FANCJ, identified as a BRCA1-interacting partner
The story of FANCJ (BACH1/BRIP1) is similar to that of the first hereditary breast cancer genes, BRCA1 and BRCA2. Initially characterized for function in DNA break repair and breast cancer suppression, their roles now extend to multiple activities in the DNA damage response and suppression of several cancers. FANCJ was the first BRCA1-interacting partner with known enzymatic activity and thus was considered a mediator of BRCA1 DNA repair function. It was thought that BRCA1 leveraged FANCJ helicase activity to unwind DNA in the vicinity of DNA damage. Consistent with this model, FANCJ was shown to function in double strand break repair in a manner dependent on BRCA1 binding 1. Moreover, FANCJ clinical mutations were identified that disrupt its helicase activity and DNA repair function 2. These findings, along with more comprehensive clinical analysis, suggested that breast cancer could arise from inherited mutations in FANCJ 3,4. However, as the cast of BRCA-interacting partners has grown, the unfolding tale is that FANCJ is one of many factors functioning in BRCA-DNA repair and tumor suppression pathway.
FANCJ is a Fanconi anemia gene
Aside from breast and ovarian cancer5,6, bi-allelic mutations in the hereditary breast cancer genes cause the rare genetic disorder Fanconi anemia (FA) that is responsible for birth anomalies, anemia, and cancer. Just two years after BRCA2 was found mutated in the FA-D1 complementation group 7, FANCJ was identified as the gene mutated in the FA-J complementation group 8–10. Joining the FA circle, BRCA1 was recently uncovered as the FA gene, FANCS 11. This link between hereditary breast cancer and FA exemplifies the fact that the hereditary breast cancer proteins are multifunctional; important not only for DNA repair and tumor suppression, but also for normal proliferation and development (for a recent review see 12). FA cells are very sensitive to agents that interfere with DNA replication, such as DNA interstrand crosslinks (ICLs). Thus, the link between BRCA proteins and FA also demonstrates that BRCA proteins function beyond DNA break repair to processing of stalled replication forks 13. More recently, additional replication stress-associated functions have been uncovered. In particular, BRCA proteins protect stalled DNA replication forks from nuclease digestion 14,15. Marching in step, FANCJ also functions at the replication fork, as outlined here, which likely contributes to genome stability and tumor suppression.
FANCJ interfaces with replication
In support of replication fork-associated functions, FANCJ is present at forks in a replication-dependent manner. Localization studies reveal that FANCJ tracks with BRCA1 not only in nuclear foci and at DNA lesions, but also to sites coincident with proliferating cell nuclear antigen (PCNA) and at DNA replication forks 1,16–21. Similar to BRCA1, FANCJ localization to DNA crosslinks or chromatin following crosslink induction is dependent on replication 22,23. An interface with replication is also suggested by proteomic analysis of proteins associated with nascent DNA. FANCJ is found directly at active-, stalled-, and collapsed replication forks 20,21. One could speculate that the BRCA1-FANCJ interaction is uniquely required for some aspect of the replication stress response. Indeed, FANCJ and BRCA1 interact most robustly during S phase 24. Moreover, not all BRCA-associated partners localize to replication forks or chromatin in a replication dependent manner. For example, BRCA1-associated CtIP weakly accumulates in chromatin following an ICL in a replication independent manner 23.
FANCJ shares functions with BRCA1 in the replication stress response
Although the localization of FANCJ to replication forks does not foretell its function, the consequence of FANCJ deficiency provides clues that, similar to BRCA1, FANCJ has function outside of break repair. While data vary from cell system analyzed, FANCJ deficient cells typically have a broad sensitivity to replication inhibitors, such as ICL-inducing agents, mitomycin C (MMC), melphalan and cisplatin as well as to hydroxyurea (HU), which depletes deoxyribonucleotide pool, and to aphidicolin, which inhibits DNA polymerases. In contrast, FANCJ deficient cells have minor sensitivity to agents that induce DNA double strand breaks (DSBs), such as ionizing radiation (IR) and camptothecin. The mild sensitivity to DSB-inducing agents and lack of sensitivity to ultraviolet light (UV) irradiation distinguishes FANCJ deficient cells from BRCA-deficient cells 8,18,25–27 (see Supplemental Table 1). Nevertheless, IR-induced DNA double strand breaks and UV-induced DNA intrastrand crosslinks are repaired with reduced kinetics in BRCA1- or FANCJ-deficient cells 17,18,28. Moreover, RPA-coating of ssDNA in S phase cells following UV irradiation and subsequent checkpoint responses are defective in these cells. Likewise UV induced mutations are enhanced by deficiency in either FANCJ or BRCA1 18,28. In the response to IR, evidence also suggests that similar to BRCA1, FANCJ promotes checkpoint activation and maintenance 16,29,30. Failure to elicit proper checkpoint responses could underlie the defective replication restart in FANCJ or BRCA1 deficient cells after release from aphidicolin treatment 31–33. Functions in the replication stress response could protect the genome beyond S phase given that both BRCA1 and FANCJ impacts centromere amplification following replication stress 34,35.
While FANCJ deficient cells resemble BRCA1 deficient cells in several respects, it remains to be determined what key functions are shared. This understanding could clarify why unlike BRCA1 deficient mice, FANCJ deficient mice are viable 36–40. In particular, it will be important to address if FANCJ functions with BRCA1 in protecting stalled forks from nuclease digestion, loading of the 9-1-1 complex, checkpoint activation, recruitment of repair factors such as FANCD2 or evicting the CMG (Cdc45, MCM2-7, and GINS) replicative DNA helicase from chromatin in ICL repair 41–43. In support of the possibility that FANCJ helicase or translocase activities could support one or more of these fork-processing activities, in FANCJ deficient cells Cdc45 remains aberrantly loaded on chromatin after IR 16, and FANCD2 foci are reduced 44,45.
FANCJ resolves secondary structures at stalled forks
It also remains unexplored as to if BRCA1 functions with FANCJ in its metabolism of replication-associated structures. In particular, FANCJ counteracts fork stalling and genomic instability by resolving secondary structures such as G-quadruplexes (G4s) that form in guanine-rich DNA 13,40,46–49. In accordance, loss of FANCJ in Xenopus egg extract system leads to persistent replication stalling at G4s 50. Likewise, cells deficient in FANCJ are sensitive to G4 binding drugs 47,51. Suggesting that G4s are abnormally accumulating even in the absence of exogenous replication stress, FANCJ deficient chicken cells have a delay in S phase progression that correlates with decreases in replication fork velocity and ssDNA induction 51. Moreover, unchallenged FANCJ null mouse or deficient human cells show slower replication fork extension rates and slower progression through S phase consistent with increased replication fork stalling 40,52. Whereas FANCJ deficient cells have an elevated S-phase accumulation in response to low dose aphidicolin, FANCJ null cells show signs of DNA break induction with high dose aphidicolin suggesting replication fork collapse 16,33. The nature of the genomic instability may be species specific given that FANCJ deficiency confers loss of G4s in C. elegans, telomere abnormalities in human cells, and microsatellite instability (MSI) in mouse and human cells 40,46,53–56. Resolving secondary structures could maintain chromatin structure given that FANCJ deficient DT40 cells have increased heterochromatin formation, localized changes in histone modifications, and gene expression 49,51. It remains to be determined how replication fork preservation functions relate to one or more of FANCJ described in vitro enzyme functions. FANCJ is an ATPase/helicase/translocase that unwinds DNA in a 5′-3′ direction and is active on a range of substrates such as 5′ flaps, forked DNA duplexes, D loops, and G4s and can displace protein/DNA interactions 2,46,47,57,58.
Loss of FANCJ-BRCA1 interaction alters the replication stress response
Despite restart defects in BRCA1 deficient cells, it is unlikely that BRCA1 binding to FANCJ is required for FANCJ function in resolving replication-blocking structures given that resistance to replication stress is enhanced in cells lacking the FANCJ-BRCA1 interaction 31,33,59. Notably, expression of a BRCA1-interaction defective mutant, FANCJS990A in FANCJ-null FA patient cells confers greater resistance to cisplatin, MMC, melphalan and UV as compared to expression of wild-type FANCJ 60. Likewise in chicken cells the FANCJ-BRCA1 interaction is not required for ICL resistance, nor appear to be conserved 25. Thus, one could speculate that in human cells, the FANCJ-BRCA1 interaction has a distinct replication stress associated function. Consistent with this idea, cells deficient in the FANCJ-BRCA1 interaction have defects in checkpoint and repair responses 59. Resembling BRCA1- or FANCJ-deficient cells, FANCJS990A cells fail to arrest DNA synthesis following IR and show reduced double strand break repair by homologous recombination (HR) 16,60. Not only are cells expressing FANCJS990A more sensitive to agents that induce DSBs, HR is reduced to the level found in cells expressing the helicase inactive mutant, FANCJK52R 60. Resistance to replication-stress inducing agents such as MMC and UV is managed through enhanced DNA damage tolerance pathways including increased reliance on the translesion synthesis (TLS) polymerase, polη 60. Thus, in human cells, the FANCJ-BRCA1 interaction could serve to coordinate the HR and TLS pathways to facilitate repair and recovery of replication with minimal error induction. While the interaction may not be required to restart replication, it will be worth considering if the FANCJ-BRCA1 interaction preserves fragile sites under replication stress given that BRCA1 and FANCJ share this function 40,61,62.
Cells lacking the FANCJ-BRCA1 interaction could have an altered replication stress response because FANCJ is not functional. With respect to replication forks, data suggest FANCJ localization is not dependent on BRCA1. In particular FANCJ localizes to UV lesions in S phase before BRCA1 and independent of BRCA1 binding. Thus, in contrast to DNA breaks, FANCJ localization to stalled forks appears to be independent of BRCA116,18. Alternatively, FANCJ could fail to be properly modified. In support of this idea, FA-J cells expressing the BRCA1-interaction defective mutant, FANCJS990A resemble FA-J cells expressing an acetylation-defective mutant, FANCJK1249R. In these cells, ICL resistance is conferred with reduced dependence on HR factors and enhanced reliance on TLS factors 63. Acetylation at FANCJ lysine K1249 promotes DNA end resection and checkpoint maintenance 63. Thus, these functions could also be promoted by BRCA1 binding. Another possibility is that when unbound to BRCA1, FANCJ fails to interact with other partners. For instance, loss of FANCJ binding to BLM or RPA could derail FANCJ’s end processing or checkpoint functions 27,64. As compared to wild-type FANCJ, FANCJS990A precipitates less of the replication checkpoint factor, TopBP1 following IR suggesting that BRCA1 binding impacts the interactions of FANCJ with other partners. The defective binding does not result from loss of a critical TopBP1 binding domain. Threonine 1133 of FANCJ is phosphorylated following DNA damage and this mediates its direct interaction with the C-terminal BRCT repeats of TopBP1. Given that FANCJ binding promotes TopBP1 stabilization at stalled forks, ATR activation and the intra-S phase checkpoint activation 65, a reduction in this interaction could switch repair processing. Likewise, loss of BRCA1 or TopBP1 binding and reduced checkpoint responses could underlie the aberrant induction of new replication in FANCJ deficient cells 44. Loss of BRCA1 binding to FANCJ could also unleash FANCJ or its helicase activity to create a gain-of-function. For example, the enhanced TLS associated with BRCA1 deficiency in response to UV could stem from an increase in unregulated FANCJ, mimicking the enhanced TLS found in cells expressing FANCJS990A 28,60. In addition, it will be important to consider if altered FANCJ function is a contributing factor to replication fork protection defects that are found in BRCA1 deficient cells or when BRCA1 is in the heterozygous state 66. If so, unregulated FANCJ could also contribute to replication-stress associated defects in breast cancers in which FANCJ is overexpressed 67.
The FANCJ-MLH1 interaction is essential for replication restart
In contrast to the finding that FANCJ binding to BRCA1 is indispensable for cells to recover from replication stress, FANCJ binding directly to the mismatch repair (MMR) protein, MLH1 is essential. Not only does expression of an MLH1-interaction defective mutant, FANCJK141/142A, in FANCJ-null patient FA-J cells fail to restore ICL resistance, but also as compared to FANCJ-null cells, ICL sensitivity is enhanced. Moreover, expression of FANCJK141/142A exacerbates the abnormal ICL-induced G2/M accumulation in FANCJ-null cells 26. Correspondingly, a hereditary colon cancer mutation MLH1L607H, which is defective in FANCJ binding, confers increased ICL sensitivity and checkpoint defects not previously detected in MLH1-null cells 68. Thus, mutations in FANCJ or MLH1 that disrupt the FANCJ-MLH1 interaction sensitize cells to ICLs to a greater extent than deficiency in FANCJ or MLH1 26,68,69. A heightened replication stress response is also detected upon release from aphidicolin. Cells lacking the FANCJ-MLH1 interaction remain arrested while FANCJ-null cells re-enter the cell cycle. The arrest correlates with γ-H2AX and DNAPKcs phosphorylation, suggesting fork breakage. Collectively, these data indicate that deficiency in the FANCJ-MLH1 interaction is worse for the restart of replication than deficiency in FANCJ or MLH1 alone.
MSH2 depletion suppresses replication restart defects in cells lacking the FANCJ–MLH1 interaction
While the underlying cause of the replication restart defect and fork collapse in cells lacking the FANCJ-MLH1 interaction is not known, it is linked to the mismatch repair protein, MSH2. Specifically, depletion of MSH2 suppresses not only MMC-induced sensitivity and chromosomal aberrations, but also aphidicolin-induced replication restart defects and break induction in cells lacking the FANCJ-MLH1 interaction 33. This finding supports a model in which FANCJ, through its MLH1 interaction, unwinds DNA structures or displaces MSH2 from DNA structures to restart stalled replication forks. MMR proteins accumulate at replication forks 20 and are poised to bind DNA mismatches or secondary structures. Therefore, if not regulated, the MMR pathway could activate checkpoints, repair, and apoptotic responses that counteract the restart of stalled replication forks. There is precedent for the regulation of DNA repair pathways by helicases, such as yeast Srs2 and bacterial UvrD (helicase II) 70,71 and FANCJ’s closest human homologue, RTEL 72. Alternatively, FANCJ could promote MMR function at stalled forks in repair or checkpoint responses. In support of this point, MMR is required for FANCJ localization to ICLs 19 and UV light-induced DNA crosslinks 18. Moreover, an interaction between FANCJ and MSH5 is important for repair of camptothecin-induced breaks 73. Furthermore, a robust checkpoint response to DNA alkylation or UV damage requires FANCJ, and the FANCJ-MLH1 interaction 18,68. Collectively, these findings suggest that FANCJ regulates the MMR response to replication stress; without FANCJ, MMR complexes could be detrimental as they block replication fork progression or fail to mount a productive response. In future studies, it will be important to understand if an ineffective MMR complex explains why similar to MMR deficiency, FANCJ deficiency confers microsatellite instability (MSI) and lymphoma 40.
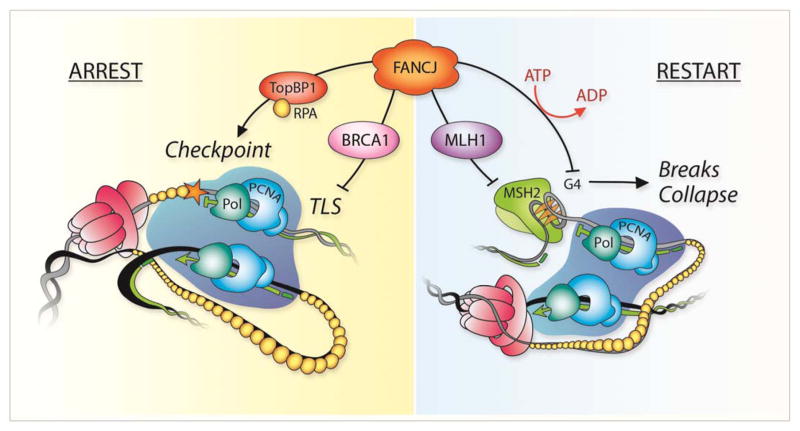
In summary, FANCJ has not receded from center stage. Rather, FANCJ is part of a BRCA network whose function in the replication stress response is an evolving story that likely involves replication stress induced arrest and restart functions (Figure 1). Building a more complete picture will be fundamental to grasp not only a full understanding of its function in DNA repair and tumor suppression pathways, but also to recognize opportunities for future therapy intervention.
Figure 1.
Potential role for FANCJ in the replication stress response: In response to replication arrest, FANCJ interactions with ToBP1 and BRCA1 promote checkpoint activation and inhibit translesion synthesis (TLS). Following repair processing, FANCJ interaction with MLH1 and helicase/translocase activities promote replication restart through displacement of MSH2 complexes and unwinding of secondary structures, such as G4s as shown.
Supplementary Material
Acknowledgments
We thank the Cantor laboratory for helpful discussions and edits. This work was supported by the National Institutes of Health (RO1 11150917) as well as charitable contributions from Mr. and Mrs. Edward T. Vitone, Jr.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Cantor SB, et al. BACH1, a novel helicase-like protein, interacts directly with BRCA1 and contributes to its DNA repair function. Cell. 2001;105:149–160. doi: 10.1016/s0092-8674(01)00304-x. [DOI] [PubMed] [Google Scholar]
- 2.Cantor S, et al. The BRCA1-associated protein BACH1 is a DNA helicase targeted by clinically relevant inactivating mutations. Proc Natl Acad Sci U S A. 2004;101:2357–2362. doi: 10.1073/pnas.0308717101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seal S, et al. Truncating mutations in the Fanconi anemia J gene BRIP1 are low-penetrance breast cancer susceptibility alleles. Nat Genet. 2006;38:1239–1241. doi: 10.1038/ng1902. [DOI] [PubMed] [Google Scholar]
- 4.Walsh T, King MC. Ten genes for inherited breast cancer. Cancer Cell. 2007;11:103–105. doi: 10.1016/j.ccr.2007.01.010. S1535-6108(07)00025-6 [pii] [DOI] [PubMed] [Google Scholar]
- 5.King MC, Marks JH, Mandell JB. Breast and ovarian cancer risks due to inherited mutations in BRCA1 and BRCA2. Science. 2003;302:643–646. doi: 10.1126/science.1088759302/5645/643. [pii] [DOI] [PubMed] [Google Scholar]
- 6.Rafnar T, et al. Mutations in BRIP1 confer high risk of ovarian cancer. Nat Genet. 2011;43:1104–1107. doi: 10.1038/ng.955. [DOI] [PubMed] [Google Scholar]
- 7.Howlett NG, et al. Biallelic inactivation of BRCA2 in Fanconi anemia. Science. 2002;297:606–609. doi: 10.1126/science.1073834. [DOI] [PubMed] [Google Scholar]
- 8.Litman R, et al. BACH1 is critical for homologous recombination and appears to be the Fanconi anemia gene product FANCJ. Cancer Cell. 2005;8:255–265. doi: 10.1016/j.ccr.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 9.Levitus M, et al. The DNA helicase BRIP1 is defective in Fanconi anemia complementation group. J Nat Genet. 2005;37:934–935. doi: 10.1038/ng1625. ng1625 [pii] [DOI] [PubMed] [Google Scholar]
- 10.Levran O, et al. The BRCA1-interacting helicase BRIP1 is deficient in Fanconi anemia. Nat Genet. 2005;37:931–933. doi: 10.1038/ng1624. [DOI] [PubMed] [Google Scholar]
- 11.Sawyer SL, et al. Biallelic mutations in BRCA1 cause a new Fanconi anemia subtype. Cancer Discov. 2015;5:135–142. doi: 10.1158/2159-8290.CD-14-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bogliolo M, Surralles J. Fanconi anemia: a model disease for studies on human genetics and advanced therapeutics. Curr Opin Genet Dev. 2015;33:32–40. doi: 10.1016/j.gde.2015.07.002. [DOI] [PubMed] [Google Scholar]
- 13.Cantor SB, Brosh RM., Jr What is wrong with Fanconi anemia cells? Cell Cycle. 2014;13:3823–3827. doi: 10.4161/15384101.2014.980633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schlacher K, et al. Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell. 2011;145:529–542. doi: 10.1016/j.cell.2011.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schlacher K, Wu H, Jasin M. A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51-BRCA1/2. Cancer cell. 2012;22:106–116. doi: 10.1016/j.ccr.2012.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Greenberg RA, et al. Multifactorial contributions to an acute DNA damage response by BRCA1/BARD1-containing complexes. Genes Dev. 2006;20:34–46. doi: 10.1101/gad.1381306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peng M, Litman R, Jin Z, Fong G, Cantor SB. BACH1 is a DNA repair protein supporting BRCA1 damage response. Oncogene. 2006;25:2245–2253. doi: 10.1038/sj.onc.1209257. 1209257 [pii] [DOI] [PubMed] [Google Scholar]
- 18.Guillemette S, et al. FANCJ localization by mismatch repair is vital to maintain genomic integrity after UV irradiation. Cancer research. 2014;74:932–944. doi: 10.1158/0008-5472.CAN-13-2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Suhasini AN, et al. Fanconi anemia group J helicase and MRE11 nuclease interact to facilitate the DNA damage response. Molecular and cellular biology. 2013;33:2212–2227. doi: 10.1128/MCB.01256-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sirbu BM, et al. Identification of proteins at active, stalled, and collapsed replication forks using isolation of proteins on nascent DNA (iPOND) coupled with mass spectrometry. J Biol Chem. 2013;288:31458–31467. doi: 10.1074/jbc.M113.511337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alabert C, et al. Nascent chromatin capture proteomics determines chromatin dynamics during DNA replication and identifies unknown fork components. Nat Cell Biol. 2014;16:281–293. doi: 10.1038/ncb2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shen X, et al. Recruitment of fanconi anemia and breast cancer proteins to DNA damage sites is differentially governed by replication. Mol Cell. 2009;35:716–723. doi: 10.1016/j.molcel.2009.06.034. S1097-2765(09)00475-4 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Raschle M, et al. DNA repair. Proteomics reveals dynamic assembly of repair complexes during bypass of DNA cross-links. Science. 2015;348:1253671. doi: 10.1126/science.1253671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu X, Chen J. DNA damage-induced cell cycle checkpoint control requires CtIP, a phosphorylation-dependent binding partner of BRCA1 C-terminal domains. Mol Cell Biol. 2004;24:9478–9486. doi: 10.1128/MCB.24.21.9478-9486.2004. 24/21/9478 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bridge WL, Vandenberg CJ, Franklin RJ, Hiom K. The BRIP1 helicase functions independently of BRCA1 in the Fanconi anemia pathway for DNA crosslink repair. Nat Genet. 2005;37:953–957. doi: 10.1038/ng1627. [DOI] [PubMed] [Google Scholar]
- 26.Peng M, et al. The FANCJ/MutLalpha interaction is required for correction of the cross-link response in FA-J cells. Embo J. 2007;26:3238–3249. doi: 10.1038/sj.emboj.7601754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Suhasini AN, et al. Interaction between the helicases genetically linked to Fanconi anemia group J and Bloom’s syndrome. The EMBO journal. 2011;30:692–705. doi: 10.1038/emboj.2010.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pathania S, et al. BRCA1 is required for postreplication repair after UV-induced DNA damage. Molecular cell. 2011;44:235–251. doi: 10.1016/j.molcel.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu B, Kim S, Kastan MB. Involvement of Brca1 in S-phase and G(2)-phase checkpoints after ionizing irradiation. Mol Cell Biol. 2001;21:3445–3450. doi: 10.1128/MCB.21.10.3445-3450.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cotta-Ramusino C, et al. A DNA damage response screen identifies RHINO, a 9-1-1 and TopBP1 interacting protein required for ATR signaling. Science. 2011;332:1313–1317. doi: 10.1126/science.1203430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yeo JE, Lee EH, Hendrickson EA, Sobeck A. CtIP mediates replication fork recovery in a FANCD2-regulated manner. Hum Mol Genet. 2014;23:3695–3705. doi: 10.1093/hmg/ddu078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Raghunandan M, Chaudhury I, Kelich SL, Hanenberg H, Sobeck A. FANCD2, FANCJ and BRCA2 cooperate to promote replication fork recovery independently of the Fanconi Anemia core complex. Cell Cycle. 2015;14:342–353. doi: 10.4161/15384101.2014.987614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peng M, Xie J, Ucher A, Stavnezer J, Cantor SB. Crosstalk between BRCA-Fanconi anemia and mismatch repair pathways prevents MSH2-dependent aberrant DNA damage responses. The EMBO journal. 2014 doi: 10.15252/embj.201387530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kais Z, Parvin JD. Regulation of centrosomes by the BRCA1-dependent ubiquitin ligase. Cancer Biol Ther. 2008;7:1540–1543. doi: 10.4161/cbt.7.10.7053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zou J, et al. FancJ regulates interstrand crosslinker induced centrosome amplification through the activation of polo-like kinase 1. Biol Open. 2013;2:1022–1031. doi: 10.1242/bio.20135801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gowen LC, Johnson BL, Latour AM, Sulik KK, Koller BH. Brca1 deficiency results in early embryonic lethality characterized by neuroepithelial abnormalities. Nat Genet. 1996;12:191–194. doi: 10.1038/ng0296-191. [DOI] [PubMed] [Google Scholar]
- 37.Hakem R, et al. The tumor suppressor gene Brca1 is required for embryonic cellular proliferation in the mouse. Cell. 1996;85:1009–1023. doi: 10.1016/s0092-8674(00)81302-1. [DOI] [PubMed] [Google Scholar]
- 38.Liu CY, Flesken-Nikitin A, Li S, Zeng Y, Lee WH. Inactivation of the mouse Brca1 gene leads to failure in the morphogenesis of the egg cylinder in early postimplantation development. Genes Dev. 1996;10:1835–1843. doi: 10.1101/gad.10.14.1835. [DOI] [PubMed] [Google Scholar]
- 39.Sun X, et al. FancJ (Brip1) loss-of-function allele results in spermatogonial cell depletion during embryogenesis and altered processing of crossover sites during meiotic prophase I in mice. Chromosoma. 2015 doi: 10.1007/s00412-015-0549-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Matsuzaki K, Borel V, Adelman CA, Schindler D, Boulton SJ. FANCJ suppresses microsatellite instability and lymphomagenesis independent of the Fanconi anemia pathway. Genes Dev. 2015;29:2532–2546. doi: 10.1101/gad.272740.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bunting SF, et al. BRCA1 Functions Independently of Homologous Recombination in DNA Interstrand Crosslink Repair. Molecular cell. 2012;46:125–135. doi: 10.1016/j.molcel.2012.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin SY, Li K, Stewart GS, Elledge SJ. Human Claspin works with BRCA1 to both positively and negatively regulate cell proliferation. Proc Natl Acad Sci U S A. 2004;101:6484–6489. doi: 10.1073/pnas.0401847101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Long DT, Joukov V, Budzowska M, Walter JC. BRCA1 promotes unloading of the CMG helicase from a stalled DNA replication fork. Mol Cell. 2014;56:174–185. doi: 10.1016/j.molcel.2014.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen X, et al. The Fanconi anemia proteins FANCD2 and FANCJ interact and regulate each other’s chromatin localization. J Biol Chem. 2014;289:25774–25782. doi: 10.1074/jbc.M114.552570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang F, Fan Q, Ren K, Auerbach AD, Andreassen PR. FANCJ/BRIP1 recruitment and regulation of FANCD2 in DNA damage responses. Chromosoma. 2010;119:637–649. doi: 10.1007/s00412-010-0285-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.London TB, et al. FANCJ is a structure-specific DNA helicase associated with the maintenance of genomic G/C tracts. J Biol Chem. 2008;283:36132–36139. doi: 10.1074/jbc.M808152200. M808152200 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu Y, Shin-Ya K, Brosh RM., Jr FANCJ Helicase Defective in Fanconia Anemia and Breast Cancer Unwinds G-Quadruplex DNA to Defend Genomic Stability. Mol Cell Biol. 2008 doi: 10.1128/MCB.02210-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bharti SK, et al. Specialization among iron-sulfur cluster helicases to resolve G-quadruplex DNA structures that threaten genomic stability. J Biol Chem. 2013;288:28217–28229. doi: 10.1074/jbc.M113.496463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sarkies P, et al. FANCJ coordinates two pathways that maintain epigenetic stability at Gquadruplex DNA. Nucleic Acids Res. 2012;40:1485–1498. doi: 10.1093/nar/gkr868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bosch PC, et al. FANCJ promotes DNA synthesis through G-quadruplex structures. Embo Journal. 2014;33:2521–2533. doi: 10.15252/embj.201488663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schwab RA, Nieminuszczy J, Shin-Ya K, Niedzwiedz W. FANCJ couples replication past natural fork barriers with maintenance of chromatin structure. The Journal of cell biology. 2013;201:33–48. doi: 10.1083/jcb.201208009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kumaraswamy E, Shiekhattar R. Activation of BRCA1/BRCA2-associated helicase BACH1 is required for timely progression through S phase. Mol Cell Biol. 2007;27:6733–6741. doi: 10.1128/MCB.00961-07. MCB.00961-07 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cheung I, Schertzer M, Rose A, Lansdorp PM. Disruption of dog-1 in Caenorhabditis elegans triggers deletions upstream of guanine-rich DNA. Nature Genetics. 2002;31:405–409. doi: 10.1038/ng928. [DOI] [PubMed] [Google Scholar]
- 54.Youds JL, et al. DOG-1 is the Caenorhabditis elegans BRIP1/FANCJ homologue and functions in interstrand cross-link repair. Mol Cell Biol. 2008;28:1470–1479. doi: 10.1128/MCB.01641-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kruisselbrink E, et al. Mutagenic capacity of endogenous G4 DNA underlies genome instability in FANCJ-defective C. elegans. Curr Biol. 2008;18:900–905. doi: 10.1016/j.cub.2008.05.013. S0960-9822(08)00610-6 [pii] [DOI] [PubMed] [Google Scholar]
- 56.Tarailo-Graovac M, et al. Spectrum of variations in dog-1/FANCJ and mdf-1/MAD1 defective Caenorhabditis elegans strains after long-term propagation. BMC Genomics. 2015;16:210. doi: 10.1186/s12864-015-1402-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gupta R, et al. Analysis of the DNA substrate specificity of the human BACH1 helicase associated with breast cancer. J Biol Chem. 2005;280:25450–25460. doi: 10.1074/jbc.M501995200. [DOI] [PubMed] [Google Scholar]
- 58.Sommers JA, et al. FANCJ uses its motor ATPase to disrupt protein-DNA complexes, unwind triplexes, and inhibit rad51 strand exchange. J Biol Chem. 2009 doi: 10.1074/jbc.M809019200. M809019200 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cantor SB, Andreassen PR. Assessing the Link between BACH1 and BRCA1 in the FA Pathway. Cell Cycle. 2006;5 doi: 10.4161/cc.5.2.2338. [DOI] [PubMed] [Google Scholar]
- 60.Xie J, et al. Targeting the FANCJ-BRCA1 interaction promotes a switch from recombination to poleta-dependent bypass. Oncogene. 2010;29:2499–2508. doi: 10.1038/onc.2010.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Turner BC, et al. The fragile histidine triad/common chromosome fragile site 3B locus and repair-deficient cancers. Cancer Res. 2002;62:4054–4060. [PubMed] [Google Scholar]
- 62.Arlt MF, et al. BRCA1 is required for common-fragile-site stability via its G2/M checkpoint function. Mol Cell Biol. 2004;24:6701–6709. doi: 10.1128/MCB.24.15.6701-6709.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xie J, et al. FANCJ/BACH1 acetylation at lysine 1249 regulates the DNA damage response. PLoS genetics. 2012;8:e1002786. doi: 10.1371/journal.pgen.1002786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Suhasini AN, et al. FANCJ helicase uniquely senses oxidative base damage in either strand of duplex DNA and is stimulated by replication protein A to unwind the damaged DNA substrate in a strand-specific manner. J Biol Chem. 2009;284:18458–18470. doi: 10.1074/jbc.M109.012229. M109.012229 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gong Z, Kim JE, Leung CC, Glover JN, Chen J. BACH1/FANCJ acts with TopBP1 and participates early in DNA replication checkpoint control. Molecular cell. 2010;37:438–446. doi: 10.1016/j.molcel.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pathania S, et al. BRCA1 haploinsufficiency for replication stress suppression in primary cells. Nature communications. 2014;5:5496. doi: 10.1038/ncomms6496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Eelen G, et al. Expression of the BRCA1-interacting protein Brip1/BACH1/FANCJ is driven by E2F and correlates with human breast cancer malignancy. Oncogene. 2008;27:4233–4241. doi: 10.1038/onc.2008.51. [DOI] [PubMed] [Google Scholar]
- 68.Xie J, et al. An MLH1 mutation links BACH1/FANCJ to colon cancer, signaling, and insight toward directed therapy. Cancer prevention research. 2010;3:1409–1416. doi: 10.1158/1940-6207.CAPR-10-0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cantor SB, Xie J. Assessing the link between BACH1/FANCJ and MLH1 in DNA crosslink repair. Environ Mol Mutagen. 2010;51:500–507. doi: 10.1002/em.20568. [DOI] [PubMed] [Google Scholar]
- 70.Papouli E, et al. Crosstalk between SUMO and ubiquitin on PCNA is mediated by recruitment of the helicase Srs2p. Mol Cell. 2005;19:123–133. doi: 10.1016/j.molcel.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 71.Hall MC, Jordan JR, Matson SW. Evidence for a physical interaction between the Escherichia coli methyl-directed mismatch repair proteins MutL and UvrD. Embo J. 1998;17:1535–1541. doi: 10.1093/emboj/17.5.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Barber LJ, et al. RTEL1 maintains genomic stability by suppressing homologous recombination. Cell. 2008;135:261–271. doi: 10.1016/j.cell.2008.08.016. S0092-8674(08)01061-1 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Xu Y, Wu X, Her C. hMSH5 Facilitates the Repair of Camptothecin-induced Double-strand Breaks through an Interaction with FANCJ. J Biol Chem. 2015;290:18545–18558. doi: 10.1074/jbc.M115.642884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Feng Z, Zhang J. A dual role of BRCA1 in two distinct homologous recombination mediated repair in response to replication arrest. Nucleic Acids Res. 2012;40:726–738. doi: 10.1093/nar/gkr748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Suhasini AN, Brosh RM., Jr Fanconi anemia and Bloom’s syndrome crosstalk through FANCJ-BLM helicase interaction. Trends in genetics : TIG. 2012;28:7–13. doi: 10.1016/j.tig.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chen X, Bosques L, Sung P, Kupfer GM. A novel role for non-ubiquitinated FANCD2 in response to hydroxyurea-induced DNA damage. Oncogene. 2016;35:22–34. doi: 10.1038/onc.2015.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Scully R, et al. Genetic analysis of BRCA1 function in a defined tumor cell line. Mol Cell. 1999;4:1093–1099. doi: 10.1016/s1097-2765(00)80238-5. [DOI] [PubMed] [Google Scholar]
- 78.Xu X, et al. Centrosome amplification and a defective G2-M cell cycle checkpoint induce genetic instability in BRCA1 exon 11 isoform-deficient cells. Mol Cell. 1999;3:389–395. doi: 10.1016/s1097-2765(00)80466-9. [DOI] [PubMed] [Google Scholar]
- 79.Zhong Q, et al. Association of BRCA1 with the hRad50-hMre11-p95 complex and the DNA damage response. Science. 1999;285:747–750. doi: 10.1126/science.285.5428.747. [DOI] [PubMed] [Google Scholar]
- 80.Gowen LC, Avrutskaya AV, Latour AM, Koller BH, Leadon SA. BRCA1 required for transcription-coupled repair of oxidative DNA damage. Science. 1998;281:1009–1012. doi: 10.1126/science.281.5379.1009. [DOI] [PubMed] [Google Scholar]
- 81.Fedier A, et al. The effect of loss of Brca1 on the sensitivity to anticancer agents in p53-deficient cells. Int J Oncol. 2003;22:1169–1173. [PubMed] [Google Scholar]
- 82.Moynahan ME, Cui TY, Jasin M. Homology-directed dna repair, mitomycin-c resistance, and chromosome stability is restored with correction of a Brca1 mutation. Cancer Res. 2001;61:4842–4850. [PubMed] [Google Scholar]
- 83.Bhattacharyya A, Ear US, Koller BH, Weichselbaum RR, Bishop DK. The breast cancer susceptibility gene BRCA1 is required for subnuclear assembly of Rad51 and survival following treatment with the DNA cross-linking agent cisplatin. J Biol Chem. 2000;275:23899–23903. doi: 10.1074/jbc.C000276200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.