Abstract
Lymphangiogenesis is supported by 2 homologous VEGFR3 ligands, VEGFC and VEGFD. VEGFC is required for lymphatic development, while VEGFD is not. VEGFC and VEGFD are proteolytically cleaved after cell secretion in vitro, and recent studies have implicated the protease a disintegrin and metalloproteinase with thrombospondin motifs 3 (ADAMTS3) and the secreted factor collagen and calcium binding EGF domains 1 (CCBE1) in this process. It is not well understood how ligand proteolysis is controlled at the molecular level or how this process regulates lymphangiogenesis, because these complex molecular interactions have been difficult to follow ex vivo and test in vivo. Here, we have developed and used biochemical and cellular tools to demonstrate that an ADAMTS3-CCBE1 complex can form independently of VEGFR3 and is required to convert VEGFC, but not VEGFD, into an active ligand. Consistent with these ex vivo findings, mouse genetic studies revealed that ADAMTS3 is required for lymphatic development in a manner that is identical to the requirement of VEGFC and CCBE1 for lymphatic development. Moreover, CCBE1 was required for in vivo lymphangiogenesis stimulated by VEGFC but not VEGFD. Together, these studies reveal that lymphangiogenesis is regulated by two distinct proteolytic mechanisms of ligand activation: one in which VEGFC activation by ADAMTS3 and CCBE1 spatially and temporally patterns developing lymphatics, and one in which VEGFD activation by a distinct proteolytic mechanism may be stimulated during inflammatory lymphatic growth.
Introduction
In all organs except the CNS, lymphatic vessels drain interstitial fluid that leaks from the high-pressure blood vessel network and provide a route by which extravasated leukocytes return to the blood (1). Lymphatics also serve specialized functions in the intestine, from which they transport absorbed fat, and in adaptive immune responses, which they coordinate by bringing antigens and antigen-presenting cells to lymph nodes, where they may contact lymphocytes (2). Loss of lymphatic function due to primary defects in lymphatic growth or secondary conditions such as filarial infection or surgical removal of lymph nodes results in edema, malabsorptive syndromes, and immune defects that may be fatal (3). Drug therapies for lymphatic insufficiency are not available, and their creation will require a detailed understanding of the molecular regulation of lymphatic vessel growth in vivo.
Critical lymphangiogenic factors have been identified through molecular cloning of ligands and receptors that are homologous to blood vessel angiogenic factors, as well as genetic studies of primary lymphatic defects in humans, mice, and, most recently, zebrafish. Such studies have identified VEGFC, a ligand for the VEGFR3 receptor that is expressed on lymphatic but not blood vascular endothelial cells (ECs), as a central player in lymphangiogenesis (4–6). Loss of VEGFC or VEGFR3 function blocks lymphatic development in fish and mice (6, 7) and underlies human primary lymphedema syndromes (8, 9). VEGFD is a second VEGFR3 ligand that is structurally and functionally homologous to VEGFC (10, 11). Loss of VEGFD does not alter lymphatic development in mice or fish (12, 13), but overexpression of either VEGFC or VEGFD drives lymphatic endothelial and vessel growth in mature animals (14–16).
The structures of VEGFC and VEGFD are distinct from those of VEGFA and VEGFB in that they contain N-terminal and C-terminal domains that are proteolytically cleaved during and after secretion from cultured cells (17, 18). Proteolytic processing of VEGFC and VEGFD is thought to regulate lymphatic vessel growth, but precisely how this processing is achieved and controlled in vivo has not been clear. Studies of mutant zebrafish that entirely lack lymphatic vessel development and rare individuals with a primary lymphedema disorder known as Hennekam syndrome have identified collagen- and calcium-binding EGF domains 1 (CCBE1) as a secreted protein that is required for lymphatic vascular development (19–21). Loss of CCBE1 completely blocks lymphatic vascular development in a manner similar to that induced by loss of VEGFC or VEGFR3 (22–24), but has no effect on blood vessel growth. Recent studies have implicated CCBE1 and a disintegrin and metalloproteinase with thrombospondin motifs 3 (ADAMTS3) in the regulation of VEGFC processing (25–27) and generated a model of lymphangiogenesis in which VEGFR3-bound VEGFC is cleaved by CCBE1 and ADAMTS3 during receptor activation (25). These studies have linked CCBE1 and ADAMTS3 to VEGFC function, but limitations in the biochemical analysis of VEGFC and VEGFD proteolytic processing, a lack of clear correlation between in vitro and in vivo studies, and a lack of in vivo evidence for the role of ADAMTS3 have limited our understanding of this complex angiogenic process.
In the present study, we use biochemical, cellular, and murine genetic approaches to dissect the relationships between CCBE1, ADAMTS3, and the lymphangiogenic factors VEGFC and VEGFD. To biochemically follow VEGFC and VEGFD proteolytic processing, we inserted an epitope tag into the mature forms of these proteins that does not interfere with proteolysis or VEGFR3 activation. To test the role of ADAMTS3 in VEGFC and VEGFD processing ex vivo, we used CRISPR-Cas9 genome editing to mutate the ADAMTS3 genes in HEK293 cells. Finally, to determine whether and to what extent these biochemical and ex vivo cellular findings predict in vivo lymphangiogenic responses, we examined lymphatic development in ADAMTS3-deficient embryos and VEGFC and VEGFD lymphangiogenic responses in adult CCBE1-deficient mice. Our studies support a mechanism in which CCBE1 functions as an activator of the ADAMTS3 protease that cleaves the N-terminus (NT) of VEGFC. We found that N-terminal cleavage of VEGFC by CCBE1 and ADAMTS3 can occur independently of the VEGFR3 receptor, but is required for VEGFC to stimulate VEGFR3 signaling. Consistent with this molecular model, embryonic ADAMTS3 deficiency resulted in a complete loss of lymphatic development in mice, a phenotype also observed with loss of either CCBE1 or VEGFC. In contrast, VEGFD processing was not regulated by either CCBE1 or ADAMTS3 in vitro, and CCBE1-deficient animals exhibited normal lymphangiogenic responses to VEGFD in vivo. We believe that these studies conclusively demonstrate distinct mechanisms of proteolytic activation for VEGFC and VEGFD that help explain their specific roles in prenatal and postnatal lymphatic vascular growth.
Results
CCBE1 is required for VEGFC to stimulate lymphatic EC proliferation in vivo.
Published studies have demonstrated that CCBE1 can enhance the lymphatic EC (LEC) response to VEGFC in mice (25) and fish (26), but direct assessment of the role of CCBE1 in VEGFC-mediated LEC responses in vivo is not possible in the developing mouse or fish, because loss of CCBE1 results in a complete absence of lymphatic vessels (20, 22). To directly address the role of CCBE1 in VEGFC-mediated LEC responses in vivo, we deleted CCBE1 in mature Ub-CreERT2 Ccbe1fl/– animals after growth and maturation of the lymphatic network were complete. Deletion of Ccbe1 in mature, 12-week-old Ub-CreERT2 Ccbe1fl/– animals (adult CCBE1-KO animals) using long-term, oral tamoxifen administration did not result in an overt phenotype. Neonatal induction of Ccbe1 deficiency compromised lymphatic growth in the ear and skin (Supplemental Figure 1, A–H; supplemental material available online with this article; doi:10.1172/JCI83967DS1), sites of postnatal lymphangiogenesis, but adult CCBE1-KO animals exhibited normal lymphatic vessel density, WT levels of LEC VEGFR3, and normal lymphatic function as measured using rhodamine dextran lymphangiography (Supplemental Figure 1, I–V). Thus, CCBE1 is required for lymphatic development and growth, but not for the maintenance or function of mature lymphatic vessels.
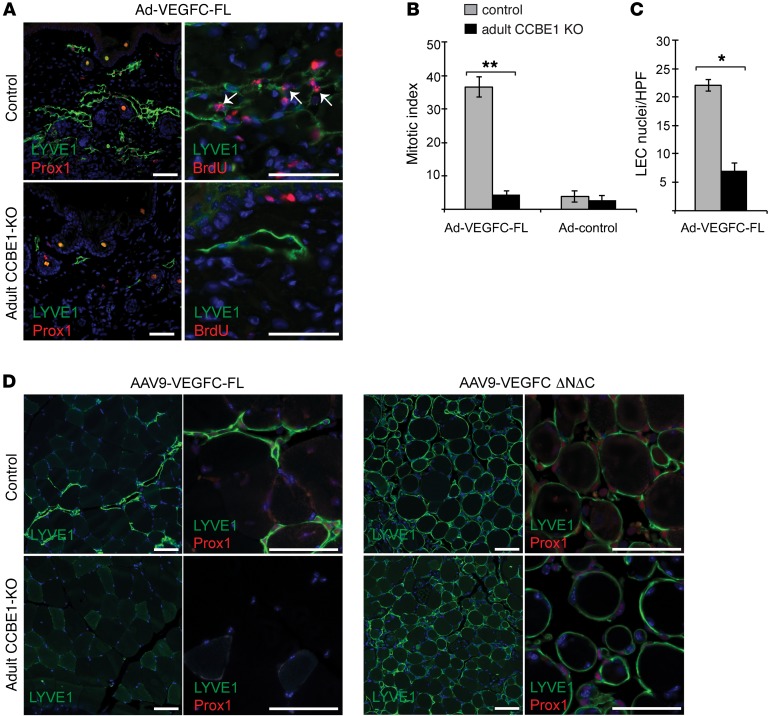
To test whether CCBE1 regulates VEGFC responses in vivo, we injected full-length VEGFC–expressing (VEGFC-FL–expressing) adenoviral vectors into the skin of adult CCBE1-KO and littermate control animals. The precise site of VEGFC expression was marked by red fluorescence generated from a small amount of coinjected adeno-Cherry (Supplemental Figure 2). LEC proliferation was measured by cell numbers and the mitotic index calculated by immunostaining for BrdU that was injected 12 hours prior to sacrifice. In control animals, adenoviral expression of VEGFC-FL stimulated strong LEC proliferation that was manifested by both the large fraction of LEC nuclei that were BrdU+ (approximately 35%–40%) and by an increase in LEC cell numbers (Figure 1, A–C). In contrast, adenoviral VEGFC-FL failed to stimulate any increase in BrdU uptake or LEC numbers in adult CCBE1-KO animals (Figure 1, A–C; n = 5 for each group, P < 0.001 for BrdU uptake and P < 0.01 for LEC numbers). To further test the role of CCBE1 in VEGFC-mediated responses in vivo, we attempted to rescue VEGFC-FL responses in adult CCBE1-KO mice by inducing expression of CCBE1-V5. Injection of adeno-associated virus expressing CCBE1-V5 into the skin of adult CCBE1-KO animals 4 weeks before the administration of VEGFC adenovirus successfully restored VEGFC-FL–induced LEC proliferation in adult CCBE1-KO animals (Supplemental Figure 3), confirming that loss of VEGFC responses is due specifically to a lack of CCBE1 and that CCBE1-V5 is biologically active in vivo. These studies demonstrate that CCBE1 is required for lymphatic endothelial responses to VEGFC in vivo, even when an intact lymphatic network is presented with very high levels of VEGFC ligand.
Figure 1. CCBE1 is required for the activity of full-length but not processed VEGFC in vivo.
(A) VEGFC-FL was not lymphangiogenic in CCBE1-deficient animals. Adenovirus expressing VEGFC-FL (Ad-VEGFC-FL) was injected into the skin of Ub-CreERT2 Ccbe1fl/– animals in which Ccbe1 had been deleted by administration of tamoxifen (adult CCBE1-KO) or of control animals. Lymphatic vessels were analyzed 72 hours after adenovirus administration and 12 hours after BrdU administration by immunostaining with the indicated Abs. Arrows indicate BrdU+ LEC nuclei. (B) Percentage of LEC nuclei that were BrdU+ (mitotic index) in the skin of adult CCBE1-KO and control animals following exposure to adenovirus expressing VEGFC-FL (Ad-VEGFC-FL). n = 5 in each group; **P < 0.001, by unpaired, 2-tailed Student’s t test. (C) LEC proliferation in the skin of adult CCBE1-KO and control animals following exposure to adenovirus expressing VEGFC-FL was calculated by counting LEC nuclei per high power field. n = 5 in each group; *P < 0.01. HPF, high-power field. (D) Proteolytically processed VEGFC ΔNΔC was lymphangiogenic in CCBE1-deficient mice. AAV expressing either VEGFC-FL (AAV9-VEGFC-FL) or the core domain of VEGFC released by proteolysis of the N and C termini (AAV9-VEGFC-ΔNΔC) was injected into the tibialis muscle of adult CCBE1-KO or control animals. Lymphatic growth was detected using LYVE1 and PROX1 immunostaining. Note that lymphatic growth stimulated by VEGFC-ΔNΔC tightly encircles muscle fibers (right) and lacks the typical branched vascular appearance associated with VEGFC-FL (left). Data shown are representative of 3 separate experiments. Scale bars: 50 μm.
CCBE1 has recently been implicated in the proteolytic processing of VEGFC (25, 26), and transgenic expression of fully proteolyzed VEGFC (VEGFC ΔNΔC) was found to rescue lymphatic growth in CCBE1 morphant zebrafish embryos (26). To more directly assess the ability of VEGFC ΔNΔC to drive lymphatic growth in CCBE1-deficient animals, we next induced expression of VEGFC ΔNΔC in adult CCBE1-KO mice. Injection of adeno-associated virus–VEGFC-FL (AAV-VEGFC-FL) that expresses VEGFC-FL stimulated a modest lymphangiogenic response in the tibialis muscle (detected as discrete foci of lymphatic vessels) that was not observed in adult CCBE1-KO animals (Figure 1D). In contrast, AAV-VEGFC ΔNΔC injection stimulated a highly robust growth of LYVE1+PROX1+ lymphatic endothelium surrounding the muscle fiber sheaths in both control and CCBE1-KO animals (Figure 1D; n = 3). These studies confirm that CCBE1 is required for VEGFC function in vivo and further implicate CCBE1 in the proteolytic processing of VEGFC in vivo. Significantly, the pattern of growth in VEGFC ΔNΔC–expressing muscle differed markedly from that driven by VEGFC-FL in WT animals, with LECs found ensheathing muscle fibers in unbranched structures, an observation consistent with a mechanism in which CCBE1 spatially regulates lymphatic growth in vivo (discussed further below).
VEGFC VHD-FLAG demonstrates that CCBE1 drives N-terminal cleavage that may be independent of both C-terminal cleavage and VEGFR3 interaction.
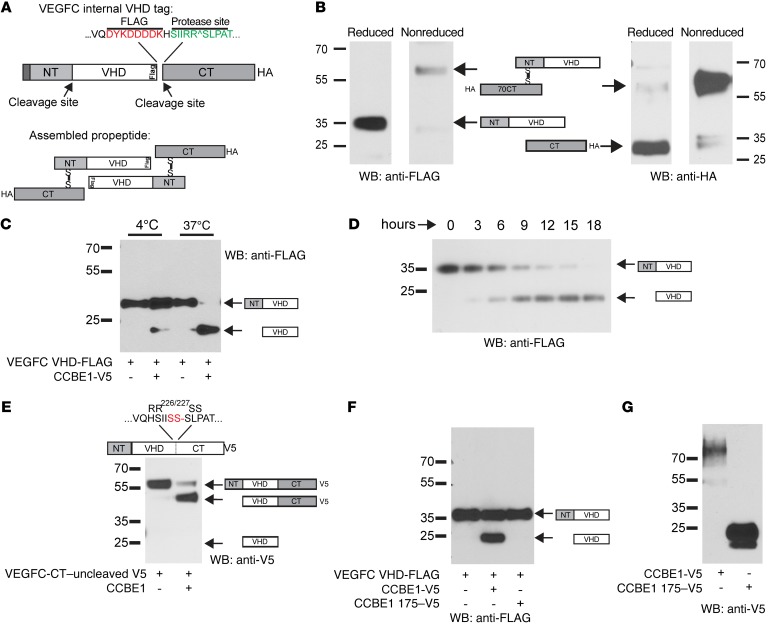
Most studies of VEGFC and VEGFD proteolytic processing have been performed using VEGFR3 ectodomain pulldown of metabolically labeled proteins to identify the different species of these secreted growth factors (17, 18). This approach has successfully demonstrated posttranslational proteolysis of these proteins, but detection based on VEGFR3 binding may bias protein detection, because VEGFC and VEGFD proteins bind more weakly to VEGFR2 and VEGFR3 receptors than do processed proteins (17, 18). VEGFC processing is believed to involve 2 distinct proteolytic events that combine to release a central peptide, termed the VEGF homology domain (VHD), that is homologous to VEGFA (17). To follow VEGFC processing in a direct and unbiased manner, we introduced a FLAG epitope into the C-terminal end of the VEGFC VHD immediately adjacent to the well-characterized site of C-terminal cleavage (Figure 2A). To follow proteolytic processing using VEGFC VHD-FLAG, we first generated a VEGFC VHD-FLAG that also contained an HA tag fused in frame to the end of the VEGFC C-terminus (CT). Immunoblot analysis for FLAG in the conditioned medium of HEK293 cells that express VEGFC VHD-FLAG CT-HA revealed a dominant band of approximately 35 kDa under reduced conditions (Figure 2B), consistent with a peptide containing the NT and VHD domains of VEGFC. Anti-FLAG immunostaining under nonreduced conditions revealed a strong 58- to 60-kDa band (Figure 2B). Anti-HA immunostaining of the same blot revealed a 28-kDa band under reduced conditions, consistent with the isolated CT, and a 58- to 60-kDa band identical to that observed with anti-FLAG Abs under nonreduced conditions (Figure 1B), indicating that the 58- to 60-kDa band most likely represents NT-VHD-FLAG and CT-HA peptides bound together by disulfide bonds (see schematic in Figure 2B).
Figure 2. VEGFC VHD-FLAG demonstrates increased VEGFC proteolysis with full-length but not truncated CCBE1.
(A) Schematic of the VEGFC VHD-FLAG protein, in which a FLAG epitope (shown in red) is inserted in frame at the C-terminal end of the VHD. The CT cleavage site is shown in green; an HA tag is placed in frame at the end of the protein CT. Disulfide bonds are predicted to link 2 full-length VEGFC VHD-FLAG molecules as shown in the bottom panel. (B) Detection of VEGFC VHD-FLAG with a C-terminal HA tag under reduced and nonreduced conditions using anti-FLAG and anti-HA Abs. The protein domains within the detected bands are illustrated schematically. (C and D) CCBE1-FL drives VEGFC proteolysis in vitro. HEK293T conditioned media containing VEGFC VHD-FLAG alone or VEGFC VHD-FLAG plus CCBE1-V5 were incubated for 24 hours at 4°C or 37°C prior to detection of FLAG by immunoblotting (C). Temporal analysis revealed slow proteolysis of VEGFC in the presence of CCBE1 (D). (E) N-terminal cleavage of VEGFC was independent of C-terminal cleavage. A VEGFC VHD-FLAG protein containing a mutation that prevents C-terminal cleavage (RR226-227SS) was incubated with CCBE1-V5 or control conditioned medium for 24 hours. (F and G) A truncated form of CCBE1 lacking its C-terminal collagen-like domain failed to drive VEGFC cleavage. VEGF VHD-FLAG was incubated with control conditioned medium, conditioned medium containing full-length CCBE1-V5, and conditioned medium containing CCBE1 175-V5 for 24 hours prior to anti-FLAG immunoblot analysis (F). Full-length CCBE1-V5 was detected as a 70- to 100-kDa smear that was expressed at lower levels than the 25-kDa CCBE1 175-V5 (G). Data shown are representative of 3 separate experiments. WB, Western blot.
To test the effect of CCBE1 on VEGFC processing, we incubated VEGFC VHD-FLAG with CCBE1-V5 by either mixing conditioned supernatants or cotransfecting a single population of HEK293T cells. CCBE1-V5 is not well expressed by HEK293T cells (22, 25), and analysis of silver-stained gels revealed a concentration of approximately 1–5 nM CCBE1 (Figure 2G and not shown). Incubation of VEGFC with CCBE1 resulted in the generation of a single 19- to 21-kDa protein containing the VEGFC VHD at 37°C but not 4°C, consistent with enzymatic digestion (Figure 2C). Timed analysis of mixed supernatants incubated at 37°C for 0 to 18 hours revealed progressive cleavage of VEGFC VHD-FLAG, with complete processing by 18 hours (Figure 2D).
Previous studies of VEGFC processing following protein expression by HEK293 and other cell types have suggested that C-terminal cleavage after aa R227 occurs before N-terminal cleavage and is highly efficient (17). To determine whether CCBE1-driven NT cleavage requires prior CT cleavage, we examined the effect of CCBE1 on VEGFC RR226/227SS-V5, a mutant in which the CT is not cleaved (17) and the protein is C-terminally tagged. VEGFC RR226/227SS-V5 was detected as a 58-kDa protein that was consistent with a VEGFC-FL protein containing the NT, VHD, and CT domains (Figure 2E). Following exposure to CCBE1, anti-V5 immunostaining identified a 45-kDa protein that was consistent with the VEGFC VHD and CT domains (Figure 1D). These findings demonstrate that CCBE1 drives N-terminal VEGFC proteolysis that is independent of its processing at the CT cleavage site. Importantly, they also demonstrate that CCBE1 regulation of VEGFC processing does not require interaction with LECs or the VEGFR3 receptor (discussed further below).
The CCBE1 N-terminal domain does not support VEGFC processing.
The CCBE1 protein is composed of an N-terminal domain containing 2 Ca++-binding EGF-like motifs and a C-terminal domain with GPP repeats resembling those in collagen (20). Since full-length CCBE1 (CCBE1-FL) is difficult to express, previous studies have investigated CCBE1 function using a truncated protein that contains only the N-terminal domain (e.g., CCBE1 175, ref. 25; and CCBE1 191, ref. 22). To directly test the function of such truncated CCBE1 proteins, we expressed VEGFC VHD-FLAG and full-length CCBE1-V5 or CCBE1 175-V5 in HEK293T cells. Mixing of conditioned media or cotransfection revealed no effect of CCBE1 175 on the processing of VEGFC under these in vitro conditions (Figure 2F), despite the presence of much higher concentrations of CCBE1 175 compared with those of CCBE1-FL (Figure 2G). Additionally, enzymatic studies revealed that the higher-than-expected molecular weight of CCBE1 and its appearance as a smear rather than a distinct band are the result of chondroitin sulfate posttranslational modification in the C-terminal domain (Supplemental Figure 4). These studies reveal that the N-terminal domain of CCBE1 is not sufficient to drive proteolytic cleavage of VEGFC and suggest that previous studies using this peptide are not likely to accurately reflect CCBE1 function (a conclusion supported by a recent in vivo structure-function analysis of CCBE1; ref. 27). Possible roles for the CCBE1 N-terminal domain in potentiating VEGFC proteolysis in vivo, e.g., by localizing the protein, will require future studies.
Activation of VEGFR3 by VEGFC requires N- and C-terminal proteolysis and is driven by CCBE1 in vitro.
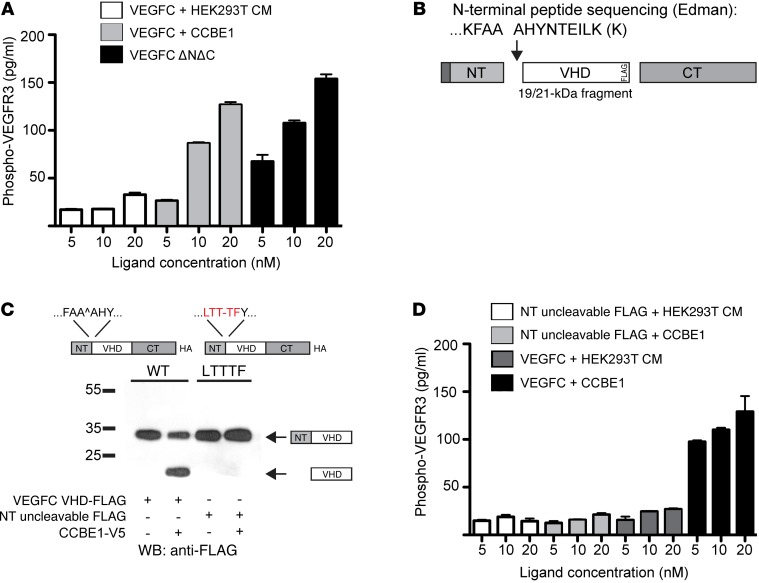
Previous studies of VEGFC processing have assessed its functional impact on VEGFC activity by measuring binding to the VEGFR3 ectodomain and common signaling endpoints such as activation of ERK1, ERK2, and AKT (17, 25). These endpoints are not ideal, because unprocessed VEGFC may bind, but not activate, the receptor (as suggested in a recent model of CCBE1 function; ref. 25), and ERK and AKT are promiscuous downstream signaling pathways that do not specifically measure VEGFR3 signaling. To better assess the functional significance of CCBE1 and VEGFC processing, we therefore measured VEGFR3 receptor phosphorylation in primary LECs using a phosphorylated VEGFR3 (phospho-VEGFR3) ELISA (28). Exposure of primary human LECs to VEGFC-containing conditioned medium without added CCBE1 resulted in little or no increase in the level of phospho-VEGFR3 as the ligand concentration was increased from 5 to 20 nM (Figure 3A). In contrast, exposure of LECs to VEGFC plus CCBE1 resulted in a dose-dependent increase in phospho-VEGFR3 that fully reproduced the response to an engineered VEGFC ΔNΔC protein (Figure 3A).
Figure 3. CCBE1-dependent proteolysis is required for VEGFC to activate VEGFR3 signaling.
(A) Phospho-VEGFR3 was measured by ELISA following LEC exposure to 5 to 20 nM VEGFC that was incubated with conditioned medium from control HEK293T cells (HEK293T CM) or CCBE1-V5 (CCBE1) or following exposure to VEGFC ΔNΔC-Fc. (B) Schematic representation of the VEGFC cleavage site generated following exposure to CCBE1 in HEK293T conditioned medium. (C) The VEGFC FAAAH109-113LTTTF mutant was not N-terminally cleaved in the presence of CCBE1. (D) N-terminally uncleavable VEGFC (NT uncleavable) was unable to activate VEGFR3. Phospho-VEGFR3 ELISA was performed as described in A using the indicated concentrations of WT and NT uncleavable VEGFC, with and without CCBE1. n = 3 for each concentration. Biochemical data shown are representative of 3 separate experiments. CM, conditioned medium.
The increase in VEGFC ligand activity conferred by CCBE1 is likely to be due to the role of CCBE1 in driving N-terminal proteolysis. To directly test this mechanism and to better characterize it at the molecular level, we isolated the 19- to 21-kDa species of VEGFC VHD-FLAG generated by exposure to CCBE1 and performed N-terminal, Edman sequencing to identify the site of proteolytic cleavage. The N-terminal sequence of this peptide was AHYNTEILK, indicating that CCBE1 drives cleavage of VEGFC following the FAA sequence ending at aa 111 (Figure 3B). Identification of the CCBE1-associated N-terminal cleavage site allowed us to next generate a VEGFC mutant that cannot not be cleaved at that site to directly test whether N-terminal proteolytic cleavage is required for VEGFC activity on LECs. Mutation of the FAA-AHY sequence in VEGFC VHD-FLAG to LTT-TFY (VEGFC-LTTTF) resulted in a protein that was well expressed by HEK293T cells but that was not N-terminally cleaved in the presence of CCBE1 (Figure 3C). VEGFC-LTTTF conferred no increase in phospho-VEGFR3 in primary LECs, with or without prior CCBE1 exposure. In contrast, WT VEGFC showed marked increases in phospho-VEGFR3 that were CCBE1 dependent (Figure 3D). These studies demonstrate that CCBE1-driven proteolysis of VEGFC is required for the ligand to activate the VEGFR3 receptor and that the effect of CCBE1 on VEGFC activity is mediated by N-terminal cleavage.
CCBE1-driven VEGFC proteolysis in HEK293T conditioned media requires ADAMTS3.
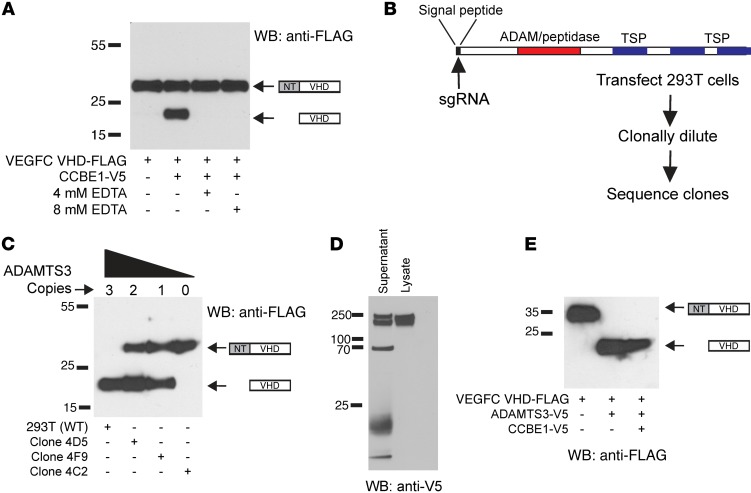
Our analysis of the N-terminal cleavage of VEGFC that is facilitated by CCBE1 revealed proteolysis at FAA-AHYN, a sequence resembling that at which collagen is processed by the ADAMTS2 and ADAMTS3 proteases (29). This finding is consistent with a previous report implicating ADAMTS3 in CCBE1-mediated VEGFC processing by HEK293 cells (25). To investigate the role of ADAMTS3, we first tested the requirement for divalent cations, as ADAMTS metalloprotease function is cation dependent. The addition of EDTA to conditioned media containing VEGFC VHD-FLAG and CCBE1 blocked VEGFC processing (Figure 4A). This result is consistent with metalloprotease activity, but the EGF-like domains of CCBE1 are also predicted to bind calcium, and this result might therefore reflect inactivation of CCBE1 function. To more definitively test the requirement for ADAMTS3, we next used CRISPR-Cas9 to mutate the sequence in the ADAMTS3 gene that encodes the protease’s signal peptide in HEK293T cells (Figure 4B). Analysis of HEK293T clonal cell populations generated after transfection with a plasmid encoding guide RNAs targeting this sequence and Cas9 revealed that HEK293T cells are triploid for the ADAMTS3 gene (consistent with previous cytogenetic studies, ref. 30). HEK293T clones with mutations predicted to disable 1, 2, or 3 ADAMTS3 alleles were identified (Figure 4C and Supplemental Table 1). Analysis of VEGFC VHD-FLAG processing following expression of VEGFC and CCBE1 in ADAMTS3-mutant HEK293T cell clones revealed complete processing in cells with 3 copies, partial processing in cells with 1 or 2 copies, and a total loss of processing in cells with no WT ADAMTS3 alleles (Figure 4C). Thus, progressive loss of ADAMTS3 alleles results in a dose-dependent reduction in CCBE1-driven VEGFC proteolysis.
Figure 4. ADAMTS3 is required for N-terminal VEGFC proteolysis in HEK293T cell supernatant.
(A) Incubation with EDTA revealed that CCBE1-dependent VEGFC proteolysis was cation dependent. (B) The ADAMTS3 gene was disrupted in HEK293T cells using CRISPR-Cas9 to generate disabling mutations in the sequence encoding the ADAMTS3 signal peptide. TSP, thrombospondin 1 motif; sgRNA, single-guide RNA. (C) Incubation of VEGFC VHD-FLAG and CCBE1-V5 generated by expression in HEK293T cell clones with 3 (WT), 2 (4D5), 1 (4F9), or 0 (4C2) WT ADAMTS3 alleles revealed a dose-dependent loss of VEGFC proteolysis, with loss of ADAMTS3. (D) Expression of ADAMTS3-V5 in HEK293T cells revealed a 220- to 240-kDa doublet in cell lysate and a number of smaller bands in conditioned supernatant, consistent with extracellular proteolysis. (E) Expression of ADAMTS3-V5 rescued proteolytic processing of VEGFC VHD-FLAG in ADAMTS3–/– HEK293T cells. Data shown are representative of 3 separate experiments.
CRISPR-Cas9 genome editing is highly efficient but may also result in off-target mutations (31). Thus, it is possible that clones exhibiting a greater loss of ADAMTS3 alleles may also harbor larger numbers of off-target mutations and that the loss of VEGFC processing may not be directly attributable to mutations in ADAMTS3. To further test the causal role for ADAMTS3 in CCBE1-directed VEGFC processing in vitro, we next expressed ADAMTS3-V5 in a HEK293T cell clone that lacked all WT ADAMTS3 genes and did not support VEGFC processing (clone 4C2). Expression of ADAMTS3-V5 revealed two 220- and 240-kDa molecular-weight bands in cell lysate and a number of smaller bands in conditioned supernatant, consistent with ADAMTS3 proteolysis in the extracellular environment (Figure 4D). Expression of ADAMTS3-V5 rescued VEGFC processing in the ADAMTS3-deficient HEK293T-mutant cell line and drove highly efficient VEGFC processing in the absence of added CCBE1 (Figure 4E). These genetic studies demonstrate that CCBE1-driven VEGFC proteolytic processing is mediated exclusively by ADAMTS3 in HEK293 cells.
ADAMTS3 is required for lymphatic development in mice.
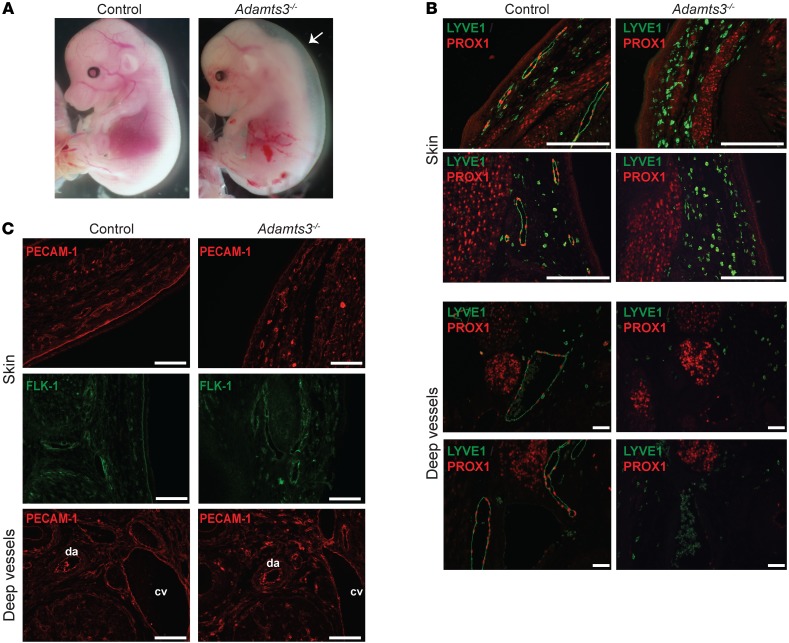
The studies described above revealed that ADAMTS3 plays a central and even necessary role in the proteolytic activation of VEGFC in HEK293T cell–conditioned medium ex vivo. However, ADAMTS3 is highly homologous to ADAMTS14 and ADAMTS2, proteases that can also cleave collagen and that are also expressed in the developing mouse embryo (32, 33). Thus, the ex vivo requirement for ADAMTS3 may merely reflect a lack of expression of related proteases by HEK293T cells and not accurately predict its in vivo lymphangiogenic role. To define the role of ADAMTS3 in vivo, we next studied lymphatic development in ADAMTS3-deficient mouse embryos. A null Adamts3 allele (Adamts3–) was generated by deletion of exon 3 in the Adamts3 gene, an event that removes the propeptidase domain and inserts multiple STOP codons immediately downstream of exon 2, resulting in an Adamts3 mRNA predicted to encode a truncated protein lacking the catalytic domain. Adamts3+/– animals were intercrossed to generate Adamts3–/– embryos and littermate controls. In E14.5 mouse embryos, the loss of ADAMTS3 resulted in severe cutaneous edema (Figure 5A), a phenotype consistent with a defect in embryonic lymphatic vascular function. Histologic examination revealed normal LYVE1+PROX1+ lymphatic vessel growth in the skin and deeper tissues of Adamts3+/– and Adamts3+/+ control littermates, but LYVE1+PROX1+ lymphatic vessels were entirely absent in embryos lacking ADAMTS3 (Figure 5B), consistent with a recent report of independently derived Adamts3–/– animals (34). PECAM and FLK1 staining for ECs revealed normal blood vessel patterning and growth in Adamts3–/– embryos (Figure 5C), a finding consistent with the observation that Adamts3–/– embryos were not growth retarded (Figure 5A). These findings closely phenocopy those previously reported for both Vegfc–/– (6) and Ccbe1–/– (22, 24, 27) embryos and reveal that ADAMTS3 plays a necessary and nonredundant role in lymphatic vascular development in mice.
Figure 5. ADAMTS3 is required for lymphatic development in mouse embryos.
(A) Loss of ADAMTS3 resulted in severe cutaneous edema at E14.5. Arrow indicates translucent space between the skin and body, indicative of edema. (B) Loss of ADAMTS3 resulted in a complete lack of PROX1+LYVE1+ LECs in the E14.5 embryo. Note the presence of smaller lymphatic vessels in the skin and larger lymphatic vessels adjacent to the dorsal aorta and cardinal vein in the WT littermate control embryo. (C) ADAMTS3 was not required for blood vessel development. Blood vessels from an ADAMTS3-deficient and control embryo were identified using anti-PECAM and anti-FLK1 immunostaining. cv, cardinal vein; da, dorsal aorta. Data shown are representative of 3 separate experiments. Scale bars: 50 μm.
VEGFD processing is independent of both CCBE1 and ADAMTS3.
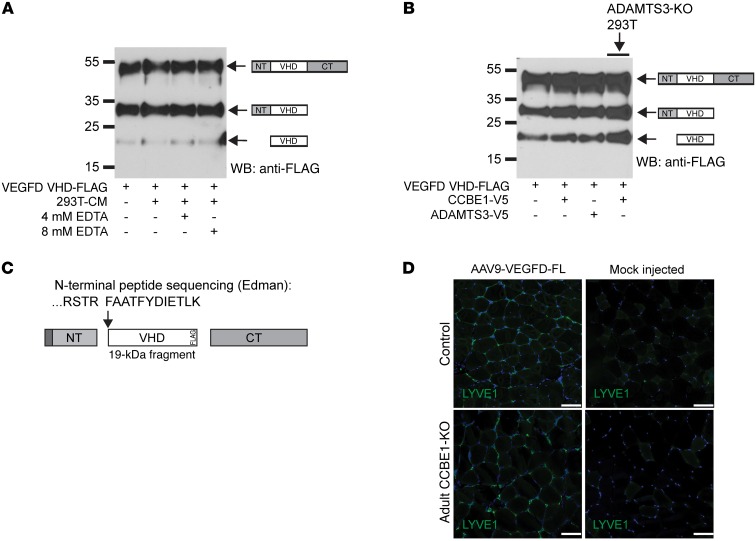
VEGFC and VEGFD are homologous proteins that activate VEGFR3, and VEGFD contains N- and C-terminal domains that are proteolytically processed in a manner similar to that observed for VEGFC (18). As for VEGFC, a number of different proteases have been implicated in the proteolytic processing of VEGFD (18, 35, 36). Zebrafish genetic studies have recently suggested that VEGFD function may be regulated by CCBE1 (13), but biochemical studies have not revealed an effect of CCBE1 or ADAMTS3 on VEGFD processing (25). To directly compare the processing of these 2 lymphangiogenic growth factors, we generated VEGFD VHD-FLAG in a manner similar to that used to generate VEGFC VHD-FLAG (Figure 6A). VEGFD VHD-FLAG secreted from HEK293T cells appeared as 3 distinct 52-, 33-, and 21-kDa bands, consistent with proteins containing NT-VHD-CT, VHD-CT, and VHD domains, respectively (Figure 6A). In contrast to VEGFC, exposure of VEGFD VHD-FLAG to either CCBE1-V5 or ADAMTS3-V5 did not increase the fraction of low-molecular-weight, fully processed protein (Figure 6B). Conversely, expression of VEGFD VHD-FLAG in ADAMTS3-deficient HEK293T cells did not reduce the fraction of low-molecular-weight, fully processed protein (Figure 6B, right). N-terminal sequencing of the 21-kDa, fully processed VEGFD VHD-FLAG protein expressed by HEK293T cells revealed cleavage after R88 rather than A91 (Figure 6C). These in vitro results are consistent with those of previous studies (18, 25) and demonstrate that despite their structural and sequence homology, VEGFC and VEGFD are processed by distinct enzymatic processes in HEK293T cell–conditioned medium.
Figure 6. VEGFD proteolysis and activity are independent of ADAMTS3 and CCBE1.
(A) Incubation with EDTA revealed that VEGFD VHD-FLAG proteolysis in HEK293T conditioned medium was cation independent. (B) VEGFD VHD-FLAG proteolysis was not increased by incubation with conditioned medium containing CCBE1 (lane 2) or ADAMTS3 (lane 3). VEGFD VHD-FLAG proteolysis was also not reduced when expressed by ADAMTS3-deficient HEK293T cells (lane 4, right). (C) N-terminal sequencing of the 21-kDa band observed in VEGFD VHD-FLAG–expressing medium revealed cleavage after R88, a site that is 3 aa N-terminal of that detected in VEGFC VHD-FLAG. frag, fragment. (D) VEGFD was lymphangiogenic in CCBE1-deficient animals. AAV expressing VEGFD-FL was injected into the tibialis muscle of adult CCBE1-KO and control animals. Lymphatic growth was detected using LYVE1 immunostaining and compared with that in mock-injected animals. Data shown are representative of 3 separate experiments. Scale bars: 50 μm.
To test whether VEGFC and VEGFD are activated through distinct mechanisms in vivo, we next expressed full-length VEGFD (VEGFD-FL) in WT and adult CCBE1-KO mouse muscle using AAV. In contrast to VEGFC, AAV-VEGFD drove lymphatic vessel growth in both WT and adult CCBE1-KO animals (Figure 6D). These findings demonstrate that, despite their strong aa and structural homology, VEGFC and VEGFD are processed by distinct proteases and are regulated independently.
VEGFC, ADAMTS3, and CCBE1 interact in a single molecular complex.
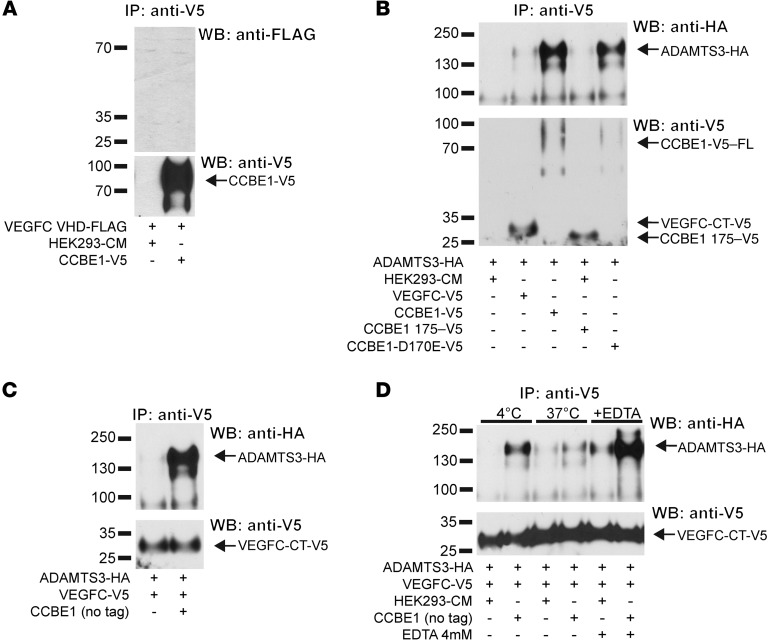
The studies described above suggested that VEGFC is cleaved by ADAMTS3 in a manner that is tightly regulated by CCBE1. To better understand the molecular interactions that govern this regulatory mechanism, we next took advantage of epitope-tagged proteins to examine the physical association of these proteins. Consistent with previously reported studies (25), VEGFC VHD-FLAG and CCBE1-V5 could not be coprecipitated from conditioned supernatant containing both proteins (Figure 7A), indicating that VEGFC and CCBE1 do not directly interact. In contrast, immunoprecipitation of CCBE1 from conditioned supernatant containing ADAMTS3-HA and CCBE1-V5 resulted in efficient coprecipitation of ADAMTS3 (Figure 7B), indicating that ADAMTS3 and CCBE1 biochemically interact in a robust manner. ADAMTS3-HA could not be coprecipitated with CCBE1 175-V5 (Figure 7B), consistent with functional studies demonstrating an inability of this truncated CCBE1 protein to drive VEGFC N-terminal cleavage (Figure 2F). Consistent with this result, CCBE1 D170E-V5, a CCBE1 protein with a mutation in the N-terminal domain that causes loss of lymphatic growth in zebrafish embryos in vivo (20), retained strong interaction with ADAMTS3 (Figure 7B). Unexpectedly, a small amount of ADAMTS3-HA could be coprecipitated with VEGFC-V5, suggesting that VEGFC may pull down some ADAMTS3 in a manner that is dependent on endogenous HEK293T CCBE1 (Figure 7B). To further explore this finding and test the possibility of a VEGFC-ADAMTS3-CCBE1 complex, we next induced expression of VEGFC-V5 and ADAMTS3 with untagged CCBE1 and repeated the VEGFC pulldown. In the presence of added CCBE1, VEGFC-V5 was able to efficiently pull down ADAMTS3-HA (Figure 7C), consistent with the formation of a trimolecular complex. To further explore the nature of this VEGFC-ADAMTS3-CCBE1 complex, we repeated this experiment at 4°C, a temperature at which ADAMTS3 activity is expected to be reduced; at 37°C, a temperature at which ADAMTS3 activity is expected to be high; and in the presence of EDTA, a known inhibitor of ADAMTS proteolytic activity. ADAMTS3 was efficiently coprecipitated with VEGFC at 4°C and in the presence of EDTA, but not at 37°C (Figure 7D). These findings support a molecular model in which ADAMTS3 binds VEGFC in a CCBE1-dependent manner to form a proteolytic complex that is required for ligand activation (Figure 8, discussed further below).
Figure 7. VEGFC, ADAMTS3, and CCBE1 form a molecular complex prior to VEGFC cleavage.
(A) VEGFC did not directly bind CCBE1. VEGFC VHD-FLAG and CCBE1-V5 conditioned media were mixed prior to immunoprecipitation with anti-V5 Abs and to immunoblot analysis with anti-FLAG Abs. Precipitated CCBE1-V5 is shown in the bottom panel. (B) ADAMTS3 coprecipitated with full-length, but not N-terminal, CCBE1. The indicated conditioned media were mixed, and CCBE1-V5 was immunoprecipitated. Coprecipitated ADAMTS3-HA is shown at the top, and precipitated CCBE1-V5 proteins are shown at the bottom. (C) VEGFC coprecipitated with ADAMTS3 in a CCBE1-dependent manner. Conditioned media containing ADAMTS3-HA and VEGFC-V5 were mixed alone and with media containing untagged CCBE1 prior to anti-V5 immunoprecipitation. (D) ADAMTS3-CCBE1 release of VEGFC required ADAMTS3 enzymatic activity. VEGFC coprecipitation with ADAMTS3 was performed in the presence of untagged CCBE1, as described in C, at 4°C and 37°C and in the presence of EDTA to inhibit metalloprotease activity. Note that VEGFC was retained at 4°C and in the presence of EDTA, conditions that block ADAMTS3 enzymatic activity. Data shown are representative of 3 separate experiments.
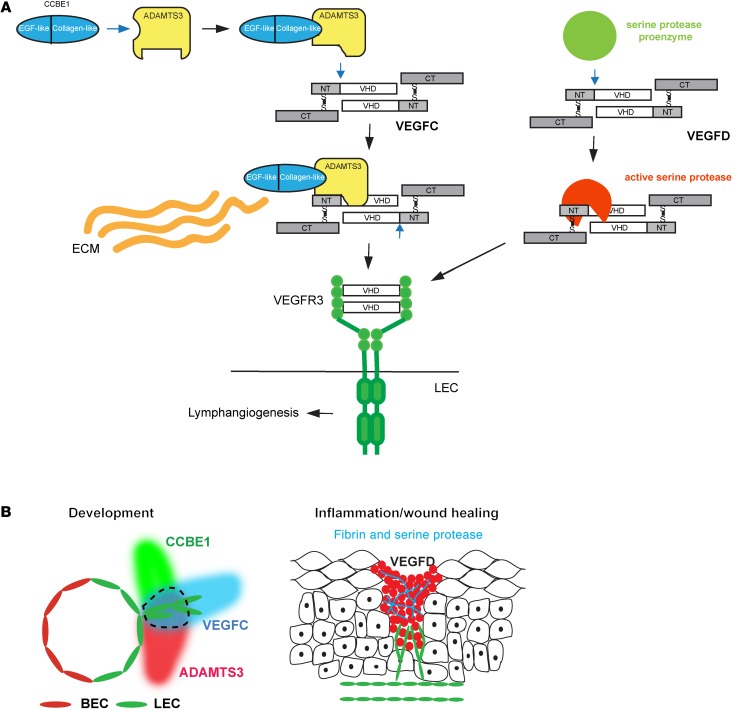
Figure 8. Distinct mechanisms of proteolytic activation support distinct biological roles for VEGFC and VEGFD.
(A) Molecular mechanisms of VEGFC and VEGFD activation. VEGFC is activated by the formation of a VEGFC-ADAMTS3-CCBE1 complex (left). CCBE1 binding to ADAMTS3 via the CCBE1 CT (left) is predicted to confer a conformational change that permits the enzyme to associate with and cleave VEGFC (middle). N-terminal cleavage of VEGFC releases the VHD that is able to activate VEGFR3 on the LEC. The CCBE1 NT may bind extracellular matrix (ECM) to localize the complex spatially during lymphatic growth. In contrast, VEGFD is activated independently of ADAMTS3 and CCBE1, most likely through a serine protease generated at sites of inflammation (right). (B) Proposed lymphangiogenic roles of VEGFC and VEGFD in vivo. VEGFC activation by ADAMTS3 and CCBE1 provides a mechanism for spatial patterning of the developing lymphatic vasculature (left). Schematic shows the mid-gestation cardinal vein with newly specified LECs that are in the process of sprouting to form the lymphatic network. The area encircled by the dotted line represents a zone of active VEGFC. VEGFD activation by inflammatory proteases such as those generated during wound healing in the skin provides a mechanism for lymphangiogenesis in mature animals with preexisting lymphatic networks (right).
Discussion
During embryonic development, lymphatic vessels arise after organs have formed and rapidly create a vascular network through a highly stereotyped, spatially controlled angiogenic program (37). VEGFC is a critical regulator of this process (6, 7), but the VEGFC homolog VEGFD appears to play no required role in lymphatic development and has been linked instead to postnatal lymphangiogenesis (12). VEGFC and VEGFD contain N- and C-terminal extensions that are not found in VEGFA. Cell culture studies performed almost 20 years ago first revealed that these extensions are proteolytically removed (17, 18), but the biological significance of these observations has only recently begun to emerge from studies connecting defects in lymphatic vascular development associated with loss of the secreted protein CCBE1 to a role for CCBE1 in VEGFC proteolysis (25–27). However, a clear molecular understanding of these events has not yet emerged because of the difficulties inherent in following VEGFC proteolysis and assessing its role in lymphatic EC responses ex vivo and in vivo and the evidence supporting a role for ADAMTS3 in lymphatic development. We addressed these issues using molecular tags to follow VEGFC and VEGFD processing without disturbing their molecular activity; genome editing to test the role of ADAMTS3 ex vivo; and mouse genetic studies to test the roles of ADAMTS3 and CCBE1 in vivo. We found that VEGFC activity is regulated by assembly of a VEGFC-ADAMTS3-CCBE1 complex and that loss of ADAMTS3 blocks lymphatic development in mice in a manner identical to that induced by loss of CCBE1 or VEGFC. In contrast, proteolysis of the closely related VEGFD, a growth factor that is dispensable during lymphatic development, is independent of ADAMTS3 and CCBE1 ex vivo, and its lymphangiogenic activity is independent of CCBE1 in vivo. These findings suggest that regulation of VEGFC activity by ADAMTS3 and CCBE1 is a molecular mechanism for programmed lymphatic growth during development, while the role of VEGFD may be connected to the lymphangiogenesis that takes place after lymphatic development is complete and that is regulated by a distinct proteolytic activation mechanism (Figure 8).
A key molecular finding in this study is that VEGFC, ADAMTS3, and CCBE1 form a trimolecular complex that is required to proteolytically activate VEGFC and that this activation mechanism can take place in the extracellular environment in a manner that does not require the LEC or the VEGFR3 receptor. Previous studies have revealed a role for CCBE1 in VEGFC proteolysis (25–27) and suggested that ADAMTS3 cleaves VEGFC (25), but a clear picture of how these secreted proteins interact and how their interaction regulates lymphatic vascular growth has not emerged. To follow VEGFC proteolytic processing, many studies have used VEGFR3 ectodomain pulldown (17, 25), an approach that necessarily inserts the VEGFR3 receptor, biases toward detection of VEGFR3-binding VEGFC proteins, and emphasizes the role of cell-surface–associated VEGFC activation. In contrast, our studies demonstrate that VEGFC can be efficiently activated independently of the VEGFR3 receptor. These findings are consistent with a model in which the spatial intersection of VEGFC, ADAMTS3, and CCBE1 proteins in the extracellular environment creates a trail of active VEGFC ligand that is followed by LECs to pattern lymphatic development in the developing embryo (Figure 8B). Such a precise spatio-temporal blueprint may be required for the lymphatic vasculature to expand as rapidly as it does in mid-gestation, e.g., from a limited number of progenitor cells to a functional network in only a few days in the mouse. Future studies that map the extracellular location of VEGFC, CCBE1, and ADAMTS3 during lymphatic development and use genetic strategies to test their cellular requirements in vivo will be required to test this model.
The finding that VEGFD proteolysis takes place independently of ADAMTS3 and CCBE1, both ex vivo and in vivo, demonstrates a remarkable difference in the regulation of these 2 lymphangiogenic factors that provides new insight into their distinct in vivo roles. Gain-of-function studies revealed that VEGFC and VEGFD have equally powerful lymphangiogenic effects in vivo (15, 38), but genetic loss-of-function studies have only demonstrated a critical role for VEGFC during lymphatic development (6, 12). If the primary role of ADAMTS3-CCBE1 processing is to lay down a roadmap of activated VEGFR3 ligand that LECs follow to create the primary lymphatic network, then this can only be accomplished by VEGFC. In contrast, lymphatic growth after wound healing or inflammatory states is a local process that may be served by the serine proteases released during hemostasis and fibrinolysis, a role consistent with activation of VEGFD by cleavage at a distinct N-terminal sequence (Figure 5 and refs. 18, 35). Thus, it is reasonable to speculate that VEGFC and VEGFD have evolved to function in distinct lymphangiogenic responses, with VEGFC serving as a factor that regulates spatially programmed lymphatic growth in the developing embryo and VEGFD serving as a factor that drives reactive, local lymphatic growth after lymphatic vessels have formed (Figure 8B). Key support for this division of lymphangiogenic labor is our finding that ADAMTS3 is required for lymphatic development in a manner similar to that previously reported for both CCBE1 and VEGFC, but not VEGFD. In contrast, previous studies have identified plasmin, a fibrinolytic serine protease that is activated at sites of tissue injury and inflammation (39), as being capable of cleaving VEGFD (35). Future studies that more precisely define the protease(s) responsible for VEGFD activation and their relationship to VEGFD function during established models of inflammatory lymphangiogenesis in vivo will help test this partitioned model of VEGFC and VEGFD lymphangiogenic function.
It is surprising that, despite its redundant function with ADAMTS2 for procollagen processing (29) and close homology to ADAMTS14 (32), ADAMTS3 plays a nonredundant role in lymphatic development in mice. The strong physical association observed between ADAMTS3 and CCBE1 in our biochemical studies suggests that the functions of these 2 proteins are tightly linked, and we found that the binding of VEGFC by this complex required the presence of ADAMTS3. Mutations in CCBE1 account for only 25% of cases of the human primary lymphedema syndrome known as Hennekam syndrome (19, 40), with another 20% attributable to mutations in the gene encoding the atypical cadherin FAT4 (41). The highly specific disruption of lymphatic development with loss of ADAMTS3 in developing mice suggests that loss-of-function mutations in ADAMTS3 may also contribute to Hennekam syndrome and that FAT4 may bind one or more of the components of the VEGFC-ADAMTS3-CCBE1 complex required to generate active VEGFC. Future studies addressing the biochemical and genetic relationships between these proteins will better define how this complex regulates lymphangiogenesis in vivo as well as its contribution to human lymphatic diseases.
There remains the molecular question of precisely how proteolysis activates the VEGFC ligand to drive lymphatic development and growth. Our findings and previous studies (17, 25) clearly demonstrate a requirement for proteolytic removal of N and C termini that are covalently linked to each other for activation of the VEGFR3 receptor by VEGFC. Several molecular mechanisms may explain this requirement. The most straightforward mechanism is that the presence of the N and C termini sterically prevents the VEGFC core ligand (i.e., the VHD) from engaging the VEGFR3 receptor to induce receptor signaling. Alternatively, the NT or CT may actively oppose VEGFC activity, e.g., by binding a distinct site in VEGFR3 or VEGFC itself. In the course of our studies, we generated a VEGFC ligand that had an uncleavable 192-aa CT: one in which the CT was replaced with a 22-aa FLAG-V5 tag and one in which there was no CT following the VHD. All of these ligands were efficiently cleaved at the NT, but we observed a progressive loss of VEGFR3 activation as the C-terminal extension grew in length from 0 to 22 to 192 aa (Supplemental Figure 5). The progressive loss of function conferred by longer C-terminal tags supports the more straightforward model of steric hindrance during the docking of VEGFC to VEGFR3 (42).
The discovery of such diverse and complex regulatory mechanisms for VEGFR3 activation reveals an unexpected complexity in the control of lymphatic vascular growth with implications for therapeutic lymphangiogenesis or lymphangiogenic blockade. How important are the stoichiometry and spatial relationships of VEGFC, ADAMTS3, and CCBE1 for productive lymphangiogenesis? The lymphatic structures generated in response to VEGFC ΔNΔC expression in the muscle of mature mice appeared highly abnormal, as these structures tightly encircled the muscle fibers that expressed that growth factor and did not form typical branching structures (Figure 1D). In contrast, VEGFC-FL stimulated the growth of branched lymphatic structures, presumably as a result of the influence of endogenous regulators such as CCBE1 and ADAMTS3. We do not yet understand the expression of these VEGFC regulators, however, and an entirely distinct set of regulators may exist for VEGFD. Conversely, it now appears that inhibition of lymphangiogenesis mediated by VEGFC may be achieved by blockade of CCBE1 or ADAMTS3. In some contexts, especially in cancer growth, the complexity of blood vessel growth has proven to be a critical barrier to antiangiogenic therapies (43); it appears that lymphatic vessel growth is no less complex and that therapeutic applications will face equally large challenges.
Methods
Mouse lines
Prox1-GFP and Ccbe1 conditional and null alleles were previously reported (24, 44). Adult VEGFC-KO and VEGFD-KO mice have been previously described (45, 46). The Ub-CreERT2 allele was obtained from The Jackson Laboratory (catalog 7001). The Adamts3-targeted mice were obtained from Adamts3tm1a(KOMP)Wtsi (clone ID: EPD0660_2_C10) sperm from the UC Davis KOMP Repository (https://www.komp.org). Adamts3-null mice were generated by crossing Adamts3tm1a(KOMP)Wtsi (clone ID: EPD0660_2_C10) and EIIa-Cre animals. Neonatal and adult deletions were induced by administration of tamoxifen as previously described (24). Adult animals were maintained on tamoxifen chow for more than 6 weeks before the in vivo studies to ensure a high-deficiency state.
Viral expression of VEGFC in vivo
Cutaneous adenoviral expression.
Adeno-VEGFC (47) or control adenovirus expressing either VEGFB or GFP (2 × 108 to 2 × 109 PFU) was injected s.c. into the dorsal skin of adult mice. Adeno-mCherry (1 × 108 PFU) was coinjected to identify the site of expression prior to skin harvesting. BrdU was injected i.p. 24 hours before harvesting to label proliferating cells. Adeno-VEGFC was provided by the Alitalo laboratory (University of Helsinki, Helsinki, Finland) and was also a gift of Calvin Kuo (Stanford University, Palo Alto, California, USA).
Muscular AAV expression.
AAV9 constructs containing murine full-length VEGFC, VEGFC ΔNΔC, or full-length VEGFD were used (38). Viral particles (9–12 × 1010) mixed with AAV9-mCherry were injected into the left tibialis anterior and/or gastrocnemius muscles. Muscles were harvested 4 weeks after injection for histologic analysis of lymphatic growth.
Histologic analysis
Paraffin-embedded tissue sections were used for all histologic studies. Immunofluorescence was performed with polyclonal anti-mouse Abs against PROX1 (Abcam; catalog 76696), LYVE1 (R&D Systems; catalog AF2125), and BrdU. Images were acquired with a Nikon Eclipse 80i microscope using a ×20/0.50 or ×40/0.75 numeric aperture (NA) dry objective.
Generation of epitope-tagged VEGFC, CCBE1, and ADAMTS3 proteins
Full-length human cDNAs for VEGFC, VEGFD, CCBE1, and ADAMTS3 were obtained from GE Dharmacon. Epitope-tagged cDNAs were generated through PCR amplification using primers containing in-frame V5, FLAG, or HA tags. Point mutations in CCBE1 and VEGFC were generated using the QuikChange II Site-Directed Mutagenesis Kit (Agilent Technologies). All final constructs were confirmed by nucleotide sequencing. The list of primers used in cloning are indicated in the Supplemental Methods.
Protein expression and characterization
Epitope-tagged cDNA constructs were cloned into pcDNA 3.1+ and transfected into WT HEK293T cells (Invitrogen) or mutant HEK 293T lines where indicated (Figure 4 and Figure 6B). Cells were plated at a density of 3 × 105 cells per 10 cm2 and transfected using Lipofectamine 2000 (Invitrogen). Unless otherwise indicated, conditioned media were collected 72 hours after transfection. Conditioned supernatants were mixed at a 1:1 ratio unless otherwise indicated and incubated for 18 hours at 37°C prior to analysis. Untransfected HEK293T conditioned media were collected and used as control supernatants. Western blotting was performed using anti-V5 HRP (Invitrogen; catalog 46-0708); anti-FLAG (Sigma-Aldrich; catalog A8592-1MG); and anti-HA (BioLegend; catalog MMS-101P-500) Abs and developed using ECL reagent (Thermo Fisher Scientific).
Phospho-VEGFR3 ELISA assay
Neonatal LECs were obtained from Lonza and used between passages 6 and 8. LECs were plated onto 6-well plates at a density of 3 × 105 cells per 10 cm2 and serum starved with 900 μl starving media (EBM2 media [Lonza] plus 0.1% BSA [Sigma-Aldrich]) for 6 hours at 37°C. Serum-starved LECs were treated with 100 μl of the indicated conditioned media (Figure 3) for 15 minutes at 37°C. The media were then aspirated and the cells washed 3 times with cold PBS before being lysed with 150 μl phospho-VEGFR3 lysis buffer (1% NP-40 Alternative, 20 mM Tris [pH 8.0], 137 mM NaCl, 10% glycerol, 2 mM EDTA, 1 mM activated sodium orthovanadate, and 10% protease inhibitors). Samples were collected and the steps followed according to the instructions in the Human Phospho-VEGFR3 ELISA Kit (R&D Systems).
Immunoprecipitation
Conditioned supernatants were harvested and mixed for 18 hours at 4°C before addition of anti–V5-tag magnetic beads (MBL). Beads were washed 3 times using Opti-MEM (Thermo Fisher Scientific) and bound proteins and then eluted in sample buffer and analyzed by Western blotting.
CRISPR-Cas9 generation of ADAMTS3-deficient HEK293T cells
Guide RNAs targeting the signal peptide of ADAMTS3 were cloned into the pSpCas9(BB)-2A-Puro (PX459) V2.0 plasmid (Addgene). The resulting plasmids were then transfected into HEK293T cells on 6-well plates using FuGENE6 reagent and standard protocols. The transfected cells were then selected by addition of puromycin to the media between 12 and 36 hours after transfection, and the selected cells were then clonally diluted onto 96-well plates. After 2 weeks of growth, wells with single colonies were identified and subsequently expanded. For each of these HEK293T clones, the targeted region of the ADAMTS3 gene was then amplified by PCR, and individual PCR products were isolated by TA cloning (T Easy vector; Promega; catalog A1360). A sample of the amplified ADAMTS3 alleles present in each cell line was obtained by sequencing at least 10 different TA-cloned PCR products from each line. Using this approach, HEK293T clones with 0, 1, 2, and 3 copies of the mutant ADAMTS3 gene were identified.
Statistics.
All error bars indicate the SEM. An unpaired, 2-tailed Student’s t test was applied to determine statistical significance. A P value of less than 0.01 was considered significant.
Study approval.
All animal protocols were approved by the IACUC of the University of Pennsylvania.
Author contributions
HMB and DE designed and performed most of the experiments and helped write the manuscript. MRR, HJN, JC, MC, YY, VD, and ET performed experiments. KJ, HZ, and RK produced recombinant CCBE1-V5 for biochemical studies. AA and KA contributed valuable reagents. MLK helped design and perform the experiments, and KA and MLK helped write the manuscript.
Supplementary Material
Acknowledgments
We thank the members of the Kahn and Alitalo laboratories for their valuable insights during the course of these studies. We would also like to thank Chari Smith and Andy Pope for their support of these studies. This work was supported by NIH grants K08AR061659 (to D. Enis); R01HL103432 (to M.L. Kahn); and R01HL111553 (to M.L. Kahn) and by the Penn Skin Disease Research Center and the Leducq Foundation (to K. Alitalo and M.L. Kahn).
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:J Clin Invest. 2016;126(6):2167–2180. doi:10.1172/JCI83967.
Contributor Information
David Enis, Email: david.enis@gmail.com.
Jennifer Cohen, Email: jencohen@mail.med.upenn.edu.
Mei Chen, Email: meichen40@yahoo.com.
Yiqing Yang, Email: yiqingy@mail.med.upenn.edu.
Veerpal Dhillon, Email: vinnzdhillon@gmail.com.
Hong Zhang, Email: Hong.2.Zhang@gsk.com.
Robert Kirkpatrick, Email: Robert.B.Kirkpatrick@gsk.com.
Elizabeth Traxler, Email: liztraxler@gmail.com.
Andrey Anisimov, Email: andrey.anisimov@helsinki.fi.
Kari Alitalo, Email: kari.alitalo@helsinki.fi.
Mark L. Kahn, Email: markkahn@mail.med.upenn.edu.
References
- 1.Tammela T, Alitalo K. Lymphangiogenesis: Molecular mechanisms and future promise. Cell. 2010;140(4):460–476. doi: 10.1016/j.cell.2010.01.045. [DOI] [PubMed] [Google Scholar]
- 2.Randolph GJ, Angeli V, Swartz MA. Dendritic-cell trafficking to lymph nodes through lymphatic vessels. Nat Rev Immunol. 2005;5(8):617–628. doi: 10.1038/nri1670. [DOI] [PubMed] [Google Scholar]
- 3.Alitalo K. The lymphatic vasculature in disease. Nat Med. 2011;17(11):1371–1380. doi: 10.1038/nm.2545. [DOI] [PubMed] [Google Scholar]
- 4.Joukov V, et al. A novel vascular endothelial growth factor, VEGF-C, is a ligand for the Flt4 (VEGFR-3) and KDR (VEGFR-2) receptor tyrosine kinases. EMBO J. 1996;15(2):290–298. [PMC free article] [PubMed] [Google Scholar]
- 5.Kaipainen A, et al. Expression of the fms-like tyrosine kinase 4 gene becomes restricted to lymphatic endothelium during development. Proc Natl Acad Sci U S A. 1995;92(8):3566–3570. doi: 10.1073/pnas.92.8.3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karkkainen MJ, et al. Vascular endothelial growth factor C is required for sprouting of the first lymphatic vessels from embryonic veins. Nat Immunol. 2004;5(1):74–80. doi: 10.1038/ni1013. [DOI] [PubMed] [Google Scholar]
- 7.Ober EA, et al. Vegfc is required for vascular development and endoderm morphogenesis in zebrafish. EMBO Rep. 2004;5(1):78–84. doi: 10.1038/sj.embor.7400047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Irrthum A, Karkkainen MJ, Devriendt K, Alitalo K, Vikkula M. Congenital hereditary lymphedema caused by a mutation that inactivates VEGFR3 tyrosine kinase. Am J Hum Genet. 2000;67(2):295–301. doi: 10.1086/303019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gordon K, et al. Mutation in vascular endothelial growth factor-C, a ligand for vascular endothelial growth factor receptor-3, is associated with autosomal dominant milroy-like primary lymphedema. Circ Res. 2013;112(6):956–960. doi: 10.1161/CIRCRESAHA.113.300350. [DOI] [PubMed] [Google Scholar]
- 10.Orlandini M, Marconcini L, Ferruzzi R, Oliviero S. Identification of a c-fos-induced gene that is related to the platelet-derived growth factor/vascular endothelial growth factor family. Proc Natl Acad Sci U S A. 1996;93(21):11675–11680. doi: 10.1073/pnas.93.21.11675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Achen MG, et al. Vascular endothelial growth factor D (VEGF-D) is a ligand for the tyrosine kinases VEGF receptor 2 (Flk1) and VEGF receptor 3 (Flt4) Proc Natl Acad Sci U S A. 1998;95(2):548–553. doi: 10.1073/pnas.95.2.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koch M, et al. VEGF-D deficiency in mice does not affect embryonic or postnatal lymphangiogenesis but reduces lymphatic metastasis. J Pathol. 2009;219(3):356–364. doi: 10.1002/path.2605. [DOI] [PubMed] [Google Scholar]
- 13.Astin JW, et al. Vegfd can compensate for loss of Vegfc in zebrafish facial lymphatic sprouting. Development. 2014;141(13):2680–2690. doi: 10.1242/dev.106591. [DOI] [PubMed] [Google Scholar]
- 14.Jeltsch M, et al. Hyperplasia of lymphatic vessels in VEGF-C transgenic mice. Science. 1997;276(5317):1423–1425. doi: 10.1126/science.276.5317.1423. [DOI] [PubMed] [Google Scholar]
- 15.Rissanen TT, et al. VEGF-D is the strongest angiogenic and lymphangiogenic effector among VEGFs delivered into skeletal muscle via adenoviruses. Circ Res. 2003;92(10):1098–1106. doi: 10.1161/01.RES.0000073584.46059.E3. [DOI] [PubMed] [Google Scholar]
- 16.Saaristo A, Karkkainen MJ, Alitalo K. Insights into the molecular pathogenesis and targeted treatment of lymphedema. Ann N Y Acad Sci. 2002;979:94–110. doi: 10.1111/j.1749-6632.2002.tb04871.x. [DOI] [PubMed] [Google Scholar]
- 17.Joukov V, et al. Proteolytic processing regulates receptor specificity and activity of VEGF-C. EMBO J. 1997;16(13):3898–3911. doi: 10.1093/emboj/16.13.3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stacker SA, et al. Biosynthesis of vascular endothelial growth factor-D involves proteolytic processing which generates non-covalent homodimers. J Biol Chem. 1999;274(45):32127–32136. doi: 10.1074/jbc.274.45.32127. [DOI] [PubMed] [Google Scholar]
- 19.Alders M, et al. Mutations in CCBE1 cause generalized lymph vessel dysplasia in humans. Nat Genet. 2009;41(12):1272–1274. doi: 10.1038/ng.484. [DOI] [PubMed] [Google Scholar]
- 20.Hogan BM, et al. Ccbe1 is required for embryonic lymphangiogenesis and venous sprouting. Nat Genet. 2009;41(4):396–398. doi: 10.1038/ng.321. [DOI] [PubMed] [Google Scholar]
- 21.Connell F, et al. Linkage and sequence analysis indicate that CCBE1 is mutated in recessively inherited generalised lymphatic dysplasia. Hum Genet. 2010;127(2):231–241. doi: 10.1007/s00439-009-0766-y. [DOI] [PubMed] [Google Scholar]
- 22.Bos FL, et al. CCBE1 is essential for mammalian lymphatic vascular development and enhances the lymphangiogenic effect of vascular endothelial growth factor-C in vivo. Circ Res. 2011;109(5):486–491. doi: 10.1161/CIRCRESAHA.111.250738. [DOI] [PubMed] [Google Scholar]
- 23.Hagerling R, et al. A novel multistep mechanism for initial lymphangiogenesis in mouse embryos based on ultramicroscopy. EMBO J. 2013;32(5):629–644. doi: 10.1038/emboj.2012.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zou Z, et al. The secreted lymphangiogenic factor CCBE1 is essential for fetal liver erythropoiesis. Blood. 2013;121(16):3228–3236. doi: 10.1182/blood-2012-10-462689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jeltsch M, et al. CCBE1 enhances lymphangiogenesis via A disintegrin and metalloprotease with thrombospondin motifs-3-mediated vascular endothelial growth factor-C activation. Circulation. 2014;129(19):1962–1971. doi: 10.1161/CIRCULATIONAHA.113.002779. [DOI] [PubMed] [Google Scholar]
- 26.Le Guen L, et al. Ccbe1 regulates Vegfc-mediated induction of Vegfr3 signaling during embryonic lymphangiogenesis. Development. 2014;141(6):1239–1249. doi: 10.1242/dev.100495. [DOI] [PubMed] [Google Scholar]
- 27.Roukens MG, et al. Functional dissection of the CCBE1 protein: a crucial requirement for the collagen repeat domain. Circ Res. 2015;116(10):1660–1669. doi: 10.1161/CIRCRESAHA.116.304949. [DOI] [PubMed] [Google Scholar]
- 28.Planas-Paz L, Strilic B, Goedecke A, Breier G, Fassler R, Lammert E. Mechanoinduction of lymph vessel expansion. EMBO J. 2012;31(4):788–804. doi: 10.1038/emboj.2011.456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Colige A, et al. Domains and maturation processes that regulate the activity of ADAMTS-2, a metalloproteinase cleaving the aminopropeptide of fibrillar procollagens types I-III and V. J Biol Chem. 2005;280(41):34397–34408. doi: 10.1074/jbc.M506458200. [DOI] [PubMed] [Google Scholar]
- 30.Bylund L, Kytölä S, Lui WO, Larsson C, Weber G. Analysis of the cytogenetic stability of the human embryonal kidney cell line 293 by cytogenetic and STR profiling approaches. Cytogenet Genome Res. 2004;106(1):28–32. doi: 10.1159/000078556. [DOI] [PubMed] [Google Scholar]
- 31.Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346(6213): doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 32.Bolz H, Ramirez A, von Brederlow B, Kubisch C. Characterization of ADAMTS14, a novel member of the ADAMTS metalloproteinase family. Biochim Biophys Acta. 2001;1522(3):221–225. doi: 10.1016/S0167-4781(01)00329-3. [DOI] [PubMed] [Google Scholar]
- 33.Le Goff C, et al. Regulation of procollagen amino-propeptide processing during mouse embryogenesis by specialization of homologous ADAMTS proteases: insights on collagen biosynthesis and dermatosparaxis. Development. 2006;133(8):1587–1596. doi: 10.1242/dev.02308. [DOI] [PubMed] [Google Scholar]
- 34.Janssen L, et al. ADAMTS3 activity is mandatory for embryonic lymphangiogenesis and regulates placental angiogenesis. Angiogenesis. 2016;19(1):53–65. doi: 10.1007/s10456-015-9488-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McColl BK, et al. Plasmin activates the lymphangiogenic growth factors VEGF-C and VEGF-D. J Exp Med. 2003;198(6):863–868. doi: 10.1084/jem.20030361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McColl BK, et al. Proprotein convertases promote processing of VEGF-D, a critical step for binding the angiogenic receptor VEGFR-2. FASEB J. 2007;21(4):1088–1098. doi: 10.1096/fj.06-7060com. [DOI] [PubMed] [Google Scholar]
- 37.Yang Y, Oliver G. Development of the mammalian lymphatic vasculature. J Clin Invest. 2014;124(3):888–897. doi: 10.1172/JCI71609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Anisimov A, et al. Activated forms of VEGF-C and VEGF-D provide improved vascular function in skeletal muscle. Circ Res. 2009;104(11):1302–1312. doi: 10.1161/CIRCRESAHA.109.197830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Romer J, et al. Impaired wound healing in mice with a disrupted plasminogen gene. Nat Med. 1996;2(3):287–292. doi: 10.1038/nm0396-287. [DOI] [PubMed] [Google Scholar]
- 40.Alders M, et al. Evaluation of clinical manifestations in patients with severe lymphedema with and without CCBE1 mutations. Mol Syndromol. 2013;4(3):107–113. doi: 10.1159/000342486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alders M, et al. Hennekam syndrome can be caused by FAT4 mutations and be allelic to Van Maldergem syndrome. Hum Genet. 2014;133(9):1161–1167. doi: 10.1007/s00439-014-1456-y. [DOI] [PubMed] [Google Scholar]
- 42.Leppanen VM, et al. Structural and mechanistic insights into VEGF receptor 3 ligand binding and activation. Proc Natl Acad Sci U S A. 2013;110(32):12960–12965. doi: 10.1073/pnas.1301415110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ebos JM, Kerbel RS. Antiangiogenic therapy: impact on invasion, disease progression, and metastasis. Nat Rev Clin Oncol. 2011;8(4):210–221. doi: 10.1038/nrclinonc.2011.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Choi I, et al. Visualization of lymphatic vessels by Prox1-promoter directed GFP reporter in a bacterial artificial chromosome-based transgenic mouse. Blood. 2011;117(1):362–365. doi: 10.1182/blood-2010-07-298562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nurmi H, Saharinen P, Zarkada G, Zheng W, Robciuc MR, Alitalo K. VEGF-C is required for intestinal lymphatic vessel maintenance and lipid absorption. EMBO Mol Med. 2015;7(11):1418–1425. doi: 10.15252/emmm.201505731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Baldwin ME, et al. Vascular endothelial growth factor D is dispensable for development of the lymphatic system. Mol Cell Biol. 2005;25(6):2441–2449. doi: 10.1128/MCB.25.6.2441-2449.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Enholm B, et al. Adenoviral expression of vascular endothelial growth factor-C induces lymphangiogenesis in the skin. Circ Res. 2001;88(6):623–629. doi: 10.1161/01.RES.88.6.623. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.