Abstract
Torsin ATPases are the only members of the AAA+ ATPase family that localize to the endoplasmic reticulum and contiguous perinuclear space. Accordingly, they are well positioned to perform essential work in these compartments, but their precise functions remain elusive. Recent studies have deciphered an unusual ATPase activation mechanism relying on Torsin-associated transmembrane cofactors, LAP1 or LULL1. These findings profoundly change our molecular view of the Torsin machinery and rationalize several human mutations in TorsinA or LAP1 leading to congenital disorders, symptoms of which have recently been recapitulated in mouse models. Here, we review these recent advances in the Torsin field and discuss the most pressing questions in relation to nuclear envelope dynamics.
Introduction
The four proteins in the Torsin ATPase family are members of the larger superfamily of AAA+ (ATPases associated with a variety of cellular activities) proteins [1]. TorsinA (TorA), the founding member and most studied Torsin ATPase, was first identified by the Breakefield laboratory nearly 20 years ago by a positional cloning approach designed to identify the gene responsible for early-onset DYT1 dystonia, a highly severe movement disorder [2]. This study mapped the disease-causing mutation to an in-frame deletion of a GAG codon resulting in the loss of a glutamate residue near the C-terminus of TorA (Fig. 1) (referred to hereafter as TorA ΔE) [2]. Accumulating evidence suggests that TorA ΔE results in a loss of function, [3-5], but Torsin's precise function is still unknown. Apparent redundancy between Torsins [6,7], early lethality in mouse models until recently [3,6], and previously poorly understood biochemical properties of Torsins have thus far complicated efforts to decipher their function.
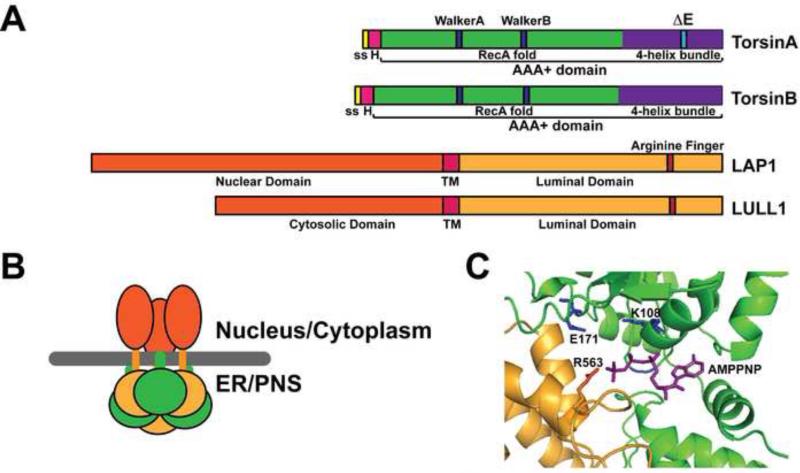
Figure 1. Structural models of Torsins and cofactors.
(A.) Domain organization of TorA, TorB, LAP1 and LULL1. ss: signal sequence; H: hydrophobic domain; ΔE: in-frame glutamate deletion in TorA that leads to DYT1 dystonia; TM: transmembrane domain (B.) Proposed mixed hexameric assembly for Torsin and cofactors. Torsin may form a mixed, alternating ring with either cofactor. Note that the Torsin assembly is highly dynamic and can likely exist in a variety of stoichiometries. Torsin is shown in green, LAP1 or LULL1 are shown in orange, the membrane is shown as grey line. Note that the precise stoichiometry of the Torsin/cofactor assembly may vary and awaits experimental validation (C.) Structural model for the composite active site at the interface of LAP1 (orange) and TorA (green). Key catalytic residues are highlighted. AMPPNP is shown in purple.
Within the AAA+ superfamily, Torsin's primary sequence is most similar to the bacterial Clp proteins [2,8], which have roles as protein unfolding or disassembly machines [9]. While early efforts in Torsin research focused on the hypothesis that Torsin may function similarly in protein quality control [10,11], a wealth of evidence now indicates that Torsins have a critical functional role at the nuclear envelope (NE), as will be discussed below. Recent structural and biochemical data revealed a much more intricate biological assembly for the Torsin core machinery than the homohexameric assembly that was originally assumed [12-14]. Instead, Torsins are essentially inactive in isolation, and strictly require the stimulation of one of two distinctly localizing transmembrane cofactors, LAP1 (lamina associated polypeptide 1) or LULL1 (luminal domain like LAP1) via their highly similar luminal domains (LDs) [4]. LAP1, an inner nuclear membrane (INM) protein, interacts with A- and B-type lamins through its N-terminal nuclear domain [15], while LULL1 localizes to the ER and has an N-terminal domain that projects into the cytoplasm [16] (Fig. 1, 2A). Rather than functioning as peripherally associating activators, these proteins are instead integral members of the Torsin core machinery (Fig. 1B) [5,17], which has significant implications for identifying and interpreting Torsin function or dysfunction. Here, we review what is known about Torsins and their cofactors, with special emphasis on recent structural and biochemical progress, as well as emerging functional implications for these essential but poorly understood ATPases.
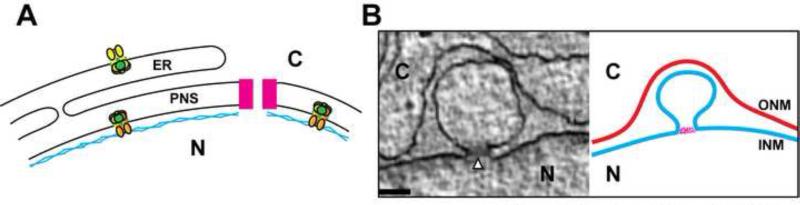
Figure 2. Torsin likely functions at the nuclear envelope.
(A.) Normal nuclear envelope and contiguous endoplasmic reticulum showing the distinct localization of Torsin (green) and cofactors. LULL1 (yellow) may form mixed rings with Torsin in the endoplasmic reticulum, while LAP1 (orange) may form mixed rings with Torsin at the inner nuclear membrane. A nuclear pore complex is shown in pink, the nuclear lamina is depicted in blue. ER: endoplasmic reticulum, PNS: perinuclear space, C: Cytoplasm, N: Nucleus (B.) Left panel: Representative TEM image of blebs seen in Torsin-deficient cells. Right panel: Graphical depiction of the blebbing phenotype. The bleb is continuous with the inner nuclear membrane (cyan). The outer nuclear membrane is red. The electron density seen at the neck (marked by an arrowhead in the left panel) of the blebs is depicted in pink. The scale bar is 100nm. C: Cytoplasm, N: Nucleus. ONM: outer nuclear membrane, INM: inner nuclear membrane.
Biochemical and structural advances
The canonical AAA+ domain consists of conserved sequence and structural motifs that enable ATP binding and hydrolysis. Most conserved is the tertiary structure, which consists of a core wedge-shaped α/ß RecA fold [18] that harbors the ATPase active site, and a C-terminal 4-helix bundle that is involved in oligomerization (Fig. 1) [1]. Key sequence motifs in the AAA+ domain include the WalkerA motif, which features a conserved lysine residue that contributes to ATP binding and the WalkerB motif, which includes a conserved glutamate residue implicated in ATP hydrolysis (corresponding to K108 and E171 for TorsinA, see Fig. 1C) [1]. Most AAA+ ATPases assemble into homohexameric ring-like structures, where the ATPase active site is formed by two adjacent subunits [1]. The WalkerA and WalkerB motifs are present on one subunit, whereas the neighboring subunit typically projects a conserved arginine residue into the active site to enable ATP hydrolysis, in part due to charge neutralization [1].
Since Torsins have significant sequence homology to many homohexameric AAA+ proteins, early efforts to develop a molecular view relied on AAA+ proteins to construct similar structural models of TorA [4,19,20]. Notably, however, Torsins lack the catalytic arginine residue that is otherwise conserved among AAA+ proteins, drawing into question whether Torsins are indeed active ATPases. In fact, when the first comprehensive in vitro analysis of purified Torsins and its previously identified [16] binding partners was conducted, Torsins were found to be completely inactive in isolation [4]. Addition of the LDs of LAP1 or LULL1, which are necessary and sufficient for Torsin binding, strongly induced ATP hydrolysis, with LULL1 being the more potent activator [4]. This stimulatory effect was attributable to a substantial stimulation of the ATP hydrolysis step that was reminiscent of the action of small GTPases by GAPs [21], suggesting that LAP1 and LULL1 serve similarly to regulate ATP hydrolysis [4]. Importantly, this activation mechanism was defective in the presence of the disease-causing TorA ΔE mutation, providing the first link between compromised enzymatic activity and disease etiology [4].
Following up on those experiments, two concurrent, complementary studies have shown that LAP1 and LULL1 are not simply peripherally associating activators but are, quite unexpectedly, core components of the ATPase machinery. This unusual property is made possible by the fact the LDs of LAP1 and LULL1 adopt folds analogous to the RecA domain of AAA+ proteins, allowing them to assume a position that is equivalent to a canonical AAA+ subunit (Fig. 1B) [5,17]. While the WalkerA and WalkerB motifs are not conserved in LAP1 or LULL1, each protein has a strictly conserved arginine residue well positioned to project into the active site of a neighboring Torsin subunit, predicting a critical “activator” interface between Torsin and the cofactor (Fig. 1C) [5,17]. Indeed, when this arginine was mutated in either cofactor, Torsin's cofactor-induced ATPase activity was strongly reduced [5,17] without changing the apparent binding affinity between Torsin and the cofactors [5]. Finally, when subjected to glutaraldehyde crosslinking, TorA and either cofactor formed a higher molecular mass complex consistent with a mixed hexamer of a 3:3 (torsin:cofactor) stoichiometry [5], and ring-shaped assemblies are observed via electron microscopy when activator and Torsin are present at an equimolar ratio [17]. It should be noted, however, that the Torsin/cofactor assembly is highly dynamic and that several, not mutually exclusive stoichiometries are possible. Indeed, it is reasonable to speculate that the dynamic properties are of functional significance in vivo.
What can we learn about the molecular defect underlying DYT1 dystonia? Given that the TorA ΔE mutation perturbs cofactor binding [4,16,20,22] and leads to a strong reduction of site-specific crosslinks at the activator interface [5], one can deduce that the glutamate deletion interferes with an orderly assembly of the activator interface, with a resulting loss of ATPase activity. This interpretation, which requires structural validation, is in close agreement with data obtained in several animal models (see below).
Importantly, these recent findings change our molecular view of the Torsin system in that we have to think about these assemblies as membrane-spanning machines capable of transducing conformational changes across biological membranes. It will thus be important to investigate nucleotide-controlled changes in conformation and stoichiometry by suitable equilibrium measurements and structural methods and to relate these findings to cell biological studies.
Mouse models and human diseases of TorA and LAP1
Early efforts to engineer mouse models of DYT1 dystonia were hampered by the early lethality of TorA and LAP1 knockout (KO) mice [3,6]. Notably, no mouse models of LULL1 or any other Torsin have been reported. However, several important insights into Torsin and cofactor function were gained from studies in several animal models. On a cellular level, the unifying hallmark phenotype that is frequently observed upon Torsin or cofactor manipulation or knockout is a “blebbing” or herniation of the INM into the perinuclear space (PNS) of the nuclear envelope (Fig. 2B). In TorA deficient mice, this phenotype is restricted to neuronal tissues and importantly, the TorA ΔE mutation fails to rescue this phenotype as well as viability [3], in excellent agreement with the aforementioned biochemical data (see above). Interestingly, in the LAP1 KO model, a highly similar but more penetrant blebbing phenotype was observed, including nonneuronal tissues. Murine embryonic fibroblasts (MEFs) from TorA KO mice exhibited normal NEs, but upon siRNA treatment against TorsinB (TorB), these cells also exhibited the blebbing phenotype, establishing functional redundancy between the two Torsins [6]. This proposed functional redundancy is consistent with LAP1's ability to activate both TorsinA and TorsinB [4].
More recently, several conditional mouse models of TorA have enabled new insights into the disease etiology of DYT1 dystonia and Torsin function [23-25]. Mice with a conditional deletion of TorA from the central nervous system (CNS) have an average lifespan of ten days, lose weight progressively, exhibit dystonic symptoms and exhibit gliosis in several sensorimotor regions [23]. Mice with one TorA ΔE allele and one allele with TorA selectively deleted in the CNS (TorAΔE/−) are viable and demonstrate INM blebbing and dystonic symptoms [23]. Interestingly, immunogold labeling from TorA conditional knockout (CKO) mice revealed ubiquitin staining in the lumen of the blebs, suggesting a link –which may be direct or indirect– to cellular protein quality control [23]. Finally, mice in which TorA was selectively deleted in embryonic progenitor cells of cholinergic and GAGAergic neurons in mouse forebrains also exhibit symptoms of dystonia [24]. These recent advances have for the first time recapitulated dystonic symptoms in a viable animal model and should prove invaluable for future studies of TorA and dystonia.
Interestingly, several recent LAP1 mouse models have also implicated this protein in muscular dystrophy and cardiomyopathy [26,27]. LAP1 was found to interact with Emerin [26], an INM protein whose loss of function in humans results in Emery-Dreifuss muscular dystrophy. Mice harboring a CKO of LAP1 in striated muscle develop muscular dystrophy, which is significantly worsened in an Emerin deficient background, even though Emerin KO mice are apparently normal. In addition, a LAP1 CKO from cardiomyocytes results in an increase in expression of cardiomyopathy-related genes and cardiac dysfunction [27]. It remains to be seen whether these LAP1-deficient phenotypes are tied with Torsins or if LAP1 has Torsin-independent functions.
Notably, there have been multiple cases of dystonia correlated with mutations in TorA [28-32] and several recent reports of dystonia, muscular dystrophy, and cardiomyopathy caused by mutations in LAP1 [33,34] (Table 1). The recent biochemical and mouse model advances described above have placed us in a position to rationalize these disease-causing mutations: the majority of these mutations map to or near the activator interface, suggesting that they likewise perturb the Torsin activation mechanism [35]. It will be interesting to test this hypothesis by biochemical means as well as in animal models, which will nurture our understanding of disease etiology.
Table 1.
Diseases Associated with TorA and LAP1
| Year first identified | Mutation | Disease | Reference |
|---|---|---|---|
| 1997 | TorA dE302/303 | Early-onset dystonia | 2 |
| 2001 | TorA dF323-Y328 | Atypical early-onset dystonia | 32 |
| 2008 | TorA R288Q | Early-onset dystonia | 31 |
| 2010 | TorA F205I | Late-onset focal dystonia | 30 |
| 2014 | TorA dA14-P15 | Early-onset dystonia | 28 |
| 2014 | TorA E121K | Early-onset dystonia | 28 |
| 2014 | TorA D194V | Early-onset segmental dystonia | 29 |
| 2014 | LAP1 E482A | Cardiomyopathy, severe dystonia, cerebellar atrophy | 33 |
| 2014 | LAP1 c. 186deiG (p.E62fsTer25) | Muscular dystrophy | 34 |
Functional perspectives: Torsins at the nuclear envelope
Despite the significant biochemical and animal model advances described above, the precise biological function of Torsin and its cofactors remains elusive. Multiple potential functions for Torsin have been put forth in the literature, including roles in ER-associated degradation [36,37] and the HSV-1 life cycle [38,39], among others. Discussing all of these potential functions is beyond the scope of this article, and these studies have been recently reviewed elsewhere [35]. We restrict this discussion to the putative role of Torsins at the NE, since it is here where the most robust phenotypes materialize on a cellular level in a wide range of animal and tissue culture models.
Accumulating evidence suggests that Torsins have a critical role at the NE. First, TorA localizes to the NE in several conditions: TorA ΔE accumulates at the NE in DYT1 patient fibroblasts [40], and a hydrolysis-deficient version of TorA (E171Q) accumulates at the NE [13,16,40]. This NE recruitment seems to be regulated in the ER at least in part by LULL1 [13,41]. As mentioned previously, the hallmark phenotype of TorA- or LAP1-deficient mice is the accumulation of perinuclear blebs at the INM [3,6,23,25]. Structures similar to these blebs have also been observed in Drosophila melanogaster and Caenorhabditis elegans upon Torsin manipulation. D. melanogaster Torsin was found to localize to the necks of large ribonucleoprotein (RNP) granules exiting the nucleus in a newly discovered nuclear pore-independent export pathway [42,43]. Upon siRNA treatment against Torsin, the number of granules with attached necks significantly increased [43]. Thus, Torsins may be required for the scission reaction that is required to pinch off vesicles from the INM (Fig. 3A) [43].
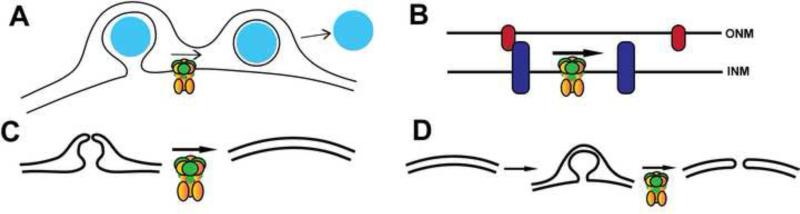
Figure 3. Models for Torsin function at the nuclear envelope.
(A.) Budding pathway through the nuclear envelope that may require Torsin and LAP1. The cargo (cyan) proceeds via a vesicular intermediate from the nucleus, into the perinuclear space and out into the cytoplasm. Torsin is proposed to mediate the scission reaction that releases the cargo into the perinuclear space. (B.) Torsin may mediate the assembly or disassembly of a protein complex in the NE, such as the LINC complex. (C.) Depiction of the nuclear envelope before and after NE reformation following mitosis or nuclear rupture. Torsin may directly or indirectly mediate this reformation. (D.) Torsin may mediate fusion between the inner and outer nuclear membranes during interphase nuclear pore complex assembly. The observed blebbing phenotype may be an otherwise transient intermediate in this assembly. Note that these models are purely hypothetical and not mutually exclusive.
Similarly in OOC-5, the C. elegans Torsin homolog, mutant worms, the nuclei exhibited a blebbing phenotype during development. These worms also displayed mislocalized nuclear pore complex components, a decrease in the rate of nuclear import, and a mislocalization of some linker of nucleoskeleton and cytoskeleton (LINC) complex components [44]. Consistently, observations from tissue culture cells also indicate that Torsin may play a role in regulating the localization of several LINC complex components [7,13,45] (Fig. 3B). These mislocalized proteins may also be an indirect result if Torsin is involved in regulating nuclear envelope reformation after mitosis or nuclear rupture (Fig. 3C).
There is still, however, uncertainty about the composition of the blebs and the role of Torsin in their formation. Since a similar phenotype is observed in several model organisms upon Torsin manipulation and similar structures have been observed in mammalian tissues during development [46-48] this is likely a phenotype that represents a “frozen intermediate” of an otherwise dynamic process. It is noteworthy that the overall dimensions and appearance of the fuzzy electron density at the neck of the blebs (Fig. 2B) is somewhat reminiscent of nuclear pore complexes (NPCs). Given the recently documented nuclear transport defect upon Torsin mutation [44], it will be critical to investigate if and which NPC components reside in those blebs, and if these structures represent NPC assembly intermediates, repurposed NPC components, or disassembly intermediates (Fig. 3D). A compositional analysis of these NE blebs, which needs to include an identification of Ubiquitin conjugates observed in the lumen of these blebs [23], will be important next steps towards relating these morphological observations to a molecular inventory. Knowing the identity of these components, and establishing their functional relationship to the Torsin/cofactor system, will enable us to define whether the observed phenotypes represent direct or indirect consequences of Torsin dysfunction. It is this conundrum that presently defines the central unresolved question in this field.
Conclusions
While much progress has been made since TorA was first discovered nearly twenty years ago, there are still several outstanding questions. Most importantly, it is imperative to define the precise biological function(s) of the Torsin ATPase/cofactor machinery. While Torsin may –not unlike other AAA+ ATPases that are often endowed with impressive functional versatility– have multiple roles throughout the ER, it is clear that Torsins have critical functions for NE integrity. Given that closely related AAA+ ATPases function by investing the energy of ATP hydrolysis to perform work on a substrate protein [1], it seems likely that Torsin targets one or several (membrane) proteins at the NE. However, it is puzzling that the ATPase activity of Torsins is significantly weaker than that of related, processively operating ATPases. This could be attributable to a yet unidentified, missing component that was not included in in vitro studies. Alternatively, Torsins may not perform processive mechanical work but could instead serve as a “timer” or holder chaperone in concert with its distinctively localizing cofactors to achieve tempo-spatial control, for example in the context of cellular transport. Understanding the structure and complex stoichiometry of Torsins and cofactors will enable us to further dissect the intricate regulation of the Torsin ATPase machine and ultimately give insights into how ATPase activation is achieved and regulated in cells.
Given the recent advances of CRISPR/Cas9 system for genome editing and our increasing understanding of the biochemical and structural properties of Torsins, this complex system is now becoming more amenable to decisive experimental attack. We can therefore anticipate a rapidly evolving understanding of this challenging system on a molecular, cellular, and organismal level, with important ramifications for developing targeted treatments for dystonia.
Acknowledgements
We apologize to the authors whose work we could not cite in this review due to size restrictions. We thank members of the C.S. laboratory for review of the manuscript. The authors are funded by NIH (DP2 OD008624-01) and EL is funded by NIH (CMB TG T32GM007223).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interest
The authors report that they have no competing interests.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
*of special interest
**of outstanding interest
- 1.Hanson PI. Whiteheart SW: AAA+ proteins: have engine, will work. Nat Rev Mol Cell Biol. 2005;6:519–529. doi: 10.1038/nrm1684. [DOI] [PubMed] [Google Scholar]
- 2.Ozelius LJ, Hewett JW, Page CE, Bressman SB, Kramer PL, Shalish C, de Leon D, Brin MF, Raymond D, Corey DP, et al. The early-onset torsion dystonia gene (DYT1) encodes an ATP-binding protein. Nat Genet. 1997;17:40–48. doi: 10.1038/ng0997-40. [DOI] [PubMed] [Google Scholar]
- 3.Goodchild RE, Kim CE, Dauer WT. Loss of the dystonia-associated protein torsinA selectively disrupts the neuronal nuclear envelope. Neuron. 2005;48:923–932. doi: 10.1016/j.neuron.2005.11.010. [DOI] [PubMed] [Google Scholar]
- 4*.Zhao C, Brown RS, Chase AR, Eisele MR, Schlieker C. Regulation of Torsin ATPases by LAP1 and LULL1. Proc Natl Acad Sci U S A. 2013;110:E1545–1554. doi: 10.1073/pnas.1300676110. [This paper showed that Torsin ATPases are inactive in isolation and strictly require ATPase activating cofactors LAP1 or LULL1 for activity. It also presents the first biochemical evidence for a loss-of-function mechanisms resulting from the DYT1 dystonia-causing mutation.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5**.Brown RS, Zhao C, Chase AR, Wang J, Schlieker C. The mechanism of Torsin ATPase activation. Proc Natl Acad Sci U S A. 2014;111:E4822–4831. doi: 10.1073/pnas.1415271111. [This paper, which is complementary to reference 17, established that LAP1 and LULL1 are integral members of the Torsin core machinery and activate Torsin via an unusual active site complementation mechanism, which is offset in the disease state. This finding established Torsin as a composite, membrane-spanning machine.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim CE, Perez A, Perkins G, Ellisman MH, Dauer WT. A molecular mechanism underlying the neural-specific defect in torsinA mutant mice. Proc Natl Acad Sci U S A. 2010;107:9861–9866. doi: 10.1073/pnas.0912877107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rose AE, Zhao C, Turner EM, Steyer AM, Schlieker C. Arresting a Torsin ATPase reshapes the endoplasmic reticulum. J Biol Chem. 2014;289:552–564. doi: 10.1074/jbc.M113.515791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ozelius LJ, Page CE, Klein C, Hewett JW, Mineta M, Leung J, Shalish C, Bressman SB, de Leon D, Brin MF, et al. The TOR1A (DYT1) gene family and its role in early onset torsion dystonia. Genomics. 1999;62:377–384. doi: 10.1006/geno.1999.6039. [DOI] [PubMed] [Google Scholar]
- 9.Sauer RT, Baker TA. AAA+ proteases: ATP-fueled machines of protein destruction. Annu Rev Biochem. 2011;80:587–612. doi: 10.1146/annurev-biochem-060408-172623. [DOI] [PubMed] [Google Scholar]
- 10.McLean PJ, Kawamata H, Shariff S, Hewett J, Sharma N, Ueda K, Breakefield XO, Hyman BT. TorsinA and heat shock proteins act as molecular chaperones: suppression of alpha-synuclein aggregation. J Neurochem. 2002;83:846–854. doi: 10.1046/j.1471-4159.2002.01190.x. [DOI] [PubMed] [Google Scholar]
- 11.Caldwell GA, Cao S, Sexton EG, Gelwix CC, Bevel JP, Caldwell KA. Suppression of polyglutamine-induced protein aggregation in Caenorhabditis elegans by torsin proteins. Hum Mol Genet. 2003;12:307–319. doi: 10.1093/hmg/ddg027. [DOI] [PubMed] [Google Scholar]
- 12.Jungwirth M, Dear ML, Brown P, Holbrook K, Goodchild R. Relative tissue expression of homologous torsinB correlates with the neuronal specific importance of DYT1 dystonia-associated torsinA. Hum Mol Genet. 2010;19:888–900. doi: 10.1093/hmg/ddp557. [DOI] [PubMed] [Google Scholar]
- 13.Vander Heyden AB, Naismith TV, Snapp EL, Hodzic D, Hanson PI. LULL1 retargets TorsinA to the nuclear envelope revealing an activity that is impaired by the DYT1 dystonia mutation. Mol Biol Cell. 2009;20:2661–2672. doi: 10.1091/mbc.E09-01-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li H, Wu HC, Liu Z, Zacchi LF, Brodsky JL, Zolkiewski M. Intracellular complexes of the early-onset torsion dystonia-associated AAA+ ATPase TorsinA. Springerplus. 2014;3:743. doi: 10.1186/2193-1801-3-743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Foisner R, Gerace L. Integral membrane proteins of the nuclear envelope interact with lamins and chromosomes, and binding is modulated by mitotic phosphorylation. Cell. 1993;73:1267–1279. doi: 10.1016/0092-8674(93)90355-t. [DOI] [PubMed] [Google Scholar]
- 16.Goodchild RE, Dauer WT. The AAA+ protein torsinA interacts with a conserved domain present in LAP1 and a novel ER protein. J Cell Biol. 2005;168:855–862. doi: 10.1083/jcb.200411026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17**.Sosa BA, Demircioglu FE, Chen JZ, Ingram J, Ploegh HL, Schwartz TU. How lamina-associated polypeptide 1 (LAP1) activates Torsin. Elife. 2014;3:e03239. doi: 10.7554/eLife.03239. [This paper, which is complementary to reference 5, established that LAP1 and LULL1 adopt AAA-like folds and activate Torsin via an unusual active site complementation mechanism, indicating that Torsin is a membrane-spanning machine.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Story RM, Weber IT, Steitz TA. The structure of the E. coli recA protein monomer and polymer. Nature. 1992;355:318–325. doi: 10.1038/355318a0. [DOI] [PubMed] [Google Scholar]
- 19.Zhu L, Wrabl JO, Hayashi AP, Rose LS, Thomas PJ. The torsin-family AAA+ protein OOC-5 contains a critical disulfide adjacent to Sensor-II that couples redox state to nucleotide binding. Mol Biol Cell. 2008;19:3599–3612. doi: 10.1091/mbc.E08-01-0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu L, Millen L, Mendoza JL, Thomas PJ. A unique redox-sensing sensor II motif in TorsinA plays a critical role in nucleotide and partner binding. J Biol Chem. 2010;285:37271–37280. doi: 10.1074/jbc.M110.123471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scheffzek K, Ahmadian MR, Wittinghofer A. GTPase-activating proteins: helping hands to complement an active site. Trends Biochem Sci. 1998;23:257–262. doi: 10.1016/s0968-0004(98)01224-9. [DOI] [PubMed] [Google Scholar]
- 22.Naismith TV, Dalal S, Hanson PI. Interaction of torsinA with its major binding partners is impaired by the dystonia-associated DeltaGAG deletion. J Biol Chem. 2009;284:27866–27874. doi: 10.1074/jbc.M109.020164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23**.Liang CC, Tanabe LM, Jou S, Chi F, Dauer WT. TorsinA hypofunction causes abnormal twisting movements and sensorimotor circuit neurodegeneration. J Clin Invest. 2014;124:3080–3092. doi: 10.1172/JCI72830. [This study established the first viable TorA mouse model that displays dystonic symptoms. These mice also exhibited accumulation of ubiquitin within the lumen of perinuclear blebs, suggesting a link between TorA and nuclear protein quality control.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24*.Pappas SS, Darr K, Holley SM, Cepeda C, Mabrouk OS, Wong JM, LeWitt TM, Paudel R, Houlden H, Kennedy RT, et al. Forebrain deletion of the dystonia protein torsinA causes dystonic-like movements and loss of striatal cholinergic neurons. Elife. 2015;4:e08352. doi: 10.7554/eLife.08352. [This study established a mouse model displaying dystonic symptoms in which TorA is selectively deleted from embryonic progenitor cells of cholinergic and GAGAergic neurons.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weisheit CE, Dauer WT. A novel conditional knock-in approach defines molecular and circuit effects of the DYT1 dystonia mutation. Hum Mol Genet. 2015;24:6459–6472. doi: 10.1093/hmg/ddv355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shin JY, Mendez-Lopez I, Wang Y, Hays AP, Tanji K, Lefkowitch JH, Schulze PC, Worman HJ, Dauer WT. Lamina-associated polypeptide-1 interacts with the muscular dystrophy protein emerin and is essential for skeletal muscle maintenance. Dev Cell. 2013;26:591–603. doi: 10.1016/j.devcel.2013.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shin JY, Le Dour C, Sera F, Iwata S, Homma S, Joseph LC, Morrow JP, Dauer WT, Worman HJ. Depletion of lamina-associated polypeptide 1 from cardiomyocytes causes cardiac dysfunction in mice. Nucleus. 2014;5:260–459. doi: 10.4161/nucl.29227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vulinovic F, Lohmann K, Rakovic A, Capetian P, Alvarez-Fischer D, Schmidt A, Weissbach A, Erogullari A, Kaiser FJ, Wiegers K, et al. Unraveling cellular phenotypes of novel TorsinA/TOR1A mutations. Hum Mutat. 2014;35:1114–1122. doi: 10.1002/humu.22604. [DOI] [PubMed] [Google Scholar]
- 29.Cheng FB, Feng JC, Ma LY, Miao J, Ott T, Wan XH, Grundmann K. Combined occurrence of a novel TOR1A and a THAP1 mutation in primary dystonia. Mov Disord. 2014;29:1079–1083. doi: 10.1002/mds.25921. [DOI] [PubMed] [Google Scholar]
- 30.Calakos N, Patel VD, Gottron M, Wang G, Tran-Viet KN, Brewington D, Beyer JL, Steffens DC, Krishnan RR, Zuchner S. Functional evidence implicating a novel TOR1A mutation in idiopathic, late-onset focal dystonia. J Med Genet. 2010;47:646–650. doi: 10.1136/jmg.2009.072082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zirn B, Grundmann K, Huppke P, Puthenparampil J, Wolburg H, Riess O, Muller U. Novel TOR1A mutation p.Arg288Gln in early-onset dystonia (DYT1). J Neurol Neurosurg Psychiatry. 2008;79:1327–1330. doi: 10.1136/jnnp.2008.148270. [DOI] [PubMed] [Google Scholar]
- 32.Leung JC, Klein C, Friedman J, Vieregge P, Jacobs H, Doheny D, Kamm C, DeLeon D, Pramstaller PP, Penney JB, et al. Novel mutation in the TOR1A (DYT1) gene in atypical early onset dystonia and polymorphisms in dystonia and early onset parkinsonism. Neurogenetics. 2001;3:133–143. doi: 10.1007/s100480100111. [DOI] [PubMed] [Google Scholar]
- 33.Dorboz I, Coutelier M, Bertrand AT, Caberg JH, Elmaleh-Berges M, Laine J, Stevanin G, Bonne G, Boespflug-Tanguy O, Servais L. Severe dystonia, cerebellar atrophy, and cardiomyopathy likely caused by a missense mutation in TOR1AIP1. Orphanet J Rare Dis. 2014;9:174. doi: 10.1186/s13023-014-0174-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kayman-Kurekci G, Talim B, Korkusuz P, Sayar N, Sarioglu T, Oncel I, Sharafi P, Gundesli H, Balci-Hayta B, Purali N, et al. Mutation in TOR1AIP1 encoding LAP1B in a form of muscular dystrophy: a novel gene related to nuclear envelopathies. Neuromuscul Disord. 2014;24:624–633. doi: 10.1016/j.nmd.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 35.Rose AE, Brown RSH, Schlieker C. Torsins: not your typical AAA+ ATPases. Critical Reviews in Biochemistry and Molecular Biology. 2015;1-18 doi: 10.3109/10409238.2015.1091804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen P, Burdette AJ, Porter JC, Ricketts JC, Fox SA, Nery FC, Hewett JW, Berkowitz LA, Breakefield XO, Caldwell KA, et al. The early-onset torsion dystonia-associated protein, torsinA, is a homeostatic regulator of endoplasmic reticulum stress response. Hum Mol Genet. 2010;19:3502–3515. doi: 10.1093/hmg/ddq266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nery FC, Armata IA, Farley JE, Cho JA, Yaqub U, Chen P, da Hora CC, Wang Q, Tagaya M, Klein C, et al. TorsinA participates in endoplasmic reticulum-associated degradation. Nat Commun. 2011;2:393. doi: 10.1038/ncomms1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maric M, Shao J, Ryan RJ, Wong CS, Gonzalez-Alegre P, Roller RJ. A functional role for TorsinA in herpes simplex virus 1 nuclear egress. J Virol. 2011;85:9667–9679. doi: 10.1128/JVI.05314-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Turner EM, Brown RS, Laudermilch E, Tsai PL, Schlieker C. The Torsin Activator LULL1 Is Required for Efficient Growth of Herpes Simplex Virus 1. J Virol. 2015;89:8444–8452. doi: 10.1128/JVI.01143-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goodchild RE, Dauer WT. Mislocalization to the nuclear envelope: an effect of the dystonia-causing torsinA mutation. Proc Natl Acad Sci U S A. 2004;101:847–852. doi: 10.1073/pnas.0304375101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goodchild RE, Buchwalter AL, Naismith TV, Holbrook K, Billion K, Dauer WT, Liang CC, Dear ML, Hanson PI. Access of torsinA to the inner nuclear membrane is activity dependent and regulated in the endoplasmic reticulum. J Cell Sci. 2015;128:2854–2865. doi: 10.1242/jcs.167452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Speese SD, Ashley J, Jokhi V, Nunnari J, Barria R, Li Y, Ataman B, Koon A, Chang YT, Li Q, et al. Nuclear envelope budding enables large ribonucleoprotein particle export during synaptic Wnt signaling. Cell. 2012;149:832–846. doi: 10.1016/j.cell.2012.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43*.Jokhi V, Ashley J, Nunnari J, Noma A, Ito N, Wakabayashi-Ito N, Moore MJ, Budnik V. Torsin mediates primary envelopment of large ribonucleoprotein granules at the nuclear envelope. Cell Rep. 2013;3:988–995. doi: 10.1016/j.celrep.2013.03.015. [This paper described a role for Torsin in a newly discovered nuclear pore- independent, vesicle driven RNA export pathway.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44*.VanGompel MJ, Nguyen KC, Hall DH, Dauer WT, Rose LS. A novel function for the Caenorhabditis elegans torsin OOC-5 in nucleoporin localization and nuclear import. Mol Biol Cell. 2015;26:1752–1763. doi: 10.1091/mbc.E14-07-1239. [This paper established the first connection between Torsins and nucleoporin localization, with defects in nuclear import kinetics upon Torsin manipulation.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nery FC, Zeng J, Niland BP, Hewett J, Farley J, Irimia D, Li Y, Wiche G, Sonnenberg A, Breakefield XO. TorsinA binds the KASH domain of nesprins and participates in linkage between nuclear envelope and cytoskeleton. J Cell Sci. 2008;121:3476–3486. doi: 10.1242/jcs.029454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Trounson A, Sathananthan AH. The application of electron microscopy in the evaluation of two- to four-cell human embryos cultured in vitro for embryo transfer. J In Vitro Fert Embryo Transf. 1984;1:153–165. doi: 10.1007/BF01139208. [DOI] [PubMed] [Google Scholar]
- 47.Szollosi MS, Szollosi D. 'Blebbing' of the nuclear envelope of mouse zygotes, early embryos and hybrid cells. J Cell Sci. 1988;91(Pt 2):257–267. doi: 10.1242/jcs.91.2.257. [DOI] [PubMed] [Google Scholar]
- 48.Kanka J, Fulka J, Jr., Fulka J, Petr J. Nuclear transplantation in bovine embryo: fine structural and autoradiographic studies. Mol Reprod Dev. 1991;29:110–116. doi: 10.1002/mrd.1080290204. [DOI] [PubMed] [Google Scholar]