Abstract
The Staphylococci comprise a diverse genus of Gram-positive, non-motile commensal organisms that inhabit the skin and mucous membranes of humans and other mammals. In general, Staphylococci are benign members of the natural flora, but many species have the capacity to be opportunistic pathogens, mainly infecting individuals who have medical device implants or are otherwise immunocompromised. S. aureus and S. epidermidis are a major source of hospital-acquired infections and are the most common causes of surgical site infections and central line-associated bloodstream infections. The ability of Staphylococci to form biofilms in vivo makes them highly resistant to chemotherapeutics and leads to chronic diseases. These biofilm infections include osteomyelitis, endocarditis, medical device implants, and persistence in the cystic fibrosis lung. Here, we provide a comprehensive analysis of our current understanding of Staphylococcal biofilm formation, with an emphasis on adhesins and regulation, while also addressing how Staphylococcal biofilms interact with the immune system. On the whole, this review will provide a thorough picture of biofilm formation of the Staphylococcus genus and how this mode of growth impacts the host.
INTRODUCTION
Bacteria from the genus Staphylococcus include a diverse group of commensals that colonize mammals on the skin or mucous membranes. Some of the best-known members of this genus, such as S. aureus and S. epidermidis, are also opportunistic pathogens and responsible for a tremendous burden on the healthcare system (1, 2). One of the reasons Staphylococci are problematic is their well-known ability to attach to surfaces and develop into recalcitrant community structures, often referred to as a “biofilm”. Generally, biofilms are defined as a community of cells encased within an exopolymeric matrix and attached to a surface, and they are recognized as being more resistant to antimicrobial therapy and host defenses (3).
The biofilm state was initially observed in studies of marine environments, in which adherent communities of bacteria were observed in natural as well as industrial aquatic environments (4). Biofilm development was subsequently found to be important in many types of infections and is now a widely accepted bacterial mode of growth. According to the NIH, as much as 80% of human infections are biofilm-based (5). Biofilm infections present a clinical challenge, as they are highly resistant to antimicrobial therapies and often occur in areas of the body that are not easily accessible for treatment (6, 7). Staphylococci in particular represent a large portion of biofilm-based infections, and are a significant burden on the healthcare system. S. aureus and the coagulase-negative Staphylococi (CoNS) are the number one and number three most common etiological agents of hospital-acquired infections in the US, respectively, including infections of medical devices and surgical wounds (8). Biofilm infections clearly are a significant burden on the healthcare system today, and the in vivo biofilm state is an important area of study.
Definition of a biofilm
The growth of Staphylococci in a biofilm has been linked to many types of infections, but one of the ongoing challenges in the field is the lack of a consensus description of the biofilm state. There is no universal agreement on what constitutes a “Staphylococcal biofilm” in terms of morphology, depth, surface coverage, regulatory state, antibiotic resistance level, or whether surface attachment is even necessary. In the field, a biofilm is defined mostly by subjective observations (i.e., it has to look like a biofilm), as well as high antibiotic resistance relative to planktonic bacteria. There have been attempts to identify biomarkers of Staphylococcal biofilm formation to provide a better definition. In one promising study, Secor et al. determined that the non-ribosomally generated peptide aureusimine (phevalin) was produced in higher levels by biofilm-grown S. aureus (9), suggesting this natural product could be a biofilm biomarker. While encouraging, there is not yet enough follow-up work on aureusimine or other potential biomarkers to reach a consensus.
The other ongoing challenge in defining biofilms is the enormity of growth states that have been linked to this term. Staphylococcal biofilm growth has been linked to foreign bodies (10), endocarditis (11), osteomyelitis (12), skin infection (13), colonization (14), cystic fibrosis (15), urinary tract infection (16), and abscess communities (17). Under such a large umbrella of different growth conditions in the host, each requiring a unique suite of bacterial factors and regulatory machinery, it is impossible to obtain a universal definition of a Staphylococcal biofilm that will be agreed upon in the field. Not surprisingly, the Staphylococcal requirements to develop infective endocarditis or a skin abscess, such as specific toxins and superantigens (18, 19), are not the same as those needed for an indwelling catheter infection that can be caused by many types of Staphylococci. Further, a much lower bacterial load (estimated at 10,000-fold lower) is needed to colonize a foreign body than to cause a skin abscess (20). The reason for this is likely the lack of vascularization at the site, and presumably a reduced presence of innate immunity factors (10). Considering this point, it seems logical that the virulence factor profile of the invading bacterial pathogen will be different in order to survive these varied host environments. As one example, S. aureus deficient in the agr quorum-sensing system are unable to properly initiate infective endocarditis or osteomyelitis (21, 22), while the same regulatory system is not essential to initiate a Staphylococcal foreign body infection (23); in fact, the agr system seems to inhibit colonization of the foreign body (24). Thus, it is increasingly important to consider the context of infection when comparing and contrasting results with other studies.
The Biofilm life cycle
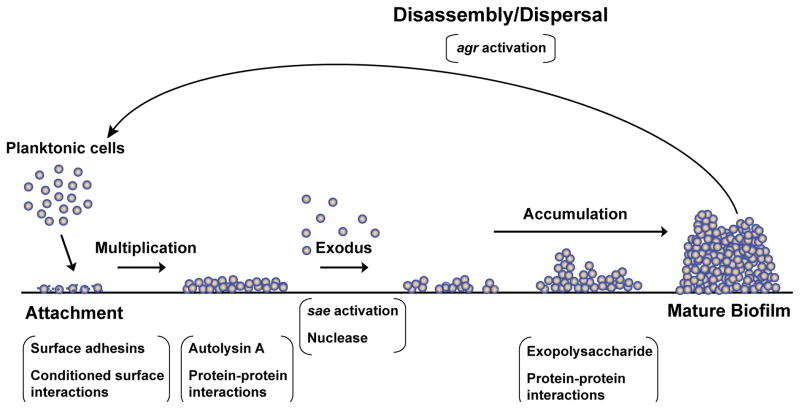
The biofilm life cycle is thought to consist of at least three stages (see Figure 1): initial attachment to an abiotic or biotic surface, maturation of the biofilm, and dispersal. Some consider “microcolony formation” to be an intermediate step between attachment and maturation, but the precise differences between a microcolony and a mature biofilm are not clearly defined. Attachment involves bacterial adhesins that can stick to the surface, while maturation is mediated by cell-cell adhesion, although some adhesins possess both properties. Dispersal or disassembly is mediated by enzymes that degrade the biofilm matrix (25–27). These enzymes may be produced by the bacteria itself or be present in the environment.
Figure 1.
The biofilm life cycle. Recent studies have identified steps present in early stages of biofilm formation. After attachment, bacteria form a lawn of growth, which undergoes an exodus period that leaves several small foci of cells. The exodus phase is mediated by the SaeRS system via nuclease enzyme activity. The foci of cells then develop into a mature biofilm, containing tower structures. Final dispersal is mediated by the agr system via secreted enzymes and PSMs.
A recent paper provided new insights into the stages of early biofilm development using a microtiter flow-based biofilm system (28). This study found that in an S. aureus biofilm, attachment and early accumulation were followed by dispersal of a portion of the cells, leaving behind small foci of biofilm growth. These foci then matured into a biofilm with tower structures. Interestingly, the early dispersal phase, termed “exodus,” was independent of the agr system, but required the sae system and was specifically modulated by the sae-regulated nuclease. These findings provide novel insight into S. aureus biofilm development and the independent roles of Staphylococcal regulatory systems in the biofilm life cycle.
Biofilm matrix
The Staphylococcal biofilm matrix has been a topic of interest in a number of reports, and various findings have demonstrated its heterogeneity and variability (29). The biofilm matrix contains eDNA, both from lysed bacteria and potentially from host neutrophil cell death, and is susceptible to dispersal by DNAses (30, 31). Proteinaceous adhesins have also been identified in the Staphylococcal biofilm matrix. These may be directly associated with bacteria in the biofilm, or free in the biofilm matrix (32). A number of cytoplasmic proteins have been identified that appear to moonlight as matrix components and undoubtedly have an important function (33). Finally, the extracellular polysaccharide intercellular adhesin (PIA) has also been identified as a major component of the Staphylococcal biofilm, especially in certain strains of S. epidermidis (34). Both proteinaceous and polysaccharide-based biofilms are susceptible to disassembly by proteases and polysaccharide-degrading agents (26, 27). Teichoic acids have also been implicated in the biofilm matrix (35), although their relative role in biofilm mechanisms has not received as much attention. Presumably, other cellular components are also present and awaiting further investigation.
Coverage of this review
This review will cover recent advances in Staphylococcal biofilm studies. We will discuss the mechanism of biofilm formation by Staphylococcal adhesins and regulatory systems, as well as the interaction of biofilms with the host immune system, with a focus on S. aureus and S. epidermidis as the model pathogens of the genus. Finally, we will discuss current knowledge on biofilm formation and virulence in other species of Staphylococci.
STAPHYLOCOCCAL ADHESINS
Staphylococci possess a number of surface-associated adhesins that mediate initial attachment of biofilm cells as well as intercellular adhesion during biofilm maturation (32, 36). The Staphylococcus aureus genome encodes more than twenty adhesins (32, 36, 37), while coagulase-negative Staphylococci (CoNS) have significantly fewer (38, 39). Staphylococcal adhesion and biofilm accumulation are mediated by covalently anchored cell wall proteins, non-covalently associated proteins, and non-protein factors. The general properties of these adhesins are presented, and their functions within biofilm development are included where information is available.
Covalently Linked Cell Wall-Anchored Proteins
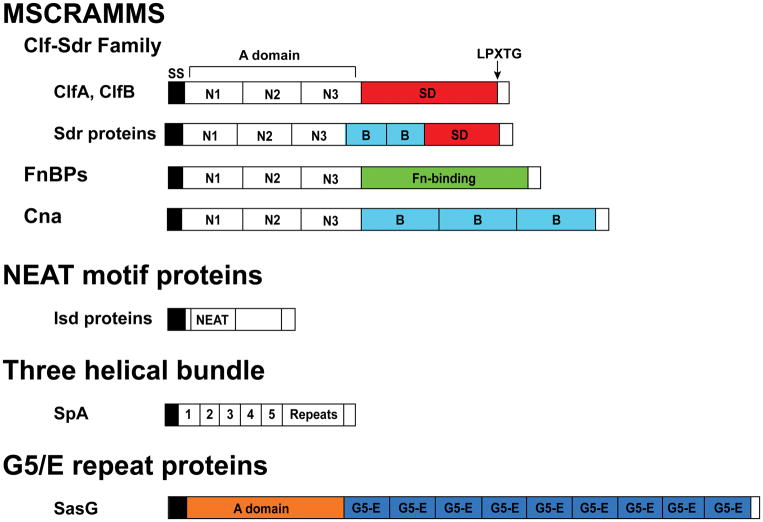
Staphylococcal cell wall-anchored (CWA) proteins are secreted by the Sec system and share a C-terminal cell wall anchoring motif, hydrophobic domain, and positively-charged domain (40). In the majority of CWA proteins, cell wall anchoring is mediated by Sortase A, which cleaves the LPXTG cell-wall anchoring motif at the threonine-glycine junction and catalyzes the covalent linkage of the CWA protein to peptidoglycan (41). Some Isd proteins in the NEAT (near iron transporter) family are instead anchored by Sortase B at the NPQT/PN/S motif (40). The Staphylococcal CWA proteins were recently discussed in a review by Foster et al., who propose to classify them into four groups (see Figure 2) based on structural motifs (40). These are the MSCRAMMs (microbial surface component recognizing adhesive matrix molecules), the NEAT motif family, the three-helical bundle family, and the G5-E repeat family. All of these types of CWA proteins are involved in biofilm formation in the Staphylococci.
Figure 2.
Cell wall-anchored adhesins. All the cell wall-anchored adhesins contain an N-terminal signal sequence (SS) and a C-terminal portion that is cleaved by Sortase A at the LPXTG sequence. MSCRAMMs contain three IgG-like folds N1, N2, and N3, followed by specific ligand-binding domains. In the Sdr protein subfamily, a variable number of B repeats is found between the IgG-like folds and the SD repeat region. SdrC is shown, which contains two of these B repeats. Similarly, the Isd proteins contain one, two, or three NEAT motifs. IsdA is shown, which has one. In SpA, there are four or five IgG-binding domains, sometimes referred to as domains E, D, A, C, and B. There follows a region containing a variable number of tandem repeats.
MSCRAMMs were originally defined as a broad category of proteins that are cell surface-associated and able to interact with the host extracellular matrix (42). The recent definition proposed by Foster et al. limits the term MSCRAMM to adhesins that contain at least two IgG-like folds and employ a ligand binding mechanism called dock, lock, and latch (40). The Staphylococcal MSCRAMMs are the Clf-Sdr family proteins, including Bbp (bone sialoprotein-bnding protein), the FnBPs (fibronectin-binding proteins), and CNA (collagen adhesion). Exposure of S. aureus to human plasma in vitro enhances both MSCRAMM expression and biofilm formation, suggesting the importance of their role in in vivo biofilm infections (43).
The Clf-Sdr family consists of Clumping factor A (ClfA), clumping factor B (ClfB), and the Sdr proteins. In addition to the IgG-like folds, their structure contains a serine-aspartate repeat domain called the SD region (40). ClfA and ClfB are fibrinogen-binding proteins in S. aureus (40, 44), and both are up-regulated in biofilm growth relative to planktonic (45). Rot and agr affect bacterial binding to fibrinogen by regulating clfB but not clfA ((46), see regulation section).
ClfA is present on the cell wall throughout the growth cycle (47) and promotes bacterial clumping in solution with fibrinogen as well as bacterial attachment to immobilized fibrinogen (48, 49). The ClfA IgG-like fold domains N2 and N3 bind at the C-terminal region of the γ-chain of fibrinogen, a region that also contains platelet binding sites (50, 51). ClfA has been shown to inhibit fibrinogen binding to platelets and fibrinogen-dependent platelet aggregation, indicating that its binding site occludes the platelet binding site (50, 52, 53). In a murine model of S. aureus septicemia, mice lacking the ClfA-binding motif of fibrinogen had better survival, suggesting that ClfA-fibrinogen interactions in the blood contribute to virulence (54). A clfA mutant in S. aureus had a decreased ability to cause vegetations in a rat endocarditis model (55) and reduced bacterial load in a murine abscess model (56).
The S. epidermidis MSCRAMM SdrG (also called Fbe) is homologous to S. aureus ClfA, although it binds to the β-chain of fibrinogen rather than the γ-chain (57, 58). SdrG is the archetypal example of the “dock, lock, and latch” mechanism of binding using its IgG-like folds. The “dock, lock, and latch” model was proposed based on the crystal structure of the SdrG-fibrinogen interaction (59, 60). SdrG mediates adherence to fibrinogen-coated surfaces in vitro (61, 62), and is required for fibrinogen-dependent platelet aggregation (63). In a rat model of central venous catheter infection, wild type S. epidermidis was more likely to cause infection and formed a more robust biofilm on the catheter in vivo than a sdrG mutant, indicating its importance in the in vivo biofilm formation of S. epidermidis (64).
The coagulase-negative species S. lugdunensis also has a ClfA homolog called Fbl (65, 66). Fbl promotes both adherence to immobilized fibrinogen and cell clumping in fibrinogen-rich solution (66). Fbl and ClfA have similar binding affinities to fibrinogen, and both interact with the C-terminus of the γ-chain (67). Fbl also is used as a species-specific detection method for S. lugdunensis (68, 69).
Like ClfA, the S. aureus ClfB IgG-like fold region binds to fibrinogen; however, it interacts with the α-chain of fibrinogen rather than the γ-chain (48, 70). ClfB promotes adherence to immobilized fibrinogen as well as S. aureus clumping in fibrinogen-rich solution (48, 70). In contrast to ClfA, ClfB becomes depleted from the cell wall beginning in late exponential phase, suggesting that it is susceptible to proteolytic degradation (48). Further studies revealed that the metalloprotease Aureolysin cleaves ClfB at two sites, resulting in the loss of fibrinogen binding (71, 72). ClfB promotes biofilm formation in vitro, and Aureolysin treatment disrupts ClfB-mediated biofilms, suggesting that Aureolysin might facilitate biofilm dispersal by processing ClfB (73). In vivo, ClfB is required for full virulence in a rat endocarditis model of infection, although the phenotype of the clfB mutant in this model was slight (74). A clfB mutant also had decreased bacterial load in a murine abscess model (56).
In addition to fibrinogen, ClfB also binds to the human epithelial proteins cytokeratin 10, cytokeratin 8, and loricrin. ClfB bound to the C-terminal tail region of purified Cytokeratin 10, as well as Cytokeratin 10 that was natively expressed in desquamated nasal epithelial cells (70, 75, 76). The ClfB IgG-like folds bind Cytokeratin 10 by the dock, lock, and latch mechanism (70). Similarly, ClfB bound immobilized Cytokeratin 8 and endogenous Cytokeratin 8 from lysates of the HaCaT keratinocyte cell line (77). ClfB also binds loricrin, the primary protein in the cornified envelope of the stratum corneum, which is present in the anterior nares. In a murine model of nasal colonization, clfB mutant S. aureus was defective for colonization compared to wild type. Colonization by both wild type and clfB mutant S. aureus was also decreased in loricrin-deficient mice, suggesting that loricrin is a critical ligand for S. aureus nasal colonization (78). In an experimental model of human nasal colonization, clfB mutant S. aureus was eliminated significantly more quickly from the nares than its wild type parent, and ClfB was required for long-term colonization (79). These results demonstrate ClfB’s versatility in ligand binding and importance for S. aureus colonization.
S. aureus Sdr proteins SdrC, SdrD, and SdrE are encoded in a single locus and have striking similarity in sequence and structural arrangement with the Clf proteins (80). SdrC can bind a host ligand as well as self-associate to promote biofilm formation. A study using a phage display peptide library found that SdrC binds to human β-neurexins, which are expressed on neuronal cells (81). Although the effect of this has not been tested in vivo, S. aureus endocarditis and sepsis are associated with polyneuropathy, meaning that SdrC interactions with neurexins could contribute to S. aureus pathogenesis. A later study showed that SdrC self-associates at the N2 domain and promotes biofilm formation, a process that is inhibited by Mn2+ (82). In contrast to SdrC, the structural stability of SdrD appears to require binding to divalent cations, specifically Ca2+ (83). Like SdrC, SdrD mediates adherence to human desquamated nasal epithelial cells (84). SdrE in S. aureus (80) induces platelet aggregation (85). SdrE also inhibits complement activation by two mechanisms. It binds the complement regulatory protein factor H, which inhibits activation of the alternative complement pathway (86). SdrE also inhibits classical complement pathway-mediated opsonization and phagocytosis by binding to the classical complement regulator C4b-binding protein (87).
SdrF in S. epidermidis binds type I collagen, mediated by its B domain repeats (88). Interestingly, SdrF was also found to bind with high affinity to Dacron, the polymeric surface of drivelines that are used in ventricular assist devices for end-stage congestive heart failure. Since S. epidermidis is a common etiological agent of medical device infections, including VAD infections, this finding is relevant to its pathogenesis. Anti-SdrF antibodies decreased infection in a murine model of S. epidermidis driveline infection, suggesting possible therapeutic interventions for these infections (89). SdrF also was found to bind other plastic materials based on ionic interactions (90). These results suggest that inhibition of SdrF binding to prosthetic devices may be a promising avenue for treatment of S. epidermidis infections.
S. epidermidis also has SdrG and SdrH, whose sequences are similar to typical SD proteins, but are not present in cell wall preparations of S. epidermidis, indicating they may be improperly sorted to the cell wall. However, antisera from patients following infection with S. epidermidis were reactive to the A domains of SdrG and SdrH, suggesting that they are expressed during infection (91).
Other coagulase-negative Staphylococci possess Sdr family proteins that have been characterized in limited detail. S. capitis SdrX has an SD repeat region, although the N-terminal domain is not strongly similar to S. aureus Sdr proteins. SdrX was reported to bind type VI collagen and mediate bacterial adherence to a type VI collagen-coated surface (92). S. saprophyticus has the Clf-Sdr family protein SdrI, which binds collagen (93) and fibronectin (94), the latter of which is a unique property among the Sdr proteins. SdrI was also found to contribute to the hydrophobicity of the S. saprophyticus surface, a property that is known to enable bacterial adherence to host epithelia (95). In a murine model of urinary tract infection, SdrI was critical for persistence but not initial colonization (96).
Bbp (bone sialoprotein-binding protein) is an Sdr-family protein in S. aureus that is considered to be an allelic variant of SdrE (87). S. aureus isolates from osteomyelitis infections were observed to bind bone sialoprotein by an unknown adhesin (97), which was eventually identified to be Bbp (98, 99). Like SdrE, Bbp binds the classical complement regulator C4b-binding protein, inhibiting classical-pathway-mediated opsonization and phagocytosis (87). Bbp appears to be associated with invasive osteomyelitis infections in particular. Of 60 patients with deep S. aureus infections following orthopedic surgery, 95% of the isolates were positive for the bbp gene (100), while of 53 S. aureus isolates from bloodstream infections, 47% were positive for bbp (101). The presence of antibodies to Bbp was also shown to be effective in distinguishing osteomyelitis from soft tissue infections in patients with diabetic foot ulcers (102).
FnBPs
S. aureus has two fibronectin-binding proteins, FnBPA and FnBPB, encoded by fnbA and fnbB, respectively (103). Like the Clf-Sdr family proteins, the FnBPs contain an N-terminal region that forms IgG-like folds; these domains bind fibrinogen and elastin (104, 105). In place of the SD repeat region, the FNBPs have a region of 10–11 tandem repeats that recognize fibronectin (106). FnBP binding to fibronectin induces bacterial invasion into epithelial cells, endothelial cells, and keratinocytes (107–109). The FnBPs have been found to affect biofilm formation and virulence. In S. aureus, a double knockout of fnbA and fnbB lost the ability to bind fibronectin and to form biofilms on microtiter plates and under shear flow conditions. Complementation of either fnbA or fnbB alone on a plasmid restored these phenotypes, as well as the ability of S. aureus to agglutinate (110). The FnBPs are thought to promote biofilm formation by a self-association mechanism that is distinct from ligand binding, making them multifunctional in the S. aureus biofilm life cycle (111, 112). The FnBPs also were shown to enhance virulence in an experimental model of endocarditis (113).
The collagen-binding adhesin CNA was initially reported to be necessary and sufficient for S. aureus binding to the collagen-rich substrate cartilage (114). CNA consists of an A domain with several collagen-binding sites, and a domain containing B repeats (114, 115). Crystal structure characterization of the CNA-collagen interaction suggested a “collagen hug” model, a variation of the “dock, lock, and latch” ligand binding scheme (115). CNA blocks activation of the classical complement pathway (116) and contributes to virulence of S. aureus keratitis (117), osteomyelitis (118), septic arthritis (119), and endocarditis (120).
S. epidermidis also has a collagen-binding protein, the GehD lipase. GehD is not LPXTG-anchored, although it is cell wall-associated, and its structure does not resemble CNA of S. aureus. However, purified GehD binds to immobilized collagen and inhibit mediate S. epidermidis binding to collagen, and therefore may contribute to colonization or pathogenesis (121).
The NEAT motif family consists of the Isd (iron-regulated surface determinant) proteins. These CWA proteins bind heme or hemoglobin, facilitating its transport into the bacterial cell, and they are up-regulated in iron limiting conditions (122). S. aureus IsdA and IsdC bind heme via their NEAT motifs (Figure 2) (123). These proteins also play a role in survival against host immune defense. S. aureus IsdA is the most abundant CWA protein in iron starvation conditions, and also decreases surface hydrophobicity, which makes S. aureus more resistant to bactericidal fatty acids and peptides in human skin (124). IsdA also is able to bind human fibrinogen and fibronectin (125). In a murine model of systemic infection, S. aureus isdB expression varied among organs to which bacteria localized in the host, and IsdB was required for colonization of the heart (126). This suggests that isd expression depends upon iron availability in each host niche of infection. In S. lugdunensis, IsdC was found to induce biofilm formation under iron-limiting conditions, and to induce attachment to polystyrene as well as self-associate to promote intercellular adhesion (127).
The sole three-helical bundle cell wall-anchored protein is Staphylococcal Protein A (SpA), which is present in all strains of S. aureus and whose sequence variation is used for strain typing (128). SpA binds the conserved Fc region of immunoglobulin IgG, which allows immune evasion (129, 130) and has also been found in the biofilm matrix in vitro (131). SpA is also released from the cell wall, and released Spa has been shown to promote bacterial survival in human blood, suggesting that free SpA contributes to disruption of the host immune response (37, 132). Presumably, free SpA could also provide adhesion in the biofilm matrix.
G5-E Repeat Family: Aap/SasG
G5-E repeats are found in cell wall-anchored adhesins in Gram-positive organisms, and are so named because of the five conserved glycine residues in each repeat. G5 domains consist of 78 residues and form six beta strands, with E domain spacers that are of similar sequence to G5, but only 50 residues (133). The S. aureus G5-E repeat protein SasG and its S. epidermidis homolog Aap have similar structures and are thought to function similarly in adhesion and biofilm formation. SasG and Aap each have an N-terminal A domain that mediates attachment to abiotic and host surfaces via unknown ligands. In S. epidermidis Aap, the A domain alone promotes attachment to polystyrene (134, 135) as well as to human corneocytes (136). S. aureus SasG promotes attached to human desquamated nasal epithelial cells via its A domain (137). Multiple studies have shown that the G5-E repeats of SasG and Aap are able to dimerize by binding to Zn2+, forming a “twisted rope” structure (133, 138). This property is thought to enable intercellular adhesion when adjacent SasG or Aap proteins dimerize via their G5-E domains. In S. epidermidis, Aap has been shown to induce biofilm formation following proteolytic removal of its A domain by exogenous proteases, although the S. epidermidis proteases that may process Aap are not identified (139). Aap has also been shown to be required for full in vitro biofilm formation in S. epidermidis, as well as for virulence in a rat catheter model of infection (134). Recently, a small 18 kDa scaffolding protein, called small basic protein or Sbp, was found in the S. epidermidis biofilm matrix and affects both PIA-dependent and Aap-dependent biofilm formation. In Aap-mediated biofilm formation, Sbp was found to interact with the B domain of Aap in the biofilm matrix, suggesting its role as a structural component of the biofilm (140).
Uncategorized CWA Proteins
The remaining uncategorized cell wall-anchored proteins are Bap and several Sas proteins, including SasA/SraP. Bap is an S. aureus cell wall-anchored protein that was identified in a transposon screen for mutants defective in biofilm formation (141). It has a unique structure, consisting of three major domains, each with sequence homology to different cell wall-anchored proteins in other bacterial genera. Bap was found to mediate attachment to an abiotic surface as well as intercellular adhesion, making it a potent enhancer of biofilm formation. In a murine model of catheter infection, the bap mutant had decreased bacterial load. Bap is also present in several coagulase-negative Staphylococci, and a mutant of the bap homolog in S. epidermidis had decreased biofilm formation relative to wild type (142). A later study demonstrated that Bap promoted adherence, but inhibited invasion, of epithelial cells in vitro by binding the Gp96 receptor (143). The authors propose that this property of Bap enhances virulence of biofilm-based infections by resisting bacterial engulfment into host cells, although this has yet to be directly tested.
S. aureus SasA/SraP (serine-rich adhesin for platelets) is a member of the serine-rich repeat family of cell wall-bound proteins found in several Gram-positive pathogens, primarily oral streptococci (144, 145). Homologs of SraP have also been identified in the coagulase-negative staphylococcal species S. epidermidis and S. haemolyticus (146, 147). SraP is a sortase-anchored, cell wall-bound adhesin that binds platelets, and it also possesses a ligand binding domain that is thought to promote intercellular adhesion and biofilm formation. A sraP mutant was reported to have decreased biofilm formation, and SraP bound to S. aureus whole cell lysates, suggesting that SraP may self-associate or bind other targets on neighboring S. aureus cells to promote biofilm development (148). The ligand binding domain was recently structurally characterized and found to contain a lectin-like module that binds N-acetylneuraminic acid (149), an abundant sugar on host glycosylated proteins. The S. haemolyticus serine-rich repeat protein UafB mediates binding to fibronectin, fibrinogen, and human uroepithelial cells (150).
SasX is another cell wall-anchored adhesin that has been shown to play an important role in virulence. SasX was linked to the spread of a MRSA epidemic in China, as its prevalence in MRSA clones increased via horizontal transfer, suggesting its importance in the pathogenic success of MRSA. SasX was also shown to be crucial for a murine model of nasal colonization, murine MRSA skin infection, and bacterial aggregation in vitro (151). SasX has recently been shown to be a promising vaccine candidate for S. aureus infection. Active or passive immunization to SasX decreased S. aureus virulence in murine models of skin infection, lung infection, and nasal colonization (152).
There are also several S. aureus Sas proteins that are poorly characterized. SasC is an LPXTG-anchored protein that contains a FIVAR domain and a domain of unknown function consisting of repeats. SasC, specifically its FIVAR domain, induced cell aggregation, binding to polystyrene, and biofilm formation (153). Genome sequence analysis of S. aureus revealed the putative cell wall-anchored adhesins SasB, SasD, SasF, SasJ, SasK, and SasL, but their structure or function have not been studied further (154).
Surface-Associated Proteinaceous Adhesins
The Autolysins AtlA and AtlE are found in S. aureus and S. epidermidis, respectively. Atl and AtlE share a similar amino acid sequence and structure, with bacteriolytic amidase and glucosoaminidase domains (155). They are known to be involved in cell wall turnover, cell division, and cell lysis (156, 157). Autolysins have two properties that could promote biofilm formation: the ability to attach to extracellular matrix materials, and to augment the biofilm matrix with eDNA by inducing cell lysis. Adhesin activity was first identified in a transposon mutant of S. epidermidis atlE that had decreased ability to adhere to polystyrene and vitronectin (155). In an S. epidermidis biofilm grown on medical device biomaterials in vitro, atlE expression decreased during the first 12 hours of biofilm growth relative to planktonic culture, but by 48 hours, expression was upregulated ten-fold (158). This may indicate that atlE is more important later in the biofilm life cycle, when autolysis is induced and eDNA is released.
Non-Proteinaceous Surface-Associated Adhesins
Wall teichoic acids and lipoteichoic acids have been shown to play a role in adhesion, colonization of host cells, and biofilm formation. Wall teichoic acids are covalently linked to the peptidoglycan and consist of alternating phosphate and ribitol, while lipoteichoic acids attach to the outer leaflet of the cell membrane and have alternating phosphate and glycerol (36). Teichoic acids are highly charged, a property that was found to be critical for S. aureus colonization of abiotic surfaces. A mutant lacking D-alanine in its wall teichoic acid lost the ability to form a biofilm in vitro. This was due to its greater net negative charge, which decreased its adherence to plastic surfaces (159). In S. epidermidis, wall teichoic acids induced adherence to immobilized fibronectin (160). Teichoic acids have been identified in the biofilm matrices of S. epidermidis and S. aureus (35, 161, 162).
The polysaccharide intercellular adhesion (PIA) is a secreted polysaccharide that is synthesized by the ica operon and has been thoroughly studied in the context of biofilm formation, immune evasion, and pathogenesis. Several reports state that PIA is required for S. epidermidis biofilm formation and virulence (163–165), and it is considered to be the most important intercellular adhesin of the Staphylococci (34). The role of PIA in Staphylococcal biofilm formation has been reviewed by O’Gara (166) and by Rohde et al. (167), and the regulation of the ica locus in the Staphylococci in (168).
SECRETED PROTEINS IN THE BIOFILM MATRIX
A number of secreted Staphylococcal proteins have been implicated in biofilm formation, the most prominent being AtlA/AtlE (discussed above as surface associated), proteases, nucleases, and phenol-soluble modulins (PSMs). The exo-enzymes and PSMs will be discussed in more detail in the regulation section below, as their role in biofilm development more relates to dispersal than accumulation. Other secreted proteins that have been linked to biofilm formation are covered here.
Alpha-toxin (Hla) is a potent cytolysin secreted by S. aureus that also affects inflammation and contributes to pathogenesis by multiple mechanisms. Many studies have shown that alpha-toxin mutants are attenuated in virulence (169). However, the contribution of this toxin to biofilm formation is less clear. In a study of in vitro biofilm formation on polystyrene, an S. aureus hla mutant had dramatically reduced biofilm in a standard microtiter plate assay and under flow conditions. Its initial attachment to the surface was also decreased, indicating that an inability to bind to the surface contributes to decreased mature biofilm (170), although the exact mechanism of this phenotype remains unclear.
S. aureus beta-toxin (Hlb) is a secreted sphingomyelinase that has hemolytic and lymphocytic activities (171, 172). Beta-toxin also has a sphingomyelinase-independent “biofilm ligase” activity, which refers to its ability to cross-link strands of DNA in the biofilm matrix. Mutants in hlb were deficient in in vitro attachment, flow-cell biofilm formation, and vegetation formation in a rabbit endocarditis model (173). Hlb is the first Staphylococcal protein identified that binds extracellular DNA (eDNA) in the biofilm matrix, providing more evidence of the importance of this matrix component.
S. aureus secretes multiple proteins that have been called Secreted Expanded Repertoire Adhesive Molecules (SERAM) (174). Two of the SERAMs, the extracellular adherence protein (Eap; also called the MHC class II analog protein or Map) and the extracellular matrix binding protein (Emp) have a demonstrated connection to biofilm formation (175, 176). Eap is a secreted adhesin that enhances S. aureus adherence to the extracellular matrix, and it has been shown to bind fibrinogen, fibronectin, vitronectin, and thrombospondin-1 with varying affinities (177–180). Eap can also self-associate to induce aggregation of S. aureus (177). Due to its ability to bind several matrix proteins, Eap is required for biofilm formation in the presence of serum (175). Regulation of the eap gene is dependent on the SaeRS two-component system (181, 182), and the gene is up-regulated under low iron conditions (176). The eap mutant biofilm phenotype is dependent on these iron-limiting conditions (176). The Eap protein has a number of known immunomodulating properties that have been summarized elsewhere (174). Less is known about Emp, but this protein can also bind matrix proteins like fibrinogen (174) and it is also SaeRS and iron regulated (176, 182). Similar to Eap, Emp is required for biofilm formation on iron limiting conditions (176).
REGULATION OF BIOFILM FORMATION
Global changes in gene regulation occur throughout the course of the Staphylococcal biofilm life cycle. Microarray studies have shown that the biofilm lifestyle requires a gene expression profile to allow tolerance of the low pH within a biofilm, as well as a metabolic quiescence that includes down-regulation of transcription, translation, and aerobic processes (183, 184). Several global regulators, such as the agr quorum sensing system, sigma factor B, and SarA, have strong connections to Staphylococcal biofilm formation in vitro and during infection and will be summarized further below. These regulators have also been examined in more acute pathogenesis mechanisms, and the focus presented here will be on the biofilm-like infections. The majority of these studies have been performed with S. aureus, but some studies on S. epidermidis are also included. It should be noted that a number of other regulators have also been linked to biofilm formation, including MgrA (185) and ArlRS (186), but these are beyond the scope of this review.
agr quorum-sensing system
The agr (accessory gene regulator) system is a peptide quorum-sensing system present in all the Staphylococci and a dominant regulator of pathogenesis and biofilm development in S. aureus. Its molecular characteristics and importance in pathogenesis have been thoroughly studied and reviewed in detail (187, 188). The agr system functions by sensing extracellular levels of an autoinducing peptide (AIP) that is produced by Staphylococci during growth. The chemical nature of this AIP signal is variable depending on the species, and can even have multiple types within a species. Briefly, the AIP is released outside the cell where it accumulates, and at a particular concentration (usually in the low nM range), the AIP binds to the surface-exposed AgrC histidine kinase, activating a two-component response. This results in phosphorylation of the response regulator AgrA, which in turn induces expression of the primary output of the system, the regulatory transcript RNAIII (189). In parallel, AgrA activates transcription of the PSMα and PSMβ transcripts (190), and also autoinduces the quorum sensing machinery. RNAIII is the main effector of the system and directly regulates production of virulence factors, and also regulates the translation of the repressor of toxins (Rot) (191). In the Staphylococcal strains where global changes in agr-dependent gene expression have been assessed (192, 193), the general dogma is that induction of the agr system leads to up-regulation of secreted enzymes and toxins, while simultaneously down-regulating adhesins.
Understanding and interpreting the literature on the agr system is a challenging task, in part due to the depth of the literature but also due to the complexities of the system. Focusing on S. aureus, one of the most overlooked issues is that the dynamic range of the agr system is tremendously variable across strains (187), meaning some strains barely produce RNAIII whereas others, like the USA300 strains, produce very high levels (194). In recent years, the molecular nature of this variability has begun to be examined (195), providing preliminary explanations for why some S. aureus strains have muted agr function, resulting in reduced RNAIII levels. The challenge becomes interpreting the results of agr mutant studies, where in a USA300 strain the mutation has a dramatic impact on many phenotypes (196–198), but in others, such as some clonal complex 30 strains (195), has little impact. Further complicating this issue, some older lab strains, such as 8325–4, have known mutations that lead to dysregulation of agr function (199). Thus, care must be taken in interpreting results of studies, especially in animal studies of infections, and unfortunately the agr function of strains used for some of these studies is not known.
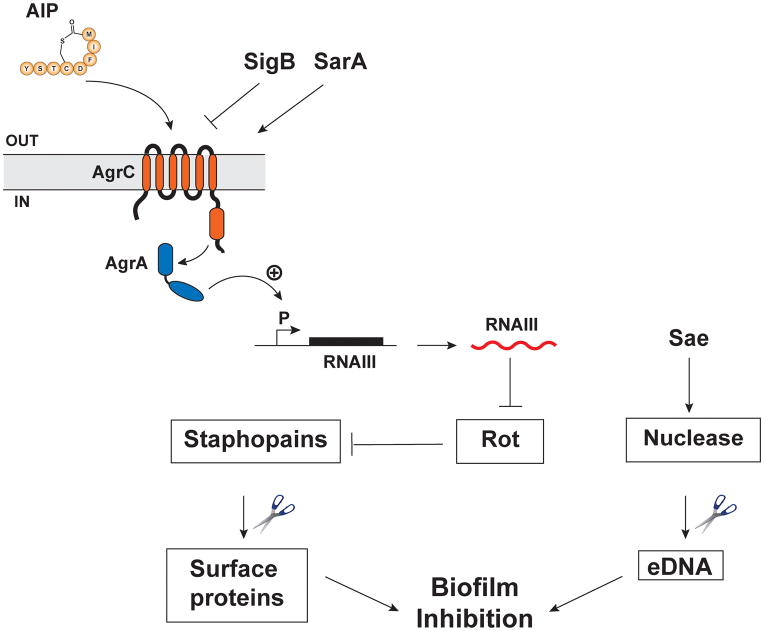
Several studies have investigated the role of quorum sensing in the biofilm life cycle. The current model is that biofilm initiation and maturation require low agr expression, while subsequent agr activation within the biofilm induces dispersion to the planktonic state. Indeed, multiple studies of S. aureus and S. epidermidis have shown that isogenic agr mutants display increased biofilm formation in vitro (24, 199–201). As the biofilm develops, small populations experience agr re-activation and disperse from the biofilm (27, 187, 202). An established S. aureus biofilm can be fully dispersed by the addition of AIP to induce agr activation (179, 203), and the dispersion process is mediated by agr-regulated proteases, most prominently the Staphopain enzymes (204). The agr regulation of the proteases is via Rot, whose transcriptional repression of the proteases is relieved when agr is induced (205, 206). A model for this regulatory pathway is shown in Figure 3. Currently, the major missing piece of this model is the specific biofilm matrix proteins that are targeted by the Staphopain enzymes, and this is a topic of ongoing investigation. The other prominent agr-regulated factors linked to biofilm dispersion are the phenol-soluble modulins (PSMs) (23). These peptides have surfactant activity that is anti-biofilm in nature. On the whole, these results support the model that under agr repressive conditions or with agr null mutants, S. aureus cells have increased biofilm capacity in vitro due to the absence of secreted dispersal factors. However, in a biofilm infection, the importance of the agr system for initiation depends on the type of infection (see below), presumably because in some tissue sites S. aureus must secrete agr-regulated immunomodulating factors to survive. Once the biofilm has been established, both in vitro and in vivo studies indicate that activation of the agr system can lead to dispersal of the cells and dissemination to new sites.
Figure 3.
Regulatory networks in biofilm formation. The agr quorum-sensing system induces expression of secreted Staphopain proteases by inhibiting translation of Rot (repressor of toxins), a negative regulator of the proteases. These proteases then degrade proteins on the Staphylococcal surface and in the biofilm matrix. The SaeRS system induces production of the nuclease enzyme that cleaves eDNA in the matrix. Sigma Factor B (SigB) inhibits agr expression, while SarA has been shown to directly enhance it.
Environmental conditions are a critical factor in controlling agr function. S. aureus can metabolize many sugars (207), and the low pH generated from excretion of short-chain fatty acids can repress agr activity (208). For development of an in vitro S. aureus biofilm, excess sugar (e.g. glucose) must be provided to trigger the pH decline and promote biofilm formation (179). The expression of agr-regulated factors, such as the Staphopains, must be repressed in order for S. aureus cells to attach and initiate biofilm development. For other Staphylococcal species, this low pH requirement is less clear. In S. epidermidis, the addition of excess glucose was not essential for promoting biofilm formation (134), but whether this is related to agr function is not known. The control of agr function is also beginning to be appreciated in the host environment. Proteins found in human serum are known to repress S. aureus agr activity (209), including apolipoprotein B (210, 211). Hemoglobin is another abundant host protein with known agr inhibitory properties (212). Environmental contaminants that accumulate in the body, like triclosan, have also been linked to enhanced biofilm formation (213). Altered environmental conditions can impact biofilm structure as well, as recently demonstrated with the conversion of PSMs into protease-resistant amyloid fibers (214).
The role of agr during infection is complex and been the focus of many studies. The contribution of agr to acute pathogenesis has been reviewed previously (187), and we will focus on chronic infections with biofilm-like properties. One of the most common models of biofilm infection is a catheter placed subcutaneously in the flank of a mouse. Staphylococci are inoculated into the lumen of the catheter and allowed to develop into a biofilm over an extended time period (215). The inoculum dose can be varied, as it is known that foreign bodies greatly reduce the bacterial load required to colonize (10). Using this model, S. aureus agr mutants have no defect in the ability to colonize the catheter and develop a biofilm (216). However, these mutants are less able to disseminate into other tissues (23), consistent with the concept that the agr system is a dispersing mechanism. Using a similar rabbit model of indwelling medical device infection, an S. epidermidis agr mutant actually had an increased ability to colonize the device (24). For both S. aureus and S. epidermidis, it has been demonstrated that PSMs are important for dispersing from a foreign-body biofilm (23, 217). It seems likely that the many exo-enzymes secreted by these pathogens are also important for dissemination through the host, and our preliminary studies confirm this hypothesis (216).
For chronic infections involving host tissue, the function of the agr system has also been assessed. S. aureus is one of the leading causes of osteomyelitis, a chronic infection of the bone that has known biofilm-like properties (12). S. aureus agr mutants are attenuated in the ability to establish osteomyelitis (22). S. aureus is also the leading cause of infective endocarditis (218), another chronic infection where bacteria attach to the heart valve and develop into a vegetation composed of matrix proteins, platelets, and immune cells. Similar to osteomyelitis, S. aureus agr mutants show a defect in the ability to establish infection (21). Interestingly, the agr system showed progressive activation within the endocarditis vegetations (219), which is in contrast to what is observed in biofilms under in vitro conditions (179). As an added complication of endocarditis, septic emboli can dislodge and enter the bloodstream, and these emboli have biofilm characteristics that make them more resistant to antibiotics than planktonic bacteria and are potentially life-threatening for the patient (220–223).
It is generally accepted that low levels of agr expression are observed in chronic infections (224–226), suggesting that the loss or decreased activity of agr is an adaptation to allow persistence in the host environment. However this is somewhat misleading, since the comparison for these claims is usually in vitro broth culture, in which agr function and RNAIII levels are extremely high. The more informative experiment is tracking agr function over time within the infection, which was performed with infective endocarditis (219), but has not been attempted more systemically in other infection types. Some of the studies on low agr expression are based on S. aureus isolates from the cystic fibrosis lung. Growth of bacterial pathogens, like Pseudomonas aeruginosa, in cystic fibrosis are known to be in biofilm state, but there is limited information on S. aureus, although link to biofilms has been suggested (15). What is clear is that agr negative mutants do frequently appear during biofilm growth in vitro (227) and during biofilm infections (226, 228). In part, this could be due to the increased fitness cost of expressing the agr system in presence of antibiotic pressure (229). When S. aureus reenters a normal state, the maintenance requirement of the agr system is restored (230).
As a global regulator, the agr quorum-sensing system has widespread effects on gene expression that can strongly impact the Staphylococcal biofilm life cycle. However, the complexity of the system and the factors that alter its expression can result in varied effects of the agr system in vivo. When studying the role of the agr system during infection, the agr expression level of the Staphylococcal strain must be considered, as well as the infection niche and the relevant host factors that may modify the importance of agr. Since inhibition of agr has begun to be investigated as possible treatment for S. aureus infections (231–233), further work needs to be done to clarify the types of in vivo conditions and infections that would benefit from this.
AI-2
AI-2, or autoinducer-2, is a second quorum-sensing molecule that is present in both Gram-positive and Gram-negative organisms, and is thought to be a bacterial interspecies signaling molecule (234, 235). AI-2 has been shown to inhibit biofilm formation and virulence in both S. aureus and S. epidermidis. In S. epidermidis, a mutant that does not produce AI-2 had greatly increased biofilm formation in vitro and increased virulence in a rat central venous catheter infection model (236). This mutant also had increased expression of the secreted polysaccharide synthesis operon ica and increased polysaccharide production, which is thought to be the mechanism for its biofilm phenotype. Similar findings were reported in S. aureus by Yu et al., in which an AI-2 mutant had increased biofilm formation in vitro and in a murine catheter infection model. AI-2 was also shown to positively regulate expression of the ica operon repressor icaR, leading to increased production of extracellular polysaccharide in the AI-2 mutant (237). An S. aureus AI-2 mutant also displayed increased survival in human blood and macrophages (238). Yu et al. also investigated the relationship between AI-2 and agr regulation in S. epidermidis biofilm formation. The two were found to have an additive effect, in which a double mutant had higher biofilm formation than either single mutant (237).
SigB
Sigma B (SigB) is an alternative sigma factor of RNA polymerase that is activated in stress response and leads to global changes in promoter specificity, and thus gene expression (239). Strains lacking SigB have tremendous changes in regulatory profiles that alter biofilm formation and virulence. S. aureus mutants in the global regulator sigB are unable to form a biofilm in vitro (199). This phenotype is mediated both by increased protease activity (240, 241) and increased nuclease activity (242). Part of the reason for increased protease activity has been linked to hyper-activation of the agr system in SigB-defective strains (199). In S. epidermidis, a sigB mutant also was reported to have decreased biofilm formation and increased expression of icaR, which represses production of the extracellular polysaccharide PIA (243, 244).
SigB also plays an important role in the response of Staphylococci to the in vivo environment. In a murine intravenous infection model with S. aureus, SigB cascade was reported to be highly transcriptionally active, although a sigB mutant had no difference in disease outcome in the mice (245). However, an S. epidermidis sigB mutant did have a defect in colonization in a rat model of foreign body infection (246). Expression of S. aureus SigB is also activated by human pulmonary surfactant and after internalization by human bronchial epithelial cells (247, 248).
Spontaneous sigB mutants arise in vitro in biofilm formation and in chronic Staphylococcal infections (249, 250). These mutants are small colony variants, which have elevated extracellular protease activity and decreased biofilm formation. They also have increased intracellular persistence and are thought to contribute to virulence of S. aureus infections (250). These results suggest that biofilm populations and infecting bacteria are heterogeneous in gene sequence as well as expression level of key virulence factors.
SarA
The sar (Staphylococcal accessory regulator) locus was discovered in a transposon mutagenesis screen for fibrinogen-binding-negative mutants (251). In addition to decreased fibrinogen binding, the sar mutant had an altered exoprotein profile, with increased protease, lipase, and α-hemolysin. The sar locus produces three transcripts from three separate promoters, all of which contain the ORF for the DNA-binding protein SarA (252, 253). Early studies showed that a sar mutant had decreased levels of the agr transcripts RNAII and RNAIII (254–256). EMSA (electrophoretic mobility shift assay) studies have shown that purified SarA directly binds to three sites within a region spanning the P2 and P3 promoters of the agr locus (253). SarA regulatory activity therefore occurs partially via its effects on agr.
SarA also directly regulates several genes that affect biofilm formation. A putative SarA binding site has been identified upstream of several SarA-regulated genes, providing a mechanism for direct regulation by DNA binding (257). SarA represses transcription of the collagen adhesion cna even in the absence of agr, and directly binds to the cna promoter region (258, 259). A microarray revealed that in addition to positively regulating fnbA and fnbB, SarA negatively regulates isaB and spa, and all of these gene promoters contain a putative SarA binding site (193). SarA also positively regulates bap, a cell wall adhesin found in bovine isolates of S. aureus, via direct binding to its promoter (260). Secreted proteases and nuclease are also up-regulated in sarA mutants (193, 261, 262). Since the opposite phenotypes are observed in agr mutants, it is apparent that this effect occurs via an agr-independent pathway.
Multiple studies have reported that in S. aureus, SarA is required for biofilm formation, both in vitro and in vivo (175, 263–265). Various mechanisms for this phenotype have been proposed. The first study to report the S. aureus sarA mutant biofilm defect also showed that this mutant had decreased ica transcription and PIA production, and suggested that this could partially account for the biofilm phenotype (264). However, a later study showed that while a S. aureus sarA mutant did not produce a biofilm, an ica operon knockout had no decrease in biofilm formation (183). These studies were performed in vitro as well as in vivo using a murine model of catheter-associated biofilm infection. The results indicate that ica regulation is not the sole factor behind the sarA knockout biofilm phenotype, and in fact that PIA production may not be critical under certain biofilm conditions.
Biofilm formation in sarA mutants is also thought to be inhibited by their increased protease and nuclease activity. Although one study found that protease inhibitor treatment did not alter the phenotype of a sarA mutant, only the initial attachment was tested rather than the endpoint of biofilm growth (264). A later report found that treatment with a cocktail of protease inhibitors for all the secreted proteases resulted in increased biofilm formation of a S. aureus sarA mutant in the clinical isolate UAMS-1 (266). This suggests that increased protease activity contributes to the sarA biofilm phenotype. The same study showed that a sarA nuc double mutant had improved biofilm formation relative to a single sarA mutant, demonstrating that the effect of SarA is also partially mediated by nuclease production. In a group of three S. aureus clinical isolates, sarA mutants had decreased biofilm formation regardless of their various levels of agr expression (267). This study also found that protease inhibitor treatment improved biofilm formation in sarA mutants of each clinical isolate, confirming the impact of secreted proteases on biofilm formation. On the whole, these results suggest that SarA-mediated effects on biofilm formation are mediated by secreted enzymes more than PIA, and are at least partially independent of agr.
There are conflicting reports of the effect of sarA mutation on S. epidermidis. In two S. epidermidis clinical isolates, sarA mutation was found to drastically increase biofilm formation (268). In the same study, S. epidermidis SarA was also found to positively regulate transcription of the ica operon and bind to the icaA promoter, indicating that SarA induces biofilm formation via PIA production. In contrast, Christner et al. showed that SarA represses biofilm formation in an S. epidermidis clinical isolate by two mechanisms (269). Mutation of sarA dramatically increased biofilm formation in the aap-negative, ica-negative S. epidermidis clinical isolate 1585 as well as in an S. epidermidis 1457 icaA mutant. The first mechanism of biofilm enhancement in the sarA mutant was increased expression of the cell wall adhesin embp (extracellular matrix binding protein). A double sarA embp mutant had decreased biofilm formation relative to the sarA mutant, indicating that the sarA phenotype was partially dependent on Embp. The second mechanism of biofilm enhancement was found to be increased protease expression in the sarA mutant, although this is contrary to previous findings in S. aureus. S. epidermidis sarA mutants have increased production of the metalloprotease SepA (270), which was shown by Christner et al. to induce processing of the autolysin AtlE, leading to increased lysis and eDNA release (269). This study shows that SarA is a positive regulator of the eDNA- and Embp-dependent biofilm.
EVASION OF THE HOST IMMUNE SYSTEM
Staphylococcal biofilms are noted for their resistance to host immune clearing, and there have been significant efforts to characterize the mechanisms that contribute to this resistance (271–273). Studies have investigated the effects of Staphylococcal biofilms on immune cell function, the host antibody response to chronic Staphylococcal infection, and the Staphylococcal transcriptional response to host innate immune cells.
In the innate immune system, PMNs (polymorphonuclear leukocytes) and macrophages are the first responders to Staphylococcal infection (17, 274–276). Although one study reported minimal PMN influx in a murine model of catheter-associated S. aureus biofilm growth (273), others have demonstrated that activated PMNs are prevalent at the site of infection in human patients with orthopedic device-associated Staphylococcal biofilm infections (277, 278). Multiple studies have reported that human PMNs in in vitro co-culture with S. aureus localize to the biofilm and can phagocytose bacteria (279, 280). In an S. epidermidis biofilm grown in vitro, PMNs were able to attach to the biofilm, release granule components from both primary and secondary granules, and phagocytose biofilm bacteria (281). These effects were observed with or without opsonization, which suggest they are mediated at least in part by bacterial components that interact with the PMNs.
PMNs can attack Staphylococcal biofilms by phagocytosis, release of toxic granule components, and production of NETs (Neutrophil Extracellular Traps), although there is evidence that these effects are dampened relative to planktonic bacteria (282). S. epidermidis is more resistant to in vitro killing by human PMNs when grown in a biofilm than grown planktonically (283). The S. epidermidis extracellular polysaccharide PIA is thought to play a role in evading PMN killing, as it has been shown that an S. epidermidis ica mutant exhibits increased susceptibility to phagocytosis and killing by human PMNs in vitro (284). Similarly, the S. aureus capsular polysaccharide inhibits opsonophagocytosis of planktonic bacteria by PMNs in vitro and is required for full virulence in a murine model of septic arthritis (285, 286). Although these studies did not directly test phagocytosis of biofilm bacteria, the results suggest that PIA and capsule in a Staphylococcal biofilm may shield the bacteria from the host immune response. The agr system may also allow the Staphylococcal biofilm to resist PMN killing. In S. aureus, an agr mutant biofilm was less cytotoxic to PMNs in co-culture than its wild type counterpart, suggesting that biofilm cells expressing agr could kill PMNs and therefore evade phagocytosis and killing by PMNs (279). This corroborates an earlier study testing interactions of planktonic wild type and agr mutant S. aureus with PMNs, which also found that the agr mutant induced decreased PMN lysis (198).
Neutrophil Extracellular Traps (NETs) were first described by Brinkmann et al., who showed that activated PMNs produce thread-like projections composed of DNA and granule components (287). NETs bind to microbes in vitro and degrade bacterial extracellular virulence factors as well as kill the bacteria. NET formation is thought to occur via a regulated cell death pathway termed NETosis that is distinct from necrosis and apoptosis (288). S. aureus extracellular nuclease has been shown to degrade NETs, thereby allowing the bacteria to resist NET-mediated killing (289, 290). Since the secreted nuclease is unique not found in all Staphylococci, speces that lack it may be more susceptible to NETs, although an additional surface-attached nuclease is conserved among the Staphylococci (291).
The interactions between Staphylococcal biofilms and host macrophages have also been investigated. These studies have led to a model (see Figure 4) in which Staphylococci promote an anti-inflammatory, pro-fibrotic environment via alternative macrophage activation (271, 273). Macrophages can undergo at least two varieties of activation, including classical and alternative (292). Classically activated (M1) macrophages are characterized by their ability to present antigen and degrade intracellular pathogens, while alternatively activated macrophages are inefficient at both of these. Alternatively activated (M2) macrophages have high arginase (Arg-1) activity, which decreases their ability to destruct intracellular pathogens (293) and promotes collagen formation, extracellular matrix proliferation, and wound healing (294). Planktonic Staphylococci have been shown to induce classical activation of macrophages and are readily phagocytosed (295). However, in a S. aureus biofilm co-culture with macrophages, Scherr et al. observed little macrophage phagocytosis of S. aureus cells, and few macrophages in close proximity to the biofilm (279). A co-culture study of S. aureus biofilms and macrophages reported that macrophages that interacted closely with the biofilm performed little phagocytosis and exhibited gene expression patterns consistent with M2 activation. Further, the same study showed that macrophage death was induced in cells that were close to the biofilm (295). Another group has also reported that clfA expression prevents macrophage phagocytosis by a mechanism that is independent of binding to fibrinogen (296). These results suggest that biofilms can promote a macrophage phenotype that favors the progression of infection and produce factors that are cytotoxic to macrophages.
Figure 4.
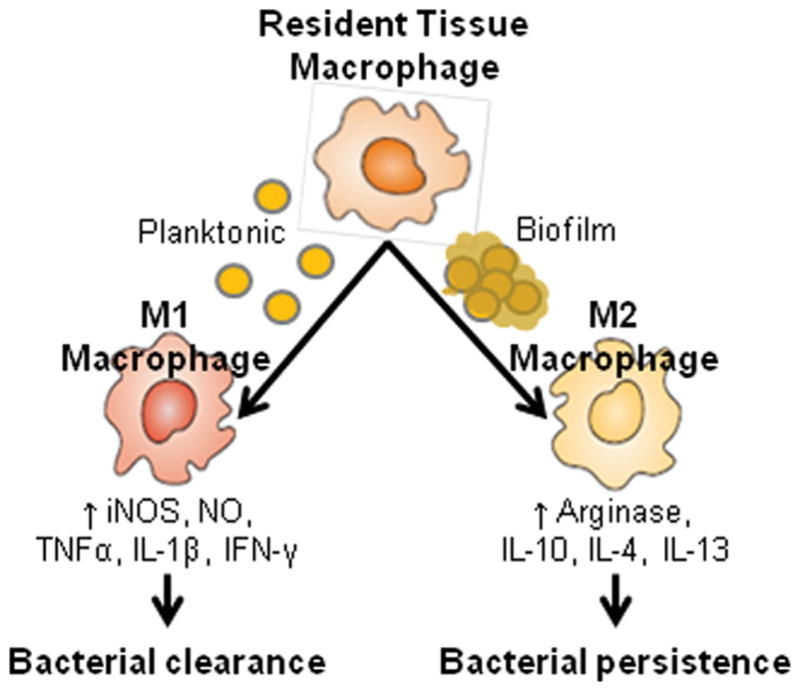
Macrophage activation pathways. Biofilm growth of Staphylococci has been shown to favor the M2 phenotype in macrophages, which is characterized by increased arginase and pro-fibrotic activity, as well as decreased antimicrobial clearance. These changes are thought to contribute to the persistence of Staphylococci in biofilm infections. This figure is a reproduction from (273).
Multiple studies have performed microarrays on S. aureus following co-culture with innate immune cells to determine global regulatory changes that facilitate Staphylococcal survival. A microarray study of S. aureus following interactions with PMNs and macrophages revealed more extensive changes in regulation after exposure to macrophages. Although various S. aureus genes were differentially regulated in response to PMNs, a striking global down regulation was observed following co-culture with macrophages. In both an immature and a mature biofilm, downregulation of hundreds of genes was observed, corresponding to a global decrease in metabolic processes (279). Another report demonstrated that following co-culture with phagocytic PMNs, S. aureus experienced widespread changes in regulation. Stress response proteins such as catalase were up-regulated, as well as virulence factors such as hemolysins and fibrinogen-binding proteins. There was also a shift in the metabolic profile relative to broth culture, and changes in global regulators. Of note, the Sae system, which positively regulates several secreted toxins and other virulence-associated proteins (196, 297–299), was up-regulated several fold after exposure to PMNs (300). The Sae system was later shown to differentially regulate its various targets in response to specific neutrophil stimuli (301), and sae mutants have decreased survival in in vitro PMN phagocytosis assays (299). These results demonstrate that S. aureus has an arsenal of tools to survive PMN and innate immune assault, and that the Sae system in particular is a crucial element.
Several studies have characterized the S. aureus proteins that are targeted by the host antibody response following infection or colonization (302–304). This group of antigens is referred to as the immune proteome, and identifies proteins that are expressed in vivo and may be important to virulence. These findings also may suggest targets for the development of a vaccine for S. aureus, which has been an area of interest in recent years.
OTHER STAPHYLOCOCCAL SPECIES
The study of Staphylococcal biofilm formation has largely focused on S. aureus and S. epidermidis. However, other Staphylococcal species are also pathogenic biofilm-formers. Staphylococci are classified based on their production of the blood-clotting enzyme coagulase, a secreted protein that induces the conversion of fibrinogen to fibrin (305, 306). The genus Staphylococcus comprises 47 species: 8 of which are coagulase-positive or coagulase-variable, 38 of which are coagulase-negative, and one, S. schleiferi, that has both a coagulase-negative and a coagulase-positive subspecies. Of the coagulase-positive Staphylococci, S. aureus is the sole species that is primarily associated with human-disease (1). However, there have been reports of other coagulase-positive Staphylococci colonizing or causing disease in human hosts who have significant contact with animals (307, 308). One example is S. pseudintermedius, a commensal of dogs that is implicated in canine opportunistic infection (309). Biofinformatic and proteomic analyses have been employed to study the cell wall adhesins of S. pseudintermedius, leading to further study of two proteins (SpsD and SpsL) to which canines have antibodies, indicating their expression in vivo (310, 311).
The coagulase-negative Staphylococci (CoNS) are the third most common cause of human healthcare-associated infections (8) and the number one cause of bovine intramammary infections (312–314). They have been reviewed thoroughly in (44) and (1). Within the CoNS, S. epidermidis is the most frequent cause of medical device-related infections, and is able to infect virtually any medical implant including catheters, central lines, ventilators, prosthetic joints, and pacemakers (8, 315–317). One reason for its high rate of infection may be its prevalence in the normal skin flora and ability to colonize many surfaces of the human body, including the anterior nares, axillae, and inguinal and perineal areas (1, 318). Other CoNS species inhabit various niches of the human body. S. lugdunensis is found particularly in the lower extremities of the body in the perineal and groin areas, as well as in the axillae (319, 320). S. haemolyticus is preferentially isolated from axillae and pubic areas (318, 321), and S. saprophyticus from the gastrointestinal tract, rectum, and urogenital tract, typically in younger individuals (1, 322, 323).
Of the coagulase-negative Staphylococci, Staphylococcus lugdunensis is considered to be the most similar to coagulase-positive Staphylococci (1). It is a skin commensal and opportunistic pathogen, responsible for 0.8%–7.8% of infectious endocarditis cases in non-drug users (324), with morality rates ranging from 38%–42% (324, 325). These high mortality rates are similar to those of S. aureus endocarditis, and are suggestive of aggressive infection. It is also implicated in infections of medical devices, such as catheters and prosthetic joints (326–328). Recent studies have also demonstrated that S. lugdunensis is a significant cause of skin and soft-tissue infections, with up to 53 per 100,000 per year (329).
For mechanisms of pathogenesis, S. lugdunensis possesses several virulence factors, including surface adhesins, that have been reviewed in (330). A mutant in the cell wall-anchoring enzyme Sortase A was attenuated in an experimental endocarditis model, confirming that like in S. aureus, adhesins contribute to S. lugdunensis-mediated endocarditis (331). S. lugdunensis also has an ica locus for PIA synthesis, although its role in biofilm formation is not clear, since S. lugdunensis biofilms in vitro were not sensitive to degradation by either of the PIA-targeting factors Na-metaperiodate or Dispersin B (332). Biofilm formation may be a significant factor in infection, as S. lugdunensis clinical isolates from prosthetic implant infections have been shown to be strong biofilm formers in vitro (333). In addition to its biofilm-forming activity, S. lugdunensis has several putative cytolysins, including the SLUSH (Staphylococcus lugdunensis synergistic hemolytic) peptides, which are similar to S. aureus delta-toxin Hld, and another Hld-like protein (330, 334). However, the activity and virulence role of these SLUSH proteins is not known.
S. haemolyticus has been implicated in a range of opportunistic infections in humans, including prosthetic device infections (335) and postoperative endophthalmitis (336). In a study of S. haemolyticus isolated from bloodstream infections, 66% of the isolates formed robust biofilms in vitro, but the same strains were all negative for PCR of the ica operon for PIA synthesis (337). This suggests that S. haemolyticus has other means of biofilm formation. The Bap adhesin was present in several biofilm-positive S. haemolyticus nosocomial infection isolates, identifying at least one PIA-independent mechanism that may contribute to S. haemolyticus biofilm formation and infection (338).
Since S. saprophyticus colonizes the rectum and urogenital tract, it is unsurprising that it is a common cause of urinary tract infections (UTIs). S. saprophyticus is the second most frequent cause of UTI in young, sexually active women, although it also causes UTIs in populations of all ages (339, 340). S. saprophyticus has several adhesins that contribute to virulence, including SdrI and the serine-rich repeat protein UafB (see adhesins section). It also secretes urease, an enzyme that hydrolyzes urea to carbon dioxide and ammonia, which raises the pH of the urinary tract and can result in calculi formation. Urease was shown to be a virulence factor in a rat model of S. saprophyticus UTI (341). Chemical inhibition of urease was able to prevent the pH change caused by S. saprophyticus growth in an artificial urine medium, although the effectiveness of this as a treatment has yet to be tested in vivo (342).
CONCLUSIONS AND FUTURE PERSPECTIVES
The goal of this review on Staphylococcal biofilms is to summarize the latest literature on the function of adhesins, regulatory cascades, and the host response to these biofilms, with a focus on the noted pathogens S. aureus and S. epidermidis, and also coverage on other CoNS biofilms. The significant body of work available indicates that there are numerous mechanisms to assemble a mature Staphylococcal biofilm, and these structures vary depending on the substratum, the adhesins particular Staphylococci express, and the matrix materials available. Within a biofilm infection, the host niche clearly also has an important role in biofilm matrix composition and impacting the regulatory pathways controlling expression of Staphylococcal biofilm factors.
A number of recent studies provide convincing evidence that the Staphylococcal biofilm cells are equipped to thwart the host immune response, making them more resistant to the host immune system than planktonic cells. Recent findings have also shown that Staphylococcal biofilms contain heterogeneous populations, with a subset of cells contributing to the development of antibiotic resistance. In several bacteria, the phenomenon of persister cells has been observed, which are a small portion of a bacterial population that remains following antibiotic treatment (343, 344). S. aureus persisters were first observed in a murine model of deep wound infection. Treatment with vancomycin killed 99% of S. aureus cells, while the remaining 1% continued to be unaffected by vancomycin even after another day of treatment, suggesting a persister population (345). Other studies have demonstrated that the appearance of heterogeneous populations can develop within a S. aureus biofilm (227), and further studies have shown these sub populations can interact to promote vancomycin-intermediate resistance phenotypes (346). Clearly, biofilms are a diverse population with varying phenotypic properties that can contribute to the progress of an infection in complex ways.
Despite all the advancements, much remains to be elucidated regarding the defined nature of the in vivo biofilm state. Although the term “biofilm” is broadly applied to various growth states ranging from benign skin colonization to endocarditis, the universal qualities among these that specifically define biofilm characteristics are not clear, making it challenging at times to compare results across studies. There have been efforts to identify universal biomarkers of a Staphylococcal biofilm (9), as well as clarify the roles of virulence determinants that are unique to certain biofilm infection types, but there is still a pressing need for more studies in this direction to standardize the field. In part, researchers themselves have created this dilemma by trying to link every Staphylococcal growth state to a biofilm without considering the limitations of such a diverse umbrella. Attempts have been made to reign in the enthusiasm by trying to keep certain growth states separate and uniquely defined, such as colonization (347), but the popularity of “biofilms” keeps this terminology at the forefront of any literature on Staphylococcal communities. Further, with the growing literature on other community states, such as synovial aggregates (348), polysaccharide aggregates (349), and fibrinogen-based clumping (350), there is a growing need for investigations to compare and contrast properties of these states with classical biofilm features.
The study of Staphylococcal biofilm development has advanced much over the past decade, and we have endeavored in this review to cover many of the recent advances. In the future, biofilm studies will need to be extended to more host relevant conditions in order to properly model and understand the in vivo biofilm state. Too often, in vitro studies leave out host factors that can impact biofilm maturation in many different ways, and these conditions need to be considered when modeling biofilm development in vitro or in vivo. We need to properly understand all these different complexities to best position therapeutic development for treating biofilm infections.
Table 1.
Adhesins
| Family | Subfamily | Adhesin | Species | Ligand(s) | Refs |
|---|---|---|---|---|---|
| MSCRAMMs | |||||
| Clf-Sdr | |||||
| ClfA | S. aureus | Fibrinogen γ-chain | 45–56 | ||
| SdrG/Fbe | S. epidermidis | Fibrinogen β-chain | 57–64 | ||
| Fbl | S. lugdunensis | Fibrinogen γ-chain | 65–69 | ||
| ClfB | S. aureus | Fibrinogen α-chain, Cytokeratin 10, Cytokeratin 8, Loricrin | 45–48; 56; 70–79 | ||
| SdrC | S. aureus | β-neurexins, self-association | 80–82 | ||
| SdrD | S. aureus | Nasal epithelial cells | 80, 83, 84 | ||
| SdrE | S. aureus | Factor H, C4b-binding protein | 80; 85–87 | ||
| Bbp | S. epidermidis | Bone sialoprotein, C4b-binding protein | 87; 97–102 | ||
| SdrF | S. epidermidis | Type I collagen, Dacron | 88–90 | ||
| SdrG | S. epidermidis | Unknown | 91 | ||
| SdrH | S. epidermidis | Unknown | 91 | ||
| SdrX | S. capitis | Type VI collagen | 92 | ||
| SdrI | S. saprophyticus | Collagen, Fibronectin | 93–96 | ||
| FnBPs | |||||
| FnBPA | S. aureus | Fibrinogen, Elastin, Fibronectin | 103–113 | ||
| FnBPB | S. aureus | Fibrinogen, Elastin, Fibronectin | 103–113 | ||
| CNA | S. aureus | Collagen | 114–120 | ||
| NEAT Motif | |||||
| IsdA | S. aureus | Heme, fibrinogen, fibronectin | 122–125 | ||
| Three Helical Bundle | |||||
| SpA | S. aureus | Fc region of IgG | 37; 128–132 | ||
| G5-E | |||||
| SasG | S. aureus | Nasal epithelial cells, self-association | 133, 137, 138 | ||
| Aap | S. epidermidis | Corneocytes, self-association | 133–136; 138–140 |
Acknowledgments
A.E.P was funded by an American Heart Association Predoctoral Fellowship 14PRE19910005. Studies in the laboratory of A.R.H are supported by grant AI083211 (Project 3) from the National Institute of Allergy and Infectious Diseases.
References
- 1.Becker K, Heilmann C, Peters G. Coagulase-negative staphylococci. Clin Microbiol Rev. 2014;27:870–926. doi: 10.1128/CMR.00109-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lowy FD. Staphylococcus aureus infections. N Engl J Med. 1998;339:520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 3.Kiedrowski MR, Horswill AR. New approaches for treating staphylococcal biofilm infections. Ann N Y Acad Sci. 2011;1241:104–121. doi: 10.1111/j.1749-6632.2011.06281.x. [DOI] [PubMed] [Google Scholar]
- 4.Costerton JW, Cheng KJ, Geesey GG, Ladd TI, Nickel JC, Dasgupta M, Marrie TJ. Bacterial biofilms in nature and disease. Annual review of microbiology. 1987;41:435–464. doi: 10.1146/annurev.mi.41.100187.002251. [DOI] [PubMed] [Google Scholar]
- 5.Davies D. Understanding biofilm resistance to antibacterial agents. Nat Rev Drug Discov. 2003;2:114–122. doi: 10.1038/nrd1008. [DOI] [PubMed] [Google Scholar]
- 6.del Pozo JL, Patel R. The challenge of treating biofilm-associated bacterial infections. Clinical pharmacology and therapeutics. 2007;82:204–209. doi: 10.1038/sj.clpt.6100247. [DOI] [PubMed] [Google Scholar]
- 7.Jacqueline C, Caillon J. Impact of bacterial biofilm on the treatment of prosthetic joint infections. The Journal of antimicrobial chemotherapy. 2014;69(Suppl 1):i37–40. doi: 10.1093/jac/dku254. [DOI] [PubMed] [Google Scholar]
- 8.Sievert DM, Ricks P, Edwards JR, Schneider A, Patel J, Srinivasan A, Kallen A, Limbago B, Fridkin S. Antimicrobial-resistant pathogens associated with healthcare-associated infections: summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2009–2010. Infection control and hospital epidemiology. 2013;34:1–14. doi: 10.1086/668770. [DOI] [PubMed] [Google Scholar]
- 9.Secor PR, Jennings LK, James GA, Kirker KR, Pulcini ED, McInnerney K, Gerlach R, Livinghouse T, Hilmer JK, Bothner B, Fleckman P, Olerud JE, Stewart PS. Phevalin (aureusimine B) production by Staphylococcus aureus biofilm and impacts on human keratinocyte gene expression. PloS one. 2012;7:e40973. doi: 10.1371/journal.pone.0040973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zimmerli W, Trampuz A, Ochsner PE. Prosthetic-joint infections. N Engl J Med. 2004;351:1645–1654. doi: 10.1056/NEJMra040181. [DOI] [PubMed] [Google Scholar]
- 11.Parsek MR, Singh PK. Bacterial biofilms: an emerging link to disease pathogenesis. Annu Rev Microbiol. 2003;57:677–701. doi: 10.1146/annurev.micro.57.030502.090720. [DOI] [PubMed] [Google Scholar]
- 12.Brady RA, Leid JG, Calhoun JH, Costerton JW, Shirtliff ME. Osteomyelitis and the role of biofilms in chronic infection. FEMS immunology and medical microbiology. 2008;52:13–22. doi: 10.1111/j.1574-695X.2007.00357.x. [DOI] [PubMed] [Google Scholar]
- 13.Percival SL, Emanuel C, Cutting KF, Williams DW. Microbiology of the skin and the role of biofilms in infection. International wound journal. 2012;9:14–32. doi: 10.1111/j.1742-481X.2011.00836.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iwase T, Uehara Y, Shinji H, Tajima A, Seo H, Takada K, Agata T, Mizunoe Y. Staphylococcus epidermidis Esp inhibits Staphylococcus aureus biofilm formation and nasal colonization. Nature. 2010;465:346–349. doi: 10.1038/nature09074. [DOI] [PubMed] [Google Scholar]
- 15.Goss CH, Muhlebach MS. Review: Staphylococcus aureus and MRSA in cystic fibrosis. Journal of cystic fibrosis : official journal of the European Cystic Fibrosis Society. 2011;10:298–306. doi: 10.1016/j.jcf.2011.06.002. [DOI] [PubMed] [Google Scholar]
- 16.Trautner BW, Darouiche RO. Role of biofilm in catheter-associated urinary tract infection. American journal of infection control. 2004;32:177–183. doi: 10.1016/j.ajic.2003.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheng AG, DeDent AC, Schneewind O, Missiakas D. A play in four acts: Staphylococcus aureus abscess formation. Trends in microbiology. 2011;19:225–232. doi: 10.1016/j.tim.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kobayashi SD, Malachowa N, Whitney AR, Braughton KR, Gardner DJ, Long D, Bubeck Wardenburg J, Schneewind O, Otto M, Deleo FR. Comparative analysis of USA300 virulence determinants in a rabbit model of skin and soft tissue infection. The Journal of infectious diseases. 2011;204:937–941. doi: 10.1093/infdis/jir441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Salgado-Pabon W, Breshears L, Spaulding AR, Merriman JA, Stach CS, Horswill AR, Peterson ML, Schlievert PM. Superantigens are critical for Staphylococcus aureus Infective endocarditis, sepsis, and acute kidney injury. mBio. 2013:4. doi: 10.1128/mBio.00494-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nowakowska J, Landmann R, Khanna N. Foreign Body Infection Models to Study Host-Pathogen Response and Antimicrobial Tolerance of Bacterial Biofilm. Antibiotics. 2014;3:378–397. doi: 10.3390/antibiotics3030378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheung AL, Eberhardt KJ, Chung E, Yeaman MR, Sullam PM, Ramos M, Bayer AS. Diminished virulence of a sar-/agr- mutant of Staphylococcus aureus in the rabbit model of endocarditis. J Clin Invest. 1994;94:1815–1822. doi: 10.1172/JCI117530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gillaspy AF, Hickmon SG, Skinner RA, Thomas JR, Nelson CL, Smeltzer MS. Role of the accessory gene regulator (agr) in pathogenesis of staphylococcal osteomyelitis. Infection and immunity. 1995;63:3373–3380. doi: 10.1128/iai.63.9.3373-3380.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Periasamy S, Joo HS, Duong AC, Bach TH, Tan VY, Chatterjee SS, Cheung GY, Otto M. How Staphylococcus aureus biofilms develop their characteristic structure. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:1281–1286. doi: 10.1073/pnas.1115006109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vuong C, Kocianova S, Yao Y, Carmody AB, Otto M. Increased colonization of indwelling medical devices by quorum-sensing mutants of Staphylococcus epidermidis in vivo. J Infect Dis. 2004;190:1498–1505. doi: 10.1086/424487. [DOI] [PubMed] [Google Scholar]
- 25.Lister JL, Horswill AR. Staphylococcus aureus biofilms: recent developments in biofilm dispersal. Frontiers in cellular and infection microbiology. 2014;4:178. doi: 10.3389/fcimb.2014.00178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaplan JB. Biofilm dispersal: mechanisms, clinical implications, and potential therapeutic uses. J Dent Res. 2010;89:205–218. doi: 10.1177/0022034509359403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boles BR, Horswill AR. Staphylococcal biofilm disassembly. Trends in microbiology. 2011;19:449–455. doi: 10.1016/j.tim.2011.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moormeier DE, Bose JL, Horswill AR, Bayles KW. Temporal and stochastic control of Staphylococcus aureus biofilm development. mBio. 2014;5:e01341–01314. doi: 10.1128/mBio.01341-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hobley L, Harkins C, MacPhee CE, Stanley-Wall NR. Giving structure to the biofilm matrix: an overview of individual strategies and emerging common themes. FEMS microbiology reviews. 2015 doi: 10.1093/femsre/fuv015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Montanaro L, Poggi A, Visai L, Ravaioli S, Campoccia D, Speziale P, Arciola CR. Extracellular DNA in biofilms. The International journal of artificial organs. 2011;34:824–831. doi: 10.5301/ijao.5000051. [DOI] [PubMed] [Google Scholar]
- 31.Mann EE, Rice KC, Boles BR, Endres JL, Ranjit D, Chandramohan L, Tsang LH, Smeltzer MS, Horswill AR, Bayles KW. Modulation of eDNA release and degradation affects Staphylococcus aureus biofilm maturation. PloS one. 2009;4:e5822. doi: 10.1371/journal.pone.0005822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Speziale P, Pietrocola G, Foster TJ, Geoghegan JA. Protein-based biofilm matrices in Staphylococci. Frontiers in cellular and infection microbiology. 2014;4:171. doi: 10.3389/fcimb.2014.00171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Foulston L, Elsholz AK, DeFrancesco AS, Losick R. The extracellular matrix of Staphylococcus aureus biofilms comprises cytoplasmic proteins that associate with the cell surface in response to decreasing pH. mBio. 2014;5:e01667–01614. doi: 10.1128/mBio.01667-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Otto M. Staphylococcal infections: mechanisms of biofilm maturation and detachment as critical determinants of pathogenicity. Annu Rev Med. 2013;64:175–188. doi: 10.1146/annurev-med-042711-140023. [DOI] [PubMed] [Google Scholar]
- 35.Jabbouri S, Sadovskaya I. Characteristics of the biofilm matrix and its role as a possible target for the detection and eradication of Staphylococcus epidermidis associated with medical implant infections. FEMS immunology and medical microbiology. 2010;59:280–291. doi: 10.1111/j.1574-695X.2010.00695.x. [DOI] [PubMed] [Google Scholar]
- 36.Heilmann C. Adhesion mechanisms of staphylococci. Advances in experimental medicine and biology. 2011;715:105–123. doi: 10.1007/978-94-007-0940-9_7. [DOI] [PubMed] [Google Scholar]
- 37.Becker S, Frankel MB, Schneewind O, Missiakas D. Release of protein A from the cell wall of Staphylococcus aureus. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:1574–1579. doi: 10.1073/pnas.1317181111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bowden MG, Chen W, Singvall J, Xu Y, Peacock SJ, Valtulina V, Speziale P, Hook M. Identification and preliminary characterization of cell-wall-anchored proteins of Staphylococcus epidermidis. Microbiology. 2005;151:1453–1464. doi: 10.1099/mic.0.27534-0. [DOI] [PubMed] [Google Scholar]
- 39.Heilbronner S, Holden MT, van Tonder A, Geoghegan JA, Foster TJ, Parkhill J, Bentley SD. Genome sequence of Staphylococcus lugdunensis N920143 allows identification of putative colonization and virulence factors. FEMS microbiology letters. 2011;322:60–67. doi: 10.1111/j.1574-6968.2011.02339.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Foster TJ, Geoghegan JA, Ganesh VK, Hook M. Adhesion, invasion and evasion: the many functions of the surface proteins of Staphylococcus aureus. Nat Rev Microbiol. 2014;12:49–62. doi: 10.1038/nrmicro3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mazmanian SK, Liu G, Ton-That H, Schneewind O. Staphylococcus aureus sortase, an enzyme that anchors surface proteins to the cell wall. Science. 1999;285:760–763. doi: 10.1126/science.285.5428.760. [DOI] [PubMed] [Google Scholar]
- 42.Patti JM, Allen BL, McGavin MJ, Hook M. MSCRAMM-mediated adherence of microorganisms to host tissues. Annual review of microbiology. 1994;48:585–617. doi: 10.1146/annurev.mi.48.100194.003101. [DOI] [PubMed] [Google Scholar]
- 43.Cardile AP, Sanchez CJ, Jr, Samberg ME, Romano DR, Hardy SK, Wenke JC, Murray CK, Akers KS. Human plasma enhances the expression of Staphylococcal microbial surface components recognizing adhesive matrix molecules promoting biofilm formation and increases antimicrobial tolerance In Vitro. BMC Res Notes. 2014;7:457. doi: 10.1186/1756-0500-7-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Otto M. Virulence factors of the coagulase-negative staphylococci. Front Biosci. 2004;9:841–863. doi: 10.2741/1295. [DOI] [PubMed] [Google Scholar]
- 45.Resch A, Rosenstein R, Nerz C, Gotz F. Differential gene expression profiling of Staphylococcus aureus cultivated under biofilm and planktonic conditions. Applied and environmental microbiology. 2005;71:2663–2676. doi: 10.1128/AEM.71.5.2663-2676.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xue T, You Y, Shang F, Sun B. Rot and Agr system modulate fibrinogen-binding ability mainly by regulating clfB expression in Staphylococcus aureus NCTC8325. Med Microbiol Immunol. 2012;201:81–92. doi: 10.1007/s00430-011-0208-z. [DOI] [PubMed] [Google Scholar]
- 47.Hartford O, Francois P, Vaudaux P, Foster TJ. The dipeptide repeat region of the fibrinogen-binding protein (clumping factor) is required for functional expression of the fibrinogen-binding domain on the Staphylococcus aureus cell surface. Molecular microbiology. 1997;25:1065–1076. doi: 10.1046/j.1365-2958.1997.5291896.x. [DOI] [PubMed] [Google Scholar]
- 48.Ni Eidhin D, Perkins S, Francois P, Vaudaux P, Hook M, Foster TJ. Clumping factor B (ClfB), a new surface-located fibrinogen-binding adhesin of Staphylococcus aureus. Molecular microbiology. 1998;30:245–257. doi: 10.1046/j.1365-2958.1998.01050.x. [DOI] [PubMed] [Google Scholar]
- 49.McDevitt D, Francois P, Vaudaux P, Foster TJ. Molecular characterization of the clumping factor (fibrinogen receptor) of Staphylococcus aureus. Molecular microbiology. 1994;11:237–248. doi: 10.1111/j.1365-2958.1994.tb00304.x. [DOI] [PubMed] [Google Scholar]
- 50.McDevitt D, Nanavaty T, House-Pompeo K, Bell E, Turner N, McIntire L, Foster T, Hook M. Characterization of the interaction between the Staphylococcus aureus clumping factor (ClfA) and fibrinogen. Eur J Biochem. 1997;247:416–424. doi: 10.1111/j.1432-1033.1997.00416.x. [DOI] [PubMed] [Google Scholar]
- 51.Ganesh VK, Rivera JJ, Smeds E, Ko YP, Bowden MG, Wann ER, Gurusiddappa S, Fitzgerald JR, Hook M. A structural model of the Staphylococcus aureus ClfA-fibrinogen interaction opens new avenues for the design of anti-staphylococcal therapeutics. PLoS pathogens. 2008;4:e1000226. doi: 10.1371/journal.ppat.1000226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu CZ, Shih MH, Tsai PJ. ClfA(221–550), a fibrinogen-binding segment of Staphylococcus aureus clumping factor A, disrupts fibrinogen function. Thrombosis and haemostasis. 2005;94:286–294. doi: 10.1160/TH05-03-0205. [DOI] [PubMed] [Google Scholar]
- 53.Liu CZ, Huang TF, Tsai PJ, Tsai PJ, Chang LY, Chang MC. A segment of Staphylococcus aureus clumping factor A with fibrinogen-binding activity (ClfA221–550) inhibits platelet-plug formation in mice. Thromb Res. 2007;121:183–191. doi: 10.1016/j.thromres.2007.03.019. [DOI] [PubMed] [Google Scholar]
- 54.Flick MJ, Du X, Prasad JM, Raghu H, Palumbo JS, Smeds E, Hook M, Degen JL. Genetic elimination of the binding motif on fibrinogen for the S. aureus virulence factor ClfA improves host survival in septicemia. Blood. 2013;121:1783–1794. doi: 10.1182/blood-2012-09-453894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Moreillon P, Entenza JM, Francioli P, McDevitt D, Foster TJ, Francois P, Vaudaux P. Role of Staphylococcus aureus coagulase and clumping factor in pathogenesis of experimental endocarditis. Infection and immunity. 1995;63:4738–4743. doi: 10.1128/iai.63.12.4738-4743.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cheng AG, Kim HK, Burts ML, Krausz T, Schneewind O, Missiakas DM. Genetic requirements for Staphylococcus aureus abscess formation and persistence in host tissues. FASEB J. 2009;23:3393–3404. doi: 10.1096/fj.09-135467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nilsson M, Frykberg L, Flock JI, Pei L, Lindberg M, Guss B. A fibrinogen-binding protein of Staphylococcus epidermidis. Infection and immunity. 1998;66:2666–2673. doi: 10.1128/iai.66.6.2666-2673.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pei L, Palma M, Nilsson M, Guss B, Flock JI. Functional studies of a fibrinogen binding protein from Staphylococcus epidermidis. Infection and immunity. 1999;67:4525–4530. doi: 10.1128/iai.67.9.4525-4530.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ponnuraj K, Bowden MG, Davis S, Gurusiddappa S, Moore D, Choe D, Xu Y, Hook M, Narayana SV. A “dock, lock, and latch” structural model for a staphylococcal adhesin binding to fibrinogen. Cell. 2003;115:217–228. doi: 10.1016/s0092-8674(03)00809-2. [DOI] [PubMed] [Google Scholar]
- 60.Bowden MG, Heuck AP, Ponnuraj K, Kolosova E, Choe D, Gurusiddappa S, Narayana SV, Johnson AE, Hook M. Evidence for the “dock, lock, and latch” ligand binding mechanism of the staphylococcal microbial surface component recognizing adhesive matrix molecules (MSCRAMM) SdrG. The Journal of biological chemistry. 2008;283:638–647. doi: 10.1074/jbc.M706252200. [DOI] [PubMed] [Google Scholar]
- 61.Pei L, Flock JI. Lack of fbe, the gene for a fibrinogen-binding protein from Staphylococcus epidermidis, reduces its adherence to fibrinogen coated surfaces. Microb Pathog. 2001;31:185–193. doi: 10.1006/mpat.2001.0462. [DOI] [PubMed] [Google Scholar]
- 62.Hartford O, O’Brien L, Schofield K, Wells J, Foster TJ. The Fbe (SdrG) protein of Staphylococcus epidermidis HB promotes bacterial adherence to fibrinogen. Microbiology. 2001;147:2545–2552. doi: 10.1099/00221287-147-9-2545. [DOI] [PubMed] [Google Scholar]
- 63.Brennan MP, Loughman A, Devocelle M, Arasu S, Chubb AJ, Foster TJ, Cox D. Elucidating the role of Staphylococcus epidermidis serine-aspartate repeat protein G in platelet activation. J Thromb Haemost. 2009;7:1364–1372. doi: 10.1111/j.1538-7836.2009.03495.x. [DOI] [PubMed] [Google Scholar]
- 64.Guo B, Zhao X, Shi Y, Zhu D, Zhang Y. Pathogenic implication of a fibrinogen-binding protein of Staphylococcus epidermidis in a rat model of intravascular-catheter-associated infection. Infection and immunity. 2007;75:2991–2995. doi: 10.1128/IAI.01741-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nilsson M, Bjerketorp J, Guss B, Frykberg L. A fibrinogen-binding protein of Staphylococcus lugdunensis. FEMS microbiology letters. 2004;241:87–93. doi: 10.1016/j.femsle.2004.10.008. [DOI] [PubMed] [Google Scholar]
- 66.Mitchell J, Tristan A, Foster TJ. Characterization of the fibrinogen-binding surface protein Fbl of Staphylococcus lugdunensis. Microbiology. 2004;150:3831–3841. doi: 10.1099/mic.0.27337-0. [DOI] [PubMed] [Google Scholar]
- 67.Geoghegan JA, Ganesh VK, Smeds E, Liang X, Hook M, Foster TJ. Molecular characterization of the interaction of staphylococcal microbial surface components recognizing adhesive matrix molecules (MSCRAMM) ClfA and Fbl with fibrinogen. The Journal of biological chemistry. 2010;285:6208–6216. doi: 10.1074/jbc.M109.062208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pereira EM, Oliveira FL, Schuenck RP, Zoletti GO, Dos Santos KR. Detection of Staphylococcus lugdunensis by a new species-specific PCR based on the fbl gene. FEMS immunology and medical microbiology. 2010;58:295–298. doi: 10.1111/j.1574-695X.2009.00626.x. [DOI] [PubMed] [Google Scholar]
- 69.Chatzigeorgiou KS, Siafakas N, Petinaki E, Zerva L. fbl gene as a species-specific target for Staphylococcus lugdunensis identification. J Clin Lab Anal. 2010;24:119–122. doi: 10.1002/jcla.20352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ganesh VK, Barbu EM, Deivanayagam CC, Le B, Anderson AS, Matsuka YV, Lin SL, Foster TJ, Narayana SV, Hook M. Structural and biochemical characterization of Staphylococcus aureus clumping factor B/ligand interactions. The Journal of biological chemistry. 2011;286:25963–25972. doi: 10.1074/jbc.M110.217414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.McAleese FM, Walsh EJ, Sieprawska M, Potempa J, Foster TJ. Loss of clumping factor B fibrinogen binding activity by Staphylococcus aureus involves cessation of transcription, shedding and cleavage by metalloprotease. The Journal of biological chemistry. 2001;276:29969–29978. doi: 10.1074/jbc.M102389200. [DOI] [PubMed] [Google Scholar]
- 72.Perkins S, Walsh EJ, Deivanayagam CC, Narayana SV, Foster TJ, Hook M. Structural organization of the fibrinogen-binding region of the clumping factor B MSCRAMM of Staphylococcus aureus. The Journal of biological chemistry. 2001;276:44721–44728. doi: 10.1074/jbc.M106741200. [DOI] [PubMed] [Google Scholar]
- 73.Abraham NM, Jefferson KK. Staphylococcus aureus clumping factor B mediates biofilm formation in the absence of calcium. Microbiology. 2012;158:1504–1512. doi: 10.1099/mic.0.057018-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Entenza JM, Foster TJ, Ni Eidhin D, Vaudaux P, Francioli P, Moreillon P. Contribution of clumping factor B to pathogenesis of experimental endocarditis due to Staphylococcus aureus. Infection and immunity. 2000;68:5443–5446. doi: 10.1128/iai.68.9.5443-5446.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.O’Brien LM, Walsh EJ, Massey RC, Peacock SJ, Foster TJ. Staphylococcus aureus clumping factor B (ClfB) promotes adherence to human type I cytokeratin 10: implications for nasal colonization. Cell Microbiol. 2002;4:759–770. doi: 10.1046/j.1462-5822.2002.00231.x. [DOI] [PubMed] [Google Scholar]
- 76.Walsh EJ, O’Brien LM, Liang X, Hook M, Foster TJ. Clumping factor B, a fibrinogen-binding MSCRAMM (microbial surface components recognizing adhesive matrix molecules) adhesin of Staphylococcus aureus, also binds to the tail region of type I cytokeratin 10. The Journal of biological chemistry. 2004;279:50691–50699. doi: 10.1074/jbc.M408713200. [DOI] [PubMed] [Google Scholar]
- 77.Haim M, Trost A, Maier CJ, Achatz G, Feichtner S, Hintner H, Bauer JW, Onder K. Cytokeratin 8 interacts with clumping factor B: a new possible virulence factor target. Microbiology. 2010;156:3710–3721. doi: 10.1099/mic.0.034413-0. [DOI] [PubMed] [Google Scholar]
- 78.Mulcahy ME, Geoghegan JA, Monk IR, O’Keeffe KM, Walsh EJ, Foster TJ, McLoughlin RM. Nasal colonisation by Staphylococcus aureus depends upon clumping factor B binding to the squamous epithelial cell envelope protein loricrin. PLoS pathogens. 2012;8:e1003092. doi: 10.1371/journal.ppat.1003092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wertheim HF, Walsh E, Choudhurry R, Melles DC, Boelens HA, Miajlovic H, Verbrugh HA, Foster T, van Belkum A. Key role for clumping factor B in Staphylococcus aureus nasal colonization of humans. PLoS Med. 2008;5:e17. doi: 10.1371/journal.pmed.0050017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Josefsson E, McCrea KW, Ni Eidhin D, O’Connell D, Cox J, Hook M, Foster TJ. Three new members of the serine-aspartate repeat protein multigene family of Staphylococcus aureus. Microbiology. 1998;144 ( Pt 12):3387–3395. doi: 10.1099/00221287-144-12-3387. [DOI] [PubMed] [Google Scholar]
- 81.Barbu EM, Ganesh VK, Gurusiddappa S, Mackenzie RC, Foster TJ, Sudhof TC, Hook M. beta-Neurexin is a ligand for the Staphylococcus aureus MSCRAMM SdrC. PLoS pathogens. 2010;6:e1000726. doi: 10.1371/journal.ppat.1000726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Barbu EM, Mackenzie C, Foster TJ, Hook M. SdrC induces staphylococcal biofilm formation through a homophilic interaction. Molecular microbiology. 2014;94:172–185. doi: 10.1111/mmi.12750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Josefsson E, O’Connell D, Foster TJ, Durussel I, Cox JA. The binding of calcium to the B-repeat segment of SdrD, a cell surface protein of Staphylococcus aureus. The Journal of biological chemistry. 1998;273:31145–31152. doi: 10.1074/jbc.273.47.31145. [DOI] [PubMed] [Google Scholar]
- 84.Corrigan RM, Miajlovic H, Foster TJ. Surface proteins that promote adherence of Staphylococcus aureus to human desquamated nasal epithelial cells. BMC Microbiol. 2009;9:22. doi: 10.1186/1471-2180-9-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.O’Brien L, Kerrigan SW, Kaw G, Hogan M, Penades J, Litt D, Fitzgerald DJ, Foster TJ, Cox D. Multiple mechanisms for the activation of human platelet aggregation by Staphylococcus aureus: roles for the clumping factors ClfA and ClfB, the serine-aspartate repeat protein SdrE and protein A. Molecular microbiology. 2002;44:1033–1044. doi: 10.1046/j.1365-2958.2002.02935.x. [DOI] [PubMed] [Google Scholar]
- 86.Sharp JA, Echague CG, Hair PS, Ward MD, Nyalwidhe JO, Geoghegan JA, Foster TJ, Cunnion KM. Staphylococcus aureus surface protein SdrE binds complement regulator factor H as an immune evasion tactic. PloS one. 2012;7:e38407. doi: 10.1371/journal.pone.0038407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hair PS, Foley CK, Krishna NK, Nyalwidhe JO, Geoghegan JA, Foster TJ, Cunnion KM. Complement regulator C4BP binds to Staphylococcus aureus surface proteins SdrE and Bbp inhibiting bacterial opsonization and killing. Results Immunol. 2013;3:114–121. doi: 10.1016/j.rinim.2013.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Arrecubieta C, Lee MH, Macey A, Foster TJ, Lowy FD. SdrF, a Staphylococcus epidermidis surface protein, binds type I collagen. The Journal of biological chemistry. 2007;282:18767–18776. doi: 10.1074/jbc.M610940200. [DOI] [PubMed] [Google Scholar]
- 89.Arrecubieta C, Toba FA, von Bayern M, Akashi H, Deng MC, Naka Y, Lowy FD. SdrF, a Staphylococcus epidermidis surface protein, contributes to the initiation of ventricular assist device driveline-related infections. PLoS pathogens. 2009;5:e1000411. doi: 10.1371/journal.ppat.1000411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Toba FA, Visai L, Trivedi S, Lowy FD. The role of ionic interactions in the adherence of the Staphylococcus epidermidis adhesin SdrF to prosthetic material. FEMS microbiology letters. 2013;338:24–30. doi: 10.1111/1574-6968.12018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.McCrea KW, Hartford O, Davis S, Eidhin DN, Lina G, Speziale P, Foster TJ, Hook M. The serine-aspartate repeat (Sdr) protein family in Staphylococcus epidermidis. Microbiology. 2000;146 ( Pt 7):1535–1546. doi: 10.1099/00221287-146-7-1535. [DOI] [PubMed] [Google Scholar]
- 92.Liu Y, Ames B, Gorovits E, Prater BD, Syribeys P, Vernachio JH, Patti JM. SdrX, a serine-aspartate repeat protein expressed by Staphylococcus capitis with collagen VI binding activity. Infection and immunity. 2004;72:6237–6244. doi: 10.1128/IAI.72.11.6237-6244.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sakinc T, Kleine B, Gatermann SG. SdrI, a serine-aspartate repeat protein identified in Staphylococcus saprophyticus strain 7108, is a collagen-binding protein. Infection and immunity. 2006;74:4615–4623. doi: 10.1128/IAI.01885-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sakinc T, Kleine B, Michalski N, Kaase M, Gatermann SG. SdrI of Staphylococcus saprophyticus is a multifunctional protein: localization of the fibronectin-binding site. FEMS microbiology letters. 2009;301:28–34. doi: 10.1111/j.1574-6968.2009.01798.x. [DOI] [PubMed] [Google Scholar]
- 95.Kleine B, Ali L, Wobser D, Sakiotanc T. The N-terminal repeat and the ligand binding domain A of SdrI protein is involved in hydrophobicity of S. saprophyticus. Microbiol Res. 2015;172:88–94. doi: 10.1016/j.micres.2014.11.003. [DOI] [PubMed] [Google Scholar]
- 96.Kline KA, Ingersoll MA, Nielsen HV, Sakinc T, Henriques-Normark B, Gatermann S, Caparon MG, Hultgren SJ. Characterization of a novel murine model of Staphylococcus saprophyticus urinary tract infection reveals roles for Ssp and SdrI in virulence. Infection and immunity. 2010;78:1943–1951. doi: 10.1128/IAI.01235-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ryden C, Yacoub AI, Maxe I, Heinegard D, Oldberg A, Franzen A, Ljungh A, Rubin K. Specific binding of bone sialoprotein to Staphylococcus aureus isolated from patients with osteomyelitis. Eur J Biochem. 1989;184:331–336. doi: 10.1111/j.1432-1033.1989.tb15023.x. [DOI] [PubMed] [Google Scholar]
- 98.Yacoub A, Lindahl P, Rubin K, Wendel M, Heinegard D, Ryden C. Purification of a bone sialoprotein-binding protein from Staphylococcus aureus. Eur J Biochem. 1994;222:919–925. doi: 10.1111/j.1432-1033.1994.tb18940.x. [DOI] [PubMed] [Google Scholar]
- 99.Tung H, Guss B, Hellman U, Persson L, Rubin K, Ryden C. A bone sialoprotein-binding protein from Staphylococcus aureus: a member of the staphylococcal Sdr family. Biochem J. 2000;345(Pt 3):611–619. [PMC free article] [PubMed] [Google Scholar]
- 100.Aamot HV, Blomfeldt A, Skramm I, Muller F, Monecke S. Molecular characterisation of methicillin-sensitive Staphylococcus aureus from deep surgical site infections in orthopaedic patients. European journal of clinical microbiology & infectious diseases : official publication of the European Society of Clinical Microbiology. 2012;31:1999–2004. doi: 10.1007/s10096-011-1532-3. [DOI] [PubMed] [Google Scholar]
- 101.Wisniewska K, Piorkowska A, Kasprzyk J, Bronk M, Swiec K. Clonal distribution of bone sialoprotein-binding protein gene among Staphylococcus aureus isolates associated with bloodstream infections. Folia Microbiol (Praha) 2014;59:465–471. doi: 10.1007/s12223-014-0321-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Persson L, Johansson C, Ryden C. Antibodies to Staphylococcus aureus bone sialoprotein-binding protein indicate infectious osteomyelitis. Clin Vaccine Immunol. 2009;16:949–952. doi: 10.1128/CVI.00442-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Jonsson K, Signas C, Muller HP, Lindberg M. Two different genes encode fibronectin binding proteins in Staphylococcus aureus. The complete nucleotide sequence and characterization of the second gene. Eur J Biochem. 1991;202:1041–1048. doi: 10.1111/j.1432-1033.1991.tb16468.x. [DOI] [PubMed] [Google Scholar]
- 104.Roche FM, Downer R, Keane F, Speziale P, Park PW, Foster TJ. The N-terminal A domain of fibronectin-binding proteins A and B promotes adhesion of Staphylococcus aureus to elastin. The Journal of biological chemistry. 2004;279:38433–38440. doi: 10.1074/jbc.M402122200. [DOI] [PubMed] [Google Scholar]
- 105.Keane FM, Loughman A, Valtulina V, Brennan M, Speziale P, Foster TJ. Fibrinogen and elastin bind to the same region within the A domain of fibronectin binding protein A, an MSCRAMM of Staphylococcus aureus. Molecular microbiology. 2007;63:711–723. doi: 10.1111/j.1365-2958.2006.05552.x. [DOI] [PubMed] [Google Scholar]
- 106.Schwarz-Linek U, Werner JM, Pickford AR, Gurusiddappa S, Kim JH, Pilka ES, Briggs JA, Gough TS, Hook M, Campbell ID, Potts JR. Pathogenic bacteria attach to human fibronectin through a tandem beta-zipper. Nature. 2003;423:177–181. doi: 10.1038/nature01589. [DOI] [PubMed] [Google Scholar]
- 107.Peacock SJ, Foster TJ, Cameron BJ, Berendt AR. Bacterial fibronectin-binding proteins and endothelial cell surface fibronectin mediate adherence of Staphylococcus aureus to resting human endothelial cells. Microbiology. 1999;145 ( Pt 12):3477–3486. doi: 10.1099/00221287-145-12-3477. [DOI] [PubMed] [Google Scholar]
- 108.Sinha B, Francois PP, Nusse O, Foti M, Hartford OM, Vaudaux P, Foster TJ, Lew DP, Herrmann M, Krause KH. Fibronectin-binding protein acts as Staphylococcus aureus invasin via fibronectin bridging to integrin alpha5beta1. Cell Microbiol. 1999;1:101–117. doi: 10.1046/j.1462-5822.1999.00011.x. [DOI] [PubMed] [Google Scholar]
- 109.Edwards AM, Potter U, Meenan NA, Potts JR, Massey RC. Staphylococcus aureus keratinocyte invasion is dependent upon multiple high-affinity fibronectin-binding repeats within FnBPA. PloS one. 2011;6:e18899. doi: 10.1371/journal.pone.0018899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.McCourt J, O’Halloran DP, McCarthy H, O’Gara JP, Geoghegan JA. Fibronectin-binding proteins are required for biofilm formation by community-associated methicillin-resistant Staphylococcus aureus strain LAC. FEMS microbiology letters. 2014;353:157–164. doi: 10.1111/1574-6968.12424. [DOI] [PubMed] [Google Scholar]
- 111.Geoghegan JA, Monk IR, O’Gara JP, Foster TJ. Subdomains N2N3 of fibronectin binding protein A mediate Staphylococcus aureus biofilm formation and adherence to fibrinogen using distinct mechanisms. Journal of bacteriology. 2013;195:2675–2683. doi: 10.1128/JB.02128-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Herman-Bausier P, El-Kirat-Chatel S, Foster TJ, Geoghegan JA, Dufrene YF. Staphylococcus aureus Fibronectin-Binding Protein A Mediates Cell-Cell Adhesion through Low-Affinity Homophilic Bonds. mBio. 2015:6. doi: 10.1128/mBio.00413-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Menzies BE. The role of fibronectin binding proteins in the pathogenesis of Staphylococcus aureus infections. Curr Opin Infect Dis. 2003;16:225–229. doi: 10.1097/00001432-200306000-00007. [DOI] [PubMed] [Google Scholar]
- 114.Patti JM, Boles JO, Hook M. Identification and biochemical characterization of the ligand binding domain of the collagen adhesin from Staphylococcus aureus. Biochemistry. 1993;32:11428–11435. doi: 10.1021/bi00093a021. [DOI] [PubMed] [Google Scholar]
- 115.Zong Y, Xu Y, Liang X, Keene DR, Hook A, Gurusiddappa S, Hook M, Narayana SV. A ‘Collagen Hug’ model for Staphylococcus aureus CNA binding to collagen. EMBO J. 2005;24:4224–4236. doi: 10.1038/sj.emboj.7600888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kang M, Ko YP, Liang X, Ross CL, Liu Q, Murray BE, Hook M. Collagen-binding microbial surface components recognizing adhesive matrix molecule (MSCRAMM) of Gram-positive bacteria inhibit complement activation via the classical pathway. The Journal of biological chemistry. 2013;288:20520–20531. doi: 10.1074/jbc.M113.454462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Rhem MN, Lech EM, Patti JM, McDevitt D, Hook M, Jones DB, Wilhelmus KR. The collagen-binding adhesin is a virulence factor in Staphylococcus aureus keratitis. Infection and immunity. 2000;68:3776–3779. doi: 10.1128/iai.68.6.3776-3779.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Elasri MO, Thomas JR, Skinner RA, Blevins JS, Beenken KE, Nelson CL, Smeltzer MS. Staphylococcus aureus collagen adhesin contributes to the pathogenesis of osteomyelitis. Bone. 2002;30:275–280. doi: 10.1016/s8756-3282(01)00632-9. [DOI] [PubMed] [Google Scholar]
- 119.Xu Y, Rivas JM, Brown EL, Liang X, Hook M. Virulence potential of the staphylococcal adhesin CNA in experimental arthritis is determined by its affinity for collagen. The Journal of infectious diseases. 2004;189:2323–2333. doi: 10.1086/420851. [DOI] [PubMed] [Google Scholar]
- 120.Hienz SA, Schennings T, Heimdahl A, Flock JI. Collagen binding of Staphylococcus aureus is a virulence factor in experimental endocarditis. The Journal of infectious diseases. 1996;174:83–88. doi: 10.1093/infdis/174.1.83. [DOI] [PubMed] [Google Scholar]
- 121.Bowden MG, Visai L, Longshaw CM, Holland KT, Speziale P, Hook M. Is the GehD lipase from Staphylococcus epidermidis a collagen binding adhesin? The Journal of biological chemistry. 2002;277:43017–43023. doi: 10.1074/jbc.M207921200. [DOI] [PubMed] [Google Scholar]
- 122.Hammer ND, Skaar EP. Molecular mechanisms of Staphylococcus aureus iron acquisition. Annual review of microbiology. 2011;65:129–147. doi: 10.1146/annurev-micro-090110-102851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Pluym M, Muryoi N, Heinrichs DE, Stillman MJ. Heme binding in the NEAT domains of IsdA and IsdC of Staphylococcus aureus. J Inorg Biochem. 2008;102:480–488. doi: 10.1016/j.jinorgbio.2007.11.011. [DOI] [PubMed] [Google Scholar]
- 124.Clarke SR, Mohamed R, Bian L, Routh AF, Kokai-Kun JF, Mond JJ, Tarkowski A, Foster SJ. The Staphylococcus aureus surface protein IsdA mediates resistance to innate defenses of human skin. Cell host & microbe. 2007;1:199–212. doi: 10.1016/j.chom.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 125.Clarke SR, Wiltshire MD, Foster SJ. IsdA of Staphylococcus aureus is a broad spectrum, iron-regulated adhesin. Molecular microbiology. 2004;51:1509–1519. doi: 10.1111/j.1365-2958.2003.03938.x. [DOI] [PubMed] [Google Scholar]
- 126.Pishchany G, Dickey SE, Skaar EP. Subcellular localization of the Staphylococcus aureus heme iron transport components IsdA and IsdB. Infection and immunity. 2009;77:2624–2634. doi: 10.1128/IAI.01531-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Missineo A, Di Poto A, Geoghegan JA, Rindi S, Heilbronner S, Gianotti V, Arciola CR, Foster TJ, Speziale P, Pietrocola G. IsdC from Staphylococcus lugdunensis induces biofilm formation under low-iron growth conditions. Infection and immunity. 2014;82:2448–2459. doi: 10.1128/IAI.01542-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Shopsin B, Gomez M, Montgomery SO, Smith DH, Waddington M, Dodge DE, Bost DA, Riehman M, Naidich S, Kreiswirth BN. Evaluation of protein A gene polymorphic region DNA sequencing for typing of Staphylococcus aureus strains. Journal of clinical microbiology. 1999;37:3556–3563. doi: 10.1128/jcm.37.11.3556-3563.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Moks T, Abrahmsen L, Nilsson B, Hellman U, Sjoquist J, Uhlen M. Staphylococcal protein A consists of five IgG-binding domains. Eur J Biochem. 1986;156:637–643. doi: 10.1111/j.1432-1033.1986.tb09625.x. [DOI] [PubMed] [Google Scholar]
- 130.Falugi F, Kim HK, Missiakas DM, Schneewind O. Role of protein A in the evasion of host adaptive immune responses by Staphylococcus aureus. mBio. 2013;4:e00575–00513. doi: 10.1128/mBio.00575-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Merino N, Toledo-Arana A, Vergara-Irigaray M, Valle J, Solano C, Calvo E, Lopez JA, Foster TJ, Penades JR, Lasa I. Protein A-mediated multicellular behavior in Staphylococcus aureus. Journal of bacteriology. 2009;191:832–843. doi: 10.1128/JB.01222-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.O’Halloran DP, Wynne K, Geoghegan JA. Protein A Is Released into the Staphylococcus aureus Culture Supernatant with an Unprocessed Sorting Signal. Infection and immunity. 2015;83:1598–1609. doi: 10.1128/IAI.03122-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Conrady DG, Wilson JJ, Herr AB. Structural basis for Zn2+-dependent intercellular adhesion in staphylococcal biofilms. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:E202–211. doi: 10.1073/pnas.1208134110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Schaeffer CR, Woods KM, Longo GM, Kiedrowski MR, Paharik AE, Buttner H, Christner M, Boissy RJ, Horswill AR, Rohde H, Fey PD. Accumulation-associated protein enhances Staphylococcus epidermidis biofilm formation under dynamic conditions and is required for infection in a rat catheter model. Infection and immunity. 2015;83:214–226. doi: 10.1128/IAI.02177-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Conlon BP, Geoghegan JA, Waters EM, McCarthy H, Rowe SE, Davies JR, Schaeffer CR, Foster TJ, Fey PD, O’Gara JP. Role for the A domain of unprocessed accumulation-associated protein (Aap) in the attachment phase of the Staphylococcus epidermidis biofilm phenotype. Journal of bacteriology. 2014;196:4268–4275. doi: 10.1128/JB.01946-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Macintosh RL, Brittan JL, Bhattacharya R, Jenkinson HF, Derrick J, Upton M, Handley PS. The terminal A domain of the fibrillar accumulation-associated protein (Aap) of Staphylococcus epidermidis mediates adhesion to human corneocytes. Journal of bacteriology. 2009;191:7007–7016. doi: 10.1128/JB.00764-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Roche FM, Meehan M, Foster TJ. The Staphylococcus aureus surface protein SasG and its homologues promote bacterial adherence to human desquamated nasal epithelial cells. Microbiology. 2003;149:2759–2767. doi: 10.1099/mic.0.26412-0. [DOI] [PubMed] [Google Scholar]
- 138.Conrady DG, Brescia CC, Horii K, Weiss AA, Hassett DJ, Herr AB. A zinc-dependent adhesion module is responsible for intercellular adhesion in staphylococcal biofilms. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:19456–19461. doi: 10.1073/pnas.0807717105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Rohde H, Burdelski C, Bartscht K, Hussain M, Buck F, Horstkotte MA, Knobloch JK, Heilmann C, Herrmann M, Mack D. Induction of Staphylococcus epidermidis biofilm formation via proteolytic processing of the accumulation-associated protein by staphylococcal and host proteases. Molecular microbiology. 2005;55:1883–1895. doi: 10.1111/j.1365-2958.2005.04515.x. [DOI] [PubMed] [Google Scholar]
- 140.Decker R, Burdelski C, Zobiak M, Buttner H, Franke G, Christner M, Sass K, Zobiak B, Henke HA, Horswill AR, Bischoff M, Bur S, Hartmann T, Schaeffer CR, Fey PD, Rohde H. An 18 kDa scaffold protein is critical for Staphylococcus epidermidis biofilm formation. PLoS pathogens. 2015;11:e1004735. doi: 10.1371/journal.ppat.1004735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Cucarella C, Solano C, Valle J, Amorena B, Lasa I, Penades JR. Bap, a Staphylococcus aureus surface protein involved in biofilm formation. Journal of bacteriology. 2001;183:2888–2896. doi: 10.1128/JB.183.9.2888-2896.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Tormo MA, Knecht E, Gotz F, Lasa I, Penades JR. Bap-dependent biofilm formation by pathogenic species of Staphylococcus: evidence of horizontal gene transfer? Microbiology. 2005;151:2465–2475. doi: 10.1099/mic.0.27865-0. [DOI] [PubMed] [Google Scholar]
- 143.Valle J, Latasa C, Gil C, Toledo-Arana A, Solano C, Penades JR, Lasa I. Bap, a biofilm matrix protein of Staphylococcus aureus prevents cellular internalization through binding to GP96 host receptor. PLoS pathogens. 2012;8:e1002843. doi: 10.1371/journal.ppat.1002843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Bensing BA, Sullam PM. An accessory sec locus of Streptococcus gordonii is required for export of the surface protein GspB and for normal levels of binding to human platelets. Molecular microbiology. 2002;44:1081–1094. doi: 10.1046/j.1365-2958.2002.02949.x. [DOI] [PubMed] [Google Scholar]
- 145.Lizcano A, Sanchez CJ, Orihuela CJ. A role for glycosylated serine-rich repeat proteins in gram-positive bacterial pathogenesis. Mol Oral Microbiol. 2012;27:257–269. doi: 10.1111/j.2041-1014.2012.00653.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Zhang YQ, Ren SX, Li HL, Wang YX, Fu G, Yang J, Qin ZQ, Miao YG, Wang WY, Chen RS, Shen Y, Chen Z, Yuan ZH, Zhao GP, Qu D, Danchin A, Wen YM. Genome-based analysis of virulence genes in a non-biofilm-forming Staphylococcus epidermidis strain (ATCC 12228) Molecular microbiology. 2003;49:1577–1593. doi: 10.1046/j.1365-2958.2003.03671.x. [DOI] [PubMed] [Google Scholar]
- 147.Takeuchi F, Watanabe S, Baba T, Yuzawa H, Ito T, Morimoto Y, Kuroda M, Cui L, Takahashi M, Ankai A, Baba S, Fukui S, Lee JC, Hiramatsu K. Whole-genome sequencing of staphylococcus haemolyticus uncovers the extreme plasticity of its genome and the evolution of human-colonizing staphylococcal species. Journal of bacteriology. 2005;187:7292–7308. doi: 10.1128/JB.187.21.7292-7308.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Sanchez CJ, Shivshankar P, Stol K, Trakhtenbroit S, Sullam PM, Sauer K, Hermans PW, Orihuela CJ. The pneumococcal serine-rich repeat protein is an intra-species bacterial adhesin that promotes bacterial aggregation in vivo and in biofilms. PLoS pathogens. 2010;6:e1001044. doi: 10.1371/journal.ppat.1001044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Yang YH, Jiang YL, Zhang J, Wang L, Bai XH, Zhang SJ, Ren YM, Li N, Zhang YH, Zhang Z, Gong Q, Mei Y, Xue T, Zhang JR, Chen Y, Zhou CZ. Structural insights into SraP-mediated Staphylococcus aureus adhesion to host cells. PLoS pathogens. 2014;10:e1004169. doi: 10.1371/journal.ppat.1004169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.King NP, Beatson SA, Totsika M, Ulett GC, Alm RA, Manning PA, Schembri MA. UafB is a serine-rich repeat adhesin of Staphylococcus saprophyticus that mediates binding to fibronectin, fibrinogen and human uroepithelial cells. Microbiology. 2011;157:1161–1175. doi: 10.1099/mic.0.047639-0. [DOI] [PubMed] [Google Scholar]
- 151.Li M, Du X, Villaruz AE, Diep BA, Wang D, Song Y, Tian Y, Hu J, Yu F, Lu Y, Otto M. MRSA epidemic linked to a quickly spreading colonization and virulence determinant. Nat Med. 2012;18:816–819. doi: 10.1038/nm.2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Liu Q, Du X, Hong X, Li T, Zheng B, He L, Wang Y, Otto M, Li M. Targeting Surface Protein SasX by Active and Passive Vaccination To Reduce Staphylococcus aureus Colonization and Infection. Infection and immunity. 2015;83:2168–2174. doi: 10.1128/IAI.02951-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Schroeder K, Jularic M, Horsburgh SM, Hirschhausen N, Neumann C, Bertling A, Schulte A, Foster S, Kehrel BE, Peters G, Heilmann C. Molecular characterization of a novel Staphylococcus aureus surface protein (SasC) involved in cell aggregation and biofilm accumulation. PloS one. 2009;4:e7567. doi: 10.1371/journal.pone.0007567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Roche FM, Massey R, Peacock SJ, Day NP, Visai L, Speziale P, Lam A, Pallen M, Foster TJ. Characterization of novel LPXTG-containing proteins of Staphylococcus aureus identified from genome sequences. Microbiology. 2003;149:643–654. doi: 10.1099/mic.0.25996-0. [DOI] [PubMed] [Google Scholar]
- 155.Heilmann C, Hussain M, Peters G, Gotz F. Evidence for autolysin-mediated primary attachment of Staphylococcus epidermidis to a polystyrene surface. Molecular microbiology. 1997;24:1013–1024. doi: 10.1046/j.1365-2958.1997.4101774.x. [DOI] [PubMed] [Google Scholar]
- 156.Takahashi J, Komatsuzawa H, Yamada S, Nishida T, Labischinski H, Fujiwara T, Ohara M, Yamagishi J, Sugai M. Molecular characterization of an atl null mutant of Staphylococcus aureus. Microbiol Immunol. 2002;46:601–612. doi: 10.1111/j.1348-0421.2002.tb02741.x. [DOI] [PubMed] [Google Scholar]
- 157.Biswas R, Voggu L, Simon UK, Hentschel P, Thumm G, Gotz F. Activity of the major staphylococcal autolysin Atl. FEMS microbiology letters. 2006;259:260–268. doi: 10.1111/j.1574-6968.2006.00281.x. [DOI] [PubMed] [Google Scholar]
- 158.Patel JD, Colton E, Ebert M, Anderson JM. Gene expression during S. epidermidis biofilm formation on biomaterials. J Biomed Mater Res A. 2012;100:2863–2869. doi: 10.1002/jbm.a.34221. [DOI] [PubMed] [Google Scholar]
- 159.Gross M, Cramton SE, Gotz F, Peschel A. Key role of teichoic acid net charge in Staphylococcus aureus colonization of artificial surfaces. Infection and immunity. 2001;69:3423–3426. doi: 10.1128/IAI.69.5.3423-3426.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Hussain M, Heilmann C, Peters G, Herrmann M. Teichoic acid enhances adhesion of Staphylococcus epidermidis to immobilized fibronectin. Microb Pathog. 2001;31:261–270. doi: 10.1006/mpat.2001.0469. [DOI] [PubMed] [Google Scholar]
- 161.Sadovskaya I, Vinogradov E, Flahaut S, Kogan G, Jabbouri S. Extracellular carbohydrate-containing polymers of a model biofilm-producing strain, Staphylococcus epidermidis RP62A. Infection and immunity. 2005;73:3007–3017. doi: 10.1128/IAI.73.5.3007-3017.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Vinogradov E, Sadovskaya I, Li J, Jabbouri S. Structural elucidation of the extracellular and cell-wall teichoic acids of Staphylococcus aureus MN8m, a biofilm forming strain. Carbohydr Res. 2006;341:738–743. doi: 10.1016/j.carres.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 163.Rupp ME, Ulphani JS, Fey PD, Bartscht K, Mack D. Characterization of the importance of polysaccharide intercellular adhesin/hemagglutinin of Staphylococcus epidermidis in the pathogenesis of biomaterial-based infection in a mouse foreign body infection model. Infection and immunity. 1999;67:2627–2632. doi: 10.1128/iai.67.5.2627-2632.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Vuong C, Kocianova S, Voyich JM, Yao Y, Fischer ER, DeLeo FR, Otto M. A crucial role for exopolysaccharide modification in bacterial biofilm formation, immune evasion, and virulence. The Journal of biological chemistry. 2004;279:54881–54886. doi: 10.1074/jbc.M411374200. [DOI] [PubMed] [Google Scholar]
- 165.Heilmann C, Schweitzer O, Gerke C, Vanittanakom N, Mack D, Gotz F. Molecular basis of intercellular adhesion in the biofilm-forming Staphylococcus epidermidis. Molecular microbiology. 1996;20:1083–1091. doi: 10.1111/j.1365-2958.1996.tb02548.x. [DOI] [PubMed] [Google Scholar]
- 166.O’Gara JP. ica and beyond: biofilm mechanisms and regulation in Staphylococcus epidermidis and Staphylococcus aureus. FEMS microbiology letters. 2007;270:179–188. doi: 10.1111/j.1574-6968.2007.00688.x. [DOI] [PubMed] [Google Scholar]
- 167.Rohde H, Frankenberger S, Zahringer U, Mack D. Structure, function and contribution of polysaccharide intercellular adhesin (PIA) to Staphylococcus epidermidis biofilm formation and pathogenesis of biomaterial-associated infections. Eur J Cell Biol. 2010;89:103–111. doi: 10.1016/j.ejcb.2009.10.005. [DOI] [PubMed] [Google Scholar]
- 168.Cue D, Lei MG, Lee CY. Genetic regulation of the intercellular adhesion locus in staphylococci. Frontiers in cellular and infection microbiology. 2012;2:38. doi: 10.3389/fcimb.2012.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Berube BJ, Bubeck Wardenburg J. Staphylococcus aureus alpha-toxin: nearly a century of intrigue. Toxins. 2013;5:1140–1166. doi: 10.3390/toxins5061140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Caiazza NC, O’Toole GA. Alpha-toxin is required for biofilm formation by Staphylococcus aureus. Journal of bacteriology. 2003;185:3214–3217. doi: 10.1128/JB.185.10.3214-3217.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Marshall MJ, Bohach GA, Boehm DF. Characterization of Staphylococcus aureus beta-toxin induced leukotoxicity. J Nat Toxins. 2000;9:125–138. [PubMed] [Google Scholar]
- 172.Huseby M, Shi K, Brown CK, Digre J, Mengistu F, Seo KS, Bohach GA, Schlievert PM, Ohlendorf DH, Earhart CA. Structure and biological activities of beta toxin from Staphylococcus aureus. Journal of bacteriology. 2007;189:8719–8726. doi: 10.1128/JB.00741-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Huseby MJ, Kruse AC, Digre J, Kohler PL, Vocke JA, Mann EE, Bayles KW, Bohach GA, Schlievert PM, Ohlendorf DH, Earhart CA. Beta toxin catalyzes formation of nucleoprotein matrix in staphylococcal biofilms. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:14407–14412. doi: 10.1073/pnas.0911032107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Chavakis T, Wiechmann K, Preissner KT, Herrmann M. Staphylococcus aureus interactions with the endothelium: the role of bacterial “secretable expanded repertoire adhesive molecules” (SERAM) in disturbing host defense systems. Thrombosis and haemostasis. 2005;94:278–285. doi: 10.1160/TH05-05-0306. [DOI] [PubMed] [Google Scholar]
- 175.Zielinska AK, Beenken KE, Mrak LN, Spencer HJ, Post GR, Skinner RA, Tackett AJ, Horswill AR, Smeltzer MS. sarA-mediated repression of protease production plays a key role in the pathogenesis of Staphylococcus aureus USA300 isolates. Molecular microbiology. 2012;86:1183–1196. doi: 10.1111/mmi.12048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Johnson M, Cockayne A, Morrissey JA. Iron-regulated biofilm formation in Staphylococcus aureus Newman requires ica and the secreted protein Emp. Infection and immunity. 2008;76:1756–1765. doi: 10.1128/IAI.01635-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Malone CL, Boles BR, Horswill AR. Biosynthesis of Staphylococcus aureus autoinducing peptides by using the synechocystis DnaB mini-intein. Applied and environmental microbiology. 2007;73:6036–6044. doi: 10.1128/AEM.00912-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Hammel M, Nemecek D, Keightley JA, Thomas GJ, Jr, Geisbrecht BV. The Staphylococcus aureus extracellular adherence protein (Eap) adopts an elongated but structured conformation in solution. Protein science : a publication of the Protein Society. 2007;16:2605–2617. doi: 10.1110/ps.073170807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Boles BR, Horswill AR. Agr-mediated dispersal of Staphylococcus aureus biofilms. PLoS pathogens. 2008;4:e1000052. doi: 10.1371/journal.ppat.1000052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Harraghy N, Hussain M, Haggar A, Chavakis T, Sinha B, Herrmann M, Flock JI. The adhesive and immunomodulating properties of the multifunctional Staphylococcus aureus protein Eap. Microbiology. 2003;149:2701–2707. doi: 10.1099/mic.0.26465-0. [DOI] [PubMed] [Google Scholar]
- 181.Harraghy N, Homerova D, Herrmann M, Kormanec J. Mapping the transcription start points of the Staphylococcus aureus eap, emp, and vwb promoters reveals a conserved octanucleotide sequence that is essential for expression of these genes. Journal of bacteriology. 2008;190:447–451. doi: 10.1128/JB.01174-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Harraghy N, Kormanec J, Wolz C, Homerova D, Goerke C, Ohlsen K, Qazi S, Hill P, Herrmann M. sae is essential for expression of the staphylococcal adhesins Eap and Emp. Microbiology. 2005;151:1789–1800. doi: 10.1099/mic.0.27902-0. [DOI] [PubMed] [Google Scholar]
- 183.Beenken KE, Dunman PM, McAleese F, Macapagal D, Murphy E, Projan SJ, Blevins JS, Smeltzer MS. Global gene expression in Staphylococcus aureus biofilms. Journal of bacteriology. 2004;186:4665–4684. doi: 10.1128/JB.186.14.4665-4684.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Yao Y, Sturdevant DE, Otto M. Genomewide analysis of gene expression in Staphylococcus epidermidis biofilms: insights into the pathophysiology of S. epidermidis biofilms and the role of phenol-soluble modulins in formation of biofilms. The Journal of infectious diseases. 2005;191:289–298. doi: 10.1086/426945. [DOI] [PubMed] [Google Scholar]
- 185.Trotonda MP, Tamber S, Memmi G, Cheung AL. MgrA represses biofilm formation in Staphylococcus aureus. Infection and immunity. 2008;76:5645–5654. doi: 10.1128/IAI.00735-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Toledo-Arana A, Merino N, Vergara-Irigaray M, Debarbouille M, Penades JR, Lasa I. Staphylococcus aureus develops an alternative, ica-independent biofilm in the absence of the arlRS two-component system. Journal of bacteriology. 2005;187:5318–5329. doi: 10.1128/JB.187.15.5318-5329.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Thoendel M, Kavanaugh JS, Flack CE, Horswill AR. Peptide signaling in the staphylococci. Chem Rev. 2011;111:117–151. doi: 10.1021/cr100370n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Novick RP, Geisinger E. Quorum sensing in staphylococci. Annu Rev Genet. 2008;42:541–564. doi: 10.1146/annurev.genet.42.110807.091640. [DOI] [PubMed] [Google Scholar]
- 189.Koenig RL, Ray JL, Maleki SJ, Smeltzer MS, Hurlburt BK. Staphylococcus aureus AgrA binding to the RNAIII-agr regulatory region. Journal of bacteriology. 2004;186:7549–7555. doi: 10.1128/JB.186.22.7549-7555.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Queck SY, Jameson-Lee M, Villaruz AE, Bach TH, Khan BA, Sturdevant DE, Ricklefs SM, Li M, Otto M. RNAIII-independent target gene control by the agr quorum-sensing system: insight into the evolution of virulence regulation in Staphylococcus aureus. Molecular cell. 2008;32:150–158. doi: 10.1016/j.molcel.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Geisinger E, Adhikari RP, Jin R, Ross HF, Novick RP. Inhibition of rot translation by RNAIII, a key feature of agr function. Molecular microbiology. 2006;61:1038–1048. doi: 10.1111/j.1365-2958.2006.05292.x. [DOI] [PubMed] [Google Scholar]
- 192.Olson ME, Todd DA, Schaeffer CR, Paharik AE, Van Dyke MJ, Buttner H, Dunman PM, Rohde H, Cech NB, Fey PD, Horswill AR. Staphylococcus epidermidis agr quorum-sensing system: signal identification, cross talk, and importance in colonization. Journal of bacteriology. 2014;196:3482–3493. doi: 10.1128/JB.01882-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Dunman PM, Murphy E, Haney S, Palacios D, Tucker-Kellogg G, Wu S, Brown EL, Zagursky RJ, Shlaes D, Projan SJ. Transcription profiling-based identification of Staphylococcus aureus genes regulated by the agr and/or sarA loci. Journal of bacteriology. 2001;183:7341–7353. doi: 10.1128/JB.183.24.7341-7353.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Li M, Diep BA, Villaruz AE, Braughton KR, Jiang X, DeLeo FR, Chambers HF, Lu Y, Otto M. Evolution of virulence in epidemic community-associated methicillin-resistant Staphylococcus aureus. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:5883–5888. doi: 10.1073/pnas.0900743106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.DeLeo FR, Kennedy AD, Chen L, Bubeck Wardenburg J, Kobayashi SD, Mathema B, Braughton KR, Whitney AR, Villaruz AE, Martens CA, Porcella SF, McGavin MJ, Otto M, Musser JM, Kreiswirth BN. Molecular differentiation of historic phage-type 80/81 and contemporary epidemic Staphylococcus aureus. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:18091–18096. doi: 10.1073/pnas.1111084108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Montgomery CP, Boyle-Vavra S, Daum RS. Importance of the global regulators Agr and SaeRS in the pathogenesis of CA-MRSA USA300 infection. PloS one. 2010;5:e15177. doi: 10.1371/journal.pone.0015177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Lauderdale KJ, Malone CL, Boles BR, Morcuende J, Horswill AR. Biofilm dispersal of community-associated methicillin-resistant Staphylococcus aureus on orthopedic implant material. Journal of orthopaedic research : official publication of the Orthopaedic Research Society. 2010;28:55–61. doi: 10.1002/jor.20943. [DOI] [PubMed] [Google Scholar]
- 198.Pang YY, Schwartz J, Thoendel M, Ackermann LW, Horswill AR, Nauseef WM. agr-Dependent interactions of Staphylococcus aureus USA300 with human polymorphonuclear neutrophils. Journal of innate immunity. 2010;2:546–559. doi: 10.1159/000319855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Lauderdale KJ, Boles BR, Cheung AL, Horswill AR. Interconnections between Sigma B, agr, and proteolytic activity in Staphylococcus aureus biofilm maturation. Infection and immunity. 2009;77:1623–1635. doi: 10.1128/IAI.01036-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Vuong C, Gerke C, Somerville GA, Fischer ER, Otto M. Quorum-sensing control of biofilm factors in Staphylococcus epidermidis. The Journal of infectious diseases. 2003;188:706–718. doi: 10.1086/377239. [DOI] [PubMed] [Google Scholar]
- 201.Batzilla CF, Rachid S, Engelmann S, Hecker M, Hacker J, Ziebuhr W. Impact of the accessory gene regulatory system (Agr) on extracellular proteins, codY expression and amino acid metabolism in Staphylococcus epidermidis. Proteomics. 2006;6:3602–3613. doi: 10.1002/pmic.200500732. [DOI] [PubMed] [Google Scholar]
- 202.Yarwood JM, Bartels DJ, Volper EM, Greenberg EP. Quorum sensing in Staphylococcus aureus biofilms. Journal of bacteriology. 2004;186:1838–1850. doi: 10.1128/JB.186.6.1838-1850.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Lauderdale KJ, Malone CL, Boles BR, Morcuende J, Horswill AR. Biofilm Dispersal of Community-Associated Methicillin-Resistant Staphylococcus aureus on Orthopedic Implant Material. J Orthop Res. 2010;28:55–61. doi: 10.1002/jor.20943. [DOI] [PubMed] [Google Scholar]
- 204.Mootz JM, Malone CL, Shaw LN, Horswill AR. Staphopains modulate Staphylococcus aureus biofilm integrity. Infection and immunity. 2013 doi: 10.1128/IAI.00377-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Mootz JM, Benson MA, Heim CE, Crosby HA, Kavanaugh JS, Dunman PM, Kielian T, Torres VJ, Horswill AR. Rot is a key regulator of Staphylococcus aureus biofilm formation. Molecular microbiology. 2015;96:388–404. doi: 10.1111/mmi.12943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Hsieh HY, Tseng CW, Stewart GC. Regulation of Rot expression in Staphylococcus aureus. Journal of bacteriology. 2008;190:546–554. doi: 10.1128/JB.00536-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Olson ME, King JM, Yahr TL, Horswill AR. Sialic acid catabolism in Staphylococcus aureus. Journal Bacteriology. 2013;195:1779–1788. doi: 10.1128/JB.02294-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Regassa LB, Novick RP, Betley MJ. Glucose and nonmaintained pH decrease expression of the accessory gene regulator (agr) in Staphylococcus aureus. Infection and immunity. 1992;60:3381–3388. doi: 10.1128/iai.60.8.3381-3388.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Yarwood JM, McCormick JK, Paustian ML, Kapur V, Schlievert PM. Repression of the Staphylococcus aureus accessory gene regulator in serum and in vivo. Journal of bacteriology. 2002;184:1095–1101. doi: 10.1128/jb.184.4.1095-1101.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Hall PR, Elmore BO, Spang CH, Alexander SM, Manifold-Wheeler BC, Castleman MJ, Daly SM, Peterson MM, Sully EK, Femling JK, Otto M, Horswill AR, Timmins GS, Gresham HD. Nox2 modification of LDL is essential for optimal apolipoprotein B-mediated control of agr type III Staphylococcus aureus quorum-sensing. PLoS pathogens. 2013;9:e1003166. doi: 10.1371/journal.ppat.1003166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Peterson MM, Mack JL, Hall PR, Alsup AA, Alexander SM, Sully EK, Sawires YS, Cheung AL, Otto M, Gresham HD. Apolipoprotein B Is an innate barrier against invasive Staphylococcus aureus infection. Cell host & microbe. 2008;4:555–566. doi: 10.1016/j.chom.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Pynnonen M, Stephenson RE, Schwartz K, Hernandez M, Boles BR. Hemoglobin promotes Staphylococcus aureus nasal colonization. PLoS pathogens. 2011;7:e1002104. doi: 10.1371/journal.ppat.1002104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Syed AK, Ghosh S, Love NG, Boles BR. Triclosan promotes Staphylococcus aureus nasal colonization. mBio. 2014;5:e01015. doi: 10.1128/mBio.01015-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Schwartz K, Syed AK, Stephenson RE, Rickard AH, Boles BR. Functional amyloids composed of phenol soluble modulins stabilize Staphylococcus aureus biofilms. PLoS pathogens. 2012;8:e1002744. doi: 10.1371/journal.ppat.1002744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Cassat JE, Lee CY, Smeltzer MS. Investigation of biofilm formation in clinical isolates of Staphylococcus aureus. Methods Mol Biol. 2007;391:127–144. doi: 10.1007/978-1-59745-468-1_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Ibberson C, Parlet C, Horswill AR. Unpublished observations [Google Scholar]
- 217.Wang R, Khan BA, Cheung GY, Bach TH, Jameson-Lee M, Kong KF, Queck SY, Otto M. Staphylococcus epidermidis surfactant peptides promote biofilm maturation and dissemination of biofilm-associated infection in mice. J Clin Invest. 2011;121:238–248. doi: 10.1172/JCI42520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Fowler VG, Jr, Miro JM, Hoen B, Cabell CH, Abrutyn E, Rubinstein E, Corey GR, Spelman D, Bradley SF, Barsic B, Pappas PA, Anstrom KJ, Wray D, Fortes CQ, Anguera I, Athan E, Jones P, van der Meer JT, Elliott TS, Levine DP, Bayer AS. Staphylococcus aureus endocarditis: a consequence of medical progress. Jama. 2005;293:3012–3021. doi: 10.1001/jama.293.24.3012. [DOI] [PubMed] [Google Scholar]
- 219.Xiong YQ, Van Wamel W, Nast CC, Yeaman MR, Cheung AL, Bayer AS. Activation and transcriptional interaction between agr RNAII and RNAIII in Staphylococcus aureus in vitro and in an experimental endocarditis model. The Journal of infectious diseases. 2002;186:668–677. doi: 10.1086/342046. [DOI] [PubMed] [Google Scholar]
- 220.Fux CA, Wilson S, Stoodley P. Detachment characteristics and oxacillin resistance of Staphyloccocus aureus biofilm emboli in an in vitro catheter infection model. Journal of bacteriology. 2004;186:4486–4491. doi: 10.1128/JB.186.14.4486-4491.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Koh TW, Brecker SJ, Layton CA. Successful treatment of Staphylococcus lugdunensis endocarditis complicated by multiple emboli: a case report and review of the literature. International journal of cardiology. 1996;55:193–197. doi: 10.1016/0167-5273(96)02679-4. [DOI] [PubMed] [Google Scholar]
- 222.Ye R, Zhao L, Wang C, Wu X, Yan H. Clinical characteristics of septic pulmonary embolism in adults: a systematic review. Respiratory medicine. 2014;108:1–8. doi: 10.1016/j.rmed.2013.10.012. [DOI] [PubMed] [Google Scholar]
- 223.Plicht B, Janosi RA, Buck T, Erbel R. Infective endocarditis as cardiovascular emergency. Der Internist. 2010;51:987–994. doi: 10.1007/s00108-009-2538-0. [DOI] [PubMed] [Google Scholar]
- 224.Goerke C, Campana S, Bayer MG, Doring G, Botzenhart K, Wolz C. Direct quantitative transcript analysis of the agr regulon of Staphylococcus aureus during human infection in comparison to the expression profile in vitro. Infection and immunity. 2000;68:1304–1311. doi: 10.1128/iai.68.3.1304-1311.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Goerke C, Wolz C. Regulatory and genomic plasticity of Staphylococcus aureus during persistent colonization and infection. International journal of medical microbiology : IJMM. 2004;294:195–202. doi: 10.1016/j.ijmm.2004.06.013. [DOI] [PubMed] [Google Scholar]
- 226.Goerke C, Wolz C. Adaptation of Staphylococcus aureus to the cystic fibrosis lung. International journal of medical microbiology : IJMM. 2010;300:520–525. doi: 10.1016/j.ijmm.2010.08.003. [DOI] [PubMed] [Google Scholar]
- 227.Yarwood JM, Paquette KM, Tikh IB, Volper EM, Greenberg EP. Generation of virulence factor variants in Staphylococcus aureus biofilms. Journal of bacteriology. 2007 doi: 10.1128/JB.00789-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Traber KE, Lee E, Benson S, Corrigan R, Cantera M, Shopsin B, Novick RP. agr function in clinical Staphylococcus aureus isolates. Microbiology. 2008;154:2265–2274. doi: 10.1099/mic.0.2007/011874-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Paulander W, Nissen Varming A, Baek KT, Haaber J, Frees D, Ingmer H. Antibiotic-mediated selection of quorum-sensing-negative Staphylococcus aureus. mBio. 2013;3:e00459–00412. doi: 10.1128/mBio.00459-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Shopsin B, Eaton C, Wasserman GA, Mathema B, Adhikari RP, Agolory S, Altman DR, Holzman RS, Kreiswirth BN, Novick RP. Mutations in agr do not persist in natural populations of methicillin-resistant Staphylococcus aureus. The Journal of infectious diseases. 2010;202:1593–1599. doi: 10.1086/656915. [DOI] [PubMed] [Google Scholar]
- 231.Daly SM, Elmore BO, Kavanaugh JS, Triplett KD, Figueroa M, Raja HA, El-Elimat T, Crosby HA, Femling JK, Cech NB, Horswill AR, Oberlies NH, Hall PR. omega-Hydroxyemodin limits staphylococcus aureus quorum sensing-mediated pathogenesis and inflammation. Antimicrobial agents and chemotherapy. 2015;59:2223–2235. doi: 10.1128/AAC.04564-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Sully EK, Malachowa N, Elmore BO, Alexander SM, Femling JK, Gray BM, DeLeo FR, Otto M, Cheung AL, Edwards BS, Sklar LA, Horswill AR, Hall PR, Gresham HD. Selective chemical inhibition of agr quorum sensing in Staphylococcus aureus promotes host defense with minimal impact on resistance. PLoS pathogens. 2014;10:e1004174. doi: 10.1371/journal.ppat.1004174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Cech NB, Horswill AR. Small-molecule quorum quenchers to prevent Staphylococcus aureus infection. Future microbiology. 2013;8:1511–1514. doi: 10.2217/fmb.13.134. [DOI] [PubMed] [Google Scholar]
- 234.Schauder S, Shokat K, Surette MG, Bassler BL. The LuxS family of bacterial autoinducers: biosynthesis of a novel quorum-sensing signal molecule. Molecular microbiology. 2001;41:463–476. doi: 10.1046/j.1365-2958.2001.02532.x. [DOI] [PubMed] [Google Scholar]
- 235.Li M, Villaruz AE, Vadyvaloo V, Sturdevant DE, Otto M. AI-2-dependent gene regulation in Staphylococcus epidermidis. BMC microbiology. 2008;8:4. doi: 10.1186/1471-2180-8-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Xu L, Li H, Vuong C, Vadyvaloo V, Wang J, Yao Y, Otto M, Gao Q. Role of the luxS quorum-sensing system in biofilm formation and virulence of Staphylococcus epidermidis. Infection and immunity. 2006;74:488–496. doi: 10.1128/IAI.74.1.488-496.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Yu D, Zhao L, Xue T, Sun B. Staphylococcus aureus autoinducer-2 quorum sensing decreases biofilm formation in an icaR-dependent manner. BMC microbiology. 2012;12:288. doi: 10.1186/1471-2180-12-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Zhao L, Xue T, Shang F, Sun H, Sun B. Staphylococcus aureus AI-2 quorum sensing associates with the KdpDE two-component system to regulate capsular polysaccharide synthesis and virulence. Infection and immunity. 2010;78:3506–3515. doi: 10.1128/IAI.00131-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Kullik II, Giachino P. The alternative sigma factor sigmaB in Staphylococcus aureus: regulation of the sigB operon in response to growth phase and heat shock. Archives of microbiology. 1997;167:151–159. doi: 10.1007/s002030050428. [DOI] [PubMed] [Google Scholar]
- 240.Mootz JM, Malone CL, Shaw LN, Horswill AR. Staphopains modulate Staphylococcus aureus biofilm integrity. Infection and immunity. 2013;81:3227–3238. doi: 10.1128/IAI.00377-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Marti M, Trotonda MP, Tormo-Mas MA, Vergara-Irigaray M, Cheung AL, Lasa I, Penades JR. Extracellular proteases inhibit protein-dependent biofilm formation in Staphylococcus aureus. Microbes and infection / Institut Pasteur. 2010;12:55–64. doi: 10.1016/j.micinf.2009.10.005. [DOI] [PubMed] [Google Scholar]
- 242.Kiedrowski MR, Kavanaugh JS, Malone CL, Mootz JM, Voyich JM, Smeltzer MS, Bayles KW, Horswill AR. Nuclease modulates biofilm formation in community-associated methicillin-resistant Staphylococcus aureus. PloS one. 2011;6:e26714. doi: 10.1371/journal.pone.0026714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Knobloch JK, Jager S, Horstkotte MA, Rohde H, Mack D. RsbU-dependent regulation of Staphylococcus epidermidis biofilm formation is mediated via the alternative sigma factor sigmaB by repression of the negative regulator gene icaR. Infection and immunity. 2004;72:3838–3848. doi: 10.1128/IAI.72.7.3838-3848.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Jager S, Jonas B, Pfanzelt D, Horstkotte MA, Rohde H, Mack D, Knobloch JK. Regulation of biofilm formation by sigma B is a common mechanism in Staphylococcus epidermidis and is not mediated by transcriptional regulation of sarA. The International journal of artificial organs. 2009;32:584–591. doi: 10.1177/039139880903200907. [DOI] [PubMed] [Google Scholar]
- 245.Depke M, Burian M, Schafer T, Broker BM, Ohlsen K, Volker U. The alternative sigma factor B modulates virulence gene expression in a murine Staphylococcus aureus infection model but does not influence kidney gene expression pattern of the host. International journal of medical microbiology : IJMM. 2012;302:33–39. doi: 10.1016/j.ijmm.2011.09.013. [DOI] [PubMed] [Google Scholar]
- 246.Pintens V, Massonet C, Merckx R, Vandecasteele S, Peetermans WE, Knobloch JK, Van Eldere J. The role of sigmaB in persistence of Staphylococcus epidermidis foreign body infection. Microbiology. 2008;154:2827–2836. doi: 10.1099/mic.0.2007/015768-0. [DOI] [PubMed] [Google Scholar]
- 247.Ishii K, Adachi T, Yasukawa J, Suzuki Y, Hamamoto H, Sekimizu K. Induction of virulence gene expression in Staphylococcus aureus by pulmonary surfactant. Infection and immunity. 2014;82:1500–1510. doi: 10.1128/IAI.01635-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Pfortner H, Burian MS, Michalik S, Depke M, Hildebrandt P, Dhople VM, Pane-Farre J, Hecker M, Schmidt F, Volker U. Activation of the alternative sigma factor SigB of Staphylococcus aureus following internalization by epithelial cells - an in vivo proteomics perspective. International journal of medical microbiology : IJMM. 2014;304:177–187. doi: 10.1016/j.ijmm.2013.11.014. [DOI] [PubMed] [Google Scholar]
- 249.Savage VJ, Chopra I, O’Neill AJ. Population diversification in Staphylococcus aureus biofilms may promote dissemination and persistence. PloS one. 2013;8:e62513. doi: 10.1371/journal.pone.0062513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Mitchell G, Fugere A, Pepin Gaudreau K, Brouillette E, Frost EH, Cantin AM, Malouin F. SigB is a dominant regulator of virulence in Staphylococcus aureus small-colony variants. PloS one. 2013;8:e65018. doi: 10.1371/journal.pone.0065018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Cheung AL, Koomey JM, Butler CA, Projan SJ, Fischetti VA. Regulation of exoprotein expression in Staphylococcus aureus by a locus (sar) distinct from agr. Proceedings of the National Academy of Sciences of the United States of America. 1992;89:6462–6466. doi: 10.1073/pnas.89.14.6462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Bayer MG, Heinrichs JH, Cheung AL. The molecular architecture of the sar locus in Staphylococcus aureus. Journal of bacteriology. 1996;178:4563–4570. doi: 10.1128/jb.178.15.4563-4570.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Rechtin TM, Gillaspy AF, Schumacher MA, Brennan RG, Smeltzer MS, Hurlburt BK. Characterization of the SarA virulence gene regulator of Staphylococcus aureus. Molecular microbiology. 1999;33:307–316. doi: 10.1046/j.1365-2958.1999.01474.x. [DOI] [PubMed] [Google Scholar]
- 254.Cheung AL, Projan SJ. Cloning and sequencing of sarA of Staphylococcus aureus, a gene required for the expression of agr. Journal of bacteriology. 1994;176:4168–4172. doi: 10.1128/jb.176.13.4168-4172.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Heinrichs JH, Bayer MG, Cheung AL. Characterization of the sar locus and its interaction with agr in Staphylococcus aureus. Journal of bacteriology. 1996;178:418–423. doi: 10.1128/jb.178.2.418-423.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Cheung AL, Bayer MG, Heinrichs JH. sar Genetic determinants necessary for transcription of RNAII and RNAIII in the agr locus of Staphylococcus aureus. Journal of bacteriology. 1997;179:3963–3971. doi: 10.1128/jb.179.12.3963-3971.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Chien Y, Manna AC, Projan SJ, Cheung AL. SarA, a global regulator of virulence determinants in Staphylococcus aureus, binds to a conserved motif essential for sar-dependent gene regulation. The Journal of biological chemistry. 1999;274:37169–37176. doi: 10.1074/jbc.274.52.37169. [DOI] [PubMed] [Google Scholar]
- 258.Gillaspy AF, Lee CY, Sau S, Cheung AL, Smeltzer MS. Factors affecting the collagen binding capacity of Staphylococcus aureus. Infection and immunity. 1998;66:3170–3178. doi: 10.1128/iai.66.7.3170-3178.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Blevins JS, Gillaspy AF, Rechtin TM, Hurlburt BK, Smeltzer MS. The Staphylococcal accessory regulator (sar) represses transcription of the Staphylococcus aureus collagen adhesin gene (cna) in an agr-independent manner. Molecular microbiology. 1999;33:317–326. doi: 10.1046/j.1365-2958.1999.01475.x. [DOI] [PubMed] [Google Scholar]
- 260.Trotonda MP, Manna AC, Cheung AL, Lasa I, Penades JR. SarA positively controls bap-dependent biofilm formation in Staphylococcus aureus. Journal of bacteriology. 2005;187:5790–5798. doi: 10.1128/JB.187.16.5790-5798.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Blevins JS, Beenken KE, Elasri MO, Hurlburt BK, Smeltzer MS. Strain-dependent differences in the regulatory roles of sarA and agr in Staphylococcus aureus. Infection and immunity. 2002;70:470–480. doi: 10.1128/IAI.70.2.470-480.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Cassat J, Dunman PM, Murphy E, Projan SJ, Beenken KE, Palm KJ, Yang SJ, Rice KC, Bayles KW, Smeltzer MS. Transcriptional profiling of a Staphylococcus aureus clinical isolate and its isogenic agr and sarA mutants reveals global differences in comparison to the laboratory strain RN6390. Microbiology. 2006;152:3075–3090. doi: 10.1099/mic.0.29033-0. [DOI] [PubMed] [Google Scholar]
- 263.Beenken KE, Blevins JS, Smeltzer MS. Mutation of sarA in Staphylococcus aureus limits biofilm formation. Infection and immunity. 2003;71:4206–4211. doi: 10.1128/IAI.71.7.4206-4211.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Valle J, Toledo-Arana A, Berasain C, Ghigo JM, Amorena B, Penades JR, Lasa I. SarA and not sigmaB is essential for biofilm development by Staphylococcus aureus. Molecular microbiology. 2003;48:1075–1087. doi: 10.1046/j.1365-2958.2003.03493.x. [DOI] [PubMed] [Google Scholar]
- 265.Snowden JN, Beaver M, Beenken K, Smeltzer M, Horswill AR, Kielian T. Staphylococcus aureus sarA regulates inflammation and colonization during central nervous system biofilm formation. PloS one. 2013;8:e84089. doi: 10.1371/journal.pone.0084089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Tsang LH, Cassat JE, Shaw LN, Beenken KE, Smeltzer MS. Factors contributing to the biofilm-deficient phenotype of Staphylococcus aureus sarA mutants. PloS one. 2008;3:e3361. doi: 10.1371/journal.pone.0003361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Beenken KE, Mrak LN, Griffin LM, Zielinska AK, Shaw LN, Rice KC, Horswill AR, Bayles KW, Smeltzer MS. Epistatic relationships between sarA and agr in Staphylococcus aureus biofilm formation. PloS one. 2010;5:e10790. doi: 10.1371/journal.pone.0010790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Tormo MA, Marti M, Valle J, Manna AC, Cheung AL, Lasa I, Penades JR. SarA is an essential positive regulator of Staphylococcus epidermidis biofilm development. Journal of bacteriology. 2005;187:2348–2356. doi: 10.1128/JB.187.7.2348-2356.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Christner M, Heinze C, Busch M, Franke G, Hentschke M, Bayard Duhring S, Buttner H, Kotasinska M, Wischnewski V, Kroll G, Buck F, Molin S, Otto M, Rohde H. sarA negatively regulates Staphylococcus epidermidis biofilm formation by modulating expression of 1 MDa extracellular matrix binding protein and autolysis-dependent release of eDNA. Molecular microbiology. 2012;86:394–410. doi: 10.1111/j.1365-2958.2012.08203.x. [DOI] [PubMed] [Google Scholar]
- 270.Lai Y, Villaruz AE, Li M, Cha DJ, Sturdevant DE, Otto M. The human anionic antimicrobial peptide dermcidin induces proteolytic defence mechanisms in staphylococci. Molecular microbiology. 2007;63:497–506. doi: 10.1111/j.1365-2958.2006.05540.x. [DOI] [PubMed] [Google Scholar]
- 271.Scherr TD, Heim CE, Morrison JM, Kielian T. Hiding in Plain Sight: Interplay between Staphylococcal Biofilms and Host Immunity. Frontiers in immunology. 2014;5:37. doi: 10.3389/fimmu.2014.00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 273.Hanke ML, Kielian T. Deciphering mechanisms of staphylococcal biofilm evasion of host immunity. Frontiers in cellular and infection microbiology. 2012;2:62. doi: 10.3389/fcimb.2012.00062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Verdrengh M, Tarkowski A. Role of neutrophils in experimental septicemia and septic arthritis induced by Staphylococcus aureus. Infection and immunity. 1997;65:2517–2521. doi: 10.1128/iai.65.7.2517-2521.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Verdrengh M, Tarkowski A. Role of macrophages in Staphylococcus aureus-induced arthritis and sepsis. Arthritis and rheumatism. 2000;43:2276–2282. doi: 10.1002/1529-0131(200010)43:10<2276::AID-ANR15>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 276.Rigby KM, DeLeo FR. Neutrophils in innate host defense against Staphylococcus aureus infections. Seminars in immunopathology. 2012;34:237–259. doi: 10.1007/s00281-011-0295-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Wagner C, Kondella K, Bernschneider T, Heppert V, Wentzensen A, Hansch GM. Post-traumatic osteomyelitis: analysis of inflammatory cells recruited into the site of infection. Shock. 2003;20:503–510. doi: 10.1097/01.shk.0000093542.78705.e3. [DOI] [PubMed] [Google Scholar]
- 278.Wagner C, Kaksa A, Muller W, Denefleh B, Heppert V, Wentzensen A, Hansch GM. Polymorphonuclear neutrophils in posttraumatic osteomyelitis: cells recovered from the inflamed site lack chemotactic activity but generate superoxides. Shock. 2004;22:108–115. doi: 10.1097/01.shk.0000132488.71875.15. [DOI] [PubMed] [Google Scholar]
- 279.Scherr TD, Roux CM, Hanke ML, Angle A, Dunman PM, Kielian T. Global transcriptome analysis of Staphylococcus aureus biofilms in response to innate immune cells. Infection and immunity. 2013;81:4363–4376. doi: 10.1128/IAI.00819-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Leid JG, Shirtliff ME, Costerton JW, Stoodley P. Human leukocytes adhere to, penetrate, and respond to Staphylococcus aureus biofilms. Infection and immunity. 2002;70:6339–6345. doi: 10.1128/IAI.70.11.6339-6345.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Meyle E, Brenner-Weiss G, Obst U, Prior B, Hansch GM. Immune defense against S. epidermidis biofilms: components of the extracellular polymeric substance activate distinct bactericidal mechanisms of phagocytic cells. The International journal of artificial organs. 2012;35:700–712. doi: 10.5301/ijao.5000151. [DOI] [PubMed] [Google Scholar]
- 282.Meyle E, Stroh P, Gunther F, Hoppy-Tichy T, Wagner C, Hansch GM. Destruction of bacterial biofilms by polymorphonuclear neutrophils: relative contribution of phagocytosis, DNA release, and degranulation. The International journal of artificial organs. 2010;33:608–620. doi: 10.1177/039139881003300906. [DOI] [PubMed] [Google Scholar]
- 283.Kristian SA, Birkenstock TA, Sauder U, Mack D, Gotz F, Landmann R. Biofilm formation induces C3a release and protects Staphylococcus epidermidis from IgG and complement deposition and from neutrophil-dependent killing. The Journal of infectious diseases. 2008;197:1028–1035. doi: 10.1086/528992. [DOI] [PubMed] [Google Scholar]
- 284.Vuong C, Voyich JM, Fischer ER, Braughton KR, Whitney AR, DeLeo FR, Otto M. Polysaccharide intercellular adhesin (PIA) protects Staphylococcus epidermidis against major components of the human innate immune system. Cellular microbiology. 2004;6:269–275. doi: 10.1046/j.1462-5822.2004.00367.x. [DOI] [PubMed] [Google Scholar]
- 285.Nilsson IM, Lee JC, Bremell T, Ryden C, Tarkowski A. The role of staphylococcal polysaccharide microcapsule expression in septicemia and septic arthritis. Infection and immunity. 1997;65:4216–4221. doi: 10.1128/iai.65.10.4216-4221.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Thakker M, Park JS, Carey V, Lee JC. Staphylococcus aureus serotype 5 capsular polysaccharide is antiphagocytic and enhances bacterial virulence in a murine bacteremia model. Infection and immunity. 1998;66:5183–5189. doi: 10.1128/iai.66.11.5183-5189.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Brinkmann V, Reichard U, Goosmann C, Fauler B, Uhlemann Y, Weiss DS, Weinrauch Y, Zychlinsky A. Neutrophil extracellular traps kill bacteria. Science. 2004;303:1532–1535. doi: 10.1126/science.1092385. [DOI] [PubMed] [Google Scholar]
- 288.Fuchs TA, Abed U, Goosmann C, Hurwitz R, Schulze I, Wahn V, Weinrauch Y, Brinkmann V, Zychlinsky A. Novel cell death program leads to neutrophil extracellular traps. The Journal of cell biology. 2007;176:231–241. doi: 10.1083/jcb.200606027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Berends ET, Horswill AR, Haste NM, Monestier M, Nizet V, von Kockritz-Blickwede M. Nuclease expression by Staphylococcus aureus facilitates escape from neutrophil extracellular traps. Journal of innate immunity. 2010;2:576–586. doi: 10.1159/000319909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Thammavongsa V, Missiakas DM, Schneewind O. Staphylococcus aureus degrades neutrophil extracellular traps to promote immune cell death. Science. 2013;342:863–866. doi: 10.1126/science.1242255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Kiedrowski MR, Crosby HA, Hernandez FJ, Malone CL, McNamara JO, 2nd, Horswill AR. Staphylococcus aureus Nuc2 is a functional, surface-attached extracellular nuclease. PloS one. 2014;9:e95574. doi: 10.1371/journal.pone.0095574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Mosser DM. The many faces of macrophage activation. Journal of leukocyte biology. 2003;73:209–212. doi: 10.1189/jlb.0602325. [DOI] [PubMed] [Google Scholar]
- 293.Hesse M, Modolell M, La Flamme AC, Schito M, Fuentes JM, Cheever AW, Pearce EJ, Wynn TA. Differential regulation of nitric oxide synthase-2 and arginase-1 by type 1/type 2 cytokines in vivo: granulomatous pathology is shaped by the pattern of L-arginine metabolism. J Immunol. 2001;167:6533–6544. doi: 10.4049/jimmunol.167.11.6533. [DOI] [PubMed] [Google Scholar]
- 294.Song E, Ouyang N, Horbelt M, Antus B, Wang M, Exton MS. Influence of alternatively and classically activated macrophages on fibrogenic activities of human fibroblasts. Cellular immunology. 2000;204:19–28. doi: 10.1006/cimm.2000.1687. [DOI] [PubMed] [Google Scholar]
- 295.Thurlow LR, Hanke ML, Fritz T, Angle A, Aldrich A, Williams SH, Engebretsen IL, Bayles KW, Horswill AR, Kielian T. Staphylococcus aureus biofilms prevent macrophage phagocytosis and attenuate inflammation in vivo. Journal of immunology. 2011;186:6585–6596. doi: 10.4049/jimmunol.1002794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Palmqvist N, Patti JM, Tarkowski A, Josefsson E. Expression of staphylococcal clumping factor A impedes macrophage phagocytosis. Microbes Infect. 2004;6:188–195. doi: 10.1016/j.micinf.2003.11.005. [DOI] [PubMed] [Google Scholar]
- 297.Olson ME, Nygaard TK, Ackermann L, Watkins RL, Zurek OW, Pallister KB, Griffith S, Kiedrowski MR, Flack CE, Kavanaugh JS, Kreiswirth BN, Horswill AR, Voyich JM. Staphylococcus aureus nuclease is an SaeRS-dependent virulence factor. Infection and immunity. 2013;81:1316–1324. doi: 10.1128/IAI.01242-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Nygaard TK, Pallister KB, Ruzevich P, Griffith S, Vuong C, Voyich JM. SaeR binds a consensus sequence within virulence gene promoters to advance USA300 pathogenesis. The Journal of infectious diseases. 2010;201:241–254. doi: 10.1086/649570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Voyich JM, Vuong C, DeWald M, Nygaard TK, Kocianova S, Griffith S, Jones J, Iverson C, Sturdevant DE, Braughton KR, Whitney AR, Otto M, DeLeo FR. The SaeR/S gene regulatory system is essential for innate immune evasion by Staphylococcus aureus. The Journal of infectious diseases. 2009;199:1698–1706. doi: 10.1086/598967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Zurek OW, Nygaard TK, Watkins RL, Pallister KB, Torres VJ, Horswill AR, Voyich JM. The role of innate immunity in promoting SaeR/S-mediated virulence in Staphylococcus aureus. Journal of innate immunity. 2014;6:21–30. doi: 10.1159/000351200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Malone CL, Boles BR, Lauderdale KJ, Thoendel M, Kavanaugh JS, Horswill AR. Fluorescent reporters for Staphylococcus aureus. Journal of microbiological methods. 2009;77:251–260. doi: 10.1016/j.mimet.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Holtfreter S, Nguyen TT, Wertheim H, Steil L, Kusch H, Truong QP, Engelmann S, Hecker M, Volker U, van Belkum A, Broker BM. Human immune proteome in experimental colonization with Staphylococcus aureus. Clin Vaccine Immunol. 2009;16:1607–1614. doi: 10.1128/CVI.00263-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Holtfreter S, Kolata J, Broker BM. Towards the immune proteome of Staphylococcus aureus - The anti-S. aureus antibody response. Int J Med Microbiol. 2010;300:176–192. doi: 10.1016/j.ijmm.2009.10.002. [DOI] [PubMed] [Google Scholar]
- 304.Broker BM, van Belkum A. Immune proteomics of Staphylococcus aureus. Proteomics. 2011;11:3221–3231. doi: 10.1002/pmic.201100010. [DOI] [PubMed] [Google Scholar]
- 305.Fairbrother RW. Coagulase production as a criterion for the classification of the staphylococci. J Pathol. 1940;50:5. [Google Scholar]
- 306.Tager M. Studies on the nature and the purification of the coagulase-reacting factor and its relation to prothrombin. J Exp Med. 1956;104:675–686. doi: 10.1084/jem.104.5.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Vandenesch F, Celard M, Arpin D, Bes M, Greenland T, Etienne J. Catheter-related bacteremia associated with coagulase-positive Staphylococcus intermedius. Journal of clinical microbiology. 1995;33:2508–2510. doi: 10.1128/jcm.33.9.2508-2510.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Hanselman BA, Kruth SA, Rousseau J, Weese JS. Coagulase positive staphylococcal colonization of humans and their household pets. Can Vet J. 2009;50:954–958. [PMC free article] [PubMed] [Google Scholar]
- 309.Bannoehr J, Guardabassi L. Staphylococcus pseudintermedius in the dog: taxonomy, diagnostics, ecology, epidemiology and pathogenicity. Vet Dermatol. 2012;23:253–266. e251–252. doi: 10.1111/j.1365-3164.2012.01046.x. [DOI] [PubMed] [Google Scholar]
- 310.Bannoehr J, Ben Zakour NL, Reglinski M, Inglis NF, Prabhakaran S, Fossum E, Smith DG, Wilson GJ, Cartwright RA, Haas J, Hook M, van den Broek AHM, Thoday KL, Fitzgerald JR. Genomic and Surface Proteomic Analysis of the Canine Pathogen Staphylococcus pseudintermedius Reveals Proteins That Mediate Adherence to the Extracellular Matrix. Infection and immunity. 2011;79:3074–3086. doi: 10.1128/IAI.00137-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Pietrocola G, Geoghegan JA, Rindi S, Di Poto A, Missineo A, Consalvi V, Foster TJ, Speziale P. Molecular Characterization of the Multiple Interactions of SpsD, a Surface Protein from Staphylococcus pseudintermedius, with Host Extracellular Matrix Proteins. PloS one. 2013:8. doi: 10.1371/journal.pone.0066901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Bergonier D, de Cremoux R, Rupp R, Lagriffoul G, Berthelot X. Mastitis of dairy small ruminants. Vet Res. 2003;34:689–716. doi: 10.1051/vetres:2003030. [DOI] [PubMed] [Google Scholar]
- 313.Piessens V, De Vliegher S, Verbist B, Braem G, Van Nuffel A, De Vuyst L, Heyndrickx M, Van Coillie E. Intra-species diversity and epidemiology varies among coagulase-negative Staphylococcus species causing bovine intramammary infections. Vet Microbiol. 2012;155:62–71. doi: 10.1016/j.vetmic.2011.08.005. [DOI] [PubMed] [Google Scholar]
- 314.Unal N, Cinar OD. Detection of stapylococcal enterotoxin, methicillin-resistant and Panton-Valentine leukocidin genes in coagulase-negative staphylococci isolated from cows and ewes with subclinical mastitis. Trop Anim Health Prod. 2012;44:369–375. doi: 10.1007/s11250-011-0032-x. [DOI] [PubMed] [Google Scholar]
- 315.Otto M. Staphylococcus epidermidis--the ‘accidental’ pathogen. Nat Rev Microbiol. 2009;7:555–567. doi: 10.1038/nrmicro2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Otto M. Molecular basis of Staphylococcus epidermidis infections. Semin Immunopathol. 2012;34:201–214. doi: 10.1007/s00281-011-0296-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Mack D, Davies AP, Harris LG, Jeeves R, Pascoe B, Knobloch JK, Rohde H, Wilkinson TS. Staphylococcus epidermidis in Biomaterial-Associated Infections. In: Moriarty F, Zaat SAJ, Busscher H, editors. Biomaterials Associated Infection: Immunological Aspects and Antimicrobial Strategies. Springer; New York: 2013. pp. 25–56. [Google Scholar]
- 318.Schleifer KH. Isolation and characterization of staphylococci from human skin. Int J Syst Bacteriol. 1975;25:17. [Google Scholar]
- 319.van der Mee-Marquet N, Achard A, Mereghetti L, Danton A, Minier M, Quentin R. Staphylococcus lugdunensis infections: high frequency of inguinal area carriage. Journal of clinical microbiology. 2003;41:1404–1409. doi: 10.1128/JCM.41.4.1404-1409.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Bieber L, Kahlmeter G. Staphylococcus lugdunensis in several niches of the normal skin flora. Clinical microbiology and infection : the official publication of the European Society of Clinical Microbiology and Infectious Diseases. 2010;16:385–388. doi: 10.1111/j.1469-0691.2009.02813.x. [DOI] [PubMed] [Google Scholar]
- 321.Kloos WE, Musselwhite MS. Distribution and persistence of Staphylococcus and Micrococcus species and other aerobic bacteria on human skin. Appl Microbiol. 1975;30:381–385. doi: 10.1128/am.30.3.381-395.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Rupp ME, Soper DE, Archer GL. Colonization of the female genital tract with Staphylococcus saprophyticus. Journal of clinical microbiology. 1992;30:2975–2979. doi: 10.1128/jcm.30.11.2975-2979.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Schneider PF, Riley TV. Staphylococcus saprophyticus urinary tract infections: epidemiological data from Western Australia. Eur J Epidemiol. 1996;12:51–54. doi: 10.1007/BF00144428. [DOI] [PubMed] [Google Scholar]
- 324.Anguera I, Del Rio A, Miro JM, Matinez-Lacasa X, Marco F, Guma JR, Quaglio G, Claramonte X, Moreno A, Mestres CA, Mauri E, Azqueta M, Benito N, Garcia-de la Maria C, Almela M, Jimenez-Exposito MJ, Sued O, De Lazzari E, Gatell JM. Staphylococcus lugdunensis infective endocarditis: description of 10 cases and analysis of native valve, prosthetic valve, and pacemaker lead endocarditis clinical profiles. Heart. 2005;91:e10. doi: 10.1136/hrt.2004.040659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Liu PY, Huang YF, Tang CW, Chen YY, Hsieh KS, Ger LP, Chen YS, Liu YC. Staphylococcus lugdunensis infective endocarditis: a literature review and analysis of risk factors. J Microbiol Immunol Infect. 2010;43:478–484. doi: 10.1016/S1684-1182(10)60074-6. [DOI] [PubMed] [Google Scholar]
- 326.Tee WS, Soh SY, Lin R, Loo LH. Staphylococcus lugdunensis carrying the mecA gene causes catheter-associated bloodstream infection in premature neonate. Journal of clinical microbiology. 2003;41:519–520. doi: 10.1128/JCM.41.1.519-520.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Choi SH, Chung JW, Lee EJ, Kim TH, Lee MS, Kang JM, Song EH, Jun JB, Kim MN, Kim YS, Woo JH, Choi SH. Incidence, characteristics, and outcomes of Staphylococcus lugdunensis bacteremia. Journal of clinical microbiology. 2010;48:3346–3349. doi: 10.1128/JCM.00609-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Sampathkumar P, Osmon DR, Cockerill FR., 3rd Prosthetic joint infection due to Staphylococcus lugdunensis. Mayo Clin Proc. 2000;75:511–512. doi: 10.4065/75.5.511. [DOI] [PubMed] [Google Scholar]
- 329.Bocher S, Tonning B, Skov RL, Prag J. Staphylococcus lugdunensis, a common cause of skin and soft tissue infections in the community. Journal of clinical microbiology. 2009;47:946–950. doi: 10.1128/JCM.01024-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Ravaioli S, Selan L, Visai L, Pirini V, Campoccia D, Maso A, Speziale P, Montanaro L, Arciola CR. Staphylococcus lugdunensis, an aggressive coagulase-negative pathogen not to be underestimated. The International journal of artificial organs. 2012;35:742–753. doi: 10.5301/ijao.5000142. [DOI] [PubMed] [Google Scholar]
- 331.Heilbronner S, Hanses F, Monk IR, Speziale P, Foster TJ. Sortase A promotes virulence in experimental Staphylococcus lugdunensis endocarditis. Microbiology. 2013;159:2141–2152. doi: 10.1099/mic.0.070292-0. [DOI] [PubMed] [Google Scholar]
- 332.Frank KL, Patel R. Poly-N-acetylglucosamine is not a major component of the extracellular matrix in biofilms formed by icaADBC-positive Staphylococcus lugdunensis isolates. Infection and immunity. 2007;75:4728–4742. doi: 10.1128/IAI.00640-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Giormezis N, Kolonitsiou F, Makri A, Vogiatzi A, Christofidou M, Anastassiou ED, Spiliopoulou I. Virulence factors among Staphylococcus lugdunensis are associated with infection sites and clonal spread. European journal of clinical microbiology & infectious diseases : official publication of the European Society of Clinical Microbiology. 2015;34:773–778. doi: 10.1007/s10096-014-2291-8. [DOI] [PubMed] [Google Scholar]
- 334.Donvito B, Etienne J, Denoroy L, Greenland T, Benito Y, Vandenesch F. Synergistic hemolytic activity of Staphylococcus lugdunensis is mediated by three peptides encoded by a non-agr genetic locus. Infection and immunity. 1997;65:95–100. doi: 10.1128/iai.65.1.95-100.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Loverix L, Timmermans P, Benit E. Successful non-surgical treatment of endocarditis caused by Staphylococcus haemolyticus following transcatheter aortic valve implantation (TAVI) Acta clinica Belgica. 2013;68:376–379. doi: 10.2143/ACB.3344. [DOI] [PubMed] [Google Scholar]
- 336.Wong RW, Rhodes KM. Endophthalmitis caused by Staphylococcus hominis and two different colonies of Staphylococcus haemolyticus after cataract surgery. Retinal cases & brief reports. 2015;9:181–184. doi: 10.1097/ICB.0000000000000133. [DOI] [PubMed] [Google Scholar]
- 337.Silva PV, Cruz RS, Keim LS, Paula GR, Carvalho BT, Coelho LR, Carvalho MC, Rosa JM, Figueiredo AM, Teixeira LA. The antimicrobial susceptibility, biofilm formation and genotypic profiles of Staphylococcus haemolyticus from bloodstream infections. Memorias do Instituto Oswaldo Cruz. 2013;108:812–813. doi: 10.1590/0074-0276108062013022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Potter A, Ceotto H, Giambiagi-Demarval M, dos Santos KR, Nes IF, do Bastos MC. The gene bap, involved in biofilm production, is present in Staphylococcus spp. strains from nosocomial infections. Journal of microbiology. 2009;47:319–326. doi: 10.1007/s12275-009-0008-y. [DOI] [PubMed] [Google Scholar]
- 339.Latham RH, Running K, Stamm WE. Urinary tract infections in young adult women caused by Staphylococcus saprophyticus. JAMA. 1983;250:3063–3066. [PubMed] [Google Scholar]
- 340.Hovelius B, Mardh PA. Staphylococcus saprophyticus as a common cause of urinary tract infections. Rev Infect Dis. 1984;6:328–337. doi: 10.1093/clinids/6.3.328. [DOI] [PubMed] [Google Scholar]
- 341.Gatermann S, John J, Marre R. Staphylococcus saprophyticus urease: characterization and contribution to uropathogenicity in unobstructed urinary tract infection of rats. Infection and immunity. 1989;57:110–116. doi: 10.1128/iai.57.1.110-116.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Loes AN, Ruyle L, Arvizu M, Gresko KE, Wilson AL, Deutch CE. Inhibition of urease activity in the urinary tract pathogen Staphylococcus saprophyticus. Lett Appl Microbiol. 2014;58:31–41. doi: 10.1111/lam.12153. [DOI] [PubMed] [Google Scholar]
- 343.Keren I, Kaldalu N, Spoering A, Wang Y, Lewis K. Persister cells and tolerance to antimicrobials. FEMS microbiology letters. 2004;230:13–18. doi: 10.1016/S0378-1097(03)00856-5. [DOI] [PubMed] [Google Scholar]
- 344.Lewis K. Riddle of biofilm resistance. Antimicrobial agents and chemotherapy. 2001;45:999–1007. doi: 10.1128/AAC.45.4.999-1007.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Conlon BP. Staphylococcus aureus chronic and relapsing infections: Evidence of a role for persister cells: An investigation of persister cells, their formation and their role in S. aureus disease. BioEssays : news and reviews in molecular, cellular and developmental biology. 2014;36:991–996. doi: 10.1002/bies.201400080. [DOI] [PubMed] [Google Scholar]
- 346.Koch G, Yepes A, Forstner KU, Wermser C, Stengel ST, Modamio J, Ohlsen K, Foster KR, Lopez D. Evolution of resistance to a last-resort antibiotic in Staphylococcus aureus via bacterial competition. Cell. 2014;158:1060–1071. doi: 10.1016/j.cell.2014.06.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Krismer B, Peschel A. Does Staphylococcus aureus nasal colonization involve biofilm formation? Future microbiology. 2011;6:489–493. doi: 10.2217/fmb.11.37. [DOI] [PubMed] [Google Scholar]
- 348.Dastgheyb S, Parvizi J, Shapiro IM, Hickok NJ, Otto M. Effect of biofilms on recalcitrance of staphylococcal joint infection to antibiotic treatment. The Journal of infectious diseases. 2015;211:641–650. doi: 10.1093/infdis/jiu514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Haaber J, Cohn MT, Frees D, Andersen TJ, Ingmer H. Planktonic aggregates of Staphylococcus aureus protect against common antibiotics. PloS one. 2012;7:e41075. doi: 10.1371/journal.pone.0041075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Walker JN, Crosby HA, Spaulding AR, Salgado-Pabon W, Malone CL, Rosenthal CB, Schlievert PM, Boyd JM, Horswill AR. The Staphylococcus aureus ArlRS Two-Component System Is a Novel Regulator of Agglutination and Pathogenesis. PLoS pathogens. 2013;9:e1003819. doi: 10.1371/journal.ppat.1003819. [DOI] [PMC free article] [PubMed] [Google Scholar]