Abstract
The histone demethylase PHF8 has been implicated in multiple pathological disorders, including X-linked mental retardation and tumorigenesis. However, it is not clear how the abundance and function of PHF8 are regulated. Here, we report that PHF8 physically associates with the deubiquitinase USP7. Specifically, we demonstrated that USP7 promotes deubiquitination and stabilization of PHF8, leading to the upregulation of a group of genes, including cyclin A2, that are critical for cell growth and proliferation. The USP7-encoding gene was also transcriptionally regulated by PHF8, via positive feedback. USP7 was overexpressed in breast carcinomas, and the level of expression positively correlated with expression of PHF8 and cyclin A2 and with the histological grade of breast cancer. We showed that USP7 promotes breast carcinogenesis by stabilizing PHF8 and upregulating cyclin A2 and that the interaction between USP7 and PHF8 is augmented during DNA damage. Moreover, USP7-promoted PHF8 stabilization conferred cellular resistance to genotoxic insults and was required for the recruitment of BLM and KU70, which are both essential for DNA double-strand break repair. Our study mechanistically links USP7 to epigenetic regulation and DNA repair. Moreover, these data support the pursuit of USP7 and PHF8 as potential targets for breast cancer intervention, especially in combination with chemo- or radiotherapies.
Introduction
Posttranslational modification of histone proteins, which is accomplished by means of a variety of enzymatic reactions, plays an important role in chromatin structure and function in eukaryotic cells (1). A well-studied type of histone modification is methylation of lysine residues, displaying 3 possible states of lysine methylation, namely, mono-, di-, and trimethylation, which are catalyzed by histone methyltransferases. Histone methylation has been implicated in a number of biological processes, including gene expression, heterochromatin formation, and genome integrity (1, 2), and aberrant histone methylation is thus linked to a number of human diseases, including various types of malignancies (3, 4).
Analogous to other reversible histone marks involved in the regulation of chromatin plasticity such as acetylation and phosphorylation, the methyl groups can be removed by histone demethylases of either the amine oxidase LSD1 or the Jumonji C-terminal–containing (JmjC) family of proteins (5). Among these demethylases, plant homeodomain finger–containing protein 8 (PHF8, also termed KDM7B) is a ubiquitously expressed nuclear protein consisting of an N-terminal plant homeodomain, which recognizes and binds histone H3 lysine 4 tri-methyl (H3K4me3) bearing nucleosomes at transcription start sites (6), and a JmjC domain catalyzing the removal of the methyl moieties from H3K9me1/2, H4K20me1, or H3K27me2 (7–10). Specifically, PHF8 interacts with PML-RARα and functions as a transcriptional coactivator in response to all-trans retinoic acid treatment (11). Physiologically, PHF8 regulates neuronal differentiation (12) and zebrafish brain and craniofacial development (10). Pathologically, mutations in the human PHF8 gene are implicated in the pathogenesis of X-linked mental retardation and/or cleft lip/cleft palate (13), and upregulation of PHF8 has been documented in several types of malignancy, including prostate cancer (14), esophagus cancer (15), laryngeal/hypopharyngeal cancer (16), and lung cancer (17). Clearly, understanding how PHF8 is regulated under physiological conditions and dysregulated in pathological settings is of great importance to understand the biological activity of this protein.
Likewise, ubiquitination of proteins is constantly opposed by deubiquitinases, which proteolytically remove polyubiquitin chains from substrates (18). Of the deubiquitinases studied to date, ubiquitin-specific protease 7 (USP7), also known as herpes virus–associated ubiquitin-specific protease (HAUSP), as it was originally identified as a herpes simplex virus type 1 Vmw110-interacting protein (19), is reported to stabilize a number of proteins thus involved in multiple cellular processes, including immune responses (20), viral replication/infection (21), mitosis progression (22), and DNA repair (23, 24). It is also found that USP7 forms a protein complex with guanosine 5′-monophosphate synthetase to catalyze the removal of H2B lysine 120 (H2BK120) ubiquitination (25). In addition, reports also implicate USP7 in several pathological states, including neurodevelopmental and neurodegenerative disorders (26), inflammation (27), dilated cardiomyopathy (28), and various types of malignancies (29–31). However, the mechanistic insights into the role of USP7 in tumor development and progression remain to be investigated.
In this study, we report that the histone demethylase PHF8 is physically associated with deubiquitinase USP7. We showed that USP7-mediated deubiquitination and stabilization of PHF8 regulate the expression of key cell cycle regulators including cyclin A2 to promote breast cancer proliferation in vitro and breast carcinogenesis in vivo. We demonstrated that the functional link between USP7 and PHF8 is augmented during DNA damage response, which contributes to DNA repair and confers cellular resistance to genotoxic insults.
Results
The histone demethylase PHF8 is physically associated with the deubiquitinase USP7.
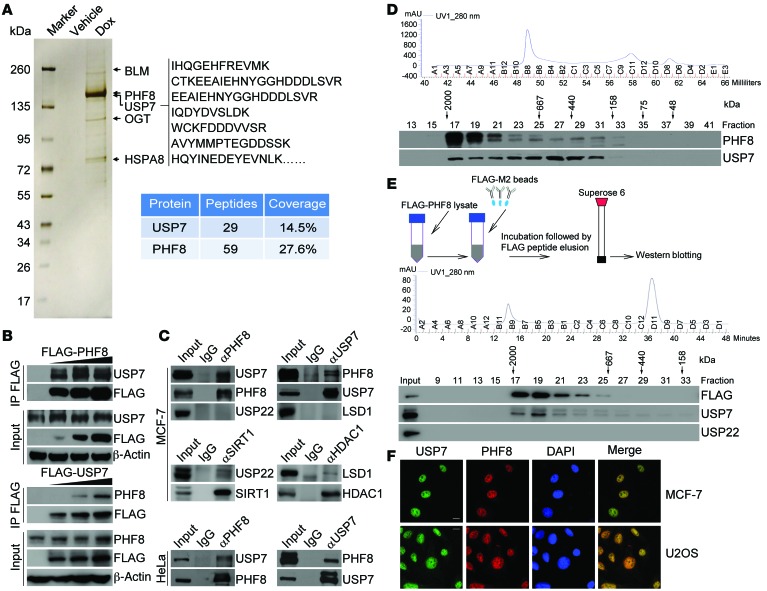
In order to further explore the biological function of PHF8 and to investigate its role in the development of cancer, we generated a mammary carcinoma MCF-7 cell line that allows doxycycline-inducible (Dox-inducible) expression of stably integrated FLAG-PHF8. Whole-cell extracts from these cells with or without Dox-inducible expression of FLAG-PHF8 were collected and subjected to affinity purification using an anti-FLAG affinity column. After extensive washing, the bound proteins were eluted with excess FLAG peptides, resolved, and then visualized by silver staining on SDS-PAGE. The protein bands on gels were recovered and analyzed by mass spectrometry. The results showed that PHF8 was associated with a number of proteins, including BLM, OGT, and HSPA8 (Figure 1A and Supplemental Table 1; supplemental material available online with this article; doi:10.1172/JCI85747DS1). Interestingly, USP7, a member of the protein deubiquitinases, was also identified in the PHF8-containing protein complex (Figure 1A).
Figure 1. The deubiquitinase USP7 is physically associated with the histone demethylase PHF8.
(A) Cellular extracts from MCF-7 cells with Dox-inducible expression of stably integrated FLAG-PHF8 were immunopurified with anti-FLAG affinity beads and eluted with FLAG peptides. The eluates were resolved on SDS-PAGE and silver-stained followed by mass spectrometry analysis. Representative peptide fragments of USP7 and peptide coverage of the indicated proteins are shown. Detailed results are provided as Supplemental Table 1. (B) Cell lysates from MCF-7 cells with Dox-inducible expression of stably integrated FLAG-PHF8 (top panel) or FLAG-USP7 (bottom panel) were immunoprecipitated (IP) and then immunoblotted with antibodies against the indicated proteins. (C) Whole cell lysates from MCF-7 cells (top panel) and HeLa cells (bottom panel) were immunoprecipitated and then immunoblotted with antibodies against the indicated proteins. (D) Cellular extracts from MCF-7 cells were fractionated on Superose 6 size exclusion columns. Chromatographic elution profiles (top panel) and Western blotting analysis (bottom panel) of the chromatographic fractions with antibodies against the indicated proteins are shown. The elution positions of calibration proteins with known molecular masses are indicated, and an equal volume from each fraction was analyzed. (E) Experiments analogous to D were performed with PHF8-containing protein complex purified from FLAG-PHF8–expressing MCF-7 cells. (F) MCF-7 cells and U2OS cells were fixed and immunostained with anti-USP7 and anti-PHF8. Scale bar: 10 μm.
To confirm the physical interaction between PHF8 and USP7, cellular extracts from MCF-7 cells expressing Dox-inducible FLAG-PHF8 were prepared, and coimmunoprecipitation experiments were performed. IP with antibodies against FLAG followed by IB with antibodies against USP7 demonstrated that USP7 was efficiently coimmunoprecipitated with FLAG-PHF8 in a Dox dose–dependent manner (Figure 1B, top panel). Analogously, coimmunoprecipitation experiments in MCF-7 cells stably integrated with Dox-inducible expression of FLAG-USP7 revealed that PHF8 was coimmunoprecipitated with FLAG-USP7 in a Dox dose–dependent fashion (Figure 1B, bottom panel). Additionally, IP in MCF-7 cells with antibodies against PHF8 followed by IB with antibodies against USP7 demonstrated that endogenous USP7, but not another protein deubiquitinase, USP22, was efficiently coimmunoprecipitated with endogenous PHF8, although USP22 could be effectively immunoprecipitated with antibodies against protein deacetylase SIRT1, as reported (ref. 32 and Figure 1C, top panel). Reciprocally, IP with antibodies against USP7 followed by IB with antibodies against PHF8 also revealed that endogenous PHF8, but not another histone demethylase, LSD1, was coimmunoprecipitated with endogenous USP7, although LSD1 could be efficiently coimmunoprecipitated with antibodies against HDAC1 (ref. 33 and Figure 1C, top panel). The interaction between endogenous PHF8 and USP7 was also detected by coimmunoprecipitation in HeLa cells (Figure 1C, bottom panel).
To gain further support of the in vivo interaction between PHF8 and USP7, protein fractionation experiments were carried out by fast protein liquid chromatography (FPLC) with Superose 6 columns and a size exclusion approach. The results indicate that native PHF8 from MCF-7 cells was eluted with an apparent molecular mass much greater than that of the monomeric protein (Figure 1D). Significantly, the elution pattern of PHF8 largely overlapped with that of USP7 (Figure 1D). Furthermore, analysis of FLAG-PHF8 affinity eluates by FPLC with Superose 6 gel filtration revealed that the majority of the purified FLAG-PHF8 existed in a multiprotein complex, which peaked in fractions 17 and 19 with USP7, but not USP22 (Figure 1E). Consistently, immunofluorescent staining with antibodies against endogenous PHF8 and USP7 showed that these 2 proteins were colocalized in the nucleus of MCF-7 and U2OS cells (Figure 1F).
To further validate the in vivo interaction between USP7 and PHF8 and to gain a molecular insight into the interaction between these 2 proteins, FLAG-tagged domain deletion mutants of PHF8 were generated and transfected into HeLa cells. IP with anti-FLAG followed by IB with anti-USP7 indicated that the C-terminal unstructured region of PHF8 was required for the association of PHF8 with USP7 (Supplemental Figure 1A). Reciprocally, domain mapping of the molecular interface of USP7 required for PHF8 binding revealed that the meprin- and TNF receptor–associated factor (TRAF) homology (MATH) domain in the N terminus of USP7 was required for the interaction of UPS7 with PHF8 (Supplemental Figure 1B). Furthermore, in vitro pull-down experiments with recombinant USP7 and full-length or truncation mutants of PHF8 that were purified from Sf9 cells demonstrated that USP7 was able to interact with full-length and C-terminal truncation mutants of PHF8 (Supplemental Figure 1C). Analogously, incubation of recombinant PHF8 with full-length or truncation mutants of USP7 purified from Sf9 cells showed that PHF8 directly interacts with full-length and MATH domain–containing truncation mutants of USP7 (Supplemental Figure 1D). Collectively, these results indicate that PHF8 interacts with USP7 through the C-terminal region of PHF8 and the MATH domain of USP7.
USP7 is functionally linked to the stabilization of PHF8.
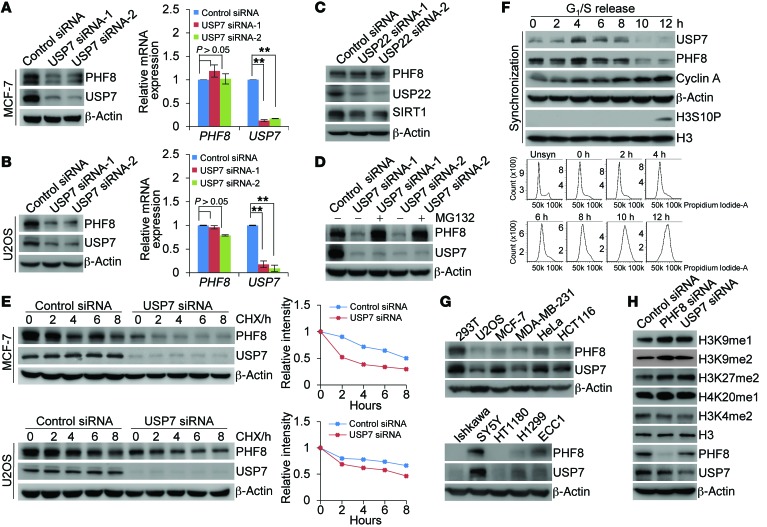
To address the functional significance of the physical interaction and spatial colocalization between USP7 and PHF8, we examined the effect of USP7 on the expression of PHF8. Western blotting analysis of cellular lysates from MCF-7 cells transfected with 2 independent sets of siRNAs targeting different regions of USP7 revealed that the level of PHF8 was significantly reduced upon USP7 depletion (Figure 2A, left panel). Similarly, knockdown of USP7 resulted in a dramatic decrease in protein level of PHF8 in U2OS cells (Figure 2B, left panel). However, knockdown of USP22, another nuclear deubiquitinase, had no evident effect on the expression level of PHF8 (Figure 2C), while USP22 knockdown affected the protein abundance of SIRT1 (32). In addition, the decreased PHF8 protein expression under USP7 depletion was not a result of reduced PHF8 mRNA, as quantitative reverse transcription PCR (qRT-PCR) measurements indicated that USP7 knockdown did not result in alterations in PHF8 mRNA level in MCF-7 cells and U2OS cells (Figure 2A, right panel, and Figure 2B, right panel). Moreover, the reduction in PHF8 protein level associated with USP7 depletion was probably through a proteasome-mediated protein degradation mechanism, as the effect could be effectively blocked by a proteasome-specific inhibitor, MG132 (Figure 2D). These observations suggest that the stability of PHF8 is regulated by USP7 and that PHF8 is a substrate of USP7.
Figure 2. USP7 promotes PHF8 stabilization.
(A) MCF-7 cells were transfected with control siRNA or different sets of USP7 siRNAs. Cellular extracts and total RNA were prepared and analyzed by Western blotting (left panel) and qRT-PCR analysis (right panel), respectively. (B) Experiments analogous to A were performed in U2OS cells. (C) MCF-7 cells were transfected with control siRNA or USP22 siRNA followed by Western blotting analysis. (D) MCF-7 cells were transfected with control siRNA or USP7 siRNA followed by treatment with DMSO or the proteasome inhibitor MG132 (10 μM). Cellular extracts were prepared and analyzed by Western blotting. (E) MCF-7 or U2OS cells transfected with control siRNA or USP7 siRNA were treated with cycloheximide (CHX; 50 μg/ml) and harvested at the indicated time points, followed by Western blotting analysis. Quantitation was done by densitometry with ImageJ software (NIH) with β-actin as a normalizer. (F) U2OS cells synchronized by double-thymidine block were released, and cellular extracts were collected at indicated time points for Western blotting analysis (top panel). Cell cycle profiles of unsynchronized (Unsyn) or synchronized cells were analyzed by FACS (bottom panel). (G) Western blotting analysis of the expression of USP7 and PHF8 in multiple cell lines. (H) Cellular extracts from MCF-7 cells transfected with control siRNA, PHF8 siRNA, or USP7 siRNA were analyzed by Western blotting. Representative images from biological triplicate experiments are shown. In A and B, each bar represents the mean ± SD for biological triplicate experiments. **P < 0.01, 1-way ANOVA.
To further support this deduction, the potential of USP7 to modulate the steady-state level of PHF8 protein was assessed by cycloheximide (CHX) chase assays. In these experiments, MCF-7 or U2OS cells transfected with control siRNA or USP7 siRNA were incubated with CHX and harvested at different time points. Western blotting analysis revealed that USP7 depletion was clearly associated with a decreased half-life of PHF8 (Figure 2E).
Then, U2OS cells were synchronized by double-thymidine block. After releasing into the cell cycle, the protein levels of USP7 and PHF8 in these cells were measured by Western blotting. Notably, the protein levels of USP7 and PHF8 oscillated in a parallel pace during cell cycle progression: both were increased in S and early G2 phases and decreased in G1 and G2–M phases (Figure 2F). In addition, examination of the expression levels of USP7 and PHF8 in multiple cell lines by Western blotting showed a correlated pattern of the expression of these 2 proteins (Figure 2G). Significantly, consistent with the reported enzymatic activity of PHF8 toward histone modifications (8, 9) and in support of our observation that PHF8 is stabilized by USP7, we found that USP7 depletion was associated with increased levels of H3K9me1, H3K9me2, H3K27me2, and H4K20me1, but not H3K4me2 (Figure 2H and Supplemental Figure 2, A and B). This effect faithfully mimicked that of PHF8 depletion (Figure 2H and Supplemental Figure 2, A and B). Together, these results strongly support the notion that USP7 controls the stability of PHF8.
USP7 deubiquitinates PHF8.
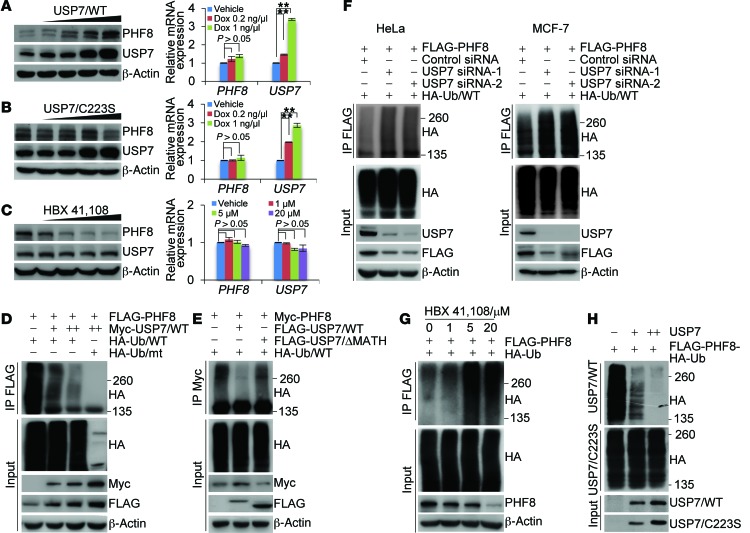
We next investigated whether USP7-promoted PHF8 stabilization is a consequence of USP7-catalyzed PHF8 deubiquitination. For this purpose, we first determined whether USP7-promoted PHF8 stabilization is dependent on the enzymatic activity of USP7. To this end, we generated 2 stable MCF-7 cell lines with Dox-inducible expression of wild-type USP7 (USP7/WT) and a catalytically inactive mutant of USP7 (USP7/C223S) (20), respectively. Western blotting analysis showed that, in cells expressing USP7/WT, the protein level of PHF8 dramatically increased in a Dox dose–dependent manner (Figure 3A, left panel), whereas in cells expressing USP7/C223S, no evident change of PHF8 protein level was detected (Figure 3B, left panel). In agreement with the notion that USP7 regulates the abundance of PHF8 at the protein level, neither USP7/WT (Figure 3A, right panel) nor USP7/C223S (Figure 3B, right panel) affects the level of PHF8 mRNA. Next, we used CRISPR/Cas9 to knock out USP7 in HeLa cells. In USP7-deficient cells, PHF8 is downregulated, and the downregulation of PHF8 in these cells could be reverted by forced expression of USP7/WT, but not USP7/C223S (Supplemental Figure 3A), while the mRNA expression level of PHF8 was essentially unchanged (Supplemental Figure 3B). Moreover, treatment of MCF-7 cells with HBX 41,108, a cyano-indenopyrazine–derived deubiquitinase inhibitor known to inhibit catalytic activity of USP7 (34), resulted in a dose-dependent reduction in the level of PHF8 protein, while it had no effect on the mRNA level of PHF8 (Figure 3C). Together, these results support the argument that USP7 regulates the stability of PHF8 through USP7 deubiquitinase activity.
Figure 3. USP7 deubiquitinates PHF8.
(A) MCF-7 cells with Dox-inducible expression of FLAG-USP7/WT were cultured in the absence or presence of increasing amounts of Dox. Cellular extracts and total RNA were collected for Western blotting (left panel) and qRT-PCR (right panel) analysis, respectively. (B) Experiments analogous to A were performed in U2OS cells with Dox-inducible expression of FLAG-USP7/C223S. (C) MCF-7 cells were cultured in the absence or presence of increasing amounts of HBX 41,108 for 2 hours as indicated. Cellular extracts and total RNA were collected for Western blotting (left panel) and qRT-PCR (right panel) analysis, respectively. (D) HeLa cells stably expressing FLAG-PHF8 were cotransfected with different amounts of Myc-USP7 and HA-Ub/WT or HA-Ub/mt as indicated. Cellular extracts were immunoprecipitated with anti-FLAG followed by IB with anti-HA. (E) HeLa cells stably expressing Myc-PHF8 were cotransfected with HA-Ub/WT and FLAG-USP7/WT or USP7/ΔMATH. Cellular extracts were immunoprecipitated with anti-Myc followed by IB with anti-HA. (F) HeLa cells (left panel) or MCF-7 cells (right panel) stably expressing FLAG-PHF8 were cotransfected with HA-Ub/WT and control siRNA or USP7 siRNAs as indicated. Cellular extracts were immunoprecipitated with anti-FLAG followed by IB with anti-HA. (G) MCF-7 cells stably expressing FLAG-PHF8 were transfected with HA-Ub/WT and cultured in the presence or absence of HBX 41,108. Cellular extracts were immunoprecipitated with anti-FLAG followed by IB with anti-HA. (H) In vitro deubiquitination assays with HA-Ub–conjugated PHF8 purified from HeLa cells using high-salt buffer and USP7/WT or USP7/C223S purified from extracts of baculovirus-infected insect cells. In A–C, each bar represents the mean ± SD for biological triplicate experiments. **P < 0.01, 1-way ANOVA.
Next, HeLa cells stably expressing FLAG-PHF8 were cotransfected with Myc-USP7 and HA-tagged wild-type ubiquitin (Ub/WT) or a ubiquitin mutant (Ub/mt) with all lysine residues replaced by arginine (35). IP of cellular lysates with anti-FLAG followed by IB with anti-HA showed that increased Myc-USP7 expression was associated with decreased levels of ubiquitinated PHF8 species (Figure 3D). Corroborating the finding that MATH domain is required for the physical association of USP7 with PHF8, expression of USP7 mutant lacking MATH domain did not affect the levels of ubiquitinated PHF8 species (Figure 3E). Meanwhile, HeLa cells stably expressing FLAG-PHF8 were cotransfected with USP7 siRNA and Ub/WT. IP of the cellular lysates with anti-FLAG followed by IB with anti-HA showed that knockdown of USP7 resulted in an increase in the level of ubiquitinated PHF8 species (Figure 3F, left panel). Similar results were also obtained in MCF-7 cells stably expressing FLAG-PHF8 (Figure 3F, right panel) and USP7 knockout HeLa cells (Supplemental Figure 3C). Consistent with the observation that enzymatic inhibition of USP7 resulted in PHF8 destabilization, treatment of MCF-7 cells with HBX 41,108 resulted in a marked increase in the level of ubiquitinated PHF8 species (Figure 3G and Supplemental Figure 3D). Furthermore, in vitro deubiquitination assays with HA-Ub–conjugated FLAG-PHF8 purified from HeLa cells and FLAG-USP7/WT or FLAG-USP7/C223S purified from Sf9 cells revealed that USP7/WT was capable of deubiquitinating PHF8, whereas USP7/C223S was not (Figure 3H). Together, these results indicate that USP7 targets PHF8 for deubiquitination, supporting a notion that the histone demethylase PHF8 is a bona fide substrate of the deubiquitinase USP7.
The biological function of USP7-promoted PHF8 stabilization.
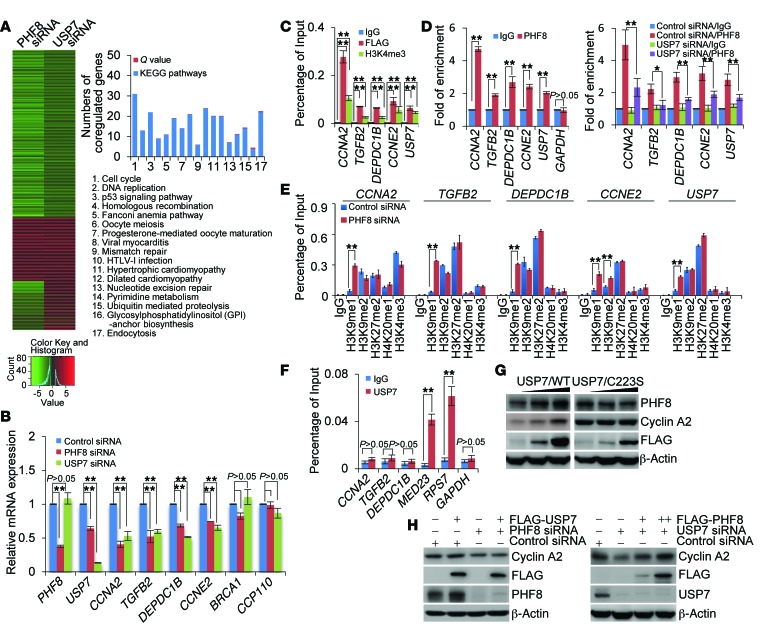
In order to understand the biological significance of USP7-promoted deubiquitination and stabilization of PHF8, we investigated the transcriptomes in PHF8- or USP7-deficient MCF-7 cells by high-throughput RNA deep sequencing (RNA-seq). In brief, total RNA was extracted from MCF-7 cells transfected with control siRNA or siRNA targeting USP7 or PHF8. RNA-seq analysis by Illumina HiSeq 2000 with a stringent cutoff (P ≤ 10–5) identified 5,680 genes whose expressions were altered upon PHF8 depletion and 1,477 genes whose expressions were changed upon USP7 knockdown. Cross-analysis of the transcriptomes from PHF8- and USP7-deficient cells identified 727 genes whose expressions were altered in both PHF8- and USP7-depleted cells, and these genes were considered as the targets that were coregulated by PHF8 and USP7 (Figure 4A). Notably, about two-thirds of these genes were clustered as targets that are transcriptionally activated by USP7 and PHF8 (Figure 4A, left panel), supporting a notion that USP7-promoted PHF8 stabilization is linked to gene activation. The genes that were coregulated by USP7 and PHF8 were then classified into various signaling pathways with a Q value (minimum FDR) cutoff of 0.5 and a P value cutoff of 0.05 by KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway analysis (Figure 4A, right panel). These signaling pathways include cell cycle, DNA replication, and p53 signaling pathways that are critically involved in cell growth and survival, consistent with the current understanding of the cellular functions of USP7 and PHF8 (9, 23).
Figure 4. USP7 and PHF8 coregulate the expression of cyclin A2.
(A) MCF-7 cells were transfected with control siRNA, PHF8 siRNA, or USP7 siRNA followed by RNA extraction and deep sequencing. Coregulated genes were clustered as indicated, and color key and histogram indicating the upregulation (red) or downregulation (green) of the targeted genes are shown (left panel). Coregulated genes were grouped and statistically analyzed according to KEGG pathways (right panel). (B) MCF-7 cells were transfected with the indicated siRNAs, followed by RNA extraction and qRT-PCR analysis of the expression of the indicated genes. (C) Soluble chromatin was immunoprecipitated from FLAG-PHF8–expressing MCF-7 cells with control IgG or antibodies against FLAG, H3K4me3, or PHF8, followed by qPCR analysis with primers for promoters of the indicated genes. (D) qChIP experiments analogous to C were performed with soluble chromatins from native MCF-7 cells (left panel) and USP7-depleted MCF-7 cells (right panel). GAPDH was used as a negative control. (E) Soluble chromatin from MCF-7 cells transfected with control siRNA or PHF8 siRNA was prepared for qChIP analysis using antibodies against the indicated histone modifications. (F) Soluble chromatin from MCF-7 cells was prepared for qChIP analysis with control IgG or anti-USP7. (G) MCF-7 cells with Dox-inducible expression of stably integrated USP7/WT (left panel) or USP7/C223S (right panel) were cultured in the absence or presence of increasing amounts of Dox. Cellular extracts were collected for Western blotting analysis. (H) MCF-7 cells with Dox-inducible expression of stably integrated FLAG-USP7 (left panel) or FLAG-PHF8 (right panel) were transfected with the indicated siRNAs and cultured in the absence or presence of Dox. Cellular extracts were prepared for Western blotting analysis. In B–F, each bar represents the mean ± SD for biological triplicate experiments. *P < 0.05 and **P < 0.01, 1-way ANOVA.
Since the cell cycle represents the most prominent pathway with the lowest Q and P values and the highest numbers of coregulated genes, we next chose 6 representative genes implicated in cell cycle regulation, CCNA2, TGFB2, DEPDC1B, CCNE2, BRCA1, and CP110, and validated their expressions in MCF-7 cells by qRT-PCR. The results indicated that the mRNA levels of CCNA2, TGFB2, DEPDC1B, and CCNE2 but not those of BRCA1 and CP110 decreased upon knockdown of either USP7 or PHF8, albeit to variable extents (Figure 4B). Interestingly, the mRNA level of USP7 itself was also decreased upon PHF8 depletion. Moreover, to determine whether these genes are directly targeted by PHF8, we examined the occupancy of PHF8 on promoters of these genes by quantitative chromatin immunoprecipitation (qChIP) assays in MCF-7 cells that were stably expressing FLAG-PHF8. Soluble chromatins were immunoprecipitated with antibodies against FLAG or H3K4me3 followed by quantitative PCR analysis of precipitated DNA. The results showed that, similarly to H3K4me3, recruitment of FLAG-PHF8 was detected in the promoter regions of CCNA2, TGFB2, DEPDC1B, CCNE2, and USP7 (Figure 4C). Furthermore, ChIP assays in MCF-7 cells also detected the occupancy of endogenous PHF8 on the promoters of these genes (Figure 4D, left panel), which was significantly weakened when USP7 was knocked down (Figure 4D, right panel). Consistently, we found that, compared with control cells, PHF8-deficient cells displayed a substantial retention of H3K9me1 in the promoters of CCNA2, TGFB2, DEPDC1B, CCNE2, and USP7, and H3K9me2 in the promoter of CCNE2 (Figure 4E). Together, these results indicate that CCNA2, TGFB2, DEPDC1B, CCNE2, and USP7 are indeed targeted by PHF8. These experiments also indicate that USP7 itself is transcriptionally regulated, via positive feedback, by PHF8.
Previous studies suggested that USP7 is a chromatin modifier that acts to remove histone H2B lysine 120 mono-ubiquitination (H2BK120ub1) (36). Therefore, it is possible that USP7, through deubiquitinating H2BK120ub1, coordinates with PHF8 to influence gene activation. To test this, qChIP experiments were performed and the recruitment of USP7 on the above-described PHF8 target genes was examined in MCF-7 cells. We did not detect the occupancy of USP7 on the promoters of CCNA2, DEPDC1B, TGFB2, and CCNE2 genes, although we did detect the presence of USP7 on the promoters of MED23 and RPS7 genes (Figure 4F), as reported by others (37). These results favor the argument that USP7 regulates the expression of PHF8 target genes through stabilizing PHF8.
To further support this proposition, MCF-7 cells stably integrated with Dox-inducible expression of USP7/WT and USP7/C223S were analyzed by Western blotting. The results showed that, in cells expressing USP7/WT but not USP7/C223S, the protein level of cyclin A2 encoded by CCNA2, a cell cycle regulator with the most evident expression changes upon USP7 or PHF8 depletion, was dramatically induced in a Dox dose–dependent manner (Figure 4G). However, when PHF8 was knocked down in USP7/WT–expressing cells, the expression of cyclin A2 was induced (Figure 4H, left panel). Meanwhile, in MCF-7 cells with Dox-inducible expression of stably integrated FLAG-PHF8, depletion of USP7 was associated with a decreased expression of cyclin A2 in the absence of Dox, the effect of which could be effectively restored by Dox-induced expression of PHF8 (Figure 4H, right panel). Together, these results support an idea that USP7 acts upstream through stabilizing PHF8 to regulate the expression of PHF8 target genes.
The USP7/PHF8/cyclin A2 axis promotes breast cancer cell proliferation in vitro and breast carcinogenesis in vivo.
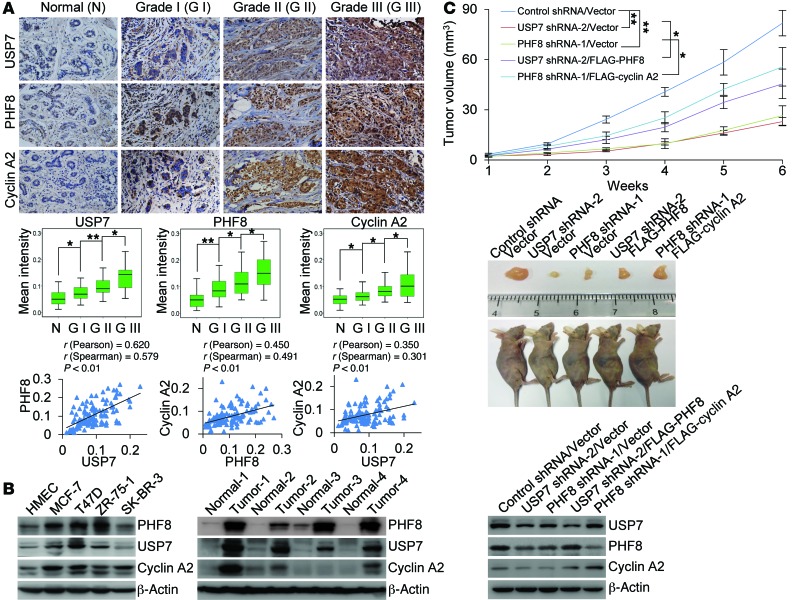
Deregulation of cyclin A2 was associated with erroneous cell proliferation and chromosomal instability (38, 39), and aberrant expression of cyclin A2 has been linked to multiple types of malignancies, including breast, liver, and lung cancers (38–40). In light of our observation that USP7-mediated deubiquitination and stabilization of PHF8 regulate the expression of cyclin A2, it is reasonable to postulate that the USP7/PHF8 signaling pathway plays a role in cell proliferation and carcinogenesis. To test this hypothesis and to extend our observations to a clinicopathologically relevant context, we first analyzed the protein expression levels of USP7, PHF8, and cyclin A2 with human tissue arrays including series of tumor samples from breast, colon, esophagus, kidney, liver, lung, rectum, and stomach, with each type of cancer having 3 malignant samples paired with adjacent normal tissues. Immunohistochemical staining showed an upregulation of all USP7, PHF8, and cyclin A2 in carcinomas derived from breast, colon, and rectum (Supplemental Figure 4). To exclude the possibility that the observed immunohistochemical signal is derived from antibody-nonspecific binding, tumor samples from the same patient were stained with isotype IgG or corresponding antibodies against these factors (Supplemental Figure 5A).
We then analyzed, by immunohistochemical staining, the expression profiles of USP7, PHF8, and cyclin A2 with human tissue arrays containing 113 samples including breast carcinoma samples and histologically normal mammary tissues. We found that, when staining was scored according to the mean intensity and nuclear staining extent of immunopositivity with Image-Pro Plus software (Media Cybernetics Inc.), USP7 and PHF8 together with cyclin A2 were highly expressed in breast carcinoma samples, and the levels of their expression were strongly correlated with each other and with the histological grades of breast cancer (Figure 5A and Supplemental Figure 5B). Consistently, measurement of the expression of USP7, PHF8, and cyclin A2 by Western blotting showed that the levels of all these proteins were substantially elevated in breast cancer cell lines (Figure 5B, left panel) as well as in breast carcinoma samples (Figure 5B, right panel). Moreover, we performed bioinformatics analysis using TCGA breast data sets from Oncomine (Thermo Fisher Scientific) (41) and found that the mRNA expression level of USP7, PHF8, and CCNA2 is upregulated in breast cancer samples (Supplemental Figure 5C), and analysis of GEO data sets GSE32646 and GSE65194 showed that the mRNA expression levels of USP7, PHF8, and CCNA2 are positively correlated with each other (Supplemental Figure 5D). These results are in agreement with the profiles of the protein expression that we observed in immunohistochemical staining and support the argument that USP7 itself is transcriptionally regulated, via positive feedback, by PHF8. Meanwhile, these results also provide a logical explanation for how the expression of USP7, PHF8, and cyclin A2 is dysregulated in breast cancer. However, the mechanism governing the elevated expression of PHF8 at the mRNA level and the contribution of USP7-promoted PHF8 stabilization at the post-transcriptional level to increased expression of PHF8 in breast carcinoma remain to be investigated. Collectively, these observations suggest that the USP7/PHF8/cyclin A2 axis plays a potential role in breast carcinogenesis.
Figure 5. USP7/PHF8/cyclin A2 signaling pathway promotes breast carcinogenesis.
(A) IHC analysis of the expression levels of USP7, PHF8, and cyclin A2 in breast tumors. Representative images (original magnification, ×200) from normal mammary tissue and breast carcinoma samples at different stages are shown (top panel). The values of the stainings were determined by Image-Pro Plus software and are presented with box plots (middle panel). *P < 0.05, **P < 0.01, 1-way ANOVA. The correlation coefficient and P values were analyzed as indicated (bottom panel). (B) Western blotting analysis of the protein levels of USP7, PHF8, and cyclin A2 in human mammary epithelial cell (HMEC) and breast cancer cell lines (left panel), or in paired samples from infiltrating ductal breast carcinomas and adjacent breast tissues (right panel). (C) MCF-7 tumors stably expressing shRNAs and genes as indicated were transplanted onto athymic mice (n = 6), and tumor volumes were measured weekly (top panel). Representative tumors and sacrificed mice are shown (middle panel). The expression levels of the indicated proteins in these tumors were examined by Western blotting (bottom panel). Each bar represents the mean ± SD. *P < 0.05 and **P < 0.01, 2-way ANOVA.
In order to investigate the role of the USP7/PHF8/cyclin A2 axis in breast carcinogenesis, we then generated MCF-7 cell lines with USP7 or PHF8 stably depleted by their specific shRNAs. We demonstrated that USP7-deficient cells (Supplemental Figure 6A) or PHF8-deficient cells (Supplemental Figure 6B) displayed a decreased expression of cyclin A2 and exhibited a much slower growth rate compared with control cells. In agreement with this observation, depletion of either USP7 or PHF8 resulted in a significant inhibition of the proliferation and thus the colony formation of MCF-7 cells (Supplemental Figure 6C). Furthermore, overexpression of PHF8 could rescue the phenotype induced by USP7 knockdown to a certain extent, the effect of which was impaired by cyclin A2 depletion (Supplemental Figure 6D). Meanwhile, overexpression of cyclin A2 could, at least partially, override the growth-inhibitory effect of PHF8 knockdown (Supplemental Figure 6E).
To further establish the role of USP7/PHF8/cyclin A2 in cell proliferation and breast carcinogenesis, we transplanted 5 types of breast tumor, developed from MCF-7 cells infected with control lentivirus, lentivirus carrying USP7 shRNA, lentivirus carrying PHF8 shRNA, lentivirus carrying USP7 shRNA and reinfected with lentivirus carrying PHF8, and lentivirus carrying PHF8 shRNA and reinfected with lentivirus carrying cyclin A2, respectively, onto the mammary fat pads of athymic mice (BALB/c; Charles River Laboratories). Tumor growth and mouse weight were monitored over 6 weeks. Notably, in athymic mice that received tumor transplants with either USP7- or PHF8-depleted cells, the tumor growth was greatly suppressed (Figure 5C). Meanwhile, overexpression of PHF8 in USP7-deficient tumors or gain of function of cyclin A2 in PHF8-deficient tumors could restore the growth of breast tumors (Figure 5C). Collectively, these results strongly support a role of the USP7/PHF8/cyclin A2 axis in promoting breast carcinogenesis.
In agreement with previous reports (42, 43), we did find that USP7 knockdown resulted in decreased expression of MDM2 (Supplemental Figure 7A), an essential regulator of the p53 pathway (42). However, PHF8 depletion had minimal effect on the expression level of MDM2 (Supplemental Figure 7A). In addition, USP7, but not PHF8, could be coimmunoprecipitated by MDM2 (Supplemental Figure 7B). Furthermore, colony formation assay demonstrated that although overexpression of MDM2 could rescue the growth inhibition phenotype resulting from USP7 depletion to a certain extent as PHF8 did, simultaneous expression of MDM2 and PHF8 showed an additive effect (Supplemental Figure 7C). The knockdown or overexpression effect of these factors was examined by Western blotting (Supplemental Figure 7D). These results suggest that PHF8 functions in breast cancer cell growth in an MDM2-independent manner.
USP7-promoted PHF8 stabilization is implemented in DNA damage response.
Cellular signal transduction is a complex process triggered by internal or external signals and involving protein-protein interactions that transmit information in a dynamic manner (2). We are interested in whether USP7-mediated PHF8 stabilization is stimulated or influenced by any internal or external stimuli. In this regard, it is interesting to note that USP7 was identified as a critical regulator of DNA damage response (DDR) in preserving genome integrity and rendering DNA damage tolerance (23, 43, 44). Therefore, we tested whether genotoxic insults could have any influence on USP7-promoted stabilization of PHF8 and whether the USP7/PHF8 signaling pathway itself is involved in DDR. To this end, MCF-7 cells were first exposed to a 6-Gy dose of x-ray irradiation (IR), and the cellular extracts were collected at different time points. Western blotting analysis showed that, remarkably, the protein level of PHF8 was increased upon IR treatment (Figure 6A, left panel). Similarly, the protein level of PHF8 was elevated when MCF-7 cells were treated with the radiomimetic DNA damage agent neocarzinostatin (NCS) (Figure 6A, right panel). However, qRT-PCR analysis showed that the levels of PHF8 mRNA were comparable in control MCF-7 cells versus cells exposed to IR or NCS (Figure 6B). Notably, IR-induced PHF8 accumulation was abrogated in USP7-deficient MCF-7 cells (Figure 6C). These results indicate that the abundance of PHF8 is regulated at the posttranscriptional level upon exposure of cells to DNA damage, a process involving USP7.
Figure 6. USP7-promoted PHF8 stabilization is involved in DNA damage response.
(A) MCF-7 cells exposed to IR (6 Gy, left panel) or NCS (0.5 μg/ml for 1 hour, right panel) were collected at the indicated time points and analyzed by Western blotting with γH2AX as a positive control. (B) Total RNA was collected from MCF-7 cells exposed to IR or NCS followed by qRT-PCR analysis of PHF8 expression. (C) MCF-7 cells transfected with control siRNA or USP7 siRNA were exposed to IR and collected at the indicated time points. The expression of the indicated proteins was examined by Western blotting. (D) MCF-7 cells (top panel) or HeLa cells (bottom panel) were exposed to IR. Two hours after IR, cellular extracts were immunoprecipitated and then immunoblotted with antibodies against the indicated proteins. (E) HeLa cells stably expressing FLAG-PHF8 were cotransfected with HA-Ub/WT and the indicated siRNAs (left panel) or expressing vectors (right panel) followed by IR treatment. Cellular extracts were immunoprecipitated with anti-FLAG and then immunoblotted with anti-HA. (F) MCF-7, T47D, or ZR-75-1 cells expressing the indicated shRNAs were cocultured with their corresponding GFP-expressing cells, exposed to IR or NCS, and analyzed by FACS after 10 days. The percentage of GFP-negative cells relative to the GFP-positive cells under IR or NCS treatment was normalized to that in untreated control mixture, and the ratio is used to reflect cellular fitness. (G) MCF-7 cells expressing the indicated shRNAs and Dox-inducible PHF8 (left panel) or USP7 (right panel) were cocultured with GFP-expressing MCF-7 cells, exposed to IR or NCS, and analyzed by FACS after 10 days. In B, F, and G, each bar represents the mean ± SD for biological triplicate experiments. *P < 0.05, **P < 0.01, 1-way ANOVA.
To support this, MCF-7 cells were exposed to IR before cellular lysates were prepared and coimmunoprecipitation experiments were performed. We found that the interaction between PHF8 and USP7 was greatly enhanced in MCF-7 cells upon IR exposure (Figure 6D, top panel). Similar results were also obtained in IR-treated HeLa cells (Figure 6D, bottom panel), suggesting that the physical association between PHF8 and USP7 is augmented by DNA damage.
To investigate whether USP7 acts to remove PHF8 polyubiquitination chains upon DNA damage, HeLa cells stably expressing FLAG-PHF8 were cotransfected with HA-Ub/WT and USP7 siRNA or Myc-USP7 followed by IR exposure. IP of cellular lysates with anti-FLAG followed by IB with anti-HA showed that IR exposure resulted in a decreased level of polyubiquitinated PHF8 species, an effect that could be abolished by USP7 knockdown (Figure 6E, left panel) and enhanced by USP7 overexpression (Figure 6E, right panel). These experiments indicate that the physical interaction and functional connection between USP7 and PHF8 are influenced by DNA damage.
In order to investigate the functional significance of DDR-enhanced stabilization of PHF8 by USP7, multicolor competition assays were performed. First, USP7-deficient MCF-7 cells were cocultured with an equal number of MCF-7 cells stably expressing GFP protein. The cell mixtures were then left untreated or exposed to IR, and analyzed by FACS after 10 days. The sensitivity to IR treatment was determined by relative fitness. Specifically, the number of GFP-negative USP7-depleted cells relative to the GFP-positive control cells under IR treatment was normalized to that in untreated control mixture. In agreement with previous reports (23), depletion of USP7 by 2 independent shRNAs rendered MCF-7 cells more sensitive to IR, although it was less severe than ATM deficiency (Figure 6F). In addition, USP7 depletion also sensitized MCF-7 cells to NCS (Figure 6F). Similar results were obtained in T47D and ZR-75-1 cells (Figure 6F). Next, we demonstrated that fitness of cells with PHF8 deficiency was also significantly compromised upon genotoxic insults (Figure 6F). Furthermore, while overexpression of PHF8 was able to desensitize USP7-deficient cells to IR or NCS exposure (Figure 6G, left panel), overexpression of USP7 failed to rescue PHF8 depletion–associated cell death upon IR or NCS exposure (Figure 6G, right panel). These results suggest that USP7-promoted PHF8 stabilization is critically implemented in DDR.
USP7-promoted PHF8 stabilization is required for double-strand break repair.
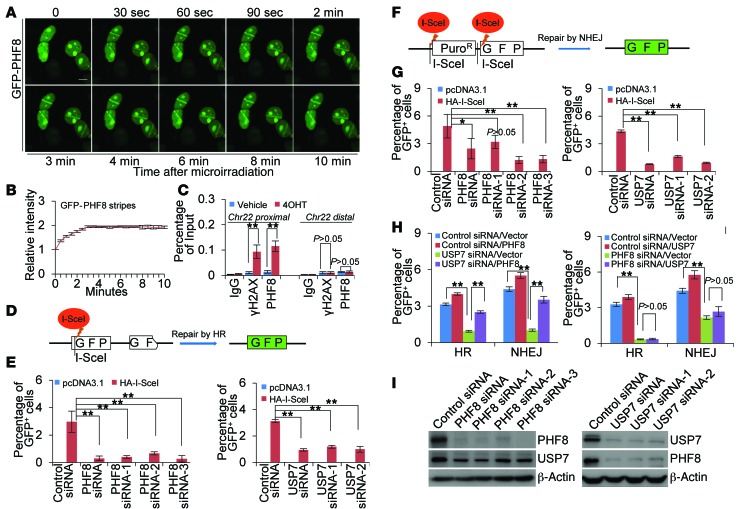
To further understand the role of USP7-promoted PHF8 stabilization in DDR, we examined whether PHF8 is directly involved in break repair. To this end, MCF-7 cells stably expressing GFP-PHF8 were subjected to laser microirradiation. Analysis of these cells by confocal microscopy indicates that PHF8 was recruited to damaged sites rapidly after laser exposure, manifested by fast accumulation of GFP signal in these sites (Figure 7, A and B). Since double-strand breaks (DSBs) are the major forms of DNA lesions induced by laser microirradiation, IR, and NCS, we next investigated the involvement of PHF8 in DSBs with an endonuclease AsiSI-based system (AsiSI is encoded by the sfaAIR gene from Sphingomonas faenia RFLI) in which endogenous sequence-specific DSBs could be generated (45). Similarly to γH2AX, we found that PHF8 was recruited to the break proximal site, but not the distal region about 2 Mb away from the break site, upon AsiSI activation by 4-hydroxyltamoxifen treatment (Figure 7C). These results suggest that PHF8 is mobilized and recruited to DSB sites upon DNA damage.
Figure 7. USP7-promoted PHF8 stabilization is required for DSB repair.
(A) MCF-7 cells stably expressing GFP-PHF8 were subjected to UV laser microirradiation and confocal microscopic analysis at the indicated time points. Scale bar: 10 μm. (B) Fluorescence intensities in microirradiated areas relative to the background signal of the undamaged regions were determined by ImageJ software. More than 20 nuclei were scored in biological triplicate experiments. Each bar represents the mean ± SD. (C) U2OS cells were transfected with HA-ER-AsiSI in the absence or presence of 0.5 μM 4-hydroxytamoxifen (4OHT). qChIP experiments were performed using anti-PHF8 or anti-γH2AX with primers that cover the DNA sequences flanking the AsiSI site and the break distal regions. (D) Assays for HR efficiency of chromosomal DSBs in DR-GFP U2OS cells. Two incomplete copies of GFP genes are integrated into chromosomal DNA. Cleavage of the I-SceI sites leads to the restoration of the GFP gene through HR. (E) HR efficiency was determined by FACS in PHF8- or USP7-deficient DR-GFP U2OS cells. (F) Assays for NHEJ of chromosomal DSBs in EJ5-U2OS cells. Excision of the 2 I-SceI sites followed by NHEJ eliminates the translation start codon of the otherwise nonsense transcript and enables the reading frame shift and subsequently expression of the GFP gene. (G) NHEJ efficiency was determined by FACS in PHF8- or USP7-deficient EJ5-U2OS cells. (H) DR-U2OS or EJ5-U2OS cells were cotransfected with siRNAs and expressing vectors as indicated. Repair efficiency was determined by FACS. (I) The knockdown effect of PHF8 and USP7 was examined by Western blotting. In C, E, G, and H, each bar represents the mean ± SD for biological triplicate experiments. *P < 0.05 and **P < 0.01, 1-way ANOVA.
To address the functional significance of the recruitment of PHF8 in DSB sites, we examined the effect of PHF8 on the repair efficiency of 2 major DSB repair pathways, homologous recombination (HR) and nonhomologous end joining (NHEJ) (46). The effect of PHF8 on HR repair of a DSB generated in chromosomal DNA was evaluated in DR-GFP U2OS cells (Figure 7D and ref. 47). As shown in the left panel of Figure 7E, depletion of PHF8 resulted in a significant reduction of HR efficiency, manifested by the decreased percentage of GFP-positive cells. This observation is consistent with the recent study reporting that the PHF8 homolog in Caenorhabditis elegans promotes DNA repair via HR (48). Meanwhile, examination of NHEJ repair efficiency by counting of the percentage of cells expressing GFP protein in U2OS cells stably integrating with an EJ5-I-SceI cassette (Figure 7F and ref. 49) revealed that depletion of PHF8 was associated with a reduced percentage of GFP-positive cells (Figure 7G, left panel). Together, these experiments indicate that PHF8 is required for efficient NHEJ and HR repair of DSBs in mammalian cells.
Next, we asked whether demethylase activity of PHF8 is required for its involvement in DNA repair. To this end, endonuclease I-SceI was cotransfected with siRNA targeting 3′-UTR of PHF8 mRNA into DR-GFP U2OS cells stably expressing wild-type PHF8 (PHF8/WT) or demethylase activity–defective PHF8 (PHF8/H247A) (8, 9). FACS analysis revealed that PHF8/WT was able to restore the decreased HR repair efficiency induced by endogenous PHF8 depletion, whereas PHF8/H247A was not (Supplemental Figure 8A, left panel). The same was true when NHEJ repair efficiency was examined in EJ5-U2OS cells with similar strategies (Supplemental Figure 8A, right panel). Meanwhile, qChIP experiments revealed that both PHF8/WT and PHF8/H247A were effectively recruited to the I-SceI–generated break region with comparable binding capacity (Supplemental Figure 8B), excluding the possibility that the loss of catalytic activity might impair the nuclear redistribution of PHF8. Although substantial retention of H3K9me1/2, H4K20me1, and H3K27me2 at the site of I-SceI–induced DSB was detected, PHF8 depletion had no evident effects on the levels of these marks (Supplemental Figure 8C). These results indicated that the demethylase activity of PHF8 is required for its activity in DSB repair, but it appears that PHF8 functions in a context-dependent manner to catalyze methylation removal of nonhistone DDR components or histone methylation marks other than H3K9me1/2, H4K20me1, and H3K27me2.
To consolidate the role of USP7-promoted PHF8 stabilization in DDR, we investigated the effect of USP7 knockdown on DSB repair by HR and NHEJ using the systems described above. We found that knockdown of USP7 greatly compromised DSB repair efficiency by both HR (Figure 7E, right panel) and NHEJ (Figure 7G, right panel), the effects of which could be largely restored by forced expression of PHF8 (Figure 7H, left panel). However, overexpression of USP7 failed to rescue the impaired DSB repair induced by PHF8 depletion (Figure 7H, right panel). The knockdown effect of PHF8 and USP7 was validated by Western blotting (Figure 7I). Taken together, these results support a notion that USP7 regulates DDR by stabilizing the histone demethylase PHF8 and that the USP7/PHF8 signaling pathway plays a critical role in the maintenance of genome integrity.
USP7-promoted PHF8 stabilization is required for the recruitment of BLM and KU70.
To gain more mechanistic insights into the role of USP7-promoted PHF8 stabilization in DDR and address the functional impact of the recruitment of PHF8 in DSB sites, we then asked whether PHF8 could influence the recruitment of key DDR proteins in break compartments. First, we analyzed the accumulation of γH2AX and MRE11, both of which act as important players in the initial processing of DSBs, to IR-induced DNA break sites, and found that PHF8 knockdown affected neither the accumulation of γH2AX nor that of MRE11 (Supplemental Figure 9).
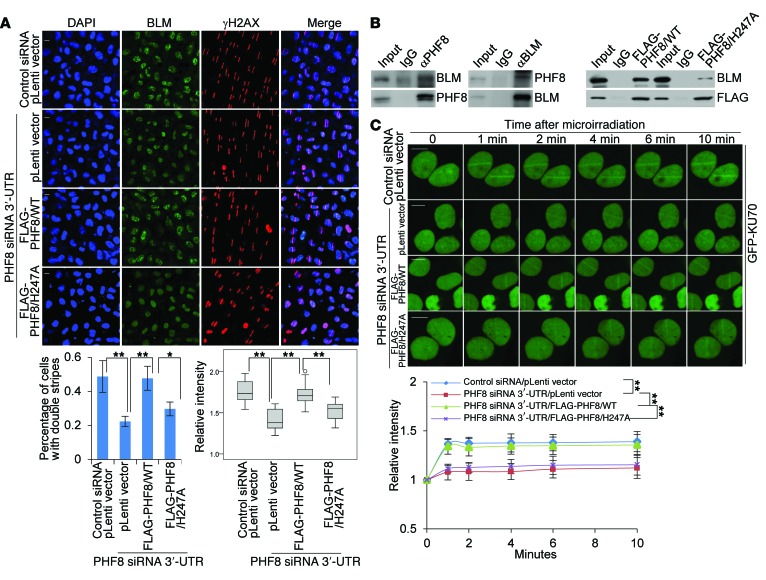
Since our mass spectrometry analysis of the PHF8 interactome revealed that BLM, an essential regulator of DNA end resection in HR repair (50, 51), is potentially associated with PHF8 (Figure 1A), we next examined whether PHF8 depletion has an effect on BLM accumulation. Indeed, we demonstrated that PHF8 deficiency was associated with an impaired recruitment of BLM to UV laser–induced DNA lesions (Figure 8A). In agreement with the finding that the catalytic activity of PHF8 is required for HR repair, forced expression of PHF8/H247A could not restore BLM accumulation as PHF8/WT did, in PHF8-depleted cells (Figure 8A). Indeed, we found that BLM could be effectively coimmunoprecipitated with endogenous PHF8 (Figure 8B, left panel), or PHF8/WT (Figure 8B, right panel), but not PHF8/H247A (Figure 8B, right panel). Thus, we deduced that the demethylase activity is required for preparation, by PHF8, of a particular DDR component, whose methylation state otherwise will impede its association with BLM and thus the engagement of BLM at DNA breaks. To corroborate the role of PHF8 in DNA end resection, we examined phosphorylation of the single-strand DNA–binding (ssDNA-binding) protein RPA and found that DNA damage–induced phosphorylation of RPA32 on Ser-4 and Ser-8 was markedly reduced upon PHF8 depletion, as shown by immunofluorescence detection of this phosphorylation event at sites of laser-induced damage (Supplemental Figure 10A) and Western blotting analysis after camptothecin (CPT) treatment (Supplemental Figure 10B, top panel). Furthermore, the resection defect in PHF8-depleted cells was overcome by overexpression of PHF8, but not that of BLM, as established by both RPA phosphorylation (Supplemental Figure 10A and Supplemental Figure 10B, bottom panel) and CPT sensitivity assays (Supplemental Figure 10C). By using a method to detect BrdU-substituted cellular DNA under nondenaturing conditions (50), we found that this direct readout of ssDNA production at DNA damage sites was also suppressed by PHF8 depletion (Supplemental Figure 10D). These data indicate that PHF8 functions to promote DNA end resection by mediating BLM recruitment.
Figure 8. PHF8 promotes BLM and KU70 accumulation at DSBs.
(A) MCF-7 cells stably expressing control vector, PHF8/WT, or PHF8/H247A were transfected with the indicated siRNAs and subjected to UV-laser microdissection. The cells were fixed and immunostained with anti-γH2AX and anti-BLM followed by microscopy analysis. Scale bar: 10 μm. Percentage of nuclei with double stripes was quantified, and each bar represents the mean ± SD for biological triplicate experiments with more than 100 nuclei. Relative intensities of BLM stripes to the uncut regions determined by ImageJ software are presented with box plots. The circle indicates the outlier value determined by SPSS software. (B) Whole-cell lysates from MCF-7 cells (left panel) or MCF-7 cells stably expressing PHF8/WT or PHF8/H247A (right panel) were immunoprecipitated and then immunoblotted with antibodies against the indicated proteins. (C) MCF-7 cells stably expressing control vector, PHF8/WT, or PHF8/H247A were cotransfected with the indicated siRNAs and GFP-KU70 followed by UV-laser microirradiation. Scale bar: 10 μm. Relative fluorescence intensities in microirradiated areas to the undamaged regions were determined by ImageJ software. More than 20 nuclei were scored in biological triplicate experiments. Each bar represents the mean ± SD. *P < 0.05 and **P < 0.01, 1-way ANOVA in A and 2-way ANOVA in C. pLenti, lentiviral plasmid.
To determine how PHF8 contributes to NHEJ repair, a UV-laser microirradiation experiment was used to monitor the accumulation kinetics of KU70, a DNA end-binding protein (52). Notably, PHF8 depletion was associated with an impaired recruitment of KU70 (Figure 8C). In line with the observation that PHF8 promotes NHEJ repair via its catalytic activity, PHF8/WT, but not PHF8/H247A, was able to rescue the phenotype of impaired KU70 recruitment in PHF8-deficient cells (Figure 8C). Meanwhile, we demonstrated that either PHF8 loss or gain of function did not result in expression alterations of BLM and KU70 (Supplemental Figure 10E), and the kinetics of laser-induced GFP-tagged PHF8/WT and PHF8/H247A accumulation at break sites exhibited a similar manner (Supplemental Figure 10F). We have thus identified PHF8 as a novel DSB repair module, which promotes the recruitment of BLM or KU70 and efficient DSB repair through an HR or NHEJ pathway, respectively.
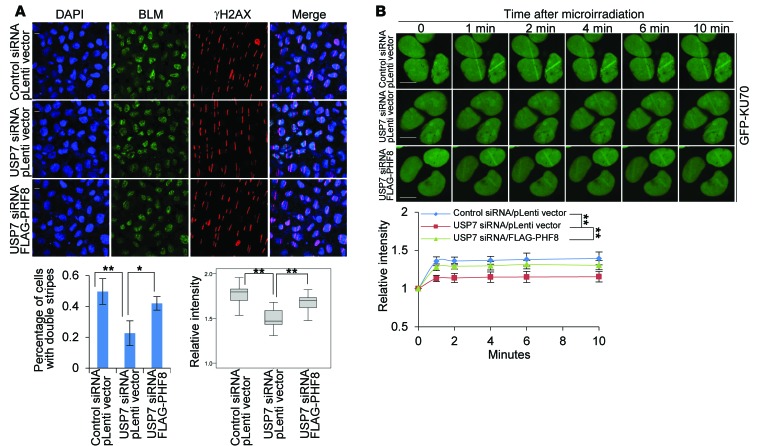
Consistent with the effects of PHF8 on DDR factor accumulation at DSBs, we found that USP7 depletion undermined the accrual of BLM (Figure 9A), RPA32 phosphorylation (Supplemental Figure 11A), and KU70 (Figure 9B), but not γH2AX and MRE11 (Supplemental Figure 9), at break sites, the effect of which could be largely reverted by forced expression of PHF8 (Figure 9A, Supplemental Figure 11A, and Figure 9B). Meanwhile, the resection defect in USP7-depleted cells was overcome by overexpression of PHF8, but not that of BLM (Supplemental Figure 11, A–C). In addition, we demonstrated that USP7 loss of function did not result in expression alterations of BLM and KU70 (Supplemental Figure 11D). Moreover, we found that the defect of NHEJ repair in PHF8-depleted or USP7-depleted cells was overcome by overexpression of PHF8, but not that of KU70, as shown by NHEJ reporter assays (Supplemental Figure 12, A and B). The expression of KU70 was monitored by Western blotting (Supplemental Figure 12C). Taken together, these results support the notion that USP7-promoted PHF8 stabilization plays critical roles in orchestrating DSB repair processes.
Figure 9. USP7-promoted PHF8 stabilization is required for BLM and KU70 accumulation.
(A) MCF-7 cells stably expressing control vector or PHF8 were transfected with the indicated siRNAs and subjected to UV-laser microdissection. The cells were fixed and immunostained with anti-γH2AX and anti-BLM followed by microscopy analysis. Scale bar: 10 μm. Percentage of nuclei with double stripes was quantified, and each bar represents the mean ± SD for biological triplicate experiments with more than 100 nuclei. Relative intensities of BLM stripes to the uncut regions determined by ImageJ software are presented with box plots. (B) MCF-7 cells stably expressing control vector or PHF8 were cotransfected with the indicated siRNAs and GFP-KU70 followed by UV-laser microirradiation. Scale bar: 10 μm. Relative fluorescent intensities in microirradiated areas to the undamaged regions were determined by ImageJ software. More than 20 nuclei were scored in biological triplicate experiments. Each bar represents the mean ± SD. *P < 0.05 and **P < 0.01, 1-way ANOVA in A and 2-way ANOVA in B.
A recent study reported that RNF168, an E3 ligase involved in ubiquitin-dependent DNA damage signaling, is a substrate of USP7 (44). Indeed, we showed that USP7 depletion led to decreased expression of RNF168 (Supplemental Figure 13A) and impaired recruitment of RNF168 to DNA lesions (Supplemental Figure 13B). However, knockdown of PHF8 did not affect the expression of RNF168 (Supplemental Figure 13A) and the recruitment of RNF168 to DNA damage sites (Supplemental Figure 13B). Analogously, the protein expression level of PHF8 was essentially unchanged upon RNF168 depletion (Supplemental Figure 13C). In addition, USP7, but not PHF8, could be coimmunoprecipitated by RNF168 (Supplemental Figure 13D). Unlike PHF8, RNF168 had no effect on the recruitment of KU70 to laser-generated DNA breaks (Supplemental Figure 13E). This finding is in line with a recent study reporting that RNF168 overexpression did not alter the recruitment of KU80, another KU protein (53). These results indicate that PHF8 functions independently of RNF168-regulated DNA damage signaling.
Discussion
It is becoming increasingly clear that dysfunction of epigenetic regulation is commonly involved in malignant transformation (4, 54). In particular, histone methylation/demethylation has been abundantly documented as a critical player in breast carcinogenesis (33, 55). In this study, we demonstrated that the histone demethylase PHF8 is physically associated with and stabilized by the deubiquitinase USP7. Importantly, we showed that USP7 is upregulated in breast cancer and its level of expression is correlated with that of PHF8. Our observations support a model in which upregulation of USP7 in breast cancer results in an elevated protein level of PHF8, which, in turn, leads to deregulation of histone modifications, thus aberrant expression of targeted genes, and eventually contributes to the development and progression of breast cancer.
As a member of the cyclins, cyclin A2 is unique in that it promotes DNA synthesis during S phase and facilitates cell cycle transition from G2 to M phase by activating CDK2 and CDK1, respectively (38). Our observations of the functional connection between USP7/PHF8 and cyclin A2 agree with recent studies reporting that PHF8 promotes cell cycle transition (9, 56). In addition, data from the Human Protein Atlas and the literature indicate that cyclin A2 is overexpressed in dozens of cancers (38, 39, 57), and inhibition of cyclin A2 complexes was shown to suppress tumorigenesis in vitro and in vivo (39, 58). Consistent with our observation that USP7-promoted PHF8 stabilization is linked to upregulation of cyclin A2, we showed that the expression level of cyclin A2 is elevated and positively associated with that of PHF8 and USP7 in clinical breast cancer samples. Moreover, we demonstrated that cyclin A2 gain of function was able to rescue PHF8 depletion–induced phenotypes both in vitro and in vivo. Together, these findings provide a molecular basis for the understanding of the dysregulation of cyclin A2 in breast cancer.
The question, then, is how USP7 is overexpressed in breast cancer. In this regard, it is interesting to note that we found that USP7 itself was also regulated by PHF8 and that PHF8 was indeed detected on the promoter region of USP7. Moreover, bioinformatics analysis indicates that the mRNA expression level of USP7 and PHF8 is upregulated in breast cancer samples, and the mRNA expression level of USP7 is positively correlated with that of PHF8. Based on these findings, we propose that a positive-feedback loop exists between USP7 and PHF8 in which USP7 deubiquitinates and stabilizes PHF8, which, in turn, contributes to the transactivation of USP7. Although the cause-and-effect relationship between USP7 overexpression and PHF8 overexpression in breast carcinomas appears to be a relationship between chicken and egg that is currently difficult to delineate, this loop underscores the importance of the deubiquitinase USP7 and the demethylase PHF8 in coordinating and orchestrating breast carcinogenesis (Supplemental Figure 14). Relevant to this, USP7 is also reported to regulate the stability of p53 and MDM2, 2 key regulators controlling cell survival and tumorigenesis (23, 42). Structural analysis indicates that both p53 and MDM2 specifically recognize the N-terminal MATH domain of USP7 in a mutually exclusive manner (59). Since the MATH domain of USP7 is also required for its interaction with PHF8, it is possible that PHF8 competes with these molecules to bind USP7. This deduction is in concert with our observation that the interaction between PHF8 and USP7 was augmented upon DNA damage, whereas USP7 and MDM2 were reported to be disassociated during DDR (43).
Significantly, we report that USP7-catalyzed PHF8 stabilization is critically involved in DSB repair. The functional involvement of PHF8 in DDR is especially appealing given that genomic instability is disclosed as a common feature in fragile X syndrome (60, 61) and PHF8 truncations (62) or mutations (62) are linked to intellectual disability. It is tempting to speculate that the role of PHF8 in DDR might represent a novel, previously unappreciated contributing factor in the development of fragile X syndrome, the understanding of which will broaden the insight into the regulation of genome/epigenome integrity and the pathogenesis of neurological diseases. Moreover, chemo- and radiotherapy are designed to eliminate cancer cells through inducing DNA damage beyond repair by DDR machinery (63). However, cancerous cells often carry abnormalities in DDR machinery, rendering these cells resistant to DNA damage–based therapy (64). In light of our observations that USP7-promoted PHF8 stabilization is required for DSB repair (Supplemental Figure 14) together with that USP7 and PHF8 are highly expressed in breast cancer, we envision that inhibition of the enzymatic activities of USP7 and/or PHF8, combined with chemo- or radiotherapy, could be potentially explored to improve the treatment of breast cancer.
The physiological significance of USP7-promoted PHF8 stabilization remains to be investigated. In addition, it will be interesting to explore the molecular mechanisms underlying the overexpression of USP7 and PHF8 and to gain more mechanistic insights into the role of PHF8 in DSB repair. Nevertheless, our study revealed that USP7 regulates the stability and thus the function of PHF8, providing a mechanistic link of the deubiquitinase USP7 to epigenetic regulation and DDR. Our experiments indicate that the USP7/PHF8/cyclin A2 axis is critically involved in breast carcinogenesis, supporting the pursuit of these molecules as potential targets for breast cancer intervention.
Methods
For a detailed description of all methods, see the Supplemental Methods.
In vitro deubiquitination assay.
HA-Ub–conjugated FLAG-PHF8 was purified from HeLa cells in high-salt and detergent buffer, and His-tagged USP7/WT or USP7/C223S was affinity purified using nickel-chelating resin from extracts of baculovirus-infected insect cells. Recombinant PHF8-Ub and USP7/WT or USP7/C223S were then incubated in DUB buffer (50 mM Tris-HCl, pH 8.0; 50 mM NaCl; 1 mM EDTA; 10 mM DTT and 5% glycerol) at 37°C for 2 hours. The reactions were stopped by boiling for 5 minutes in 5× SDS-PAGE loading buffer, and the boiled protein complexes were subjected to SDS-PAGE followed by IB with appropriate antibodies.
Sequences of shRNAs and primers.
Sequences of shRNAs and sequences of primers used in quantitative PCR are provided in Supplemental Tables 2–4.
RNA sequencing.
The transcriptome data sets are available at the NCBI Sequence Read Archive with accession number SRP066280, and the analyzed results with cutoff (P value ≤ 10–5, FDR ≤0.001, and |log2 ratio| ≥1) are provided in Supplemental Table 5.
Statistics.
Data from biological triplicate experiments are presented with error bars as mean ± SD. ANOVA with Bonferroni’s correction was used to compare multiple groups of data. A P value of less than 0.05 was considered significant. All of the statistical testing results were determined by SPSS 20.0 software.
Study approval.
All procedures involving animals were approved by the Ethics Committee of Tianjin Medical University and followed the NIH Guide for the Care and Use of Laboratory Animals (8th ed. National Academies Press. 2011). All studies associated with patient samples were approved by the Ethics Committee of Tianjin Medical University, and informed consent was obtained from all patients.
Author contributions
QW, SM, ZY, and LS (Lei Shi) designed the research studies. QW, SM, NS, XL (Xin Li), and LL conducted experiments. QW, SM, NS, XL (Xin Li), LL, SY, XD, LS (Lin Shan), XZ, DS, YW, QZ, XL (Xinhua Liu), NY, and KZ acquired data. QW, SM, XL (Xinhua Liu), ZY, and LS (Lei Shi) analyzed data. QW, SM, ZY, and LS (Lei Shi) wrote the manuscript.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (81272284 and 91219102 to L. Shi, 81572882 to Z. Yao, and 91219201, 81530073, and 81130048 to Y. Shang). We thank Xingzhi Xu (Capital Normal University, Beijing, China) and Bin Peng (Capital Normal University, Beijing, China) for providing help on the UV-laser micropoint experiment. We thank Tao Sun (Nankai University, Tianjin, China) for providing help on the IHC experiment.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:J Clin Invest. 2016;126(6):2205–2220. doi:10.1172/JCI85747.
Contributor Information
Qian Wang, Email: wangqian@tmu.edu.cn.
Shuai Ma, Email: yjs_mashuai@tmu.edu.cn.
Nan Song, Email: songnan@tmu.edu.cn.
Xin Li, Email: lixinangla@tmu.edu.cn.
Ling Liu, Email: lingliu@tmu.edu.cn.
Shangda Yang, Email: yangshangda@tmu.edu.cn.
Xiang Ding, Email: xding@moon.ibp.ac.cn.
Lin Shan, Email: shanlin@tmu.edu.cn.
Xing Zhou, Email: zhouxing@tmu.edu.cn.
Dongxue Su, Email: yjs_sudongxue@tmu.edu.cn.
Yue Wang, Email: yjs_wangyue@tmu.edu.cn.
Qi Zhang, Email: Tjzhangqi@tmu.edu.cn.
Xinhua Liu, Email: liuxinhua@tmu.edu.cn.
Na Yu, Email: na_yuna2001@163.com.
Kai Zhang, Email: zhangkai@nankai.edu.cn.
Yongfeng Shang, Email: yshang@tmu.edu.cn.
Zhi Yao, Email: yaozhi@tmu.edu.cn.
Lei Shi, Email: shilei@tmu.edu.cn.
References
- 1.Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447(7143):407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- 2.Groth A, Rocha W, Verreault A, Almouzni G. Chromatin challenges during DNA replication and repair. Cell. 2007;128(4):721–733. doi: 10.1016/j.cell.2007.01.030. [DOI] [PubMed] [Google Scholar]
- 3.Schneider R, Bannister AJ, Kouzarides T. Unsafe SETs: histone lysine methyltransferases and cancer. Trends Biochem Sci. 2002;27(8):396–402. doi: 10.1016/S0968-0004(02)02141-2. [DOI] [PubMed] [Google Scholar]
- 4.Hojfeldt JW, Agger K, Helin K. Histone lysine demethylases as targets for anticancer therapy. Nat Rev Drug Discov. 2013;12(12):917–930. doi: 10.1038/nrd4154. [DOI] [PubMed] [Google Scholar]
- 5.Shi Y, Whetstine JR. Dynamic regulation of histone lysine methylation by demethylases. Mol Cell. 2007;25(1):1–14. doi: 10.1016/j.molcel.2006.12.010. [DOI] [PubMed] [Google Scholar]
- 6.Fortschegger K, de Graaf P, Outchkourov NS, van Schaik FM, Timmers HT, Shiekhattar R. PHF8 targets histone methylation and RNA polymerase II to activate transcription. Mol Cell Biol. 2010;30(13):3286–3298. doi: 10.1128/MCB.01520-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kleine-Kohlbrecher D, et al. A functional link between the histone demethylase PHF8 and the transcription factor ZNF711 in X-linked mental retardation. Mol Cell. 2010;38(2):165–178. doi: 10.1016/j.molcel.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Suganuma T, Workman JL. Features of the PHF8/KIAA1718 histone demethylase. Cell Res. 2010;20(8):861–862. doi: 10.1038/cr.2010.110. [DOI] [PubMed] [Google Scholar]
- 9.Liu W, et al. PHF8 mediates histone H4 lysine 20 demethylation events involved in cell cycle progression. Nature. 2010;466(7305):508–512. doi: 10.1038/nature09272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qi HH, et al. Histone H4K20/H3K9 demethylase PHF8 regulates zebrafish brain and craniofacial development. Nature. 2010;466(7305):503–507. doi: 10.1038/nature09261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arteaga MF, et al. The histone demethylase PHF8 governs retinoic acid response in acute promyelocytic leukemia. Cancer Cell. 2013;23(3):376–389. doi: 10.1016/j.ccr.2013.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qiu J, et al. The X-linked mental retardation gene PHF8 is a histone demethylase involved in neuronal differentiation. Cell Res. 2010;20(8):908–918. doi: 10.1038/cr.2010.81. [DOI] [PubMed] [Google Scholar]
- 13.Abidi F, Miano M, Murray J, Schwartz C. A novel mutation in the PHF8 gene is associated with X-linked mental retardation with cleft lip/cleft palate. Clin Genet. 2007;72(1):19–22. doi: 10.1111/j.1399-0004.2007.00817.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma Q, Chen Z, Jia G, Lu X, Xie X, Jin W. The histone demethylase PHF8 promotes prostate cancer cell growth by activating the oncomiR miR-125b. Onco Targets Ther. 2015;8:1979–1988. doi: 10.2147/OTT.S85443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun X, et al. Oncogenic features of PHF8 histone demethylase in esophageal squamous cell carcinoma. PLoS One. 2013;8(10): doi: 10.1371/journal.pone.0077353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu G, et al. Elevated expression of histone demethylase PHF8 associates with adverse prognosis in patients of laryngeal and hypopharyngeal squamous cell carcinoma. Epigenomics. 2015;7(2):143–153. doi: 10.2217/epi.14.82. [DOI] [PubMed] [Google Scholar]
- 17.Shen Y, Pan X, Zhao H. The histone demethylase PHF8 is an oncogenic protein in human non-small cell lung cancer. Biochem Biophys Res Commun. 2014;451(1):119–125. doi: 10.1016/j.bbrc.2014.07.076. [DOI] [PubMed] [Google Scholar]
- 18.Komander D, Clague MJ, Urbe S. Breaking the chains: structure and function of the deubiquitinases. Nat Rev Mol Cell Biol. 2009;10(8):550–563. doi: 10.1038/nrm2731. [DOI] [PubMed] [Google Scholar]
- 19.Everett RD, Meredith M, Orr A, Cross A, Kathoria M, Parkinson J. A novel ubiquitin-specific protease is dynamically associated with the PML nuclear domain and binds to a herpesvirus regulatory protein. EMBO J. 1997;16(7):1519–1530. doi: 10.1093/emboj/16.7.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Daubeuf S, et al. HSV ICP0 recruits USP7 to modulate TLR-mediated innate response. Blood. 2009;113(14):3264–3275. doi: 10.1182/blood-2008-07-168203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee HR, et al. Bilateral inhibition of HAUSP deubiquitinase by a viral interferon regulatory factor protein. Nat Struct Mol Biol. 2011;18(12):1336–1344. doi: 10.1038/nsmb.2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Giovinazzi S, Morozov VM, Summers MK, Reinhold WC, Ishov AM. USP7 and Daxx regulate mitosis progression and taxane sensitivity by affecting stability of Aurora-A kinase. Cell Death Differ. 2013;20(5):721–731. doi: 10.1038/cdd.2012.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khoronenkova SV. ATM-dependent downregulation of USP7/HAUSP by PPM1G activates p53 response to DNA damage. Mol Cell. 2012;45(6):801–813. doi: 10.1016/j.molcel.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schwertman P, et al. UV-sensitive syndrome protein UVSSA recruits USP7 to regulate transcription-coupled repair. Nat Genet. 2012;44(5):598–602. doi: 10.1038/ng.2230. [DOI] [PubMed] [Google Scholar]
- 25.van der Knaap JA, et al. GMP synthetase stimulates histone H2B deubiquitylation by the epigenetic silencer USP7. Mol Cell. 2005;17(5):695–707. doi: 10.1016/j.molcel.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 26.Hao YH, et al. USP7 acts as a molecular rheostat to promote WASH-dependent endosomal protein recycling and is mutated in a human neurodevelopmental disorder. Mol Cell. 2015;59(6):956–969. doi: 10.1016/j.molcel.2015.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Loosdregt J, et al. Stabilization of the transcription factor Foxp3 by the deubiquitinase USP7 increases Treg-cell-suppressive capacity. Immunity. 2013;39(2):259–271. doi: 10.1016/j.immuni.2013.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Birks EJ, et al. Elevated p53 expression is associated with dysregulation of the ubiquitin-proteasome system in dilated cardiomyopathy. Cardiovasc Res. 2008;79(3):472–480. doi: 10.1093/cvr/cvn083. [DOI] [PubMed] [Google Scholar]
- 29.Fan YH, et al. USP7 inhibitor P22077 inhibits neuroblastoma growth via inducing p53-mediated apoptosis. Cell Death Dis. 2013;4: doi: 10.1038/cddis.2013.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chauhan D, et al. A small molecule inhibitor of ubiquitin-specific protease-7 induces apoptosis in multiple myeloma cells and overcomes bortezomib resistance. Cancer Cell. 2012;22(3):345–358. doi: 10.1016/j.ccr.2012.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Epping MT, Meijer LA, Krijgsman O, Bos JL, Pandolfi PP, Bernards R. TSPYL5 suppresses p53 levels and function by physical interaction with USP7. Nat Cell Biol. 2011;13(1):102–108. doi: 10.1038/ncb2142. [DOI] [PubMed] [Google Scholar]
- 32.Lin Z, et al. USP22 antagonizes p53 transcriptional activation by deubiquitinating Sirt1 to suppress cell apoptosis and is required for mouse embryonic development. Mol Cell. 2012;46(4):484–494. doi: 10.1016/j.molcel.2012.03.024. [DOI] [PubMed] [Google Scholar]
- 33.Wang Y, et al. LSD1 is a subunit of the NuRD complex and targets the metastasis programs in breast cancer. Cell. 2009;138(4):660–672. doi: 10.1016/j.cell.2009.05.050. [DOI] [PubMed] [Google Scholar]
- 34.Reverdy C, et al. Discovery of specific inhibitors of human USP7/HAUSP deubiquitinating enzyme. Chem Biol. 2012;19(4):467–477. doi: 10.1016/j.chembiol.2012.02.007. [DOI] [PubMed] [Google Scholar]
- 35.Li M, Brooks CL, Wu-Baer F, Chen D, Baer R, Gu W. Mono- versus polyubiquitination: differential control of p53 fate by Mdm2. Science. 2003;302(5652):1972–1975. doi: 10.1126/science.1091362. [DOI] [PubMed] [Google Scholar]
- 36.Sarkari F, Sanchez-Alcaraz T, Wang S, Holowaty MN, Sheng Y, Frappier L. EBNA1-mediated recruitment of a histone H2B deubiquitylating complex to the Epstein-Barr virus latent origin of DNA replication. PLoS Pathog. 2009;5(10): doi: 10.1371/journal.ppat.1000624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lecona E, Narendra V, Reinberg D. USP7 cooperates with SCML2 to regulate the activity of PRC1. Mol Cell Biol. 2015;35(7):1157–1168. doi: 10.1128/MCB.01197-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Malumbres M, Barbacid M. Cell cycle, CDKs and cancer: a changing paradigm. Nat Rev Cancer. 2009;9(3):153–166. doi: 10.1038/nrc2602. [DOI] [PubMed] [Google Scholar]
- 39.Gopinathan L, Tan SL, Padmakumar VC, Coppola V, Tessarollo L, Kaldis P. Loss of Cdk2 and cyclin A2 impairs cell proliferation and tumorigenesis. Cancer Res. 2014;74(14):3870–3879. doi: 10.1158/0008-5472.CAN-13-3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang F, Hu Y, Liu HX, Wan YJ. MiR-22-silenced cyclin A expression in colon and liver cancer cells is regulated by bile acid receptor. J Biol Chem. 2015;290(10):6507–6515. doi: 10.1074/jbc.M114.620369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rhodes DR, et al. Oncomine 3. Neoplasia. 2007;9(2):166–180. doi: 10.1593/neo.07112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li M, Brooks CL, Kon N, Gu W. A dynamic role of HAUSP in the p53-Mdm2 pathway. Mol Cell. 2004;13(6):879–886. doi: 10.1016/S1097-2765(04)00157-1. [DOI] [PubMed] [Google Scholar]
- 43.Meulmeester E, et al. Loss of HAUSP-mediated deubiquitination contributes to DNA damage-induced destabilization of Hdmx and Hdm2. Mol Cell. 2005;18(5):565–576. doi: 10.1016/j.molcel.2005.04.024. [DOI] [PubMed] [Google Scholar]
- 44.Zhu Q, Sharma N, He J, Wani G, Wani AA. USP7 deubiquitinase promotes ubiquitin-dependent DNA damage signaling by stabilizing RNF168. Cell Cycle. 2015;14(9):1413–1425. doi: 10.1080/15384101.2015.1007785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Iacovoni JS, et al. High-resolution profiling of gammaH2AX around DNA double strand breaks in the mammalian genome. EMBO J. 2010;29(8):1446–1457. doi: 10.1038/emboj.2010.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bunting SF, et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell. 2010;141(2):243–254. doi: 10.1016/j.cell.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pierce AJ, Johnson RD, Thompson LH, Jasin M. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 1999;13(20):2633–2638. doi: 10.1101/gad.13.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee C, Hong S, Lee MH, Koo HS. A PHF8 homolog in C. PLoS One. 2015;10(4): doi: 10.1371/journal.pone.0123865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li X, et al. Histone demethylase KDM5B is a key regulator of genome stability. Proc Natl Acad Sci U S A. 2014;111(19):7096–7101. doi: 10.1073/pnas.1324036111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Polo SE, et al. Regulation of DNA-end resection by hnRNPU-like proteins promotes DNA double-strand break signaling and repair. Mol Cell. 2012;45(4):505–516. doi: 10.1016/j.molcel.2011.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gravel S, Chapman JR, Magill C, Jackson SP. DNA helicases Sgs1 and BLM promote DNA double-strand break resection. Genes Dev. 2008;22(20):2767–2772. doi: 10.1101/gad.503108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Spagnolo L, Rivera-Calzada A, Pearl LH, Llorca O. Three-dimensional structure of the human DNA-PKcs/Ku70/Ku80 complex assembled on DNA and its implications for DNA DSB repair. Mol Cell. 2006;22(4):511–519. doi: 10.1016/j.molcel.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 53.Feng L, Chen J. The E3 ligase RNF8 regulates KU80 removal and NHEJ repair. Nat Struct Mol Biol. 2012;19(2):201–206. doi: 10.1038/nsmb.2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dawson MA, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell. 2012;150(1):12–27. doi: 10.1016/j.cell.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 55.Kim JH, et al. UTX and MLL4 coordinately regulate transcriptional programs for cell proliferation and invasiveness in breast cancer cells. Cancer Res. 2014;74(6):1705–1717. doi: 10.1158/0008-5472.CAN-13-1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lim HJ, Dimova NV, Tan MK, Sigoillot FD, King RW, Shi Y. The G2/M regulator histone demethylase PHF8 is targeted for degradation by the anaphase-promoting complex containing CDC20. Mol Cell Biol. 2013;33(21):4166–4180. doi: 10.1128/MCB.00689-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Uhlen M, et al. Towards a knowledge-based Human Protein Atlas. Nat Biotechnol. 2010;28(12):1248–1250. doi: 10.1038/nbt1210-1248. [DOI] [PubMed] [Google Scholar]
- 58.Chen W, Lee J, Cho SY, Fine HA. Proteasome-mediated destruction of the cyclin a/cyclin-dependent kinase 2 complex suppresses tumor cell growth in vitro and in vivo. Cancer Res. 2004;64(11):3949–3957. doi: 10.1158/0008-5472.CAN-03-3906. [DOI] [PubMed] [Google Scholar]
- 59.Hu M, Gu L, Li M, Jeffrey PD, Gu W, Shi Y. Structural basis of competitive recognition of p53 and MDM2 by HAUSP/USP7: implications for the regulation of the p53-MDM2 pathway. PLoS Biol. 2006;4(2): doi: 10.1371/journal.pbio.0040027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jenkins EC, et al. Reduced telomere length in individuals with FMR1 premutations and full mutations. Am J Med Genet A. 2012;158A(5):1060–1065. doi: 10.1002/ajmg.a.35275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Entezam A, Usdin K. ATM and ATR protect the genome against two different types of tandem repeat instability in Fragile X premutation mice. Nucleic Acids Res. 2009;37(19):6371–6377. doi: 10.1093/nar/gkp666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Laumonnier F, et al. Mutations in PHF8 are associated with X linked mental retardation and cleft lip/cleft palate. J Med Genet. 2005;42(10):780–786. doi: 10.1136/jmg.2004.029439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ummat A, et al. Structural basis for cisplatin DNA damage tolerance by human polymerase eta during cancer chemotherapy. Nat Struct Mol Biol. 2012;19(6):628–632. doi: 10.1038/nsmb.2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang B, Hurov K, Hofmann K, Elledge SJ. NBA1, a new player in the Brca1 A complex, is required for DNA damage resistance and checkpoint control. Genes Dev. 2009;23(6):729–739. doi: 10.1101/gad.1770309. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.