Abstract
Drug-induced kidney injury (DIKI) is a severe complication in hospitalized patients associated with higher probabilities of developing progressive chronic kidney disease or end-stage renal diseases. Furthermore, DIKI is a problem during preclinical and clinical phases of drug development leading to high rates of project terminations. Understanding the molecular perturbations caused by DIKI would pave the way for a new class of therapeutics to mitigate the damage. Yet, another approach to ameliorate DIKI is identifying sensitive and specific translational biomarkers that outperform the current diagnostic analytes like serum creatinine and facilitate early diagnosis. MicroRNAs (miRNAs), a class of non-coding RNAs, are increasingly being recognized to have a two-pronged approach towards DIKI management: 1) miRNAs have a regulatory role in gene expression and signaling pathways thereby making them novel interventional targets and 2) miRNAs enable diagnosis and prognosis of DIKI because of their stable presence in biofluids. In this review, apart from summarizing the literature on miRNAs in DIKI, we report small RNA sequencing results showing miRNA expression profiles at baseline in normal kidney samples from mice and humans. Additionally, we also compared the miRNA expression in biopsies of normal human kidneys to patients with acute tubular necrosis, and found 76 miRNAs significantly downregulated and 47 miRNAs upregulated (FDR adjusted p<0.05, +/−2-fold change).
In summary, we highlight the transformative potential of miRNAs in therapeutics and translational medicine with a focus on drug-induced kidney damage.
Keywords: microRNA, Kidney, Kidney toxicity, Acute kidney injury, Biomarker, Therapeutic targets
1. Introduction
Drug-induced kidney injury
The high susceptibility of the kidney to toxicity is mainly due to its function of eliminating endogenous waste products as well as xenobiotics. These substances can induce toxic responses due to a high local concentration and/or transformation into reactive metabolites (Kahl, Schmuck, & Vohr, 2010). Commonly prescribed drugs (Table 1) are known to cause acute kidney injury (AKI) that is a severe condition associated with high probabilities of developing progressive chronic kidney disease or end-stage renal diseases, thus leading to high mortality rates (Chawla, Eggers, Star, & Kimmel, 2014). In fact, the incidence of dialysis-requiring AKI increased from 222 to 533 cases per million person-years from 2000 to 2009 in the US (Hsu, McCulloch, Dudley, Lo, & Hsu, 2013). Epidemiological studies show that drugs are the cause of 18–27% of hospitalizations and 19% of intensive care unit patients within the group of AKI patients (Taber & Pasko, 2008; Uchino, et al., 2005). Taking into account that treating patients with end stage renal diseases accounted for over $40 billion in public and private US funds in 2009 (niddk.nih.gov, 2016), drug-induced AKI (DIKI) is a major public health concern. Additionally, DIKI accounts for approximately 10% of the failures in the preclinical and clinical stages (Cook, Hansen, Siu, & Abdul Razak, 2015) thus having a high relevance and a significant economic impact in drug development.
Table 1.
Commonly used drugs with nephrotoxic side effects
| Name | Pharmacological Class | Name | Pharmacological Class |
|---|---|---|---|
| Capreomycin | aminoglycosides antibiotics | 5-Fluoruracil | antineoplasic |
| Gentamicin | aminoglycosides antibiotics | Arsenic Trioxide | antineoplasic |
| Kanamycin | aminoglycosides antibiotics | Camptothecin | antineoplasic |
| Neomycin | aminoglycosides antibiotics | Carmustine | antineoplastic |
| Streptomycin | aminoglycosides antibiotics | Cisplatin | antineoplasic |
| Tobramycin | aminoglycosides antibiotics | Doxorubicin | antineoplasic |
| Acetaminophen | analgesic | Idarubicin | antineoplasic |
| Bacitracin | antibiotics | Mitomycin C | antineoplasic |
| Ciprofloxacin | antibiotics | Paclitaxel | antineoplasic |
| Demeclocycline | antibiotics | Puromycin | antineoplasic |
| Imipenem | antibiotics | Aldesleukin | antineoplasic, immunomudulating |
| Methoxyflurane | analgesic | Ifosfamide | antineoplasic, immunosuppressive |
| Polymyxins | antibiotics | Methotrexate | antineoplastic, antimetabolite, immunosuppressant |
| Rifampicin | antibiotics | Pentamidine | antiprotozoal |
| Streptozocin | antibiotics | Acyclovir | antiviral |
| Sulfamethoxazole | antibiotics | Cidofovir | antiviral |
| Tetracyclines | antibiotics | Foscarnet | antiviral |
| Trimethoprim | antibiotics | Tenofovir | antiviral |
| Vancomycin | antibiotics | Deferoxamine | chelating agent |
| Sulfonamides | antibiotics, anti-diabetics, diuretics | EDTA | chelating agent |
| Pentamidine isethionate | antifungal | Sodium aurothiomalat | immunosuppressive, anti-rheumatic |
| Amphotericin B | antifungal, antiprotozoal | Cyclosporine A | immunsupressive |
| Chlorpropamide | anti-hyperglycemic | Penicillamine | immunsupressive |
| Gallium nitrate | anti-hypercalcemia | Tacrolimus | immunsupressive |
| Pamidronate | anti-hypercalcemia | Lithium | psychiatric medication |
| Aspirin | anti-inflammatory, analgesic, antipyretic | Diatrizoate | radiocontrast |
| Ibuprofen | anti-inflammatory, analgesic, antipyretic | Iodipamide | radiocontrast |
Kidney injury in humans is measured using functional biomarkers like blood urea nitrogen and/or serum creatinine. Although theses biomarkers are considered to be the standard diagnostic analytes in routine care, they are known to be modified by nutrition, muscle mass, age, sex, muscle injury, and aggressive fluid resuscitation (Waikar, Betensky, Emerson, & Bonventre, 2012). Furthermore, they increase only when glomerular filtration rate decreases by more than 50% and they do not reflect dynamic changes in filtration rates (Uchino, 2010). Novel sensitive and specific biomarkers are urgently needed to provide for cost-effective and non-invasive methods of detecting and treating early stage kidney injury. Early diagnostic and predictive biomarkers would also allow for stratification of patients that may be susceptible to develop AKI thereby facilitating clinical trials. Currently, in the absence of any therapeutics for AKI, renal replacement therapy remains the only option (Bellomo, 2015) for severe AKI, leading to an indispensable need for improved kidney injury management, i.e. detection as well as improved therapy.
In the last two decades, due to significant advances in understanding the molecular pathogenesis of AKI using state-of-the-art genome sequencing technologies, microRNAs (miRNAS) have emerged as novel therapeutic targets as well as biomarker candidates for AKI.
microRNA Biogenesis, Function and Extracellular Features
MiRNAs are approximately 20–25 nucleotides long, non-coding and evolutionary conserved small RNAs. MiRNAs were first discovered in C. elegans (R. C. Lee, Feinbaum, & Ambros, 1993; Wightman, Ha, & Ruvkun, 1993) followed by the recognition of their conservation in a wide range of species (Pasquinelli, et al., 2000), leading to the current status of 788 known miRNAs in rats, 1899 in mice and 2585 in humans (miRBase, 2014).
In the cell, miRNAs regulate gene expression at the post-transcriptional level. As part of a ribonucleoprotein complex called miRISC (miRNA-induced silencing complex) they bind to complementary sequences in the 3′-untranslated regions of target mRNAs thus inhibiting mRNA translation. The process of miRNA maturation, miRISC incorporation and subsequent mRNA binding is relatively well explored and reviewed in detail in several review articles (Desvignes, et al., 2015; Garcia-Lopez, Brieno-Enriquez, & Del Mazo, 2013; Krol, Loedige, & Filipowicz, 2010). The complementarity between miRNA and mRNA does not have to be perfect for translational inhibition, therefore one miRNA regulates several hundred mRNAs and likewise, one mRNA is regulated by several miRNAs (Filipowicz, Bhattacharyya, & Sonenberg, 2008). In fact, it is estimated that over 50% of all protein-coding genes are regulated by miRNAs in mammals (Krol, et al., 2010) revealing their overall involvement in diverse physiological as well as pathological processes (Ceman & Saugstad, 2011; T. Li, et al., 2011; Szabo & Bala, 2013; Visone & Croce, 2009; Y. Wang & Lee, 2009; Wiemer, 2007). Many miRNAs are found to be highly enriched in particular organs or at a particular stage of development or disease progression in human body (Kriegel, et al., 2013; Landgraf, et al., 2007) – for instance the liver specific miR-122 (Lagos-Quintana, et al., 2002), kidney cortex enriched miR-192 (Tian, Greene, Pietrusz, Matus, & Liang, 2008), skeletal muscle enriched miR-133a and -b (Sempere, et al., 2004), or the cardiomyocyte specific miR-208a (van Rooij, et al., 2007). The expression of the miR-17~92 cluster, consisting of miR-17, -18a, 19a, -20a, -19b1 and -92a1, seems to be essential for normal nephrogenesis since ablation of the cluster in a mouse model resulted in reduced numbers of nephrons (Marrone, et al., 2014). Furthermore, miR-21 and miR-150 were found highly enriched in kidney cysts of patients with polycystic kidney disease and kidney biopsies from patients with lupus nephritis, respectively (Lakhia, et al., 2015; H. Zhou, et al., 2013).
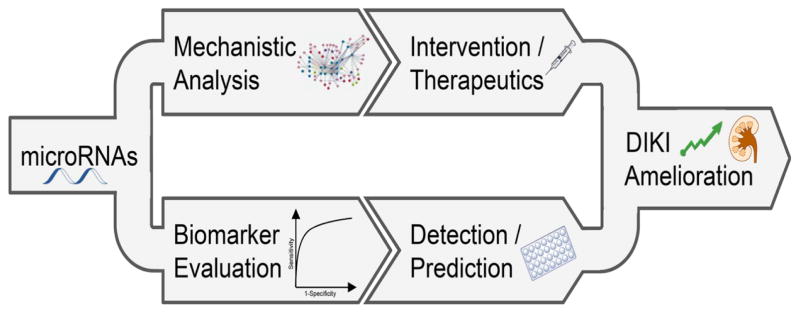
Outside the cell, miRNAs were discovered for the first time in serum/plasma from cancer patients (X. Chen, et al., 2008; Mitchell, et al., 2008) and afterwards in other body fluids like urine, breast milk, saliva and cerebral fluid (Weber, et al., 2010). Extracellular miRNAs are very stable and resistant to degradation even with long-time storage at room temperature, pH variability and multiple freeze-thaw cycles (Y. Li, et al., 2011; J. S. McDonald, Milosevic, Reddi, Grebe, & Algeciras-Schimnich, 2011; Mitchell, et al., 2008; Mraz, Malinova, Mayer, & Pospisilova, 2009). Their stability is probably due to an association with RNA-binding proteins or being packed into vesicles (Arroyo, et al., 2011; Vickers, Palmisano, Shoucri, Shamburek, & Remaley, 2011; Vickers & Remaley, 2012; K. Wang, Zhang, Weber, Baxter, & Galas, 2010; Xu, Yang, & Ai, 2013). The function of extracellular miRNAs is not yet understood and some studies suggest that they might be active signaling molecules (Melo, et al., 2014; Y. Zhang, et al., 2010). However, unique features of extracellular miRNAs have made them promising biomarker candidates. Consequently, due to their intracellular function and their stable presence in biofluids, miRNAs could significantly contribute to improved kidney injury management as both potential biomarkers and interventional targets (Figure 1).
Figure 1. MicroRNAs ameliorate DIKI via a two-pronged approach: identifying interventional strategies and facilitating early diagnosis.
Due to the intracellular function of miRNAs and their stable presence in biofluids, miRNAs could significantly contribute to ameliorate kidney injury outcomes as interventional targets and potential biomarkers.
2. Mechanistic Role of microRNAs in Drug-induced Kidney Injury
MicroRNA Expression in the Kidney
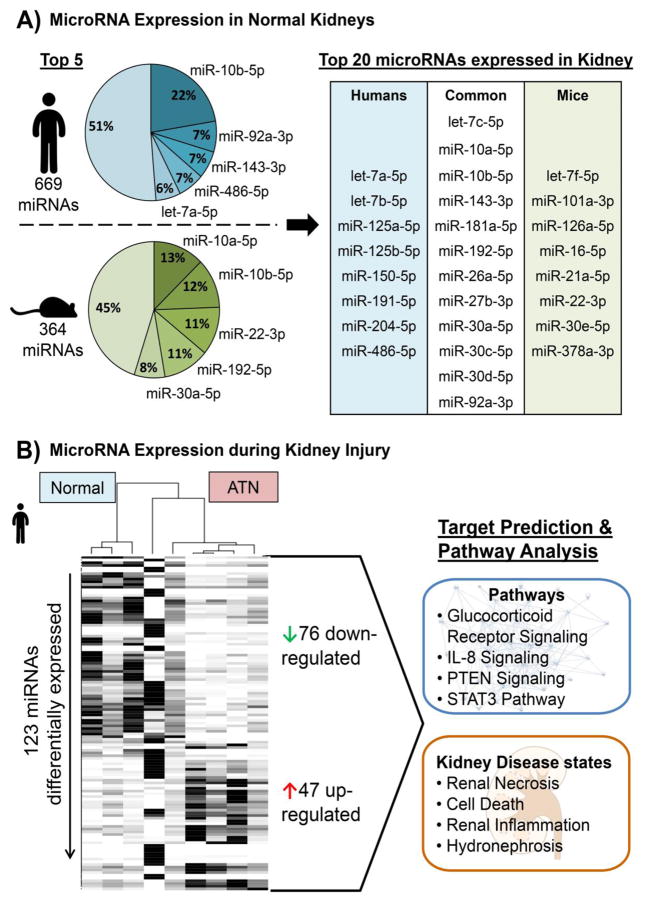
By functioning as regulators of gene expression miRNAs play a crucial role in a variety of molecular processes in multiple organs including the kidney. To gain insight into kidney miRNA expression at baseline, we conducted small RNA sequencing in normal human and mouse kidney samples. After ranking the normalized read counts, the top expressed miRNAs were compared between human and mouse kidneys (Figure 2A). Sixty percent of the top 20 miRNAs were overlapping in human and mouse kidney, of which for example miR-10b was in the top five of both. This confirms the high conservation between species and leads to the assumption that these kidney-enriched miRNAs are involved in normal kidney homeostasis. Less is known about miR-10b in kidney physiology, but deregulation was reported in the context of clear cell renal cell carcinoma and acute allograft rejection (Fritz, Lindgren, Ljungberg, Axelson, & Dahlback, 2014; X. Liu, et al., 2015). However, the role of other kidney-enriched miRNAs like miR-192 or the miR-30 family are better elucidated. MiR-192 targets the β1 subunit of the Na+/K+-ATPase and by inhibiting the expression of this subunit it negatively affects the enzyme activity thereby contributing to renal handling of fluid balance, whereas the miR-30 family is involved in the nephron development and glomerular integrity (Agrawal, Tran, & Wessely, 2009; Mladinov, Liu, Mattson, & Liang, 2013; Wu, et al., 2014). Overall, several studies have demonstrated miRNA involvement in kidney physiology and pathophysiology (Bhatt, Kato, & Natarajan, 2016). When we conducted small RNA sequencing comparing the miRNA expression in biopsies of normal human kidneys to patients with acute tubular necrosis (ATN), we found 76 miRNAs significantly downregulated and 47 miRNAs upregulated (FDR adjusted p<0.05, +/-2-fold change; Figure 2B and Table 2). Using target prediction software, the miRNA expression profile revealed affected pathways in the ATN as well as associated kidney disease states that highlight the importance of miRNA analysis for understanding the pathogenesis of DIKI.
Figure 2. MicroRNA Expression in Healthy and Injured Kidneys.
A) To measure the baseline expression levels of microRNAs in the kidney total RNA was extracted from normal human (n=5) and mice (n=3) kidney samples followed by a small RNA sequencing using the Illumina platform. The normalized mean read counts were ranked by defining read counts >10 as being expressed. The proportions of the top expressed microRNAs were compared within each species as well as between human and mouse and revealed a big overlap of highly abundant microRNAs in the kidneys. B) Total RNA was also extracted and small RNAs were sequenced from kidney samples of patients with acute tubular necrosis (ATN; n=5). Differentially expressed microRNAs between normal and ATN kidney were identified using the DEseq2 algorithm including an adjustment for age and gender of the patients. This resulted in 47 up- and 76 down-regulated microRNAs in ATN vs. normal (FDR adjusted p<0.05 and fold-change cut-off +/− 2). Subjecting the 123 total deregulated microRNAs to a target prediction and pathway analysis utilizing Ingenuity Pathway Analysis demonstrated that these miRNAs are associated with specific pathways as well as kidney disease states.
Table 2. MicroRNA expression changes in kidney injury.
List of 123 differentially expressed microRNAs between normal human kidneys and kidneys from patients with acute tubular necrosis (ATN). Data corresponds to Figure 2B; FDR adjusted p-value.
| 76 downregulated microRNAs | 47 upregulated microRNAs | |||||||
|---|---|---|---|---|---|---|---|---|
| microRNA | fold change | adjusted p-value | microRNA | fold change | adjusted p-value | microRNA | fold change | adjusted p-value |
| miR-124-3p | −13.45 | <0.001 | miR-424-5p | −3.19 | 0.004 | miR-339-5p | 2.03 | 0.0240 |
| miR-206 | −12.90 | <0.001 | miR-204-3p | −3.18 | 0.020 | miR-130b-5p | 2.05 | 0.0159 |
| miR-582-3p | −12.74 | <0.001 | miR-10a-3p | −3.18 | 0.002 | miR-15b-5p | 2.10 | 0.0251 |
| miR-892a | −10.28 | <0.001 | miR-26a-1-3p | −3.16 | 0.049 | miR-574-3p | 2.17 | 0.0355 |
| miR-184 | −9.72 | <0.001 | miR-500b-5p | −3.14 | 0.003 | miR-4742-3p | 2.18 | 0.0283 |
| miR-4482-3p | −9.12 | 0.002 | miR-10b-3p | −3.08 | 0.004 | miR-361-3p | 2.19 | 0.0209 |
| miR-135a-5p | −8.80 | 0.001 | miR-218-5p | −3.06 | 0.028 | miR-3173-5p | 2.26 | 0.0388 |
| miR-20b-5p | −7.58 | <0.001 | miR-10a-5p | −3.01 | 0.005 | miR-125b-1-3p | 2.27 | 0.0390 |
| miR-190a-5p | −7.25 | <0.001 | miR-99a-3p | −2.97 | 0.010 | miR-25-5p | 2.34 | 0.0190 |
| miR-194-5p | −7.19 | <0.001 | miR-548q | −2.90 | 0.048 | miR-5701 | 2.36 | 0.0082 |
| miR-1251-5p | −7.10 | 0.006 | miR-4510 | −2.89 | 0.025 | miR-4301 | 2.36 | 0.0040 |
| miR-891a-5p | −7.09 | 0.001 | miR-30d-3p | −2.88 | 0.020 | miR-423-5p | 2.51 | 0.0235 |
| miR-106a-5p | −7.06 | <0.001 | miR-30b-3p | −2.86 | 0.013 | miR-6747-3p | 2.54 | 0.0090 |
| miR-216b-5p | −6.79 | 0.006 | miR-30a-5p | −2.85 | 0.001 | miR-342-5p | 2.62 | 0.0101 |
| miR-192-5p | −6.63 | 0.001 | miR-30a-3p | −2.82 | 0.029 | miR-7977 | 2.62 | 0.0010 |
| miR-3065-3p | −6.56 | 0.001 | miR-501-5p | −2.78 | 0.001 | miR-663b | 2.64 | 0.0290 |
| miR-9-5p | −6.45 | <0.001 | miR-338-3p | −2.70 | 0.039 | miR-342-3p | 2.70 | 0.0081 |
| miR-6500-3p | −5.71 | 0.012 | miR-629-5p | −2.70 | 0.016 | miR-941 | 2.70 | 0.0355 |
| miR-26a-2-3p | −5.54 | 0.003 | miR-22-5p | −2.63 | 0.016 | miR-330-3p | 2.72 | <0.0019 |
| miR-3065-5p | −5.40 | 0.006 | miR-1270 | −2.60 | 0.034 | miR-4286 | 2.94 | 0.0034 |
| miR-10b-5p | −5.38 | <0.001 | miR-500a-3p | −2.55 | 0.002 | miR-197-5p | 2.96 | 0.0284 |
| miR-362-5p | −5.37 | <0.001 | miR-17-5p | −2.47 | 0.011 | let-7i-3p | 3.00 | 0.0284 |
| miR-187-3p | −5.14 | 0.002 | miR-30e-3p | −2.42 | 0.044 | miR-629-3p | 3.07 | 0.0049 |
| miR-20a-5p | −5.11 | <0.001 | miR-30c-2-3p | −2.40 | 0.033 | miR-642a-3p | 3.15 | 0.0420 |
| miR-577 | −5.07 | 0.003 | miR-6511b-5p | −2.38 | 0.021 | miR-197-3p | 3.17 | 0.0012 |
| miR-1269a | −4.97 | 0.008 | miR-200c-3p | −2.31 | 0.016 | miR-4767 | 3.18 | 0.0156 |
| miR-138-2-3p | −4.93 | 0.016 | miR-664a-5p | −2.19 | 0.044 | miR-4516 | 3.27 | 0.0137 |
| miR-204-5p | −4.90 | 0.001 | miR-152-3p | −2.18 | 0.034 | miR-6812-3p | 3.27 | 0.0165 |
| miR-211-5p | −4.77 | 0.001 | miR-151a-3p | −2.12 | 0.003 | miR-5010-5p | 3.29 | 0.0242 |
| miR-5588-5p | −4.45 | 0.014 | miR-766-3p | 3.51 | <0.0016 | |||
| miR-4461 | −4.33 | 0.014 | miR-6769b-3p | 3.53 | 0.0137 | |||
| let-7f-5p | −4.19 | 0.005 | miR-34b-3p | 3.59 | 0.0110 | |||
| miR-660-5p | −4.18 | 0.001 | miR-935 | 3.66 | 0.0157 | |||
| miR-338-5p | −4.12 | 0.008 | miR-483-3p | 3.69 | 0.0290 | |||
| miR-6723-5p | −4.05 | 0.008 | miR-3960 | 3.71 | 0.0375 | |||
| miR-194-3p | −3.94 | 0.021 | miR-142-3p | 3.96 | 0.0269 | |||
| miR-532-5p | −3.84 | <0.001 | miR-3690 | 3.99 | 0.0206 | |||
| miR-502-5p | −3.71 | 0.006 | miR-150-3p | 4.10 | 0.0027 | |||
| miR-379-5p | −3.67 | 0.029 | miR-4792 | 4.26 | 0.0075 | |||
| miR-26b-5p | −3.67 | 0.008 | miR-150-5p | 4.36 | 0.0034 | |||
| miR-98-5p | −3.61 | 0.002 | miR-3195 | 4.54 | 0.0128 | |||
| miR-138-5p | −3.60 | 0.028 | miR-642a-5p | 5.39 | <0.0017 | |||
| miR-374b-5p | −3.56 | 0.002 | miR-4448 | 6.16 | 0.0027 | |||
| miR-188-5p | −3.43 | 0.008 | miR-3168 | 6.36 | 0.0049 | |||
| let-7e-5p | −3.34 | 0.001 | miR-6731-3p | 6.43 | 0.0017 | |||
| miR-107 | −3.27 | 0.003 | miR-4492 | 6.67 | 0.0030 | |||
| miR-196a-5p | −3.26 | 0.025 | miR-4508 | 7.20 | <0.0019 | |||
In line with these findings as well as based on results from other disciplines, miRNAs have emerged as promising therapeutic targets either by restoring or more commonly by inhibiting their function with synthetic miRNA mimics and anti-miRs, respectively.
Wei and colleagues (2010) showed for the first time the essential involvement of miRNAs in AKI by using a proximal tubule specific Dicer knockout (a miRNA processing enzyme). They reported a differential miRNA expression profile at baseline (80% and 16% of miRNAs were decreased and increased, respectively) due to the knockout which resulted in a significant protection against ischemic AKI (Wei, et al., 2010). The majority of published studies have focused on understanding the regulation of miRNAs during ischemia/reperfusion (I/R)-induced AKI. For example, miR-24 was shown to be increased in mouse kidneys after I/R injury in human allograft biopsies as well as in primary human proximal tubule cells after anoxia/hypoxia (Lorenzen, et al., 2014). Further, heme oxygenase 1 and H2A histone family, member X, were verified as miR-24 targets and silencing miR-24 resulted in decreased injury in cells and in mouse kidneys probably via apoptosis inhibition. Similarly, miR-687 was found to be up-regulated in mouse kidneys after ischemic injury and again anti-miR-687 treatment led to significantly less injury (Bhatt, et al., 2015). PTEN was identified as a direct target and thus blocking of miR-687 preserved PTEN expression and attenuated cell cycle activation and apoptosis.
The potential use of miRNAs for treating kidney injury is an extremely exciting area of ongoing investigation and since there are distinct as well as common mechanisms between different kinds of AKI, results for I/R injury also provide new insights and innovative targets for DIKI.
MicroRNAs in Kidney Toxicity
The involvement of specific miRNAs in DIKI is overall less well explored. The currently known miRNAs with the respective DIKI models are summarized in Table 3. One of the earliest reports showed miR-34a, a p53 target, to be increased in mouse proximal tubular epithelial cells after cisplatin treatment (Bhatt, et al., 2010). Silencing of miR-34a increased cisplatin toxicity leading to the conclusion that miR-34a has a protective role during kidney injury. Although miR-34a was also increased in an I/R mouse model, its inhibition was associated with decreased autophagy and thus aggravated injury (X. J. Liu, et al., 2015). However, miR-155 was shown to be significantly upregulated in rats with kidney injury either induced by gentamicin administration or following I/R and, subsequently, miR-155 KO-mice exhibited significantly enhanced kidney toxicity in response to cisplatin administration (Pellegrini, et al., 2014; Saikumar, et al., 2012). This finding suggests a protective role of miR-155 during DIKI but also an involvement in a common mechanism between I/R and drug-induced AKI. A comparable protective effect was also seen for miR-125b, which was increased in in vivo and in vitro models of cisplatin-induced kidney injury and was suggested to be part of the Nrf2 pathway (Joo, Lee, Koo, & Kim, 2013). There are some published reports that show the contribution of miRNA deregulation in the onset and progression of injury. Treating the immortalized human proximal tubule epithelial cell line HK-2 with cisplatin increased the expression of miR-181a which subsequently inhibited its known target BCL-2 resulting in apoptosis (G. Chen, et al., 2010; Zhu, et al., 2012) and correspondingly less apoptosis was detected when cells were treated with anti-miR-181a. In mouse kidneys and in a rat kidney cell line, miR-122 was decreased after cisplatin, gentamicin and doxorubicin treatment (C. G. Lee, et al., 2014). Since FOXO3 is a verified target of miR-122, FOXO3 stimulated downstream activation of p53 in the absence of miR-122 and resulted in progression of apoptosis and kidney injury. Similar findings were observed in a mouse toxicity model with doxorubicin where miR-133a was increased which in turn was found in other models to directly inhibit the multidrug resistance-associated protein 2 (MRP2; (Loeser, et al., 2015)). MRP2 is one of the numerous transporters in proximal tubule epithelial cells located on the apical membrane and is known to be decreased in injured proximal tubules and thereby further augmenting kidney toxicity (Wen, et al., 2014).
Table 3.
MicroRNAs involved in drug-induced kidney injury (DIKI)
| microRNA | DIKI Models | in vitro Models | in vivo Models | Expression | Upstream Regulator | Targetsa | Proposed Effect | Reference |
|---|---|---|---|---|---|---|---|---|
| miR-122 | Cisplatin, Gentamicin, Doxorubicin | Mouse | Decrease | FOXO3 | p53 Activation | (C. G. Lee, et al., 2014) | ||
| miR-124 | Cyclosporin A | HPTEC | Mouse | Increase | (J. Chen, et al., 2015) | |||
| miR-133a | Doxorubicin | HPTEC c | Mouse, Human kidney biopsies | Increase | ET-1/ ET-B Receptor | MRP2 | Increasing Injury | (Loeser, et al., 2015) |
| miR-146b | Cisplatin | HPTEC | Mousec | Decrease | (Pellegrini, et al., 2015) | |||
| miR-155 | Cisplatin, Gentamicin | Mouse, Rat | Increase | c-Fosb | (Saikumar, et al., 2012) (Pellegrini, et al., 2014) |
|||
| miR-181a | Cisplatin | HK-2 | Increase | BCL-2 | Apoptosis | (Zhu, et al., 2012) | ||
| miR-18a | Cisplatin | HPTEC | Mousec | Decrease | (Pellegrini, et al., 2015) | |||
| miR-21 | Cyclosporin A | HPTEC | Mouse | Increase | AKT/PTEN Pathway | EMT | (J. Chen, et al., 2015) | |
| miR-21 | Gentamicin | Mouse | Increase | (Saikumar, et al., 2012) | ||||
| miR-21 | Gentamicin | Rat | Increase | (Jia, et al., 2013) | ||||
| miR-34a | Cisplatin | BUMPT- 306 | Mouse | Increase | p53 | Cytoprotection | (Bhatt, et al., 2010) | |
| miR-34a | Cisplatin | Rat | Increase | p53b | (Pavkovic, et al., 2014) | |||
| miR-34a | Cisplatin, Gentamicin, Doxorubicin | Mouse | Increase | Sirt1b | p53 Activation via FOXO3 Acetylation | (C. G. Lee, et al., 2014) | ||
| miR-494 | Cyclosporin A | HK-2 | Mouse | Increase | PTEN | EMT | (Yuan, et al., 2015) |
in the context of DIKI;
no direct interaction shown in this study;
kidney injury model but not DIKI; EMT, Epithelial-Mesechymal Transition
Besides the kidney toxicity of cytostatic drugs including cisplatin or doxorubicin and aminoglycoside antibiotics like gentamicin, miRNAs have been studied in cyclosporine A toxicity. Cyclosporine A is an immunosuppressive agent commonly given to transplant or autoimmune disease patients but it is known for its long-term kidney toxicity characterized by severe renal tubulointerstitial fibrosis. Treating human proximal tubular epithelial cells in vitro with cyclosporine A deregulated 46 miRNAs (J. Chen, Zmijewska, Zhi, & Mannon, 2015). One of the few increased miRNAs was miR-21 (~5.5-fold), which is widely explored in the context of kidney disease and injury (Li, et al., 2013; T. B. Zhou & Jiang, 2014). In the cyclosporine A model, miR-21 up-regulation was associated with AKT activation, PTEN decrease and the increase of several markers of epithelial-mesenchymal transition (EMT) including vimentin and αsmooth muscle actin. Epithelial to mesenchymal transition has been shown to play an important role in kidney fibrosis (Lovisa, et al., 2015) and also in the context of cyclosporine A toxicity (Slattery, Campbell, McMorrow, & Ryan, 2005). These results correspond with a therapeutic study on kidney fibrosis, demonstrating that inhibition of miR-21 was protective against TGF-β-induced fibrogenesis in a mouse model of Alport nephropathy (Gomez, et al., 2015). Further, Yuan et al. (2015) found another miRNA, miR-494, to be involved in cyclosporine A induced EMT (Yuan, Benway, Bagley, & Iacomini, 2015). MiR-494 was increased approximately 2-fold in mouse kidneys as well as in HK-2 cells after cyclosporine A treatment. Again a PTEN decrease was observed (~4-fold), which was identified as a direct target of miR-494. Counteracting a PTEN inhibition by using anti-miR-494 prevented cyclosporine A induced EMT.
In summary, the further exploration of the miRNA role in the pathogenesis of DIKI could lead to the development of miRNA-based therapeutics and this seems promising based on the current findings and results seen in other related disciplines.
3. MicroRNAs as Biomarkers for Drug-induced Kidney Injury
Biomarker Candidates
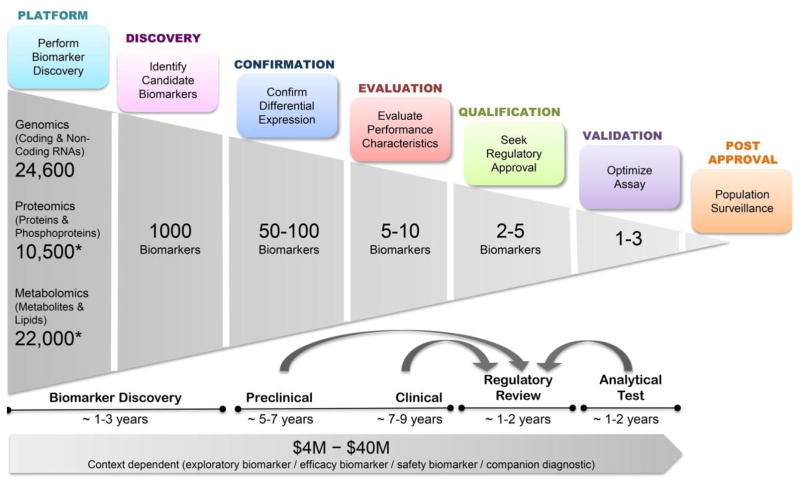
With the 21st century advances in omics technologies, biomarker science has emerged as a very exciting multidisciplinary approach to understand and classify disease pathogenesis. However, the biomarker science pipeline that involves carrying the biomarker from discovery to confirmation, evaluation, qualification and validation steps requires a significant commitment of resources and time (Figure 3). Based on the platform of choice for a biomarker discovery effort, starting with >10,000 candidates, one could identify a handful of successfully qualified and validated biomarkers that enables improved patient care as well as a more efficient drug development. Over a decade ago, this path was formed and successfully taken, culminating in 2008 with a qualification of seven urinary protein biomarkers by the US Food and Drug Administration (FDA) and European Medicines Agency for the assessment of DIKI in preclinical studies. In spite of this breakthrough work, implementation of these pre-clinically qualified biomarkers into clinical practice is still awaited (Dieterle, et al., 2010; EMA, 2009; Murray, et al., 2014). Protein-based biomarker evaluation is often challenging because of the high diversity of proteins and their post-translational modifications and the resulting issues in assay development.
Figure 3. Biomarker Development Pipeline.
The overview is based on estimations and the experiences gained during the Critical Path’s Predictive Safety Testing Consortium’s work on protein biomarkers for kidney injury. *Dependent on platforms used as well as type of biospecimen.
On the contrary, referring to the FDA’s description of an ideal biomarker (Wallace, et al., 2001), extracellular miRNAs fit all the criteria: 1) easy accessibility in diverse body fluids, 2) stability, 3) conservation across species, 4) association with particular tissues or pathological states and 5) a sensitive measurement method. Due to these unique features miRNAs have shown great promise as non-invasive biomarkers. They were detected in almost all body fluids including the clinically most relevant blood and urine (Weber, et al., 2010). Although high concentrations of RNA degrading enzymes are present in the extracellular space, miRNAs are found to be remarkably stable (McDonald, Milosevic, Reddi, Grebe, & Algeciras-Schimnich, 2011), which is probably due to their packing into microvesicles and exosomes or their association with proteins and high-density lipoproteins (Arroyo, et al., 2011; Hoy & Buck, 2012; Turchinovich, Weiz, Langheinz, & Burwinkel, 2011). Furthermore, miRNAs not only show organ specificity many times but are also highly conserved in sequence across species (Landgraf, et al. 2007; Sun, et al. 2004). Commonly, they are measured by real-time PCR which is a sensitive and well-established method for nucleic acids. To date, several hundred studies have evaluated the potential of miRNAs as biomarkers for various pathological conditions including cancers, cardiovascular and neurodegenerative diseases. For circulating miRNAs i.e. miRNAs from blood, only, 35 different clinical studies are currently registered assessing their performance as biomarkers for human diseases (clinicaltrials.gov, 2015); several of these are also associated with kidney diseases like autosomal dominant polycystic kidney disease, renal cell carcinoma and AKI after cardiac surgery.
In terms of kidney toxicity biomarkers, work with miRNAs has thus far been focused on urine as it is non-invasive, directly derived from kidneys and has been shown to contain miRNAs. Our laboratory was amongst the first few laboratories to demonstrate the isolation of miRNAs from urinary supernatants and showed the differential expression of miR-21 and miR-155 in the urines of rats with AKI or gentamicin-induced AKI (Saikumar, et al., 2012). These results were confirmed by a precompetitive consortium of pharmaceutical industries, ILSI Health and Environmental Sciences Institute. In spite of using a different cisplatin doses, fasting/ feeding conditions and different rat strains, approximately 20 miRNA candidates were found to be the same in both studies (Kanki, et al., 2014; Pavkovic, Riefke, & Ellinger-Ziegelbauer, 2014). Increases of specific miRNAs were also measured in urine from rats treated with gentamicin or doxorubicin (Church, et al., 2014; Nassirpour, et al., 2014). All miRNA levels correlated with histopathological changes as well as the qualified protein biomarker Kim-1 and were increased before serum creatinine and blood urea nitrogen. Almost all these studies were conducted in rats, (Table 4), but for example, miR-21 was also found in urine from human AKI patients from the intensive care unit (Ramachandran, et al., 2013) as well as in patients with acetaminophen or cisplatin induced AKI (Pavkovic, Robinson-Cohen, et al., 2015) thereby strengthening the translational potential of miRNAs in DIKI settings. A recent study evaluated miRNAs in plasma as biomarkers for contrast-induced kidney injury (Gutierrez-Escolano, Santacruz-Vazquez, & Gomez-Perez, 2015). First, the miR-30 family (miR-30a, -c, and -e) was found to be increased in rat plasma after the administration of contrast agent and these results were validated in a patient cohort where especially miR-30a performed very well in differentiating patients with contrast-induced nephropathy from those without. Even though the number and diversity of miRNA biomarker studies specifically for DIKI are small, the data is promising and results from other kidney diseases including immunoglobulin A nephropathy, ischemic AKI, glomerulonephritis or focal segmental glomerulosclerosis (Ichii, et al., 2014; Pavkovic, Riefke, Frisk, Groticke, & Ellinger-Ziegelbauer, 2015; G. Wang, et al., 2011; J. F. Wang, et al., 2014; W. Zhang, et al., 2014) support the value of further exploration.
Table 4.
MicroRNA biomarker candidates for drug-induced kidney injury (DIKI)
| microRNARNA | DIKI (Model) | Species | Biofluid | Direction | Reference |
|---|---|---|---|---|---|
| let-7g, miR-93, -191, -195 | Cisplatin | Rat | Urine | UP | (Kanki, et al., 2014) |
| miR-15, -16, -20a, -192, -193, -210 | Cisplatin | Rat | Urine | UP | (Pavkovic, et al., 2014) |
| miR-21, -200c, -423 | Cisplatin, Acetaminophen | Patients | Urine | UP | (Pavkovic, Robinson- Cohen, et al., 2015) |
| miR-203a, let-7d | Gentamicin | Rat | Urine | DOWN | (Nassirpour, et al., 2014) |
| miR-21, miR-155 | Gentamicin | Rat | Urine | DOWN | (Saikumar, et al., 2012) |
| miR-30a,c,e | Contrast agent* | Rat Patients | Plasma | UP | (Gutierrez-Escolano, et al., 2015) |
| miR-320 | Gentamicin | Rat | Urine | UP | (Nassirpour, et al., 2014) |
| miR-34c | Doxorubicin | Rat | Urine | UP | (Church, et al., 2014) |
iohexhol + furosemide + indomethacin were used for rats
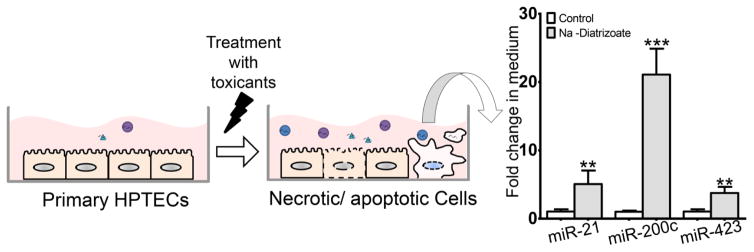
In addition, extracellular miRNA biomarkers could have more advantages in drug development. Since they are conserved and ubiquitously expressed, they could not only be implemented in preclinical and clinical studies but also in vitro studies for screening of compounds with potential toxic effects. The presence of miRNAs in cell culture medium was mostly demonstrated while exploring their potential role in inter-cellular communication (M. K. McDonald, Capasso, & Ajit, 2013; Rani, 2014; Y. Zhou, et al., 2014). However, assuming that miRNAs can be actively as well as passively released from injured kidney cells, cell culture media of treated and untreated cells could have different miRNA profiles. We had previously found miR-21, -200c and -423 to be increased in patients with AKI and here we show that when primary human proximal tubular epithelial cells are treated with the contrast agent sodium diatrizoate all three miRNAs, miR-21, -200c and -423 significantly increased in medium of cells (Figure 5). Furthermore, due to their intracellular function, extracellular miRNAs could mirror the events taking place in the injured kidney, thus being not only indicative of the injury itself but also of the affected pathways. For instance, cisplatin induces miR-34a expression in rat kidneys which itself is involved in the p53-mediated apoptosis pathway and increased miR-34a levels were also measured in urine of the same cisplatin treated rats (Pavkovic, et al., 2014).
All findings and hypotheses around miRNA biomarkers for kidney toxicity seem promising and valuable; nonetheless more replication and validation of the biomarkers are needed in large multi-centered cohorts to confirm the reproducibility and performance.
Challenges of microRNAs as Biomarkers
Despite all the promising results with miRNA biomarkers it is worth mentioning that the field is facing several basic challenges that hinder a smooth process of biomarker qualification and validation.
One of the biggest challenges in clinical miRNA biomarker studies is the comparison with serum creatinine (SCr) as a gold standard definition for acute kidney injury. Although in preclinical studies renal histopathological examination is the gold standard for DIKI diagnosis, in clinical assessments SCr remains widely used. In fact, moderate performances of new AKI biomarker candidates are frequently seen in clinical studies, where AKI is mostly defined based on increased SCr levels (Pavkovic, Robinson-Cohen, et al., 2015). A potential solution for DIKI studies could be to use the treatment with the nephrotoxic drug per se for comparison.
Yet, another challenge is the lack of standardization in not only miRNA isolation and measurement approaches but also sample collection, handling and storage conditions. The most common methods of miRNA isolation are phenol/chloroform-based techniques including silica column purification, but these have also been shown to vary greatly depending on the vendor (El-Khoury, Pierson, Kaoma, Bernardin, & Berchem, 2016; Martinez-Fernandez, Paramio, & Duenas, 2016). In terms of miRNA measurement, qRT-PCR is widely used especially from biospecimens having overall low RNA yields like serum or urine. The issue here is the absence of generally agreed endogenous control miRNAs. The reported normalization strategies differ greatly, including normalization with 1.) synthetic miRNAs that were spiked in during RNA isolation (Argyropoulos, et al., 2013; J. F. Wang, et al., 2010), 2.) small RNAs like RNU48 or U6 (G. Wang, et al., 2012; N. Wang, et al., 2012) or 3.) invariant miRNAs identified within the specific study (Hanke, et al., 2010; Yang, et al., 2012). All three approaches have disadvantages: normalization to spike-ins does not account for a true biological variability but rather accounts for a technical variation; normalizing to small RNAs could bias the results because the small RNA itself could have been differentially expressed during injury or the isolation and transcription efficiency is different for small RNAs vs. miRNAs (e.g. U6 ~106 nucleotides vs. miRNAs 20–25 nucleotides); finally the use of invariant miRNAs for normalization, although popular, seems not to be universal and only limited to specific studies or even specific datasets. Furthermore, urine as a biospecimen has its own challenges regarding normalization, which is necessary to account for variations in urine flow rate/concentrations due to hydration or diuresis. Yet, urine is very interesting since it is directly associated with the kidneys and thereby serves as a relevant and most-proximal non-invasive matrix to perform miRNA biomarker discovery for AKI. Normalizing biomarker levels with urinary creatinine as is usually done for protein biomarkers was suggested in a recent rat study (Pavkovic, et al., 2014). This approach, however, has several limitations due to potential changes of urinary creatinine induced by factors like variations in diurnal production, physical activity, diet, muscle mass (Greenblatt, et al., 1976; Heymsfield, Arteaga, McManus, Smith, & Moffitt, 1983; Waikar, Sabbisetti, & Bonventre, 2010). Furthermore, simulations on creatinine kinetics revealed that protein biomarker performance is actually affected by this way of normalization (Waikar, et al., 2010) which could also be the case for miRNAs.
In general the harmonization of miRNA biomarker evaluation is important to enable the establishment of reference ranges accounting for potential differences in strain, species, fasting/feeding conditions in preclinical studies and age, sex, ethnicity, comorbid conditions in clinical studies. Expression analyses in kidneys during the life span of F344 rats have shown that miRNA levels were sex and age dependent (Kwekel, Vijay, Desai, Moland, & Fuscoe, 2015), therefore also miRNA levels in biofluids could be affected by these factors. Thus, a systematic effort towards standardization could accelerate qualification of these miRNAs and improve the interpretation of miRNA biomarker candidates from different studies.
4. Conclusion
DIKI is still a large problem in clinical as well as non-clinical settings. The detection of DIKI is hindered by poorly performing standard diagnostic analytes and once AKI is diagnosed therapeutic approaches are nearly nonexistent. On the one hand, miRNAs regulate over 50% of all protein-coding genes at the post-transcriptional level. Thus miRNAs are involved in almost all physiological as well as pathological processes, which make them interesting therapeutic targets. On the other hand, extracellular miRNAs are remarkably stable and found to be valuable biomarker candidates in diverse disease settings. Therefore, due to the intracellular function of miRNAs and their presence in biofluids, miRNAs could significantly contribute to improved management of DIKI as interventional targets but also as potential biomarkers. Positive results from other disciplines and the limited yet promising data in the field of kidney medicine hold great promise for a successful application of miRNA-based interventions in the context of DIKI.
Figure 4. MicroRNAs as Biomarkers for in vitro Toxicity Testing.
Normal cells have been shown to release miRNA-containing vesicles. Likewise, injured cells also release miRNAs actively as well as passively (apoptosis/necrosis) which then can be measured in different biofluids. In in vitro studies, this would correspond to miRNA profiles in the supernatant of untreated and treated cultured cells. Differential levels could enable an evaluation of these extracellular miRNAs as biomarker candidates for screening of potential kidney toxic agents. To test this hypothesis, primary human proximal tubular epithelial cells (HPTECs) were cultured and treated with the contrast agent sodium diatrizoate (200 mM) for 24h. Total RNA was isolated from 200 μl medium and miR-21, -200c and -423 were measured by qRT-PCR as these miRNAs have been reported to increase following kidney injury in human urines (Ramachandran, et al., 2013). All three miRNAs were found to be significantly increased in the medium of treated HPTECs as compared to untreated cells. Data is presented as mean fold changes with standard deviation (n=4) relative to the untreated cells (2−ΔΔCt with internal reference). 1-way ANOVA with Dunnett’s test was used for p-value calculation: ** p<0.01 and *** p<0.001.
Acknowledgments
MP is a recipient of a research fellowship from the Deutsche Forschungsgesellschaft (DFG). We thank Cory Gerlach for scientific and linguistic support. Work in the Vaidya laboratory was supported by Outstanding New Environmental Sciences (ONES) award from NIH/NIEHS (ES017543), Innovation in Regulatory Science Award from Burroughs Wellcome Fund (BWF-1012518) and a collaborative research agreement with Biogen (A24378).
Abbreviations
- AKI
acute kidney injury
- ATN
acute tubular necrosis
- DIKI
drug-induced kidney injury
- EMT
epithelial to mesenchymal transition
- FDR
false discovery rate
- I/R
ischemia/reperfusion
- miRNA
microRNA
- qRT-PCR
quantitative real time PCR
Footnotes
Conflict of Interest
The authors declare that there are no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agrawal R, Tran U, Wessely O. The miR-30 miRNA family regulates Xenopus pronephros development and targets the transcription factor Xlim1/Lhx1. Development. 2009;136:3927–3936. doi: 10.1242/dev.037432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argyropoulos C, Wang K, McClarty S, Huang D, Bernardo J, Ellis D, Orchard T, Galas D, Johnson J. Urinary microRNA profiling in the nephropathy of type 1 diabetes. PLoS One. 2013;8:e54662. doi: 10.1371/journal.pone.0054662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, Gibson DF, Mitchell PS, Bennett CF, Pogosova-Agadjanyan EL, Stirewalt DL, Tait JF, Tewari M. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci U S A. 2011;108:5003–5008. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellomo R. Decade in review--acute kidney injury: Acute kidney injury--a decade of progress. Nat Rev Nephrol. 2015;11:636–637. doi: 10.1038/nrneph.2015.147. [DOI] [PubMed] [Google Scholar]
- Bhatt K, Kato M, Natarajan R. Mini-review: emerging roles of microRNAs in the pathophysiology of renal diseases. Am J Physiol Renal Physiol. 2016;310:F109–118. doi: 10.1152/ajprenal.00387.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatt K, Wei Q, Pabla N, Dong G, Mi QS, Liang M, Mei C, Dong Z. MicroRNA-687 Induced by Hypoxia-Inducible Factor-1 Targets Phosphatase and Tensin Homolog in Renal Ischemia-Reperfusion Injury. J Am Soc Nephrol. 2015;26:1588–1596. doi: 10.1681/ASN.2014050463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatt K, Zhou L, Mi QS, Huang S, She JX, Dong Z. MicroRNA-34a is induced via p53 during cisplatin nephrotoxicity and contributes to cell survival. Mol Med. 2010;16:409–416. doi: 10.2119/molmed.2010.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceman S, Saugstad J. MicroRNAs: Meta-controllers of gene expression in synaptic activity emerge as genetic and diagnostic markers of human disease. Pharmacol Ther. 2011;130:26–37. doi: 10.1016/j.pharmthera.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chawla LS, Eggers PW, Star RA, Kimmel PL. Acute kidney injury and chronic kidney disease as interconnected syndromes. N Engl J Med. 2014;371:58–66. doi: 10.1056/NEJMra1214243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Zhu W, Shi D, Lv L, Zhang C, Liu P, Hu W. MicroRNA-181a sensitizes human malignant glioma U87MG cells to radiation by targeting Bcl-2. Oncol Rep. 2010;23:997–1003. doi: 10.3892/or_00000725. [DOI] [PubMed] [Google Scholar]
- Chen J, Zmijewska A, Zhi D, Mannon RB. Cyclosporine-mediated allograft fibrosis is associated with micro-RNA-21 through AKT signaling. Transpl Int. 2015;28:232–245. doi: 10.1111/tri.12471. [DOI] [PubMed] [Google Scholar]
- Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, Guo J, Zhang Y, Chen J, Guo X, Li Q, Li X, Wang W, Wang J, Jiang X, Xiang Y, Xu C, Zheng P, Zhang J, Li R, Zhang H, Shang X, Gong T, Ning G, Zen K, Zhang CY. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- Church RJ, McDuffie JE, Sonee M, Otieno M, Ma JY, Liu X, Watkinsa PB, Harrilla AH. MicroRNA-34c-3p is an early predictive biomarker for doxorubicin-induced glomerular injury progression in male Sprague-Dawley rats. Toxicology Research. 2014;3:384–394. [Google Scholar]
- clinicaltrials.gov. A service of the US National Institutes of Health. 2015. [Google Scholar]
- Cook N, Hansen AR, Siu LL, Abdul Razak AR. Early phase clinical trials to identify optimal dosing and safety. Mol Oncol. 2015;9:997–1007. doi: 10.1016/j.molonc.2014.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desvignes T, Batzel P, Berezikov E, Eilbeck K, Eppig JT, McAndrews MS, Singer A, Postlethwait JH. miRNA Nomenclature: A View Incorporating Genetic Origins, Biosynthetic Pathways, and Sequence Variants. Trends Genet. 2015;31:613–626. doi: 10.1016/j.tig.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieterle F, Perentes E, Cordier A, Roth DR, Verdes P, Grenet O, Pantano S, Moulin P, Wahl D, Mahl A, End P, Steadler F, Legay F, Carl K, Laurie D, Chibout S, Vonderscher J, Maurer G. Qualification of urinary clusterin to detect drug-induced tubular injury and of urinary cystatin C, β2-microglobulin and total protein to detect drug-induced glomerular injury. Nat Biotechnol. 2010;28 doi: 10.1038/nbt.1622. In Press. [DOI] [PubMed] [Google Scholar]
- El-Khoury V, Pierson S, Kaoma T, Bernardin F, Berchem G. Assessing cellular and circulating miRNA recovery: the impact of the RNA isolation method and the quantity of input material. Sci Rep. 2016;6:19529. doi: 10.1038/srep19529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EMA. Final conclusions of the pilot joint EMEA/FDA VXDA experience on qualification of nephrotoxicity biomarkers. 2009 www.emea.europa.eu, Doc.ref. EMEA/679719/2008 Rev. 1.
- Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nature reviews Genetics. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- Fritz HK, Lindgren D, Ljungberg B, Axelson H, Dahlback B. The miR(21/10b) ratio as a prognostic marker in clear cell renal cell carcinoma. Eur J Cancer. 2014;50:1758–1765. doi: 10.1016/j.ejca.2014.03.281. [DOI] [PubMed] [Google Scholar]
- Garcia-Lopez J, Brieno-Enriquez MA, Del Mazo J. MicroRNA biogenesis and variability. Biomol Concepts. 2013;4:367–380. doi: 10.1515/bmc-2013-0015. [DOI] [PubMed] [Google Scholar]
- Gomez IG, MacKenna DA, Johnson BG, Kaimal V, Roach AM, Ren S, Nakagawa N, Xin C, Newitt R, Pandya S, Xia TH, Liu X, Borza DB, Grafals M, Shankland SJ, Himmelfarb J, Portilla D, Liu S, Chau BN, Duffield JS. Anti-microRNA-21 oligonucleotides prevent Alport nephropathy progression by stimulating metabolic pathways. J Clin Invest. 2015;125:141–156. doi: 10.1172/JCI75852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenblatt DJ, Ransil BJ, Harmatz JS, Smith TW, Duhme DW, Koch-Weser J. Variability of 24-hour urinary creatinine excretion by normal subjects. J Clin Pharmacol. 1976;16:321–328. doi: 10.1002/j.1552-4604.1976.tb01527.x. [DOI] [PubMed] [Google Scholar]
- Gutierrez-Escolano A, Santacruz-Vazquez E, Gomez-Perez F. Dysregulated microRNAs involved in contrast-induced acute kidney injury in rat and human. Ren Fail. 2015;37:1498–1506. doi: 10.3109/0886022X.2015.1077322. [DOI] [PubMed] [Google Scholar]
- Hanke M, Hoefig K, Merz H, Feller AC, Kausch I, Jocham D, Warnecke JM, Sczakiel G. A robust methodology to study urine microRNA as tumor marker: microRNA-126 and microRNA-182 are related to urinary bladder cancer. Urol Oncol. 2010;28:655–661. doi: 10.1016/j.urolonc.2009.01.027. [DOI] [PubMed] [Google Scholar]
- Heymsfield SB, Arteaga C, McManus C, Smith J, Moffitt S. Measurement of muscle mass in humans: validity of the 24-hour urinary creatinine method. Am J Clin Nutr. 1983;37:478–494. doi: 10.1093/ajcn/37.3.478. [DOI] [PubMed] [Google Scholar]
- Hoy AM, Buck AH. Extracellular small RNAs: what, where, why? Biochem Soc Trans. 2012;40:886–890. doi: 10.1042/BST20120019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu R, McCulloch C, Dudley R, Lo L, Hsu CY. Temporal Changes in Incidence of Dialysis-Requiring AKI. Journal of the American Society of Nephrology : JASN. 2013;24:37–42. doi: 10.1681/ASN.2012080800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichii O, Otsuka-Kanazawa S, Horino T, Kimura J, Nakamura T, Matsumoto M, Toi M, Kon Y. Decreased miR-26a expression correlates with the progression of podocyte injury in autoimmune glomerulonephritis. PLoS One. 2014;9:e110383. doi: 10.1371/journal.pone.0110383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia P, Teng J, Zou J, Fang Y, Zhang X, Bosnjak ZJ, Liang M, Ding X. miR-21 Contributes to Xenon-conferred Amelioration of Renal Ischemia-Reperfusion Injury in Mice. Anesthesiology. 2013 doi: 10.1097/ALN.0b013e318298e5f1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo MS, Lee CG, Koo JH, Kim SG. miR-125b transcriptionally increased by Nrf2 inhibits AhR repressor, which protects kidney from cisplatin-induced injury. Cell Death Dis. 2013;4:e899. doi: 10.1038/cddis.2013.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahl R, Schmuck G, Vohr H-W. Toxikologie der Organe und Organsysteme. In: Vohr H-W, editor. Toxikologie Band 1: Grundlagen der Toxikologie. 1. Weinheim: WILEY-VCH Verlag; 2010. pp. 95–106. [Google Scholar]
- Kanki M, Moriguchi A, Sasaki D, Mitori H, Yamada A, Unami A, Miyamae Y. Identification of urinary miRNA biomarkers for detecting cisplatin-induced proximal tubular injury in rats. Toxicology. 2014;324:158–168. doi: 10.1016/j.tox.2014.05.004. [DOI] [PubMed] [Google Scholar]
- Kriegel AJ, Liu Y, Liu P, Baker MA, Hodges MR, Hua X, Liang M. Characteristics of microRNAs enriched in specific cell types and primary tissue types in solid organs. Physiol Genomics. 2013;45:1144–1156. doi: 10.1152/physiolgenomics.00090.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nature reviews Genetics. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- Kwekel JC, Vijay V, Desai VG, Moland CL, Fuscoe JC. Age and sex differences in kidney microRNA expression during the life span of F344 rats. Biol Sex Differ. 2015;6:1. doi: 10.1186/s13293-014-0019-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- Lakhia R, Hajarnis S, Williams D, Aboudehen K, Yheskel M, Xing C, Hatley ME, Torres VE, Wallace DP, Patel V. MicroRNA-21 Aggravates Cyst Growth in a Model of Polycystic Kidney Disease. J Am Soc Nephrol. 2015 doi: 10.1681/ASN.2015060634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, Lin C, Socci ND, Hermida L, Fulci V, Chiaretti S, Foa R, Schliwka J, Fuchs U, Novosel A, Muller RU, Schermer B, Bissels U, Inman J, Phan Q, Chien M, Weir DB, Choksi R, De Vita G, Frezzetti D, Trompeter HI, Hornung V, Teng G, Hartmann G, Palkovits M, Di Lauro R, Wernet P, Macino G, Rogler CE, Nagle JW, Ju J, Papavasiliou FN, Benzing T, Lichter P, Tam W, Brownstein MJ, Bosio A, Borkhardt A, Russo JJ, Sander C, Zavolan M, Tuschl T. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CG, Kim JG, Kim HJ, Kwon HK, Cho IJ, Choi DW, Lee WH, Kim WD, Hwang SJ, Choi S, Kim SG. Discovery of an integrative network of microRNAs and transcriptomics changes for acute kidney injury. Kidney Int. 2014;86:943–953. doi: 10.1038/ki.2014.117. [DOI] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Li T, Cao H, Zhuang J, Wan J, Guan M, Yu B, Li X, Zhang W. Identification of miR-130a, miR-27b and miR-210 as serum biomarkers for atherosclerosis obliterans. Clin Chim Acta. 2011;412:66–70. doi: 10.1016/j.cca.2010.09.029. [DOI] [PubMed] [Google Scholar]
- Li Y, Jiang Z, Xu L, Yao H, Guo J, Ding X. Stability analysis of liver cancer-related microRNAs. Acta Biochim Biophys Sin (Shanghai) 2011;43:69–78. doi: 10.1093/abbs/gmq114. [DOI] [PubMed] [Google Scholar]
- Li YF, Jing Y, Hao J, Frankfort NC, Zhou X, Shen B, Liu X, Wang L, Li R. MicroRNA-21 in the pathogenesis of acute kidney injury. Protein Cell. 2013;4:813–819. doi: 10.1007/s13238-013-3085-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Dong C, Jiang Z, Wu WK, Chan MT, Zhang J, Li H, Qin K, Sun X. MicroRNA-10b downregulation mediates acute rejection of renal allografts by derepressing BCL2L11. Exp Cell Res. 2015;333:155–163. doi: 10.1016/j.yexcr.2015.01.018. [DOI] [PubMed] [Google Scholar]
- Liu XJ, Hong Q, Wang Z, Yu YY, Zou X, Xu LH. MicroRNA-34a Suppresses Autophagy in Tubular Epithelial Cells in Acute Kidney Injury. Am J Nephrol. 2015;42:168–175. doi: 10.1159/000439185. [DOI] [PubMed] [Google Scholar]
- Loeser H, von Brandenstein M, Herschung A, Schlosser M, Buttner R, Fries JW. ET-1 Induced Downregulation of MRP2 via miRNA 133a - A Marker for Tubular Nephrotoxicity? Am J Nephrol. 2015;41:191–199. doi: 10.1159/000381272. [DOI] [PubMed] [Google Scholar]
- Lorenzen JM, Kaucsar T, Schauerte C, Schmitt R, Rong S, Hubner A, Scherf K, Fiedler J, Martino F, Kumarswamy R, Kolling M, Sorensen I, Hinz H, Heineke J, van Rooij E, Haller H, Thum T. MicroRNA-24 antagonism prevents renal ischemia reperfusion injury. J Am Soc Nephrol. 2014;25:2717–2729. doi: 10.1681/ASN.2013121329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovisa S, LeBleu VS, Tampe B, Sugimoto H, Vadnagara K, Carstens JL, Wu CC, Hagos Y, Burckhardt BC, Pentcheva-Hoang T, Nischal H, Allison JP, Zeisberg M, Kalluri R. Epithelial-to-mesenchymal transition induces cell cycle arrest and parenchymal damage in renal fibrosis. Nat Med. 2015;21:998–1009. doi: 10.1038/nm.3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrone AK, Stolz DB, Bastacky SI, Kostka D, Bodnar AJ, Ho J. MicroRNA-17~92 is required for nephrogenesis and renal function. J Am Soc Nephrol. 2014;25:1440–1452. doi: 10.1681/ASN.2013040390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Fernandez M, Paramio JM, Duenas M. RNA Detection in Urine: From RNA Extraction to Good Normalizer Molecules. J Mol Diagn. 2016;18:15–22. doi: 10.1016/j.jmoldx.2015.07.008. [DOI] [PubMed] [Google Scholar]
- McDonald JS, Milosevic D, Reddi HV, Grebe SK, Algeciras-Schimnich A. Analysis of circulating microRNA: preanalytical and analytical challenges. Clin Chem. 2011;57:833–840. doi: 10.1373/clinchem.2010.157198. [DOI] [PubMed] [Google Scholar]
- McDonald MK, Capasso KE, Ajit SK. Purification and microRNA profiling of exosomes derived from blood and culture media. J Vis Exp. 2013:e50294. doi: 10.3791/50294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo SA, Sugimoto H, O’Connell JT, Kato N, Villanueva A, Vidal A, Qiu L, Vitkin E, Perelman LT, Melo CA, Lucci A, Ivan C, Calin GA, Kalluri R. Cancer exosomes perform cell-independent microRNA biogenesis and promote tumorigenesis. Cancer Cell. 2014;26:707–721. doi: 10.1016/j.ccell.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- miRBase. 2014;21 http://mirbase.org/ Faculty of Life Sciences at the University of Manchester with funding from the BBSRC, and was previously hosted and supported by the Wellcome Trust Sanger Institute. [Google Scholar]
- Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O’Briant KC, Allen A, Lin DW, Urban N, Drescher CW, Knudsen BS, Stirewalt DL, Gentleman R, Vessella RL, Nelson PS, Martin DB, Tewari M. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mladinov D, Liu Y, Mattson DL, Liang M. MicroRNAs contribute to the maintenance of cell-type-specific physiological characteristics: miR-192 targets Na+/K+-ATPase beta1. Nucleic Acids Res. 2013;41:1273–1283. doi: 10.1093/nar/gks1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mraz M, Malinova K, Mayer J, Pospisilova S. MicroRNA isolation and stability in stored RNA samples. Biochem Biophys Res Commun. 2009;390:1–4. doi: 10.1016/j.bbrc.2009.09.061. [DOI] [PubMed] [Google Scholar]
- Murray PT, Mehta RL, Shaw A, Ronco C, Endre Z, Kellum JA, Chawla LS, Cruz D, Ince C, Okusa MD workgroup A. Potential use of biomarkers in acute kidney injury: report and summary of recommendations from the 10th Acute Dialysis Quality Initiative consensus conference. Kidney Int. 2014;85:513–521. doi: 10.1038/ki.2013.374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassirpour R, Mathur S, Gosink MM, Li Y, Shoieb AM, Wood J, O’Neil SP, Homer BL, Whiteley LO. Identification of tubular injury microRNA biomarkers in urine: comparison of next-generation sequencing and qPCR-based profiling platforms. BMC Genomics. 2014;15:485. doi: 10.1186/1471-2164-15-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- niddk.nih.gov. The National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) 2016. [Google Scholar]
- Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B, Hayward DC, Ball EE, Degnan B, Muller P, Spring J, Srinivasan A, Fishman M, Finnerty J, Corbo J, Levine M, Leahy P, Davidson E, Ruvkun G. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:86–89. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- Pavkovic M, Riefke B, Ellinger-Ziegelbauer H. Urinary microRNA profiling for identification of biomarkers after cisplatin-induced kidney injury. Toxicology. 2014;324:147–157. doi: 10.1016/j.tox.2014.05.005. [DOI] [PubMed] [Google Scholar]
- Pavkovic M, Riefke B, Frisk AL, Groticke I, Ellinger-Ziegelbauer H. Glomerulonephritis-Induced Changes in Urinary and Kidney MicroRNA Profiles in Rats. Toxicol Sci. 2015;145:348–359. doi: 10.1093/toxsci/kfv053. [DOI] [PubMed] [Google Scholar]
- Pavkovic M, Robinson-Cohen C, Nicoara O, Antoine D, Himmelfarb J, Waikar SS, Vaidya SV. Early Detection of Kidney Toxicity in Humans Using Urinary microRNA-21, -200c and -423. The Toxicologist: Supplement to Toxicological Sciences. 2015;144 Abstract no. 2297. [Google Scholar]
- Pellegrini KL, Gerlach CV, Craciun FL, Ramachandran K, Bijol V, Kissick HT, Vaidya VS. Application of small RNA sequencing to identify microRNAs in acute kidney injury and fibrosis. Toxicol Appl Pharmacol. 2015 doi: 10.1016/j.taap.2015.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrini KL, Han T, Bijol V, Saikumar J, Craciun FL, Chen WW, Fuscoe JC, Vaidya VS. MicroRNA-155 deficient mice experience heightened kidney toxicity when dosed with cisplatin. Toxicol Sci. 2014;141:484–492. doi: 10.1093/toxsci/kfu143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran K, Saikumar J, Bijol V, Koyner JL, Qian J, Betensky RA, Waikar SS, Vaidya VS. Human miRNome profiling identifies microRNAs differentially present in the urine after kidney injury. Clin Chem. 2013;59:1742–1752. doi: 10.1373/clinchem.2013.210245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rani S. MicroRNA profiling of exosomes isolated from biofluids and conditioned media. Methods Mol Biol. 2014;1182:131–144. doi: 10.1007/978-1-4939-1062-5_12. [DOI] [PubMed] [Google Scholar]
- Saikumar J, Hoffmann D, Kim TM, Gonzalez VR, Zhang Q, Goering PL, Brown RP, Bijol V, Park PJ, Waikar SS, Vaidya VS. Expression, circulation, and excretion profile of microRNA-21, -155, and -18a following acute kidney injury. Toxicol Sci. 2012;129:256–267. doi: 10.1093/toxsci/kfs210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sempere LF, Freemantle S, Pitha-Rowe I, Moss E, Dmitrovsky E, Ambros V. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 2004;5:R13. doi: 10.1186/gb-2004-5-3-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slattery C, Campbell E, McMorrow T, Ryan MP. Cyclosporine A-induced renal fibrosis: a role for epithelial-mesenchymal transition. Am J Pathol. 2005;167:395–407. doi: 10.1016/S0002-9440(10)62984-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Koo S, White N, Peralta E, Esau C, Dean NM, Perera RJ. Development of a micro-array to detect human and mouse microRNAs and characterization of expression in human organs. Nucleic Acids Res. 2004;32:e188. doi: 10.1093/nar/gnh186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo G, Bala S. MicroRNAs in liver disease. Nat Rev Gastroenterol Hepatol. 2013 doi: 10.1038/nrgastro.2013.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taber SS, Pasko DA. The epidemiology of drug-induced disorders: the kidney. Expert Opin Drug Saf. 2008;7:679–690. doi: 10.1517/14740330802410462. [DOI] [PubMed] [Google Scholar]
- Tian Z, Greene AS, Pietrusz JL, Matus IR, Liang M. MicroRNA-target pairs in the rat kidney identified by microRNA microarray, proteomic, and bioinformatic analysis. Genome research. 2008;18:404–411. doi: 10.1101/gr.6587008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchino S. Creatinine. Curr Opin Crit Care. 2010;16:562–567. doi: 10.1097/MCC.0b013e32833ea7f3. [DOI] [PubMed] [Google Scholar]
- Uchino S, Kellum JA, Bellomo R, Doig GS, Morimatsu H, Morgera S, Schetz M, Tan I, Bouman C, Macedo E, Gibney N, Tolwani A, Ronco C. Acute renal failure in critically ill patients: a multinational, multicenter study. JAMA. 2005;294:813–818. doi: 10.1001/jama.294.7.813. [DOI] [PubMed] [Google Scholar]
- van Rooij E, Sutherland LB, Qi X, Richardson JA, Hill J, Olson EN. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science. 2007;316:575–579. doi: 10.1126/science.1139089. [DOI] [PubMed] [Google Scholar]
- Vickers KC, Palmisano BT, Shoucri BM, Shamburek RD, Remaley AT. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol. 2011;13:423–433. doi: 10.1038/ncb2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vickers KC, Remaley AT. Lipid-based carriers of microRNAs and intercellular communication. Curr Opin Lipidol. 2012;23:91–97. doi: 10.1097/MOL.0b013e328350a425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visone R, Croce CM. MiRNAs and cancer. Am J Pathol. 2009;174:1131–1138. doi: 10.2353/ajpath.2009.080794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waikar SS, Betensky RA, Emerson SC, Bonventre JV. Imperfect gold standards for kidney injury biomarker evaluation. J Am Soc Nephrol. 2012;23:13–21. doi: 10.1681/ASN.2010111124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waikar SS, Sabbisetti VS, Bonventre JV. Normalization of urinary biomarkers to creatinine during changes in glomerular filtration rate. Kidney Int. 2010;78:486–494. doi: 10.1038/ki.2010.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace KB, Murphy E, Rosenblum IY, Herman G, Metz AL, Rosen MR, York MJ, Holt G, MacGregor JT, Hausner E, Essayan DM. NCSS Fact Finding Cardiotoxicity Expert Working Group. FDA; 2001. pp. PTT slides. [Google Scholar]
- Wang G, Kwan BC, Lai FM, Chow KM, Li PK, Szeto CC. Elevated levels of miR-146a and miR-155 in kidney biopsy and urine from patients with IgA nephropathy. Dis Markers. 2011;30:171–179. doi: 10.3233/DMA-2011-0766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Kwan BC, Lai FM, Chow KM, Li PK, Szeto CC. Urinary miR-21, miR-29, and miR-93: novel biomarkers of fibrosis. Am J Nephrol. 2012;36:412–418. doi: 10.1159/000343452. [DOI] [PubMed] [Google Scholar]
- Wang JF, Yu ML, Yu G, Bian JJ, Deng XM, Wan XJ, Zhu KM. Serum miR-146a and miR-223 as potential new biomarkers for sepsis. Biochem Biophys Res Commun. 2010;394:184–188. doi: 10.1016/j.bbrc.2010.02.145. [DOI] [PubMed] [Google Scholar]
- Wang JF, Zha YF, Li HW, Wang F, Bian Q, Lai XL, Yu G. Screening plasma miRNAs as biomarkers for renal ischemia-reperfusion injury in rats. Med Sci Monit. 2014;20:283–289. doi: 10.12659/MSM.889937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Zhang S, Weber J, Baxter D, Galas DJ. Export of microRNAs and microRNA-protective protein by mammalian cells. Nucleic Acids Res. 2010;38:7248–7259. doi: 10.1093/nar/gkq601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N, Zhou Y, Jiang L, Li D, Yang J, Zhang CY, Zen K. Urinary MicroRNA-10a and MicroRNA-30d Serve as Novel, Sensitive and Specific Biomarkers for Kidney Injury. PLoS One. 2012;7:e51140. doi: 10.1371/journal.pone.0051140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Lee CG. MicroRNA and cancer--focus on apoptosis. J Cell Mol Med. 2009;13:12–23. doi: 10.1111/j.1582-4934.2008.00510.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber JA, Baxter DH, Zhang S, Huang DY, Huang KH, Lee MJ, Galas DJ, Wang K. The microRNA spectrum in 12 body fluids. Clin Chem. 2010;56:1733–1741. doi: 10.1373/clinchem.2010.147405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Q, Bhatt K, He HZ, Mi QS, Haase VH, Dong Z. Targeted deletion of Dicer from proximal tubules protects against renal ischemia-reperfusion injury. J Am Soc Nephrol. 2010;21:756–761. doi: 10.1681/ASN.2009070718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen X, Buckley B, McCandlish E, Goedken MJ, Syed S, Pelis R, Manautou JE, Aleksunes LM. Transgenic expression of the human MRP2 transporter reduces cisplatin accumulation and nephrotoxicity in Mrp2-null mice. Am J Pathol. 2014;184:1299–1308. doi: 10.1016/j.ajpath.2014.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiemer EA. The role of microRNAs in cancer: no small matter. Eur J Cancer. 2007;43:1529–1544. doi: 10.1016/j.ejca.2007.04.002. [DOI] [PubMed] [Google Scholar]
- Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- Wu J, Zheng C, Fan Y, Zeng C, Chen Z, Qin W, Zhang C, Zhang W, Wang X, Zhu X, Zhang M, Zen K, Liu Z. Downregulation of microRNA-30 facilitates podocyte injury and is prevented by glucocorticoids. J Am Soc Nephrol. 2014;25:92–104. doi: 10.1681/ASN.2012111101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Yang BF, Ai J. MicroRNA transport: a new way in cell communication. J Cell Physiol. 2013;228:1713–1719. doi: 10.1002/jcp.24344. [DOI] [PubMed] [Google Scholar]
- Yang X, Greenhaw J, Shi Q, Su Z, Qian F, Davis K, Mendrick D, Salminen W. Identification of urinary microRNA profiles in rats that may diagnose hepatotoxicity. Toxicological sciences : an official journal of the Society of Toxicology. 2012;125:335–379. doi: 10.1093/toxsci/kfr321. [DOI] [PubMed] [Google Scholar]
- Yuan J, Benway CJ, Bagley J, Iacomini J. MicroRNA-494 promotes cyclosporine-induced nephrotoxicity and epithelial to mesenchymal transition by inhibiting PTEN. Am J Transplant. 2015;15:1682–1691. doi: 10.1111/ajt.13161. [DOI] [PubMed] [Google Scholar]
- Zhang W, Zhang C, Chen H, Li L, Tu Y, Liu C, Shi S, Zen K, Liu Z. Evaluation of microRNAs miR-196a, miR-30a-5P, and miR-490 as biomarkers of disease activity among patients with FSGS. Clin J Am Soc Nephrol. 2014;9:1545–1552. doi: 10.2215/CJN.11561113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu D, Chen X, Li J, Li L, Bian Z, Sun F, Lu J, Yin Y, Cai X, Sun Q, Wang K, Ba Y, Wang Q, Wang D, Yang J, Liu P, Xu T, Yan Q, Zhang J, Zen K, Zhang CY. Secreted monocytic miR-150 enhances targeted endothelial cell migration. Mol Cell. 2010;39:133–144. doi: 10.1016/j.molcel.2010.06.010. [DOI] [PubMed] [Google Scholar]
- Zhou H, Hasni SA, Perez P, Tandon M, Jang SI, Zheng C, Kopp JB, Austin H, 3rd, Balow JE, Alevizos I, Illei GG. miR-150 promotes renal fibrosis in lupus nephritis by downregulating SOCS1. J Am Soc Nephrol. 2013;24:1073–1087. doi: 10.1681/ASN.2012080849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou TB, Jiang ZP. Role of miR-21 and its signaling pathways in renal diseases. J Recept Signal Transduct Res. 2014;34:335–337. doi: 10.3109/10799893.2014.896382. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Xiong M, Niu J, Sun Q, Su W, Zen K, Dai C, Yang J. Secreted fibroblast-derived miR-34a induces tubular cell apoptosis in fibrotic kidney. J Cell Sci. 2014;127:4494–4506. doi: 10.1242/jcs.155523. [DOI] [PubMed] [Google Scholar]
- Zhu HY, Liu MY, Hong Q, Zhang D, Geng WJ, Xie YS, Chen XM. Role of microRNA-181a in the apoptosis of tubular epithelial cell induced by cisplatin. Chin Med J (Engl) 2012;125:523–526. [PubMed] [Google Scholar]