Abstract
The linear sequence of eukaryotic genomes is arranged in a specific manner within the three-dimensional nuclear space. Interactions between distant sites partition the genome into domains of highly associating chromatin. Interaction domains are found in many organisms, but their properties and the principles governing their establishment vary between different species. Topologically associating domains (TADs) extending over large genomic regions are found in mammals and D. melanogaster, whereas other types of contact domains exist in lower eukaryotes. Here we review recent studies that explore the mechanisms by which long distance chromatin interactions determine the 3D organization of the genome and the relationship between this organization and the establishment of specific patterns of gene expression.
Keywords: chromatin, epigenetics, CTCF, cohesin, architectural proteins, transcription
Introduction: A Three-dimensional Genome
The eukaryotic nucleus is a complex three-dimensional environment in which genome function depends not only on the linear arrangement of regulatory sequence elements but also on their spatial organization for effective control of gene expression [1,2]. Modulation of transcription occurs in part through spatial proximity of regulatory elements and gene promoters [1,2]. These interactions are essential for organismal development and response to environmental stimuli [2–5] in eukaryotes, including yeast, worms, plants, flies, and mammals [6–12]. Analysis of the role of chromatin 3D organization in gene expression is progressing rapidly, largely due to the development of chromosome conformation capture methods such as Hi-C [13]. Studies of long-range chromatin interactions have highlighted principles of three-dimensional genome organization, and whole genome chromatin contact maps have provided significant insights into how the 3D organization of the genome relates to gene expression [1,2]. Due to these advances we are beginning to understand overall chromosomal organization in the nucleus, how this organization is established, and how it can modulate gene expression. Here we discuss recent work that has helped answer important questions about the establishment and role of chromatin organization in genome regulation.
Units of Organization
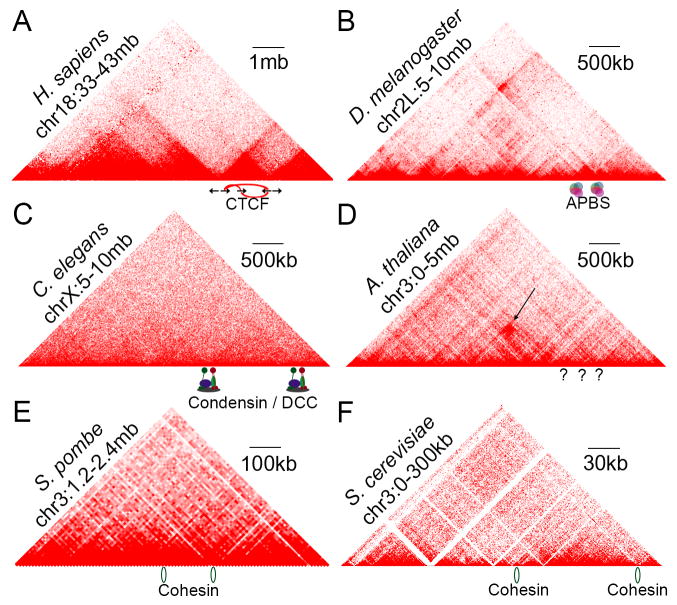
Whole-genome chromatin conformation capture (involving ligation and sequencing of spatially proximal DNA fragments - Hi-C as described in [13]) has been performed in several organisms and the results indicate that some features of chromatin 3D organization are consistent between some species (Figure 1). In many species and tissue types there are easily observable features of chromatin contact maps consisting of large genomic regions organized as contact domains [1,2]. Sequences within these Topologically Associated Domains (TADs) interact more frequently with sites inside than outside the domain. TADs with a median size of 880 kb have been found in mammals (Figure 1A) whereas Drosophila TADs have a median size of 107 kb [1] (Figure 1B). TADs in C. elegans are fairly weak, with the more easily defined domains located in the X chromosome of hermaphrodites [12] (Figure 1C). In S. pombe TAD-like contact domains (termed globules) are present at sizes ranging from 50 to 100 kb [14] (Figure 1E). However, large TAD-like structures are not as easily identifiable in some model organisms such as S. cerevisiae and A. thaliana [6–9] (Figure 1DF). This poses the question of whether TADs are truly conserved features of chromatin organization in eukaryotes.
Figure 1. Chromosome Organization in Eukaryotes.
Chromatin interactions detected by Hi-C displayed as heatmaps for human [47] (A); D. melanogaster [4] (B); C. elegans [12] (C); A. thaliana [9] (D); S. pombe [14] (E); and S. cerevisiae [15] (F).
A) Large TADs in mammals occur with CTCF in reverse – forward orientation at domain borders. Arrows indicate CTCF motif orientation connected by chromatin interactions (red arcs).
B) Clustering of architectural proteins at domain borders. High occupancy architectural binding sites (APBS) correlate with domain borders in D. melanogaster.
C) The Dosage Compensation Complex (DCC) binds domain borders in C. elegans hermaphrodites. Interaction domains are stronger on the X chromosome in C. elegans and correlate with condensin-containing DCC binding sites.
D) Architectural proteins in A. thaliana are unknown. A. thaliana has distinct chromatin interaction structures (indicated by arrow).
E) Globule domains in S. pombe. Cohesin (displayed as rings) sites correlate with edges of globule domains.
F) Small domains in S. cerevisiae. Micro-C allows high resolution contact maps and identification of short chromatin interaction domains. Cohesin (displayed as rings) loader Scc2 correlates with domain borders.
TADs vary in size throughout an individual genome and are overall shorter in the smaller genome of Drosophila (Figure 1A, 1B). Since TAD borders form at sites of active transcription and in regions with high gene density, it is likely that these features occur more often in smaller genomes [15]. To search for TADs in S. cerevisiae, the Hi-C protocol was modified to increase resolution by using micrococcal nuclease (MNase) digestion in lieu of restriction fragmentation (Micro-C) [15]. Using this method high-frequency contact domains were observed in S. cerevisiae [15] (Figure 1F). These domains are indeed smaller than those of mammals and Drosophila, between 2–10 kb in size, and contain only a few genes each [15]. It is unknown whether these small domains are similar to TADs, especially compared to the relatively large domains found in S. pombe [14] (Figure 1E). However, cohesin which is thought to be an important contributor to TAD border formation in many organisms, is enriched at domain borders in S. pombe as is the cohesin loader Scc2 in S. cerevisiae [14,15] (Figure 1E, 1F). It is therefore likely that the difference in domain size is related to the distribution of active genes and architectural proteins across the respective genomes [14,15].
It is clear from these studies that even simple eukaryotes contain structured contact domains and it is likely that chromatin domain organization is a conserved principle of eukaryotic life. However, contact maps of Arabidopsis do not display large TAD structures despite a genome size similar to that of Drosophila [7–9] (Figure 1D). The Arabidopsis genome does contain more genes, and thus gene density may contribute to smaller domains not as easily visible at the resolutions used. Indeed, a recent study using higher resolution contact maps has described small domain-like structures that are not as large or distinct as TADs in other organisms [9]. Another factor that may be confounding clear TAD detection is that samples used for Arabidopsis Hi-C have been from whole tissue and thus represent a population of different cell types [7–9]. While almost nothing is known about the genome-wide distribution of architectural proteins in Arabidopsis, several features of chromatin organization stand out from Hi-C experiments carried out in this organism. Most prominent are inter-centromeric interactions and heterochromatic knot structures defined by high local and inter-chromosomal interactions [8] (Figure 1D). These organizational structures indicate that, despite a lack of knowledge on architectural proteins, Arabidopsis has distinct features of chromatin organization.
Although it is tempting to think of TADs as well defined discrete genomic units, in reality TAD structure is more complex. While sharp boundaries between TADs can sometimes be seen, many boundaries are fuzzy and/or have multiple possible locations (Figure 1). In addition, smaller structures can be seen within TADs corresponding to domains with even higher contact frequencies [10,16]. It is not clear how these smaller domains are different from TADs or if they simply represent similar organizational features at a finer scale. The complexity of Hi-C contact maps makes domain calling difficult and algorithms for this purpose vary and can produce either low resolution or differing boundary calls [11,17,18]. The complexity of TAD organization could suggest dynamic chromatin interactions and/or borders in which structures seen by Hi-C represent average or usual contact points in a population. At a finer scale than domains, point to point chromatin contacts such as enhancer-promoter interactions, represent important features of chromatin organization [1,2]. These contacts can be somewhat difficult to see by Hi-C due to the high frequency of interactions within TADs and many algorithms seek to optimally choose significant contacts [19,20]. More directly capturing interactions with promoter probes (Capture Hi-C/HiCap) has allowed these interactions to be explored in more detail [21,22]. These studies (along with Pol II precipitated interactions) found that promoter-promoter interactions are fairly frequent and form multigene complexes [22,23]. Although no total interaction bias was seen for inactive vs active promoters, the level of expression between contact points was correlated, which implies the existence of a matrix of expression regulation [21,22]. Promoter contacts as well as the complex point to point interaction matrix contribute to overall domain organization. Exploration of these and other fine scale organization principles is an important area of future research and may help to explain the formation and function of contact domains as discussed below.
Establishment of long-range interactions
How two specific distal sites can find each other in the three-dimensional space is an open question. It is known that architectural proteins such as CTCF and cohesin play a major role in long-range contact formation [1,24]. CTCF has not been identified in plants, yeast, or C. elegans [25,26] which may account for the smaller or weaker organizational units observed in these organisms (Figure 1C–F). However, the high abundance of detectable long-distance interactions in these organisms [6–9] and the actual presence of contact domains in yeast [14,15] suggest that other factors can function similar to CTCF in genome organization. In support of this idea, while no known CTCF protein has been found in C. elegans, large TAD-like domains exist [12] (Figure 1C). While these domains are relatively weak on autosomes, they have better defined borders on the X chromosome of hermaphrodites [12]. Interestingly, these borders coincide with the Dosage Compensation Complex (DCC), which contains condensin [12] (Figure 1C). Depletion of the DCC reduces the strength of TAD borders [12], thus the DCC (condensin) acts as a border defining complex without the necessity of CTCF. It is likely that other factors can function similarly in other organisms either for the establishment of domain borders or long-distance chromatin interactions.
Even in organisms that have CTCF, other architectural proteins contribute to genome organization [1,27]. In Drosophila, CTCF and other architectural proteins are present at thousands of sites throughout the genome called Architectural Protein Binding Sites (APBS) [1]. Clustering of APBSs occurs at TAD borders and is thought to effectively insulate chromatin contacts from crossing these sites and/or promote long distance contacts solely in one direction [1] (Figure 1B). In mammals, CTCF motifs often cluster in the genome, but they usually lie too close together to resolve distinct peaks using ChIP-seq. Analysis of the distribution of BORIS and CTCF in the same cells enabled examination of occupancy at clustered sites [28]. The BORIS protein binds the same motif as CTCF, but it is expressed only in the male germ line and in tumor cells. In BORIS positive cells, CTCF and BORIS often bind next to each other at 2X CTCF sites containing two CTCF motifs separated by 30–50 bp. In cells that do not express BORIS, these sites are thought to be occupied by two CTCF proteins, as detected by DNase-seq [28]. These 2X CTCF sites are enriched at active promoters and enhancers, suggesting that clustered CTCF sites mediate interactions involved in transcription activation [28]. CTCF sites have also been shown to cluster with TFIIIC and Prdm5 at TAD borders [29]. It is therefore tempting to speculate that clustering of architectural proteins may play a role in the formation of TAD borders in both mammals and Drosophila [29].
CTCF is found not only at TAD borders, but also inside TADs, suggesting that the mere presence of this protein is not sufficient for TAD border formation [16,30]. Chromatin 3D organization in mammals seems to rely in part on the orientation of CTCF motifs with respect to each other in order to establish contacts between specific regions in the genome, such that interactions tend not to extend beyond CTCF motifs found in reverse – forward orientation [16,24,31,32] (Figure 1A). Recently, it was shown that deletion of reverse – forward oriented CTCF motifs at looping boundaries results in merging of domain/loop structures and causes interactions to extend beyond the deleted site [31,33]. This was also true if the CTCF motif orientation was disrupted solely by inversion [31,33]. Interestingly, inversion/deletion of only one CTCF motif in the reverse - forward pair does not merge the domains (i.e. the border is not lost) but does allow more interactions to occur between domains [33]. This is probably because the remaining CTCF site is still able to insulate interactions from one direction, but not the other. Interestingly, it was found that even palindromic motifs are recognized by CTCF in only one orientation and that inversion of even these sites results in reverse CTCF binding orientation and loss of cohesin binding [31]. Other studies hint at a more complex situation in the establishment of interactions between CTCF sites. Analysis of CTCF-mediated interactions in mouse embryonic stem cells (ESCs) and neural progenitor cells (NPCs) using 4C indicates that contacts between CTCF sites, even over distances of 5.8 Mb, show a preference in directionality, with 65% of loops forming between CTCF sites in convergent, 1% divergent, and 34% in the same orientation [34]. Although deletion of specific CTCF sites results in disruption of loops between convergent sites, inversion of a site does not result in the formation of new loops with sites now arranged in a convergent orientation. Further work will be necessary to resolve these discrepancies, which may be the result of the particular genomic contexts being analyzed in different studies.
Another recent study demonstrated the significance of CTCF orientation by inserting RAG recombinase sites into different regions of the genome [35]. At insertion sites, recombination events correlated with Hi-C interaction intensity and were restricted to domains marked by forward-reverse oriented CTCF motifs [35]. This was dramatically exemplified at the IgH locus where forward-reverse CTCF sites at IGCR1 act as a domain boundary [35]. The function of this boundary was shown by deletion of these sites, which resulted in aberrations in recombination [35]. The importance of CTCF motif orientation has been documented by several groups [16,24,31,32] giving rise to models where linear sequence determines three-dimensional genome structure [33,36,37]. In these models, cohesin tracks along the chromosome such that DNA is pulled through a cohesin ring into a loop until appropriately oriented CTCF motifs are reached [33,36,37].
Functional Aspects of Chromatin Organization
One role of 3D chromatin organization may be to enable enhancer-promoter contacts over long linear distances [2,22]. Proper regulation of transcription requires interactions between enhancers and promoters via long distance contacts [1,2]. This strategy can complicate the interpretation of GWAS analyses because SNPs affecting specific traits are not necessarily located proximally to the affected genes. Indeed it was recently shown that QTLs are more accurately identified if long distance contacts are accounted for [38]. Individuals with differential histone modifications in regulatory regions had corresponding SNPs at distally interacting transcription factor motifs. These differences in histone modifications among individuals correlate with differences in gene expression between non-adjacent genes that interact in the 3D space of the nucleus [38]. Similarly, another study identified variable chromatin modules (VCMs) composed of enhancer marks and genes with coordinated expression [39]. Coordinated variability in chromatin occurs mostly within topological domains and vcmQTLs are enriched for transcription factor binding sites and changes in gene expression [39]. These studies also indicate that genes may share regulatory regions and that changes at single sites may affect chromatin state and gene expression at multiple locations [38,39]. Thus TAD organization and finer scale contacts are important features to consider when evaluating factors affecting gene expression. In fact the overall function of TADs in controlling gene expression could be the creation of local genomic environments in which enhancer-promoter interactions occur [10,16,21,22], and it is possible that TADs are simply low resolution views of clusters of enhancer-promoter matrices.
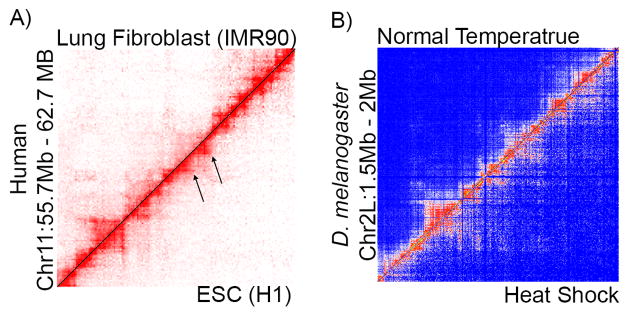
Consistent with the idea that TADs represent matrices of contacts among regulatory regions, interactions within TADs are different between cell types and are influenced by cellular differentiation as well as environmental conditions [4,40,41]. Individual domain contact strength is different between lineage specific cell types while widespread changes occur in histone marks and CTCF binding [40]. The changes to chromatin interactions are best correlated with the enhancer mark H3K4me1 such that increased interactions correspond to increased active enhancer density [40]. This suggests that interactions can change to connect regulatory regions being actively used [41]. In support of this idea, differences in TAD organization can easily be seen when comparing different cell types. For example, specific domains that are well defined in embryonic cell lines are missing in lung fibroblasts (Figure 2A). This indicates that TAD structure is variable among cell types. Furthermore, TAD organization in Drosophila is altered during the heat-shock response [4] (Figure 2B). Architectural proteins are re-distributed from TAD borders to inside TADs, resulting in a decrease in TAD border strength and an increase in inter-TAD interactions [4] (Figure 2B). These changes in 3D organization may allow long-range interactions between Polycomb (Pc) bound enhancers and promoters, forming new Pc bodies and causing general transcription silencing; a characteristic of the stress response to temperature elevation [4].
Figure 2. Changes to Domain Organization.
A) Hi-C of human embryonic stem cells (H1 ESC – bottom right) compared to lung fibroblast cells (IMR90 – top left) [47]. Arrows indicate TAD structure changes.
B) Hi-C of D. melanogaster under heat shock (bottom right) compared to normal temperature (top left) [4].
While the effect of transcription inhibition on TAD organization appears to be small [4,10], RNA may play an important role in the 3D organization of the genome. Enhancers often contain lncRNAs, which may be important for recruitment of transcription factors or architectural proteins, or for other means of establishing long-range contacts [42–45]. Studies of individual loci have found that elimination of specific lncRNAs do in fact alter chromatin interactions [42,44,45]. However, lncRNAs are also involved in several aspects of chromatin structure, including histone modifications and nucleosome positioning [43,46], and thus the effect on long range interactions may be indirect. This is probably not the case at the HOXA locus, as depletion of blincRNA did not affect CTCF binding, but drastically altered local chromatin interactions [44]. The relationship between chromatin organization and lncRNAs is likely not one-sided, and just as long-distance interactions influence gene expression, the same may be true for lncRNA transcription.
Overall the chromatin environment in the eukaryotic nucleus involves long-distance contacts that form higher-order domains. The effects on chromatin interaction structures likely depends on communication among several features such as covalent histone modifications, nucleosome position, transcription factor binding, lncRNAs, and gene expression. Teasing apart independent roles in long-range contact formation may be difficult as each component must take part in the overall chromatin community.
Summary
While our knowledge of the three-dimensional genome has advanced significantly, we still do not understand much about the specificity underlying the establishment of long-range interactions. From a wide perspective it seems that chromatin association domains exist in most species tested [4,12,14,15,47] (Figure 1), but the functional significance of these domains is only partially understood. Several layers of genome organization exist [1,2,16,47] and the contributions of each to the control of gene expression is an important direction for future studies. Understanding this relationship will require knowledge of how these domains form and, more specifically, how two sites in the genome are chosen for contact. Research on proteins or lncRNA with undiscovered architectural function may provide answers to these questions [29,42,44]. Truly, the study of genome organization is an exciting field at the cusp of really understanding the basic mechanisms that control gene expression.
Acknowledgments
Work in the authors’ lab was supported by U.S. Public Health Service Award R01GM035463. MJR was supported by award F32GM113570 from the National Institutes of Health. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Cubeñas-Potts C, Corces VG. Architectural proteins, transcription, and the three-dimensional organization of the genome. FEBS Lett. 2015;589:2923–2930. doi: 10.1016/j.febslet.2015.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dekker J, Misteli T. Long-Range Chromatin Interactions. Cold Spring Harb Perspect Biol. 2015:7. doi: 10.1101/cshperspect.a019356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3**.Lupiáñez DG, Kraft K, Heinrich V, Krawitz P, Brancati F, Klopocki E, Horn D, Kayserili H, Opitz JM, Laxova R, et al. Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell. 2015;161:1012–1025. doi: 10.1016/j.cell.2015.04.004. This study examined chromatin interactions in patients with limb malformations and found that each was associated with disruption of a TAD boundary. Deletion of TAD borders resulted in increased interactions from genes to neighboring TADs containing enhancer elements. This suggested that enhancers act past the previous TAD border which results in misregulation of gene expression. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4*.Li L, Lyu X, Hou C, Takenaka N, Nguyen HQ, Ong C-T, Cubeñas-Potts C, Hu M, Lei EP, Bosco G, et al. Widespread rearrangement of 3D chromatin organization underlies polycomb-mediated stress-induced silencing. Mol Cell. 2015;58:216–231. doi: 10.1016/j.molcel.2015.02.023. Through comparison of Hi-C contact maps in normal and heat-shocked Drosophila cells it was shown that TAD organization and architectural protein localization can change drastically in response to environmental stimuli. This study also explored effects of transcription inhibition on long-range chromatin interactions, as well as the knockdown of key aarchitectural proteins. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Apostolou E, Ferrari F, Walsh RM, Bar-Nur O, Stadtfeld M, Cheloufi S, Stuart HT, Polo JM, Ohsumi TK, Borowsky ML, et al. Genome-wide chromatin interactions of the Nanog locus in pluripotency, differentiation, and reprogramming. Cell Stem Cell. 2013;12:699–712. doi: 10.1016/j.stem.2013.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duan Z, Andronescu M, Schutz K, McIlwain S, Kim YJ, Lee C, Shendure J, Fields S, Blau CA, Noble WS. A three-dimensional model of the yeast genome. Nature. 2010;465:363–367. doi: 10.1038/nature08973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feng S, Cokus SJ, Schubert V, Zhai J, Pellegrini M, Jacobsen SE. Genome-wide Hi-C Analyses in Wild-Type and Mutants Reveal High-Resolution Chromatin Interactions in Arabidopsis. Mol Cell. 2014;55:694–707. doi: 10.1016/j.molcel.2014.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grob S, Schmid MW, Grossniklaus U. Hi-C Analysis in Arabidopsis Identifies the KNOT, a Structure with Similarities to the flamenco Locus of Drosophila. Mol Cell. 2014;55:678–693. doi: 10.1016/j.molcel.2014.07.009. [DOI] [PubMed] [Google Scholar]
- 9.Wang C, Liu C, Roqueiro D, Grimm D, Schwab R, Becker C, Lanz C, Weigel D. Genome-wide analysis of local chromatin packing in Arabidopsis thaliana. Genome Res. 2015;25:246–256. doi: 10.1101/gr.170332.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jin F, Li Y, Dixon JR, Selvaraj S, Ye Z, Lee AY, Yen C-A, Schmitt AD, Espinoza CA, Ren B. A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature. 2013 doi: 10.1038/nature12644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sexton T, Yaffe E, Kenigsberg E, Bantignies F, Leblanc B, Hoichman M, Parrinello H, Tanay A, Cavalli G. Three-Dimensional Folding and Functional Organization Principles of the Drosophila Genome. Cell. 2012;148:458–472. doi: 10.1016/j.cell.2012.01.010. [DOI] [PubMed] [Google Scholar]
- 12*.Crane E, Bian Q, McCord RP, Lajoie BR, Wheeler BS, Ralston EJ, Uzawa S, Dekker J, Meyer BJ. Condensin-driven remodelling of X chromosome topology during dosage compensation. Nature. 2015;523:240–244. doi: 10.1038/nature14450. This represents the first high-resolution view of chromatin interactions in C. elegans and the data suggest that differences in domain organization may exist between sexes. A role for the Dosage Compensation Complex is implicated in domain border formation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fraser J, Williamson I, Bickmore WA, Dostie J. An Overview of Genome Organization and How We Got There: from FISH to Hi-C. Microbiol Mol Biol Rev MMBR. 2015;79:347–372. doi: 10.1128/MMBR.00006-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14*.Mizuguchi T, Fudenberg G, Mehta S, Belton J-M, Taneja N, Folco HD, FitzGerald P, Dekker J, Mirny L, Barrowman J, et al. Cohesin-dependent globules and heterochromatin shape 3D genome architecture in S. pombe Nature. 2014;516:432–435. doi: 10.1038/nature13833. This study found that domain organization exists in yeast as chromatin globules. It showed that not only is Cohesin an important factor in this organization, but that heterochromatin factor Clr4 was shown to play a role in domain formation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15*.Hsieh T-HS, Weiner A, Lajoie B, Dekker J, Friedman N, Rando OJ. Mapping Nucleosome Resolution Chromosome Folding in Yeast by Micro-C. Cell. 2015;162:108–119. doi: 10.1016/j.cell.2015.05.048. Using a novel approach to Hi-C by using MNase-digestion, a high resolution map of S. cerevisiae was created allowing visualization of previously undiscovered chromatin organization domains. This study suggests that domain size may scale with gene density. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16**.Rao SSP, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, Sanborn AL, Machol I, Omer AD, Lander ES, et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159:1665–1680. doi: 10.1016/j.cell.2014.11.021. By developing in-situ Hi-C and obtaining close to five billion contacts in one line and performing Hi-C in hundreds of lines, this study mapped the human genome contact matrix at unprecedented resolution. Using these maps and sophisticated algorithms they were able to identify chromatin loops enriched for CTCF binding sites that were oriented convergently. These loops are at the edge of contact domains, and are fairly conserved across cell lines. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Filippova D, Patro R, Duggal G, Kingsford C. Identification of alternative topological domains in chromatin. Algorithms Mol Biol. 2014;9:14. doi: 10.1186/1748-7188-9-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weinreb C, Raphael BJ. Identification of hierarchical chromatin domains. Bioinforma Oxf Engl. 2015 doi: 10.1093/bioinformatics/btv485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mifsud B, Martincorena I, Darbo E, Sugar R, Schoenfelder S, Fraser P, Luscombe N. GOTHiC, a simple probabilistic model to resolve complex biases and to identify real interactions in Hi-C data. 2015 doi: 10.1371/journal.pone.0174744. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ay F, Bailey TL, Noble WS. Statistical confidence estimation for Hi-C data reveals regulatory chromatin contacts. Genome Res. 2014;24:999–1011. doi: 10.1101/gr.160374.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21*.Mifsud B, Tavares-Cadete F, Young AN, Sugar R, Schoenfelder S, Ferreira L, Wingett SW, Andrews S, Grey W, Ewels PA, et al. Mapping long-range promoter contacts in human cells with high-resolution capture Hi-C. Nat Genet. 2015;47:598–606. doi: 10.1038/ng.3286. Using capture Hi-C, this study describes features of promoter interactions and found that promoters interact with many other promoters. The number of interactions per promoter was independent of transcriptional activity, but interactions tend to occur more with the gene body than upstream. Comparison of cell lines indicate that genes with differential expression also have differential promoter interactions. [DOI] [PubMed] [Google Scholar]
- 22.Sahlén P, Abdullayev I, Ramsköld D, Matskova L, Rilakovic N, Lötstedt B, Albert TJ, Lundeberg J, Sandberg R. Genome-wide mapping of promoter-anchored interactions with close to single-enhancer resolution. Genome Biol. 2015:16. doi: 10.1186/s13059-015-0727-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li G, Ruan X, Auerbach RK, Sandhu KS, Zheng M, Wang P, Poh HM, Goh Y, Lim J, Zhang J, et al. Extensive Promoter-Centered Chromatin Interactions Provide a Topological Basis for Transcription Regulation. Cell. 2012;148:84–98. doi: 10.1016/j.cell.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24*.Vietri Rudan M, Barrington C, Henderson S, Ernst C, Odom DT, Tanay A, Hadjur S. Comparative Hi-C reveals that CTCF underlies evolution of chromosomal domain architecture. Cell Rep. 2015;10:1297–1309. doi: 10.1016/j.celrep.2015.02.004. Comparison of Hi-C maps of several species showed that domain organization is fairly conserved. CTCF motifs that were diverged usually corresponded to weaker interactions, occurred within domains, and corresponded to increased local insulation. This study also showed that TAD structure is maintained through chromosomal rearrangements and that these events typically occur at domain borders. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heger P, Marin B, Schierenberg E. Loss of the insulator protein CTCF during nematode evolution. BMC Mol Biol. 2009;10:84. doi: 10.1186/1471-2199-10-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ong C-T, Corces VG. Insulators as mediators of intra- and inter-chromosomal interactions: a common evolutionary theme. J Biol. 2009;8:73. doi: 10.1186/jbiol165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maksimenko O, Bartkuhn M, Stakhov V, Herold M, Zolotarev N, Jox T, Buxa MK, Kirsch R, Bonchuk A, Fedotova A, et al. Two new insulator proteins, Pita and ZIPIC, target CP190 to chromatin. Genome Res. 2015;25:89–99. doi: 10.1101/gr.174169.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pugacheva EM, Rivero-Hinojosa S, Espinoza CA, Méndez-Catalá CF, Kang S, Suzuki T, Kosaka-Suzuki N, Robinson S, Nagarajan V, Ye Z, et al. Comparative analyses of CTCF and BORIS occupancies uncover two distinct classes of CTCF binding genomic regions. Genome Biol. 2015:16. doi: 10.1186/s13059-015-0736-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Van Bortle K, Nichols MH, Li L, Ong C-T, Takenaka N, Qin ZS, Corces VG. Insulator function and topological domain border strength scale with architectural protein occupancy. Genome Biol. 2014;15:R82. doi: 10.1186/gb-2014-15-5-r82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ong C-T, Corces VG. CTCF: an architectural protein bridging genome topology and function. Nat Rev Genet. 2014;15:234–246. doi: 10.1038/nrg3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31**.Guo Y, Xu Q, Canzio D, Shou J, Li J, Gorkin DU, Jung I, Wu H, Zhai Y, Tang Y, et al. CRISPR Inversion of CTCF Sites Alters Genome Topology and Enhancer/Promoter Function. Cell. 2015;162:900–910. doi: 10.1016/j.cell.2015.07.038. This study showed that CTCF recognizes its motif in only one direction and that CTCF mediated interactions usually occur between motifs in forward - reverse orientation. Inversion of CTCF sites resulted in a switch in the direction that interactions occur. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gómez-Marín C, Tena JJ, Acemel RD, López-Mayorga M, Naranjo S, de la Calle-Mustienes E, Maeso I, Beccari L, Aneas I, Vielmas E, et al. Evolutionary comparison reveals that diverging CTCF sites are signatures of ancestral topological associating domains borders. Proc Natl Acad Sci. 2015;112:7542–7547. doi: 10.1073/pnas.1505463112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33**.Sanborn AL, Rao SSP, Huang S-C, Durand NC, Huntley MH, Jewett AI, Bochkov ID, Chinnappan D, Cutkosky A, Li J, et al. Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc Natl Acad Sci U S A. 2015 doi: 10.1073/pnas.1518552112. Different models of chromatin organization are compared and it was found that chromatin extrusion best fits actual Hi-C data. Normal Hi-C data was compared to simulated data, as was data where CTCF sites at borders were systematically inverted or deleted. This effectively explores the role of CTCF motif orientation in domain organization. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de Wit E, Vos ESM, Holwerda SJB, Valdes-Quezada C, Verstegen MJAM, Teunissen H, Splinter E, Wijchers PJ, Krijger PHL, de Laat W. CTCF Binding Polarity Determines Chromatin Looping. Mol Cell. 2015;60:676–684. doi: 10.1016/j.molcel.2015.09.023. [DOI] [PubMed] [Google Scholar]
- 35.Hu J, Zhang Y, Zhao L, Frock RL, Du Z, Meyers RM, Meng F, Schatz DG, Alt FW. Chromosomal Loop Domains Direct the Recombination of Antigen Receptor Genes. Cell. 2015 doi: 10.1016/j.cell.2015.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nichols MH, Corces VG. A CTCF Code for 3D Genome Architecture. Cell. 2015;162:703–705. doi: 10.1016/j.cell.2015.07.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fudenberg G, Imakaev M, Lu C, Goloborodko A, Abdennur N, Mirny LA. Formation of Chromosomal Domains by Loop Extrusion. 2015 doi: 10.1016/j.celrep.2016.04.085. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38**.Grubert F, Zaugg JB, Kasowski M, Ursu O, Spacek DV, Martin AR, Greenside P, Srivas R, Phanstiel DH, Pekowska A, et al. Genetic Control of Chromatin States in Humans Involves Local and Distal Chromosomal Interactions. Cell. 2015;162:1051–1065. doi: 10.1016/j.cell.2015.07.048. This study indicates that SNPs and chromatin variation can influence gene/trait expression over long distances. These variations could affect distal enhancers, transcription factors, or chromatin modifications that interact with several different genomic regions. It suggests that accounting for Hi-C contacts improves the power of genome-wide association studies. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39**.Waszak SM, Delaneau O, Gschwind AR, Kilpinen H, Raghav SK, Witwicki RM, Orioli A, Wiederkehr M, Panousis NI, Yurovsky A, et al. Population Variation and Genetic Control of Modular Chromatin Architecture in Humans. Cell. 2015;162:1039–1050. doi: 10.1016/j.cell.2015.08.001. By comparing 47 individual genome-wide maps of chromatin factors this study found regions of the genome where variations in chromatin correlate (termed variable chromatin modules). These modules generally occur within contact domains and are associated with transcription factor binding sites. This indicates that changes within domains can alter multiple sites connected by long-range interactions. [DOI] [PubMed] [Google Scholar]
- 40.Dixon JR, Jung I, Selvaraj S, Shen Y, Antosiewicz-Bourget JE, Lee AY, Ye Z, Kim A, Rajagopal N, Xie W, et al. Chromatin architecture reorganization during stem cell differentiation. Nature. 2015;518:331–336. doi: 10.1038/nature14222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Heidari N, Phanstiel DH, He C, Grubert F, Jahanbani F, Kasowski M, Zhang MQ, Snyder MP. Genome-wide map of regulatory interactions in the human genome. Genome Res. 2014;24:1905–1917. doi: 10.1101/gr.176586.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang H, Zeitz MJ, Wang H, Niu B, Ge S, Li W, Cui J, Wang G, Qian G, Higgins MJ, et al. Long noncoding RNA-mediated intrachromosomal interactions promote imprinting at the Kcnq1 locus. J Cell Biol. 2014;204:61–75. doi: 10.1083/jcb.201304152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Quinodoz S, Guttman M. Long noncoding RNAs: an emerging link between gene regulation and nuclear organization. Trends Cell Biol. 2014;24:651–663. doi: 10.1016/j.tcb.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nwigwe IJ, Kim YJ, Wacker DA, Kim TH. Boundary Associated Long Noncoding RNA Mediates Long-Range Chromosomal Interactions. PloS One. 2015;10:e0136104. doi: 10.1371/journal.pone.0136104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ariel F, Jegu T, Latrasse D, Romero-Barrios N, Christ A, Benhamed M, Crespi M. Noncoding Transcription by Alternative RNA Polymerases Dynamically Regulates an Auxin-Driven Chromatin Loop. Mol Cell. 2014;55:383–396. doi: 10.1016/j.molcel.2014.06.011. [DOI] [PubMed] [Google Scholar]
- 46.Böhmdorfer G, Wierzbicki AT. Control of Chromatin Structure by Long Noncoding RNA. Trends Cell Biol. 2015;25:623–632. doi: 10.1016/j.tcb.2015.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]