Abstract
The sequence of genomic alterations acquired by cancer cells during tumor progression and metastasis is poorly understood. Focal adhesion kinase (FAK) is a non-receptor tyrosine kinase that integrates cytoskeleton remodeling, mitogenic signaling and cell survival. FAK has previously been reported to undergo nuclear localization during cell migration, cell differentiation and apoptosis. However, the mechanism behind FAK nuclear accumulation and its contribution to tumor progression has remained elusive. We report that amplification of FAK and the SUMO E3 ligase PIAS1 gene loci frequently co-occur in non-small cell lung cancer (NSCLC) cells, and that both gene products are enriched in a subset of primary NSCLCs. We demonstrate that endogenous FAK and PIAS1 proteins interact in the cytoplasm and the cell nucleus of NSCLC cells. Ectopic expression of PIAS1 promotes proteolytic cleavage of the FAK C-terminus, focal adhesion maturation and FAK nuclear localization. Silencing of PIAS1 deregulates focal adhesion turnover, increases susceptibility to apoptosis in vitro and impairs tumor xenograft formation in vivo. Nuclear FAK in turn stimulates gene transcription favoring DNA repair, cell metabolism and cytoskeleton regulation. Consistently, ablation of FAK by CRISPR/Cas9 editing, results in basal DNA damage, susceptibility to ionizing radiation and impaired oxidative phosphorylation. Our findings provide insight into a mechanism regulating FAK cytoplasm-nuclear distribution and demonstrate that FAK activity in the nucleus promotes NSCLC survival and progression by increasing cell-ECM interaction and DNA repair regulation.
Introduction
Protein inhibitor of activated STAT1 (PIAS1) is a SUMO E3 ligase implicated in the regulation of several oncogenes and tumor suppressors such as AKT, BRCA1, BRCA2, PML and PML-RARA [1], [2], [3], [4]. In addition, PIAS1 is over-expressed in prostate and lung cancers [4], [5]. Moreover, increase in PIAS1 protein levels has recently been linked to breast cancer tumorigenesis, albeit reports disagree as to the relevance of PIAS1 to tumorigenesis and metastasis [6], [7]. Thus, we decided to characterize the relevance of PIAS1 in non-small cell lung cancer (NSCLC) progression and metastasis. We also investigated PIAS1 downstream targets that could account for the phenotype observed and potentially serve as a therapeutic target in NSCLC.
Lung cancer metastasis is an indicator of poor prognosis and a main determinant of cancer-related mortality. Consequently, targeting and prevention of cancer cell metastasis is among the biggest hurdles in clinical oncology [8]. During metastasis, cancer cells rely heavily on cell-extracellular matrix (ECM) interactions, cytoskeleton remodeling and gene transcription. An important player in these processes is focal adhesion kinase (FAK). FAK is a non-receptor tyrosine kinase that contributes to almost every aspect of metastasis; from ECM sensing, cytoskeleton remodeling to gene transcription [9], [10], [11], [12]. The FAK gene is rarely mutated in human lung cancers, but the FAK locus (chromosome 8q) is frequently amplified in lung, colon, breast and gastric tumors [13], [14], [15], [16].
FAK controls cytoskeleton remodeling by transducing signals from integrin receptors to ERK/MAPK, PI3K, RAC1 and RHOA [10], [17], [18], [19]. Importantly, FAK promotes integrin β1 (ITGβ1) gene expression, which in turn, increases the survival of cancer cells [20]. FAK has also been linked to transcriptional activation of SNAIL, TWIST, ZEB1, and ZEB2 genes, which are essential for epithelial to mesenchymal (EMT) reprogramming in epithelial cells [21], [22], [23]. However, whether FAK is involved in transcriptional regulation is still a matter of debate because FAK resides mainly in the cytoplasm where it is associated with the plasma membrane. However, FAK protein can relocate to the cell nucleus during cell differentiation or cancer progression [24], [25]. Despite several studies reporting FAK protein nuclear localization and involvement in gene transcription, no unifying mechanism exists to explain the nuclear accumulation of FAK and the potential implications of nuclear FAK for tumorigenesis and metastasis.
Small ubiquitin-like modifiers (SUMO) have recently gained attention because of their participation in the covalent modification of target protein substrates, a process referred to as SUMOylation. This process consists of an enzymatic cascade whereby SUMO proteins are added onto target substrates with the involvement of E1, E2 and a limited number of SUMO E3 ligases. Typically, only a small fraction of a given protein is SUMOylated [26]. SUMOylation has been implicated in several cellular processes that include the regulation of nuclear import, DNA damage repair and signal transduction, however its role in tumorigenesis is still incompletely understood [27].
Using single nucleotide polymorphism (SNPs) data, we discovered that PIAS1 and FAK are frequently co-amplified in lung cancer specimens. We found a positive correlation between increased gene copy number and FAK and PIAS1 protein levels in a subset of NSCLC cell lines in vitro, human lung tumor samples in vivo and in a mouse model of tumor metastasis. Herein, we report an interaction between FAK-PIAS1 leads to FAK nuclear relocation, which is crucial for the regulation of the turnover of focal adhesions, and cell survival during oncogenic stress.
Materials and Methods
Gene Copy Number Analysis
SNP was performed as previously described [4]. Briefly, SNP profile was obtained using Illumina DNA analysis Bead Chip (Illumina, Inc.). PIAS1 and FAK gene copy number was extrapolated from their relative probe intensity compared with diploid controls.
Histochemistry and Immunofluorescence
Formalin-fixed, paraffin-embedded NSCLC specimens were obtained from the human tumor tissue bank at UT Southwestern Medical Center. Tissues and cells were processed for immunohistochemistry (IHC) or immunofluorescence (IF) using standard protocols [19]. See also supplementary methods.
Cell Lines and Tissue Culture
Human NSCLC cells and human bronchoalveolar epithelial cells (HBECs) were from the Hamon Center Cell Line Repository and were a gift of Dr. John Minna (UT Southwestern Medical Center) [28], [29]. All cell lines were DNA-fingerprinted for provenance (PowerPlex 1.2 Kit; Promega) and Mycoplasma-free (e-Myco Kit; Boca Scientific). NSCLC cells were cultured in RPMI supplemented with 5% serum and antibiotics. HBEC cells were grown in keratinocyte growth media with supplements (Invitrogen, No.: 17005-042). NIH 3T3 were grown in 5% DMEM. All cells were maintained in humidified incubator with 5% CO2 at 37°C. See also supplementary methods.
Western Blotting and Immunoprecipitation
We followed standard protocols [19].
Soft Agar Colony Formation, Transwell Migration and Scratch Assays
We performed these assays following established procedures [11], [30].
Subcellular Fractionation
Cellular fractionation was carried out as previously described [31]. Cells were scraped with ice-cold PBS, pelleted by centrifugation at 600 g for 5 minutes, suspended in hypotonic lysis buffer (10 mM Tris-base pH 7.5, 10 mM KCl, 1.5 mM MgCl2) with protease/phosphatase inhibitors for 10 minutes. Cells were then lysed by Dounce homogenization and centrifuged at 3000 × g to separate cytoplasmic (supernatant) from nuclear (pellet) fractions. To extract nuclear proteins, the nuclear pellet was resuspended in high salt buffer (50 mM Tris-base, pH 7.9, 0.5 M NaCl) 2 mM EDTA, 10% sucrose, 10% glycerol) with protease and phosphatase inhibitors.
RNAi Interference
Stable lentiviral transduction was performed with pGIPZ Lentiviral vectors containing shRNAs targeting PIAS1 and non-targeting scrambled shRNA controls (Dharmacon). siRNA (siGenome) targeting PIAS1, or non-targeting siRNA controls were purchased from Dharmacon. See also supplementary methods.
Oxygen Consumption Rate Measurement
This assay was performed with a Seahorse XF analyzer following the manufacturer's protocols (Seahorse Bioscience).
Mouse Xenografts
Xenografts were performed as described previously [32] and were approved by the institutional IACUC guidelines.
Results
FAK and PIAS1 Genes are Frequently Co-Amplified in a Subset of NSCLCs
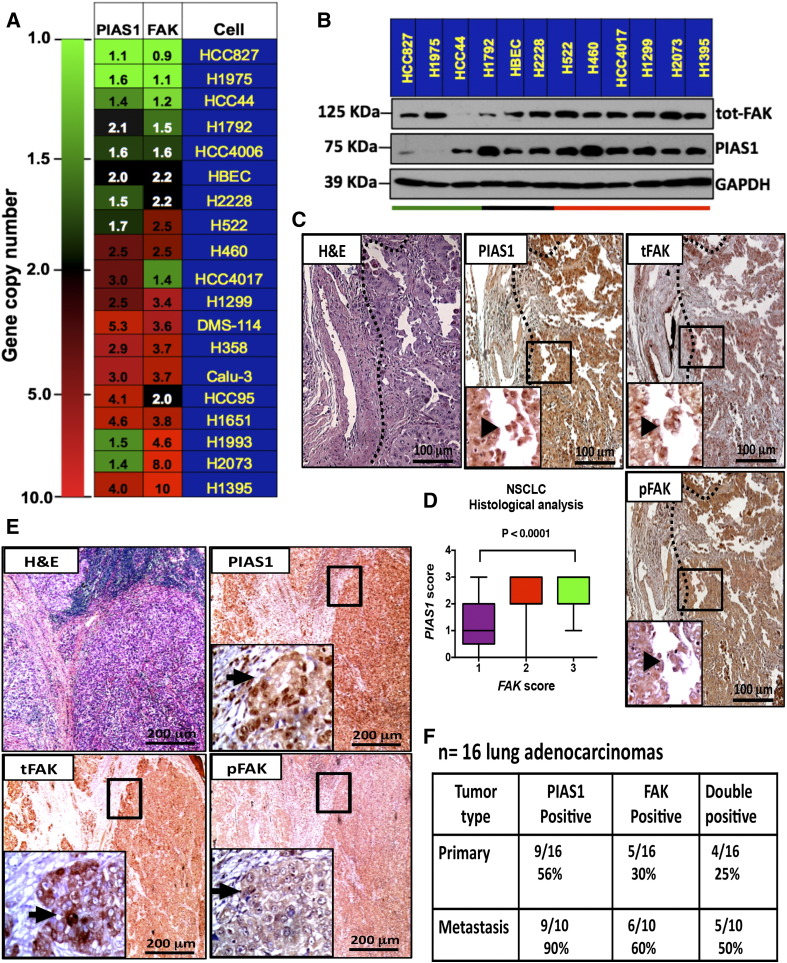
Using SNPs gene copy number analysis we looked for novel genomic cooperating alterations in a panel of 108 non-small cell lung cancer (NSCLC) cells. This analysis revealed that approximately 8% (ie, 12 cell lines) of NSCLC cells had amplification of the FAK and/or PIAS1 genes (Figure 1A).
Figure 1.
FAK and PIAS1 protein levels are correlated in NSCLC. (A) Heatmap shows single nucleotide polymorphism (SNPs) gene copy number analysis for FAK and PIAS1 genes in the indicated lung cancer cell lines. Green, black and red indicate copy number loss, no change or gene copy number gain, respectively. (B) Immunoblot confirms increment in FAK and PIAS1 at the protein level in a representative panel of cells with gene amplification. Green, black and red horizontal lines indicate NSCLC cells with loss, no change or gene copy number gain of PIAS1 and FAK. (C) Hematoxylin and eosin (H&E) and IHC stain of PIAS1, phospho-FAK (pFAK) and total FAK in primary NSCLC samples. Insert shows cytoplasmic and nuclear positivity for both proteins (black arrowheads). Scale bar: 100 μm. (D) IHC positivity score analysis of FAK and PIAS1 in human NSCLC tumor tissue microarrays samples (n = 330). Graph shows positive correlation between increase positivity in FAK stain and PIAS1. Score = 1 low intensity; 2 = moderate intensity; 3 = high intensity. Statistical analysis was done using a Mann-Whitney U test. (E) H&E and IHC stain of PIAS1, phospho-FAK (pFAK) and total FAK in metastatic NSCLC. Insert shows positivity for cytoplasmic and nuclear stain for both proteins. Scale bar: 200 μm. (F) Contingency table shows positive correlation between FAK and PIAS1 IHC stain positivity in primary vs metastatic samples. The number of samples is indicated.
To investigate the relationship between gene copy number gain and gene expression we detected FAK and PIAS1 by immunoblot in cell lines representative of our larger panel of NSCLC cells. We found that cell lines with increased PIAS1 gene copy number display concordant increase in PIAS1 protein (Figure 1B). Since gene amplification/deletions may result from adaptive responses to selective pressures of tissue culture, we tested primary NSCLC specimens for FAK and PIAS1 proteins using IHC. We found that primary NSCLC samples exhibited elevated total FAK (tot-FAK), phospho-FAK (p-FAK; Y397) and PIAS1 positivity compared to normal surrounding tissue (Figure 1C, arrowhead). Using lung tumor tissue microarrays we further confirmed that samples with high tot-FAK positivity score had a concomitant increase in PIAS1 positivity, but not p-FAK positivity (Figure 1D). Because FAK is associated with cell migration [9], [33], we tested paired primary and metastatic NSCLC samples for FAK and PIAS1 protein levels. As observed in primary lung tumor samples, we found a positive correlation with FAK and PIAS1 protein levels in lymph node metastasis, but not with p-FAK protein levels (Figure 1, E and F). These observations suggest that PIAS1 and total-FAK protein levels correlate with tumor progression, independent of FAK phosphorylation.
The KrasG12D/p53R172H mouse model (KP hereafter) faithfully recapitulates aggressive human lung adenocarcinoma, developing advanced lung tumors that are highly metastatic [34], [35]. Metastatic lung tumors in this mouse model have been fully characterized and show elevated expression of the ZEB1 transcription factor and activation of Notch1 and Jagged2 signaling pathways, recapitulating the EMT profiles associated with human metastatic NSCLC [36], [37]. As in human NSCLC, we found that KP lung tumors are positive for total-FAK and PIAS1, but not for p-FAK (Supplementary Fig. 1A). Next we took advantage of 393P and 344SQ lung cancer lines, which are a pair of lung cancer cell lines derived from a KP lung specimen. 393P cells, were derived from a primary lung tumor, are epithelial and have low metastatic potential; while 344SQ cells were derived from a metastatic KP lung tumor, are mesenchymal and are highly metastatic [34]. In this experimental system we found that PIAS1 is significantly up-regulated in vivo and metastatic cell line 344SQ (Supplementary Fig. 1B).
In view of these findings, we concluded that co-amplification of FAK and PIAS1 genes occurs in a subset of primary human NSCLC samples. Moreover, subsets of primary and metastatic NSCLC samples have elevated PIAS1 and tot-FAK protein levels.
PIAS1 and FAK Proteins Physically Interact in NSCLC
We tested whether FAK and PIAS1 physically interact with co-IP assays using NIH-3T3 cells transiently transfected with FAK or PIAS1 cDNAs. We successfully co-immunoprecipitated FAK with PIAS1 from transfected NIH-3T3 cells (Figure 2A). Furthermore, we co-immunoprecipitated endogenous FAK and PIAS1 proteins in several NSCLC cells, which co-amplify FAK and PIAS1 genes (Figure 2B).
Figure 2.
PIAS1 and FAK physically interact in human NSCLC cell lines. (A) NIH 3T3 cells were transiently transfected with the indicated plasmids and FAK and PIAS1 protein interaction was analyzed by co-IP followed by immunoblotting. (B) Immunoblot showing endogenous PIAS1 and FAK protein co-IP in the indicated NSCLC cell lines. (C) Subcellular fractionation and immunoblotting of endogenous FAK and PIAS1 proteins was performed in serum starved lung cancer cells H460 following treatment with: DMSO; EGF; protein kinase C inhibitor Calphostin (PKCi) and 10% FBS. Fractions: C; cytosolic; M; matrix; N: nuclear. Note significant cytoplasmic FAK and PIAS1 protein accumulation in PKCi and serum treatment, but not with EGF. (D) IF and confocal micrograph of FAK (red) and PIAS1 (green) localization at nuclear periphery in H460 NSCLC cells. Scale bar: 10um. (E) Immunoblot following Flag-Pias1 and Fak overexpression in NIH-3T3. Blot shows no change in total FAK or pFAK (Y397), but note the appearance of low and higher molecular weight species of FAK protein in cells overexpressing Flag-Pias1 or Flag-Pias1-SUMO1 overexpression, which correspond to SUMOylated-FAK and cleaved FAK. (F) Cells were transfected with mCherry-Fakwt or mCherry-FAKV744G, a cleavage resistant mutation, in combination with Flag-Pias1 and analyzed with immunoblot as indicated. Note that Flag-Pias1 promotes the cleavage of mCherry-Fakwt as expected, but cleavage of C-terminus mutant FAKV744G is impaired.
To gain insights into the location and stimulus required for FAK and PIAS1 interaction, we performed cellular co-fractionation of endogenous proteins during serum starvation, apoptosis or mitogenic stimulation. To our surprise, we discovered that FAK and PIAS1 mostly co-purified in the cytosolic fractions, when cells were treated with protein kinase C inhibitor (PKCi: Calphostin-C, a proapoptotic agent), or serum for 2-4 hours post starvation, but not after stimulation with epithelial growth factor (EGF) (Figure 2C lanes 5, 7 and 10). Using IF and confocal microscopy we found significant co-localization of endogenous FAK and PIAS1 at the nuclear periphery of serum treated cells (Figure 2D). We subsequently performed immunoblot and confocal co-localization studies with well-characterized organelle markers following serum stimulation. Although partial localization of PIAS1 and FAK can be seen with the endoplasmic reticulum protein Calnexin, the dominant sites of interaction contain the RAB11 and LAMP1, endosome associated proteins. These findings suggest that internalization of growth factor receptors complexes promote the association of FAK and PIAS1 proteins in endosomes localized in the cytoplasm and nuclear periphery (Supplementary Fig. 2A-C).
To begin testing the biological significance of the interaction between FAK and PIAS1, we determined the effects of Flag-Pias1 and or SUMO overexpression on FAK proteins. We found that overexpression of Flag-Pias1 or SUMO-1 did not promote a significant change in tot-FAK or p-FAK (Figure 2E). However, expression of Flag-Pias1 or the combination of Flag-Pias1 and SUMO1 resulted in the appearance of a lower molecular weight species of FAK (Figure 2E arrowheads).
FAK was previously reported to undergo proteolytic cleavage by Calpain proteases on its C-terminus to allow disengagement from focal adhesions [38]. To test whether PIAS1 contributes to FAK C-terminal cleavage we used a mutant (FakV744G) that abolishes FAK protein C-terminal cleavage. Indeed, we found that the FakV744G is resistant to PIAS1-induced FAK C-terminal proteolysis (Figure 2F).
Thus, we concluded that mitogenic signaling promotes FAK and PIAS1 physical interaction, which correlates with PIAS1-induced FAK C-terminal cleavage. Importantly, this phenotype is rescued by using a FAK mutant resistant to proteolytic cleavage.
PIAS1-FAK Interaction Regulates Focal Adhesion Dynamics
Endogenous PIAS1 is mostly nuclear, but can also be found scattered in the cytoplasm of lung cancer cells in vivo and in vitro (Figures 1C and 2D). We hypothesized that PIAS1-FAK interaction may affect focal adhesion dynamics and how cells integrate extracellular signaling. To test this hypothesis, we used RNAi to determine how PIAS1 affects F-Actin stress fiber formation. We found that PIAS1 silencing in lung cancer cells destabilizes F-Actin fibers and reduces Vinculin (VCL) localization to focal adhesions (Figure 3A). Then we tested if the opposite was true using HBECs, which are immortalized, but not transformed, by transduction of CDK4 and human Telomerase Reverse Transcriptase (hTERT) [29]. Indeed, we found that ectopic expression of PIAS1 (Flag-Pias1) in HBECs increases VCL puncta at focal adhesions, while also promoting the nuclear localization of GFP-FAK (GFP-Fak) (Figure 3B). We further tested for changes in VCL puncta formation in live cells using mCherry labeled VCL (mCherry-VCL). We measured the formation, duration and turnover of mCherry-VCL focal adhesion, as inverse correlation to FAK-dependent turnover of focal adhesions [11], [38]. Compared to control shRNA, PIAS1 silencing increased the turnover of mCherry-VCL puncta; along the leading and lagging edge of cells (Figure 3, C–F). Furthermore, we found that Flag-Pias1 gene expression induces membrane ruffle formation in fibroblasts and rescues endogenous VCL puncta loss in GFP-Fak overexpressing cells (Supplementary Fig. 3A, B). Finally, induction of membrane ruffles by PIAS1 correlates with increases in Rac1-GTPase and ROCK-1 protein levels without an increase in pFAK levels (Supplementary Fig. 3C).
Figure 3.
PIAS1 silencing reduces stable stress fiber formation. (A) IF image of filamentous Actin (F-Actin) fiber formation in H460 NSCLC cells. White arrows indicate F-Actin fibers in control siRNA treated cells (Ctrl siRNA) or following PIAS1 gene silencing; yellow arrows indicates Vinculin (VCL) protein localization to F-Actin fibers, which is lost in cells with PIAS1 silencing. Scale bar: 10 μm. (B) IF image of VCL and GFP-FAK co-localization at F-Actin fibers in HBECs cells following ectopic Flag-Pias1 expression. Top row shows HBECs cells expressing ectopic GFP-Fak and endogenous VCL. Bottom row image shows HBECs expressing ectopically expressed Flag-Pias1 and GFP-FAK stained as indicated. Note FAK nuclear accumulation and increase in VCL puncta at focal adhesions (yellow arrows). Pink arrows indicate co-localization of GFP-FAK and VCL proteins. Scale bar: 10 μm. (C) Confocal live cell imaging of H460 NSCLC cells examining mCherry-VCL puncta formation dynamics in cells expressing the indicated shRNAs. Yellow arrows indicate sites of rapid focal adhesion turnover over a time lapse of 25 minutes. (D-F) Histograms show the average time for mCherry-VCL puncta formation, duration and turnover. Values represent an average of n = 3 cells.
In view of these findings we concluded that PIAS1 regulates focal adhesion dynamics by promoting FAK nuclear localization and directional cell motility.
PIAS1 Silencing Impairs the Ability of NSCLC Cells to Form Colonies on Soft Agar
To elucidate how PIAS1-FAK interaction can modulate lung cancer progression we tested the effect of PIAS1 inhibition on cell proliferation and survival. We found that PIAS1 silencing with a short hairpin RNA (shRNA) had a modest effect on cell growth on plastic for most NSCLC cell lines (Figure 4A). Next, we examined how PIAS1 inhibition would affect growth of NSCLC cells in soft agar. Indeed, following PIAS1 inhibition, we observed a marked reduction in colony formation in soft agar assays (Figure 4, B–D). Next we tested whether PIAS1 inhibition may result in defective p-FAK activation, which could explain the inability of NSCLC cells with silenced PIAS1 to establish colonies in soft agar assays. Surprisingly, p-FAK levels did not correlate with soft agar growth impairment in lung cancer cells (Figure 4E). Notably, the degree of knockdown achieved by shRNA A and B correlates with their effect on cell viability (Figure 4A). However, PIAS1 silencing promotes increase baseline levels of pro-apoptotic protein BIM in several lung cancer cells including: H2228 and H1395 (Figure 4F). This finding suggests that PIAS1 silencing does not directly affect p-FAK or total-FAK protein levels in NSCLC, but promotes changes in FAK subcellular localization that are independent on FAK phosphorylation. Furthermore, these findings indicate that PIAS1 silencing lowers the threshold for apoptosis in NSCLC cells.
Figure 4.
PIAS1 silencing impairs tumor progression in NSCLC cells with FAK gene amplification. (A) Histogram shows cell viability following PIAS1 gene silencing. FAK gene copy number status is also indicated. (B) Representative soft agar colonies of NSCLC cells expressing the indicated shRNA. Scale bar: 500 μm. (C) Quantification of soft agar colonies of H2228 NSCLC cells 3 weeks post plating. (D) Quantification of soft agar colonies of H460 NSCLC cells 10 days post plating. (E) H2228, H522, H460 and H1395 NSCLC cells were treated with the indicated shRNAs and analyzed by Immunoblot with the indicated antibodies. (F) Immunoblot following PIAS1 shRNA knockdown in H2228, H460 and H1395 NSCLC cells shows BIM pro-apoptotic protein upregulation. (G) Representative image of a Transwell migration assay performed with H460 cells expressing the indicated shRNAs. (H) Histogram shows the absolute number of migrating cells in triplicate wells following PIAS1 knockdown in lung cancer cell line H460. Student's t test = ***P < .001. (I) Representative image of a Transwell migration assay performed with H1395 cells expressing the indicated shRNAs. (J) Histogram shows the absolute number of migrating cells in triplicate wells following PIAS1 knockdown in H1395 cells. Student's t test = **P < .01.
Our analysis of human NSCLC samples suggested that PIAS1 and FAK proteins are elevated in a subset of highly invasive tumors. Thus, we tested whether PIAS1 silencing affects NSCLC cell invasion and migration potential in vitro. To do this, we performed Transwell migration and scratch assays in NSCLC cells with gene amplification of FAK and PIAS1 [21], [30], [39]. In Transwell assays, H460 cells expressing the control shRNA completed migration after 16 hours (16 h) [40], whereas cells expressing a shRNA targeting PIAS1 showed reduced migration through Transwell membranes (Figure 4, G–H). These findings were replicated in H1395 cells, another NSCLC cell line with gene amplification of FAK and PIAS1 (Figure 4, I–J).
These results suggest that PIAS1 suppression significantly reduce the migration capacity of NSCLC cells. Thus, we performed scratch migration assays using NSCLC cells: H1792, H522 and H460 as representative examples of NSCLC cells from our panel. We found that PIAS1 suppression did not have an effect on H1972, which has a loss of FAK gene copy number, but reduced the migration of H522 and H460, which have substantially higher levels of FAK and PIAS1 protein (Supplementary Figure 4, A–F). We assessed for changes in the cytoskeleton following scratch formation in H460 cells by staining for F-Actin and cells polarity with the GM130 Golgi maker. After 8 hours, cells transfected with the siRNA control were oriented towards the scratch site, whereas cells with PIAS1 silencing were not, suggesting reduced ability to polarize towards the scratch site (Supplementary Figure 4, G–H).
In view of our findings we conclude that PIAS1 suppression is associated with reduced anchorage-independent growth. In addition, PIAS1 silencing reduces cell polarization during stimulus-driven migration in NSCLC cells with FAK and PIAS1 gene amplification.
PIAS1-FAK Interaction Regulates Gene Transcription
We determined that PIAS1 promotes focal adhesion maturation and FAK protein nuclear accumulation. This phenotype was conserved in NSCLC cells, NIH-3T3 fibroblasts and HBECs (Figure 5, A–B). GFP-FAK nuclear accumulation also occurs in HBECs cells harboring the oncogenic KRASG12D mutation and p53 knockdown, suggesting a positive correlation with cancer progression (Figure 5B).
Figure 5.
PIAS1 promotes FAK protein nuclear localization and gene transcription. (A) Immunoblot of immortalized HBECs transduced/transfected with the indicated plasmids. Arrow indicates cleaved FAK. (B) Confocal microscopy of HBECs transduced as indicated. Note the nuclear relocation of GFP-FAK after either ectopic Flag-Pias1 overexpression in HBECs or cells harboring KRASG12D mutations (green and red arrows indicate cytoplasmic and nuclear FAK, respectively). Scale bar: 20 μm. (C-D) Heatmap and gene ontology analysis of HBECs expressing GFP-Fak alone or in combination with Flag-Pias1. (E) Immunoblot of FAK wild type (FAK+) or FAK deleted (FAK-) H460 NSCLC cells treated with IR and harvested at the indicated time points. Note upregulation of p-CHK2 and g-H2AX in null cells, indicating DNA damage hypersensitivity following FAK loss. (F) Histogram shows the change in ATP productions (pmol/min) and oxygen consumption rate (OCR) in H460 NSCLC following FAK gene deletion. Analysis of mitochondrial function shows reduced ATP production and oxygen consumption in FAK- H460 cells.
To test whether PIAS1-FAK interaction and FAK nuclear accumulation are directly associated with a pro-tumorigenic gene transcription program, we analyzed HBECs ectopically expressing GFP-Fak and Flag-Pias1. We analyzed mRNA-transcript linearized data with the Benjamini-Hochberg statistics (log ratio 0.6; and p value < 0.3) and subtracted values obtained from HBECs expressing GFP-Fak alone or Flag-Pias1 alone to obtain transcripts that change only when both genes are co-expressed in HBECs. We identified 473 differentially up/down-regulated transcripts, which we used for further characterization (GEO accession ID: GSE73280). Using gene ontology analysis we uncovered that co-expression of GFP-FAK and PIAS1 correlates with activation of several transcriptional programs that include DNA damage repair genes, oxidative phosphorylation genes and a pancreatic adenocarcinoma signature (Figure 5D). This finding suggests that nuclear FAK may participate in DNA repair and cell cycle progression, a hypothesis consistent with the known function of PIAS1 in these processes [1], [5].
To test whether FAK is involved in DNA repair or mitochondrial metabolism, we targeted its deletion in NSCLC cells by CRISPR/Cas9 gene editing. First, we tested for changes in γH2AX and pCHK2, which are well-known DNA damage response genes [31], [41]. We found that in addition to having baseline activation of γH2AX and phospho-Chk2 (pCHK2), surrogate markers of DNA damage, FAK null (FAK-) H460 NSCLC cells were hypersensitive to ionizing radiation (IR) (Figure 5E). This result suggests a link between FAK nuclear localization and DNA damage response in NSCLC cells. We also tested the effect of FAK loss on oxidative phosphorylation by assessing ATP production and oxygen consumption rate. We found that in agreement with the perturbation in oxidative phosphorylation signatures during PIAS1-FAK overexpression, FAK protein depletion by CRISPR/Cas9 led to a reduced mitochondria ATP production and oxygen consumption rates (Figure 5, F and G).
Taken together these results suggest that PIAS1, by promoting FAK nuclear localization, promotes tumor progression by engaging transcriptional programs that regulate DNA damage repair, and oxidative phosphorylation.
PIAS1 Silencing is Detrimental for Xenograft Tumor Growth In Vivo.
Lastly, we examined whether PIAS1 was required for tumor engraftment in vivo by performing xenograft experiments. We compared H460 cells growth rate in vivo following stable viral transduction of control shRNA or a PIAS1 shRNA. Xenografts expressing PIAS1 shRNA showed a significant reduction in growth 15 days post implantation as compared to controls (P < .05) (Figure 6A). Furthermore, mice in the control group were euthanized on average after 37-days due to tumor burden; whereas the survival of mice in the PIAS1 shRNA group averaged 55 days (Figure 6B). We performed postmortem histological analysis of xenograft tumors and found no significant difference in cell morphology or vascularity (Figure 6C). We then examined the contribution of shRNA harboring cells to the xenograft using the GFP reporter in the shRNA vector backbone and discovered a significant underrepresentation of GFP positive cells in the PIAS1-shRNA treatment group as compared to shRNA controls (**P < .05) (Figure 6, D–E).
Figure 6.
PIAS1 gene silencing impairs xenograft tumor growth in vivo. (A) The histogram shows the volume of xenografts from H460 cells grown subcutaneously in NOD-SCID mice following transduction with scrambled shRNA (shScr; black line) or PIAS1 shRNA (shPIAS1; red line). Mice per group N = 7. (B) Kaplan–Meier curve of mice carrying xenografts shown in panel A. Mice were sacrificed when the tumors reached 2000 mm3. (C) H&E histological analysis of xenografts expressing shScr or PIAS1 shRNA at the end point of the experiment. Squares indicate the sections analyzed for the GFP reporter in the shRNA construct. (D) Anti-GFP IF shows loss of GFP positive cells in the PIAS1 shRNA xenograft group compared to the scramble shRNA group. (E) Quantification of GFP positive cells in the indicated xenograft sections. Student's t test = **P < .01.
In view of these findings, we concluded that PIAS1 is required for the growth of tumor xenografts in vivo. Furthermore, PIAS1 inhibition is selected against during tumor growth as demonstrated by the underrepresentation of PIAS1 shRNA expressing cells at the experiment endpoint.
Discussion
Metastasis accounts for more than 90% of cancer related deaths worldwide [34], [35], [42], [43]. Consequently, identification of novel biomarkers and potential therapeutic targets is of paramount importance. FAK nuclear localization has been reported to be important for disease progression in various cancer types and a requirement during embryogenesis and tissue homeostasis [40], [44], [45]. However, the mechanism that mediates FAK relocation to the cell nucleus and if and how this enhances cell survival, metastasis and tissue development has remained unclear [25], [46].
Using SNPs data, we discovered that the SUMO-E3 ligase PIAS1 and FAK genes are co-amplified in a subset of NSCLC specimens. Furthermore, we show a positive correlation between gene copy number gain and increase in total FAK and PIAS1 protein levels, using NSCLC cell lines in vitro and human lung tumor samples in vivo. Interestingly, we found that FAK and PIAS1 protein are elevated in a subset of primary and invasive human lung tissues and in a bona fide mouse model of NSCLC metastasis [35].
It was previously reported that FAK and PIAS1 protein interact in a yeast two-hybrid screen and in transfected HEK-293T cells. In this setting PIAS1 promotes FAK phosphorylation at tyrosine 397 (Y397), a key event for FAK activation [47]. We confirmed that endogenous PIAS1 and FAK interact in NSCLC cells and this interaction is observed at the nuclear periphery or within the cell nucleus. However, our findings indicate that PIAS1 expression does not affect P-FAK or total FAK protein levels in NSCLC, instead our data support the conclusion that PIAS1 promotes changes in FAK subcellular localization that are independent on its phosphorylation. Indeed, PIAS1 expression, or the combined expression of PIAS1 and SUMO-1, results in the appearance of a ~ 87 KDa form of FAK, previously reported as a Calpain-mediated FAK cleavage product [38]. Indeed we found that using a point mutant form of FAK protein (FakV744G), that renders FAK resistant to Calpain proteolysis, the ~ 87KDa form of FAK was significantly reduced. Because FAK cleavage is part of its negative regulation at focal adhesions, we conclude that PIAS1 may be involved in the regulation of FAK and focal adhesion dynamics.
Recently, inhibition of PIAS1 was found to reduce breast cancer tumorigenesis by increasing genomic instability and loss of stem cell potential [7]. Silencing of PIAS1 in a subset of NSCLC cells with FAK amplification results in deregulated F-Actin formation and increase in focal adhesion turnover. We found that PIAS1 expression can promote GFP-FAK nuclear accumulation and rescues the accumulation of VCL puncta at focal adhesions. The latter correlates with an increase in the activation of integrin downstream targets (RAC-1 and ROCK-1), but not pFAK activation, as it is no longer at the cell membrane where its phosphorylation is known to occur [48], [49].
Although FAK is widely associated with metastasis [10], [11], [25], it is still unclear whether PIAS1 or SUMOylation negates or contributes to tumor progression [5], [50]. PIAS1 was reported to repress TGF-β and reduced EMT transition in breast cancer cells by repression of N-cadherin [6]. However, other reports suggest that PIAS1 is necessary for breast cancer progression via activation of WNT5A, regulation of the estrogen receptor (ER) signaling and promoting tumor growth in vivo [7]. In our study, we found that PIAS1 silencing in NSCLC did not have an effect on EMT genes including N-Cadherin or E-Cadherin (JDC and PPS, unpublished data). However, PIAS1 silencing in NSCLC reduced cell polarization and migration, suggesting that PIAS1 contributes to NSCLC stimulus-driven migration. Specifically, we observed deficient lamellipodia formation and GM130 leading edge orientation following migration stimulus in vitro, which may have contributed to cells inability to integrate extracellular signaling and properly migrate. We propose that FAK recruitment to the nucleus by means of increase interaction with PIAS1, allows for focal adhesion maturation and increase integrin signaling, as demonstrated by the appearance of lamellipodia projections in Flag-Pias1 expressing cells. In contrast, lamellipodia are decreased or absent in migrating cells after PIAS1 knockdown. PIAS1 gene silencing in NSCLC cells also reduced cell viability, independently of P-FAK activation. Following PIAS1 gene silencing we observed a concomitant increase in BIM protein levels. In addition, we also observed a significant decrease in soft agar growth following PIAS1 knockdown.
An unexpected result in our study was that PIAS1-induced FAK nuclear recruitment promotes DNA repair transcription program. Because FAK deletion led to DNA-damage hypersensitivity, shown by γH2AX and pCHK2 activation, we speculate that the co-amplification of FAK and PIAS1 promotes DNA damage repair, providing a survival advantage to genomically unstable tumors. In fact, FAK was found to be a negative regulator of p53 tumor suppressor in immortalized fibroblasts, and FAK inhibition radiozensitizes head and neck carcinoma [46], [51]. Work is already in progress to characterize in better detail the involvement FAK in DNA damage repair and possible applications for NSCLC radiotherapy. Another unexpected finding was that silencing of FAK is associated with decrease of oxygen consumption and ATP production, an observation that underscores the importance of FAK in the maintenance of FAK-dependent NSCLC cells. Work is currently in progress to further characterize this phenotype.
In agreement with previous findings in breast cancer, PIAS1 silencing affects NSCLC cells growth in soft agar in vitro and tumor xenograft growth in vivo. We speculate DNA damage regulation in NSCLC may provide the prosurvival signaling required for tumor progression in PIAS1/FAK overexpressing cancers. It will be of interest to identify the transcriptional modulators interacting with FAK while in the nucleus, as they represent potential tumor biomarkers or therapeutic targets.
Finally, our results show that a subset of NSCLCs has co-amplification of FAK and PIAS1 and that these proteins are enriched in metastatic NSCLC. We conclude that PIAS1 is oncogenic and that, at least in part, this activity depends on its ability to promote FAK nuclear accumulation, integrin signaling activation and DNA damage repair. We conclude that the FAK-PIAS1 signaling axis is a novel regulator of NSCLC progression, integrating extracellular cues that regulate cell survival, migration and the DNA damage response. We propose that FAK-PIAS1 status would serve as a biomarker for the selection of patients undergoing personalized cancer treatment protocols likely to respond to FAK inhibitors currently in clinical trials.
Conflict of Interest
Authors declare no conflict of interest.
Author Contribution
JDC and PPS conceived the project, designed experiments and wrote the manuscript. JDC, KT, SM, HL and MM performed experiments; XT, JRC and IW performed and interpreted tumor microarray histochemistry experiments.
Acknowledgments
We thank Lisa Sowels, for administrative assistance and the Genomics core facility at UT Southwestern for assistance with microarray experiments. We would like to thank John Minna, Luc Girard (UT Southeastern Medical Center) for providing NSCLC cells and access to SNP bioinformatics data and Jon Kurie (MD Anderson Cancer Center) for providing KP lung tumors and 393P and 344Q cell lines. We are grateful to Dr. Hongtao Yu, Dr. Jerry W. Shay and Dr. Rolf Brekken for helpful discussions of this manuscript. We would like to acknowledge funding support from NCI grant #1F31CA180689-01 (to JDC), NIH grants #1R01CA137195, American Cancer Society Scholar Award 13-068-01-TBG, CDMRP LCRP Grant LC110229, UT Southwestern Friends of the Comprehensive Cancer Center, the Gibson Foundation and Texas 4000 (to PPS), Science and Technology Program of Guangzhou grant # 2012J5100031 (to KJT), NCI Cancer Center support grant #1P30 CA 142543-01 (Harold C. Simmons Cancer Center), NCI grant #2P30CA016672, MD Anderson Cancer Center Institutional Tissue Bank (IIW).
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.neo.2016.03.003.
Appendix A. Supplementary data
Supplementary figures
Supplementary material.
References
- 1.Galanty Y, Belotserkovskaya R, Coates J, Polo S, Miller KM, Jackson SP. Mammalian SUMO E3-ligases PIAS1 and PIAS4 promote responses to DNA double-strand breaks. Nature. 2009;462:935–939. doi: 10.1038/nature08657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li R, Wei J, Jiang C, Liu D, Deng L, Zhang K, Wang P. Akt SUMOylation regulates cell proliferation and tumorigenesis. Cancer Res. 2013;73:5742–5753. doi: 10.1158/0008-5472.CAN-13-0538. [DOI] [PubMed] [Google Scholar]
- 3.Morris JR, Boutell C, Keppler M, Densham R, Weekes D, Alamshah A, Butler L, Galanty Y, Pangon L, Kiuchi T. The SUMO modification pathway is involved in the BRCA1 response to genotoxic stress. Nature. 2009;462:886–890. doi: 10.1038/nature08593. [DOI] [PubMed] [Google Scholar]
- 4.Rabellino A, Carter B, Konstantinidou G, Wu SY, Rimessi A, Byers LA, Heymach JV, Girard L, Chiang CM, Teruya-Feldstein J. The SUMO E3-ligase PIAS1 regulates the tumor suppressor PML and its oncogenic counterpart PML-RARA. Cancer Res. 2012;72:2275–2284. doi: 10.1158/0008-5472.CAN-11-3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoefer J, Schafer G, Klocker H, Erb HH, Mills IG, Hengst L, Puhr M, Culig Z. PIAS1 is increased in human prostate cancer and enhances proliferation through inhibition of p21. Am J Pathol. 2012;180:2097–2107. doi: 10.1016/j.ajpath.2012.01.026. [DOI] [PubMed] [Google Scholar]
- 6.Dadakhujaev S, Salazar-Arcila C, Netherton SJ, Chandhoke AS, Singla AK, Jirik FR, Bonni S. A novel role for the SUMO E3 ligase PIAS1 in cancer metastasis. Oncoscience. 2014;1:229–240. doi: 10.18632/oncoscience.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu B, Tahk S, Yee KM, Yang R, Yang Y, Mackie R, Hsu C, Chernishof V, O'Brien N, Jin Y. PIAS1 regulates breast tumorigenesis through selective epigenetic gene silencing. PLoS One. 2014;9:e89464. doi: 10.1371/journal.pone.0089464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frisch SM, Schaller M, Cieply B. Mechanisms that link the oncogenic epithelial-mesenchymal transition to suppression of anoikis. J Cell Sci. 2013;126:21–29. doi: 10.1242/jcs.120907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McKean DM, Sisbarro L, Ilic D, Kaplan-Alburquerque N, Nemenoff R, Weiser-Evans M, Kern MJ, Jones PL. FAK induces expression of Prx1 to promote tenascin-C-dependent fibroblast migration. J Cell Biol. 2003;161:393–402. doi: 10.1083/jcb.jcb.200302126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pylayeva Y, Gillen KM, Gerald W, Beggs HE, Reichardt LF, Giancotti FG. Ras- and PI3K-dependent breast tumorigenesis in mice and humans requires focal adhesion kinase signaling. J Clin Invest. 2009;119:252–266. doi: 10.1172/JCI37160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sieg DJ, Hauck CR, Schlaepfer DD. Required role of focal adhesion kinase (FAK) for integrin-stimulated cell migration. J Cell Sci. 1999;112(Pt 16):2677–2691. doi: 10.1242/jcs.112.16.2677. [DOI] [PubMed] [Google Scholar]
- 12.Wilson C, Nicholes K, Bustos D, Lin E, Song Q, Stephan JP, Kirkpatrick DS, Settleman J. Overcoming EMT-associated resistance to anti-cancer drugs via Src/FAK pathway inhibition. Oncotarget. 2014;5:7328–7341. doi: 10.18632/oncotarget.2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Agochiya M, Brunton VG, Owens DW, Parkinson EK, Paraskeva C, Keith WN, Frame MC. Increased dosage and amplification of the focal adhesion kinase gene in human cancer cells. Oncogene. 1999;18:5646–5653. doi: 10.1038/sj.onc.1202957. [DOI] [PubMed] [Google Scholar]
- 14.Golubovskaya VM, Conway-Dorsey K, Edmiston SN, Tse CK, Lark AA, Livasy CA, Moore D, Millikan RC, Cance WG. FAK overexpression and p53 mutations are highly correlated in human breast cancer. Int J Cancer. 2009;125:1735–1738. doi: 10.1002/ijc.24486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Park JH, Lee BL, Yoon J, Kim J, Kim MA, Yang HK, Kim WH. Focal adhesion kinase (FAK) gene amplification and its clinical implications in gastric cancer. Hum Pathol. 2010;41:1664–1673. doi: 10.1016/j.humpath.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 16.Rodenhuis S, Slebos RJ, Boot AJ, Evers SG, Mooi WJ, Wagenaar SS, van Bodegom PC, Bos JL. Incidence and possible clinical significance of K-ras oncogene activation in adenocarcinoma of the human lung. Cancer Res. 1988;48:5738–5741. [PubMed] [Google Scholar]
- 17.Carr HS, Zuo Y, Oh W, Frost JA. Regulation of focal adhesion kinase activation, breast cancer cell motility, and amoeboid invasion by the RhoA guanine nucleotide exchange factor Net1. Mol Cell Biol. 2013;33:2773–2786. doi: 10.1128/MCB.00175-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chang F, Lemmon CA, Park D, Romer LH. FAK potentiates Rac1 activation and localization to matrix adhesion sites: a role for betaPIX. Mol Biol Cell. 2007;18:253–264. doi: 10.1091/mbc.E06-03-0207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Konstantinidou G, Ramadori G, Torti F, Kangasniemi K, Ramirez RE, Cai Y, Behrens C, Dellinger MT, Brekken RA, Wistuba II. RHOA-FAK is a required signaling axis for the maintenance of KRAS-driven lung adenocarcinomas. Cancer Discov. 2013;3:444–457. doi: 10.1158/2159-8290.CD-12-0388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eke I, Deuse Y, Hehlgans S, Gurtner K, Krause M, Baumann M, Shevchenko A, Sandfort V, Cordes N. beta(1)Integrin/FAK/cortactin signaling is essential for human head and neck cancer resistance to radiotherapy. J Clin Invest. 2012;122:1529–1540. doi: 10.1172/JCI61350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen L, Gibbons DL, Goswami S, Cortez MA, Ahn YH, Byers LA, Zhang X, Yi X, Dwyer D, Lin W. Metastasis is regulated via microRNA-200/ZEB1 axis control of tumour cell PD-L1 expression and intratumoral immunosuppression. Nat Commun. 2014;5 doi: 10.1038/ncomms6241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li CW, Xia W, Huo L, Lim SO, Wu Y, Hsu JL, Chao CH, Yamaguchi H, Yang NK, Ding Q. Epithelial-mesenchymal transition induced by TNF-alpha requires NF-kappaB-mediated transcriptional upregulation of Twist1. Cancer Res. 2012;72:1290–1300. doi: 10.1158/0008-5472.CAN-11-3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li XY, Zhou X, Rowe RG, Hu Y, Schlaepfer DD, Ilic D, Dressler G, Park A, Guan JL, Weiss SJ. Snail1 controls epithelial-mesenchymal lineage commitment in focal adhesion kinase-null embryonic cells. J Cell Biol. 2011;195:729–738. doi: 10.1083/jcb.201105103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luo SW, Zhang C, Zhang B, Kim CH, Qiu YZ, Du QS, Mei L, Xiong WC. Regulation of heterochromatin remodelling and myogenin expression during muscle differentiation by FAK interaction with MBD2. EMBO J. 2009;28:2568–2582. doi: 10.1038/emboj.2009.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Serrels A, Lund T, Serrels B, Byron A, McPherson RC, von Kriegsheim A, Gomez-Cuadrado L, Canel M, Muir M, Ring JE. Nuclear FAK Controls Chemokine Transcription, Tregs, and Evasion of Anti-tumor Immunity. Cell. 2015;163:160–173. doi: 10.1016/j.cell.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Flotho A, Melchior F. Sumoylation: a regulatory protein modification in health and disease. Annu Rev Biochem. 2013;82:357–385. doi: 10.1146/annurev-biochem-061909-093311. [DOI] [PubMed] [Google Scholar]
- 27.Bettermann K, Benesch M, Weis S, Haybaeck J. SUMOylation in carcinogenesis. Cancer Lett. 2012;316:113–125. doi: 10.1016/j.canlet.2011.10.036. [DOI] [PubMed] [Google Scholar]
- 28.Gazdar AF, Girard L, Lockwood WW, Lam WL, Minna JD. Lung cancer cell lines as tools for biomedical discovery and research. J Natl Cancer Inst. 2010;102:1310–1321. doi: 10.1093/jnci/djq279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ramirez RD, Sheridan S, Girard L, Sato M, Kim Y, Pollack J, Peyton M, Zou Y, Kurie JM, Dimaio JM. Immortalization of human bronchial epithelial cells in the absence of viral oncoproteins. Cancer Res. 2004;64:9027–9034. doi: 10.1158/0008-5472.CAN-04-3703. [DOI] [PubMed] [Google Scholar]
- 30.Zhang Y, Gong Y, Hu D, Zhu P, Wang N, Zhang Q, Wang M, Aldeewan A, Xia H, Qu X. Nuclear SIPA1 activates integrin beta1 promoter and promotes invasion of breast cancer cells. Oncogene. 2015;34:1451–1462. doi: 10.1038/onc.2014.36. [DOI] [PubMed] [Google Scholar]
- 31.Gil del Alcazar CR, Hardebeck MC, Mukherjee B, Tomimatsu N, Gao X, Yan J, Xie XJ, Bachoo R, Li L, Habib AA. Inhibition of DNA double-strand break repair by the dual PI3K/mTOR inhibitor NVP-BEZ235 as a strategy for radiosensitization of glioblastoma. Clin Cancer Res. 2014;20:1235–1248. doi: 10.1158/1078-0432.CCR-13-1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Konstantinidou G, Bey EA, Rabellino A, Schuster K, Maira MS, Gazdar AF, Amici A, Boothman DA, Scaglioni PP. Dual phosphoinositide 3-kinase/mammalian target of rapamycin blockade is an effective radiosensitizing strategy for the treatment of non-small cell lung cancer harboring K-RAS mutations. Cancer Res. 2009;69:7644–7652. doi: 10.1158/0008-5472.CAN-09-0823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen YJ, Shie MY, Hung CJ, Wu BC, Liu SL, Huang TH, Kao CT. Activation of focal adhesion kinase induces extracellular signal-regulated kinase-mediated osteogenesis in tensile force-subjected periodontal ligament fibroblasts but not in osteoblasts. J Bone Miner Metab. 2014;32:671–682. doi: 10.1007/s00774-013-0549-3. [DOI] [PubMed] [Google Scholar]
- 34.Gibbons DL, Lin W, Creighton CJ, Zheng S, Berel D, Yang Y, Raso MG, Liu DD, Wistuba II, Lozano G. Expression signatures of metastatic capacity in a genetic mouse model of lung adenocarcinoma. PLoS One. 2009;4 doi: 10.1371/journal.pone.0005401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zheng S, El-Naggar AK, Kim ES, Kurie JM, Lozano G. A genetic mouse model for metastatic lung cancer with gender differences in survival. Oncogene. 2007;26:6896–6904. doi: 10.1038/sj.onc.1210493. [DOI] [PubMed] [Google Scholar]
- 36.Yang Y, Ahn YH, Chen Y, Tan X, Guo L, Gibbons DL, Ungewiss C, Peng DH, Liu X, Lin SH. ZEB1 sensitizes lung adenocarcinoma to metastasis suppression by PI3K antagonism. J Clin Invest. 2014;124:2696–2708. doi: 10.1172/JCI72171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang Y, Ahn YH, Gibbons DL, Zang Y, Lin W, Thilaganathan N, Alvarez CA, Moreira DC, Creighton CJ, Gregory PA. The Notch ligand Jagged2 promotes lung adenocarcinoma metastasis through a miR-200-dependent pathway in mice. J Clin Invest. 2011;121:1373–1385. doi: 10.1172/JCI42579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chan KT, Bennin DA, Huttenlocher A. Regulation of adhesion dynamics by calpain-mediated proteolysis of focal adhesion kinase (FAK) J Biol Chem. 2010;285:11418–11426. doi: 10.1074/jbc.M109.090746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen Q, Xu R, Zeng C, Lu Q, Huang D, Shi C, Zhang W, Deng L, Yan R, Rao H. Down-regulation of Gli transcription factor leads to the inhibition of migration and invasion of ovarian cancer cells via integrin beta4-mediated FAK signaling. PLoS One. 2014;9 doi: 10.1371/journal.pone.0088386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ward KK, Tancioni I, Lawson C, Miller NL, Jean C, Chen XL, Uryu S, Kim J, Tarin D, Stupack DG. Inhibition of focal adhesion kinase (FAK) activity prevents anchorage-independent ovarian carcinoma cell growth and tumor progression. Clin Exp Metastasis. 2013;30:579–594. doi: 10.1007/s10585-012-9562-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Camacho CV, Mukherjee B, McEllin B, Ding LH, Hu B, Habib AA, Xie XJ, Nirodi CS, Saha D, Story MD. Loss of p15/Ink4b accompanies tumorigenesis triggered by complex DNA double-strand breaks. Carcinogenesis. 2010;31:1889–1896. doi: 10.1093/carcin/bgq153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cho JH, Robinson JP, Arave RA, Burnett WJ, Kircher DA, Chen G, Davies MA, Grossmann AH, VanBrocklin MW, McMahon M. AKT1 Activation Promotes Development of Melanoma Metastases. Cell Rep. 2015;13:898–905. doi: 10.1016/j.celrep.2015.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guan X. Cancer metastases: challenges and opportunities. Acta Pharm Sin B. 2015;5:402–418. doi: 10.1016/j.apsb.2015.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Albasri A, Fadhil W, Scholefield JH, Durrant LG, Ilyas M. Nuclear expression of phosphorylated focal adhesion kinase is associated with poor prognosis in human colorectal cancer. Anticancer Res. 2014;34:3969–3974. [PubMed] [Google Scholar]
- 45.Lim ST, Miller NL, Chen XL, Tancioni I, Walsh CT, Lawson C, Uryu S, Weis SM, Cheresh DA, Schlaepfer DD. Nuclear-localized focal adhesion kinase regulates inflammatory VCAM-1 expression. J Cell Biol. 2012;197:907–919. doi: 10.1083/jcb.201109067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lim ST, Chen XL, Lim Y, Hanson DA, Vo TT, Howerton K, Larocque N, Fisher SJ, Schlaepfer DD, Ilic D. Nuclear FAK promotes cell proliferation and survival through FERM-enhanced p53 degradation. Mol Cell. 2008;29:9–22. doi: 10.1016/j.molcel.2007.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kadare G, Toutant M, Formstecher E, Corvol JC, Carnaud M, Boutterin MC, Girault JA. PIAS1-mediated sumoylation of focal adhesion kinase activates its autophosphorylation. J Biol Chem. 2003;278:47434–47440. doi: 10.1074/jbc.M308562200. [DOI] [PubMed] [Google Scholar]
- 48.Schaller MD, Hildebrand JD, Shannon JD, Fox JW, Vines RR, Parsons JT. Autophosphorylation of the focal adhesion kinase, pp125FAK, directs SH2-dependent binding of pp60src. Mol Cell Biol. 1994;14:1680–1688. doi: 10.1128/mcb.14.3.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fincham VJ, Frame MC. The catalytic activity of Src is dispensable for translocation to focal adhesions but controls the turnover of these structures during cell motility. EMBO J. 1998;17:81–92. doi: 10.1093/emboj/17.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Netherton SJ, Bonni S. Suppression of TGFbeta-induced epithelial-mesenchymal transition like phenotype by a PIAS1 regulated sumoylation pathway in NMuMG epithelial cells. PLoS One. 2010;5 doi: 10.1371/journal.pone.0013971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lim ST, Mikolon D, Stupack DG, Schlaepfer DD. FERM control of FAK function: implications for cancer therapy. Cell Cycle. 2008;7:2306–2314. doi: 10.4161/cc.6367. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures
Supplementary material.