Abstract
DNA replication is essential for faithful transmission of genetic information and is intimately tied to chromosome structure and function. Genome duplication occurs in a defined temporal order known as the replication-timing (RT) program, which is regulated during the cell cycle and development in discrete units corresponding to topologically-associating domains (TADs) that are spatially compartmentalized in the nucleus. Correlations of RT to chromatin organization and gene regulation have been known for decades but causal and mechanistic links remain unknown. The complete elucidation of these intriguing liaisons is critical to understand the connection between the three-dimensional organization of the nucleus and cellular function. Here, we discuss emerging evidence providing new insights into regulation of chromosome architecture, transcription and its connections with RT.
Introduction
Two decades ago studies of DNA replication discovered the presence of stable submegabase units of chromosome structure and their spatio-temporal compartmentalization in the nucleus. Pulse-labeling followed by in situ detection revealed punctate sites of DNA synthesis (“replication foci”) that segregated into distinct spatial compartments depending upon the time of S phase labeling (Figure 1A) [1–5]. When chased for multiple generations, replication foci persisted as stable chromosome units [6–8], each estimated to contain 0.5–1Mb of DNA replicated by clusters of several synchronously firing replicons [9]. Early replicated foci located at the nuclear interior while late replicating foci were more tightly clustered and associated with the periphery of the nucleus and nucleolus (Figure 1A), as well as other sites of heterochromatin [4,5,10*]. The development of genome-wide methods to map RT found that during cell fate commitment half of the genome changes RT, coinciding with changes in gene expression and select examples of re-localization within the nucleus (Figure 2) [11,12]. These RT changes occurred in units of 400–800 kilobases termed replication domains (RDs) that are likely to be molecular equivalents of replication foci.
Figure 1.
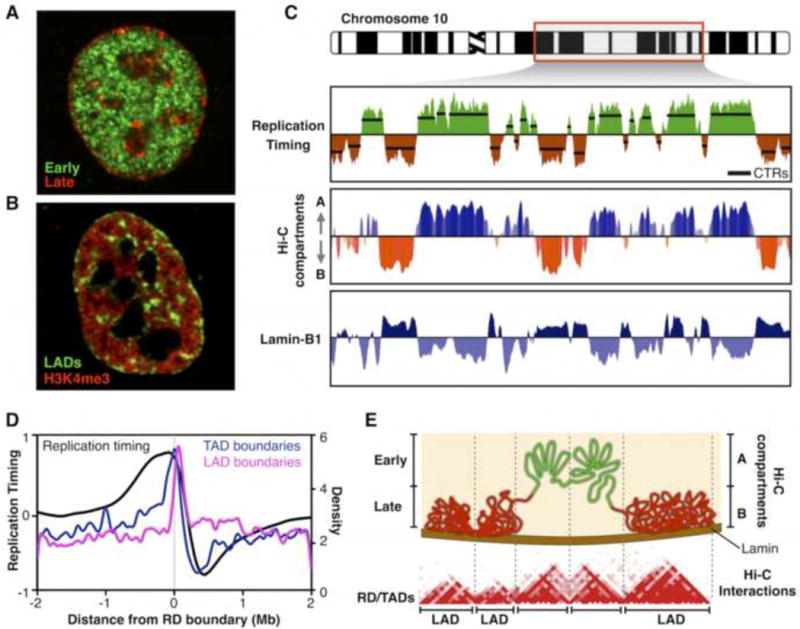
Replication timing reflects genome organization. A) Cells were pulse-labeled with CldU, chased for 3 hours, pulse-labeled with IdU, and fixed and immunofluorescently stained for CldU (green) and IdU (red) [29]. Shown is a nucleus from a cell labeled with the early replicating compartments labeled in green and the late replicating compartment in red. B) LADs (green) labeled at the nuclear periphery during one cell cycle re-distribute in the next cell cycle to locations that resemble the late replicating foci. H3K4me3 labels active chromatin [19]. C) Exemplary RT profile of 50 Mb of human Chr10 from IMR90 fibroblasts, segments of chromosomes with uniform RT (CTRs) aligned with Hi-C interaction compartments (Eigenvector display) and LaminB1 contact maps (DamID). D) RD boundaries align with the boundaries of both topologically associating domains (TADs) and lamin-associated domains (LADs). E) Replication domain model. Early and late replicating regions correspond to the Hi-C A/B compartments and RDs are equivalent to TADs, while late RDs and RDs in the passively replicated transitional regions between early and late RDs are associated with the nuclear periphery (LADs). The graphs were plotted using public data: RT from [29], HiC from [28], lamin-B1 from [16] and boundaries alignment from [26**].
Figure 2.
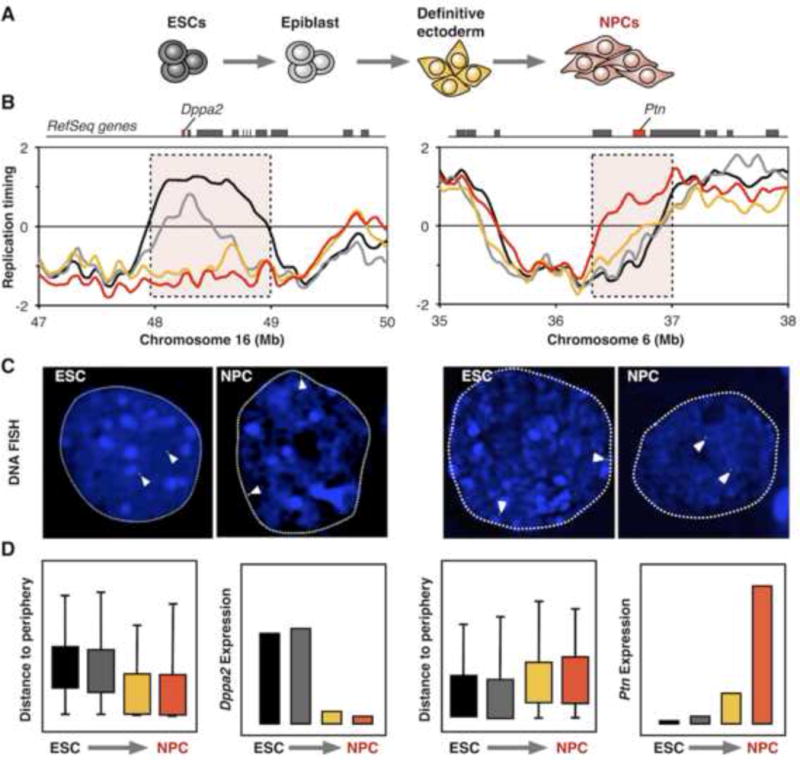
RT changes are associated with nuclear re-localization and gene expression. A) Schematic depiction of neural differentiation (NPCs) from mouse embryonic stem cells (ESCs). B) RT changes during neural differentiation. Location of Dppa2 and Ptn genes are shown at the top. C) Nuclear re-localization of Dppa2 and Ptn domains during neural differentiation visualized by DNA FISH. D) Measurements of radial nuclear positioning (distance to the nuclear periphery) and gene expression of Dppa2 and Ptn genes. Colors in the graphs represent the distinct differentiation stages as shown in (A). Data and FISH images were obtained from [12].
Each 7 years, Current Opinions has asked us to review the enigmatic relationships between replication timing, sub-nuclear chromatin organization and transcriptional regulation [13,14]. 14 years ago, we had little more than anecdotal observations from cytogenetics and comparisons of individual gene behavior in several cell lines to suggest a mutually reinforcing relationship between RT and chromatin structure for which teasing out causality resembled the chicken and egg conundrum (Figure 1 of [13]). 7 years ago genomics had emerged to fill in gaps from anecdotal studies into complete descriptions of the complexity of RT and transcription, suggesting strong correlative links to 3D genome organization (Figure 1 of [14]). In the past 7 years, developments in stem cell differentiation have facilitated extensive mapping of regions of the genome subject to developmental control of RT and 3D organization, and identified unique properties of constitutive vs. developmental RDs that challenge longstanding correlations previously presumed to be clues to causal linkages between these complex phenomena. Mapping interactions with the nuclear lamina revealed chromosome domains physically associated with the nuclear periphery (lamina-associated domains; LADs) [15–18]. Intriguingly, live single-cell tracking of LADs from one cell cycle to the next revealed that a significant proportion of peripheral chromatin redistributed to locations throughout the nucleus that resemble the spatial distribution of late replicating foci (Figure 1A) [19], and in situ hybridization suggests that late replicating segments can associate with any of these locations [20*] suggesting that these varied locations that replicate simultaneously constitute a single functionally equivalent sub-nuclear compartment. The explosion of chromatin conformation capture methods (Hi-C) has permitted genome-wide mapping of early and late replicating spatial compartments, and has identified stable structural units of topologically self-associating chromatin (TADs) that correspond to RDs we discussed 7 years ago and likely to foci we discussed 14 years ago. Here we summarize the most recent findings, emphasizing structure-function relationships in the nucleus, and we predict that the next 7-year cycle will be dominated by genome editing methods that may finally get us “beyond cause and effect”.
Replication timing reflects the spatial organization of the genome
The biggest conceptual leap for RT in the last 7 years is the molecular elucidation of 3D genome conformation by Hi-C, providing genome map coordinates that indisputably align the spatio-temporal relationships revealed through cytogenetics to those mapped by RT functional genomics. RT profiles are generally displayed as the ratio of early/late replication along the length of each chromosome [21–24*] and reveal chromosome segments with uniform replication timing, known as constant timing regions or CTRs (Figure 1B), which often consist of several adjacent RDs that replicate within 1–2 hours [25–27]. Intriguingly, chromatin conformation methods (Hi-C) detect not only chromatin partitioning into TADs but also folding of TADs into multi-megabase compartments of active/open (A-compartments) or inactive/closed (B-compartments) chromatin [28]. Consistent with the cytogenetic studies, comparison of RT profiles and Hi-C compartments shows remarkable alignment of RT to physical compartmentalization of the genome (Figure 1B, [29–32]). Moreover, by mapping RT transitions in multiple cell types the majority of RD boundaries could be mapped revealing a near one-to-one alignment to TAD boundaries [26**] (Figure 1C). Hence, the “replication domain model” [26**,27,33] proposes that RD/TADs are stable chromosome units segregated into spatially distinct and coordinately replicated nuclear compartments (Figure 1D). Indeed, tracking the formation of TADs and 3D compartments through the cell cycle revealed that 3D structure is dismantled during mitosis [34,35**], but both TAD structure and interaction compartments were re-established coincident with the re-establishment of RT in a very early window during G1 referred as the timing decision point (TDP) [35**], demonstrating a convincing intimate co-appearance of structure and function. However, TADs and compartments persist into G2 phase, when RT function is lost [36], suggesting that 3D structure is not sufficient for RT, but may provide a necessary scaffold on which cell cycle regulated factors operate to maintain the RT program [35**].
In fact, several factors have recently been implicated in regulating replication timing in the context of chromosome 3D organization, all of which act by antagonizing or recruiting the essential replication initiation cell division cycle 7 kinase (Cdc7). In budding yeast, forkhead proteins (Fkh1 and Fkh2) are thought to organize early replication origins into clusters that permit the concentration of Cdc7 activity [37]. In yeast and mammals, the large multi-functional rap1 interacting factor (Rif1) has been shown to localize to late replicating regions of the genome [38], and in yeast Rif1 recruits phosphatase PP1 to antagonize Cdc7 [38–41]. Rif1 depletion appears to disrupt 3D chromatin organization in mammals [42,43**], leading to speculation that Rif1 regulates RT through establishing 3D structure. However, there is still contradictory data as to whether Rif1 is removed from chromatin during replication [42,44], so whether Rif1 is directing 3D structure necessary but not sufficient for RT, or whether Rif1 interacts with the 3D scaffold in a cell cycle regulated fashion to influence RT is still a matter of debate.
Intrinsic and extrinsic variation in RT and large-scale chromosome architecture
An important gap in our understanding of RT is the degree of stochastic variation and cell-to-cell heterogeneity. Recent studies of budding and fission yeast replicon structure using isolated DNA fibers demonstrate a great deal of heterogeneity in the sites and timing of initiation for replication [45,46*]. However, current DNA fiber methods do not retain cell of origin information to distinguish extrinsic (cell to cell) and intrinsic (homologue to homologue) variation. Moreover, compelling cytogenetic evidence in mammalian cells suggests that the same cohort of replication foci labeled in one cell cycle are also labeled coordinately in the subsequent cell cycle [47], implying a great degree of coordination. It is now imperative to develop methods to probe how structure and function co-vary within single cells. Although single-cell RT has not been developed, great progress in single-cell isolation and whole genome amplification techniques have paved the way for transcriptome and epigenome analyses of individual cells [48,49]. The first glimpses of single-cell Hi-C suggest stability in the structure of TADs but considerable variability in long range interactions that nonetheless remained within their respective interaction compartment [50] while single-cell LAD mapping has confirmed cell to cell heterogeneity in chromatin interaction with the nuclear periphery [51*] predicted from the earlier single cell studies [19]. However, both studies examined haploid chromosomes, precluding measurements of intrinsic variation. In mammals, RT profiles may soon emerge from single-cell copy number measurements [52*], and high-throughput methods to analyze individual DNA fibers are becoming state-of-the-art [53,54*], suggesting measurements of origin firing and RT variability are on the horizon.
While we do not yet have good assessments of the degree of stochastic intrinsic variation, there is evidence for deterministic influences on intrinsic RT variation. Genomic imprinting in mammals is associated with silencing and delayed replication of the imprinted allele [55,56] and mono-allelically expressed genes are also generally replicated earlier when active [57,58]. Moreover, during chromosome X inactivation in female mammals, the inactive chromosome X (Xi) replicates later than the active X (Xa) [59,60]. Intriguingly, the Xi is depleted of TADs [61–63] and replicated by a rapid and synchronous firing of origins throughout the whole chromosome [64,65*]. Analysis of RT by deep sequencing of phased genomes (i.e. with all allelic variants mapped to distinguish the parental origin of each haplotype) permits correlations between allelic DNA sequence variation and RT. In general, homologues replicate highly synchronously with very few regions showing allelic differences in RT or in origin usage related to DNA sequence variation, although some reach statistical or disease-associated significance worth further investigation [24*,66*,67,68*,69*]. An elegant series of recent papers has revealed a new class of cis-acting elements involved in the regulation of RT, mitotic condensation and chromosome stability [70–72**]. These elements consist of monoallelically expressed long non-coding RNAs that appear to be present on each mammalian chromosome, and interact in cis to regulate RT, monoallelic gene expression and structural stability of the entire chromosome [72**]. Overall, these results uncover specific mechanisms that control intrinsic allelic variation in RT that are certain to be the subject of much investigation in the coming years.
Clinging onto Proteus’s neck
Strong correlations between genome organization, RT and gene regulation are found in all multicellular organisms studied [11,12,22,73–80]. Half of the genome changes RT during development in units of 400–800 kilobases (entire RD/TADs) closely coordinated with transcriptional competence (Figure 2) [11,12,77,81–83]. Just as holding the mythical Proteus throughout all his transformations was necessary to learn his secrets, understanding these intriguing liaisons requires careful observation of their order of events during cell fate transitions. However, until recently, systems capable of eliciting change found that the changes in RT, gene activity and nuclear positioning were too synchronous to temporally separate them [12]. By taking advantage of new human embryonic stem cell (hESC) differentiation systems that allow highly synchronous derivation of distinct cell types [84–86**], we were able to track changes in RT and gene expression through multiple intermediate steps in lineage specification [86**]. Surprisingly, our study revealed that the strong correlation between early replication and active transcription is restricted to RT-constitutive genes (i.e. genes that do not change their RT program), while the RT-switching genes have a much weaker correlation that is further diminished during differentiation. Moreover, some RT-switching genes are expressed only when replicated early (E-class), while the majority (C-class) can be strongly expressed in one or more cell types while late replicating, demonstrating that transcription is not sufficient for early replication (Figure 3). Additionally, a rare category of genes (L-class) were expressed exclusively when replicated late. Tracking C-class gene activity through cell fate transitions showed that this class of genes commonly reach high expression levels in the same lineage where an RT switch occurs, but preceding the change to early RT by one or two intermediate stages of differentiation, while down-regulation often followed changes to late RT (Figure 3) [86**]. These results suggest that some aspects of transcriptional control could indirectly influence RT. One possibility is that RT responds to aspects of transcriptional regulatory circuitry, but is not directly related to transcription itself.
Figure 3.
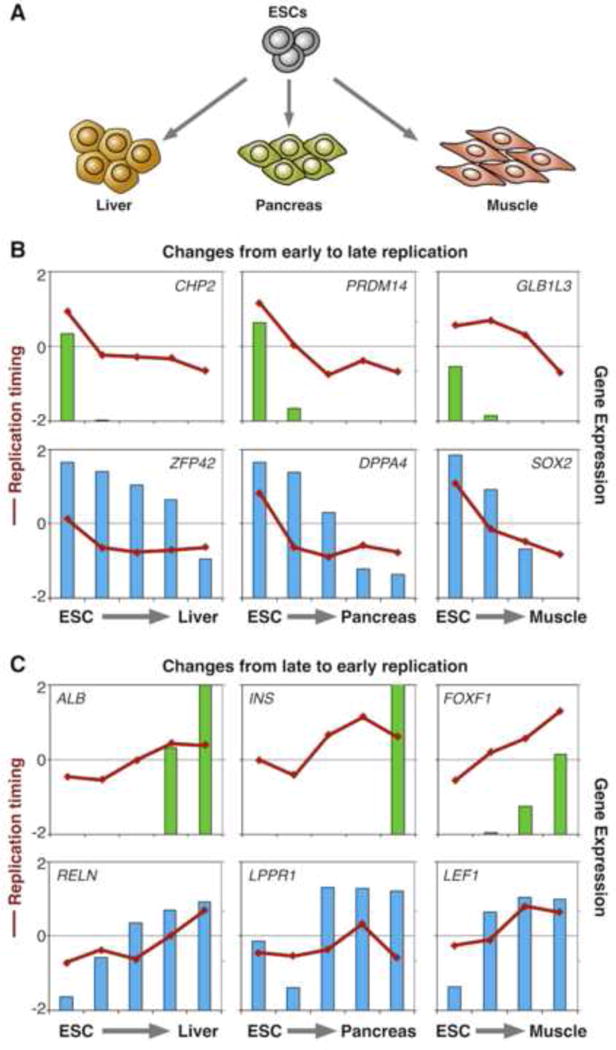
Transcription changes precede or follow RT changes for different classes of genes. RT changes and gene regulation during differentiation of human ESCs to liver, pancreas and muscle (Schematized in A). Exemplary genes switching from early to late (B) and from late to early (C) replication are shown. Lines represent the RT and bars the expression values at distinct cell fate transitions. E-class genes (top panels in B and C) are transcribed only when replicate early. C-class genes (bottom panels in B and C) down-regulation follows changes to late replication while their induction precedes changes to early replication. Specific details of differentiation stages and original data from [86**].
Studies of RT changes during differentiation also revealed that many other longstanding correlations to RT, in addition to transcription, apply only to the RT-constitutive domains and that RT-switching or “developmental domains” have independent organizational principles that challenge many of the assumptions that guided our hypotheses over the last few decades. In addition to their distinct sequence composition (see [86**] and Figure 1 of [14]), they are highly nuclease insensitive, depleted of replication origins [87,88], less confined to A/B Hi-C compartments and chromatin states correlated to RT in constitutive domains are much less correlated to RT in developmental domains [35**]. Hence, chromosome domains fall into categories that reflect their developmental control of DNA RT. Importantly, it is not known whether the lack of correlations in the RT-switching half of the genome is due to intermediate properties of these domains, increased cell-to-cell heterogeneity, or structural instability within a single cell cycle. It is now imperative to tease out what properties actually do correlate with RT within the half of the genome that changes RT and 3D organization during differentiation.
Twisting the lion’s tail
Understanding causal relationships between RT, chromosome architecture and gene regulation requires experimental manipulation or ‘twisting the lion’s tail’ [89]. Several studies expressing artificial proteins consisting of sequence-specific DNA binding domains fused to domains that can target to sub-nuclear domains or strongly remodel chromatin have revealed some important principles. Rapid re-localization (1–2 hrs) of a chromosome site has been observed from the periphery to the nuclear interior upon robust transcriptional induction [90] or towards RNA processing sites following heat shock [91*]. Conversely, artificial anchoring of distinct genes to the nuclear lamina leads to their reversible repression [92–94], but this re-positioning occurs only after a passage through mitosis [93]. Tethering to the periphery is sufficient to suppress the expression of some but not all genes [92,95,96], implying the existence of different sensitivity classes of genes, as was found with the RT studies discussed above. Although RT was not measured in these studies, since the periphery is a late replicating compartment such analogies are reasonable to expect. Additionally, mechanisms tethering LADs to the nuclear periphery remain unknown and some evidence is contradictory: mES cells depleted of nuclear lamins preserved normal chromatin interactions with the nuclear periphery [97], while other studies suggest that lamin A/C is required to anchor chromatin to the nuclear periphery [98,99].
Several earlier studies suggested that targeting strong artificial transcriptional activators is sufficient to induce a change from late to early replication of some specific loci [100–103]. More recently, targeting an exceptionally strong transactivator induced changes in both nuclear repositioning and RT [104*], while targeting a mutated transactivator with chromatin unfolding but not transcription activity [105] elicited repositioning but not a change in RT, prompting the authors to conclude the transcription is sufficient for the RT switch [104*]. This is clearly not a generalized conclusion, since genome-wide studies reveal numerous examples of transcriptional upregulation without alterations in RT and approximately 20% of late replicating genes are expressed [11,12,73,86**]. Moreover, RT changes in response to the artificial transactivator were less robust than changes observed at this same locus during differentiation [104*] and some evidence suggests that remodeling of chromatin during DNA replication precedes and is necessary for gene activation during differentiation at other loci [106]. Hence, we are beginning to witness a transition in the field from correlative to more manipulative studies that are providing glimpses of causality, but also complexity. It is possible that transcription, RT, chromatin structure and chromosome organization all influence each other in a contextually or quantitatively dependent manner. Clearly there is much exciting work ahead to establish the governing principles relating large-scale structure and function in the nucleus.
Conclusions and future directions
There is an intimate connection between, on the one hand, fundamental chromosome functions of RT and transcription and, on the other hand, the structural organization of chromosomes into TADs and their 3D organization in the nucleus. A critical issue to be resolved is the extent to which genome structure is cause or consequence of genome function. From the intriguing equivalence between RDs and TADs it is tempting to speculate that RT reflects the organization of the genome, but to date it is not clear how RT is regulated while TAD organization is not sufficient to dictate RT [35**]. Moreover, studies of RT have revealed that domains with developmentally plastic RT lack or have measurably weaker associations of RT to chromatin structure and transcriptional control as compared to the RT-constitutive domains. Transcriptional activity appears to be capable of influencing both gene position and RT, but only in as yet poorly defined contexts. Moreover, developmental domains can be just as early or late replicating as constitutive domains, yet they do not follow the correlations between RT and chromatin properties identified over the last 30 years. Figure 4 presents a hypothetical model in which chromosomes are partitioned into distinct RD/TADs that agglomerate to form early and late compartments. Dynamic changes in nuclear organization, initiated by the transcriptional induction of C-class genes, elicit a compartment switch in the following mitosis that changes RT, creating a new stable state reinforced by chromatin assembly at a different time and location in the nucleus. Newly emerging approaches may finally be capable of teasing out cause and effect in these relationships. For example, combining chromosome “domain engineering” with evaluating effects during cell fate transitions [107,108] will identify necessary and sufficient cis-acting elements of chromosome structure and function, while recently developed methodologies for analysis of thousands of randomly integrated reporters [109] will provide much needed insight into influence of large scale architecture on functional outputs.
Figure 4.
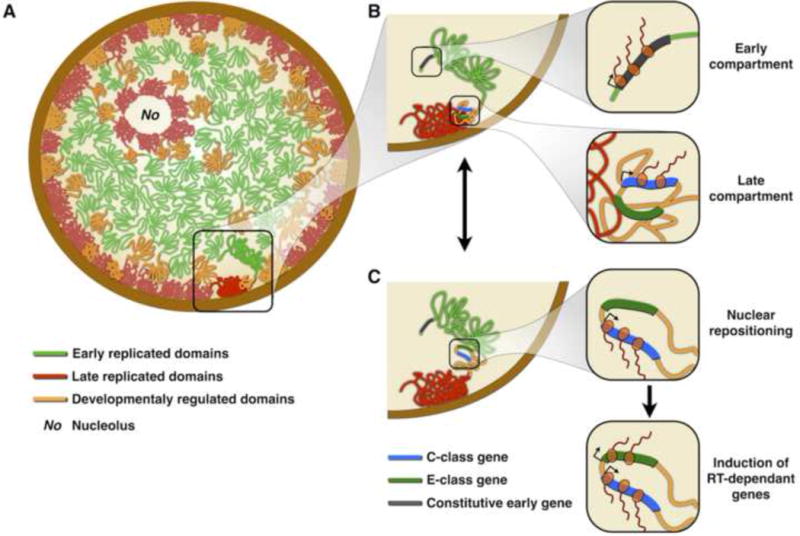
Genome organization, RT and gene regulation in the nucleus. A) Organization of RDs at the nuclear interior. Late replicating domains are located at the nuclear and nucleolar periphery (and other regions as shown in Figure 1A), early replicating domains are located at the nuclear interior and developmental regulated domains are less well compartmentalized [35**]. B) Early replicating compartments contain open/active chromatin at the nuclear interior where genes can be highly expressed. Developmentally regulated RDs contain distinct classes of genes: E-class genes are silenced while C-class genes can be expressed despite being late replicating and close to the periphery [86**]. C) RT changes and nuclear repositioning are commonly accompanied by an increase in expression of C-class genes, while E-class genes are induced only after the RT change.
Acknowledgments
We would like to thank C. Trevilla-Garcia for critical reading of the manuscript and Karen Reddy for helpful discussions. This work was supported by National Institutes of Health (NIH) grants GM083337 and GM085354 (D.M.G.).
References
Papers of particular interest, published within the period of review, have been highlighted as:
*of special interest
**of outstanding interest
- 1.Nakamura H, Morita T, Sato C. Structural organizations of replicon domains during DNA synthetic phase in the mammalian nucleus. Exp Cell Res. 1986;165:291–297. doi: 10.1016/0014-4827(86)90583-5. [DOI] [PubMed] [Google Scholar]
- 2.Nakayasu H. Mapping replicational sites in the eucaryotic cell nucleus. The Journal of Cell Biology. 1989;108:1–11. doi: 10.1083/jcb.108.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.O’Keefe RT, Henderson SC, Spector DL. Dynamic organization of DNA replication in mammalian cell nuclei: spatially and temporally defined replication of chromosome-specific alpha-satellite DNA sequences. The Journal of Cell Biology. 1992;116:1095–1110. doi: 10.1083/jcb.116.5.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Visser AE, Eils R, Jauch A, Little G, Bakker PJ, Cremer T, Aten JA. Spatial distributions of early and late replicating chromatin in interphase chromosome territories. Exp Cell Res. 1998;243:398–407. doi: 10.1006/excr.1998.4144. [DOI] [PubMed] [Google Scholar]
- 5.Zink D. The temporal program of DNA replication: new insights into old questions. Chromosoma. 2006;115:273–287. doi: 10.1007/s00412-006-0062-8. [DOI] [PubMed] [Google Scholar]
- 6.Sparvoli E, Levi M, Rossi E. Replicon clusters may form structurally stable complexes of chromatin and chromosomes. J Cell Sci. 1994;107:3097–3103. doi: 10.1242/jcs.107.11.3097. [DOI] [PubMed] [Google Scholar]
- 7.Ferreira J. Spatial Organization of Large-Scale Chromatin Domains in the Nucleus: A Magnified View of Single Chromosome Territories. The Journal of Cell Biology. 1997;139:1597–1610. doi: 10.1083/jcb.139.7.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jackson DA, Pombo A. Replicon clusters are stable units of chromosome structure: evidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells. The Journal of Cell Biology. 1998;140:1285–1295. doi: 10.1083/jcb.140.6.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ma H, Samarabandu J, Devdhar RS, Acharya R, Cheng PC, Meng C, Berezney R. Spatial and temporal dynamics of DNA replication sites in mammalian cells. The Journal of Cell Biology. 1998;143:1415–1425. doi: 10.1083/jcb.143.6.1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10*.Bass HW, Hoffman GG, Lee T-J, Wear EE, Joseph SR, Allen GC, Hanley-Bowdoin L, Thompson WF. Defining multiple, distinct, and shared spatiotemporal patterns of DNA replication and endoreduplication from 3D image analysis of developing maize (Zea mays L.) root tip nuclei. Plant Mol Biol. 2015;89:339–351. doi: 10.1007/s11103-015-0364-4. While replication foci have been well characterized in multiple animal cell culture systems, this study constitutes the first 3D characterization of replication foci during development in plants. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hiratani I, Ryba T, Itoh M, Yokochi T, Schwaiger M, Chang C-W, Lyou Y, Townes TM, Schübeler D, Gilbert DM. Global reorganization of replication domains during embryonic stem cell differentiation. PLoS Biol. 2008;6:e245. doi: 10.1371/journal.pbio.0060245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hiratani I, Ryba T, Itoh M, Rathjen J, Kulik M, Papp B, Fussner E, Bazett-Jones DP, Plath K, Dalton S, et al. Genome-wide dynamics of replication timing revealed by in vitro models of mouse embryogenesis. Genome Res. 2010;20:155–169. doi: 10.1101/gr.099796.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gilbert DM. Replication timing and transcriptional control: beyond cause and effect. Curr Opin Cell Biol. 2002;14:377–383. doi: 10.1016/s0955-0674(02)00326-5. [DOI] [PubMed] [Google Scholar]
- 14.Hiratani I, Takebayashi S-I, Lu J, Gilbert DM. Replication timing and transcriptional control: beyond cause and effect–part II. Curr Opin Genet Dev. 2009;19:142–149. doi: 10.1016/j.gde.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vogel MJ, Peric-Hupkes D, van Steensel B. Detection of in vivo protein-DNA interactions using DamID in mammalian cells. Nat Protoc. 2007;2:1467–1478. doi: 10.1038/nprot.2007.148. [DOI] [PubMed] [Google Scholar]
- 16.Guelen L, Pagie L, Brasset E, Meuleman W, Faza MB, Talhout W, Eussen BH, de Klein A, Wessels L, de Laat W, et al. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 2008;453:948–951. doi: 10.1038/nature06947. [DOI] [PubMed] [Google Scholar]
- 17.Amendola M, van Steensel B. Mechanisms and dynamics of nuclear lamina-genome interactions. Curr Opin Cell Biol. 2014;28:61–68. doi: 10.1016/j.ceb.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 18.Gordon MR, Pope BD, Sima J, Gilbert DM. Many paths lead chromatin to the nuclear periphery. Bioessays. 2015;37:862–866. doi: 10.1002/bies.201500034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kind J, Pagie L, Ortabozkoyun H, Boyle S, de Vries SS, Janssen H, Amendola M, Nolen LD, Bickmore WA, van Steensel B. Single-cell dynamics of genome-nuclear lamina interactions. Cell. 2013;153:178–192. doi: 10.1016/j.cell.2013.02.028. [DOI] [PubMed] [Google Scholar]
- 20*.Ragoczy T, Telling A, Scalzo D, Kooperberg C, Groudine M. Functional redundancy in the nuclear compartmentalization of the late-replicating genome. Nucleus. 2014;5:626–635. doi: 10.4161/19491034.2014.990863. This study uses in situ hybridization to show that the same late replicating DNA loci can localize either at the nuclear periphery, nucleolar periphery or pericentromeric heterochromatin, suggesting functional equivalence of these spatially varied late replicating compartments. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ryba T, Battaglia D, Pope BD, Hiratani I, Gilbert DM. Genome-scale analysis of replication timing: from bench to bioinformatics. Nat Protoc. 2011;6:870–895. doi: 10.1038/nprot.2011.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Woodfine K, Fiegler H, Beare DM, Collins JE, McCann OT, Young BD, Debernardi S, Mott R, Dunham I, Carter NP. Replication timing of the human genome. Hum Mol Genet. 2004;13:191–202. doi: 10.1093/hmg/ddh016. [DOI] [PubMed] [Google Scholar]
- 23.Gilbert DM. Evaluating genome-scale approaches to eukaryotic DNA replication. Nat Rev Genet. 2010;11:673–684. doi: 10.1038/nrg2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24*.Koren A, Handsaker RE, Kamitaki N, Karlić R, Ghosh S, Polak P, Eggan K, McCarroll SA. Genetic variation in human DNA replication timing [Internet] Cell. 2014;159:1015–1026. doi: 10.1016/j.cell.2014.10.025. This study shows that clusters of SNPs and small indels correlate with RT variations between individuals or between homologues, suggesting that small variations in DNA sequence may influence RT (albeit not replication origin specification as they conclude; see reference 28). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rhind N, Gilbert DM. DNA replication timing. Cold Spring Harb Perspect Biol. 2013;5 doi: 10.1101/cshperspect.a010132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26**.Pope BD, Ryba T, Dileep V, Yue F, Wu W, Denas O, Vera DL, Wang Y, Hansen RS, Canfield TK, et al. Topologically-associating domains are stable units of replication-timing regulation [Internet] Nature. 2014;515:402–405. doi: 10.1038/nature13986. Mapping RT transitions across multiple cell types demonstrates a near one to one correspondence of RDs and TADs and finds that LADs associate with both late replicating RDs and the temporal transition regions (TTRs) between early and late RDs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dileep V, Rivera-Mulia JC, Sima J, Gilbert DM. Large-Scale Chromatin Structure-Function Relationships during the Cell Cycle and Development: Insights from Replication Timing. [Internet] Cold Spring Harb Symp Quant Biol. 2015 doi: 10.1101/sqb.2015.80.027284. [DOI] [PubMed] [Google Scholar]
- 28.Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ryba T, Hiratani I, Lu J, Itoh M, Kulik M, Zhang J, Schulz TC, Robins AJ, Dalton S, Gilbert DM. Evolutionarily conserved replication timing profiles predict long-range chromatin interactions and distinguish closely related cell types. Genome Res. 2010;20:761–770. doi: 10.1101/gr.099655.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yaffe E, Farkash-Amar S, Polten A, Yakhini Z, Tanay A, Simon I. Comparative Analysis of DNA Replication Timing Reveals Conserved Large-Scale Chromosomal Architecture. PLoS Genet. 2010;6:e1001011. doi: 10.1371/journal.pgen.1001011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moindrot B, Audit B, Klous P, Baker A, Thermes C, de Laat W, Bouvet P, Mongelard F, Arneodo A. 3D chromatin conformation correlates with replication timing and is conserved in resting cells. 2012;40:9470–9481. doi: 10.1093/nar/gks736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pope BD, Aparicio OM, Gilbert DM. SnapShot: Replication timing. Cell. 2013;152:1390–1390.e1. doi: 10.1016/j.cell.2013.02.038. [DOI] [PubMed] [Google Scholar]
- 34.Naumova N, Imakaev M, Fudenberg G, Zhan Y, Lajoie BR, Mirny LA, Dekker J. Organization of the Mitotic Chromosome. Science. 2013;342:948–953. doi: 10.1126/science.1236083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35**.Dileep V, Ay F, Sima J, Vera DL, Noble WS, Gilbert DM. Topologically associating domains and their long-range contacts are established during early G1 coincident with the establishment of the replication-timing program. Genome Res. 2015;25:1104–1113. doi: 10.1101/gr.183699.114. Using 4C-seq to track chromatin interactions during the cell cycle this study demonstrates that both TADs and 3D interaction compartments are re-established coincident with the re-establishment of the RT program during early G1 phase. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lu J, Li F, Murphy CS, Davidson MW, Gilbert DM. G2 phase chromatin lacks determinants of replication timing. The Journal of Cell Biology. 2010;189:967–980. doi: 10.1083/jcb.201002002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Knott SRV, Peace JM, Ostrow AZ, Gan Y, Rex AE, Viggiani CJ, Tavaré S, Aparicio OM. Forkhead transcription factors establish origin timing and long-range clustering in S. cerevisiae. Cell. 2012;148:99–111. doi: 10.1016/j.cell.2011.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Davé A, Cooley C, Garg M, Bianchi A. Protein Phosphatase 1 Recruitment by Rif1 Regulates DNA Replication Origin Firing by Counteracting DDK Activity. Cell Reports. 2014;7:53–61. doi: 10.1016/j.celrep.2014.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hiraga SI, Alvino GM, Chang F, Lian HY, Sridhar A, Kubota T, Brewer BJ, Weinreich M, Raghuraman MK, Donaldson AD. Rif1 controls DNA replication by directing Protein Phosphatase 1 to reverse Cdc7-mediated phosphorylation of the MCM complex. Genes & Development. 2014;28:372–383. doi: 10.1101/gad.231258.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mattarocci S, Shyian M, Lemmens L, Damay P, Altintas DM, Shi T, Bartholomew CR, Thomä NH, Hardy CFJ, Shore D. Rif1 Controls DNA Replication Timing in Yeast through the PP1 Phosphatase Glc7. Cell Reports. 2014;7:62–69. doi: 10.1016/j.celrep.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 41.Peace JM, Ter-Zakarian A, Aparicio OM. Rif1 Regulates Initiation Timing of Late Replication Origins throughout the S. cerevisiae Genome. 2014;9:e98501. doi: 10.1371/journal.pone.0098501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamazaki S, Ishii A, Kanoh Y, Oda M, Nishito Y, Masai H. Rif1 regulates the replication timing domains on the human genome. EMBO J. 2012;31:3667–3677. doi: 10.1038/emboj.2012.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43**.Foti R, Gnan S, Cornacchia D, Dileep V, Bulut-Karslioglu A, Diehl S, Buness A, Klein FA, Huber W, Johnstone E, et al. Nuclear Architecture Organized by Rif1 Underpins the Replication-Timing Program. Mol Cell. 2016;61:260–273. doi: 10.1016/j.molcel.2015.12.001. ChIP-seq genome-wide analysis of Rif1 shows that late replicating domains are marked by Rif1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cornacchia D, Dileep V, Quivy J-P, Foti R, Tili F, Santarella-Mellwig R, Anthony C, Almouzni G, Gilbert DM, Buonomo SBC. Mouse Rif1 is a key regulator of the replication-timing programme in mammalian cells. EMBO J. 2012 doi: 10.1038/emboj.2012.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bechhoefer J, Rhind N. Replication timing and its emergence from stochastic processes. Trends in Genetics. 2012;28:374–381. doi: 10.1016/j.tig.2012.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46*.Kaykov A, Nurse P. The spatial and temporal organization of origin firing during the S-phase of fission yeast. Genome Res. 2015;25:391–401. doi: 10.1101/gr.180372.114. Methods to preserve DNA fibers at near chromosome length in fission yeast were developed to map positions and timing of replication initiation, revealing a strong stochastic component to origin firing throughout S phase. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sadoni N, Cardoso MC, Stelzer EHK, Leonhardt H, Zink D. Stable chromosomal units determine the spatial and temporal organization of DNA replication. J Cell Sci. 2004;117:5353–5365. doi: 10.1242/jcs.01412. [DOI] [PubMed] [Google Scholar]
- 48.Macaulay IC, Voet T. Single Cell Genomics: Advances and Future Perspectives. PLoS Genet. 2014;10:e1004126. doi: 10.1371/journal.pgen.1004126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schwartzman O, Tanay A. Single-cell epigenomics: techniques and emerging applications. Nat Rev Genet. 2015 doi: 10.1038/nrg3980. [DOI] [PubMed] [Google Scholar]
- 50.Nagano T, Lubling Y, Stevens TJ, Schoenfelder S, Yaffe E, Dean W, Laue ED, Tanay A, Fraser P. Single-cell Hi-C reveals cell-to-cell variability in chromosome structure. Nature. 2013;502:59–64. doi: 10.1038/nature12593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51*.Kind J, Pagie L, de Vries SS, Nahidiazar L, Dey SS, Bienko M, Zhan Y, Lajoie B, de Graaf CA, Amendola M, et al. Genome-wide Maps of Nuclear Lamina Interactions in Single Human Cells. Cell. 2015;163:134–147. doi: 10.1016/j.cell.2015.08.040. By adapting DamID technology to single cells, this study mapped genome-wide the interactions to the nuclear lamina identifying domains that varied from being at the periphery in only some to most or all cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52*.Van der Aa N, Cheng J, Mateiu L, Esteki MZ, Kumar P, Dimitriadou E, Vanneste E, Moreau Y, Vermeesch JR, Voet T. Genome-wide copy number profiling of single cells in S-phase reveals DNA-replication domains. 2013;41:e66–e66. doi: 10.1093/nar/gks1352. Using genome-wide CNV, the authors were able to see RT patterns in single cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lam ET, Hastie A, Lin C, Ehrlich D, Das SK, Austin MD, Deshpande P, Cao H, Nagarajan N, Xiao M, et al. Genome mapping on nanochannel arrays for structural variation analysis and sequence assembly. Nat Biotechnol. 2012;30:771–776. doi: 10.1038/nbt.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54*.Mak ACY, Lai YYY, Lam ET, Kwok T-P, Leung AKY, Poon A, Mostovoy Y, Hastie AR, Stedman W, Anantharaman T, et al. Genome-Wide Structural Variation Detection by Genome Mapping on Nanochannel Arrays. Genetics. 2015 doi: 10.1534/genetics.115.183483. In this study structural variants in the human genome were mapped using high-throughput methods to analyze individual long DNA fibers that may someday be adapted to study genome organization at single molecule resolution. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kitsberg D, Selig S, Brandeis M, Simon I, Keshet I, Driscoll DJ, Nicholls RD, Cedar H. Allele-specific replication timing of imprinted gene regions. Nature. 1993;364:459–463. doi: 10.1038/364459a0. [DOI] [PubMed] [Google Scholar]
- 56.Simon I, Tenzen T, Reubinoff BE, Hillman D, McCarrey JR, Cedar H. Asynchronous replication of imprinted genes is established in the gametes and maintained during development [Internet] 1999;401:929–932. doi: 10.1038/44866. [DOI] [PubMed] [Google Scholar]
- 57.Mostoslavsky R, Singh N, Tenzen T, Goldmit M, Gabay C, Elizur S, Qi P, Reubinoff BE, Chess A, Cedar H, et al. Asynchronous replication and allelic exclusion in the immune system. Nature. 2001;414:221–225. doi: 10.1038/35102606. [DOI] [PubMed] [Google Scholar]
- 58.Farago M, Rosenbluh C, Tevlin M, Fraenkel S, Schlesinger S, Masika H, Gouzman M, Teng G, Schatz D, Rais Y, et al. Clonal allelic predetermination of immunoglobulin-κ rearrangement. Nature. 2012;490:561–565. doi: 10.1038/nature11496. [DOI] [PubMed] [Google Scholar]
- 59.Avner P, Avner P, Heard E. X-chromosome inactivation: counting, choice and initiation. Nat Rev Genet. 2001;2:59–67. doi: 10.1038/35047580. [DOI] [PubMed] [Google Scholar]
- 60.Galupa R, Heard E. X-chromosome inactivation: new insights into cis and trans regulation. Curr Opin Genet Dev. 2015;31:57–66. doi: 10.1016/j.gde.2015.04.002. [DOI] [PubMed] [Google Scholar]
- 61.Splinter E, de Wit E, Nora EP, Klous P, van de Werken HJG, Zhu Y, Kaaij LJT, van Ijcken W, Gribnau J, Heard E, et al. The inactive X chromosome adopts a unique three-dimensional conformation that is dependent on Xist RNA. Genes & Development. 2011;25:1371–1383. doi: 10.1101/gad.633311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, Servant N, Piolot T, van Berkum NL, Meisig J, Sedat J, et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012 doi: 10.1038/nature11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Deng X, Ma W, Ramani V, Hill A, Yang F, Ay F, Berletch JB, Blau CA, Shendure J, Duan Z, et al. Bipartite structure of the inactive mouse X chromosome. Genome Biol. 2015;16:67. doi: 10.1186/s13059-015-0728-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Casas-Delucchi CS, Brero A, Rahn H-P, Solovei I, Wutz A, Cremer T, Leonhardt H, Cardoso MC. Histone acetylation controls the inactive X chromosome replication dynamics. Nat Comms. 2011;2:222. doi: 10.1038/ncomms1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65*.Koren A, McCarroll SA. Random replication of the inactive X chromosome. Genome Res. 2014;24:64–69. doi: 10.1101/gr.161828.113. This study used SNPs to establish allele-specific RT of the active and inactive X chromosomes. The results confirmed dramatic difference in RT of X homologues, and the highly synchronous late replication of the entire inactive X, previously shown by cytogenetic methods in ref: 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66*.Mukhopadhyay R, Lajugie J, Fourel N, Selzer A, Schizas M, Bartholdy B, Mar J, Lin CM, Martin MM, Ryan M, et al. Allele-specific genome-wide profiling in human primary erythroblasts reveal replication program organization. PLoS Genet. 2014;10:e1004319. doi: 10.1371/journal.pgen.1004319. By using SNPs to establish allele-specific RT this study shows that <10 % of the autosomal genome varies between homologues with large structural variants, but not SNPs, associated with RT changes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gerhardt J, Tomishima MJ, Zaninovic N, Colak D, Yan Z, Zhan Q, Rosenwaks Z, Jaffrey SR, Schildkraut CL. The DNA replication program is altered at the FMR1 locus in fragile X embryonic stem cells. Mol Cell. 2014;53:19–31. doi: 10.1016/j.molcel.2013.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68*.Gerhardt J, Zaninovic N, Zhan Q, Madireddy A, Nolin SL, Ersalesi N, Yan Z, Rosenwaks Z, Schildkraut CL. Cis-acting DNA sequence at a replication origin promotes repeat expansion to fragile X full mutation [Internet] The Journal of Cell Biology. 2014;206:599–607. doi: 10.1083/jcb.201404157. This study and ref 67 together demonstrate that a single SNP upstream of the FMR1 gene linked to Fragile X syndrome is associated with loss of a local origin activity, alteration in replication timing of the locus, and the disease phenotype. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69*.Bartholdy B, Mukhopadhyay R, Lajugie J, Aladjem MI, Bouhassira EE. Allele-specific analysis of DNA replication origins in mammalian cells. Nat Comms. 2015;6:7051–4. doi: 10.1038/ncomms8051. By deep sequencing of nascent strands and SNPs identification of individual homologous chromosomes, this study showed that RT differences associated with structural variations shown in ref 66 result from differences in efficiency of DNA replication origins, rather than silencing or de novo activation of origins. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stoffregen EP, Donley N, Stauffer D, Smith L, Thayer MJ. An autosomal locus that controls chromosome-wide replication timing and mono-allelic expression. Hum Mol Genet. 2011;20:2366–2378. doi: 10.1093/hmg/ddr138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Donley N, Stoffregen EP, Smith L, Montagna C, Thayer MJ. Asynchronous Replication, Mono-Allelic Expression, and Long Range Cis-Effects of ASAR6. PLoS Genet. 2013;9:e1003423. doi: 10.1371/journal.pgen.1003423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72**.Donley N, Smith L, Thayer MJ. ASAR15, A cis-Acting Locus that Controls Chromosome-Wide Replication Timing and Stability of Human Chromosome 15. PLoS Genet. 2015;11:e1004923. doi: 10.1371/journal.pgen.1004923. In this study a new long ncRNA involved in the control of RT, monoallelic gene expression and chromosomal stability was identified. As other ncRNAs with similar properties have been described for other chromosomes, this study proposed that these cis-acting elements might be present in all mammalian chromosomes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schübeler D, Scalzo D, Kooperberg C, van Steensel B, Delrow J, Groudine M. Genome-wide DNA replication profile for Drosophila melanogaster: a link between transcription and replication timing. Nat Genet. 2002;32:438–442. doi: 10.1038/ng1005. [DOI] [PubMed] [Google Scholar]
- 74.Schübeler D, MacAlpine DM, Scalzo D, Wirbelauer C, Kooperberg C, van Leeuwen F, Gottschling DE, O’Neill LP, Turner BM, Delrow J, et al. The histone modification pattern of active genes revealed through genome-wide chromatin analysis of a higher eukaryote. Genes & Development. 2004;18:1263–1271. doi: 10.1101/gad.1198204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.MacAlpine DM, Rodríguez HK, Bell SP. Coordination of replication and transcription along a Drosophila chromosome. Genes & Development. 2004;18:3094–3105. doi: 10.1101/gad.1246404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Huvet M, Nicolay S, Touchon M, Audit B, d’Aubenton-Carafa Y, Arneodo A, Thermes C. Human gene organization driven by the coordination of replication and transcription. Genome Res. 2007;17:1278–1285. doi: 10.1101/gr.6533407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Desprat R, Thierry-Mieg D, Lailler N, Lajugie J, Schildkraut CL, Thierry-Mieg J, Bouhassira EE. Predictable dynamic program of timing of DNA replication in human cells. Genome Res. 2009;19:2288–2299. doi: 10.1101/gr.094060.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schwaiger M, Stadler MB, Bell O, Bell O, Kohler H, Oakeley EJ, Schubeler D. Chromatin state marks cell-type- and gender-specific replication of the Drosophila genome. Genes & Development. 2009;23:589–601. doi: 10.1101/gad.511809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Maric C, Prioleau M-N. Interplay between DNA replication and gene expression: a harmonious coexistence. Curr Opin Cell Biol. 2010;22:277–283. doi: 10.1016/j.ceb.2010.03.007. [DOI] [PubMed] [Google Scholar]
- 80.Lubelsky Y, Prinz JA, DeNapoli L, Li Y, Belsky JA, Belsky JA, MacAlpine DM. DNA replication and transcription programs respond to the same chromatin cues. Genome Res. 2014;24:1102–1114. doi: 10.1101/gr.160010.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhou J, Ermakova OV, Riblet R, Birshtein BK, Schildkraut CL. Replication and subnuclear location dynamics of the immunoglobulin heavy-chain locus in B-lineage cells. Mol Cell Biol. 2002;22:4876–4889. doi: 10.1128/MCB.22.13.4876-4889.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Schultz SS, Desbordes SC, Du Z, Kosiyatrakul S, Lipchina I, Studer L, Schildkraut CL. Single-molecule analysis reveals changes in the DNA replication program for the POU5F1 locus upon human embryonic stem cell differentiation. Mol Cell Biol. 2010;30:4521–4534. doi: 10.1128/MCB.00380-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yue F, Cheng Y, Breschi A, Vierstra J, Wu W, Ryba T, Sandstrom R, Ma Z, Davis C, Pope BD, et al. A comparative encyclopedia of DNA elements in the mouse genome [Internet] Nature. 2014;515:355–364. doi: 10.1038/nature13992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Schulz TC, Young HY, Agulnick AD, Babin MJ, Baetge EE, Bang AG, Bhoumik A, Cepa I, Cesario RM, Haakmeester C, et al. A Scalable System for Production of Functional Pancreatic Progenitors from Human Embryonic Stem Cells. 2012;7:e37004. doi: 10.1371/journal.pone.0037004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Menendez L, Kulik MJ, Page AT, Park SS, Lauderdale JD, Cunningham ML, Dalton S. Directed differentiation of human pluripotent cells to neural crest stem cells. Nat Protoc. 2013;8:203–212. doi: 10.1038/nprot.2012.156. [DOI] [PubMed] [Google Scholar]
- 86**.Rivera-Mulia JC, Buckley Q, Sasaki T, Zimmerman J, Didier RA, Nazor K, Loring JF, Lian Z, Weissman S, Robins AJ, et al. Dynamic changes in replication timing and gene expression during lineage specification of human pluripotent stem cells [Internet] Genome Res. 2015;25:1091–1103. doi: 10.1101/gr.187989.114. By taking advantage of highly synchronous methods of human embryonic stem cell differentiation this study analyzed the dynamic changes in RT at specific cell fate transitions and its coordination with gene expression. In contrast with all previous literature, this study shows that correlation between early replication and gene expression is not maintained within developmentally regulated RDs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Takebayashi S-I, Ryba T, Gilbert DM. Developmental control of replication timing defines a new breed of chromosomal domains with a novel mechanism of chromatin unfolding. Nucleus. 2012;3:500–507. doi: 10.4161/nucl.22318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Besnard E, Babled A, Lapasset L, Milhavet O, Parrinello H, Dantec C, Marin J-M, Lemaitre JM. Unraveling cell type-specific and reprogrammable human replication origin signatures associated with G-quadruplex consensus motifs. Nat Struct Mol Biol. 2012;19:837–844. doi: 10.1038/nsmb.2339. [DOI] [PubMed] [Google Scholar]
- 89.Kuhn TS. Mathematical vs experimental traditions in the development of physical science. The Journal of Interdisciplinary History. 1976;7:1–31. [Google Scholar]
- 90.Chuang C-H, Carpenter AE, Fuchsova B, Johnson T, de Lanerolle P, Belmont AS. Long-range directional movement of an interphase chromosome site. Curr Biol. 2006;16:825–831. doi: 10.1016/j.cub.2006.03.059. [DOI] [PubMed] [Google Scholar]
- 91*.Khanna N, Hu Y, Belmont AS. HSP70 transgene directed motion to nuclear speckles facilitates heat shock activation. Curr Biol. 2014;24:1138–1144. doi: 10.1016/j.cub.2014.03.053. This study characterized the rapid movements of heat shock protein 70 (HSP70) loci from the nuclear periphery towards speckles upon induction of transcription by heat shock. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Finlan LE, Sproul D, Thomson I, Boyle S, Kerr E, Perry P, Ylstra B, Chubb JR, Bickmore WA. Recruitment to the Nuclear Periphery Can Alter Expression of Genes in Human Cells. PLoS Genet. 2008;4:e1000039. doi: 10.1371/journal.pgen.1000039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Reddy KL, Zullo JM, Bertolino E, Singh H. Transcriptional repression mediated by repositioning of genes to the nuclear lamina. Nature. 2008;452:243–247. doi: 10.1038/nature06727. [DOI] [PubMed] [Google Scholar]
- 94.Zullo JM, Demarco IA, Piqué-Regi R, Gaffney DJ, Epstein CB, Spooner CJ, Luperchio TR, Bernstein BE, Pritchard JK, Reddy KL, et al. DNA sequence-dependent compartmentalization and silencing of chromatin at the nuclear lamina. Cell. 2012;149:1474–1487. doi: 10.1016/j.cell.2012.04.035. [DOI] [PubMed] [Google Scholar]
- 95.Kumaran RI, Spector DL. A genetic locus targeted to the nuclear periphery in living cells maintains its transcriptional competence. The Journal of Cell Biology. 2008;180:51–65. doi: 10.1083/jcb.200706060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Jost KL, Bertulat B, Rapp A, Brero A, Hardt T, Domaing P, Gösele C, Schulz H, Hübner N, Cardoso MC. Gene repositioning within the cell nucleus is not random and is determined by its genomic neighborhood. Epigenetics & Chromatin. 2015;8:1050. doi: 10.1186/s13072-015-0025-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Amendola M, Amendola M, van Steensel B. Nuclear lamins are not required for lamina-associated domain organization in mouse embryonic stem cells. EMBO reports. 2015 doi: 10.15252/embr.201439789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Solovei I, Wang AS, Thanisch K, Schmidt CS, Krebs S, Zwerger M, Cohen TV, Devys D, Foisner R, Peichl L, et al. LBR and Lamin A/C Sequentially Tether Peripheral Heterochromatin and Inversely Regulate Differentiation. Cell. 2013;152:584–598. doi: 10.1016/j.cell.2013.01.009. [DOI] [PubMed] [Google Scholar]
- 99.Harr JC, Luperchio TR, Wong X, Cohen E, Wheelan SJ, Reddy KL. Directed targeting of chromatin to the nuclear lamina is mediated by chromatin state and A-type lamins. The Journal of Cell Biology. 2015;208:33–52. doi: 10.1083/jcb.201405110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Simon I. Developmental regulation of DNA replication timing at the human beta globin locus. EMBO J. 2001;20:6150–6157. doi: 10.1093/emboj/20.21.6150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lin CM, Fu H, Martinovsky M, Bouhassira EE, Bouhassira E, Aladjem MI. Dynamic alterations of replication timing in mammalian cells. Curr Biol. 2003;13:1019–1028. doi: 10.1016/s0960-9822(03)00382-8. [DOI] [PubMed] [Google Scholar]
- 102.Hassan-Zadeh V, Chilaka S, Cadoret J-C, Ma MK-W, Boggetto N, West AG, Prioleau M-N. USF binding sequences from the HS4 insulator element impose early replication timing on a vertebrate replicator. PLoS Biol. 2012;10:e1001277. doi: 10.1371/journal.pbio.1001277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Koryakov DE, Pokholkova GV, Maksimov DA, Belyakin SN, Belyaeva ES, Zhimulev IF. Induced transcription results in local changes in chromatin structure, replication timing, and DNA polytenization in a site of intercalary heterochromatin. Chromosoma. 2012 doi: 10.1007/s00412-012-0382-9. [DOI] [PubMed] [Google Scholar]
- 104*.Therizols P, Illingworth RS, Courilleau C, Boyle S, Wood AJ, Bickmore WA. Chromatin decondensation is sufficient to alter nuclear organization in embryonic stem cells. Science. 2014;346:1238–1242. doi: 10.1126/science.1259587. In this study a viral transactivator was employed to artificially induce gene expression, resulting in changes in nuclear repositioning and a moderate advance in RT. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Carpenter AE, Memedula S, Plutz MJ, Belmont AS. Common Effects of Acidic Activators on Large-Scale Chromatin Structure and Transcription. Mol Cell Biol. 2005;25:958–968. doi: 10.1128/MCB.25.3.958-968.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Pop R, Shearstone JR, Shen Q, Liu Y, Hallstrom K, Koulnis M, Gribnau J, Socolovsky M. A key commitment step in erythropoiesis is synchronized with the cell cycle clock through mutual inhibition between PU.1 and S-phase progression. PLoS Biol. 2010;8 doi: 10.1371/journal.pbio.1000484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Gilbert DM. Cell fate transitions and the replication timing decision point. The Journal of Cell Biology. 2010;191:899–903. doi: 10.1083/jcb.201007125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Cavalli G, Misteli T. Functional implications of genome topology. Nat Struct Mol Biol. 2013;20:290–299. doi: 10.1038/nsmb.2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Akhtar W, de Jong J, Pindyurin AV, Pagie L, Meuleman W, de Ridder J, Berns A, Wessels LFA, van Lohuizen M, van Steensel B. Chromatin position effects assayed by thousands of reporters integrated in parallel. Cell. 2013;154:914–927. doi: 10.1016/j.cell.2013.07.018. [DOI] [PubMed] [Google Scholar]


