Abstract
The TWIST1 embryonic transcription factor displays biphasic functions during the course of carcinogenesis. It facilitates the escape of cells from oncogene-induced fail-safe programs (senescence, apoptosis) and their consequent neoplastic transformation. Additionally, it promotes the epithelial-to-mesenchymal transition and the initiation of the metastatic spread of cancer cells. Interestingly, cancer cells recurrently remain dependent on TWIST1 for their survival and/or proliferation, making TWIST1 their Achilles’ heel. TWIST1 has been reported to form either homodimeric or heterodimeric complexes mainly in association with the E bHLH class I proteins. These complexes display distinct, sometimes even antagonistic, functions during development and unequal prometastatic functions in prostate cancer cells. Using a tethered dimer strategy, we successively assessed the ability of TWIST1 dimers to cooperate with an activated version of RAS in human mammary epithelial cell transformation, to provide mice with the ability to spontaneously develop breast tumors, and lastly to maintain a senescence program at a latent state in several breast cancer cell lines. We demonstrate that the TWIST1-E12 complex, unlike the homodimer, is an oncogenic form of TWIST1 in mammary epithelial cells and that efficient binding of both partners is a prerequisite for its activity. The detection of the heterodimer in human premalignant lesions by a proximity ligation assay, at a stage preceding the initiation of the metastatic cascade, is coherent with such an oncogenic function. TWIST1-E protein heterodimeric complexes may thus constitute the main active forms of TWIST1 with regard to senescence inhibition over the time course of breast tumorigenesis.
Introduction
The TWIST1 gene is aberrantly reactivated in a large set of solid cancer types, including a variety of carcinomas, melanomas, sarcomas, and neuroblastomas [1]. As a reminiscence of its embryonic functions, this transcription factor was originally shown to promote the metastatic dissemination of cancer cells through its ability to trigger an epithelial-to-mesenchymal transition [2]. We and others additionally highlighted its ability to alleviate the induction of fail-safe programs (senescence, apoptosis) in response to oncogenic activations, thereby cooperating with mitogenic oncoproteins in promoting cell neoplastic transformation in vitro and tumor initiation in vivo [3], [4], [5], [6], [7], [8], [9]. Furthermore, cancer cells often remained dependent on TWIST1 functions to protect them from latent senescence or apoptosis [3], [4], [8], [9], [10], [11]. Its oncogenic properties are mostly related to its ability to downmodulate the activity of p53 [6], [9], [11], [12] and to dampen the expression of numerous cyclin-kinase inhibitors (p15INK4B, p16INK4A p21CIP1), the relative contribution of these activities varying according to the cellular context [3], [4].
Basic-helix-loop-helix (bHLH) transcription factors were originally classified into different categories based on their expression pattern, partners, and structural features [13]. The TWIST1 protein belongs to class II, which encompasses tissue-specific bHLH, such as MyoD. These proteins heterodimerize with the ubiquitously expressed class I bHLH proteins, termed E proteins [13]. They include the two splice variants of the TCF3/E2A protein E12 and E47, the TCF4/ITF2, and the TCF12/HEB transcription factors. All TWIST1-E heterodimers have been shown to display similar functional properties [14]. The TWIST1 protein also interacts with the class II bHLH transcription factors HAND1 and HAND2 and additionally displays the property to constitute functional homodimers, as originally demonstrated in Drosophila [15]. The TWIST1 protein preferentially heterodimerizes, providing a rationale for the similar limb formation defects triggered by the enforced expression of the TWIST1-E12 heterodimer or of the TWIST1 monomer [16], [17], [18]. Nonetheless, homodimerization is privileged at low protein concentration (e.g., TWIST1 haploinsufficiency in Saethre-Chotzen patients), in the presence of ID HLH proteins which titrate E proteins, or following the phosphorylation of residues located in the helix I of TWIST1 (i.e., Thr121 and Ser123) by protein kinase A [14], [19], [20]. Point mutations preventing the posttranslational modification of these residues (TS121-123AA) or mimicking their constitutive phosphorylation (TS121-123ED) were demonstrated to functionally mimic the TWIST1 homodimer and heterodimer, respectively [16]. The phosphoregulation of TWIST1 was additionally shown to influence its affinity for E-boxes in a cis-element–dependent manner [20]. Posttranslational modifications of TWIST1 thus have an impact on dimer choice and on the downstream activation of targeted genes [21]. The contribution of these complexes to the embryonic TWIST1 functions has been largely explored using tethered dimers, the reliability of this approach being unquestionably established through successful in vivo complementation assays [15], [16], [19]. The validity of this strategy was also supported by the demonstration that the tethered TWIST1~E12 dimer binds DNA and transactivates reporter genes similarly to when TWIST1 and E12 are expressed as separated polypeptides [15], [20]. Exploiting this tethered dimer strategy unveiled differential and even antagonistic properties of the TWIST1 complexes during embryonic development, demonstrating the key role of the TWIST1 partner in determining and regulating TWIST1 functions. Whether the balance between these dimers also modulates TWIST1 functions during tumorigenesis remains relatively unexplored. Enforced expression of tethered TWIST1~TWIST1 or TWIST1~E12 dimers in prostate cancer cell lines led to the conclusion that the prometastatic properties of TWIST1 are allotted to the heterodimeric TWIST1 complex [22]. Their contribution to the malignant transformation nonetheless still remains to be determined. Moreover, identifying the TWIST1 complex implicated in the escape from fail-safe program and upon which cancer cells are dependent for their proliferation and survival constitutes an essential step in the development of novel therapeutic strategies aiming at eradicating cancer cells through TWIST1 inactivation. To tackle this question, we employed this tethered dimer strategy to assess the oncogenic potentials of both TWIST dimers in vitro and in vivo.
Material and Methods
DNA Constructs
The EcoR pBabe Zeo, ER-H-RASG12V pLNCX2-Neo, and H-RASG12V pbabe-Puro retroviral construct and the VIM1-luciferase reporter have been previously described [23], [24], [25], [26]. The tethered dimers were generated by polymerase chain reaction (PCR) using FLAG-human TWIST1 [3] and E12 expression constructs [27] as templates. The strategy employed to generate the tethered dimers is described in detail in the Supplementary Information section. The shRNA TWIST1 A pLKO.1 lentiviral construct was previously described [3]. The shRNA TWIST1 B pSIREN (5′-CTCTGGAGCTGGATAACTAAA-3′) retroviral construct was kindly provided by Patrice Lassus (IGMM, Montpellier). The shRNA TCF3 pLKO.1 was supplied by Sigma.
Mouse Strains
Animal maintenance and experiments were performed in a specific pathogen-free animal facility, “Anican,” in accordance with the animal care guidelines of the European Union and French laws and were validated by the local Animal Ethic Committee (CECAAP). The CAG-LSL-Twist1-Twist1 or CAG-LSL-Twist1-E12 (FVB/NJ genetic background) [19] was crossed with the WAP-Cre (whey acidic protein promoter [28]) mouse strain [B6.Cg-Tg(Wap-Cre)11738 Mam, obtained from the mouse models of Human Cancers Consortium and back-crossed to the FVB/NJ genetic background]. Cohorts of multiparous female WAP-Cre; Twist1-Twist1 and WAP-Cre; Twist1-E12 mice (with two rounds of lactation) were generated. Genotyping was performed on genomic DNA from tails using the 5′-AATGAAATGGAGAGCTTGGGCGAC-3′ and 5′-CATCACTCGTTGCATCGACC-3′ primer pair for the WAP-Cre transgene and 5′-GCAAGCGCGGCAAGAAATCTG-3′ and 5′-CCCGTTCAAGTCCTCTTCAGAAATGAC-3′ primer pair for the Twist1 transgene as described previously [19], [28]. Mice were monitored twice a week for tumor incidence. End points were based on tumor diameter (up to 1.5 cm) unless natural death occurred. Mice were euthanized by CO2 inhalation. Tumors were fixed in 10% phosphate-buffered formalin for 24 hours and embedded in paraffin. Mammary gland, lung, liver, kidney, and spleen were collected either to check for malignant lesions or to assess potential metastatic dissemination.
Cell Culture
HMEC-hTERT cells and derived cells were cultured as described previously [7]. The Hs578T, MDA-MB-436, MDA-MB-231, and BT-474 breast cancer cell lines and HEK293T were provided by the ATCC and cultured according to the supplier’s instructions.
Protein Analysis and Immunohistochemistry
Cells extracts were performed in radioimmunoprecipitation assay (RIPA) buffer [100 mM NaCl, 1% NP40, 0.1% sodium dodecyl sulfate (SDS), 50 mM Tris pH 8] supplemented with protease (Roche) and phosphatase inhibitor cocktails (Sigma) and clarified by centrifugation. Proteins were analyzed by Western blot using the monoclonal anti-TWIST1 2C1a (Abcam), anti-p21CIP1 clone SX118 (Dako), and anti-HEB A9 (Santa Cruz Biotechnology) and the polyclonal anti-TWIST2 (proteintech), anti-E2A V18, anti-ITF2 K12, anti-H-RAS C20, anti-p15INK4B C20, anti-p16INK4A H156, p-histone H3 (Ser10)-R (Santa Cruz Biotechnology), and anti-GAPDH Abs16 (Millipore) antibodies and horseradish peroxidase–conjugated secondary antibodies (Dako). Antigen-antibody complexes were revealed with the Western-blotting Luminol reagent (Santa Cruz Biotechnology).
Analysis of TWIST1 by immunohistochemistry in mouse breast carcinomas was performed as described previously [7] using the monoclonal anti-TWIST1 2C1a (Abcam).
Gene Expression Analysis
RNA preparation and reverse transcription were performed as described previously [29]. Real-time PCR intron-spanning primers were designed with the primer3 software. The HPRT1 housekeeping gene was used for normalization. The combinations of primers used are listed in the Supplementary Information section.
Retroviral Infections, SA-β-Galactosidase, Growth Curves, and Soft-Agar Colony Assays
Enforced expression of H-RASG12V and/or TWIST1 proteins was performed through retroviral infections as described previously [29]. Briefly, cells were “murinized” by expressing the ecotropic receptor [24], [30] before being infected with retroviral expression constructs 48 hours later. Selection was initiated 24 hours post–second infection with puromycin (0.5 μg/ml) or neomycin (100 μg/ml).
shRNA TWIST1 lentiviral particles were generated through the co-transfection of 293T cells with pLKO.1, pCMV ΔR8.91 (gag-pop-Tat-Rev) [31], and phCMVG-VSVG (env) [32] expression constructs, and shRNA TWIST1 retroviral particles were generated through the transfection of HEK293GP cells with pSIREN and phCMVG-VSVG (env) expression constructs using the calcium phosphate precipitation technique. TWIST1 knockdown was achieved via double retroviral (shRNA TWIST1 B pSIREN) or lentiviral (shRNA TWIST1 A pLKO.1) infections at 48-hour intervals. Over 90% of cells were infected. Consequences of TWIST depletion on cell survival and proliferation were assessed in the absence of selection. Activation of ER-RASG12V with 4-OH-tamoxifen (4-OHT, 625 nM, Sigma Aldrich) was specifically performed in medium supplemented with 1 mM D-glucose. Three days later, cells were washed twice with PBS, either fixed in 3% formaldehyde and stained with crystal violet or tested with a SA-β-galactosidase assay [33]. Transformation assays were performed 1 week postinfection (TWIST1 transduction) or post–second infection (RAS + TWIST transduction), as described previously [29].
Immunofluorescence
A total of 104 cells were seeded on a 12-well IBIDI-chamber slide, fixed in 4% formaldehyde (Sigma) for 15 minutes, washed with 1% bovine serum albumin (BSA) in PBS buffer, and permeabilized in 0.1% Triton 100 × (Sigma) and 1% BSA in PBS buffer at room temperature for 10 minutes. The cells were then washed three times with 1% BSA in PBS buffer and incubated with the anti-TWIST1 2C1a primary antibody (Abcam, diluted 1/100 in 0.5% BSA in PBS buffer) overnight at 4°C. Following extensive washing in 1% BSA in PBS buffer, cells were incubated with fluorescein isothiocyanate–conjugated Alexa goat anti-mouse antibody (Life Technology, 1/600 in 0.5% BSA in PBS buffer) for 30 minutes at room temperature. Nuclei were stained with Hoechst 5 mg/ml in 0.5% BSA in PBS buffer for 10 minutes and mounted with Fluoromount-G (SouthernBiotech). Images were taken on an immunofluorescence microscope (Leica) at identical exposure times.
Immunoprecipitation
HEK293T cells were transfected with wild-type or mutant FLAG-tagged TWIST1 and/or MYC-tagged E12 expression constructs using the calcium-phosphate technique. Cells were lysed 36 hours posttransfection in a 50-mM Tris-HCl pH 7.4, 150-mM NaCl, 1-mM EDTA, and 1% Triton buffer supplemented with protease inhibitors (Roche), cleared by centrifugation, and incubated with an anti-FLAG M2 resin (Sigma-Aldrich). After intensive washing to eliminate nonspecific binding, the resin was resuspended in Laemmli buffer and boiled for 3 minutes. After elimination of the beads by centrifugation, samples were reduced by adding β-mercaptoethanol and separated on SDS–polyacrylamide gel electrophoresis. Proteins were analyzed with the murine monoclonal anti-TWIST1 C2a antibody (Abcam) and the rabbit polyclonal V18 E2A antibody (Santa Cruz Biotechnology).
Immunoprecipitation of endogenous E12 protein by ectopically expressed FLAG-TWIST1 was performed in HMEC-hTERT cells. A total of 2 × 106 cells were resuspended in an extraction buffer (20 mM Tris pH 8, 125 mM NaCl, 1 mM EDTA, 0.5% NP40) supplemented with protease inhibitors (Roche), sonicated, and cleared by centrifugation. Protein extracts were diluted in a final 20-mM Tris pH 8 buffer containing 125 mM NaCl, 1 mM EDTA, NP40 0.125%, and glycerol 10% and successively incubated in the presence of 4 μg of M2 α-FLAG antibody (Sigma) for 4 hours at 4°C and protein-A sepharose beads (protein A sepharose 4B, InVitrogen) for 1 hour. After intensive washing in a 20-mM Tris pH 8 buffer containing 125 mM NaCl, 1 mM EDTA, NP40 0.1%, and glycerol 10%, beads were resuspended in Laemmli buffer (in absence of β-mercaptoethanol) boiled at 95°C for 3 minutes. After elimination of the beads by centrifugation, samples were reduced by adding β-mercaptoethanol and separated on SDS–polyacrylamide gel electrophoresis.
Proximity Ligation Assay
Human ductal in situ carcinoma (DCIS) samples were obtained through the Biological Resource Center of the Centre Léon Bérard with the agreement of the ethical review board of the Centre Léon Bérard. Samples were used with the patient’s written informed consent. The present study was approved by the ethical review board of the Centre Léon Bérard. DCIS samples were selected on the basis of TWIST1 protein detection, as previously reported [7].
After deparaffinization and rehydratation, tissue sections were boiled in a 10-mM pH 6 citrate buffer for 40 minutes. After saturating unspecific binding sites for 20 minutes with a 1% BSA PBS buffer, sections were incubated overnight with the mouse monoclonal anti-TWIST1 Twist2C1a (Abcam) and the rabbit polyclonal anti-E2A N-649 (Santa Cruz) antibodies in a 0.5% BSA in PBS buffer. Incubation with PLA probes, ligation, and amplification steps were performed according to the manufacturer instructions (Duolink In Situ kit, Sigma). The specificity of the signal was confirmed using paraffin-embedded MDA-MB-436, Hs578T (TWIST1 positive, E12 positive), and HMEC-hTERT (TWIST1 negative, E12 positive) mammary cells in which TWIST1 or TCF3 (the two E2A protein variants E12 and E47 encoding gene) were specifically knocked down through RNA interference. Number of dots per cell was assessed using the ImageJ software.
Results
Design of Wild-Type and Mutant Tethered TWIST1 Dimers
Because both HAND1 and HAND2 genes were found to be transcriptionally inactive in breast cancer cells and because the TCF3/E2A proteins are the predominant E proteins expressed in the breast cancer cell lines employed in this study, as assessed by Western blot (Figure S1A), the experiments were performed either with homodimeric (TWIST1-TWIST1, hereafter named T~T) or heterodimeric (TWIST1-E2A E12, hereafter named T~E) tethered dimers. Following previous studies [15], [19], [20], [22], the E2A splice variant E12 was employed as the model for TCF3/E2A proteins. Tethered dimers were generated by linking the two partners with a flexible glycine-serine polylinker, also used in previous studies [15], [19], [20], [22], and the TWIST1 protein was tagged at the N-terminus with a single FLAG peptide, a position that we previously confirmed does not interfere with TWIST1 oncogenic functions [3] (Figure S1B). The production and correct nuclear localization of the resulting tethered dimers were validated (Figure S1C). Noticeably, C-terminal truncated subproducts (revealed with both anti-TWIST1 and anti-FLAG antibodies) were detected. Very similar subproducts were revealed with the functional murine MYC-tagged T~T and MYC-tagged T~E forced dimers generated in the laboratory of D. Spicer [14] (Figure S1D). In particular, the detection with the T~T dimer of a predominant subproduct with a molecular size close to the monomer likely reflects a difference in the accessibility of a consensus caspase 3/7 cleavage site present in the C-terminal of TWIST1 within the two tethered dimers. Cleavage of TWIST1 by caspase 3 was originally reported in apoptotic cells and was quickly followed by its proteasome-mediated degradation [34]. It is, therefore, very likely that the cleavage occurs during protein extraction.
As the study progressed, additional mutants were generated including the T R154P~E heterodimer (hereafter named T RP~E) to confirm the need of the interaction between both partners to generate an active tethered heterodimer. The R154P mutation was previously reported to disrupt the TWIST1-E2A protein interaction [35], and this was herein confirmed by performing a co-immunoprecipitation assay (Figure S2A). Insertion of this mutation into the T~E tethered dimer (T RP~E) did not have an impact on its nuclear sublocalization (Figure S2B) but, as anticipated, abrogated the ability of the fusion protein to activate the TWIST1-targeted vimentin (VIM) gene in a reporter assay (Figure S2C). A TWIST1 K145E-TWIST1 K145E (T KE~T KE) tethered dimer was also generated as an inactive version of the T~T tethered dimer. The Lys145 residue plays a determining role in stabilizing TWIST1 complexes on the DNA by contributing to the establishment of the interhelical loops and by directly interacting with oxygen atoms of DNA bases [36], [37]. The K145E mutation thus affects the global structure of TWIST1 complexes and abrogates TWIST1 DNA binding properties [38].
The TWIST1 Dimers Unequally Alleviate Oncogene-Induced Senescence in Human Mammary Epithelial Cells
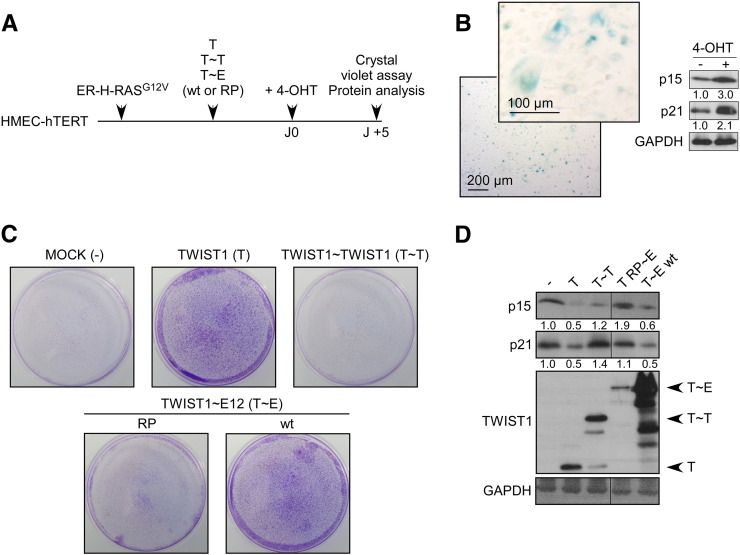
To compare the ability of both TWIST1 dimers in preventing oncogene-induced senescence (OIS), immortalized and nontransformed human mammary epithelial cells (HMEC-hTERT) were sequentially infected with retroviral vectors encoding either for an inducible activated version of HRAS (ER-H-RASG12V) or for TWIST1 (monomer T, T~T or T~E dimer) (Figure 1A). Activation of the ER-H-RASG12V fusion protein by 4-hydroxyl-tamoxifen (+ 4-OHT) triggered a senescence program, as revealed by the permanent proliferation arrest and the detection of a SA-β-galactosidase activity (Figure 1B). While the CDKN2A-ARF locus is methylated in HMEC-hTERT cells [39], [40], [41], commitment into senescence was associated with an accumulation of p15INK4B (p15) and p21CIP1/WAF1 (p21) (Figure 1B). In line with our previous observations [3], enforced expression of the TWIST1 monomer successfully prevented the activation of cyclin-kinase inhibitors and sustained cell proliferation, as assessed in a crystal violet coloration assay (Figure 1, C and D). The heterodimer turned out to be functionally active in this assay and the homodimer inactive (Figure 1, C and D), suggesting that the dimerization between TWIST1 and E protein partners is a prerequisite to carry out this function. To support this hypothesis, we observed that the disruption of the partner interaction through the insertion of the R154P point mutation in TWIST1 abolished T~E activity (Figure 1, C and D). Collectively, this assay demonstrated that both monomeric and tethered heterodimers similarly prevent OIS, supporting the concept that ectopically expressed TWIST1 can form heterodimers with endogenous E proteins [17], [18]. Indeed, ectopic TWIST1 was confirmed to immunoprecipitate the endogenous E12 protein (Figure S3). Of note, in light of the absence of activity of the TWIST1 homodimeric complex, the functionality of the fusion protein was further controlled by assessing its ability to transactivate a set of TWIST1 targeted genes, selected on the basis of previous studies [7], [42]. As shown in Figure S4, the homodimer was confirmed to be functional. As an internal control, the insertion of the K145E mutation (T KE~T KE tethered dimer), described to abolish the TWIST1 DNA binding capability [38], annihilated its transactivation potential.
Figure 1.
Homo- and heterodimeric TWIST1 complexes unequally avoid RAS-induced senescence in immortalized human mammary epithelial cells.
(A) HMEC-hTERT cells were sequentially infected with the inducible ER-H-RASG12V fusion protein and with monomeric TWIST1, the homodimeric TWIST1~TWIST1 tethered dimer, or the wild-type (wt) or mutant (RP) TWIST1~E12 heterodimeric tethered dimer. (B) In the absence of TWIST1, the activation of the ER-H-RASG12V protein, using 4-OHT, was associated with the induction of senescence as shown in the SA-β-galactosidase assay and the accumulation of the cyclin-kinase inhibitors p15INK4B (p15) and p21CIP1 (p21) as assessed by Western blot. (C and D) Five days postactivation of ER-H-RASG12V using 4-OHT, cell proliferation was assessed through a crystal violet staining (C), and p21CIP1 (p21), p15INK4B and TWIST1 proteins were analyzed by Western blot (D). Relative signal quantification with respect to MOCK cells is indicated.
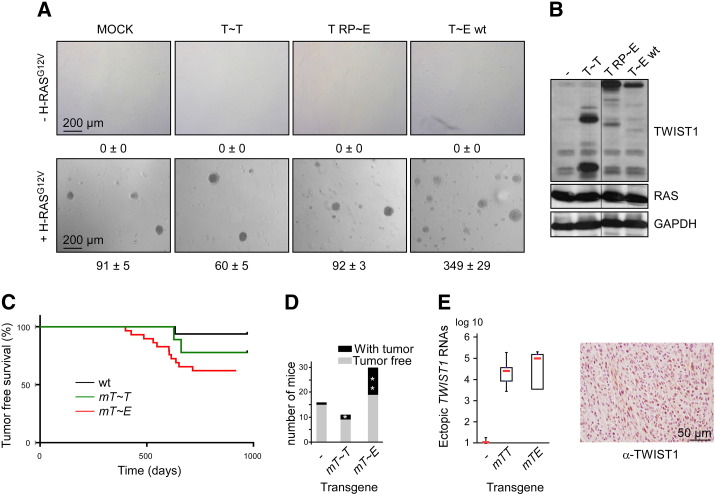
To strengthen the differences observed between the two TWIST1 complexes, we next assessed their ability to cooperate with the H-RASG12V protein in promoting the neoplastic transformation of HMEC-hTERT cells. To this end, cells were sequentially infected with TWIST1 and constitutive H-RASG12V retroviral expression constructs. Whereas the sole expression of the TWIST1 tethered dimers failed to transform cells, concomitant production of H-RASG12V and the active heterodimeric complex successfully transformed cells, as assessed in a colony formation assay (Figure 2, A and B). Neither the homodimeric T~T complex nor the inactive form of the T~E complex (T RP ~ E) was found to be functional in this second assay.
Figure 2.
The heterodimeric TWIST1 complex displays oncogenic properties in vitro and in vivo.
(A) HMEC-hTERT cells were sequentially infected with TWIST1 (T~T, wt, or mutant T~E) and H-RASG12V retroviral expression vectors. The transformation potential of the established cell lines was assessed in a soft-agar colony formation assay. Mean of numbered colonies and SD of one experiment performed in triplicates are shown. (B) Analysis of TWIST1 and RAS proteins by Western blot. (C–E) Assessment of the TWIST oncogenic potential in vivo. The enforced expression of Twist1 in luminal committed mammary epithelial cells was achieved by crossing either CAG-LSL-Twist~ Twist1 or CAG-LSL-Twist1~ E12 with WAP-Cre mice. (C) Kaplan-Meier tumor-free survival. (D) Histogram indicating the number of mice spontaneously developing breast carcinoma during their lifespan. Stars mark mice with two breast tumors. (E, left panel) Plot boxes indicating the expression of ectopic TWIST1 transcripts in tumors as assessed by quantitative reverse transcription (qRT)–PCR. Levels were expressed relative to the housekeeping HPRT1 gene transcript and were normalized against wild-type mice–derived breast carcinoma. (E, right panel) Example of TWIST1 staining in a metaplastic spindle cell carcinoma.
The enforced Expression of the T~E Tethered Dimer Induces the Spontaneous Development of Breast Carcinoma in Mice
Next, we validated the different oncogenic activities observed between the two TWIST1 complexes in vivo. We previously reported that the combined production of either the murine monomeric TWIST1 or the T~E fusion protein with an activated version of K-RAS (K-RASG12D) in luminal committed epithelial cells induced breast tumor development in the first 5 months with a complete penetrance [7]. However, not a single one of the three mice examined, coexpressing the homodimer T~T and K-RASG12D, developed breast tumors within the same period of time, suggesting that the homodimer failed to cooperate with RAS in promoting tumor initiation (G.W. Hinkal, unpublished data). To strengthen this result, we examined whether the ectopic expression of T~T or T~E in the absence of an oncogenic insult was sufficient to promote breast carcinogenesis, with an anticipated lower penetrance and extended latency to accumulate requested secondary events. T~T or T~E production was enforced in luminal committed cells through the generation of WAP: Twist1~ Twist1 and WAP: Twist1~ E12 mice (producing the murine T~T or T~E fusion proteins under the control of the whey acidic protein promoter) by crossing either the CAG-LSL-Twist1~ Twist1 or CAG-LSL-Twist1~ E12 mouse strain with the WAP-Cre mouse strain [19], [28]. Full activation of the WAP promoter was assured by two consecutive lactations. As shown in Figure 2, C–E, T~E-producing mice were found to develop breast carcinomas with a higher frequency (37.9%, n = 29) and a shorter latency than T~T-producing mice (18.1%, n = 14) and control littermates (6.2%, n = 16). Tumor phenotypes were found to be heterogeneous and classified [43] as (i) adenosquamous, (ii) metaplastic spindle cell, (iii) papillary and cribriform, (iv) solid and cribriform, and (v) adenosquamous carcinomas (Table S1). No secondary tumor site was detected. Collectively, these observations support our conclusion that the heterodimeric complexes are the oncogenic forms of TWIST1 in mammary epithelial cells.
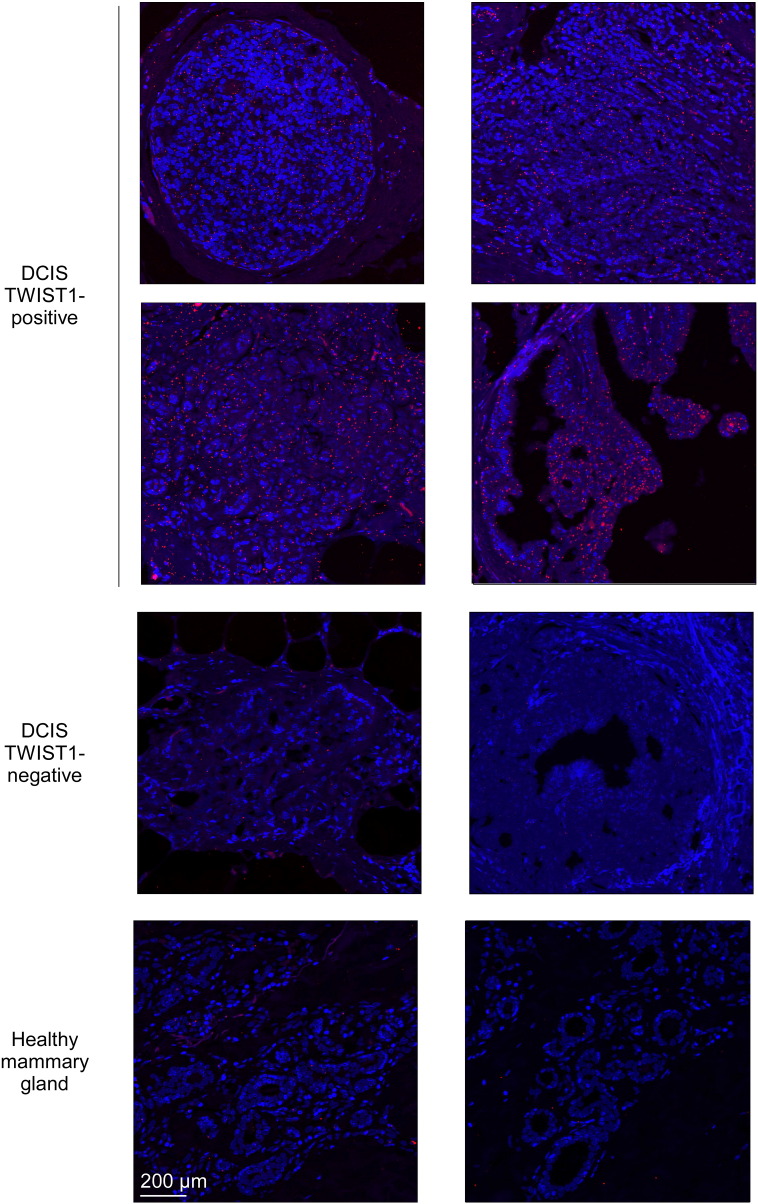
The Heterodimeric TWIST1-E12 Complex Is Detected in Human Breast Premalignant Lesions
If the assumption that the heterodimeric TWIST1 complex plays a role in malignant transformation, breast cancer cell survival, and proliferation is correct, we would expect that this complex would already be detectable in DCIS before the initiation of the metastatic cascade. We thus assessed TE complex formation through a proximity ligation assay (PLA), selecting cases previously defined as TWIST1-positive or -negative by immunohistochemistry [7]. Experimental conditions were established in mammary carcinoma cell lines, and the specificity of the signal was confirmed by turning down the expression of either the endogenous TWIST1 or TCF3 (the E12/E47 proteins-encoding gene) through RNA interference. Depletion in the TWIST1 and E2A proteins significantly reduced the number of dots detectable in the TWIST1-positive MDA-MB436 (8.8 ± 1.1 dots/cell to 4.9 ± 1.0 and 1.6 ± 1.5 dots/cell, respectively) and Hs578T (9.8 ± 3.1 dots/cell to 3.1 ± 1.9 and 2.2 ± 1.7 dots/cell, respectively), whereas the number of dots/cell remained < 1 in TWIST1-negative HMEC-hTERT cells (Figure S5). As shown in Figure 3, the TE complex was invariably detected in all TWIST1-positive DCIS, whereas no signal was detected in the control samples, namely, TWIST1-negative DCIS and normal mammary glands. The detection of the TE complex in premalignant lesions is thus compatible with its role in the inhibition of a fail-safe program.
Figure 3.
The heterodimeric complex is detectable in breast premalignant lesions, as assessed in a PLA.
Assessment of the TWIST1-E12 heterodimeric complex in a PLA in TWIST1-positive and TWIST1-negative DCIS [7]. Healthy mammary gland was used as a control. Each single dot corresponds to the detection of a TE complex.
The TWIST1-E12 Heterodimeric Complex, Unlike the Homodimeric TWIST1 Complex, Complements the Loss of the Endogenous TWIST1 Protein in Sustaining Mammary Epithelial Cancer Cell Proliferation
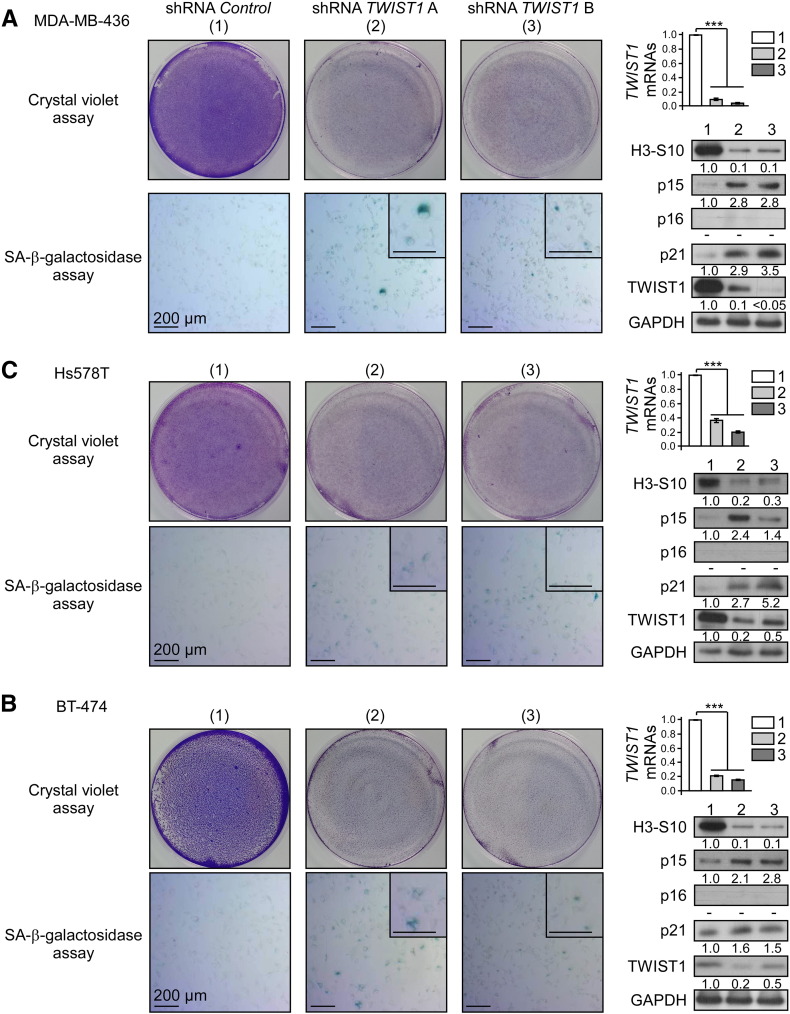
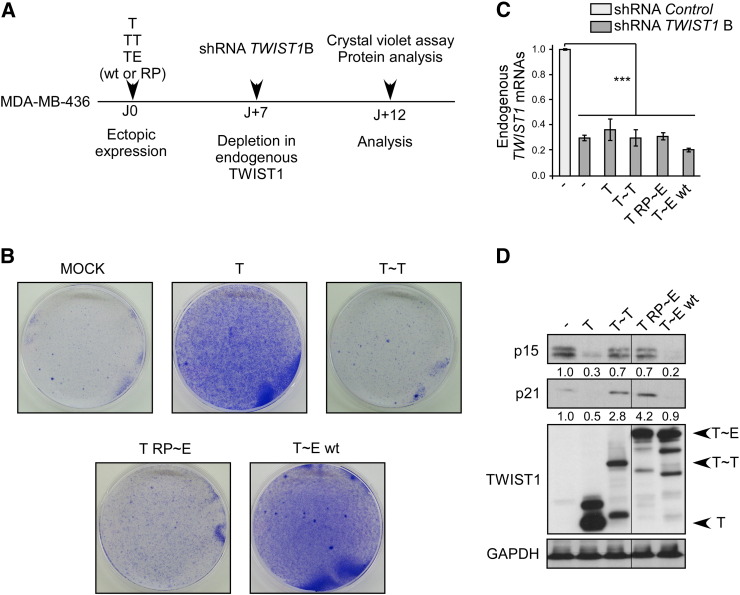
By lowering the activity of oncosuppressive pathways and of the cell cycle control machinery, the TWIST1 protein not only favors the cell neoplastic transformation but also protects cancer cells from latent OIS and/or apoptosis [3], [4], [5], [8], [9], [10]. To assess the contribution of TWIST1 dimers in these secondary activities, we selected a shRNA targeting the 3′-untranslated sequences of the endogenous TWIST1 RNA, which are absent from the heterodimer cDNAs, to inhibit endogenous TWIST1 expression and performed complementation assays with the tethered dimers. We confirmed that this shRNA (shRNA TWIST1 B), similarly to the shRNA used in our previous studies (shRNA TWIST1A [3]), induced a proliferation arrest (as assessed by a crystal violet coloration assay and the amount of phospho-Ser10 histone H3) and triggered a senescence program, as revealed by the detection of a SA-β-galactosidase activity (Figure 4), in all three breast cell lines tested (basal-B/claudin-low MDA-MB-436 and Hs578T cell lines and the luminal B BT-474 cell line). The permanent growth arrest was invariably associated with an accumulation of p15INK4B (p15) and p21CIP1 (p21) cyclin-dependent kinase inhibitors, p16INK4A (p16) remaining undetectable (Figure 4). No trace of cleaved caspases-3 or PARP fragments was detected, excluding a concomitant apoptosis induction (data not shown). To evaluate the contribution of both the homodimeric and heterodimeric TWIST1 complexes, MDA-MB-436 cells were next sequentially infected with TWIST1-expressing constructs (using a monomeric TWIST1 cDNA lacking 3′-untranslated sequence as an internal positive control) and depleted in endogenous TWIST1 through RNA interference. As shown in Figure 5, similarly to the ectopically expressed TWIST1 monomer, the heterodimeric T~E complex avoided cyclin-kinase inhibitor accumulation and sustained cell proliferation. As anticipated, the inactive T~E variant (T RP ~ E) failed to do so. The T~T homodimer also failed to complement the endogenous TWIST1 functions. Similar results were obtained in the two additional Hs578T and BT-474 cell lines (Figure S6). These data unambiguously demonstrated that the T~E heterodimeric complex is necessary to prevent OIS and to maintain the senescence program in a latent state in mammary epithelial cells.
Figure 4.
Depletion in TWIST1 reactivates latent OIS in breast cancer cells.
Breast cancer cells (A, MDA-MB-436; B, Hs578T; C, BT-474) were depleted in TWIST1 through RNA interference (shRNA TWIST1 A or B). (Left panels) Two days postinfection, cells were seeded at low density and stained after 3 days with crystal violet or assessed for their SA-β-galactosidase activity. (Right upper panels) Analysis of the level of the endogenous TWIST1 transcripts by qRT-PCR. Levels were expressed relative to the housekeeping HPRT1 gene transcript and were normalized against uninfected cells. Mean, SD and Student’s t test of one experiment performed in triplicate are shown. (Right lower panels) Analysis of Ser10-histone H3, p15INK4B (p15), p16INK4A (p16), p21CIP1 (p21), and TWIST1 by Western blotting (relative signal quantification with respect to shRNA control infected cells is indicated). “−” means below the threshold.
Figure 5.
A fully active TWIST1-E12 heterodimeric complex is required to complement the endogenous TWIST1 function in sustaining MDA-MB-436 breast cancer cell proliferation.
(A) MDA-MB-436 breast cancer cells were sequentially infected with T, T~T, or T~E (either wild-type or RP mutant) retroviral expression vectors and depleted in endogenous TWIST1 through RNA interference (shRNA TWIST1B). Of note, the shRNA targets 3′-untranslated sequences absent from ectopically expressed TWIST1 constructs and thus specifically turns down the endogenous TWIST1 expression. (B) Cells were seeded at low density 48 hours post–second infection and stained with crystal violet 3 days later. (C) Analysis of the level of expression of endogenous TWIST1 by qRT-PCR. Levels were expressed relative to the housekeeping HPRT1 gene transcript and were normalized against uninfected MDA-MB-436 cells. Mean, SD, and Student’s t test of one experiment performed in triplicate are shown. (D) Analysis of p15INK4B (p15), p21CIP1 (p21), and TWIST1 proteins by Western blot (relative signal quantification in respect to MOCK cells is indicated).
Discussion
The embryonic transcription factor TWIST1 displays pleiotropic functions during carcinogenesis, promoting the neoplastic transformation of cells and tumor initiation, on the one hand, and the metastatic spread of cancer cells through EMT induction, on the other hand [1]. Determining whether these functions rely on the activities on various TWIST1 complexes or whether the oncogenic and prometastatic properties are conferred upon a single complex is essential to further understand the TWIST1 functions as well as to design strategies to inactivate the transcription factor. By using a tethered dimer approach, previously proven to delineate the TWIST1 complex functions in vitro as well as in vivo, we demonstrated that the TWIST1-E12 heterodimer efficiently avoided OIS in mammary epithelial cells. Furthermore, its enforced expression complements the activity of the endogenous TWIST1 protein in keeping cyclin-kinase inhibitors under a threshold compatible with breast cancer cell proliferation. In comparison, the homodimeric complex failed to do so, highlighting the pivotal role of the TWIST1 partner in regulating its oncogenic functions. To support this conclusion, the monomeric TWIST1 protein and the heterodimeric TWIST1 complex were found to efficiently cooperate with RAS in promoting breast carcinogenesis in vivo [7]. Despite the limited number of mice examined, the absence of pathology observed when the homodimeric TWIST1 complex was combined with RAS demonstrated that T~T was less efficient than T~E in promoting tumor initiation in this context (G.W. Hinkal, unpublished results). The tendency of mice, ectopically expressing T~E in mammary epithelial cells, to spontaneously develop breast carcinoma with a higher frequency than their T~T counterparts and control littermates further supports this conclusion.
Several bHLH transcription factors are known to heterodimerize with TWIST1, including the TCF3/E2A splice variants E12 and E47, the TCF4/ITF2, the TCF12/HEB, and the HAND proteins. As the HAND genes were found to be transcriptionally inactive in the examined breast cancer cells (Figure S1), we focused on E proteins, using E12 as a prototype. Considering the degree of homology in their bHLH domains (81.8% identity with TCF3/E2A E47 splice variant, 89.1% identity with TCF4/E2-2, and 92.7% identity with TCF12/HEB), it is likely that all of these complexes display similar oncogenic properties. E2A proteins are the predominant E proteins expressed in the breast cancer cell lines employed in the present study, as assessed by Western blot (Figure S1A). Unfortunately, we cannot exclude that additional TWIST1 bHLH protein partners also contribute to its oncogenic activity. For example, the TWIST1-related TWIST2 protein is expressed in the two basal-B/claudin-low breast cancer cell lines examined (Figure S1A). Although coexpression of both transcription factors at the cellular level remains to be demonstrated, the two proteins are able to heterodimerize [44]. It is worth noting that the heterodimeric TWIST1-E12 complex was additionally shown to drive the prometastatic activity of TWIST1 in prostate cancer cells [22], suggesting that the TWIST1-E complexes are the active forms of TWIST1 with regard to tumorigenesis, promoting both tumor initiation and progression. Using a PLA, we confirmed that the heterodimer is detectable in DCIS before the metastatic cascade is initiated (Figures 3 and S5).
Because the ectopic expression of the tethered dimer T~T did not affect breast cancer cell proliferation, it is possible that the homodimeric complex does not display transdominant negative properties, at least with regard to the functions and cellular models considered in this study. The balance between the two complexes rather spatially and temporally controls the amount of heterodimeric complex and, thereby, the oncogenic and prometastatic properties of TWIST1. To support this hypothesis, ID HLH proteins which tilt the balance in favor of the TWIST1 homodimer were shown to suppress the TWIST1-driven invasive properties and to favor secondary site colonization through the induction of a mesenchymal-to-epithelial transition [45], [46]. Interestingly, in both Caenorhabditis elegans and mouse development, respectively, the HLH8 (the C. elegans TWIST ortholog) and HAND1 homodimeric complexes were found to play a key role, and the heterodimers’ functions were restricted to that of counterbalancing their activity [47], [48]. The relative contribution of bHLH complexes during embryonic development and senescence inhibition over the time course of breast carcinogenesis may thus be inverted. Nonetheless, the ability of the homodimer to modulate downstream targeted genes suggests that it might also display unrelated and yet-uncharacterized protumoral properties.
In numerous tumor models, cancer cells were shown to remain dependent on TWIST1 to sustain proliferation [3], [4], [8], [9], [10], [11]. We now extend this observation to EMT-committed basal-B breast cancer cells (such as MDA-MB-436 and Hs578T), which express a large spectrum of EMT inducers including ZEB1 and TWIST2. ZEB1, similarly to TWIST1, has been demonstrated to downregulate the expression of INK4B and CIP1 and to prevent EGFR-driven OIS in lung cancer cells [49], [50]. TWIST1 and TWIST2 were also shown to similarly downregulate RB and p53 pathways [3]. The observed lack of complementation between the proteins in these breast cancer cells either suggests cell type–specific regulations (e.g., posttranslational regulations) or unveils yet-unidentified TWIST1-dependent properties. The key regulators leading to the specificity of the genetic program driven by TWIST1 complexes remain to be defined. Of note, a tandem E-box separated by a 5-nucleotide spacer was recently shown to be specifically recognized by the TWIST1-E2A complexes (similarly with both E2A splice variants), a tetrameric complex formed and stabilized through the interaction between the α-helixes of the WR domain of TWIST1 proteins [51]. Enforced expression of the T~E heterodimer in HMEC-hTERT cells affected the expression of a limited number of genes (data not shown), suggesting that the tethered dimer strategy avoids titration of bHLH factors. This experimental approach may lead the way to defining the TWIST1-E complex downstream targeted genes and to exploring their contribution to the oncogenic activity of TWIST1.
Funding
This work was supported by the Labellisation program of the Ligue Nationale contre le Cancer and by institutional grants from the LabEX DEVweCAN (ANR-10-LABX-61) and from the LyRIC (Lyon Recherche Intégrée en Cancérologie, Institut National contre le Cancer, INCa-4664). L. J., G. R., and C. B. were recipients of fellowships from the Ligue Nationale contre le Cancer and the Foundation ARC pour la Recherche sur le Cancer.
Author Contributions
Conceptualization and design: L. J., L. P., A. T., S. A.; development of methodology: L. J., G. R., M. D., J. C., A. T., S. A.; material support: D. B. S.; analysis and interpretation of data: L. J., C. B., L. P., A. P., A. T., S. A.; writing: S. A.; revision: G. C. and S. A.
Acknowledgements
We are grateful to the Biological Resource Center of the Centre Léon Bérard for providing the cohort of human breast samples. We are thankful to Prof. Cornelis Murre (University of California, San Diego, USA) for providing the human E12 expression construct, Prof. William C. Hahn for providing the EcoR retroviral expressing construct (Harvard University, USA), Dr. Patrice Lassus (IGMM, Montpellier, France) for the shRNA TWIST1 directed against its 3′UTR (shRNA TWIST1 B), Prof. Jesus Gil (MRC, London, UK) for the ER-RAS construct, Dr. Ivan Mikaelian (CRCL, Lyon, France) for the VIM1-luciferase reporter, and Dr. Didier Negre (INSERMU111/CIRI, Ecole Normale Supérieure de Lyon, France) for providing the pCMVΔR8.91 and phCMVG-VSVG vectors. The authors are thankful to Dr. Isabelle Treilleux, Audrey Pierrot, Sophie Léon, and Nicolas Gado for their technical help and to Dr. Louise Hill and Dr. Brigitte Manship for critical reading of the manuscript.
Footnotes
This work was supported by the Labellisation program of the Ligue Nationale contre le Cancer and institutional grants from the LabEX DEVweCAN (ANR-10-LABX-61) and from the LyRIC (Lyon Recherche Intégrée en Cancérologie, Institut National contre le Cancer, INCa-4664).
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.neo.2016.03.007.
Appendix A. Supplementary data
Supplementary material.
References
- 1.Ansieau S, Morel AP, Hinkal G, Bastid J, Puisieux A. TWISTing an embryonic transcription factor into an oncoprotein. Oncogene. 2010;29:3173–3184. doi: 10.1038/onc.2010.92. [DOI] [PubMed] [Google Scholar]
- 2.Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C, Savagner P, Gitelman I, Richardson A, Weinberg RA. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117:927–939. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 3.Ansieau S, Bastid J, Doreau A, Morel AP, Bouchet BP, Thomas C, Fauvet F, Puisieux I, Doglioni C, Piccinin S. Induction of EMT by twist proteins as a collateral effect of tumor-promoting inactivation of premature senescence. Cancer Cell. 2008;14:79–89. doi: 10.1016/j.ccr.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 4.Burns TF, Dobromilskaya I, Murphy SC, Gajula RP, Thiyagarajan S, Chatley SN, Aziz K, Cho YJ, Tran PT, Rudin CM. Inhibition of TWIST1 leads to activation of oncogene-induced senescence in oncogene-driven non–small cell lung cancer. Mol Cancer Res. 2013;11:329–338. doi: 10.1158/1541-7786.MCR-12-0456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kwok WK, Ling MT, Yuen HF, Wong YC, Wang X. Role of p14ARF in TWIST-mediated senescence in prostate epithelial cells. Carcinogenesis. 2007;28:2467–2475. doi: 10.1093/carcin/bgm185. [DOI] [PubMed] [Google Scholar]
- 6.Maestro R, Dei Tos AP, Hamamori Y, Krasnokutsky S, Sartorelli V, Kedes L, Doglioni C, Beach DH, Hannon GJ. Twist is a potential oncogene that inhibits apoptosis. Genes Dev. 1999;13:2207–2217. doi: 10.1101/gad.13.17.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morel AP, Hinkal GW, Thomas C, Fauvet F, Courtois-Cox S, Wierinckx A, Devouassoux-Shisheboran M, Treilleux I, Tissier A, Gras B. EMT inducers catalyze malignant transformation of mammary epithelial cells and drive tumorigenesis towards claudin-low tumors in transgenic mice. PLoS Genet. 2012;8:e1002723. doi: 10.1371/journal.pgen.1002723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tran PT, Shroff EH, Burns TF, Thiyagarajan S, Das ST, Zabuawala T, Chen J, Cho YJ, Luong R, Tamayo P. Twist1 suppresses senescence programs and thereby accelerates and maintains mutant Kras-induced lung tumorigenesis. PLoS Genet. 2012;8:e1002650. doi: 10.1371/journal.pgen.1002650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Valsesia-Wittmann S, Magdeleine M, Dupasquier S, Garin E, Jallas AC, Combaret V, Krause A, Leissner P, Puisieux A. Oncogenic cooperation between H-Twist and N-Myc overrides failsafe programs in cancer cells. Cancer Cell. 2004;6:625–630. doi: 10.1016/j.ccr.2004.09.033. [DOI] [PubMed] [Google Scholar]
- 10.Beck B, Lapouge G, Rorive S, Drogat B, Desaedelaere K, Delafaille S, Dubois C, Salmon I, Willekens K, Marine JC. Different levels of Twist1 regulate skin tumor initiation, stemness, and progression. Cell Stem Cell. 2015;16:67–79. doi: 10.1016/j.stem.2014.12.002. [DOI] [PubMed] [Google Scholar]
- 11.Piccinin S, Tonin E, Sessa S, Demontis S, Rossi S, Pecciarini L, Zanatta L, Pivetta F, Grizzo A, Sonego M. A "twist box" code of p53 inactivation: twist box: p53 interaction promotes p53 degradation. Cancer Cell. 2012;22:404–415. doi: 10.1016/j.ccr.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 12.Stasinopoulos IA, Mironchik Y, Raman A, Wildes F, Winnard P, Jr., Raman V. HOXA5-twist interaction alters p53 homeostasis in breast cancer cells. J Biol Chem. 2005;280:2294–2299. doi: 10.1074/jbc.M411018200. [DOI] [PubMed] [Google Scholar]
- 13.Massari ME, Murre C. Helix-loop-helix proteins: regulators of transcription in eucaryotic organisms. Mol Cell Biol. 2000;20:429–440. doi: 10.1128/mcb.20.2.429-440.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Connerney J, Andreeva V, Leshem Y, Muentener C, Mercado MA, Spicer DB. Twist1 dimer selection regulates cranial suture patterning and fusion. Dev Dyn. 2006;235:1345–1357. doi: 10.1002/dvdy.20717. [DOI] [PubMed] [Google Scholar]
- 15.Castanon I, Von SS, Kass J, Baylies MK. Dimerization partners determine the activity of the Twist bHLH protein during Drosophila mesoderm development. Development. 2001;128:3145–3159. doi: 10.1242/dev.128.16.3145. [DOI] [PubMed] [Google Scholar]
- 16.Firulli BA, Krawchuk D, Centonze VE, Vargesson N, Virshup DM, Conway SJ, Cserjesi P, Laufer E, Firulli AB. Altered Twist1 and Hand2 dimerization is associated with Saethre-Chotzen syndrome and limb abnormalities. Nat Genet. 2005;37:373–381. doi: 10.1038/ng1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laursen KB, Mielke E, Iannaccone P, Fuchtbauer EM. Mechanism of transcriptional activation by the proto-oncogene Twist1. J Biol Chem. 2007;282:34623–34633. doi: 10.1074/jbc.M707085200. [DOI] [PubMed] [Google Scholar]
- 18.Villavicencio EH, Yoon JW, Frank DJ, Fuchtbauer EM, Walterhouse DO, Iannaccone PM. Cooperative E-box regulation of human GLI1 by TWIST and USF. Genesis. 2002;32:247–258. doi: 10.1002/gene.10078. [DOI] [PubMed] [Google Scholar]
- 19.Connerney J, Andreeva V, Leshem Y, Mercado MA, Dowell K, Yang X, Lindner V, Friesel RE, Spicer DB. Twist1 homodimers enhance FGF responsiveness of the cranial sutures and promote suture closure. Dev Biol. 2008;318:323–334. doi: 10.1016/j.ydbio.2008.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Firulli BA, Redick BA, Conway SJ, Firulli AB. Mutations within helix I of Twist1 result in distinct limb defects and variation of DNA binding affinities. J Biol Chem. 2007;282:27536–27546. doi: 10.1074/jbc.M702613200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Firulli AB, Conway SJ. Phosphoregulation of Twist1 provides a mechanism of cell fate control. Curr Med Chem. 2008;15:2641–2647. doi: 10.2174/092986708785908987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gajula RP, Chettiar ST, Williams RD, Nugent K, Kato Y, Wang H, Malek R, Taparra K, Cades J, Annadanam A. Structure-function studies of the bHLH phosphorylation domain of TWIST1 in prostate cancer cells. Neoplasia. 2015;17:16–31. doi: 10.1016/j.neo.2014.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barradas M, Anderton E, Acosta JC, Li S, Banito A, Rodriguez-Niedenfuhr M, Maertens G, Banck M, Zhou MM, Walsh MJ. Histone demethylase JMJD3 contributes to epigenetic control of INK4a/ARF by oncogenic RAS. Genes Dev. 2009;23:1177–1182. doi: 10.1101/gad.511109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boehm JS, Hession MT, Bulmer SE, Hahn WC. Transformation of human and murine fibroblasts without viral oncoproteins. Mol Cell Biol. 2005;25:6464–6474. doi: 10.1128/MCB.25.15.6464-6474.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Le MS, Fromigue O, Marie PJ. Sp1/Sp3 and the myeloid zinc finger gene MZF1 regulate the human N-cadherin promoter in osteoblasts. Exp Cell Res. 2005;302:129–142. doi: 10.1016/j.yexcr.2004.08.028. [DOI] [PubMed] [Google Scholar]
- 26.Mikaelian I, Malek M, Gadet R, Viallet J, Garcia A, Girard-Gagnepain A, Hesling C, Gillet G, Gonzalo P, Rimokh R. Genetic and pharmacologic inhibition of mTORC1 promotes EMT by a TGF-beta-independent mechanism. Cancer Res. 2013;73:6621–6631. doi: 10.1158/0008-5472.CAN-13-0560. [DOI] [PubMed] [Google Scholar]
- 27.Murre C, Voronova A, Baltimore D. B-cell– and myocyte-specific E2-box-binding factors contain E12/E47-like subunits. Mol Cell Biol. 1991;11:1156–1160. doi: 10.1128/mcb.11.2.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wagner KU, Wall RJ, St-Onge L, Gruss P, Wynshaw-Boris A, Garrett L, Li M, Furth PA, Hennighausen L. Cre-mediated gene deletion in the mammary gland. Nucleic Acids Res. 1997;25:4323–4330. doi: 10.1093/nar/25.21.4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gras B, Jacqueroud L, Wierinckx A, Lamblot C, Fauvet F, Lachuer J, Puisieux A, Ansieau S. Snail family members unequally trigger EMT and thereby differ in their ability to promote the neoplastic transformation of mammary epithelial cells. PLoS One. 2014;9:e92254. doi: 10.1371/journal.pone.0092254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baker BW, Boettiger D, Spooncer E, Norton JD. Efficient retroviral-mediated gene transfer into human B lymphoblastoid cells expressing mouse ecotropic viral receptor. Nucleic Acids Res. 1992;20:5234. doi: 10.1093/nar/20.19.5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zufferey R, Nagy D, Mandel RJ, Naldini L, Trono D. Multiply attenuated lentiviral vector achieves efficient gene delivery in vivo. Nat Biotechnol. 1997;15:871–875. doi: 10.1038/nbt0997-871. [DOI] [PubMed] [Google Scholar]
- 32.Burns JC, Friedmann T, Driever W, Burrascano M, Yee JK. Vesicular stomatitis virus G glycoprotein pseudotyped retroviral vectors: concentration to very high titer and efficient gene transfer into mammalian and nonmammalian cells. Proc Natl Acad Sci U S A. 1993;90:8033–8037. doi: 10.1073/pnas.90.17.8033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dimri GP, Lee X, Basile G, Acosta M, Scott G, Roskelley C, Medrano EE, Linskens M, Rubelj I, Pereira-Smith O. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci U S A. 1995;92:9363–9367. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Demontis S, Rigo C, Piccinin S, Mizzau M, Sonego M, Fabris M, Brancolini C, Maestro R. Twist is substrate for caspase cleavage and proteasome-mediated degradation. Cell Death Differ. 2006;13:335–345. doi: 10.1038/sj.cdd.4401744. [DOI] [PubMed] [Google Scholar]
- 35.Spicer DB, Rhee J, Cheung WL, Lassar AB. Inhibition of myogenic bHLH and MEF2 transcription factors by the bHLH protein Twist. Science. 1996;272:1476–1480. doi: 10.1126/science.272.5267.1476. [DOI] [PubMed] [Google Scholar]
- 36.Bouard C, Terreux R, Hope J, Chemelle JA, Puisieux A, Ansieau S, Payen L. Interhelical loops within the bHLH domain are determinant in maintaining TWIST1-DNA complexes. J Biomol Struct Dyn. 2014;32:226–241. doi: 10.1080/07391102.2012.762722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maia AM, da Silva JH, Mencalha AL, Caffarena ER, Abdelhay E. Computational modeling of the bHLH domain of the transcription factor TWIST1 and R118C, S144R and K145E mutants. BMC Bioinformatics. 2012;13:184. doi: 10.1186/1471-2105-13-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.El Ghouzzi V, Legeai-Mallet L, Benoist-Lasselin C, Lajeunie E, Renier D, Munnich A, Bonaventure J. Mutations in the basic domain and the loop-helix II junction of TWIST abolish DNA binding in Saethre-Chotzen syndrome. FEBS Lett. 2001;492:112–118. doi: 10.1016/s0014-5793(01)02238-4. [DOI] [PubMed] [Google Scholar]
- 39.Brenner AJ, Stampfer MR, Aldaz CM. Increased p16 expression with first senescence arrest in human mammary epithelial cells and extended growth capacity with p16 inactivation. Oncogene. 1998;17:199–205. doi: 10.1038/sj.onc.1201919. [DOI] [PubMed] [Google Scholar]
- 40.Huschtscha LI, Noble JR, Neumann AA, Moy EL, Barry P, Melki JR, Clark SJ, Reddel RR. Loss of p16INK4 expression by methylation is associated with lifespan extension of human mammary epithelial cells. Cancer Res. 1998;58:3508–3512. [PubMed] [Google Scholar]
- 41.Kiyono T, Foster SA, Koop JI, McDougall JK, Galloway DA, Klingelhutz AJ. Both Rb/p16INK4a inactivation and telomerase activity are required to immortalize human epithelial cells. Nature. 1998;396:84–88. doi: 10.1038/23962. [DOI] [PubMed] [Google Scholar]
- 42.Taube JH, Herschkowitz JI, Komurov K, Zhou AY, Gupta S, Yang J, Hartwell K, Onder TT, Gupta PB, Evans KW. Core epithelial-to-mesenchymal transition interactome gene-expression signature is associated with claudin-low and metaplastic breast cancer subtypes. Proc Natl Acad Sci U S A. 2010;107:15449–15454. doi: 10.1073/pnas.1004900107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cardiff RD. Validity of mouse mammary tumour models for human breast cancer: comparative pathology. Microsc Res Tech. 2001;52:224–230. doi: 10.1002/1097-0029(20010115)52:2<224::AID-JEMT1007>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 44.Fu J, Qin L, He T, Qin J, Hong J, Wong J, Liao L, Xu J. The TWIST/Mi2/NuRD protein complex and its essential role in cancer metastasis. Cell Res. 2011;21:275–289. doi: 10.1038/cr.2010.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rahme GJ, Israel MA. Id4 suppresses MMP2-mediated invasion of glioblastoma-derived cells by direct inactivation of Twist1 function. Oncogene. 2015;34:53–62. doi: 10.1038/onc.2013.531. [DOI] [PubMed] [Google Scholar]
- 46.Stankic M, Pavlovic S, Chin Y, Brogi E, Padua D, Norton L, Massague J, Benezra R. TGF-beta-Id1 signaling opposes Twist1 and promotes metastatic colonization via a mesenchymal-to-epithelial transition. Cell Rep. 2013;5:1228–1242. doi: 10.1016/j.celrep.2013.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hu D, Scott IC, Snider F, Geary-Joo C, Zhao X, Simmons DG, Cross JC. The basic helix-loop-helix transcription factor Hand1 regulates mouse development as a homodimer. Dev Biol. 2013;382:470–481. doi: 10.1016/j.ydbio.2013.07.025. [DOI] [PubMed] [Google Scholar]
- 48.Philogene MC, Small SG, Wang P, Corsi AK. Distinct Caenorhabditis elegans HLH-8/twist-containing dimers function in the mesoderm. Dev Dyn. 2012;241:481–492. doi: 10.1002/dvdy.23734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu Y, El-Naggar S, Darling DS, Higashi Y, Dean DC. Zeb1 links epithelial-mesenchymal transition and cellular senescence. Development. 2008;135:579–588. doi: 10.1242/dev.007047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ohashi S, Natsuizaka M, Wong GS, Michaylira CZ, Grugan KD, Stairs DB, Kalabis J, Vega ME, Kalman RA, Nakagawa M. Epidermal growth factor receptor and mutant p53 expand an esophageal cellular subpopulation capable of epithelial-to-mesenchymal transition through ZEB transcription factors. Cancer Res. 2010;70:4174–4184. doi: 10.1158/0008-5472.CAN-09-4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chang AT, Liu Y, Ayyanathan K, Benner C, Jiang Y, Prokop JW, Paz H, Wang D, Li HR, Fu XD. An evolutionarily conserved DNA architecture determines target specificity of the TWIST family bHLH transcription factors. Genes Dev. 2015;29:603–616. doi: 10.1101/gad.242842.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material.