Abstract
The thyroid hormone–inactivating (TH-inactivating) enzyme type 3 iodothyronine deiodinase (D3) is an oncofetal protein that is rarely expressed in adult life but has been shown to be reactivated in the context of proliferation and neoplasms. D3 terminates TH action within the tumor microenvironment, thereby enhancing cancer cell proliferation. However, the pathological role of D3 and the contribution of TH metabolism in cancer have yet to be fully explored. Here, we describe a reciprocal regulation between TH action and the cancer-associated microRNA-21 (miR21) in basal cell carcinoma (BCC) skin tumors. We found that, besides being negatively regulated by TH at the transcriptional level, miR21 attenuates the TH signal by increasing D3 levels. The ability of miR21 to positively regulate D3 was mediated by the tumor suppressor gene GRHL3, a hitherto unrecognized D3 transcriptional inhibitor. Finally, in a BCC mouse model, keratinocyte-specific D3 depletion markedly reduced tumor growth. Together, our results establish TH action as a critical hub of multiple oncogenic pathways and provide functional and mechanistic evidence of the involvement of TH metabolism in BCC tumorigenesis. Moreover, our results identify a miR21/GRHL3/D3 axis that reduces TH in the tumor microenvironment and has potential to be targeted as a therapeutic approach to BCC.
Introduction
Basal cell carcinoma (BCC) is the most frequently diagnosed human cancer and accounts for approximately 80% of all nonmelanoma skin cancers. BCC formation is based on a combination of inherited and environmental factors, mainly UV irradiation and long-term sun exposure. Proliferation and survival of BCC cells is physiologically regulated by the hedgehog (Hh) pathway, which plays a pivotal role in embryogenic development and tumorigenesis (1–4). Sporadic BCCs in humans are predominantly due to inactivation of PTCH1, an inhibitor of Hh signaling, and the consequent activation of Gli transcription factors (5, 6). Therapeutics targeting Sonic hedgehog (SHH) signaling are now being used in patients and represent an important nonsurgical approach to interfere with BCC occurrence and growth (7).
Type 3 iodothyronine deiodinase (D3, encoded by the DIO3 gene) is an oncofetal protein commonly deregulated in a wide range of adult tissues and re-expressed in cancer (8). D3 belongs to the deiodinase family of enzymes, which regulate thyroid hormone (TH) availability in target cells, thereby allowing the intracellular adaptation of the TH signal (9). The major product of the thyroid gland is T4, while the only active TH (T3) derives from the conversion of T4 into T3 catalyzed by the deiodinases D1 and D2. D3 degrades T4 and T3 into inactive metabolites and is thus the major physiological inactivator of TH (9). D3 is overexpressed in BCCs of both mouse and human origin, and is under the control of SHH, which increases D3 expression by acting via a conserved GLI2 binding site on the human DIO3 promoter (10). By decreasing TH action in the BCC microenvironment, D3 promotes BCC tumorigenesis. In addition, GLI2 and the SHH pathway are negative targets of TH, which thus attenuates cancer formation (11).
Among all cancer-related microRNAs (miRNAs), miR21 has been related to the pathogenesis of various malignant tumors, including prostate, gastric, colon, breast, and lung cancer (12–20). Apart from being involved in cancerogenesis, miR21 plays a crucial role in a plethora of biological functions and diseases, namely development, cardiovascular diseases, and inflammation (21). miR21 is also expressed in skin (22–24), and its potential role as a regulator of skin homeostasis is demonstrated by its upregulation in such diseases as psoriasis, atopic dermatitis (25, 26), melanoma, and squamous cell carcinoma (SCC) (27–29). However, nothing is known about miR21 expression in BCC.
GRHL3 is a transcription factor expressed in the skin in the differentiated suprabasal layers, and is essential for epidermal differentiation and barrier formation at the end of mouse embryogenesis (30–33). Grhl3 KO mice exhibit impaired epidermal differentiation and decreased expression of several genes involved in barrier formation (32). Accordingly, in humans, dominant-negative GRHL3 mutations are associated with defective periderm development in Van der Woude syndrome (34). Moreover, GRHL3 was found to be a potent tumor suppressor in mouse and human SCC, and an upstream regulator of PTEN (31).
In this study we demonstrate that miR21 is a negative TH target. Moreover, besides being regulated by TH, miR21 also modulates TH metabolism by inducing D3 expression. Importantly, we found that a miR21 target gene, GRHL3, which is essential for keratinocyte differentiation, is a novel suppressor of D3 to our knowledge and accounts for the positive regulation of miR21 on D3 levels.
Our data demonstrate the existence of a miR21/GRHL3/D3 regulatory circuit that leads to a reduction of the TH signal in the tumor microenvironment. Disruption of this regulation in vivo drastically attenuates the oncogenic potential of BCC cells. Indeed, targeted mutagenesis of D3 potently reduces the tumorigenic ability of BCC cells and of miR21-overexpressing BCC cells in a xenograft mouse model. Furthermore, genetic D3 depletion in the keratinocyte compartment drastically reduced formation of BCC-like tumors in a GLI2-driven mouse model.
Together, our results provide what we believe to be novel insight into the function of TH in regulating the skin tumorigenic program and reveal a general mode of TH downregulation by oncogenic signals.
Results
TH regulates the expression of multiple miRNAs and, in particular, downregulates miR21 in BCC.
We previously showed that TH regulates the proliferative potential of BCC cells in vitro and in vivo (10, 11). To explore whether TH action in BCC involves the regulation of miRNAs, we analyzed the differential expression of mature miRNAs (miChip) in D3-silenced mouse BCC cells. We used a microarray platform of miRNA, the Exiqon platform (miRCURY LNA Arrays; Exiqon, Denmark), to compare the expression of miRNAs in BCC cells transfected with a control scrambled shRNA oligo versus the D3-targeting shRNA oligo (10). This analysis showed that 46 miRNAs were differentially expressed (Supplemental Table 1; supplemental material available online with this article; doi:10.1172/JCI84465DS1). Among them, we selected miR21 for further studies, since it is overexpressed in a wide variety of cancers and has been causally linked to cell proliferation, apoptosis, and migration (12, 13), and, importantly, it is the most sensitive target upon D3 depletion (variation of 6.62-fold; Figure 1A and Supplemental Table 1).
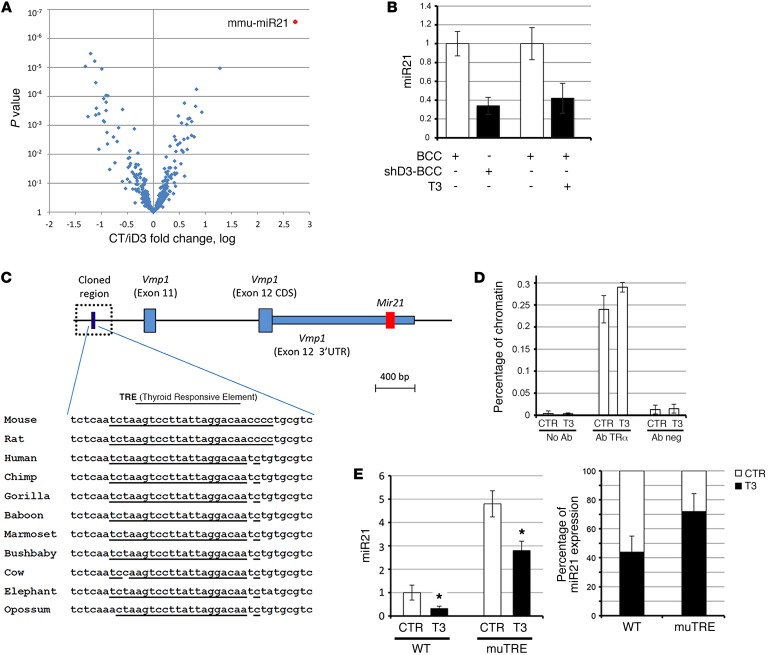
Figure 1. TH represses miR21 in mouse and human BCC.
(A) Genome-wide miRNA expression analysis in D3-depleted and control BCC cells. All the deregulated miRNAs in shD3 relative to control cells are plotted against the P value. mmu-miR21, murine miR-21. (B) miR21 expression was measured by TaqMan real-time PCR in cDNAs from control BCC, shD3, and BCC cells treated with 30 nM T3 for 48 hours. (C) Schematic structural organization of the miR21 gene (red) and the miR21 enhancer region. The expression of the thyroid hormone–responsive element (TRE) and its comparison between species are indicated below. (D) ChIP analysis of the interaction between TRα and the miR21 enhancer region. Chromatin extracted from BCC cells was immunoprecipitated using the indicated antibodies. (E) miR21 expression was measured by TaqMan PCR in BCC cells carrying a mutated TRE element in the miR21 region (mutTRE) and control BCC cells (WT) treated or not with 30 nM T3 for 48 hours. Right: Reduction of miR21 in T3-treated cells versus nontreated cells in WT cells and CRISPR/Cas9–mediated TRE-mutated cells. *P < 0.05. Data represent the mean of at least 3 independent experiments in duplicate. SDs are indicated. Differences between samples were assessed by Student’s 2-tailed t test for independent samples.
We validated miR21 as a negative TH target gene by measuring the mature miR21 transcript by TaqMan RT-PCR and by Northern blotting of BCC cells treated with TH (30 nM T3), and in D3-depleted BCC cells (shD3) (10), in which the TH signal was amplified (Figure 1B and Supplemental Figure 1A). Accordingly, while repressed by TH treatment and D3 depletion, miR21 was upregulated by D3 overexpression and by TH deprivation in serum (Supplemental Figure 1, B and C). To explore the possibility that TH directly inhibits miR21 transcription, we analyzed the sequence of the miR21 enhancer region (35). In silico analysis revealed a conserved thyroid hormone–responsive element–binding (TRE-binding) site within the enhancer (Figure 1C). We cloned 389 bp of the enhancer within the pGL3-basic vector to perform functional studies in the human embryonic kidney (HEK) 293 cell line and in mouse BCC cells. As shown in Supplemental Figure 2, A and B, the miR21 enhancer was efficiently downregulated by TH and by double thyroid receptor transfection/TH treatment in both cell lines, thus demonstrating that TH represses the transcription of the pre-miR21. ChIP analysis revealed that the thyroid receptor (TR) physically binds to the newly identified TRE (Figure 1D). Furthermore, we mutagenized the core motif of the TRE within the miR21 enhancer region using CRISPR/Cas9 technology in the BCC cells. Effective mutagenesis was confirmed by sequencing of the DNA of clones containing the mutated TRE. One of the positive clones was treated with T3, and miR21 expression was measured using TaqMan PCR. As shown in Figure 1E, the endogenous miR21 was expressed at higher levels in cells with mutated TRE when compared with the control, WT BCC cells. Furthermore, the repression of miR21 by T3 was partially reduced in cells with mutated TRE, which demonstrates that mutation of the TRE attenuates the effect of T3 on miR21 (Figure 1E, right). The fact that TRE mutation does not completely abolish the regulation of T3 on miR21 suggests the existence of multiple TREs in other parts of the miR21 regulatory elements. Together, these results point to a negative regulation of T3 on the miR21 transcription via a direct T3-TR binding to the miR21 enhancer.
miR21 positively regulates D3 expression.
Next, we explored the potential of miR21 to modulate the TH signal in BCC cells. To this aim, we measured the expression of deiodinases D2 and D3 and other TH signaling modulators (TRα and TRβ, MCT8 and MCT10) in BCC cells and in BCC cells transiently overexpressing miR21. While DIO2 mRNA, the TH receptors, and the transporters were not modified by miR21 expression (Supplemental Figure 3), overexpression of miR21 strongly over induced D3 levels in a time-dependent manner (Figure 2A and Supplemental Figure 3).
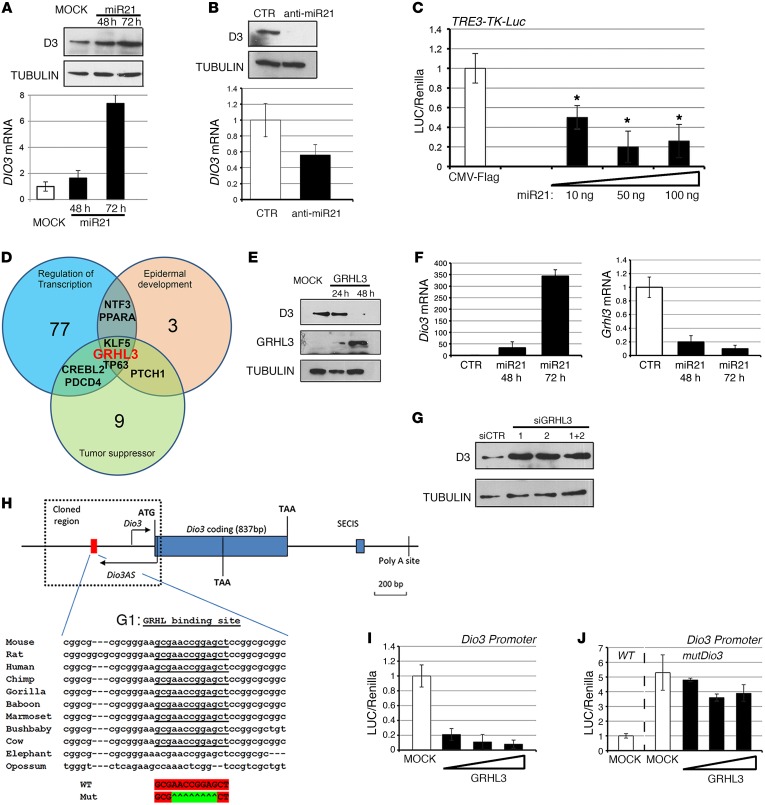
Figure 2. miR21 upregulates D3 expression by targeting GRHL3.
(A) BCC cells were transiently transfected with miR21 expression plasmid, and cells were collected at the indicated times. D3 mRNA and protein were measured by real-time PCR and Western blot (see complete unedited blots in the supplemental material). (B) LNA anti-miR21 or a control LNA-scrambled oligo was transfected in BCC cells, and D3 mRNA and protein were measured as in A. (C) BCC cells transfected with the artificial T3-responsive promoter TRE3-TK-Luc and CMV-renilla as internal control. Increasing amounts of miR21 were cotransfected, and cells were analyzed for luciferase activity. The results are shown as means ± SD of the luciferase/renilla (LUC/Renilla) ratios from at least 3 separate experiments, performed in duplicate. (D) miR21 target genes were classified for functional classification with the DAVID program (36). (E) D3 protein levels were measured by Western blot in BCC cells transfected with Flag-GRHL3 and harvested at the indicated times. GRHL3 (anti-Flag) and tubulin levels were measured as control. (F) Primary cultures of keratinocytes were transfected with miR21, and cells were harvested at 48 and 72 hours. Dio3 and Grhl3 mRNAs were measured by real-time PCR. (G) BCC cells were transfected with 2 different siRNAs targeting endogenous Grhl3 oligos or a combination of them, and D3 protein was measured by Western blot analysis. (H) Schematic representation of the Dio3 locus, the Dio3 promoter, and the G1 site (red) responsive to GRHL. (I and J) BCC cells were transiently transfected with Dio3-Luc promoter (I) or Dio3-Luc and mutDio3-Luc promoters (J) and with increasing amounts of the GRHL3 plasmid. Cells were harvested 48 hours after transfection and analyzed for luciferase activity. CMV-renilla was cotransfected as internal control. The results are shown as means ± SD of the LUC/renilla ratios from at least 3 separate experiments, performed in duplicate. *P < 0.05.
We then inhibited miR21 in BCC cells by transfecting the LNA oligonucleotide complementary to miR21 (anti-miR21). In accordance with its role as an inducer of D3 expression, miR21 inhibition potently reduced D3 expression at the mRNA and protein levels (Figure 2B). Importantly, overexpression of miR21 also attenuated intracellular TH action as indicated by the cotransfection of increasing amounts of miR21 together with an artificial T3-responsive promoter (Figure 2C).
miR21 induces D3 expression by targeting the tumor suppressor GRHL3.
miRNAs often inhibit mRNA expression at the posttranscriptional level. Since D3 is induced by miR21, we tested the hypothesis that a direct miR21 target, and putative suppressor of D3, might mediate this effect. To this aim, we searched for miR21 downstream targets that could potentially inhibit D3 expression. By using the computational algorithm TargetScan (http://www.targetscan.org/) together with published and/or validated miR21 target genes, we obtained a list of genes potentially controlled by miR21 (Supplemental Table 2). This list was further analyzed with the DAVID program for functional classification (36). By selecting the categories of “Regulation of transcription,” “Epidermal development,” and “Tumor suppressor,” we found only a few genes that were present in all these 3 categories (Figure 2D). We tested the ability of 3 different genes belonging to these categories, TP63, KLF5, and GRHL3, to affect D3 expression. Among these, only 1 gene repressed D3 expression, namely GRHL3 (Figure 2E and Supplemental Figure 4).
GRHL3 is a tumor suppressor functionally related to SCC formation (31, 33). We first evaluated the expression of GRHL3 in BCC and normal skin, and, as shown in Supplemental Figure 5A, we observed that GRHL3 is highly expressed in normal skin and in WT keratinocytes, while it is repressed in BCC tissues and cells. We also tested whether GRHL3 is a miR21 downstream target in the BCC cells, as already demonstrated in other cell contexts (31). BCC cells transfected with miR21 or with anti-miR21 LNA (shown in Figure 2, A and B) were analyzed for GRHL3 mRNA and protein expression. miR21 overexpression reduced GRHL3, while miR21 silencing increased GRHL3 levels, thus validating GRHL3 as a miR21 target in the BCC cell context (Supplemental Figure 6). Importantly, the same effects were observed in primary cultures of keratinocytes, in which, thanks to the very low level of endogenous miR21, overexpression of miR21 changed DIO3 and GRHL3 to a much greater extent than it did in BCC cells (Figure 2F).
To determine whether GRHL3 indeed negatively regulates D3, we knocked down the endogenous protein using 2 different siRNA oligos (siGRHL3-1 and siGRHL3-2). D3 levels were significantly higher in BCC cells transfected with 1 or more siGRHL3 oligos than in BCC cells transfected with scrambled oligos (siCTR). This result confirms that GRHL3 is a repressor of D3 expression (Figure 2G and Supplemental Figure 5B).
We next tested whether GRHL3 directly binds to the Dio3 promoter. In silico analysis revealed a putative GRHL3 binding site within the promoter region of the Dio3 gene (G1: GCGAACCGGAGCT), which is highly conserved, and located 436 bp from the ATG codon (Figure 2H). Functional studies demonstrated that GRHL3 binds to the G1 site and represses the activity of the Dio3 promoter. Indeed, cotransfection of the mouse Dio3 promoter (pmD3Luc) with increasing amounts of GRHL3 showed that the Dio3 promoter is potently inhibited by GRHL3 (Figure 2I). Mutation of the G1 site within the Dio3 promoter revealed that, while the basal activity of the mutated promoter was increased as expected, the GRHL3-induced repression was abolished (Figure 2, I and J, and Supplemental Figure 6D). These data indicate that the tumor suppressor GRHL3 is a direct repressor of D3.
To assess the contribution of GRHL3 to the miR21-dependent D3 upregulation, we transfected miR21 or GRHL3 or both in BCC cells (Figure 3A). The forced expression of GRHL3 downregulated D3 expression and, most importantly, abolished the miR21-mediated induction of D3 (Figure 3A). This observation was confirmed by a different approach, cotransfection of the Dio3 promoter with miR21 alone and with miR21 plus GRHL3. Also in this context, GRHL3 antagonized the effects induced by miR21 on D3 (Figure 3C). Interestingly, concomitant miR21 overexpression and GRHL3 downregulation by siRNA did not result in synergistic D3 upregulation, which reinforces the concept that suppression of GRHL3 is the mechanism by which miR21 enhances D3 (Figure 3, B and D). Notably, miR21 and GRHL3 expression ultimately decreased and increased, respectively, the effect of TH in the nucleus (Figure 3E). Furthermore, when the G1 site in the Dio3 promoter was mutated, miR21 failed to regulate D3 expression, thus reinforcing the finding that GRHL3 is the only gene mediating miR21-dependent D3 regulation (Supplemental Figure 6D). The picture that emerges from these results is that miR21 upregulates D3 expression by inhibiting the D3 repressor GRHL3.
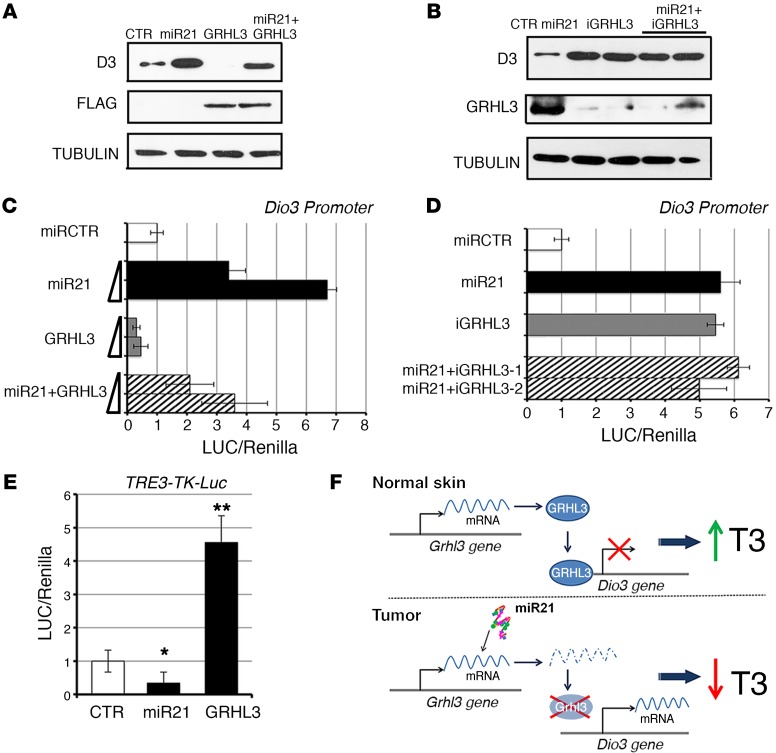
Figure 3. The miR21/GRHL3 axis regulates D3 expression and TH intracellular concentration.
(A) D3 protein levels were measured by Western blot from BCC cells transfected with miR21, GRHL3, and the 2 expression plasmids together. Lysates were also probed with anti-Flag antibody as control of Flag-GRHL3 transfection efficiency. (B) BCC cells were transfected with miR21 and miR21 plus 2 different siRNAs for GRHL3 knockdown. Anti-GRHL3 antibody was used as internal control of GRHL3 knockdown. (C and D) The Dio3 promoter was cotransfected with the indicated constructs together with CMV-renilla as internal control. Luciferase activity was measured and reported as means ± SD of the LUC/renilla ratios from at least 3 separate experiments, performed in duplicate. (E) The T3-responsive promoter TRE3-TK-Luc was cotransfected in BCC cells with the indicated plasmids. Luciferase activity was measured as in C and D. *P < 0.05, **P < 0.01. (F) The miR21/GRHL3/D3 axis regulates intracellular T3 availability.
Since TH negatively regulates miR21, we tested the possibility that TH treatment could in turn increase GRHL3 expression. Indeed, we observed that T3 treatment positively regulates GRHL3 and that this effect is mediated by miR21, as demonstrated by the results of T3 treatment and miR21 silencing shown in Supplemental Figure 7.
The identified miR21/GRHL3/D3 axis and its ability to modulate TH activity are depicted in Figure 3F.
BCC tumorigenesis requires an intact miR21/GRHL3/D3 axis.
To assess the functional role of the newly identified network, we sought to disrupt the miR21/D3 axis by depleting D3 or by inducing miR21 in a genetic Dio3-null background in vitro and in vivo. We used the CRISPR/Cas9 technology to genetically deplete D3 in BCC cells (D3KO clones). We also stably overexpressed miR21 in BCC cells (miR cells). One representative D3KO clone was subsequently stably transfected with the miR21 plasmid, and double D3KO-miR21-overexpressing cells were selected (D3KO-miR cells). We first confirmed D3 depletion in D3KO clones (Supplemental Figure 8, A−C), and miR21 and D3 expression in miR cells (Supplemental Figure 8, D and E) and in double D3KO-miR cells (Supplemental Figure 8, F and G). Because the level of miR21 was higher in BCC tissue than in normal skin (Figure 1, F and G), we tested whether the increase of miR21 exceeded its basal levels of transcription. Indeed, we observed that miR21 is 10-fold overexpressed in miR clones versus control BCC cells (Supplemental Figure 8D). GRHL3 expression was very low in all generated cells and was further suppressed by miR21 overexpression (Supplemental Figure 8, H and I).
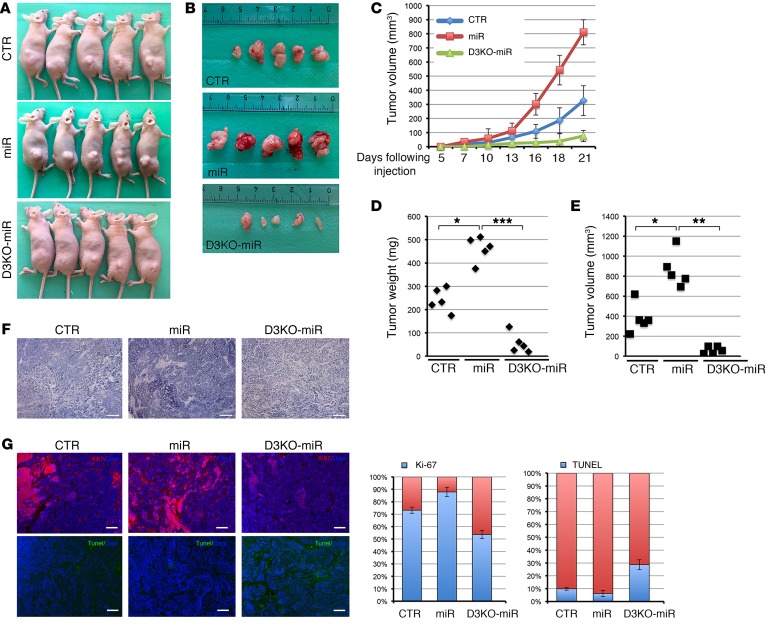
We next subcutaneously injected the cells generated into athymic (nude) mice and examined the weight and morphology of the resulting subcutaneous xenograft tumors. Tumors originating from miR cells grew rapidly, and were palpable 2 weeks after injection (Figure 4, A and B). D3KO-miR cells generated much smaller tumors, which become palpable 3 weeks after injection, while D3KO cells did not generate tumors (Figure 4, A–C). Importantly, miR21 overexpression exacerbated tumorigenesis versus control BCC cells (Figure 4, A–C). On the contrary, D3 ablation almost completely abolished the tumorigenic advantage acquired upon miR21 overexpression; consequently, D3 depletion results in a potent antitumoral effect even in the presence of an excess of miR21. We found a 3.54-fold reduction (D3KO-miR) and a 2.04-fold increase (miR21) in tumor weight, and a 4.2-fold reduction (D3KO-miR) and a 2.2-fold increase (miR21) in tumor volume, compared with control cell–generated tumors (Figure 4, D and E). Accordingly, H&E, Ki-67, and TUNEL staining showed that control and miR-derived tumors have a high mitotic grade, while D3KO-miR–derived tumors showed multiple areas of cell death (Figure 4, F and G). Consistent with increased D3 expression in miR cells versus control cells, D3 mRNA and protein were upregulated in tumors overexpressing miR21 (Supplemental Figure 9). Furthermore, none of the generated tumors expressed GRHL3, thus confirming the tumor-suppressor role of this transcriptional factor (Supplemental Figure 10).
Figure 4. Targeted D3 depletion drastically reduces the tumorigenic potential of BCC cells in nude mice.
(A and B) 1 × 106 per 100 μl of control CRISPR-CTR cells (CTR), stable miR21-overexpressing (miR) BCC cells, CRISPR/Cas9-D3KO cells (D3KO), and double D3KO-miR cells were injected s.c. into the flanks of nude mice. Mice were collected 3 weeks after injection, and tumors were measured as indicated in Methods. (C) Tumor volume was measured with a caliper for 3 weeks in CTR, miR, and D3KO-miR cells deriving tumors. (D and E) Tumor weight and volume measured 3 weeks after cell injection. (F and G) Tumor sections were analyzed by H&E staining and by immunofluorescence for Ki-67 and TUNEL expression in the indicated tumors. Scale bars: 100 μm. Right: Percentage of Ki-67–positive and TUNEL-positive cells. *P < 0.05, **P < 0.01, ***P < 0.001.
Genetic D3 depletion potently reduces BCC-like tumor growth in vivo.
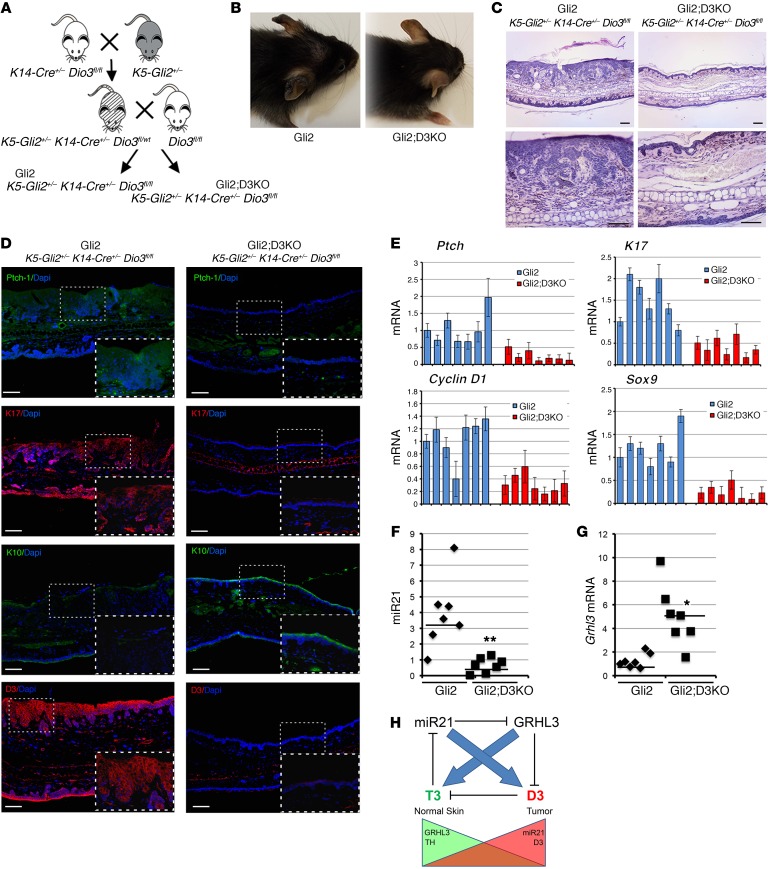
Having shown that D3 depletion drastically attenuates BCC tumorigenesis and miR21 oncogenic potential in xenografts, we evaluated the role of D3 depletion in genetically induced BCC tumors in vivo. To this aim, we generated a keratinocyte-specific conditional D3-null mouse (K14-Cre+/– Dio3fl/fl) in a mouse model developing BCC-like tumors (K5-Gli2) (37), thereby generating a spontaneously forming mouse BCC-like model in an epidermal-specific D3-null background, namely, K5-Gli2 K14-Cre+/– Dio3fl/fl, henceforth referred to as “Gli2;D3KO” (Figure 5A), while K5-Gli2 K14-Cre–/– Dio3fl/fl mice (referred to as “Gli2”) served as controls. Constitutive activation of GLI2 in keratinocytes of Gli2 mice resulted in epidermal hyperplasia, BCC, and basaloid follicular hematoma with conspicuous down-growth, arising mostly on the ears of mice at 6 months of age (Figure 5, B and C, and Supplemental Figure 6). Strikingly, Dio3 deletion in Gli2;D3KO mice reduced the number and size of tumors (Figure 5, B and C, and Supplemental Figure 6). The few lesions that developed on the ears of Gli2;D3KO mice were much smaller than those found in Gli2 mice. Expression of the BCC markers K17 and PTCH1 and SOX9 was drastically reduced in D3-depleted mice as measured by immunofluorescence and RT-PCR (Figure 5, D and E). Additionally, K10, a marker of the differentiating layers of the epidermis, was significantly downregulated in Gli2 mice, while it was well preserved in Gli2;D3KO mice (Figure 5D).
Figure 5. Epidermal D3 depletion significantly reduces Gli2-driven BCC-like tumor formation and inhibits miR21 expression.
(A) Genetic D3 depletion was specifically induced in keratinocytes by crossing of K5-Gli2 K14-Cre–/+ with Dio3fl/fl mice. (B) Mice were analyzed at 6 months of age when BCC-like formations were abundant and readily visible on the ears of control (Gli2) mice (n = 11). (C) Histological analysis of ears from Gli2 and Gli2;D3KO mice. H&E stainings were performed on sections of ears from Gli2 and Gli2;D3KO mice. Scale bars: 100 μm. (D) Immunofluorescent staining for PTCH1, K17, K10, and D3 expression. Scale bars: 100 μm. (E) Real-time PCR analysis to measure the expression of the indicated genes in Gli2 versus Gli2;D3KO mice (n = 7). (F and G) miR21 and Grhl3 expression was measured in the ears from Gli2 and Gli2;D3KO mice (n = 7). (H) Schematic representation of the mutual, negative-feedback loop by which TH regulates miR21 and miR21 controls intracellular TH through GRHL3. *P < 0.05, **P < 0.01.
We next evaluated whether, by enhancing TH levels, D3 ablation in skin could affect the expression of miR21 and GRHL3 in the tumor. As shown in Figure 5, F and G, miR21 was drastically reduced upon D3 depletion, while GRHL3 was upregulated in the absence of D3.
The data reported in this section indicate a functional requirement of D3 for growth in Hh pathway–driven skin tumors. These data also suggest that effective D3 loss and consequent TH activation lead to a remarkable attenuation of miR21 expression in vivo in BCC and a corresponding increase in the miR target GRHL3, and functionally validate our model of a reciprocal regulation of TH on miR21 and miR21 over TH metabolism in skin. The TH-miR21 interplay is schematically represented in Figure 5H.
The miR21/GRHL3/D3 axis is recapitulated in human BCC.
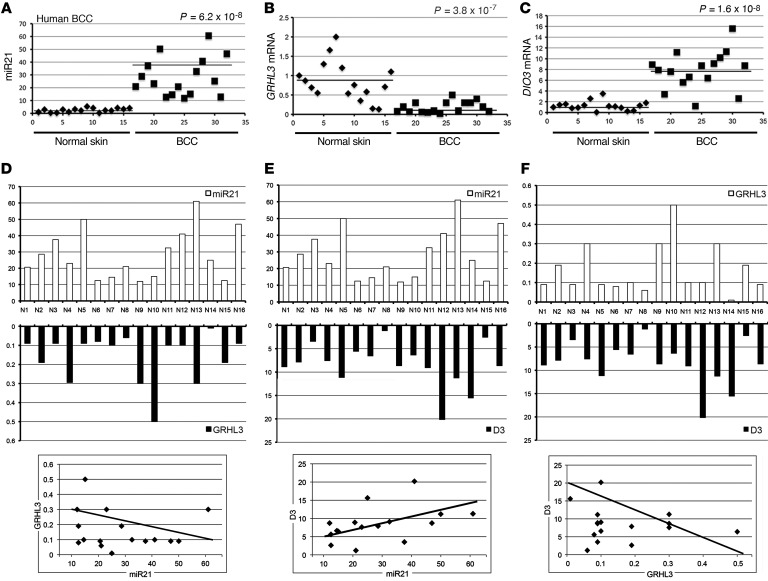
Lastly, we sought to address whether the regulation of T3 on miR21 and GRHL3 is recapitulated also in human BCC. To this aim, we evaluated miR21 levels in 16 human BCC tissues versus samples of healthy skin from the same patients by TaqMan real-time RT-PCR. In accordance with the concept that miR21 is a marker of tumorigenesis, miR21 was expressed at significantly higher levels in BCC tissue than in the healthy skin samples (12.05-fold, P = 6.2 × 10−8; Figure 6A). Similar miR21 overexpression in tumors occurred in mouse BCC cells and tumors (Supplemental Figure 11). We also measured GRHL3 and DIO3 expression in the human samples (Figure 6, B and C). As expected, GRHL3 was highly expressed in normal skin and drastically repressed in most of the BCC samples. D3 expression correlated with miR21 expression (Figure 6C).
Figure 6. miR21 inversely correlates with GRHL3 and is positively correlated with D3 in human BCCs.
(A–C) The expression of miR21, GRHL3, and DIO3 was measured in 16 human BCC tumors compared with normal skin counterparts. All samples were run in triplicate and referred to normal skin in sample 1 set arbitrarily as 1. P values were calculated by 1-way ANOVA. (D–F) The same data as in A–C were reported as scale bars, and correlations between miR21/GRHL3, miR21/D3, and D3/GRHL3 were calculated by Pearson’s correlation analysis (bottom).
Considering that miR21 and D3 are upregulated in human BCC, and GRHL3 is remarkably lower in BCC than in nontumor tissues, we performed a correlation analysis between miR21/GRHL3, miR21/D3, and GRHL3/D3 expression in BCC tissues. Using Pearson’s correlation analysis, we observed a significant inverse correlation between miR21 and GRHL3 and between D3 and GRHL3; conversely, miR21 expression and D3 expression were linked by a positive correlation (Figure 6, D–F; miR21 vs. GRHL3: R = –0.35, P = 0.048; miR21 vs. D3: R = 0.5, P = 0.0049; GRHL3 vs. D3: R = –0.35, P = 0.048). The inverse correlation between miR21 and GRHL3 is in line with the demonstration by Bhandari et al. that miR21 negatively regulates GRHL3 in skin (33). Taken together, these results confirm the model established in our study and suggest that targeting D3 and increasing the TH signal might represent a therapeutic approach to BCC.
Discussion
Here we describe what we believe to be a novel network by which TH signaling, by modulating miRNA expression, interacts with oncogenic pathways to regulate the growth of BCC tumors. T3 suppresses miR21 expression, thus attenuating BCC, and is contextually regulated by miR21 through the control of the TH-inactivating enzyme D3. These conclusions are supported by genome-wide, biochemical, and expression data coupled with functional studies in vivo in xenografts and in a BCC mouse model.
TH deregulation is frequent in human tumors, but it is not known whether it is a bystander event or whether it might drive or control cancer growth in vivo. Notably, the TH-inactivating enzyme D3 is upregulated in many human tumors and cancer cell lines (8). Determining the role of TH in cancer is challenging, mostly because TH plays pleiotropic roles in the body and it has been shown to trigger distinct, even opposing, functions in different cellular districts. On the basis of in vitro findings and studies conducted with a xenograft model, we previously reported that TH induces apoptosis and inhibits cell-cycle progression in BCC (10, 11). Our present findings complement these previous reports and extend the function of TH in BCC to the control of miR21-dependent oncogenic action. Importantly, using a unique skin-specific Dio3 KO mouse model, we provide the first in vivo genetic evidence that D3 depletion, which is equivalent to TH treatment, drastically reduces BCC formation.
TH signaling, miR21, and cancer.
The classical mode of TH action is mainly mediated by nuclear receptors, the TRs, which are ligand-dependent transcription factors that bind to TREs in the promoter of target genes. In the absence of the ligand, TRs are associated with corepressors and with histone deacetylases, thereby preventing transcription. Upon T3 binding, the repressive complex is destabilized and corepressors are replaced by coactivators that ultimately recruit RNA polymerase II in transcription of target genes (38). Many cell cycle–related and proliferation-controlling genes are TH targets, and, as such, TH positively or negatively controls cell proliferation and differentiation in various contexts (8). Moreover, TH signaling is often compromised in carcinogenesis as demonstrated by the reduced expression or deletion of TR genes in human cancers, which suggests that TRs could function as tumor suppressors. The inverse association between TH and cancer was confirmed by the findings that the v-erb-a oncogene, which is the avian erythroblastosis gene, is a mutated form of the THRA gene (coding for the TRα protein) (39), and that v-erb-a antagonizes T3 function in avian erythroblastosis virus–transformed erythroleukemic cells, thereby blocking T3-dependent cell differentiation (40).
In this perspective, our finding that TH exerts negative control over miR21 expression is particularly relevant. miR21 is one of a handful of key miRNAs that promote human and mouse oncogenesis (12–20). Here, we provide what is to our knowledge the first evidence that miR21 is overexpressed in human and mouse BCC, while it is barely detectable in normal skin (Figure 6 and Supplemental Figure 11). Our data demonstrate that miR21 is suppressed by TH at the transcriptional level by directly binding to a TRE located within the miR21 enhancer region. Decreased expression of D3 or TH treatment of BCC cells contributes to downregulation of miR21 levels (Figure 1 and Supplemental Figure 1). Furthermore, our finding that miR21 is also an upstream regulator of D3 (Figure 2) highlights the existence of a reciprocal TH-miR21 interplay, which regulates skin tumorigenesis.
Besides in cancer, there are many other contexts in which miR21 functions, D3 expression, and TH metabolism are linked. For instance, miR21 is implicated in many types of cardiac stress, including the myocardial remodeling that occurs after infarction, in which both D3 and miR21 are induced (41, 42). Moreover, miR21 is induced during immune cell activation (21, 43). The analogy of miR21 with D3 expression in multiple settings suggests that the miR21/D3 axis is a potential therapeutic target.
Although thyroid dysfunctions affect the skin phenotype and many people with hypothyroidism and hyperthyroidism have skin-related conditions (44), there is no evidence of an association between thyroid dysfunctions and BCC formation. This apparent lack of an association may be explained in 1 of 2 ways. Firstly, BCCs grow very slowly, while alterations of TH circulation at plasma levels are acute conditions that generally do not become chronic. Secondly, the deiodinase family represents a potent homeostatic mechanism when thyroid dysfunction induces a minimal variation of TH, and is thus able to preserve intracellular “normal” T3 concentrations even in the presence of altered thyroid status at plasma level. Conversely, D3 upregulation in the BCC might not alter circulating TH concentrations.
The tumor suppressor GRHL3 is an inhibitor of D3.
Our data demonstrate that the developmental factor and tumor suppressor GRHL3 represses DIO3 transcription and mediates the positive regulation of D3 expression by miR21. Although GRHL3 has been shown to be a miR21 target gene in SCCs (31, 33), this regulation appears to be context-dependent, since it was not among the miR21 targets in tissues in which miR21 was modified (45). Here we demonstrate that GRHL3 is downregulated by miR21 in BCC (Figures 2 And 3). Accordingly, GRHL3 inhibition increases the expression of D3 in BCC cells. This is consistent with the poor expression of D3 in adult tissues and its reactivation in many tumoral contexts (8). Remarkably, forced GRHL3 expression prevents miR21-dependent D3 transcription, thus providing genetic evidence for a miR21/GRHL3/D3 epistatic cascade (Figure 3, A–D).
The discovery of a miR21/GRHL3/D3 axis prompts some considerations regarding the role of TH metabolism in the skin. First, both miR21 and GRHL3 (which are functionally related to D3 expression) play critical roles in different skin cancer subtypes (33, 46). These observations reinforce the concept that D3 expression and TH metabolism play a critical role in skin cancer. Second, we previously demonstrated that D3 overexpression amplifies keratinocyte proliferation (10), and while its expression is very low in adult tissues, D3 is re-expressed in many hyperproliferative conditions (8). The inverse correlation between D3 and GRHL3 in BCC emphasizes the opposite roles played by the 2 genes in BCC tumorigenesis and, most importantly, suggests that the negative control exerted by GRHL3 on D3 expression might be extended to such other pathological conditions as psoriasis and wound healing in which GRHL3 plays a regulatory role (47, 48). Third, GRHL3 is critical for keratinocyte differentiation and skin barrier formation, and Grhl3–/– mice exhibit impaired epidermal differentiation and decreased expression of multiple differentiation genes (47). This implies that D3 suppression and increased TH signaling are also part of the differentiation program of keratinocytes. Should this be the case, it would be clinically relevant, since alterations of TH levels in patients are associated with cutaneous manifestations, and generalized myxedema (increased glycosaminoglycan deposition in skin, which appears cold, dry, and pale, with abundant hair loss) is the classical clinical sign of hypothyroid patients (44). Further studies are needed to determine whether T3 directly affects skin barrier formation and hair follicle cycle regulation.
D3 depletion drastically reduces BCC tumorigenesis.
To assess the pathological role of the newly identified circuit in vivo, we inhibited D3 by CRISPR/Cas9 technology in BCC cells and in miR21-overexpressing BCC cells. These experiments demonstrated that blockade of D3 potently attenuates BCC cell proliferation and in vivo xenograft tumorigenesis. miR21 overexpression stimulated the oncogenic potential of BCC cells to a greater extent than control cells (Figure 4). Notably, although BCC cells were transduced with miR21, their ability to form tumors was significantly hampered by D3 ablation, which strongly supports the concept that the TH inactivation pathway plays a pivotal role in tumor growth.
Intracellular TH regulation by D3 appears to be critical for tumorigenic potential in an in vivo model of BCC-like tumor formation. Indeed, we demonstrate that skin-specific D3 depletion drastically reduces tumor occurrence in mice with spontaneously forming BCC tumors, and prevents miR21 overexpression in these tumors. Thus, our data demonstrate that blocking TH degradation at the cellular level is a new modality with which to interfere with skin tumorigenesis. The few tumors that developed in our D3-depleted mice contained very low levels of miR21, but substantially increased GRHL3 expression. D3 depletion potently attenuates BCC growth driven by the Shh/Gli2 pathway and by miR21 overexpression. However, although in a drastically attenuated fashion, BCC cells still generate tumors when miR21 is overexpressed and D3 is depleted, thus suggesting that increasing the intracellular TH signaling is not sufficient to eradicate BCC tumorigenesis. Collectively, these data are consistent with a strong link between miR21 expression and D3 overexpression, and an inverse correlation between D3 and GRHL3. Moreover, they suggest the existence of a common route by which tumor cells attenuate the TH signal in the tumor microenvironment (Figure 5H).
In conclusion, our findings demonstrate that TH functions as a suppressor of BCC tumors by regulating multiple pathways, irrespective of miR21, and identify the TH-inactivating enzyme D3 as a potential target of anticancer therapy for human BCC.
Methods
Cell cultures and transfections.
BCC cells were derived from transgenic mice expressing a constitutively active form of Gli2 under the control of the keratin 5 promoter, as previously described (49). BCC cells were cultured under low-calcium conditions with 8% Ca2+-chelated fetal bovine serum (FBS) and keratinocyte growth factor (Sigma-Aldrich). HEK-293 cells were cultured in DMEM with 10% FBS, 2 mM l-glutamine, and 10 U/ml penicillin/streptomycin. All transient transfections for BCC cells were performed using Lipofectamine 2000 (Life Technologies) according to the manufacturer’s instructions. HEK-293 cells were transfected using IBAfect reagent (IBA BioTAGnology). Mouse primary keratinocytes were isolated from C57BL/6 newborn mice and cultured under low-calcium conditions (0.05 mM) in the presence of 4% calcium-chelated FBS (Invitrogen) and EGF (Invitrogen) as previously described (50).
Plasmids, siRNA, and reagents.
Two luciferase-driving constructs (pmD3Luc and miR21-Enhancer-Luc) were generated as follows: For the pmD3Luc (Dio3 promoter) a genomic mouse DNA was used as a PCR template; the oligonucleotides pmD3U and pmD3L (Supplemental Table 3) generated a 750-bp fragment containing the mouse Dio3 5′ flanking region. For the miR21-Enhancer-Luc plasmid, we used the oligos mMiR21U and mMiR21L to amplify 389 bp of the miR21 5′ flanking region (35). The amplified regions were cloned into the NheI/BglII (pmD3Luc) or SacI/XhoI (miR21-Enhancer-Luc) sites of a pGL3-basic vector (Promega Corp.). The mutated plasmid pmD3Luc-del was obtained from the pmD3Luc by recombinant PCR with 2 sets of oligonucleotides. Briefly, the 2 PCR products, pmD3U/pmD3mGRHLL2 and pmD3mGRHLU/pmD3L to obtain pmD3Luc-del, were combined by PCR using outside oligonucleotides, and the final PCR products were reinserted into the pGL3-basic vector (Promega Corp.). The Flag-GRHL3 plasmid was generated by PCR on mouse normal skin cDNA; the coding sequence of mouse Grhl3 was amplified by PCR with specific oligonucleotides (mGRHL3clonU and mGRHL3clonL); the PCR product was inserted into the HindIII/EcoRI sites of the pFLAG-CMV-2 vector (Sigma-Aldrich). GRHL3 silencing was performed with siRNA technology: 2 siRNAs targeting GRHL3 were purchased from Life Technologies (catalog s106644, s106646). Stealth RNAi negative control duplex (Invitrogen) served as control. Effective GRHL3 silencing was confirmed by measurement of GRHL3 protein levels in transfected cells (Supplemental Figure 4B). The primer sequence is reported in Supplemental Table 3. The miR21-overexpressing plasmid was provided by G. De Vita (University of Naples Federico II, Naples, Italy). Knockdown of miR21 was performed by transient transfection of anti-miR21 LNA (ID mh10206; Ambion). LNA-scrambled CTR served as control. Western blot analysis was performed with the following antibodies: anti-D3 (D3-718; ref. 51), anti–cyclin D1 (sc-246; Santa Cruz Biotechnology Inc.), and anti-tubulin (sc-8035; Santa Cruz Biotechnology Inc.). Anti-K17 (NCL-CK17; Novocastra), anti-K10 (PRB-159P), anti-Sox9 (ab26414; Abcam), and anti-PTCH1 (2113; Novus Biologicals) antibodies were used for immunofluorescence experiments.
Genome editing of the Dio3 locus using the CRISPR/Cas9 system.
Targeted depletion of D3 in mouse BCC cells was obtained with CRISPR/Cas9 technology using the CRISPR/Cas9 system from Santa Cruz Biotechnology Inc. (Dio3 CRISPR/Cas9 KO Plasmid, catalog sc-430733) and control plasmid (Control CRISPR/Cas9 Plasmid, catalog sc-418922). The D3 CRISPR/Cas9 KO plasmid consists of a pool of 3 plasmids, each encoding the Cas9 nuclease and a target-specific 20-nucleotide guide RNA. CRISPR/Cas9–guided mutagenesis in this case resulted in a deletion in the Dio3 coding region. Briefly, BCC cells were transfected with 500 ng of the D3 CRISPR/Cas9 KO plasmid or control plasmid with Lipofectamine 2000 (Life Technologies). Three days after transfection, the cells were FACS-sorted for GFP expression, and single clones were selected and analyzed by PCR to verify D3 coding sequence integrity using specific oligonucleotides (gdio3U: CGACCCAAGATTTCTGGGGCA; and gdio3L: AGCAGAGTCTCAAGTTAGCCAGAC; Supplemental Figure 4). Effective D3 depletion was confirmed by Western blot analysis (Supplemental Figure 4).
Genome editing of the TRE in the miR21 enhancer using the CRISPR/Cas9 system.
Targeted mutagenesis of the TRE site within the miR21 enhancer region was performed with CRISPR/Cas9 technology and the CRISPR/Cas9 system from ADDGENE [pSpCas9(BB)-2A-GFP]. We used 2 custom-designed synthetic single guide RNAs (sgRNAs) targeting the genomic region flanking the TRE site. Each sgRNA was cloned into the pX458 [pSpCas9(BB)-2A-GFP] vector, which also encodes the Cas9 nuclease (52). To obtain the mutated template, recombinant PCR was performed on the genomic mouse DNA (Supplemental Figure 2) using 2 sets of oligos (mHmir21dU and mMIR21mutL; mMIR21mutU and mHmir21dL), which generated a 2-kb fragment containing the mutation TCCTAATAAGGA to TttTAATAAaaA. The vector and the TRE-mutated sequence were transfected into BCC cells, and cells expressing GFP were sorted by FACS into individual wells of 96-well plates and expanded in culture. Successful homology directed repair–mediated mutation was assessed by PCR and Sanger sequencing.
Luciferase assays.
The Luc plasmids (pmD3Luc, miR21-Enhancer-Luc, or TRE3-TK-Luc) and CMV-renilla reporters were cotransfected into BCC and HEK-293 cells, and treated with 30 nM T3 as indicated. Luciferase activities were measured 48 hours after transfection using the Dual Luciferase Reporter Assay System (Promega Corp.), and differences in transfection efficiency were corrected relative to the level of renilla activity. Each construct was studied in triplicate in at least 3 separate transfection experiments.
Western blot analysis.
Total protein extracts from BCC cells were run on a 10% SDS-PAGE gel and transferred on Immobilon-P transfer membrane (Millipore). The membrane was then blocked with 5% nonfat dry milk in PBS, probed with anti-D3 polyclonal antibodies (1:500), anti-Flag (1:1,000, M2; Sigma-Aldrich), or GRHL3 antibody (1:500, sc-398839; Santa Cruz Biotechnology Inc.) for 2 hours, washed and incubated with HRP-conjugated donkey anti-rabbit IgG secondary antibody (1:3,000), and detected by chemiluminescence (Millipore). After extensive washing, the membrane was incubated with anti-tubulin-specific antibodies (sc-8035, 1:10,000; Santa Cruz Biotechnology Inc.) as loading control. All Western blots were run in triplicate, and bands were quantitated in 1 representative gel.
miRNA Northern blot.
Total RNA was isolated from BCC cells treated or not with T3, using TRIzol (Invitrogen) and quantitated at 260 nm using a standard spectrometry. Ten micrograms of total RNA was resuspended in 2′ Tris/borate/EDTA (TBE) urea sample buffer (Invitrogen) to a final volume of 10 μl. Samples were heated at 70°C for 5 minutes and loaded onto a 15% polyacrylamide gel containing TBE urea. Electrophoresis was performed in 1′ TBE running buffer at 120 V with a run time between 60 and 90 minutes. After electrophoresis, the gel was rinsed in deionized water, followed by a 5-minute wash in 1′ TBE. RNA was transferred onto a Hybond-N+ membrane for 30 minutes at 20 V using a Bio-Rad Miniprotean II apparatus. The nylon membrane was placed in the Stratagene UV Crosslinker and RNA cross-linked at 1,200 kJ. As miR21 probe, we designed the oligonucleotide MiR21probe (Supplemental Table 3) containing the mature miRNA sequence. The oligo was hybridized to the OprobAAA: 5′-TTTTTTTTTTCTCTGTCC-3′, and elongated with Klenow exo– (Life Technologies) and ATP α-32P (PerkinElmer Inc.) to generate a tail of radioactive adenine. To normalize the RNA, we used the 5S subunit of rRNA (Supplemental Table 3), which was also hybridized to the OprobAAA and radiolabeled.
Animals, histology, and immunostaining.
K5-Gli2 K14-Cre+/– Dio3fl/fl (Gli2;D3KO) mice were obtained by crossing of a keratinocyte-specific conditional D3-null mouse (K14-Cre+/– D3fl/fl) with the mouse model K5-Gli2 (37). Female 5-week-old BALB/c (nude) mice were purchased from Charles River Laboratories. All animal experiments and mouse husbandry were done in the animal facility at CEINGE-Biotecnologie Avanzate, Naples, Italy, in accordance with institutional guidelines. For immunofluorescence and histology, ears from Gli2;D3KO and control xenograft tumors of mice at 6 months of age were embedded in paraffin, cut into 7-μm sections, and H&E-stained. Slides were baked at 60°C, deparaffinized by xylenes, dehydrated with ethanol, rehydrated in PBS, and permeabilized by 0.5% Triton X-100 in PBS. Antigen retrieval was performed by incubation in 0.01 M citrate buffer (pH 6.0) or 0.5 M Tris buffer (pH 8.0) at 95°C for 15 minutes. Sections were blocked in 1% BSA/0.05% Tween/PBS for 30 minutes at 37°C. Primary antibody incubation was carried out overnight at 4°C in blocking buffer followed by washing in 0.2% Tween/PBS. Secondary antibody incubation was carried out at room temperature for 1 hour, followed by washing in 0.2% Tween/PBS. Images were acquired with an IX51 Olympus microscope and the Cell*F Olympus Imaging Software.
Real-time RT-PCR.
mRNAs were extracted with TRIzol reagent (Life Technologies). cDNAs were prepared with SuperScript VILO Master Mix (Life Technologies) as indicated by the manufacturer. The cDNAs were amplified by PCR in an iQ5 Multicolor Real-Time Detector System (Bio-Rad) with the fluorescent double-stranded DNA-binding dye SYBR Green (Applied Biosystems). Specific primers for each gene were designed to work under the same cycling conditions (95°C for 10 minutes followed by 40 cycles at 95°C for 15 seconds and 60°C for 1 minute), generating products of comparable sizes (~200 bp for each amplification). Primer combinations were positioned whenever possible to span an exon-exon junction and the RNA digested with deoxyribonuclease to avoid genomic DNA interference. For each reaction, standard curves for reference genes were constructed based on six 4-fold serial dilutions of cDNA. All samples were run in triplicate. The template concentration was calculated from the cycle number when the amount of PCR product passed a threshold established in the exponential phase of the PCR. The relative amounts of gene expression were calculated with cyclophilin A expression as an internal standard (calibrator). The results, expressed as n-fold differences in target gene expression, were determined as follows: n × target = 2(ΔCtsample – ΔCtcalibrator).
miRNA microarray analysis.
miRNA microarray analysis was performed in BCC cells transfected with shD3 or shCTR stealth oligos. Briefly, total RNA was harvested using the TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. The samples were hybridized to the miRCURY LNA Array (version 9.2; Exiqon). Data have been submitted to the Gene Expression Omnibus database (GEO GSE78265).
TaqMan miRNA assay.
miR21, RNU6B, and Sno234 were analyzed using 2-step real-time PCR protocols with TaqMan MicroRNA Assays: hsa-miR-21 catalog 000397; RNU6B catalog 001093; SnoRNA234 catalog 001234 (Applied Biosystems). Reverse transcriptase reactions were performed using the TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems) in a reaction mix of 15 μl total volume with 7 μl of Master Mix containing 1′ RT buffer, 1.0 mM of total dNTPs, 50 U MultiScribe Reverse Transcriptase Enzyme, and 0.25 U of RNase inhibitor; 3 μl of RT primers (Applied Biosystems); and 5 μl of RNA sample. The amplification conditions were as follows: annealing at 16°C for 30 minutes, extension at 42°C for 30 minutes, and RT inactivation at 85°C for 10 minutes. Quantitative PCRs (qPCRs) were performed on Applied Biosystems Step One Plus Real-Time PCR System with a mixture of 20 μl final volume containing 1′ TaqMan Universal PCR Master Mix (Applied Biosystems), 1 MicroRNA Assay (Applied Biosystems), and 3 μl cDNA (RT product diluted 1:5). Thermal cycling conditions were 10 minutes at 95°C followed by 40 cycles of 15 seconds at 95°C and 1 minute at 60°C. The end point of qPCR data is the threshold cycle (Ct), which represents the fractional cycle number at which the fluorescence reaches the fixed threshold. All qPCR reactions were run in duplicate. The relative quantification of miRNA expression was analyzed using the 2−ΔΔCt method.
Tumor xenografts.
To evaluate in vivo tumorigenesis, a BCC xenografting mouse model was established. After resuspension in PBS, 1 × 106 in 100 μl of CRISPR-CTR BCC cells (control), 1 D3KO clone (D3KO n.12, Supplemental Figure 4), and miR21 and D3KO-miR21 cells were injected s.c. into the flanks of 5-week-old female BALB/c-nu mice. Mice were examined twice a week, and the tumor was measured with a caliper and then weighed 3 weeks after injection. All animals were maintained in a sterile environment on a daily 12-hour light/12-hour dark cycle. Tumor volume was calculated according to the formula: volume (mm3) = 4/3 × π × (length/2) × (width/2) × (height/2). Tumor xenografts were harvested, weighed, and snap-frozen for immunofluorescence analysis.
Human samples.
Human BCC samples were collected at the Istituto Nazionale Tumori Ospedale Pascale in Naples.
Statistics.
The results are shown as means ± SD throughout. Differences between samples were assessed by Student’s 2-tailed t test for independent samples. In all experiments, differences were considered significant when P was less than 0.05. Asterisks indicate significance at *P < 0.05, **P < 0.01, and ***P < 0.001 throughout. Differences of mRNA levels in human BBC samples versus normal skin samples were analyzed using a 1-way ANOVA, and P less than 0.05 was considered significant. Relative mRNA levels (in which the control sample was arbitrarily set as 1) are reported as results of real-time PCR, in which expression of cyclophilin A was used as a housekeeping gene. The relationship between miR21/GRHL3, miR21/D3, and GRHL3/D3 expression was analyzed using Pearson’s correlation.
Study approval.
This work was approved by the appropriately constituted research ethics committee of the University of Naples Federico II.
Author contributions
DDG, GM, TP, CL, and EDC performed the in vitro and in vivo experiments and prepared the figures. RA generated plasmids and mouse models. MADS performed histochemistry and immunofluorescence. GS performed FACS sorting studies. DS, CM, LDV, AC, and AAD provided observations and scientific interpretations. DS contributed to experiment supervision and interpretation. MD designed the overall study, supervised the experiments, analyzed the results, and wrote the paper. All authors discussed the results and provided input on the manuscript.
Supplementary Material
Acknowledgments
This work was supported by grants from the European Research Council under the European Union’s Horizon 2020 Programme–ERC-StG-2014 (STARS–639548) and by Italian Association for Cancer Research grants to M. Dentice (IG 13065), D. Salvatore (IG 11362), and C. Missero (IG 17079). We thank Jean Ann Gilder (Scientific Communication Srl., Naples, Italy) for writing assistance.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:J Clin Invest. 2016;126(6):2308–2320. doi:10.1172/JCI84465.
Contributor Information
Daniela Di Girolamo, Email: danieladigirolamo1988@gmail.com.
Giuseppina Mancino, Email: man.giusy@gmail.com.
Tommaso Porcelli, Email: tommasoporcelli@gmail.com.
Cristina Luongo, Email: cristina.luongo@libero.it.
Emery Di Cicco, Email: emery2304@gmail.com.
Giulia Scalia, Email: scalia@ceinge.unina.it.
Luigi Del Vecchio, Email: luigi.delvecchio@unina.it.
Annamaria Colao, Email: colao@unina.it.
Caterina Missero, Email: missero@ceinge.unina.it.
Domenico Salvatore, Email: domsalva@unina.it.
Monica Dentice, Email: monica.dentice@unina.it.
References
- 1.Gailani MR, et al. The role of the human homologue of Drosophila patched in sporadic basal cell carcinomas. Nat Genet. 1996;14(1):78–81. doi: 10.1038/ng0996-78. [DOI] [PubMed] [Google Scholar]
- 2.Kasper M, Jaks V, Hohl D, Toftgard R. Basal cell carcinoma — molecular biology and potential new therapies. J Clin Invest. 2012;122(2):455–463. doi: 10.1172/JCI58779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oro AE, Higgins KM, Hu Z, Bonifas JM, Epstein EH., Jr Scott MP. Basal cell carcinomas in mice overexpressing sonic hedgehog. Science. 1997;276(5313):817–821. doi: 10.1126/science.276.5313.817. [DOI] [PubMed] [Google Scholar]
- 4.Xie J, et al. Activating Smoothened mutations in sporadic basal-cell carcinoma. Nature. 1998;391(6662):90–92. doi: 10.1038/34201. [DOI] [PubMed] [Google Scholar]
- 5.Johnson RL, et al. Human homolog of patched, a candidate gene for the basal cell nevus syndrome. Science. 1996;272(5268):1668–1671. doi: 10.1126/science.272.5268.1668. [DOI] [PubMed] [Google Scholar]
- 6.Hahn H, et al. Mutations of the human homolog of Drosophila patched in the nevoid basal cell carcinoma syndrome. Cell. 1996;85(6):841–851. doi: 10.1016/S0092-8674(00)81268-4. [DOI] [PubMed] [Google Scholar]
- 7.Wong SY, Dlugosz AA. Basal cell carcinoma, Hedgehog signaling, and targeted therapeutics: the long and winding road. J Invest Dermatol. 2014;134(e1):E18–E22. doi: 10.1038/skinbio.2014.4. [DOI] [PubMed] [Google Scholar]
- 8.Dentice M, Antonini D, Salvatore D. Type 3 deiodinase and solid tumors: an intriguing pair. Expert Opin Ther Targets. 2013;17(11):1369–1379. doi: 10.1517/14728222.2013.833189. [DOI] [PubMed] [Google Scholar]
- 9.Gereben B, et al. Cellular and molecular basis of deiodinase-regulated thyroid hormone signaling. Endocr Rev. 2008;29(7):898–938. doi: 10.1210/er.2008-0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dentice M, et al. Sonic hedgehog-induced type 3 deiodinase blocks thyroid hormone action enhancing proliferation of normal and malignant keratinocytes. Proc Natl Acad Sci U S A. 2007;104(36):14466–14471. doi: 10.1073/pnas.0706754104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luongo C, Ambrosio R, Salzano S, Dlugosz AA, Missero C, Dentice M. The sonic hedgehog-induced type 3 deiodinase facilitates tumorigenesis of basal cell carcinoma by reducing Gli2 inactivation. Endocrinology. 2014;155(6):2077–2088. doi: 10.1210/en.2013-2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Volinia S, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103(7):2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65(14):6029–6033. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- 14.Xu LF, Wu ZP, Chen Y, Zhu QS, Hamidi S, Navab R. MicroRNA-21 (miR-21) regulates cellular proliferation, invasion, migration, and apoptosis by targeting PTEN, RECK and Bcl-2 in lung squamous carcinoma, Gejiu City, China. PLoS One. 2014;9(8): doi: 10.1371/journal.pone.0103698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lv L, et al. MicroRNA-21 is overexpressed in renal cell carcinoma. Int J Biol Markers. 2013;28(2):201–207. doi: 10.5301/JBM.2013.10831. [DOI] [PubMed] [Google Scholar]
- 16.Hiyoshi Y, et al. MicroRNA-21 regulates the proliferation and invasion in esophageal squamous cell carcinoma. Clin Cancer Res. 2009;15(6):1915–1922. doi: 10.1158/1078-0432.CCR-08-2545. [DOI] [PubMed] [Google Scholar]
- 17.Li X, Huang K, Yu J. Inhibition of microRNA-21 upregulates the expression of programmed cell death 4 and phosphatase tensin homologue in the A431 squamous cell carcinoma cell line. Oncol Lett. 2014;8(1):203–207. doi: 10.3892/ol.2014.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ren W, et al. MiR-21 modulates chemosensitivity of tongue squamous cell carcinoma cells to cisplatin by targeting PDCD4. Mol Cell Biochem. 2014;390(1–2):253–262. doi: 10.1007/s11010-014-1976-8. [DOI] [PubMed] [Google Scholar]
- 19.Asangani IA, et al. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008;27(15):2128–2136. doi: 10.1038/sj.onc.1210856. [DOI] [PubMed] [Google Scholar]
- 20.Cao Z, Yoon JH, Nam SW, Lee JY, Park WS. PDCD4 expression inversely correlated with miR-21 levels in gastric cancers. J Cancer Res Clin Oncol. 2012;138(4):611–619. doi: 10.1007/s00432-011-1140-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morrisey EE. The magic and mystery of miR-21. J Clin Invest. 2010;120(11):3817–3819. doi: 10.1172/JCI44596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hildebrand J, et al. A comprehensive analysis of microRNA expression during human keratinocyte differentiation in vitro and in vivo. J Invest Dermatol. 2011;131(1):20–29. doi: 10.1038/jid.2010.268. [DOI] [PubMed] [Google Scholar]
- 23.Yi R, et al. Morphogenesis in skin is governed by discrete sets of differentially expressed microRNAs. Nat Genet. 2006;38(3):356–362. doi: 10.1038/ng1744. [DOI] [PubMed] [Google Scholar]
- 24.Ahmed MI, Mardaryev AN, Lewis CJ, Sharov AA, Botchkareva NV. MicroRNA-21 is an important downstream component of BMP signalling in epidermal keratinocytes. J Cell Sci. 2011;124(pt 20):3399–3404. doi: 10.1242/jcs.086710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Joyce CE, et al. Deep sequencing of small RNAs from human skin reveals major alterations in the psoriasis miRNAome. Hum Mol Genet. 2011;20(20):4025–4040. doi: 10.1093/hmg/ddr331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zibert JR, Lovendorf MB, Litman T, Olsen J, Kaczkowski B, Skov L. MicroRNAs and potential target interactions in psoriasis. J Dermatol Sci. 2010;58(3):177–185. doi: 10.1016/j.jdermsci.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 27.Dziunycz P, et al. Squamous cell carcinoma of the skin shows a distinct microRNA profile modulated by UV radiation. J Invest Dermatol. 2010;130(11):2686–2689. doi: 10.1038/jid.2010.169. [DOI] [PubMed] [Google Scholar]
- 28.Akagi I, et al. Relationship between altered expression levels of MIR21, MIR143, MIR145, and MIR205 and clinicopathologic features of esophageal squamous cell carcinoma. Dis Esophagus. 2011;24(7):523–530. doi: 10.1111/j.1442-2050.2011.01177.x. [DOI] [PubMed] [Google Scholar]
- 29.Satzger I, et al. microRNA-21 is upregulated in malignant melanoma and influences apoptosis of melanocytic cells. Exp Dermatol. 2012;21(7):509–514. doi: 10.1111/j.1600-0625.2012.01510.x. [DOI] [PubMed] [Google Scholar]
- 30.Stramer B, Martin P. Cell biology: master regulators of sealing and healing. Curr Biol. 2005;15(11):R425–R427. doi: 10.1016/j.cub.2005.05.034. [DOI] [PubMed] [Google Scholar]
- 31.Darido C, et al. Targeting of the tumor suppressor GRHL3 by a miR-21-dependent proto-oncogenic network results in PTEN loss and tumorigenesis. Cancer Cell. 2011;20(5):635–648. doi: 10.1016/j.ccr.2011.10.014. [DOI] [PubMed] [Google Scholar]
- 32.Yu Z, et al. The Grainyhead-like epithelial transactivator Get-1/Grhl3 regulates epidermal terminal differentiation and interacts functionally with LMO4. Dev Biol. 2006;299(1):122–136. doi: 10.1016/j.ydbio.2006.07.015. [DOI] [PubMed] [Google Scholar]
- 33.Bhandari A, et al. The Grainyhead transcription factor Grhl3/Get1 suppresses miR-21 expression and tumorigenesis in skin: modulation of the miR-21 target MSH2 by RNA-binding protein DND1. Oncogene. 2013;32(12):1497–1507. doi: 10.1038/onc.2012.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peyrard-Janvid M, et al. Dominant mutations in GRHL3 cause Van der Woude Syndrome and disrupt oral periderm development. Am J Hum Genet. 2014;94(1):23–32. doi: 10.1016/j.ajhg.2013.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fujita S, et al. miR-21 Gene expression triggered by AP-1 is sustained through a double-negative feedback mechanism. J Mol Biol. 2008;378(3):492–504. doi: 10.1016/j.jmb.2008.03.015. [DOI] [PubMed] [Google Scholar]
- 36.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 37.Grachtchouk M, et al. Basal cell carcinomas in mice overexpressing Gli2 in skin. Nat Genet. 2000;24(3):216–217. doi: 10.1038/73417. [DOI] [PubMed] [Google Scholar]
- 38.Ortiga-Carvalho TM, Sidhaye AR, Wondisford FE. Thyroid hormone receptors and resistance to thyroid hormone disorders. Nat Rev Endocrinol. 2014;10(10):582–591. doi: 10.1038/nrendo.2014.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weinberger C, Thompson CC, Ong ES, Lebo R, Gruol DJ, Evans RM. The c-erb-A gene encodes a thyroid hormone receptor. Nature. 1986;324(6098):641–646. doi: 10.1038/324641a0. [DOI] [PubMed] [Google Scholar]
- 40.Gandrillon O, Ferrand N, Michaille JJ, Roze L, Zile MH, Samarut J. c-erbA alpha/T3R and RARs control commitment of hematopoietic self-renewing progenitor cells to apoptosis or differentiation and are antagonized by the v-erbA oncogene. Oncogene. 1994;9(3):749–758. [PubMed] [Google Scholar]
- 41.van Rooij E, et al. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc Natl Acad Sci U S A. 2006;103(48):18255–18260. doi: 10.1073/pnas.0608791103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tatsuguchi M, et al. Expression of microRNAs is dynamically regulated during cardiomyocyte hypertrophy. J Mol Cell Cardiol. 2007;42(6):1137–1141. doi: 10.1016/j.yjmcc.2007.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lu TX, Munitz A, Rothenberg ME. MicroRNA-21 is up-regulated in allergic airway inflammation and regulates IL-12p35 expression. J Immunol. 2009;182(8):4994–5002. doi: 10.4049/jimmunol.0803560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Antonini D, Sibilio A, Dentice M, Missero C. An intimate relationship between thyroid hormone and skin: regulation of gene expression. Front Endocrinol (Lausanne) 2013;4: doi: 10.3389/fendo.2013.00104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ma X, et al. Loss of the miR-21 allele elevates the expression of its target genes and reduces tumorigenesis. Proc Natl Acad Sci U S A. 2011;108(25):10144–10149. doi: 10.1073/pnas.1103735108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Medina PP, Nolde M, Slack FJ. OncomiR addiction in an in vivo model of microRNA-21-induced pre-B-cell lymphoma. Nature. 2010;467(7311):86–90. doi: 10.1038/nature09284. [DOI] [PubMed] [Google Scholar]
- 47.Ting SB, et al. A homolog of Drosophila grainy head is essential for epidermal integrity in mice. Science. 2005;308(5720):411–413. doi: 10.1126/science.1107511. [DOI] [PubMed] [Google Scholar]
- 48.Gordon WM, et al. A GRHL3-regulated repair pathway suppresses immune-mediated epidermal hyperplasia. J Clin Invest. 2014;124(12):5205–5218. doi: 10.1172/JCI77138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sheng H, et al. Dissecting the oncogenic potential of Gli2: deletion of an NH(2)-terminal fragment alters skin tumor phenotype. Cancer Res. 2002;62(18):5308–5316. [PubMed] [Google Scholar]
- 50.Antonini D, et al. An autoregulatory loop directs the tissue-specific expression of p63 through a long-range evolutionarily conserved enhancer. Mol Cell Biol. 2006;26(8):3308–3318. doi: 10.1128/MCB.26.8.3308-3318.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huang SA, et al. Transforming growth factor–beta promotes inactivation of extracellular thyroid hormones via transcriptional stimulation of type 3 iodothyronine deiodinase. Mol Endocrinol. 2005;19(12):3126–3136. doi: 10.1210/me.2005-0173. [DOI] [PubMed] [Google Scholar]
- 52.Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8(11):2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.