Abstract
The hypothesis that the recent rapid evolution of primate cytochromes c, which primarily involves residues in the least stable Ω-loop (Ω-loop C, residues 40 – 57), stabilizes the heme crevice of cytochrome c relative to other mammals, is tested. To accomplish this goal, we have compared the properties of human and spider monkey cytochrome c and a set of four variants produced in the process of converting human cytochrome c into spider monkey cytochrome c. The global stability of all variants has been measured by guanidine hydrochloride denaturation. The stability of the heme crevice has been assessed with the alkaline conformational transition. Structural insight into the effects of the five amino acid substitutions needed to convert human cytochrome c into spider monkey cytochrome c is provide by a 1.15 Å resolution structure of spider monkey cytochrome c. The global stability for all variants is near 9.0 kcal/mol at 25 °C and pH 7, which is higher than that observed for other mammalian cytochromes c. The heme crevice stability is more sensitive to the substitutions required to produce spider monkey cytochrome c with decreases of up to 0.5 units in the apparent pKa of the alkaline conformational transition relative to human cytochrome c. The structure of spider monkey cytochrome c indicates that the Y46F substitution destabilizes the heme crevice by disrupting an extensive hydrogen bond network that connects three surface loops including Ω-loop D (residues 70–85), which contains the Met80 heme ligand.
Keywords: Spider Monkey Cytochrome c, Global stability, Alkaline conformational transition, Heme crevice stability, Hydrogen bond network
Graphical Abstract
Destabilization of the heme crevice during the rapid evolution of primate cytochromes c is primarily due to a Y46F substitution, which disrupts of a hydrogen bond network that knits together three surface Ω-loops.
1. Introduction
Cytochrome c (Cytc) resides in the intermembrane space of mitochondria. Its function as a component of the electron transport chain has been known for many years [1, 2]. About 20 years ago, it was discovered that release of Cytc from mitochondria, followed by its interaction with Apaf-1 to form the apoptosome [3, 4], is a critical signaling event in the intrinsic pathway of apoptosis [5, 6]. Furthermore, peroxidative activity, particularly directed at the inner mitochondrial membrane lipid, cardiolipin (CL), is believed to be the earliest signal in the intrinsic pathway of apoptosis [7]. More recently it has been postulated that the peroxidase activity of Cytc may provide a rich source of CL-based lipid signaling agents [8, 9]. The peroxidase activity of Cytc is linked to loss of the Met80-heme bond [5, 6]. Thus, there has been considerable interest recently in the factors that control the stability of the heme crevice [10, 11].
It has been noted recently that there has been a rapid evolution of Cytc in the primate lineage [12, 13]. It has been postulated that this evolution may be coupled to evolution of cytochrome c oxidase. However, many of the mutations in the evolution of primate cytochromes c are found in a surface loop, Ω-loop C (residues 40–57), which is believed to modulate the opening of the heme crevice in the alkaline conformational transition [14]. Human Cytc (Hu Cytc) and spider monkey Cytc (SM Cytc) are on different branches of the evolutionary tree of primate cytochromes c. Furthermore, it has been postulated that the A50E substitution, found in SM Cytc relative Hu Cytc, may destabilize the heme crevice by providing an electrostatic interaction that can compete with the hydrogen bond between the ε-amino group of Lys79 and the main chain carbonyl of residue 47 [15].
As an initial test of the hypothesis that the rapid evolution of primate cytochromes c may also affect the stability of the heme crevice and thus could be important for the peroxidase activity of Cytc in apoptosis, we have prepared SM Cytc from Hu Cytc in a stepwise manner. There are only five substitutions in the sequence of SM Cytc relative to Hu Cytc [15]. However, four of these substitutions are in Ω-loop C. Here, we characterize the global stability of Hu Cytc, SM Cytc and the four intermediate variants used to produce SM Cytc from Hu Cytc. To characterize the effects of these substitutions on the stability of the heme crevice, we have measured the alkaline conformational transition of the set of six cytochromes c. We also report a 1.15 Å structure of SM Cytc that provides high resolution structural insight into the effects of these mutations on heme crevice stability.
We find that the effects of the five substitutions required to produce SM Cytc from Hu Cytc on the global stability of Cytc are modest. However, the apparent pKa of the alkaline transition is decreased by up to 0.5 units. The Y46F mutation has the largest effect on the alkaline transition. This observation is supported by comparison of our SM Cytc structure with that of Hu Cytc, which shows that the Y46F mutation compromises the integrity a hydrogen bond network that holds together three surface loops in the structure of Cytc.
2. Experimentals
2.1. Preparation of the spider monkey cytochrome c expression vector
The pBTR(HumanCc) plasmid for expression of Hu Cytc in Escherichia coli was provided by Gary Pielak at the University of North Carolina [3]. The pBTR(HumanCc) plasmid is a derivative of the pBTR1 plasmid [16, 17] produced by replacing the yeast iso-1-Cytc gene (CYC1) with a synthetic Hu Cytc gene [3, 18]. Thus, the plasmid co-expresses the yeast heme lyase gene (CYC3) permitting covalent attachment of heme in the cytoplasm of E. coli. SM Cytc differs at five sequence positions from Hu Cytc [15]. The necessary K8R, P44S, Y46F, S47T and A50E mutations were introduced sequentially starting at the N-terminus of the protein using the QuikChange PCR-based mutagenesis protocol (Agilent) producing the set of variants K8R, K8R/P44S, K8R/P44S/Y46F, K8R/P44S/Y46F/S47T and finally SM Cytc (K8R/P44S/Y46F/S47T/A50E). The final SM Cytc plasmid pBTR(SMCytc) is available through Addgene (https://www.addgene.org/). Primers used for mutagenesis are listed in Table S1 of the Supplementary Data. Plasmid DNA for each variant was prepared from TG-1 E. coli cells transformed with DNA produced by the QuikChange protocol using the Qiagen miniprep procedure (Qiagen). The entire coding region of the Cytc gene was sequenced at the Murdock DNA Sequencing facility at the University of Montana to confirm the identity of each variant.
2.2. Expression of Cytc variants
Cytc variants were expressed after transformation of the expression plasmid into competent E. coli BL21 DE3 cells (EdgeBio). Transformed cells on agar plates were re-suspended in sterile L-broth (4 mL) and used to inoculate 1 L of sterilize 2xYT broth (1 L deionized H2O, 16 g tryptone, 10 g yeast extract, 5 g NaCl) in a Fernbach flask. 2 mL of sterile ampicillin (100 mg/mL) was added to each flask. All flasks were grown in an orbital shaker incubator (Thermo Forma 435) at 30 °C or 37 °C for 24 h.
After growth, the cells were pelleted using a Sorvall RC 5C+ centrifuge (Thermo) with a GS3 rotor (5,000 rpm) and stored at −80 °C. Protein was isolated from cell pellets using methods similar to those reported for yeast iso-1-cytochrome c (iso-1-Cytc) [19]. In brief, before mechanically lysing the cells, they were subjected to three freeze-thaw cycles at −80 °C (3 h) and 4 °C (2 h), respectively. Cell pellets were then re-suspended in 2 mL of lysis buffer (100 mM Tris, 1 mM NaEDTA, 500 mM NaCl, pH 8.0) per gram of cells. A small amount of lyophilized DNase and RNase powder was added along with the protease inhibitor PMSF to a final concentration of 3 mM. The resuspended cells were then passed through a French Pressure Cell (SLM Aminco) a minimum of four times. The lysed cells were spun down at 10,000 rpm (GSA rotor) in a Sorvall RC 5C+ centrifuge for 30 min and the supernatant was passed through a filter cloth. The supernatant was brought to 50% saturation with ammonium sulfate and stirred overnight at 4 °C, followed by centrifugation at 10,000 rpm (GSA rotor) to remove precipitated protein impurities. The supernatant was passed through filter cloth and dialyzed against 12.5 mM sodium phosphate, pH 7.2, 1.0 mM EDTA, 2.0 mM β-mercaptoethanol. The dialyzed protein was batch loaded onto CM Sepharose fast-flow beads (GE Healthcare) and the resin packed into glass column (3.2 cm × 18.4 cm) followed by elution of Cytc with a 200 mL linear gradient from 0 to 0.8 M NaCl in 50 mM sodium phosphate, pH 7.2, 1mM EDTA, 2 mM β-mercaptoethanol. Cytc containing fractions were exchanged into 50 mM sodium phosphate at pH 7 (HPLC buffer A) using an Amicon stirred ultrafiltration cell and then concentrated to ~1.6 mg/mL using centrifuge ultrafiltration. Aliquots of 1.5 mL were flash frozen and stored at −80°C freezer for later use. Yields of SM Cytc are 9 – 10 mg per liter of 2x YT culture.
Aliquots were thawed and purified by HPLC using a Bio-Rad UNO S6 column immediately in advance of experiments. The following gradient at a flow rate of 3.0 mL/min was used for purification: 0 % B from 0 to 7 min, increase to 30 % B from 7 to 34 min, maintain 30 % B from 34 to 40 min., increase to 100 % B from 40 to 43 min, maintain 100 % B from 43 to 50 min, decrease to 0 % B from 50 to 53 min, maintain 0 % B from 53 to 63 min. HPLC buffer B is HPLC buffer A with 1.0 M NaCl. Pure Cytc fractions were concentrated by ultrafiltration.
The molecular weight of the SM Cytc was measured to be 12,307.7 g/mol (M+H+, expected M+H+ = 12,309.1 g/mol) using a Bruker microflex MALDI-ToF mass spectrometer. Protein Calibration Standard I (Bruker Part No. 206355) was used to calibrate the mass spectrometer.
2.3 Oxidization of Cytc
All experiments were performed on the oxidized state, Fe(III)heme, of Cytc. Potassium ferricyanide was used to oxidize the protein using 5 mg of ferricyanide per mg of protein. After 15 min at 4 °C, the Cytc solution was spun in a microcentrifuge for 5 min and loaded on a column packed with Sephadex G-25 superfine size-exclusion resin. The sephadex G-25 resin was equilibrated and run with the buffer to be used in the experiment.
2.4 Measurement of global stability by guanidine hydrochloride denaturation
Circular dichroism (CD) spectroscopy was used to monitor guanidine hydrochloride (GdnHCl) unfolding of Cytc variants. A 4 µM Cytc sample was prepared in CD Buffer (20 mM Tris, 40 mM NaCl adjusted to pH 7.0). A 6 M GdnHCl solution in CD Buffer, also containing 4 µM Cytc, was used to titrate the Cytc sample in CD buffer. Titrations were carried out at 25 °C using a Hamilton Microlab 500 titrator interfaced with an Applied Photophysics Chirascan CD spectrophotometer. Data were collected at 222 nm using 250 nm as background to account for instrumental drift during the titration.
Plots of background corrected ellipticity at 222 nm, θ222corr (θ222corr = θ222 – θ250), versus GdnHCl concentration were fit to a linear free energy relationship between the free energy of unfolding, ΔGu, and GdnHCl concentration using nonlinear least squares methods (SigmaPlot 7), and Eq. (1) [20], where θN is the ellipticity of the native state of Cytc, θD is the ellipticity of the
| (1) |
denatured state extrapolated to 0 M GdnHCl, mD is the slope of the denatured state baseline as a function of GdnHCl concentration, ΔGu°(H2O) is ΔGu extrapolated to 0 M GdnHCl and m is the slope of ΔGu as a function GdnHCl concentration.
2.5 Measurement of local stability via the alkaline conformational transition
The alkaline conformation transition of the Cytc variants was measured by pH titration monitored at 695 nm, an absorbance band sensitive to Met80-hemeFe(III) ligation [2, 21, 22]. A 400 µM protein in 200 mM NaCl solution (2x-stock) was mixed 1:1 with deionized water to make the titration solution (200 µM Cytc in 100 mM NaCl). An equal volume of HCl solution and the 2x-stock was added to adjust the titration solution to about pH 6.3. For each step of the titration, equal volumes of an NaOH solution of appropriate concentration and of the 2x-stock solution were added to raise the pH of the titration solution by 0.15–0.20 pH units. The pH was recorded with a semimicro calomel electrode and the spectrum was recorded from 750 to 500 nm at each step. The titration was continued until a pH of 11 was achieved. Titrations were carried out at room temperature, 23 ± 1 °C.
The absorbance at 750 nm, A750, was subtracted from the absorbance at 695 nm, A695, to correct for instrumental drift during the titration. The corrected absorbance at 695 nm, A695corr (A695corr = A695 – A750), was plotted against pH and fit to a modified form of the Henderson-Hasselbalch equation, Eq. (2), which accounts for the number of protons linked to the alkaline transition, n.
| (2) |
Fits were carried out using nonlinear least squares methods (SigmaPlot 7) yielding, n and the apparent pKa, pKapp, of the alkaline conformational transition.
2.6 Ionic strength dependence of the alkaline conformational transition of SM Cytc
The methods of Osheroff et al. [15] were used to carry out measurement of the alkaline conformational transition as a function of ionic strength. A 25 mM Tris/acetate buffer, pH 7.0, stock buffer was prepared by titrating acetic acid with 1 M Tris base solution until a pH of 7 was achieved. The buffer was adjusted to its final volume in a volumetric flask such that the concentration of acetate was 25 mM. For pH titrations with NaOH, each TrisH+ titrated by NaOH is replace by Na+, so the ionic strength does not change with pH. This buffer was diluted to 10 mM for oxidation of SM Cytc and separation of oxidized protein from ferricyanide by Sephadex G25 chromatography. To increase the ionic strength of the solution, NaCl was added to create the desired solution ionic strength. Titrations were carried out and data were analyzed as described in section 2.5.
2.7. Crystallization, data collection, and structure determination of SM Cytc
The isolated SM Cytc protein was concentrated to 37 mg/ml by centrifuge ultrafiltration in a buffer of 50 mM Tris·HCl, pH 7. The initial crystallization condition was obtained by commercial crystallization screening trials. Additional vapor diffusion crystallization experiments were set up in grid screening by expanding the pH range and precipitant concentrations in 24-well VDX plates. After 2 to 4 weeks of equilibration at 20 °C, large cubic-shaped crystals were obtained by mixing 1 µL of protein with an equal volume of 0.1 M LiCl, 20% PEG 6000, 0.1 M HEPES sodium salt, pH 8.2, reservoir solution. For cryoprotection, crystals were transferred into reservoir solution containing 30% (v/v) PEG 400 and flash-frozen in a 100 K nitrogen gas stream.
Diffraction data were collected at the Stanford Synchrotron Radiation Lightsource SSRL-SMB 14-1 beamline using a MAR325 CCD detector at an incident beam wavelength of 1.22 Å. Images were indexed, integrated, and scaled using HKL2000 [23]. The initial phasing map was determined by molecular replacement with Phaser, integrated in the PHENIX software suite [24], using the coordinates of yeast iso-1-Cytc (PDB 2YCC) as a search model, against 8 to 2 Å experimental data. A continuous electron density map with a clearly defined protein-solvent boundary was observed. The initial model was first subjected to one cycle of rigid-body refinement using PHENIX. Subsequently, the model was further refined by iterative model rebuilding with COOT [25] and refinement of atomic positions, real space, occupancy, and isotropic B-factor parameters with PHENIX using a subset of reflections for calculation of Rfree (Table 1). At a later stage of refinement, the refined structure was improved to a final resolution of 1.15 Å and the coordinates of hydrogen atoms were included in the subsequent refinement cycles. Data collection and refinement statistics of the final model are shown in Table 1. The coordinates and structure factors for spider monkey Cytc have been deposited in the Protein Data Bank (PDB) under the ID code: 5DFS.
Table 1.
X-ray crystallography data collection and refinement statistics.
| Beamline | 14.1 |
| Wavelength (Å) | 1.22 |
| Resolution range (Å) | 15.0 – 1.15 (1.19 – 1.15)a |
| Space group | P 21 |
| Unit cell dimensions | |
| a, b, c (Å) | 31.5, 42.7, 65.7 |
| α, β, γ (°) | 90, 94.5, 90 |
| Total reflections | 172044 |
| Unique reflections | 62439 (6168) |
| Multiplicity | 2.8 (2.7)a |
| Completeness (%) | 97.8 (97.6)a |
| Mean I/σ(I) | 15.4 (2.05)a |
| Wilson B-factor | 9.30 |
| Rsymb | 0.065 (0.615)a |
| Refinement | |
| Rworkc | 0.159 (0.241)a |
| Rfreec | 0.182 (0.249)a |
| Number of total atoms | 2099 |
| protein molecule | 1679 |
| Ligands | 112 |
| Water | 308 |
| Total protein residues | 208 |
| RMS (bonds, Å) | 0.018 |
| RMS (angles, °) | 1.64 |
| Ramachandran favored (%)d | 98 |
| Rotamer outliers (%) | 0 |
| Average B-factor | 14.50 |
| macromolecules | 13.10 |
| Ligands | 11.10 |
| Solvent | 23.40 |
Data for highest resolution shell are given in brackets.
Rsym =∑hkl ∑i | Ii (hkl)- 〈I (hkl)〉|/∑hkl ∑i Ii (hkl) where Ii (hkl) is the ith observation of the intensity of the reflection hkl.
Rwork=∑hkl || Fobs|-|Fcalc||/∑hkl |Fobs|, where Fobs and Fcalc are the observed and calculated structure-factor amplitudes for each reflection hkl. Rfree was calculated with 6% of the diffraction data that were selected randomly and excluded from refinement.
Calculated using MolProbity (Chen et al., 2010).
3. Results and Discussion
3.1 Global Stability of Human and Spider Monkey Cytc
Spider monkey cytochrome c, SM Cytc, has five substitutions relative to Human Cytc, Hu Cytc. Most of these substitutions occur in the Ω-loop that runs from residues 40 to 57 (Ω-loop C). The sole substitution outside of this region is in the N-terminal helix, a K8R substitution. We made this conservative mutation first expecting that its effect on stability would be modest. The remaining four substitutions occur in a short sequence segment from residues 44 to 50. Based on native-state hydrogen-deuterium exchange (NSHX) studies on equine Cytc and limited proteolysis on yeast iso-1-Cytc, this Ω-loop is the least stable part of mitochondrial Cytc [26, 27]. Thus, changes in stability are expected to propagate upward affecting the global stability of Hu Cytc versus SM Cytc [28, 29]. Mutations were made sequentially starting at position 44 and ending with the position 50 substitution (A50E).
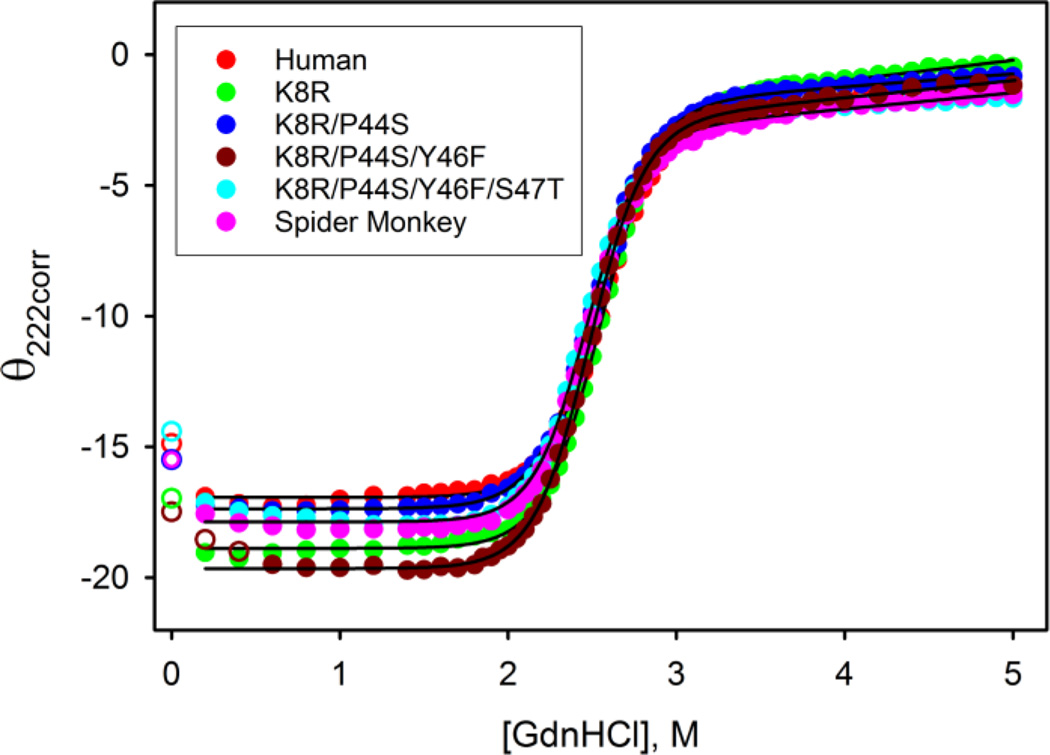
To assess the effect of each mutation on global stability we carried out GdnHCl denaturation of each protein. The unfolding titration curves for Hu Cyt, SM Cytc and the four intervening variants are shown in Fig. 1. It is evident from the GdnHCl denaturation data that the unfolding midpoint concentration, Cm, for all six sequence variants of Cytc is similar. The ΔGu°(H2O) of 8.9 ± 0.2 kcal/mol (Table 2) that we observe for the Hu Cytc, is similar to global stability of ~ 8.7 kcal/mol at 298 K reported for Hu Cytc based on thermal denaturation data [3]. Our GdnHCl data also indicate that Hu Cytc is more stable that equine or bovine Cytc as determined by GdnHCl denaturation methods (Table 2). Thermal denaturation data also show that Hu Cytc is more stable than equine Cytc (stability ~6.6 kcal/mol at 298 K, see ref. [3]). Table 2 shows that SM Cytc and the set of variants used to produce SM Cytc from Hu Cytc all have global stability similar to Hu Cytc.
Fig. 1.
Observed changes in corrected ellipticity, θ222corr, as a function of GdnHCl concentration for Hu Cytc, SM Cytc and the set of variants required to convert Hu Cytc into SM Cytc. The solid lines are fits to Eq. (1) in section 2.4 of the Experimentals. Parameters from the fits to Eq. (1) are collected in Table 2. All data were obtained at 25 °C in 20 mM Tris, pH 7, 40 mM NaCl.
Table 2.
Global stability parameters from GdnHCl denaturation of Cytc variants at 25 °C and pH 7.
| Variant | ΔGu°(H2O) | m | Cm |
|---|---|---|---|
| Human | 8.9 ± 0.2 | 3.51 ± 0.06 | 2.54 ± 0.01 |
| K8R | 9.0 ± 0.1 | 3.48 ± 0.04 | 2.58 ± 0.04 |
| K8R/P44S | 8.8 ± 0.2 | 3.52 ± 0.11 | 2.51 ± 0.02 |
| K8R/P44S/Y46F | 8.7 ± 0.2 | 3.44 ± 0.17 | 2.52 ± 0.06 |
| K8R/P44S/Y46F/S47T | 9.0 ± 0.1 | 3.66 ± 0.03 | 2.45 ± 0.01 |
| K8R/P44S/Y46F/S47T/A50E (Spider Monkey) |
8.7 ± 0.4 | 3.49 ± 0.19 | 2.48 ± 0.02 |
| Equinea | 7.27 | 3.01 | 2.42 |
| Bovinea | 8.38 | 3.19 | 2.62 |
| Yeastb | 5.77 ± 0.4 | 5.11 ± 0.36 | 1.13 ± 0.02 |
Hu Cytc and SM Cytc and the set of variants used to convert Hu Cytc to SM Cytc also have similar denaturant m-values of ~3.5 kcal mol−1M−1. These values are higher than observed for equine and bovine Cytc but smaller than observed for yeast iso-1-Cytc. Analysis of the denaturant dependence of the stability of the substructures of equine Cytc derived from NSHX studies shows that there is a significant population of intermediates in the transition region for GdnHCl unfolding of equine Cytc [30]. The lower GdnHCl m-value for equine Cytc (Table 1, see ref [31]) versus Hu and SM Cytc is consistent with a greater population of intermediates during global unfolding of equine Cytc than for primate cytochromes c. Limited proteolysis studies on yeast iso-1-Cytc show that the least stable substructure is close to the global stability of iso-1-Cytc [27]. The m-value of iso-1-Cytc [32] is larger than for Hu and SM Cytc, indicating that population of intermediates is less during global unfolding of yeast than for the primate cytochromes c. Thus, the intermediate magnitude of the m-value for Hu and SM Cytc relative to equine and yeast suggests that the free energy required to unfold the least stable substructure of Hu and SM Cytc is greater than for equine Cytc.
3.2 Alkaline conformational transition of Hu and SM Cytc
The alkaline conformational transition of oxidized, Fe(III)heme, Cytc occurs at mildly alkaline pH [2, 33]. It involves replacement of the native state heme ligand, Met80, with lysine side chains from Ω-loop D (residues 70–85), altering the structure of this Ω-loop [34, 35]. The alkaline conformational transition correlates well with the stability of the unstable substructure of Cytc corresponding to Ω-loop D [36] and thus is often used as a proxy for the stability of the heme crevice. Hydrogen exchange methods indicate that the rate of the alkaline conformational transition is limited by opening of Ω-loop C (residues 40–57) the least stable substructure of mitochondrial cytochromes c [26, 27]. Thus, the substitutions in this region required to convert Hu to SM Cytc might be expected to affect the alkaline conformational transition.
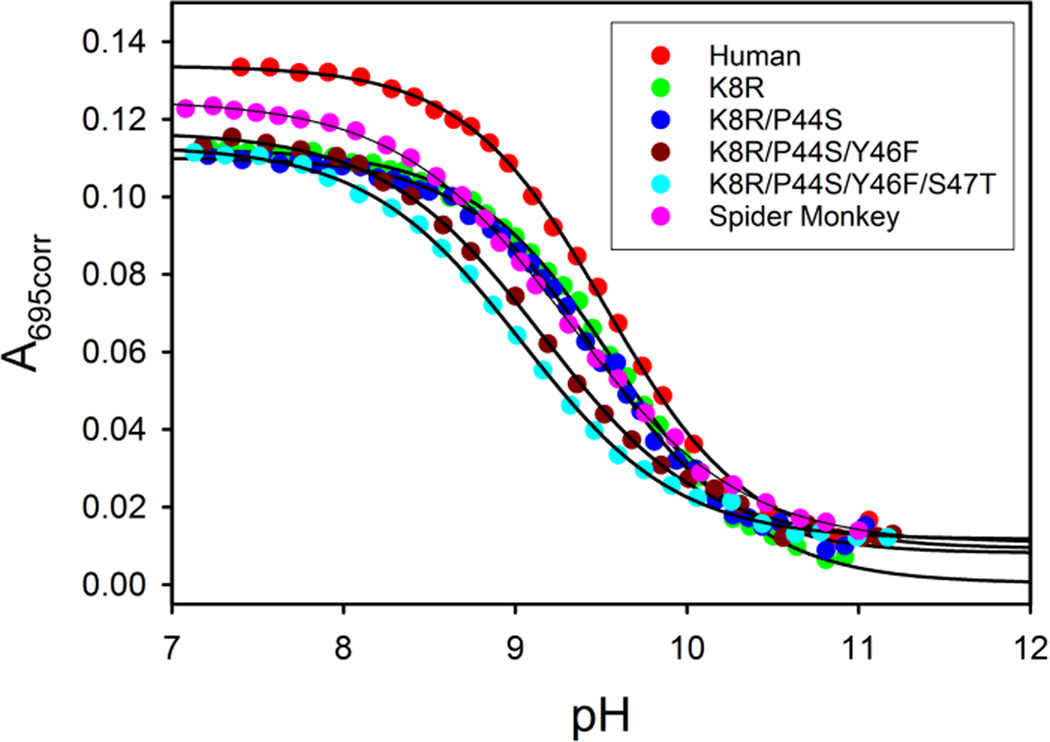
Fig. 2 shows that, unlike global stability, the local stability of Ω-loop D is sensitive to mutations in Ω-loop C. The magnitude of pKapp for Hu Cytc and the K8R and K8R/P44S variants is within error unchanged (Table 3). However, addition of the Y46F mutation leads to a significant decrease of 0.4 units in pKapp relative to Hu Cytc. There is a small additional decrease in pKapp when the S47T mutation is added. The combined effect these two mutations leads to a destabilization of 0.66 kcal/mol of the native state of Hu Cytc relative to its alkaline conformer. The final mutation (A50E), producing SM Cytc, partially reverses the destabilizing effect of the Y46F and S47T mutations on the native state relative to the alkaline state of Cytc. Thus, the net destabilization of the native state relative to the alkaline state for SM versus Hu Cytc is only ~0.3 kcal/mol (ΔΔGalk = ln(10)RTΔpKapp).
Fig. 2.
Alkaline conformational transition for Hu Cytc, SM Cytc and variants used to convert Hu to SM Cytc. Plots of A695corr versus pH are fit to Eq. (2) in the Experimental (solid curves). Parameters from the fits to Eq. (2) are collected in Table 3. Data were obtained in 100 mM NaCl at ~200 µM protein concentration and at 23 ± 1 °C.
Table 3.
Parameters for the alkaline conformational transition of Cytc at 23 ± 1 °C in 0.1 M NaCl solution.
| Variant | pKapp | n |
|---|---|---|
| Human | 9.54 ± 0.03 | 1.03 ± 0.02 |
| K8R | 9.63 ± 0.07 | 1.02 ± 0.06 |
| K8R/P44S | 9.51 ± 0.05 | 1.06 ± 0.06 |
| K8R/P44S/Y46F | 9.15 ± 0.02 | 0.93 ± 0.02 |
| K8R/P44S/Y46F/S47T | 9.05 ± 0.01 | 0.90 ± 0.05 |
| Spider Monkey | 9.32 ± 0.03 | 0.87 ± 0.01 |
There are a number of measurements of the pKapp for the alkaline conformational transition of Hu Cytc available in the literature. Margoliash and coworkers reported the pKapp of Hu Cytc isolated from human heart (with N-terminal acetylation) as 9.50 at 10 mM ionic strength rising to 9.85 at 250 and 500 mM ionic strength. Our pKapp of 9.54 obtained at ~100 mM ionic strength falls in the middle of this range as would be expected. Under similar conditions, equine Cytc has a pKapp ~ 0.4 to 0.45 units lower [15]. Thus, the lower global stability of equine Cytc relative to Hu Cytc is correlated with lower stability of the heme crevice as measured by the alkaline conformational transition. Other reported values for the pKapp of Hu Cytc are for E. coli-expressed Hu Cytc (not N-terminally acetylated) and range from 9.0 to 9.9 [37–41]. The pKapp of Cytc is sensitive to ionic strength and specific ion interactions [15, 42, 43]. Some of this observed range likely results from specific anion binding effects due to phosphate and citrate in the titration buffers used.
For Hu Cytc, the pKapp for a Y46F variant has been reported. Relative to the value reported for Hu Cytc, the pKapp decreases by ~0.6 units for the Y46F variant, similar the ~0.4 unit decrease in pKapp we observe when the Y46F mutation is added to the K8R/P44S variant (Table 3).
Hu Cytc and the K8R and K8R/P44S all have alkaline conformational transitions consistent with the release of a single proton (n = 1, see Table 3) being linked to the conformational change. After addition of the Y46F mutation, n drops to ~0.9. The decrease in n may reflect a change in coupling of protonation equilibria to the alkaline conformational transition, which have been shown to be complex in this pH regime [44–46]. It may also reflect mutation-induced changes in the pKa of one of the lysine ligands contributing to the alkaline state leading to a broadening of the transition [47, 48].
3.3 Ionic strength dependence of pKapp
The pKapp of 9.32 ± 0.03 we observed for SM Cytc in 0.1 M NaCl is similar to the pKapp of 9.3 observed by Margoliash and coworkers in the presence of 0.25 M sodium acetate [15]. However, at low ionic strength (10 mM Tris acetate pH 7) the pKapp observed by Margoliash and coworkers decreased to 8.70 [15]. They proposed that the large decrease in the pKapp observed for SM Cytc was due to an electrostatic interaction between Glu50 and Lys79 competing with the heme crevice stabilizing hydrogen bond between the carbonyl-O of residue 47 and the ε-amino group of Lys79. The observation that the A50E mutation partially reverses the effect of the Y46F and S47T mutations appears at odds with this proposal. Therefore, we studied the ionic strength dependence of the pKapp of SM Cytc.
In 10 mM Tris acetate, we find that the pKapp of E. coli-expressed SM Cytc is similar to that observed in 100 mM NaCl and about 0.5 units higher that observed for SM Cytc obtained from its natural source (Table 4). Addition of NaCl to 10 mM Tris acetate to raise the ionic strength to 50 mM and 250 mM has only modest effects on the pKapp. The large difference in the pKapp in 10 mM Tris acetate for SM Cytc expressed from E. coli versus from its natural source, points to the N-terminal acetylation of SM Cytc from its native source as a possible cause for the observed difference.
Table 4.
Parameters for the alkaline conformational transition of SM Cytc at 23 ± 1 °C as a function of solution ionic strength.
| Ionic Strengtha | Spider Monkey (E. Coli-expressed) |
Spider Monkey (Natural Source)b |
|
|---|---|---|---|
| pKapp | n | pKapp | |
| 10 mMc | 9.21 ± 0.03 | 0.79 ± 0.04 | 8.70 ± 0.05 |
| 50 mM | 9.15 ± 0.05 | 0.78 ± 0.01 | _ |
| 250 mM | 9.22 ± 0.02 | 0.89 ± 0.05 | 9.30 ± 0.05 |
Ionic strength adjusted with NaCl in the presence of 10 mM Tris acetate
Values from ref. [15]. Values of n not reported individually, a range of 0.8 to 1.1 was indicated. 10 mM value was obtained in the presence of Tris acetate. Ionic strength for 250 mM was adjusted with sodium acetate.
10 mM Tris acetate, no NaCl added.
It has been shown that phosphate, but not acetate, can strongly increase the intensity of the 695 nm absorbance band suggesting that phosphate can strengthen the heme-Met80 bond [49]. While acetate binds only weakly to cytochrome c [42], both phosphate and chloride [50] bind specifically. Thus, as a control experiment the effect of increasing Cl− concentration on the 695 nm absorbance band of SM Cytc was monitored. In contrast to phosphate binding to horse Cytc [49], no significant effect is observed from 0 to 250 mM Cl− on the magnitude of the extinction coefficient at 695 nm (Fig. S1). Thus, Cl− does not appear to directly affect the strength of the heme-Met80 bond for SM Cytc.
Interestingly, there is a drop in the number of protons, n, linked to the alkaline conformational transition at ionic strengths of 10 mM and 50 mM by ~0.1 relative to the n observed in 100 mM NaCl and at an ionic strength of 250 mM (Tables 3 and 4). It has also been shown that the alkaline transition is less cooperative at low ionic strength for horse Cytc [45]. Loss of cooperativity can be achieved if the alkaline transition involves two ionizable ligands, where one of the ionizable ligands has a significantly lower pKa than the other ligand but does not drive the alkaline transition to completion [51]. Fig. S2 demonstrates that an equally good fit to the data as with Eq. (2) can be achieved at low ionic strength with this model assuming that there are two lysine ligands for the alkaline state with significantly different pKa values [47]. The N-terminal amino group of E. coli-expressed horse Cytc can participate in the alkaline conformational transition [36]. Thus, we cannot rule out the N-terminal amino group as a ligand in the alkaline state.
3.4 High resolution X-ray structure of SM Cytc
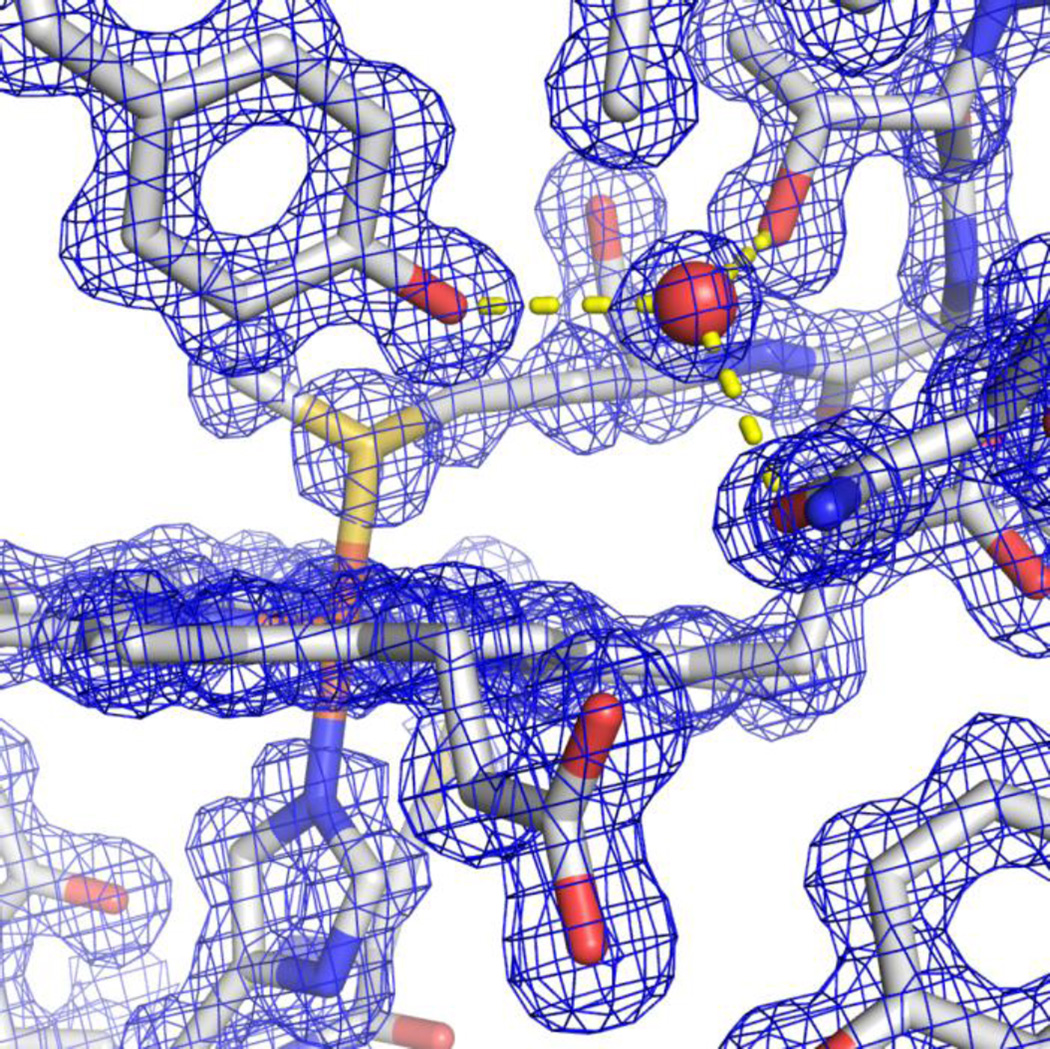
To gain insight into the differences in the alkaline conformational transition between Hu Cytc and SM Cytc we prepared crystals of SM Cytc. The crystals diffracted to 1.15 Å resolution providing a high quality structural model for SM Cytc (Rwork = 0.159; Rfree = 0.182; see Table 1). Fig. 3 shows the electron density around the heme including the hydrogen bonding network that includes Tyr67, Asn52, Thr78 and a buried water molecule, showing the high quality of the electron density map and its fit to the model. There are two molecules of SM Cytc (chains A and B) in the asymmetric unit, which align with a Cα RMS deviation of 0.385 Å. There are different conformers for 17 side chains in the two molecules of the asymmetric unit. Most of these are at solvent exposed sites. Two side chains in chain A and four in chain B were fit to two rotamers. Only Arg91 has two rotamers in both chains. Several of these alternate side chain conformers will be highlighted in the discussion that follows.
Fig. 3.
Electron density around the heme of chain B of SM Cytc (pdb code: 5DFS) showing buried water (W 332 of chain B) with hydrogen bonds (yellow dashed lines) to the −OH of Tyr67 (2.78 Å), the side chain hydroxyl of Thr78 (2.67 Å) and the carbonyl of the primary amide side chain of Asn52 (2.72 Å).
Buried water molecules are characteristic of mitochondrial cytochromes c [52]. There are three buried water molecules in chain A and six in chain B. Two of the additional buried waters in chain B are near the surface and result from alternate conformers of the His33 and Asn103 side chains in chain A versus B. One buried water molecule (B-factor = 10.35 Å2, chain A) is hydrogen bonded to Asn52, Tyr67 and Thr78 and is observed for both chains (Fig. 3 and Fig. 4). This water has been implicated in redox function [52, 53]. A second buried water molecule (B-factor = 10.84 Å2, chain A) is hydrogen bonded to heme propionate A, main chain atoms of Lys39 and Gln42 and the side chain of Arg38 (Fig. 4). This water is believed to be important for holding Arg38 against the heme, allowing Arg38 to regulate the heme redox potential by charge-charge interactions [54]. Recent structural studies on an alternate conformer of K72A iso-1-Cytc show that the Arg38 side chain can gate access of water to the heme crevice, perhaps mediating general acid/base catalysis during peroxidase activity associated with apoptosis [11]. These two buried waters are broadly conserved in mitochondrial cytochromes c and as indicated above may be important for function [52, 53]. A third buried water (B-factor = 9.29 Å2, chain A) hydrogen bonded to main atoms of Thr19, Lys25, Lys27, and Gly29 and the side chain of Thr19 sits near the heme ligand His18 and may help to stabilize a tight turn [55]. The remaining buried water is only observed in chain B and is hydrogen bonded to heme propionate A, the main chain of Asn31 and the side of Arg38. This water is observed in yeast iso-1-Cytc [52, 56, 57], but not in horse [53, 55] and human [41] Cytc (Fig. S3).
Fig. 4.
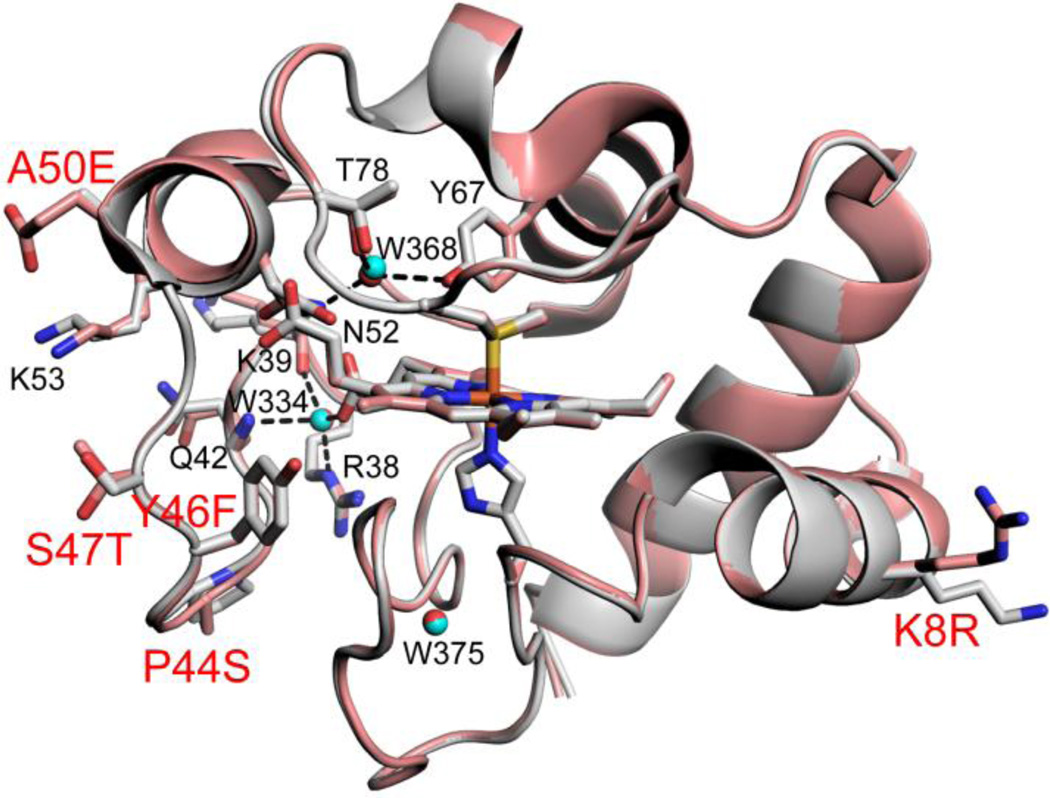
Overlay of chain A of Hu Cytc (gray, pdb code: 3ZCF, [41]) with chain A of SM Cytc (salmon, pdb code: 5DFS) showing the five side chain substitutions (stick models with red labels, for Lys8 of Hu Cytc, only one of two fitted side chain rotamers is shown) and the three buried water molecules (cyan spheres for Hu Cytc, red spheres for SM Cytc). The heme and the His18 and Met80 ligands and the side chains of Arg 38, Lys39, Gln42, Asn52, Lys53, Tyr67 and Thr78 are also shown as stick models (black labels). Buried waters are denoted with numbering from the pdb file for chain A of SM Cytc (black labels). Hydrogen bonds from the buried water (W 368) to Tyr67 (2.84 Å), Thr78 (2.80 Å) and Asn52 (3.03 Å) and buried water (W334) to Arg38-δN (2.91 Å), Lys39 carbonyl-O (2.70 Å), Gln42 amide-NH (3.03 Å), and heme propionate A (2.66 Å), are shown with black dashed lines.
As can be seen in Fig. 4, the structures of SM Cytc and Hu Cytc overlay well (Cα RMSD of 0.390 Å, chain A versus chain A). The main chain overlays nearly perfectly with small deviations near the P44S substitution, in the single turn of helix running from residues 49 to 56 and near Gly77 in Ω-loop D. The rotamers of the side chains at the positions of the substituted side chains P44S (in chain B a different rotamer is observed, Fig. S3), Y46F and S47T are surprisingly similar. The surface substitutions, K8R and A50E, have similar Cβ positions. The small main chain deviation in the short 50’s helix leads to an alternate rotamer for Asn52 (Fig. 4) that significantly lengthens the hydrogen bonds to the buried water adjacent to Met80 relative to those observed in chain B where the same rotamer of Asn52 as for Hu Cytc is observed (Fig. 4 versus Fig. S3). For chain B of SM Cytc, the main chain deviations near the P44S substitution are smaller relative to Hu Cytc, and similar in the short 50’s helix. However, the displacement of Ω-loop D in chain B of SM Cytc relative to Hu Cytc occurs primarily after Met80 (Fig. S3). Thus, while different in detail, both molecules of SM Cytc, show structural deviations that might affect the stability of Ω-loop D.
The A50E mutation has been proposed to provide an electrostatic interaction with Lys79 which competes with a hydrogen bond from the ε-amino group of Lys79 to the main chain carbonyl of residue 47. This hydrogen bond is believed to stabilize the heme crevice and thus the A50E mutation in SM Cytc is believed to explain the low pKapp for SM Cytc relative to Hu Cytc. Our structural data show that Glu50 forms an electrostatic contact with Lys53 (Fig. 4). Thus, the effect of the A50E mutation on heme crevice stability may be only indirect, leading instead to subtle adjustments to the main chain of the 50’s helix as described above.
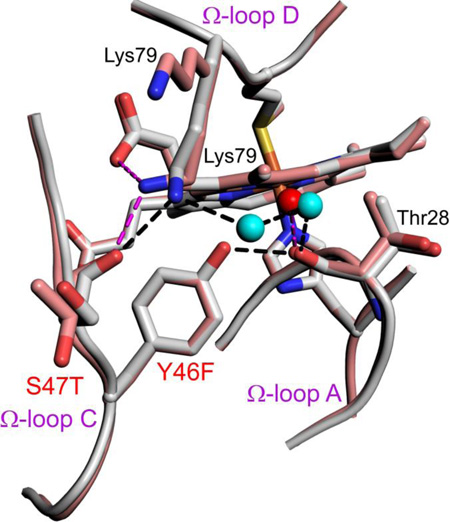
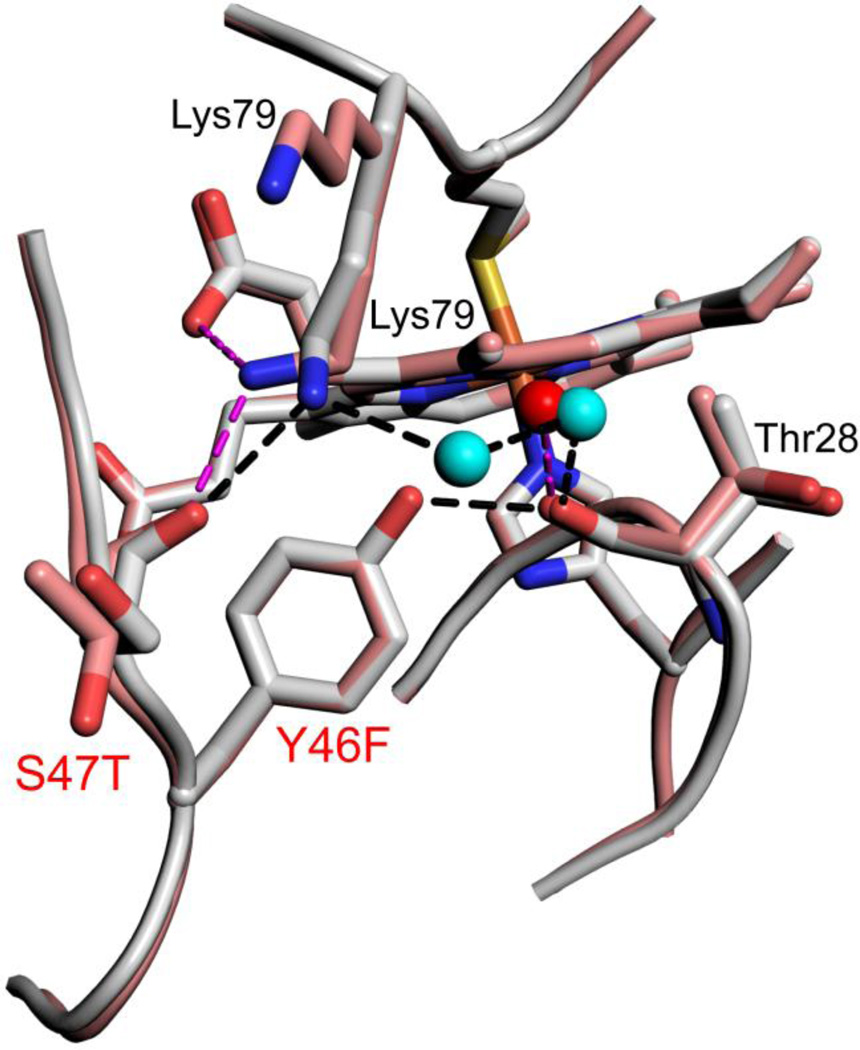
The Y46F mutation introduces the largest decrease in the pKapp of the alkaline transition as Hu Cytc is converted toward SM Cytc (Table 2). In Hu Cytc, Tyr46 is part of a hydrogen bond network that knits together three different surface Ω-loops (Fig. 5). The hydroxyl group of Tyr46 is hydrogen bonded to the carbonyl-O of Thr28. The ε-amino group of Lys79 is hydrogen bonded to the main chain carbonyl-O of Ser47 and to the Thr28 carbonyl-O through two bridging water molecules. In SM Cytc, the Y46F substitution causes loss of the hydrogen bond to the Thr28 carbonyl-O. Loss of this contact destabilizes the Lys79 ε-amino group to Thr28 carbonyl-O water-bridge causing Lys79 to adopt two conformations in chain A (Fig. 5). In one conformer (44% occupancy), the Lys79 ε-amino group to Thr47 carbonyl-O interaction is maintained and a new hydrogen bond to heme propionate D is formed. In the other conformer (56% occupancy), Lys79 projects into the solvent. In Chain B of SM Cytc, the conformation of Lys79 is similar to that in Hu Cytc. However, loss of the Tyr46 hydroxyl to Thr28 carbonyl-O hydrogen bond causes the Lys79 to Thr28 carbonyl-O water-bridge to be less optimal (Fig. S4). Inspection of the structure of the Y46F variant of Hu Cytc [41], shows that the water bridge from the ε-amino group of Lys79 to the Thr28 carbonyl-O is disrupted, in addition to the loss of the Tyr46 hydroxyl to Thr28 carbonyl-O hydrogen bond (Fig. S5). Thus, the Y46F substitution observed in SM Cytc appears to have a general destabilizing effect on the hydrogen bond network connecting the Ω-loops of Cytc.
Fig. 5.
Close-up of changes in the hydrogen bond network around position 46 for Hu Cytc (Tyr 46, gray, chain A, pdb code: 3ZCF, [41]) and SM Cytc (Phe46, salmon, chain A, pdb code: 5DFS). Sites of sequence substitutions are labeled red. Other residues involved in the hydrogen bond network have black labels. Both positions of Lys79 in SM Cytc are labeled. Waters in the hydrogen bond network are shown as cyan (Hu Cytc) and red (SM Cytc) spheres. Hydrogen bonds (black dashed lines) for Hu Cytc have the following lengths: Lys79 εN to Ser47 carbonyl-O, 2.84 Å; Lys79 εN to water 2100, 2.76 Å; W2100 to W2043, 2.63 Å; W2043 to Thr28 carbonyl-O, 2.74 Å; Tyr46-OH to Thr28 carbonyl-O, 2.63 Å. Hydrogen bonds (magenta dashed lines) for SM Cytc have the following lengths: Lys79 εN to Thr47 carbonyl-O, 2.76 Å; Lys79 εN to heme propionate D, 3.05 Å; water 362 to Thr28 carbonyl-O, 2.79 Å.
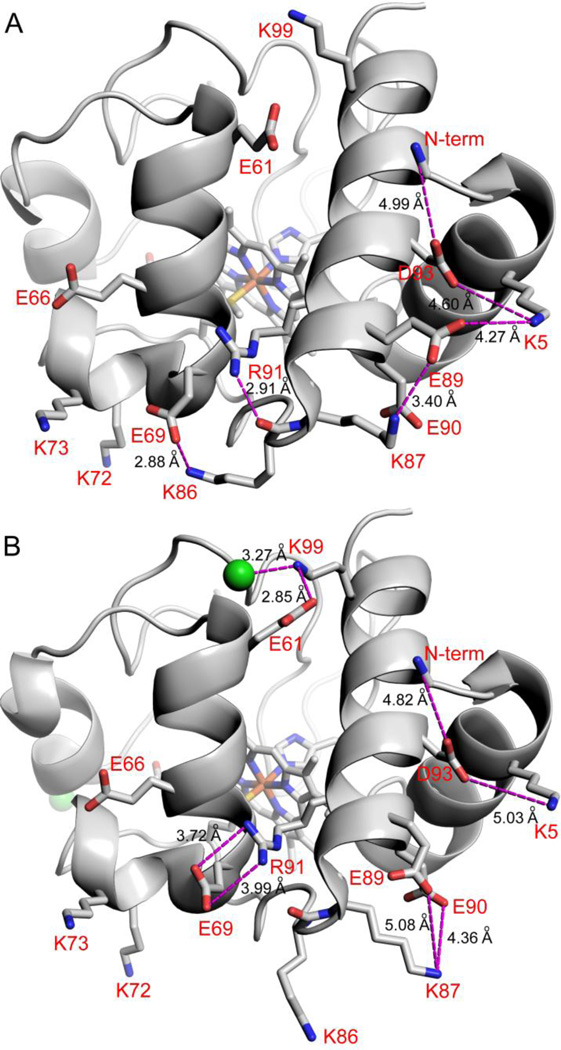
We also noted differences in anion binding for the two molecules in the asymmetric unit of SM Cytc. Chain B has no chloride bound (Fig. 6A). Chain A has two chlorides bond, one at the N-terminal end of the 60’s helix and the other at the N-terminal end of the short 50’s helix (Fig. 6B). Both presumably interact with the positive end of the helix dipole of these helices. There are no electrostatic interactions at distances of ~5 Å or less for the Cl− near the short 50’s helix. The presence of these two different molecules in the asymmetric unit of our structure allows comparisons that may aid in understanding the lack of dependence of pKapp for the alkaline transition of E. coli-expressed SM Cytc on NaCl concentration. Interpretation of the position of surface side chains, necessary for the following analysis, must be approached with caution because often electron density is missing for outer atoms of long side chains at the protein surface and the effects of packing constraints are difficult to assess. The high resolution of the current structure provides a high quality electron density map (Fig. 3). Good electron density is observed for the surface Glu and Asp side chains discussed below. The surface Lys side chains, discussed below, have good electron density out to at least the γ-carbon. Still, the limitations of the following discussion should be kept in mind.
Fig. 6.
Effect of Cl− binding on electrostatic networks in SM Cytc. (A) Molecule B of SM Cytc (pdb code: 5DFS) showing electrostatic contacts (magenta dashed lines) of ~5 Å or less (distances given in black type) in the absence of Cl− binding. The dominant conformer (63% occupancy) of Arg91 is shown. The other conformer of Arg91 forms a shorter hydrogen bond of 2.72 Å to the main chain carbonyl of Lys86. (B) Molecule A of SM Cytc (pdb code: 5DFS) showing electrostatic contacts (magenta dashed lines) of ~5 Å or less (distances given in black type) in the presence of Cl− binding. Cl− ions are shown as green spheres. The dominant conformer of Arg91 (83% occupancy) is shown. The minor conformer of Arg91 forms a 2.78 Å hydrogen bond with the main chain carbonyl of Lys86. In both panels residues are labeled in red using one letter amino acids abbreviations and the N-terminal amino group is labeled N-term.
The Cl− at the N-terminus of the 60’s helix of chain B of SM Cytc forms a triad of close electrostatic interactions with Lys99 and Glu61 (Fig. 6B). Although Glu61 is 6.7 (chain B, with Cl−) to 7.0 Å (chain A, no Cl−) from the N-terminal amino group, the binding of Cl− at the N-terminus of the 60’s helix appears to affect the balance of electrostatic interactions around the N-terminal amino group. The main result is a substantial shift in the position of the side chain of Glu89. In the absence of Cl−, Glu89 is part of a five-residue salt bridge network with the N-terminal amino group, Asp93, Lys5 and Lys87. In the presence of Cl−, Glu89 forms a three-way salt bridge with Lys87 and Glu90. The change in the position of Lys87 caused by this three-way salt bridge moves the main chain sufficiently that a hydrogen bond between the carbonyl of Lys86 and the side chain of Arg91, present in the absence of Cl− (Fig. 6A), is broken (Fig. 6B, see also Fig. 6, caption). This small change in turn affects the electrostatic interactions of Glu69. In the absence of Cl−, Glu69 forms a salt bridge with Lys86 instead of Arg91. The change in the position of Glu69 in the absence of Cl− places it 5.6 Å from Lys73 (2.1 Å closer than in the presence of Cl−).
The binding of Cl− should stabilize the native state of SM Cytc. Since Ω-loop C controls the opening of Ω-loop D in the alkaline transition [14], stabilization of Ω-loop C by Cl− binding to the short 50’s helix should increase the pKapp of the alkaline transition as Cl− concentration increases. However, the increase in the distance of Glu69 from Lys73 when Cl− binds should decrease the pKa of the alkaline state ligand, Lys73, which should decrease pKapp. These opposing effects of Cl− binding to SM Cytc may explain the insensitivity of the pKapp of E. coli-expressed SM Cytc to increasing NaCl concentration.
The electrostatic network mediated by the N-terminal amino group of E. coli-expressed SM Cytc in the absence of Cl−, places Lys73 in a more negative charge environment that would raise its pKa and thus the pKapp of the Lys73-mediated alkaline transition. This effect of the N-terminal amino group could provide an explanation for the higher pKapp at low ionic strength observed for E. coli-expressed SM Cytc relative to SM Cytc isolated from its natural source with an acetylated N-terminus (Table 4). In the five-residue salt bridge network in Fig. 6A, Glu89 and Asp93 are within 4.4 Å of each other. An acetylated N-terminal amino group could not provide a positive charge to help mitigate the close approach of Glu89 and Asp93. Thus, the position of Glu89 in the five-residue salt bridge network observed in the absence of Cl− would be expected to be considerably less favorable with an acetylated N-terminal amino group. Thus, an acetylated N-terminus might favor the less negative electrostatic environment for Lys73 (Fig. 6B), depressing its pKa at lower ionic strength and thus contributing to the lower pKapp of the alkaline transition of SM Cytc from its natural source (Table 4).
4. Conclusion
The recent rapid evolution of primate cytochromes c has been attributed primarily to co-evolution with cytochrome c oxidase [12, 13]. In this study, we have shown that the stability of the heme crevice may also be affected on different branches of the primate evolutionary tree. This destabilization may have implications for the peroxidase function of Cytc early in apoptosis. Previous work has suggested that the A50E substitution in SM Cytc versus Hu Cytc destabilizes the heme crevice by causing competition for the heme crevice stabilizing hydrogen bond between the ε-amino group of Lys79 and main chain carbonyl-O of residue 47. Our work shows that the primary effect on heme crevice destabilization is due to the Y46F substitution in SM Cytc versus Hu Cytc. Our high resolution structure of SM Cytc suggests that loss of the Tyr46 hydroxyl to Thr28 carbonyl-O hydrogen bond and of a water bridge that connects the ε-amino group of Lys79 to the Thr28 carbonyl-O are the primary structural factors which lead to heme crevice destabilization.
Supplementary Material
Highlights.
The rapid evolution of primate cytochromes c affects heme crevice stability
The heme crevice stability of spider monkey and human cytochrome c is compared
The Y46F substitution is the primary destabilizer of the heme crevice
A 1.15 Å resolution structure of spider monkey cytochrome c is presented
The Y46F substitution disrupts a hydrogen bond network connecting 3 surface Ω-loops
Acknowledgments
This research was supported by National Science Foundation grants, CHE-0910616 and CHE-1306903 (B.E.B.). B.E.B. acknowledges support from CoBRE grant P20GM103546 from NIGMS. The Bruker microflex MALDI-ToF mass spectrometer was purchased with support from NSF Major Research Instrumentation Grant CHE-1039814.
Abbreviations
- Cytc
cytochrome c
- Hu Cytc
Human cytochrome c
- iso-1-Cytc
yeast iso-1-cytochrome c
- SM Cytc
spider monkey cytochrome c
- GdnHCl
guanidine hydrochloride
- NSHX
native-state hydrogen-deuterium exchange
- pKapp
apparent pKa of the alkaline conformational transition
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Winge DR. Mol. Cell. Biol. 2012;32:2647–2652. doi: 10.1128/MCB.00573-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moore GR, Pettigrew GW. Cytochromes c: Evolutionary, Structural and Physicochemical Aspects. 1990 [Google Scholar]
- 3.Olteanu A, Patel CN, Dedmon MM, Kennedy S, Linhoff MW, Minder CM, Potts PR, Deshmukh M, Pielak GJ. Biochem. Biophys. Res. Commun. 2003;312:733–740. doi: 10.1016/j.bbrc.2003.10.182. [DOI] [PubMed] [Google Scholar]
- 4.Yu T, Wang X, Purring-Koch C, Wei Y, McLendon GL. J. Biol. Chem. 2001;276:13034–13038. doi: 10.1074/jbc.M009773200. [DOI] [PubMed] [Google Scholar]
- 5.Ow YP, Green DR, Hao Z, Mak TW. Nat. Rev. Mol. Cell Biol. 2008;9:532–542. doi: 10.1038/nrm2434. [DOI] [PubMed] [Google Scholar]
- 6.Kagan VE, Bayir HA, Belikova NA, Kapralov O, Tyurina YY, Tyurin VA, Jiang J, Stoyanovsky DA, Wipf P, Kochanek PM, Greenberger JS, Pitt B, Shvedova AA, Borisenko G. Free Radical Biol. Med. 2009;46:1439–1453. doi: 10.1016/j.freeradbiomed.2009.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kagan VE, Tyurin VA, Jiang J, Tyurina YY, Ritov VB, Amoscato AA, Osipov AN, Belikova NA, Kapralov AA, Kini V, Vlasova II, Zhao Q, Zou M, Di P, Svistunenko DA, Kurnikov IV, Borisenko GG. Nat. Chem. Biol. 2005;1:223–232. doi: 10.1038/nchembio727. [DOI] [PubMed] [Google Scholar]
- 8.Kagan VE, Chu CT, Tyurina YY, Cheikhi A, Bayira H. Chem. Phys. Lipids. 2014;179:64–69. doi: 10.1016/j.chemphyslip.2013.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tyurina YY, Poloyac SM, Tyurin VA, Kapralov AA, Jiang J, Anthonymuthu TS, Kapralova VI, Vikulina AS, Jung M-Y, Epperly MW, Mohammadyani D, Klein-Seetharaman J, Jackson TC, Kochanek PM, Pitt BR, Greenberger JS, Vladimirov YA, Bayır H, Kagan VE. Nature Chemistry. 2014;6:542–552. doi: 10.1038/nchem.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bandi S, Bowler BE. Biochemistry. 2015;54:1729–1742. doi: 10.1021/bi501252z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McClelland LJ, Mou T-C, Jeakins-Cooley ME, Sprang SR, Bowler BE. Proc. Natl. Acad. Sci. U.S.A. 2014;111:6648–6653. doi: 10.1073/pnas.1323828111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pierron D, Wildman DE, Hütteman M, Letellier T, Grossman LI. Adv. Exp. Med. Biol. 2012;748:185–213. doi: 10.1007/978-1-4614-3573-0_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pierron D, Opazo JC, Heiske M, Papper Z, Uddin M, Chand G, Wildman DE, Romero R, Goodman M, Grossman LI. PLoS ONE. 2011;6:e26269. doi: 10.1371/journal.pone.0026269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoang L, Maity H, Krishna MM, Lin Y, Englander SW. J. Mol. Biol. 2003;331:37–43. doi: 10.1016/s0022-2836(03)00698-3. [DOI] [PubMed] [Google Scholar]
- 15.Osheroff N, Borden D, Koppenol WH, Margoliash E. J. Biol. Chem. 1980;255:1689–1697. [PubMed] [Google Scholar]
- 16.Rosell FI, G Mauk A. Biochemistry. 2002;41:7811–7818. doi: 10.1021/bi016060e. [DOI] [PubMed] [Google Scholar]
- 17.Pollock WB, Rosell FI, Twitchett MB, Dumont ME, Mauk AG. Biochemistry. 1998;37:6124–6131. doi: 10.1021/bi972188d. [DOI] [PubMed] [Google Scholar]
- 18.Patel CN, Lind MC, Pielak GJ. Protein Expression Purif. 2001;22:220–224. doi: 10.1006/prep.2001.1438. [DOI] [PubMed] [Google Scholar]
- 19.Cherney MM, Junior C, Bowler BE. Biochemistry. 2013;52:837–846. doi: 10.1021/bi301599g. [DOI] [PubMed] [Google Scholar]
- 20.Hammack BN, Smith CR, Bowler BE. J. Mol. Biol. 2001;311:1091–1104. doi: 10.1006/jmbi.2001.4909. [DOI] [PubMed] [Google Scholar]
- 21.Eaton WA, Hochstrasser RM. J. Chem. Phys. 1967;46:2533–2539. doi: 10.1063/1.1841081. [DOI] [PubMed] [Google Scholar]
- 22.Dragomir I, Hagarman A, Wallace C, Schweitzer-Stenner R. Biophys. J. 2007;92:989–998. doi: 10.1529/biophysj.106.095976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Otwinowski Z, Minor W. Methods Enzymol. 1997;276A:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 24.Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L-W, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Emsley P, Lohkamp B, Scott WG, Cowtan K. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krishna MM, Lin Y, Rumbley JN, Englander SW. J. Mol. Biol. 2003;331:29–36. doi: 10.1016/s0022-2836(03)00697-1. [DOI] [PubMed] [Google Scholar]
- 27.Duncan MG, Williams MD, Bowler BE. Protein Sci. 2009;18:1155–1164. doi: 10.1002/pro.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu Y, Mayne L, Englander SW. Nat. Struct. Biol. 1998;5:774–778. doi: 10.1038/1810. [DOI] [PubMed] [Google Scholar]
- 29.Krishna MM, Maity H, Rumbley JN, Lin Y, Englander SW. J. Mol. Biol. 2006;359:1410–1419. doi: 10.1016/j.jmb.2006.04.035. [DOI] [PubMed] [Google Scholar]
- 30.Mayne L, Englander SW. Protein Sci. 2000;9:1873–1877. doi: 10.1110/ps.9.10.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Knapp JA, Pace CN. Biochemistry. 1974;13:1289–1294. doi: 10.1021/bi00703a036. [DOI] [PubMed] [Google Scholar]
- 32.Godbole S, Hammack B, Bowler BE. J. Mol. Biol. 2000;296:217–228. doi: 10.1006/jmbi.1999.3454. [DOI] [PubMed] [Google Scholar]
- 33.Cherney MM, Bowler BE. Coord. Chem. Rev. 2011;255:664–677. [Google Scholar]
- 34.Assfalg M, Bertini I, Dolfi A, Turano P, Mauk AG, Rosell FI, Gray HB. J. Am. Chem. Soc. 2003;125:2913–2922. doi: 10.1021/ja027180s. [DOI] [PubMed] [Google Scholar]
- 35.Amacher JF, Zhong F, Lisi GP, Zhu MQ, Alden SL, Hoke KR, Madden DR, Pletneva EV. J. Am. Chem. Soc. 2015;137:8435–8449. doi: 10.1021/jacs.5b01493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maity H, Rumbley JN, Englander SW. Proteins. 2006;63:349–355. doi: 10.1002/prot.20757. [DOI] [PubMed] [Google Scholar]
- 37.García-Heredia JM, Díaz-Quintana A, Salzano M, Orzáez M, Pérez-Payá E, Teixeira M, Rosa MADl, Díaz-Moreno I. J. Biol. Inorg. Chem. 2011;16:1155–1168. doi: 10.1007/s00775-011-0804-9. [DOI] [PubMed] [Google Scholar]
- 38.García-Heredia JM, Díaz-Moreno I, Nieto PM, Orzáez M, Kocanis S, Teixeira M, Pérez-Payá E, Díaz-Quintana A, Rosa MADl. Biochim. Biophys. Acta. 2010;1797:981–993. doi: 10.1016/j.bbabio.2010.03.009. [DOI] [PubMed] [Google Scholar]
- 39.Ying T, Zhong F, Xie J, Feng Y, Wang Z-H, Huang Z-X, Tan X. J. Bioenerg. Biomembr. 2009;41:251–257. doi: 10.1007/s10863-009-9223-9. [DOI] [PubMed] [Google Scholar]
- 40.Josephs TM, Liptak MD, Hughes G, Lo A, Smith RM, Wilbanks SM, Bren KL, Ledgerwood EC. J. Biol. Inorg. Chem. 2013;18:289–297. doi: 10.1007/s00775-012-0973-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rajagopal BS, Edzuma AN, Hough MA, Blundell KLIM, Kagan VE, Kapralov AA, Fraser LA, Butt JN, Silkstone GG, Wilson MT, Svistunenko DA, Worrall JAR. Biochem. J. 2013;456:441–452. doi: 10.1042/BJ20130758. [DOI] [PubMed] [Google Scholar]
- 42.Battistuzzi G, Borsari M, Ranieri A, Sola M. Arch. Biochem. Biophys. 2001;386:117–122. doi: 10.1006/abbi.2000.2183. [DOI] [PubMed] [Google Scholar]
- 43.Battistuzzi G, Borsari M, Loschi L, Martinelli A, Sola M. Biochemistry. 1999;38:7900–7907. doi: 10.1021/bi983060e. [DOI] [PubMed] [Google Scholar]
- 44.Martinez RE, Bowler BE. J. Am. Chem. Soc. 2004;126:6751–6758. doi: 10.1021/ja0494454. [DOI] [PubMed] [Google Scholar]
- 45.Verbaro D, Hagarman A, Soffer J, Schweitzer-Stenner R. Biochemistry. 2009;48:2990–2996. doi: 10.1021/bi802208f. [DOI] [PubMed] [Google Scholar]
- 46.Weinkam P, Zimmermann J, Sagle LB, Matsuda S, Dawson PE, Wolynes PG, Romesberg FE. Biochemistry. 2008;47:13470–13480. doi: 10.1021/bi801223n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Battistuzzi G, Borsari M, De Rienzo F, Di Rocco G, Ranieri A, Sola M. Biochemistry. 2007;46:1694–1702. doi: 10.1021/bi061961e. [DOI] [PubMed] [Google Scholar]
- 48.Blouin C, Guillemette JG, Wallace CJA. Biophys. J. 2001;81:2331–2338. doi: 10.1016/S0006-3495(01)75879-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shah R, Schweitzer-Stenner R. Biochemistry. 2008;47:5250–5257. doi: 10.1021/bi702492n. [DOI] [PubMed] [Google Scholar]
- 50.Hagihara Y, Aimoto S, Fink A, Goto Y. J. Mol. Biol. 1993;231:180–184. doi: 10.1006/jmbi.1993.1272. [DOI] [PubMed] [Google Scholar]
- 51.Nelson CJ, Bowler BE. Biochemistry. 2000;39:13584–13594. doi: 10.1021/bi0017778. [DOI] [PubMed] [Google Scholar]
- 52.Brayer GD, Murphy MEP. In: Cytochrome c. A Multidisciplinary Approach. Scott RA, Mauk AG, editors. Sausalito, California: University Science Books; 1996. pp. 103–166. [Google Scholar]
- 53.Bushnell GW, Louie GV, Brayer GD. J. Mol. Biol. 1990;214:585–595. doi: 10.1016/0022-2836(90)90200-6. [DOI] [PubMed] [Google Scholar]
- 54.Lo TP, Komar-Panucci S, Sherman F, McLendon G, Brayer GD. Biochemistry. 1995;34:5259–5268. doi: 10.1021/bi00015a041. [DOI] [PubMed] [Google Scholar]
- 55.Sanishvili R, Volz KW, Westbrook EM, Margoliash E. Structure. 1995;3:707–717. doi: 10.1016/s0969-2126(01)00205-2. [DOI] [PubMed] [Google Scholar]
- 56.Berghuis AM, Brayer GD. J. Mol. Biol. 1992;223:959–976. doi: 10.1016/0022-2836(92)90255-i. [DOI] [PubMed] [Google Scholar]
- 57.Louie GV, Brayer GD. J. Mol. Biol. 1990;214:527–555. doi: 10.1016/0022-2836(90)90197-T. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.