Although genetics and genomics play an increasingly large role in the practice of medicine, the clinical care of patients suffering from cardiovascular disease or stroke has not been significantly affected. This is despite the tremendous strides being made to understand the genetic basis of both rare and common cardiovascular and stroke disorders through techniques such as genome-wide association studies (GWASs) and next-generation sequencing studies. Much of this knowledge remains to be translated to the clinic and must be subjected to clinical trials to ensure patient safety and a meaningful impact on clinical outcomes. However, even if this knowledge were to be successfully implemented into clinical practice, a potential barrier to widespread adoption is a lack of familiarity with basic concepts of genetics and genomics. Another concern is the possibility of the emergence of a significant gap in clinical care provided by practitioners who are informed about the clinical use of genetics and genomics knowledge and those who are not. Thus, there is a critical need to foster genetics/genomics literacy among all involved in the care of cardiovascular and stroke patients because it can be expected that these topics will transform the way medicine is practiced.
The purpose of this document is to serve as a resource for practitioners in cardiovascular and stroke medicine on the application of genetics and genomics to patient care. Although not exhaustive, it contains an overview of the field written specifically to be accessible and relevant to practitioners. It also refers to additional educational materials available in the literature, in textbooks, and on the Internet. (Because this article is intended to be primarily educational in nature, rather than providing a review of the literature, citations are limited to a small number of research articles and reviews of exceptional interest.) It recommends a core knowledge base with which practitioners and especially trainees in cardiovascular and stroke clinical care should be familiar. Finally, it is intended to be a companion to the American Heart Association's Council on Functional Genomics and Translational Biology Online Educational Series, in which online modules covering the topics outlined in this document are discussed in greater depth and are accessible to members of the cardiovascular and stroke clinical communities.
Primer on Genetics and Genomics
Basics of Molecular Biology
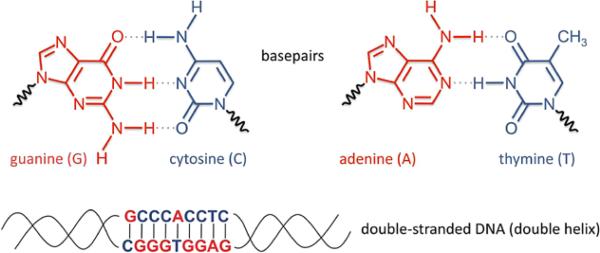
Deoxyribonucleic acid (DNA) is a molecule with 2 strands that are wrapped around each other in a helical formation, hence its description as a double helix (Figure 1). The outer portion of the helix contains the sugar and phosphate backbone; the inner portion contains the coding bases: adenine (A), cytosine (C), guanine (G), and thymine (T). The genetic information of an organism is determined by the order of the sequence of the bases; with 4 bases available, the number of potential sequences is almost infinite. The versatility of DNA results from the obligatory pairing of bases in the 2 strands, forming base pairs. An adenine in 1 strand is always matched up with a thymine in the other strand, and cytosine is always paired with guanine. Thus, the 2 strands contain redundant information, and each can serve as a template on which a new complementary strand can be synthesized. This allows easy duplication of the DNA so that, when a cell divides into 2 cells, each descendant cell receives the same genetic information as the original cell.
Figure 1.
The structure of DNA. Each DNA strand has a sugar-phosphate backbone (not shown in detail) with a sequence of bases that come in 4 versions: adenine (A), cytosine (C), guanine (G), and thymine (T). Two DNA strands can combine to form a double helix, the stable form of DNA found in chromosomes. Holding the strands together are base pairs: Guanine on 1 strand binds to cytosine on the other strand, and adenine on 1 strand binds to thymine on the other strand. Thus, the 2 strands are complementary and contain redundant information.
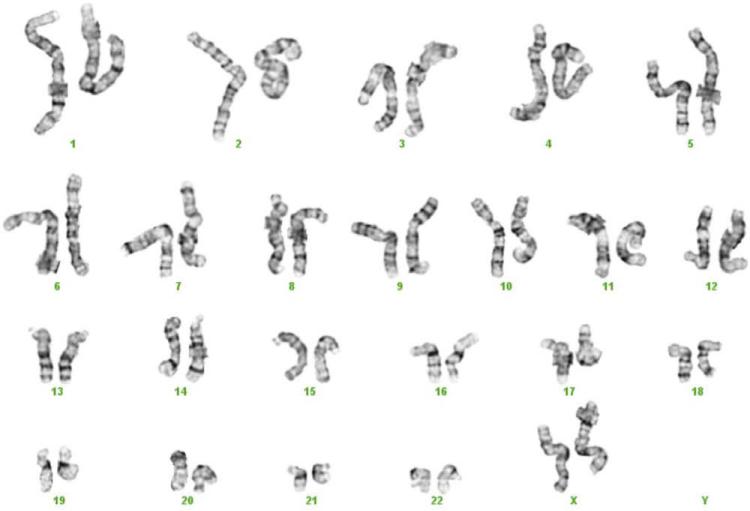
The DNA of an organism is organized into extremely long strands that are packaged by a large complex of supporting proteins into chromosomes. Humans have 23 pairs of chromosomes, including the pair that determines sex, which in women comprises 2 X chromosomes and in men 1 X and 1 Y chromosome (Figure 2). For each chromosome pair, 1 chromosome was inherited from the mother and 1 from the father. The full set of chromosomes is collectively called the genome. The human genome is largely contained within the nucleus of each cell, where it is separated from the rest of the cell functions. However, a small amount of DNA exists outside the nucleus in the mitochondria and is considered to be part of the human genome.
Figure 2.
The human genome. As seen in this karyotypic spread, the typical human cell has 46 chromosomes with 22 pairs of autosomes (numbered 1–22) and a pair of sex chromosomes, either XX or XY.
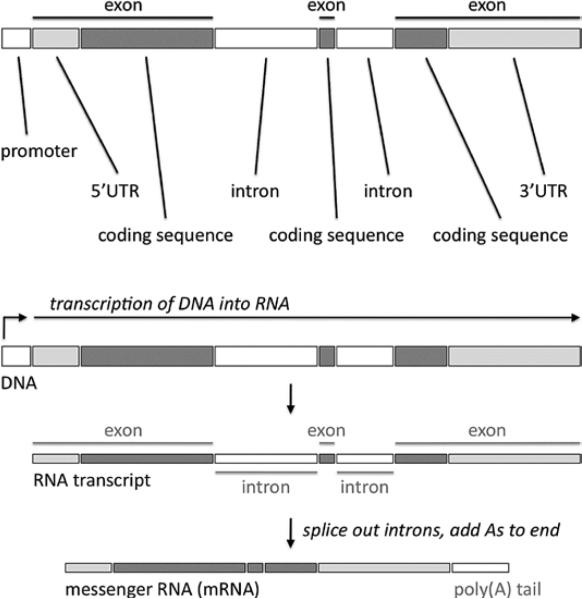
In general, the genome is characterized by vast regions of noncoding DNA sequence punctuated by small areas of coding DNA, also called genes, that contain the instructions needed by cells to perform their functions. Coding DNA is transcribed into a single-stranded molecule called ribonucleic acid (RNA) by a collection of specialized enzymes (Figure 3). RNA is structurally similar to a DNA strand and contains 4 types of bases, including adenine, cytosine, and guanine (in RNA, uracil [U] is substituted for thymine [T] in DNA). The transcription enzymes have “proofreading” functions that ensure that the sequence of the RNA molecule faithfully matches the sequence of the DNA template from which it was synthesized. RNA is more flexible and mobile than DNA and is transported out of the nucleus of the cell into the outer compartment, the cytoplasm. Thus, RNA is the mechanism by which genetic information is expressed and relayed from the central repository (DNA) to the rest of the cell, where it directs cellular functions.
Figure 3.
The structure of a gene and its transcription into an mRNA. Genes have exons and introns. The exons are coding sequences (with the exception of 2 noncoding elements at the beginning and end of the gene called 5′ untranslated region [UTR] and 3′ UTR, the significance which is beyond the scope of this document). The introns are noncoding sequences. Transcription enzymes, starting at the noncoding promoter, produce an RNA transcript from the DNA gene. This RNA transcript is processed by splicing factors to remove the introns (and a “poly(A) tail” is added, the significance of which is beyond the scope of this document).
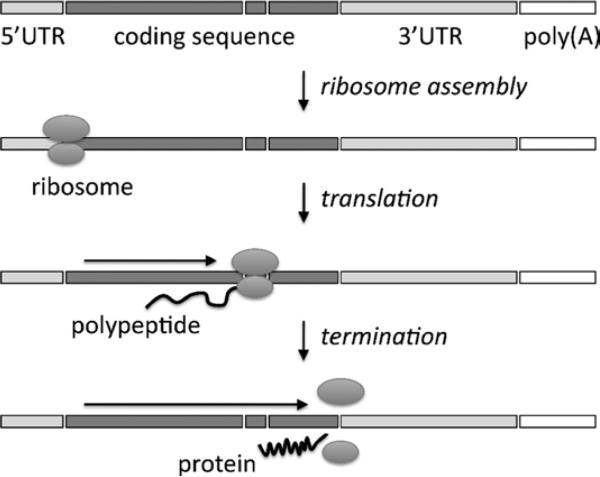
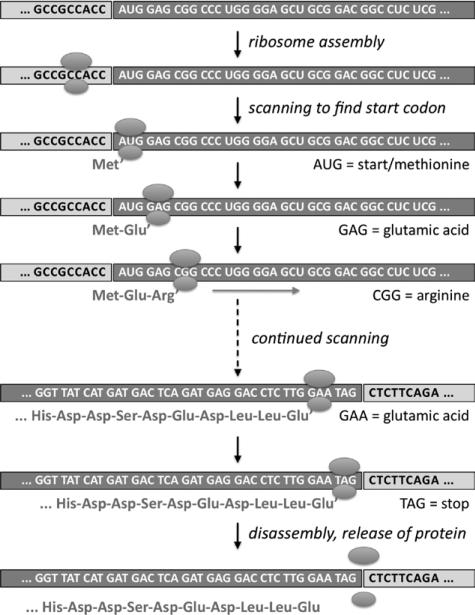
Although some RNAs have specialized functions, for example, serving as structural components of certain parts of the cell, most RNAs take the form of messenger RNAs (mRNAs), which are translated by ribosomes into proteins (Figures 4 and 5). The ribosome reads from the beginning of the mRNA and uses it as a coding template to build proteins, with each nonoverlapping set of 3 consecutive bases (codons) serving to specify a particular amino acid. With 4 available bases, there are 64 possible codon combinations; with some redundancy, these codons are translated into any of 20 different amino acids or into a stop signal. The RNA sequence is converted into an amino acid sequence until a stop signal is reached that prompts the ribosome to finish and release the protein. The protein is then processed by the cell and deployed to its purpose (as an enzyme, secreted hormone, etc).
Figure 4.
Translation of an mRNA into a protein. After splicing, the coding sequence lies in 1 contiguous block, and a ribosome scans through the coding sequence to produce the protein. UTR indicates untranslated region.
Figure 5.
From RNA codons to amino acids. Groups of 3 RNA bases, or codons, are translated by the ribosome into amino acids. AUG (which encodes the amino acid methionine) is the standard start codon. As it scans the mRNA, the ribosome adds amino acids to an increasingly long chain. When it reaches a stop codon, the completed protein is released.
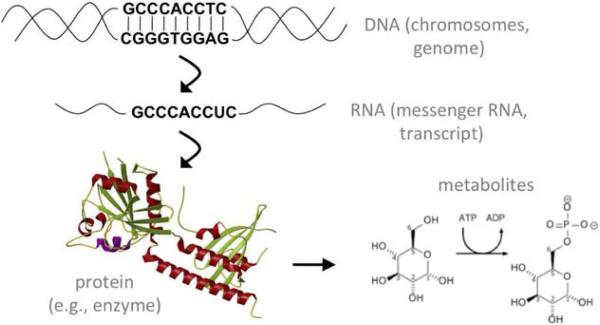
This organized progression from DNA to transcribed RNA to translated protein is known as the central dogma of molecular biology (Figure 6), and although there are exceptions to this sequence of events, the central dogma explains the vast majority of cellular processes. In humans, these processes combine with environmental influences to determine each person's individual characteristics, susceptibility to diseases, and responses to medications. New technology is now available to study the cellular processes at any step of the central dogma. When an investigation occurs at the level of DNA, it is called genetics if it deals primarily with 1 gene. The term genomics is used if it deals with the interactions among multiple genes or all of the genes in the genome. When at the level of mRNAs and proteins, the terms transcriptomics and proteomics, respectively, are used. Processed proteins or other products of enzymatic reactions are called metabolites, the study of which is called metabolomics. Together, the effects of DNA, RNA, proteins, and metabolites, when combined with environmental factors, result in phenotypes. Phenotypes can refer to phenomena occurring within a single cell or in an entire organism. Phenotypes include individual characteristics (eg, hair color), clinical traits (eg, blood cholesterol levels), or diseases (eg, myocardial infarction). In this document, we focus primarily on disease phenotypes.
Figure 6.
The central dogma. With a few exceptions, genetic information flows in 1 direction.
Basic Characteristics of the Genome
The human genome is roughly 6 billion DNA base pairs in size, spanning the 23 chromosome pairs, and represents virtually the entire list of coded instructions needed to create a human being. There are an estimated 20 000 genes in the human genome, most of which encode proteins or components of proteins. What makes each person unique is a large number of DNA variants distributed throughout the genome. Some people have particular DNA variants that can predispose them to cardiovascular disease or stroke. These variants often require the presence of environmental factors (eg, smoking and obesity) to trigger disease. Less commonly, certain variants have such a strong effect that they can cause disease outright. Other variants may determine how well or poorly patients respond to particular medications.
One reason that some people are more susceptible to getting a disease than other people or respond differently to medications is that their DNA variants affect the function of genes. There are rare variants that have a large effect on the function of a gene by either significantly increasing or decreasing the activity of the gene; these are the kind of variants that cause disease in many members of a single family and are known as mutations. Classic examples include hypertrophic cardiomyopathy and Marfan syndrome. There are common variants (>1% of the general population) that have a small effect on the function of a gene. These variants do not change gene activity enough to cause disease by themselves but instead need to be combined with other variants in other genes or with environmental factors for disease to occur. This is the case with most cardiovascular disorders for which there are many contributing factors, for example, hypercholesterolemia, myocardial infarction, and ischemic stroke.
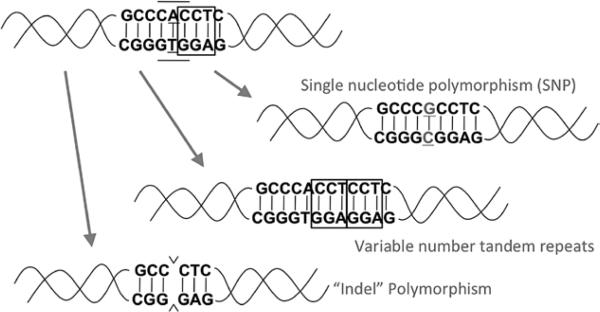
All of these differences at the DNA level are called polymorphisms, of which there are several types (Figure 7). Single-nucleotide polymorphisms (SNPs) occur when a single base in the DNA differs from the usual base at that position. Variable-number tandem repeats are polymorphisms in which the number of repeats of a short DNA sequence at a location varies from person to person; when the length of the repeat ranges from 2 to 6 base pairs, other names for this type of polymorphism include microsatellites, single-sequence repeats, and short tandem repeats. A copy number variation (CNV) is a polymorphism in which the number of repeats of a large DNA sequence (>1000 base pairs) at a location varies from person to person, with the number typically ranging from zero copies (deletion of the sequence) up to a few copies. An indel (short for insertion-deletion) is a polymorphism in which a DNA sequence of any size is either present or absent at a location, varying from person to person. An indel can be characterized as either a variable-number tandem repeat or a CNV, depending on the size of the involved sequence.
Figure 7.
Three types of polymorphisms. The top left DNA molecule is the reference sequence; each of the other 3 DNA molecules demonstrates 1 type of DNA variant. “Indel” indicates insertion-deletion; SNP, single-nucleotide polymorphism.
SNPs are the most common and best characterized of the polymorphisms, with tens of millions SNPs now identified across the human genome (they are cataloged in a database called dbSNP, http://www.ncbi.nlm.nih.gov/SNP/). On average, they occur every few hundred base pairs. SNPs are a large contributor to the genomic variation that distinguishes each individual person. Much of genomics research has focused on understanding how SNPs are distributed in different populations, how they affect gene function, and how they contribute to disease. Most GWASs (see below) have largely focused on discovering associations between SNPs (rather than variable-number tandem repeats or CNVs) and diseases.
Coding and Noncoding DNA Variants
As mentioned, the genome can be divided into coding and noncoding DNA. Coding DNA, which makes up just 1% of the genome, contains the gene sequences that are transcribed into mRNAs and then translated into proteins. The coding DNA of a single gene is usually not present as a single continuous block but rather is split into a number of distinct blocks called exons that are separated by stretches of noncoding DNA called introns (Figure 3). When a gene is transcribed, it begins with a change in the balance of regulatory proteins called transcription factors that are associated with an upstream region of noncoding DNA called the promoter. Specific transcription factors can either enhance or repress this process, so that transcription is initiated when the balance of transcriptional enhancers outweighs translational repressors. Immediately downstream of the promoter is the first exon of the gene, followed by an intron, followed by the next exon, followed by another intron, etc. The entire region of DNA (including both the exons and introns of the gene but not the promoter) is transcribed into RNA.
After the full RNA is transcribed, it is processed in the nucleus with the help of proteins called splicing factors. Introns are excised and the ends of the exons are joined, thereby creating an mRNA with all of the exons now forming a continuous sequence (Figure 3). In some cases, alternative splicing occurs. Depending on circumstances, a particular exon may be either included or excluded from the final mRNA, or a choice may be made between 2 adjacent exons, resulting in either 1 or the other exon being included in the final mRNA. Alternative splicing can thereby result in the creation of a heterogeneous pool of mRNAs transcribed from a single gene, resulting in a heterogeneous mix of slightly different proteins, called isoforms. In different situations, the pool of mRNAs from a gene may be dominated by some splice forms versus other splice forms, allowing an extra level of regulation of gene function.
When SNPs fall in the midst of coding DNA, a variety of consequences for gene function can occur, despite a change of just 1 DNA base. Nonsynonymous variants are SNPs that alter a codon in a way that changes the amino acid that is encoded by the codon. One type of nonsynonymous variant, called a missense variant, results in a single amino acid being changed in the protein product that is translated from the gene. This is because of the codon being switched from 1 type of amino acid to another. For example, a change in a codon from AAG to AAC would result in the substitution of the amino acid asparagine for the amino acid lysine in the protein. Another type of nonsynonymous variant, called a nonsense variant, results in the protein being prematurely truncated at that position as a result of the codon being changed to a stop signal. An example is a change in a codon from AAG, which encodes the amino acid lysine, to UAG, a stop codon. Typically, although not always, a nonsense variant will have greater consequences for gene function than a missense variant. Many SNPs are synonymous variants, which change a DNA base without changing the amino acid specified by the codon. This can occur as a result of the redundancy of the genetic code. Because there are 64 possible codons that encode only 20 different amino acids, most of the amino acids are encoded by multiple codons that are very similar; for example, they may vary only in the third base of the codon. For example, the amino acid lysine is encoded by the codons AAA and AAG; the amino acid glycine is encoded by the codons GGA, GGC, GGG, and GGT. Thus, a single base change may not ultimately affect the protein.
Although most synonymous variants are not thought to affect gene function in any way, there can be exceptions. For example, if the variant occurs at the very beginning or end of any exon, it can potentially interfere with splicing of that exon and the adjacent intron. Splice-site variants can affect alternative splicing of exons or, in some scenarios, can cause introns to be inappropriately included in mRNAs, with deleterious consequences for the translated protein products.
Small indels that cause the insertion or deletions of a few base pairs of coding DNA can result in the disruption of gene function. Frameshift variants can result in the frame of an mRNA being placed out of register so that the ribosome is no longer reading the appropriate codons. For example, because codons are read as groups of 3 bases, deletion (or insertion) of 1 base would result in each of the subsequent codons being misread by the ribosome. The same would occur with deletion (or insertion) of 2 bases. This usually, but not always, results in a premature stop signal occurring soon after the site of the variant, causing a dysfunctional truncated protein to be made. (Thus, functionally, frameshift variants and nonsense variants are similar.) The deletion (or insertion) of multiples of 3 bases would have different effects and would therefore not constitute frameshift variants. In this case, ≥1 amino acids would be missing from (or extra amino acids would be present in) the final protein, but because the subsequent codons would still be in the correct frame, the remainder of the protein would be normally translated and therefore would be intact. The missing (or extra) amino acids may or may not affect the activity of the protein, depending on where they fall in the protein.
Noncoding DNA variants occur either within a gene (promoter, introns) or outside a gene. Although noncoding DNA variants do not affect codons, they can nevertheless affect the final protein products of genes. Variants within introns can affect the splicing of nearby exons, thereby affecting which protein isoforms are produced. Variants within promoters can directly affect gene transcription, resulting in higher or lower levels of mRNAs being produced, which in turn results in higher or lower levels of protein being produced. Even when far away from genes, variants can affect their transcription. Noncoding DNA elements called transcriptional enhancers and transcriptional repressors can affect the expression of genes from large distances, as many as thousands of bases (kilobases) or even millions of bases (megabases), through 3-dimensional interactions between different regions of a chromosome, that is, folding of a chromosome resulting in 2 remote sites being brought into proximity, with transcription factors bridging between the 2 sites. Variants in these transcriptional elements can thereby modulate gene expression.
Finally, various classes of RNAs exist that are transcribed from noncoding DNA and therefore do not code for proteins but can nevertheless affect the functions of other genes. MicroRNAs (miRNAs) are small noncoding RNAs ≈22 nucleotides in size that match complementary sequences within mRNA molecules. By forming base pairs with an mRNA sequence, an miRNA can regulate the amount of protein produced by the mRNA. This can occur by blocking of translation of the mRNA, which directly reduces the yield of protein, or by inducing the degradation of the mRNA, which indirectly reduces the yield of protein. In some cases, an miRNA may enhance transcription of a gene or translation of an mRNA, thereby increasing the level of the protein product. Many miRNAs are contained in the introns of coding genes, with the others lying in regions between genes.
Long noncoding RNAs (lncRNAs) are transcripts longer than 200 nucleotides. They can play a number of different roles in regulating gene expression and protein production. Some lncRNAs can bind to and modulate the activity of specific transcription factors, thereby affecting the transcription of certain genes. Other lncRNAs regulate the basic enzymes involved in the transcription of all genes, thereby causing global changes in the cell, or act to silence genes in large portions of or even entire chromosomes. Yet other lncRNAs are involved in the regulation of translation of mRNAs, often via the formation of base pairs with a complementary sequence in an mRNA, similar to the mechanism by which miRNAs act, or in the regulation of mRNA splicing. Thus, for both miRNAs and lncRNAs, noncoding DNA variants that fall within the sequences encoding these RNAs can potentially have important functional consequences.
Genotyping and Sequencing to Determine the Identity of DNA Variants
In most cases, each person has 2 copies of each DNA sequence, called alleles, because of the pairing of chromosomes; the exceptions are DNA sequences on the X or Y chromosome in men, who have only 1 of each chromosome. A person's genotype at the site of a polymorphism is the identity of the DNA sequence for each of the 2 alleles on the paired chromosomes. For an SNP, a genotype is typically designated as 2 letters corresponding to the identities of the bases at the SNP position (eg, AA versus AG versus GG). For a variable-number tandem repeat or CNV, a genotype is typically designated as 2 numbers corresponding to the copy numbers of the 2 alleles. A haplotype is a combination of SNPs at multiple locations on a chromosome, often within kilobases of each other, that are usually transmitted as a group from parents to offspring.
There are 2 methods to determine the genotypes of a polymorphism. First, there are assays that allow the direct genotyping of a polymorphism. Although a description of the technical details of these assays is beyond the scope of this document, the assays have the advantages of being relatively inexpensive (compared with sequencing, as described below) and can be combined into a high-throughput format, usually in a genotyping array or chip format, that can ascertain the genotypes of up to millions of polymorphisms in a person's genomic DNA sample in a single experiment. This is the technique used by commercial DNA testing services. Such services extract genomic DNA from the cells in a person's saliva sample and then apply the DNA to a genotyping chip to determine the genotypes of a large number of SNPs and CNVs distributed across the genome. One disadvantage of this methodology is that it can ascertain the genotypes of only predetermined polymorphisms. It cannot interrogate any other DNA bases in the genome and, importantly, cannot discover new polymorphisms.
The second method to determine the genotypes of polymorphisms entails DNA sequencing. DNA sequencing techniques date back to the 1970s, when it could take days to determine the identity of the bases in a sequence of a few dozen DNA bases. In the 1990s, improved DNA sequencing techniques were developed that allowed an international consortium to sequence the entirety of the human genome, the Human Genome Project, in ≈12 years at a cost of US $3 billion.1 The 2000s saw the invention of next-generation sequencing techniques, which enormously decreased the time and costs required to sequence increasingly large stretches of DNA. In 2009, the first reports of whole-exome sequencing of DNA samples from patients were published.2,3 The exome comprises the entirety of the coding portions of the genome, that is, all of the exons of the ≈20 000 genes, which together constitute ≈1% of the genome. Shortly thereafter, whole-genome sequencing of DNA samples from patients was reported.4 As expected, whole-exome sequencing remains cheaper than whole-genome sequencing, but further advances in next-generation sequencing technology have made it possible to sequence a patient's genome in a single day for a few thousand US dollars.
Because known polymorphisms affect only a small proportion of the DNA bases in the genome, it remains more expensive to sequence the entire genome than to genotype polymorphisms; thus, direct genotyping assays remain in common use. However, a significant advantage of whole-exome and whole-genome sequencing is the ability to discover new DNA variants, especially rare DNA variants that are unique to particular individuals or families. As sequencing technologies become even cheaper, it can be expected that whole-genome sequencing will eventually supplant direct genotyping.
Monogenic Cardiovascular and Stroke Disorders
Rare DNA Variants and Monogenic (Mendelian) Disorders
Classic genetics focused largely on monogenic, or mendelian, diseases, that is, those that follow the Mendel laws of inheritance. In these diseases, a DNA variant or variants in a single gene are responsible for causing disease. Perforce, these variants must have large effects on gene function because they are able to singlehandedly induce disease. Typically, these variants are quite rare in a given population because they are unique to a patient or a family and thus are called mutations. The reason for the rarity of these mutations is natural selection: If the mutations result in disorders that decrease health and reproductive fitness, they will eventually be eliminated from a population. In exceptional cases, mutations may cause both beneficial and detrimental consequences, resulting in opposing forces of positive selection and negative selection that may cause the mutations to be preserved at nonrare frequencies in a population. For example, the HbS mutation in the HBB gene (which produces the β subunit of hemoglobin) causes sickle cell disease when present in both alleles, a detrimental consequence, but protects against malaria when present in 1 allele, a beneficial consequence, ensuring that the mutation persists in populations in areas of the world where malaria is endemic.
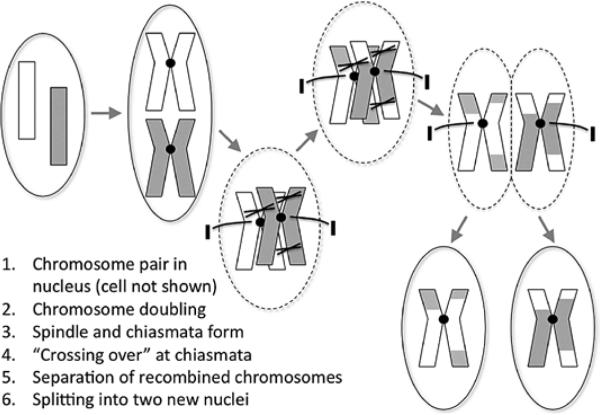
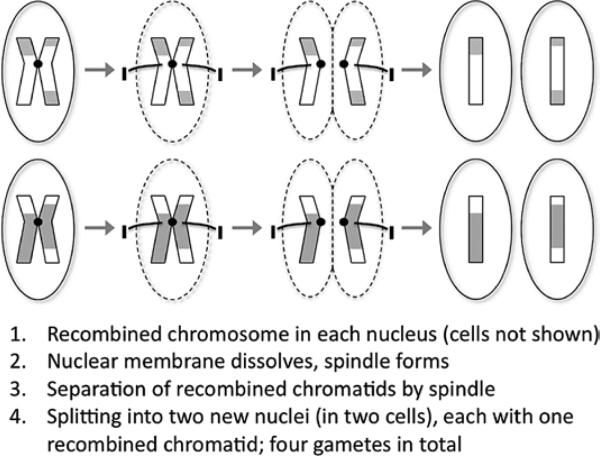
Genes are passed from parents to offspring via the process of meiosis by which gametes, the egg cells in the mother and the sperm cells in the father, are generated. Ordinarily, each cell has 23 pairs of chromosomes; the gametes have 23 unpaired chromosomes. In meiosis, the 23 pairs are split so that each gamete receives 1 chromosome from each pair (Figures 8 and 9). Two gametes (egg and sperm) ultimately join into a single cell, the zygote, which has the full complement of 23 chromosome pairs restored. If all goes well, the zygote gives rise to a live offspring.
Figure 8.
Meiosis, part 1. Before the first cell division, meiotic recombination (crossing over) between a chromosome pair occurs.
Figure 9.
Meiosis, part 2. The second cell division yields gametes, which have only half of the complete genome (unpaired chromosomes). Two gametes subsequently fuse (fertilization) to create a zygote that has a complete genome and can give rise to an organism.
The Mendel Laws: Segregation and Independent Assortment
Both of the Mendel laws pertain directly to the process of meiosis. The first Mendel law, the law of segregation, states that each parent passes a randomly selected allele for a given DNA base to an offspring. Stated another way, the chance of a gamete receiving 1 or the other chromosome of a pair is 50%. Thus, neither chromosome of a pair, and, by extension, any particular allele of a polymorphism, is favored during the process of meiosis. The second Mendel law, the law of independent assortment, states that 2 separate genes (or 2 alleles of 2 polymorphisms) are passed independently of one another from a parent to an offspring. This can be rationalized as being the result of chromosomes of different pairs being distributed into gametes independently; which specific chromosome of 1 pair ends up in a gamete is entirely unconnected to which chromosome of another pair ends up in the same gamete.
The second law can be violated because, if 2 genes or 2 polymorphisms are on the same chromosome, they should be passed together via the single chromosome from a parent to offspring 100% of the time; there is no longer independent assortment. However, this violation is somewhat attenuated because, during the process of meiosis, crossing over, also called recombination, can occur between each pair of chromosomes wherein pieces of the chromosomes are swapped before they are separated into the gametes (Figure 6). The consequence of recombination is that the alleles of 2 polymorphisms that were previously linked together in a haplotype on a single chromosome may end up on the 2 different chromosomes of a pair and thus end up in different gametes. This breaking of the haplotype is more likely to occur if the polymorphisms lie far apart on a chromosome than if they lie close together on a chromosome (recombination between the 2 polymorphisms will occur more frequently in the former scenario than the latter scenario). This phenomenon has important implications for both linkage studies and GWASs (see below).
Mendelian Transmission of Disease
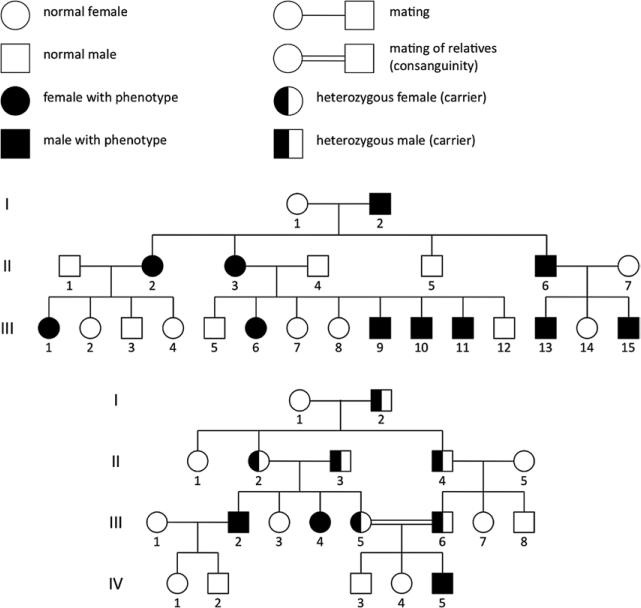
In classic genetics, there are 5 major modes of inheritance: autosomal dominant, autosomal recessive, X-linked dominant, X-linked recessive, and maternal (or extranuclear). The mode of inheritance of a given monogenic disorder depends on the nature of the mutation and the chromosome where the mutation is located. Family trees called pedigrees can be useful in determining the mode of inheritance of a disease (Figures 10 and 11).
Figure 10.
Pedigrees of families with monogenic disorders, part 1. Symbols commonly used in pedigrees are shown at the top. The 2 pedigrees show a set of parents and successive generations of offspring. In order, the modes of inheritance are autosomal dominant and autosomal recessive.
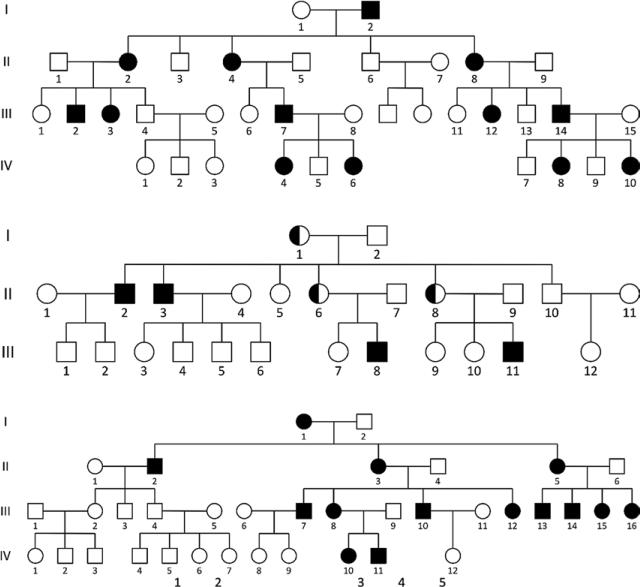
Figure 11.
Pedigrees of families with monogenic disorders, part 2. In order, the modes of inheritance are X-linked dominant, X-linked recessive, and maternal (extranuclear) inheritance.
Some mutations have such a large effect on gene function that having the mutation in just 1 copy of the gene is sufficient to cause disease, even if the other copy of the gene is normal, a condition called heterozygosity. In this scenario, an offspring needs to inherit only 1 mutation, from either the mother or the father, to manifest the disease. These mutations are considered to be dominant. Other mutations will have a large effect on gene function only if both copies of the gene are mutated, which can occur if the same mutation is present in both copies, called homozygosity. When 2 different mutations are present in the 2 copies, it is called compound heterozygosity. In either case, 1 mutation is inherited from the mother and the other from the father. These mutations are considered to be recessive and will not cause significant or clinically detectable disease if there is a normal copy of the gene present. Other mutations are considered to be codominant or additive, although in most cases, these types of mutations are relevant to a quantitative trait (eg, blood cholesterol level) rather than a disease. With these mutations, there is an increasing effect as the number of mutant gene copies increases. Two mutations have a greater effect than 1 mutation, which in turn has an effect not observed in individuals with no mutations.
Autosomal Dominant and Autosomal Recessive Inheritance
All chromosomes other than the X and Y chromosomes are called autosomes and are present in pairs. If a parent has a dominant mutation in a gene on an autosome, that parent should have the disease and moreover by the first Mendel law has a 50% chance of passing the mutation to an offspring, who in turn will have the disease. This is autosomal dominant inheritance (Figure 10). A classic example of an autosomal dominant disorder is hypertrophic cardiomyopathy (see below for a more detailed description of the genetics of this disorder). If a parent has a recessive mutation in a gene on an autosome and the other parent does not, there is a 50% chance the first parent will pass the mutation to an offspring, but that person will not have the disease because he or she will inherit a normal gene copy from the other parent. Rather, the offspring will be a carrier of the mutation, with the possibility of passing on the mutation to future generations. If both parents have recessive mutations in a gene on an autosome, then it is possible for an offspring to inherit 2 mutated gene copies and thereby develop the disease. This is autosomal recessive inheritance (Figure 10). A distinctive feature of recessive inheritance is that a disease can skip generations of people and then re-emerge in a later generation if one mutation carrier should happen to mate with another mutation carrier. Classic examples of autosomal recessive disorders include cystic fibrosis and sickle cell anemia.
X-Linked Inheritance
More complicated patterns emerge if a disease mutation is present in a gene on the X chromosome. If a mutation is dominant, then a mother with the mutation (who herself should have the disease) has a 50% chance of passing the mutation to an offspring, who in turn will have the disease. In contrast, a father with the mutation (who himself should have the disease) has a 50% chance of passing the mutation to a daughter because he passes an X chromosome to her, but he cannot transmit the mutation to a son because he passes a Y chromosome to him. Thus, the inheritance of disease depends on sex. This is X-linked dominant inheritance (Figure 11). An example of an X-linked dominant disorder is Rett syndrome. If a mutation is recessive, then a mother with the mutation (who should be a healthy carrier) has a 50% chance of passing the mutation to an offspring. A daughter who inherits the mutation will be a carrier, whereas a son who inherits the mutation will have the disease because he has only a single X chromosome and has no normal gene copy to counteract the mutant gene copy. A father with the mutation (who should have the disease) has a 50% chance of passing the mutation to a daughter, who will be a carrier, but cannot transmit the mutation to a son. The only way a daughter can have the disease is if she inherits mutant gene copies from both parents. This is X-linked recessive inheritance (Figure 11). Such diseases are much more likely to affect men than women. Classic examples of X-linked recessive disorders include red-green color blindness and hemophilia.
Maternal Inheritance
In the fifth mode of inheritance, the disease mutation lies not on a chromosome in the nucleus but rather in mitochondrial DNA outside the nucleus. Mitochondria are inherited exclusively from an offspring's mother; because of this phenomenon, the mutation and thus the disease can be passed only from a mother to her offspring. This is maternal inheritance, also known as extranuclear inheritance (Figure 11). Representative disorders include various mitochondrial myopathies.
An important initial step in studying a monogenic disease in a family is to determine the mode of inheritance at work. Although the patterns described above may seem straightforward, there can be complicating circumstances that make it difficult to ascertain the mode of inheritance. This usually results from an unaffected individual, one who appears to be healthy, having the mutation but not manifesting the disease or from an affected individual, one who appears to have the disease, not actually having the mutation(s) but manifesting the disease for some other reason. One scenario in which this occurs is when a mutation sometimes fails to produce disease as a result of balancing genetic or environmental factors. This is known as incomplete penetrance. In another scenario, the mutation causes disease to manifest at a late age, resulting in young people being categorized as unaffected when in fact they will become affected in the future.
Linkage Studies
It is possible to use genetic information from both affected and unaffected family members to map the location of the mutation(s) responsible for a monogenic disease, that is, to determine which region of which chromosome harbors the variant. This is done by genotyping a number of marker polymorphisms throughout the genome, typically microsatellites, although SNPs also can be used as well, and then assessing whether any particular marker is linked to the disease. For a marker that is in close proximity to the disease gene, a particular allele of the marker should be present in the family members with disease and absent in the family members without disease. Such analyses are known as linkage studies.
If the mutation and the marker are on different chromosomes, then by the second Mendel law (the law of independent assortment), there should be no relationship at all between the mutation and an allele of the marker. Whether one ends up in a gamete has nothing to do with whether the other ends up in the same gamete. In contrast, if the mutation and the marker allele are close together on the same chromosome, they should be tightly linked and therefore violate the second Mendel law. There is a high probability that they will end up in the same gamete or, alternatively, that neither will end up in a gamete. If either of these possibilities occurs, the gamete is considered to be nonrecombinant. In other words, no recombination has occurred between the mutation and the marker allele. If the mutation ends up in the gamete but the marker allele does not or if the marker allele ends up in the gamete but the mutation does not, then the gamete is considered to be recombinant; that is, recombination must have occurred between the mutation and the marker allele (assuming that they are on the same chromosome).
A linkage study assesses the number of nonrecombinant versus recombinant gametes generated within a family for any of the genotyped markers. The numbers of each type of gamete can be inferred directly from the relationships between each set of parents and offspring within a family. In principle, the higher the ratio of nonrecombinant gametes to recombinant gametes for a marker is, the closer the marker must lie to the mutation. Perfect linkage would mean all nonrecombinant gametes and no recombinant gametes. Statistical methods can be used to formalize the degree of linkage in a metric called the logarithm of the odds (LOD) score; the higher the LOD score is, the more likely it is that the marker is near the mutation. The LOD score depends in part on the size of the family or, in some cases, the number of families (because it is possible to combine data from multiple families under the assumption that they have mutations in the same gene). Thus, all else being equal, a linkage study with a large number of people is more likely to be successful than a linkage study with few people. A marker with an LOD score >3.0 is generally regarded as being a statistically significant result.
Even with a successful linkage study, the disease mutation will not be directly identified. Rather, the region of the chromosome harboring the mutation is identified. Because of inherent limitations of the analysis, a linkage study will at best define an interval of about a million bases (1 megabase), somewhere in which the mutation lies. Further work is required to pinpoint the exact location of the mutation and its consequences for gene function. A common follow-up study entails the sequencing of all of the genes in the linkage interval in affected family members, with the hope of discovering the mutation in 1 of the coding regions of 1 of the genes. (This assumes that the mutation is in fact a coding variant, which is not a given.) This approach can be prohibitive if the linkage interval in question contains tens or even hundreds of genes, which is often the case. The scientific literature has many examples in which a linkage study was successful (ie, an LOD score >3.0) but no follow-up report of the discovery of the disease mutation was subsequently published.
Next-Generation Sequencing Studies
An alternative approach to identifying mutations responsible for monogenic disorders in families has been made possible by the advent of next-generation sequencing technologies. It is now feasible to perform whole-exome sequencing of DNA samples from a few affected family members and to search for the mutation(s) linked to the disease. In principle, this should yield the full set of coding variants in each of the sequenced individuals, with the disease mutation being among those variants. For a family with a linkage interval that has already been defined with a linkage study, the list of coding variants can be pared down to just those in the linkage interval, making the number of candidate mutations much smaller. (In a sense, whole-exome sequencing is a brute-force approach to dealing with a linkage interval that contains tens or hundreds of genes.)
Even if no linkage study has been performed for a family, it is still quite feasible to discover the disease mutation(s). This is particularly true if the genetic disorder is recessive because this means that both copies of the disease gene must have mutations. For example, if DNA samples from 2 affected siblings are subjected to whole-exome sequencing, the list of candidate mutations can be winnowed down by the following:
Eliminating any variants that are not shared by the 2 siblings
Eliminating variants that have already been found in humans (this assumes that the family has unique, extremely rare mutations that have not already been cataloged)
Eliminating any variants that are unlikely to affect gene function, that is, synonymous variants
Accepting only variants in those genes that are either homozygous for a variant or are compound heterozygous for 2 different variants (meaning the gene must be mutated in both copies)
The number of gene variants meeting all of these criteria is likely to be very small, perhaps limited to a single gene. Genetic disorders that are dominant in nature are more difficult (although by no means impossible) to elucidate in this way because only 1 copy of the disease gene needs to be affected, so the final list of variants may nominate dozens of genes, requiring further studies to determine which is the disease gene.
A particular challenge that has emerged from next-generation sequencing studies is the difficulty in determining whether a newly discovered DNA variant affects gene function. In general, synonymous variants can be assumed to have no effect on gene function, although there are exceptions. Conversely, nonsense variants, as well as frameshift variants, typically will have significant effects on gene function if they result in significantly truncated protein products. Missense variants are more difficult to predict, given that only a single amino acid is altered in a protein product. If the amino acid is in a critical part of the protein, it can inactivate the protein (eg, by impairing the active site of an enzyme or by causing the protein to unfold or fall apart), increase the activity of the protein, or even confer some entirely new function on the protein. If the amino acid is in an unimportant part of the protein, it may have no effect. The precise effect can depend on the identity of the altered amino acid. If the amino acid change is conservative, the new amino acid may act very similarly to the original amino acid and thus result in no change in function. If the new amino acid has biochemical properties that are different from those of the original amino acid, it may have a profound effect.
Unfortunately, there is no reliable method for determining the effect of a missense variant without performing experiments using in vitro or in vivo models, which can be prohibitive if trying to analyze numerous coding variants from a next-generation sequencing experiment. Computational techniques are being used to make predictions about the effect of variants on protein structure and folding, but they remain a work in progress. Another means of predicting whether a particular amino acid is important for gene function is to compare the sequences of the gene from a variety of species across the evolutionary spectrum, from unicellular organisms to humans. If the amino acid has remained identical or similar in all versions of the gene, that is, is conserved across species, it argues for that specific amino acid being critical for the function of the gene, so any missense variant affecting that amino acid is more likely (but not certain) to have a disruptive effect. Recent advances in the ability to generate stem cells from specific patients with specific mutations and in vitro genome-editing technologies that allow one to rapidly and efficiently insert a mutation into the genome of a normal cell are providing new options for exploring the functional effects of newly identified variants.5
Novel noncoding variants are even more challenging because they can fall anywhere in the 99% of the genome that does not encode genes. There is almost no way to know a priori how they might affect nearby or faraway genes. Hence, the notion of sequencing a person's genome and being able to accurately predict the person's lifelong health and disabilities remains a fantasy, notwithstanding the enormous contribution of environmental influences, because of the inability to reliably predict the functional consequences of any given rare, novel DNA variant discovered in that person.
Monogenic Disorders
Although numerous monogenic cardiovascular disorders have been defined, a few well-known classic examples are discussed here.
Familial Hypercholesterolemia
Familial hypercholesterolemia is an inherited condition in which patients have extremely high blood levels of low-density lipoprotein (LDL) cholesterol, which results in abnormal deposition of cholesterol in various parts of the body and a dramatically increased risk of cardiovascular disease, which often manifests at an early age. Several genes have been implicated in this disorder. Mutations in LDLR, which encodes the LDL receptor, can affect the synthesis, structure, and function of the LDL receptor in a variety of ways,6 resulting in the impaired ability of cells to remove cholesterol-carrying LDL particles from the bloodstream and thus the accumulation of LDL cholesterol in the blood. Although familial hypercholesterolemia is often regarded as an autosomal dominant disorder, LDLR mutations have an additive (codominant) effect such that patients who have 2 LDLR mutations have higher blood LDL cholesterol levels and experience earlier cardiovascular disease (as early as childhood) compared with patients with 1 LDLR mutation. Mutations in the APOB gene, which encodes the apolipoprotein B protein, which is a core protein of LDL particles and facilitates their removal from the bloodstream, can mimic the effects of LDLR mutations and result in familial hypercholesterolemia.7 Finally, mutations in 2 other genes that encode proteins that affect the function of the LDL receptor, PCSK9 and LDLRAP1, can also result in familial hypercholesterolemia.8,9 Unlike the other 3 genes, LDLRAP1 mutations are recessive and thus are required to affect both copies of the gene for patients to manifest disease.
Long-QT Syndrome
In long-QT syndrome (LQTS), delayed repolarization of the heart after contraction predisposes to ventricular arrhythmias. Different forms of the condition are inherited in either an auto-somal dominant or autosomal recessive fashion. Mutations in more than a dozen genes have been linked to LQTS, typically affecting the function of potassium, sodium, or calcium channels in cardiac myocytes.10 Clinical gene sequencing is available to assess for mutations in many of these genes; the most commonly affected genes are KCNQ1 (type 1), KCNQ2/HERG (type 2), and SCNA5 (type 3). (See below for a discussion of how the use of gene sequencing may be useful for patient management.) The Brugada syndrome is another inherited cause of ventricular arrhythmias, with mutations in at least 8 genes linked to the syndrome.11 Interestingly, the most commonly affected gene is SCNA5, which is the same gene involved in LQTS type 3; it is notable that different mutations in this gene can give rise to different inherited disorders.
Hypertrophic Cardiomyopathy
Hypertrophic cardiomyopathy (HCM), which is defined by a thickening of the cardiac left ventricle and septum in the absence of any identifiable cause, is the leading cause of sudden cardiac death in young populations. This autosomal dominant disorder has been linked to mutations in more than a dozen genes, most of which encode proteins that are components of the sarcomere, the basic contractile unit of the cardiac myocyte12; >900 distinct mutations in these genes have been identified, presumably leading to disease by interfering with the normal function of the sarcomere. Clinical gene sequencing is now available to assess for mutations in most of these genes. (See below for a discussion of how the use of gene sequencing may be useful for patient management.) HCM is notable in that the first manifestation of disease is often sudden cardiac death; indeed, it has been responsible for a number of high-profile cases in which adolescent or young adult athletes suddenly died while playing sports. Unlike many genetic disorders in which the clinical consequences are apparent at birth, infancy, or childhood, HCM typically does not come to clinical attention until later in life.
Arrhythmogenic Right Ventricular Dysplasia
Arrhythmogenic right ventricular dysplasia is an autosomal dominant cardiomyopathy characterized by fibrofatty replacement of myocardium, primarily in the right ventricle. As with HCM, arrhythmogenic right ventricular dysplasia often first comes to clinical attention in young, healthy people who experience sudden cardiac death during physical activity. It has been linked to mutations in at least 7 genes that encode desmosomal proteins in cardiac myocytes,13 although the pathogenesis of the disease remains unclear.
Polygenic Cardiovascular and Stroke Disorders Polygenic Disorders
In contrast to monogenic disorders, most diseases are complex, that is, they reflect contributions from multiple genes and additional influences such as lifestyle and environmental factors. The contribution of genetics to the development of a disease is reflected in the heritability of the disease. The methods used to calculate heritability are beyond the scope of this document, but it is typically expressed as a number between 0 (no genetic component) and 1 (completely genetically determined). Most cardiovascular disorders are complex in nature and accordingly have heritability estimates in the middle range between 0 and 1.
If DNA variants in multiple genes may contribute to the development of disease, the disease is considered to be polygenic. Single DNA variants will not have a large enough effect to produce disease on their own. Multiple variants with small effects typically combine, possibly along with nongenetic factors, for a person to develop the disease. Accordingly, the disease will not be observed to follow straightforward mendelian modes of inheritance (dominant transmission, recessive transmission, etc). Consequently, family-based study designs (eg, linkage studies) are poorly suited to investigate complex diseases. A different approach is needed to detect the small effects contributed by each of the individual DNA variants.
Common DNA Variants and Linkage Disequilibrium
In contrast to family-based studies, GWASs use large numbers (as many as hundreds of thousands) of unrelated individuals in a population to detect associations between particular SNP markers and diseases. Typically, an SNP marker will have 2 different alleles in a given population, with the more common allele called the major allele and the less common allele called the minor allele. The minor allele frequency (MAF) of an SNP can vary widely between different populations (eg, different ethnic groups); it is used as the criterion by which to judge whether an SNP is common (MAF >5%), low frequency (0.5%<MAF<5%), or rare (MAF <0.5%) in a given population.
Linkage
Linkage equilibrium occurs when there is no linkage between 2 SNPs. This is certainly the case when the 2 SNPs lie on different chromosomes because they will segregate independently during meiosis; the allele of 1 SNP inherited by an offspring from a parent will have no correlation with the inherited allele of the other SNP. This will also occur when 2 SNPs are some distance apart on the same chromosome; the reason is that meiotic recombination within chromosomes occurs at particular recombination hotspots distributed widely across chromosomes, typically on the order of tens to hundreds of kilobases apart. These hotspots define discrete chromosomal regions or loci; SNPs that are separated by 1 hotspot will have a low degree of linkage (even if they lie just across the hotspot from each other), and SNPs that are separated by multiple hotspots will be in linkage equilibrium, that is, have no linkage. In contrast, SNPs that lie within the same locus, that is, not separated by hotspots, will be in linkage disequilibrium (LD); they will have a high degree of linkage because they will be inherited together by the offspring of a parent.
In a scenario in which 2 SNPs are in perfect linkage, the minor allele of the first SNP is always found with the minor allele of the second SNP, and the major allele of the first SNP is always found with the major allele of the second SNP. They will segregate together during meiosis, so the alleles of the 2 SNPs will always be inherited together by an offspring from a parent. By definition, the SNPs will have the same MAFs in a given population. Moreover, 1 SNP can act as a perfect proxy for the other SNP. If one knows the allele at the first SNP, one can reliably predict the identity of the allele at the second SNP without having to directly genotype it (a process called imputation).
Two SNPs may be within the same locus but in only partial linkage. In this scenario, the minor allele of the first SNP may always be found with the minor allele of the second SNP, but the minor allele of the second SNP may occur either with the minor or major allele of the first SNP. This situation arises when the 2 SNPs have different histories; that is, the first SNP arose at a different time in the past from the second SNP. The MAFs of these 2 SNPs will be different.
The metric r2 is commonly used as a gauge of the degree of LD. r2 ranges from 0, which indicates no linkage, to 1, which indicates perfect linkage. Intermediate values indicate partial linkage. It is important to understand that 2 SNPs with r2>0, particularly if r2>0.5, are considered to “tag” each other; that is, they have some degree of correlation. Thus, if 1 SNP is associated with a disease, the other SNP will also be associated with disease to some degree, whether more strongly or more weakly.
By mapping all of the SNPs that have significant linkage with a tag SNP, one can determine the locations of the recombination hotspots that define the boundaries of the locus, which is critical for undertaking a GWAS. Of note, LD patterns differ among different ethnic groups, so a single tag SNP may define quite different loci in different populations. For this reason, each GWAS is typically restricted to data from people of a single ethnicity.
Genome-Wide Association Studies
In essence, a GWAS asks whether for a given SNP marker the MAF differs between a group of individuals with disease (cases) and a group of individuals without disease (controls). For the vast majority of SNPs in the genome, there will be no such difference. In a successful GWAS, there will be at least a handful of SNPs that display a statistically significant difference in MAF between the cases and controls. As with linkage studies, the markers identified by GWASs serve primarily to define an interval in the genome within which lies the causal DNA variant, that is, the DNA variant that causes or contributes to the pathogenesis of a disease. As a rule, the intervals defined by GWASs are much smaller than those defined by linkage studies (tens to hundreds of kilobases versus megabases). This is attributable to the existence of recombination hotspots and the phenomenon of LD, as described above.
In performing GWASs, researchers take advantage of LD by choosing 1 or a few tag SNPs in a given locus, rather than genotyping every single known SNP in the locus. They can thereby do a full study by genotyping a minimum of ≈300 000 tag SNPs across the genome, rather than genotyping the tens of millions of known SNPs in the genome. The genotyping can be done quite efficiently with the use of an array or chip that determines the genotypes of hundreds of thousands or even millions of SNPs in a DNA sample at once.
An important statistical consideration is that testing each of the tag SNPs for an association with disease constitutes an independent experiment. If 1 million SNPs are tested, then 1 million experiments are being performed, and if one accepts the standard threshold of P<0.05 as the criterion for statistical significance, then 5% of the 1 million SNPs, or 50 000 SNPs, will meet that criterion by chance, representing 50 000 false positives, which would swamp out the handful of true positives. One means to address this problem is to adjust the statistical significance threshold using the Bonferroni correction, which simply divides the P value threshold by the number of experiments. In this case, the correction yields a threshold of P<5×10−8, which has become the accepted criterion for statistical significance among GWAS researchers. The need to achieve such a stringent P value, along with the small effects on polygenic disease typically conferred by each individual causal DNA variants (see discussion below), explains why GWASs must often include tens of thousands or even hundreds of thousands of people to have sufficient power to establish SNP associations with disease. It also explains why GWASs are best suited to find common DNA variants that contribute to disease; rare DNA variants, by definition, are found so infrequently in a study population that it is difficult to ascertain their effects on disease with statistical robustness.
A tag SNP found to have significant statistical association with disease may not be (and probably is not) the causal DNA variant itself but rather is in some degree of LD with the causal DNA variant, which is to be found somewhere in the locus. Thus, the tag SNP serves as a signpost around which one must do a finer search. The causal DNA variant may be a coding variant in a gene and thereby contribute to disease by directly altering the function of the gene. Alternatively, the causal DNA variant may be a noncoding variant that influences the expression of a gene, splicing, or other characteristic; in some cases, the causal DNA variant may be as far as hundreds of kilobases away from the causal gene.
GWASs on Cardiovascular Disorders
GWASs have been performed for virtually every cardiovascular disease and trait. Particularly large studies, with study populations numbering as high as the hundreds of thousands, have been performed for coronary artery disease (CAD),14 stroke,15 atrial fibrillation,16 QRS interval,17 blood pressure,18 and blood lipid levels.19 We discuss the studies that have been performed for CAD and stroke in more detail as illustrative examples.
Among the very first GWASs to be performed for any disease were 3 studies for CAD.20–22 Each had a similar design: collecting DNA samples from several thousand patients who had suffered heart attacks and control individuals (who had not had heart attacks but were otherwise similar to the patients), genotyping up to hundreds of thousands of SNPs with gene arrays or chips, and performing statistical tests for association for each of the SNPs. All of the studies identified tag SNPs in the same locus on chromosome 9p21 as being convincingly associated with CAD. Individuals with 1 copy of the at-risk allele of the tag SNP (ie, the allele associated with increased risk of CAD) had a 20% to 40% increase in disease risk compared with individuals with no copies of the at-risk allele; individuals with 2 copies of the at-risk allele had a 30% to 70% increase in risk.
GWASs on CAD
Subsequent GWASs on CAD took advantage of the fact that it is straightforward to combine genetic data from multiple study populations into a single meta-analysis, as long as the populations are of the same ethnicity. Such studies have tremendously increased power to detect SNPs with significant statistical associations. Early meta-analyses identified a total of 13 loci association with CAD or myocardial infarction (listed in Table 1). Two larger meta-analyses used data from tens of thousands of CAD patients and control individuals and identified or confirmed some 2 dozen CAD-associated loci.27,28 In the largest meta-analysis to date, which included data from >60 000 patients with CAD and 130 000 control individuals, a total of 46 CAD-associated loci were identified or confirmed,14 demonstrating the power of increasingly larger sample sizes. However, for many of these new loci, the tag SNPs have rather small effects on CAD risk, with the at-risk alleles conferring increases of only a few percent each.
Table 1.
Tag SNPs in Loci Associated With CAD or Myocardial Infarction
| Unique Locus | Chromosome | SNP | Risk Allele Frequency in Europeans, % | Odds Ratio of Disease (95% CI) per risk allele | Gene(s) of Interest Within or Near the Associated Interval | Functional Effect | Reference |
|---|---|---|---|---|---|---|---|
| 1 | 9p21 | rs4977574 | 56 | 1.29 (1.25–1.34) | CDKN2A-CDKN2B-ANRIL | Cellular proliferation? | 23 |
| 2 | 1p13 | rs646776 | 81 | 1.19 (1.13–1.26) | SORT1 | Blood lipids | 23 |
| 3 | 21q22 | rs9982601 | 13 | 1.20 (1.14–1.27) | SLC5A3-MRPS6-KCNE2 | ? | 23 |
| 4 | 1q41 | rs17465637 | 72 | 1.14 (1.10–1.19) | MIA3 | ? | 23 |
| 5 | 10q11 | rs1746048 | 84 | 1.17 (1.11–1.24) | CXCL12 | ? | 23 |
| 6 | 6p24 | rs12526453 | 65 | 1.12 (1.08–1.17) | PHACTR1 | ? | 23 |
| 7 | 19p13 | rs1122608 | 75 | 1.15 (1.10–1.20) | LDLR | Blood lipids | 23 |
| 8 | 2q33 | rs6725887 | 14 | 1.17 (1.11–1.23) | WDR12 | ? | 23 |
| 9 | 1p32 | rs11206510 | 81 | 1.15 (1.10–1.21) | PCSK9 | Blood lipids | 23 |
| 10 | 12q24 | rs2259816 | 37 | 1.08 (1.05–1.11) | HNF1A | Blood lipids, diabetes mellitus | 24 |
| 11 | 12q24 | rs3184504 | 40 | 1.13 (1.08–1.18) | SH2B3 | ? | 25 |
| 12 | 3q22 | rs9818870 | 15 | 1.15 (1.11–1.19) | MRAS | ? | 24 |
| 13 | 6q26 | rs3798220 | 2 | 1.47 (1.35–1.60) | LPA | Blood lipids | 26 |
CAD indicates coronary artery disease; CI, confidence interval; and SNP, single-nucleotide polymorphism.
Modified from the Myocardial Infarction Genetics Consortium23 with permission from Macmillan Publishers Ltd, copyright © 2009, Rights Managed by Nature Publishing Group; from Erdmann et al24 with permission from Macmillan Publishers Ltd, copyright © 2009, Rights Managed by Nature Publishing Group; from Gudbjartsson et al25 with permission from Macmillan Publishers Ltd, copyright © 2009, Rights Managed by Nature Publishing Group; and from Clarke et al26 with permission from Massachusetts Medical Society, copyright © 2009, Massachusetts Medical Society.
GWASs on Stroke
Initial GWASs on stroke were less informative. This may have been attributable in part to issues of study design. Because stroke is not a single clinical entity but rather multiple disorders of varying subtypes and severity, each of which may have its own unique genetic factors, a GWAS that aggregates stroke cases may include a fairly heterogeneous mix of patients, thereby weakening the power of the study to detect a causal DNA variant involved in any one stroke subtype. Once again demonstrating the power of numbers, subsequent meta-analyses of stroke GWASs that included data from much larger numbers of patients of defined subtypes have turned up a handful of statistically significant associations.
The largest stroke GWAS to date, a meta-analysis of genetic data from >12 000 patients with ischemic stroke and 60 000 control individuals, has confirmed tag SNPs in 3 loci with statistically significant associations with disease.15 Two of the loci, which harbor the PITX2 and ZFHX3 genes, have previously been shown in GWASs to be associated with atrial fibrillation.16 Subgroup analyses show that the 2 loci are specifically associated with cardioembolic stroke, with at-risk alleles conferring a 20% to 40% increase in risk of this type of stroke. A plausible mechanism is that the at-risk alleles of the causal DNA variants directly increase the risk of atrial fibrillation, which in turn increases the risk of stroke. The third locus, harboring the HDAC9 gene, is specifically associated with large-vessel ischemic stroke, with the at-risk allele conferring a 40% increase in risk. The mere presence of the PITX2, ZFHX3, and HDAC9 genes near the tag SNPs does not confirm that these are the causal genes underlying stroke risk (just as the tag SNPs are not necessarily the causal DNA variants); rather, experiments in cellular or animal models are needed to establish that they are in fact the causal genes, as well as the molecular mechanisms by which the genes contribute to stroke.
Cardiovascular and Stroke Risk Prediction
Use of Genetics and Genomics for Risk Prediction
In principle, a patient's genetic data could be used to help forecast the risk of developing a disease (assuming there is a genetic component to the disease) and the outcomes of the disease course. This could be in the context of a monogenic disorder, in which the effect of the causal gene or even the causal mutation may be well understood by virtue of studying the outcomes for many patients with the gene/mutation. Alternatively, this could be related to a polygenic disorder, in which determination of the genotypes of variants in the involved genes may be useful. (When variants in multiple genes rather than a single gene are considered together, it should be considered a genomics application.)
With respect to a complex disease, after GWASs identify a number of tag SNPs at different chromosomal loci across the genome that are convincingly associated with that disease, one can use these SNPs to calculate a genomic risk score for the disease. One simple version of a genomic risk score entails cataloging for each SNP: Does the patient have 2, 1, or 0 copies of the at-risk variant of the SNP? Risk points are assigned, depending on the genotype at the SNP. These points are summed for all of the SNPs, yielding a total risk score. This risk score, especially when combined with a traditional risk score that accounts for lifestyle and environmental factors, might be useful in predicting the likelihood of developing the disease. Providers would be able to test for this specific panel of SNPs versus using whole-genome data that have already been obtained to calculate a risk score that would help guide patient management. Risk scores are already offered as part of commercial genotyping services, and patients may seek interpretations of these risk scores from their providers.
Challenges in Clinical Practice
For a provider presented with this type of genomic information, it will be a challenge to meaningfully integrate it into clinical practice. The relative risks associated with SNP variants are typically small, with at-risk alleles individually conferring between 1.0 to 1.2 times the risk of developing the disease. It has been estimated that a person would need to have dozens or even hundreds of at-risk alleles to have double or triple the risk of a complex disease.29 Thus, any useful clinical applications involving SNP panels would require broad testing of a large number of informative SNPs, and informatics solutions would be required to appropriately analyze and interpret the data, to properly classify patients, and to guide providers in managing the patients. This is in contrast to the typical laboratory test, for which a provider sees the result and quickly interprets it as being normal or abnormal.
There are other important limitations of SNP panels. The SNP panels do not include rare DNA variants with large effects that cause disease or protect against disease and thus may outweigh the small effects of the common variants aggregated in genomic risk scores. Because most GWASs to date have been performed in populations of European ancestry, SNP panels derived from those GWASs may not be relevant to individuals of other racial and ethnic backgrounds. Finally, it should not be overlooked by either providers or patients that many old-fashioned preventive health practices (good diet, weight control, exercise, smoking cessation, etc) can have a far larger impact on one's risk of getting a disease than any genetic influences that one may learn about from genetic testing.
Examples of Risk Prediction
Long-QT Syndrome
As an example of a monogenic disorder, for patients with LQTS, identification of the responsible gene can be useful in predicting the incidence and triggers of a ventricular arrhythmia.30,31 Arrhythmias in LQTS type 1, which is caused by mutations in KCNQ1, are triggered by exercise, particularly swimming, and lifestyle modification can reduce their incidence. Male children more commonly experience cardiac events than female children, whereas adult women have more cardiac events than adult men. In contrast, arrhythmias in LQTS type 2, which is caused by mutations in KCNQ2/HERG, are triggered by emotional or auditory stimuli; there is no sex predilection for cardiac events. Cardiac events in LQTS type 3, which is caused by mutations in SCNA5, commonly occur during rest or sleep; the sex predilection is similar to that found in LQTS type 1. Although events occur less frequently in LQTS type 3 compared with types 1 and 2, the events are more likely to be fatal (because they are more likely to occur during sleep). Thus, identification of the causal mutation in a LQTS patient can potentially help guide patient management, whether that entails counseling lifestyle changes, prescribing antiarrhythmic medications, or counseling reluctant patients who otherwise meet guidelines for placement of an implantable cardioverter-defibrillator to undergo device placement.
CAD and Stroke
There is significant interest in using genomics to improve risk prediction for complex disorders such as CAD and stroke, but thus far, this application has not been straightforward. To date, the paucity of SNPs that have been found to be convincingly associated with stroke has prevented the formulation and testing of a genomic risk score for stroke. A study of a genomic risk score comprising 13 tag SNPs from GWASs on CAD or myocardial infarction (from the 13 loci listed in Table 1) found a 66% increase in risk for incident CAD events for individuals in the top quintile of the score compared with individuals in the bottom quintile of the score.32 However, when the genomic risk score was added to a risk prediction algorithm incorporating traditional cardiovascular risk factors (age, blood pressure, cholesterol levels, tobacco use, etc), there was no improvement in the C statistic, a metric by which the ability of risk prediction algorithms to distinguish high-risk subjects from low-risk subjects is judged. Interestingly, the association of the genomic risk score with incident disease was not affected by adjustment for family history of cardiovascular disease. This may signify that patients with a strong family history of disease have inherited rare DNA variants with large effects (variants that are not included in the genomic risk score). Accordingly, the genomic risk score and the family history may be assessing 2 independent influences on disease.
The modest (66%) increase in risk for CAD signified by a high versus low genomic risk score in this example suggests that risk scores will not affect the management of most patients, particularly patients who are already judged to be at low risk or at high risk for disease. There may be a role for genomic risk scores in reclassifying patients at intermediate risk who are “on the fence”; that is, it is unclear whether to be aggressive or conservative in their management. Another possibility is that genomic risk scores may prove useful in children or young individuals to gauge lifetime cardiovascular risk and to guide early interventions (so-called primordial prevention), although such a strategy must be validated by clinical studies.
Pharmacogenetics
Use of Genetic Information to Predict Response to Medications or Therapies
Pharmacogenetics entails the use of genetic information to predict a patient's response to therapy, that is, its efficacy and toxicity, with the ultimate objective being to safely deliver the right therapy at the right dose for the right patient. The DNA variants used in pharmacogenetic tests are identified in 1 of 2 ways: through analysis of DNA variants in candidate genes with biological links to drug activity or in an unbiased GWAS to find SNPs that are associated with a particular drug response or adverse effect. For a DNA variant to be useful, patients with different genotypes of the variant should display significantly different responses to the therapy, whether a positive response or a negative response.
The earliest, and still most common, examples are ones in which the tested DNA variant is located in or near a gene that encodes a transporter or enzyme that metabolizes the medication. One allele may result in increased or decreased activity of the enzyme compared with the alternate allele, resulting in varied blood levels of the original medication or of an active metabolite. Another common situation occurs when the DNA variant is in or around the drug target (eg, VKORC1 with warfarin; see below) or in related downstream pathways. In some cases in which the full spectrum of drug effects is not known, there may be no known biological link between the DNA variant and the medication, only a statistical association between the variant and the patient response to the medication.
One scenario for the application of pharmacogenetics is the use of a genetic test to identify patients who are at risk for adverse side effects from a therapy (increased toxicity) or who are unlikely to respond to the therapy (decreased efficacy). A patient presenting to medical attention with a particular condition would undergo the test to identify the genotype of a relevant polymorphism or set of polymorphisms. (Alternatively, the genotype may already be available if the patient has previously undergone whole-genome analysis.) The genotype information would be used to determine whether the patient's condition is likely to improve from the therapy, how much of the therapy should be given, or whether the therapy poses an unacceptable risk and should be avoided altogether. If the last is true, an alternative therapy may be chosen.
Examples of Pharmacogenetic Applications
Pharmacogenetics of Clopidogrel
No cardiovascular pharmacogenetic application has yet been fully validated or widely adopted. One application of significant interest involves the antiplatelet agent clopidogrel, which is widely used in patients who have acute coronary syndrome, particularly after percutaneous coronary intervention (PCI). Patients display variable responses to clopidogrel therapy because clopidogrel is not itself an active drug but must be converted into an active metabolite by the hepatic cytochrome P-450 2C19 enzyme. There are a number of identified DNA variants in the CYP2C19 gene that reduce the activity of this enzyme, called reduced-function variants, and thereby result in lower levels of the active metabolite in the bloodstream. There are also CYP2C19 variants that increase the activity of the enzyme, although these variants have not been studied as extensively.
Three large studies of mostly post–acute coronary syndrome or post-PCI patients on clopidogrel therapy found that carriers of reduced-function CYP2C19 variants experienced significantly higher rates of cardiovascular death, myocardial infarction, and stroke.33-35 Subsequent studies with mostly lower-risk patients who did not undergo PCI did not find a difference in the effects of clopidogrel on reduced-function variant carriers versus noncarriers.36 Meta-analyses of numerous studies have reached conflicting conclusions over whether reduced-function variant carriers are disadvantaged when taking clopidogrel.37-39 This was perhaps foreseeable because constituent studies comprise patients with widely varying levels of cardiovascular risk.
In aggregate, the available data suggest that patients at highest risk for cardiovascular events (those who have undergone PCI and are in the acute period after the procedure) may have worse outcomes on clopidogrel if they are reduced-function variant carriers. This has led a few institutions to explore the possibility of point-of-care CYP2C19 genotype testing before PCI. Patients who are noncarriers of reduced-function variants would be prescribed clopidogrel as per usual clinical practice, whereas those who are carriers would be prescribed an alternative medication of the same drug class that is not metabolized by cytochrome P-450 2C19, for example, prasugrel or ticagrelor. To date, no clinical trials assessing the utility of a CYP2C19 genotype test to guide and tailor therapy in a way that leads to improved patient outcomes have been published (although such clinical trials are underway), so any use of such a test must be considered experimental at the present time and cannot be recommended for routine use. Also being explored is the use of platelet function testing as a surrogate marker for the reduced-function genotype, although outcomes trials have so far been negative.40,41
Pharmacogenetics of Warfarin
Another cardiovascular pharmacogenetic application involves the use of the anticoagulant warfarin. Patients receiving warfarin require frequent monitoring of blood clotting activity as measured by the prothrombin time/international normalized ratio; there is risk of either thromboembolism, if the warfarin dose is too low, or bleeding, if the dose is too high. DNA variants in 2 genes, CYP2C9 and VKORC1, account for much of the person-to-person variation in stable therapeutic dosing of warfarin, and algorithms to determine the optimal initiation dosing now include these genetic data.
In 2 randomized trials comparing a pharmacogenetic algorithm with a clinical algorithm for determining the initiation dosing of warfarin, there were no significant differences in the percentage of time in the therapeutic range during the respective study periods (4 and 12 weeks after initiation).42,43 However, there were trends toward reduced clinically significant bleeds with the use of pharmacogenetic algorithms. In a separate randomized trial comparing a pharmacogenetic algorithm with a nonalgorithmic, loading-dose regimen (intended to represent standard clinical care), the pharmacogenetic algorithm yielded a significantly greater percentage of time in the therapeutic range, a significantly shorter median time to therapeutic international normalized ratio, and significantly fewer supratherapeutic international normalized ratio measurements.44 Building on these early findings, additional clinical studies of warfarin pharmacogenetics are underway.
Clinical, Social, and Ethical Implications
There are 2 methods by which genetic testing can occur: specific gene sequencing or genotyping arranged by providers for patients with clinical diagnoses for which the likelihood of a genetic cause is high (ie, risk prediction) or for patients for whom the appropriateness of a specific treatment is being evaluated (ie, pharmacogenetics), and direct-to-consumer genome-wide SNP genotyping services. In either case, although there may be no immediate physical harm for a patient in undergoing genetic testing, which typically involves only swabbing of the inside of a cheek, collection of saliva, or drawing of a blood sample, there are important long-term consequences to consider.
Specific gene testing often occurs at the discretion of the provider rather than the patient (although it should not occur without the patient's permission). Such testing may be informative because the presence of particular mutations may have diagnostic and therapeutic implications. For example, the finding of a BRCA1 or BRCA2 mutation that indicates increased risk of breast cancer may result in a management plan (made jointly by the provider and patient) in which the patient chooses to undergo prophylactic mastectomy. The finding of a mutation that augurs heightened risk of sudden cardiac death in a cardiomyopathy patient may result in the provider and patient opting for the placement of an implantable cardioverter-defibrillator. Typically, these sorts of decisions are driven by the presence of mutations that, on the basis of prior research, are likely to have large clinical effects. However, this is not always the case, and the premature use of a genetic test may carry risks. In 1 example, a company marketed a test for a variant in the KIF6 gene that initial research studies had found to predict patient response to statin therapy. Many providers used the test, presumably to help decide whether to prescribe statins to patients. Subsequent larger studies failed to replicate the KIF6 association with statin response, undermining the validity of the indication for the marketed test and suggesting that use of the test may have adversely affected patient management (if a provider had chosen not to prescribe a statin to a patient who otherwise met guidelines for statin therapy).
Commercial Genetic Testing
The use of a commercial SNP genotyping service is undertaken by the patient because the service is provided directly to consumers without the need for provider approval. Typically, the patient is healthy and is simply seeking information about the future risk of disease. Although some patients may take the initiative to avail themselves of the SNP genotyping service first and then bring genetic information to providers for interpretation, others will approach their providers first and ask whether the testing is advisable. At the present time, such providers are in a difficult position because it is not yet clear whether such testing is of any benefit or how they should best interpret the results of testing, given that few clinical studies have been done. The SNPs included in the testing are mostly common variants with small effects on risk; even when aggregated into genomic risk scores, the SNPs rarely signify more than a doubling or halving of risk. Importantly, rare variants with large effects on risk are not included in the genotype testing. It is possible that a patient with a genomic risk score indicating a modestly reduced risk of a disease may in fact have an undetected rare variant that greatly increases the risk or, conversely, a genomic risk score indicating a modestly reduced risk and a rare variant that greatly decreases the risk. Such a patient, with knowledge only of the genomic risk score, will come away with an inappropriate impression of the true risk of disease.
Thus, there may be significant harm for patients who overanalyze the results of their tests on the basis of, for example, misleading information available on the Internet. One possibility is that a patient may be falsely reassured by hearing that his or her genetic risk for a particular condition is low. The patient may decline to make lifestyle changes that would reduce the risk of disease even more than the protection offered by the presumptive favorable genomic profile (which may itself be misleading because it fails to account for rare variants of large effect). Conversely, learning of an increased genetic risk for a disorder may cause undue worry and even strain family relations. For example, learning that one is more likely to develop a serious illness may affect one's relationship with one's spouse, as well as relationships with parents and potential offspring. Arranging for a patient and family members to meet a genetic counselor is advisable if there is the potential for this type of situation to arise.
The Challenge of Interpreting Genetic Information
The difficulty for providers and patients in appropriately interpreting and acting on the results of genetic testing will only be exacerbated by the “thousand-dollar genome” that will become feasible in the near future. It can be anticipated that whole-genome sequencing for patients will rapidly become routine, either through clinical settings or through direct-to-consumer commercial services. It may be feasible to accurately assess the effects of common variants (by virtue of large population studies in which many people with the variants can be compared with people without the variants), although these effects are usually quite small. In contrast, the interpretation of rare variants that have never before been observed and are unique in an individual or a family will be an enormous challenge. When called on by patients to explain such variants of unknown significance found in their genomes, providers will be in the unenviable position of having to plead ignorance and, at the same time, warn patients not to overanalyze information available from other sources (eg, a Web site with a prediction algorithm that calls a variant “possibly damaging” or “probably damaging,” which refers only to the likelihood of the variant affecting the structure and function of the protein rather than its relevance to disease).
It is worth considering potential situations that providers may soon face and whether genetic information would be useful and would affect patient management. Consider the case of a young asymptomatic adult with no family history of heart disease who undergoes whole-genome sequencing through a commercial service. Among the large number of unique variants found in her genome, the vast majority having no clinical consequence, is a missense variant in the TTN gene, which encodes titin, a component of the sarcomere in cardiac myocytes. Researching the gene on the Internet, she discovers that the gene has been linked to cardiomyopathy; moreover, she finds a Web site with prediction software that calls her variant possibly damaging. Concerned that she may have or may develop a heart condition, she seeks advice from her provider. With the lack of data on this particular variant, it would be hard to argue that the patient should be aggressively managed, whether through lifestyle changes (eg, avoiding competitive sports), intensive monitoring (eg, annual echocardiograms), or intervention (eg, prescription of β-blocker or angiotensin-converting enzyme inhibitor). Is reassurance sufficient, or is further evaluation (eg, a 1-time echocardiogram) warranted, recognizing that such an evaluation might uncover “incidentalomas” that prompt further testing, incur significant healthcare expenses, and expose the patient to unnecessary risks? Clearly, such questions need to be intensively studied as genetic testing becomes more commonplace.
Consider the rather different situation of a patient who faints while exercising and is diagnosed with HCM by echocardiography. The pretest probability of detecting a pathogenic mutation will be far higher in this patient compared with the asymptomatic individual discussed above. In this case, targeted sequencing of TTN and other HCM genes (or whole-genome sequencing, which will soon be able to provide the same gene sequences at less cost) might yield useful diagnostic and prognostic information for the patient. Furthermore, identification of a variant in 1 of these genes would prompt screening for the same variant in family members. It is even possible that whole-genome sequencing of this patient may discover a variant in a gene not previously implicated in cardiomyopathy, prompting research studies on the gene and ultimately advancing the scientific understanding of cardiomyopathy.
Privacy Issues
Finally, privacy issues should be seriously considered when the use of genetic testing is contemplated, especially with respect to whole-genome sequencing of healthy people. It is an unanswered question under what circumstances, to what extent, and by what means genetic data should be incorporated into the medical record. Although easy access to such data could be helpful to providers in improving patient care, it remains to be seen how other parties (eg, insurance companies) might act on the data in ways that do not benefit patients. The US Congress acted to prohibit discrimination by employers and health insurers on the basis of genetic testing with the Genetic Information Nondiscrimination Act in 2008, but further safeguards will undoubtedly be needed as the health implications of genetic data become clearer.
Educational Resources
Although a comprehensive list of educational resources available on the Internet and via other sources is beyond the scope of this document, we wish to highlight a few reliable, curated resources with which providers can begin a search for genetics and genomics information (Table 2).
Table 2.
Relevant Resources
| American Heart Association (AHA) | http://www.heart.org/ |
| Centers for Disease Control and Prevention (CDC) | http://www.cdc.gov/ |
| European Stroke Organisation (ESO) | http://www.eso-stroke.org/ |
| Genetics/Genomics Competency Center (G2C2) | http://www.g-2-c-2.org/ |
| GeneReviews | http://www.ncbi.nlm.nih.gov/books/NBK1116/ |
| GeneTests | http://www.genetests.org/ |
| Heart Rhythm Society (HRS) | http://www.hrsonline.org/ |
| Hypertrophic Cardiomyopathy Association (HCMA) | http://www.4hcm.org/ |
| International Society of Nurses in Genetics (ISONG) | http://www.isong.org/ |
| National Stroke Association | http://www.stroke.org/ |
| National Institutes of Health (NIH) | http://www.nih.gov/ |
| Sudden Arrhythmia Death Syndromes (SADS) Foundation | http://www.sads.org/ |
Appendix
Glossary
The following terms appeared in this document. Some of these definitions were adapted from References 45 and 46.
Additive—A quality found in the relationship between 2 alleles of a polymorphism or gene. In an additive relationship, 1 of the alleles will partly contribute to a phenotype such that 2 copies of the allele will have a larger effect than 1 copy of the allele, which in turn will have a larger effect than 0 copies of the allele.
Affected–A family member who has disease.
Allele—One of ≥2 versions of a polymorphism or gene. An individual typically inherits 2 alleles for each polymorphism, 1 from each parent. If the 2 alleles are the same, the individual is homozygous for that polymorphism. If the alleles are different, the individual is heterozygous. Although the term allele was originally used to describe variation among genes, it now also refers to variation among noncoding DNA sequences.
Alternative splicing—A regulatory mechanism by which variations in the incorporation of the exons of a gene, or coding regions, into mRNA lead to the production of >1 related protein or isoform.
Amino acid—A single unit of a protein.
Array—A technology used to study many genes or polymorphisms at once. Also known as a chip.
At-risk allele—The allele of a polymorphism that is associated with increased risk of disease. An allele associated with decreased risk of disease is a protective allele.
Autosomal dominant—A mendelian pattern of inheritance characteristic of some genetic diseases. “Autosomal” means that the gene in question is located on an autosome. “Dominant” means that a single copy of the mutant gene is enough to cause the disease.
Autosomal recessive—A mendelian pattern of inheritance characteristic of some genetic diseases. “Autosomal” means that the gene in question is located on an autosome. “Recessive” means that 2 copies of the mutant gene are needed to cause the disease.
Autosome—One of the numbered, or nonsex, chromosomes (1 through 22). X and Y are the sex chromosomes.
Base—A single unit of a DNA or an RNA strand. May also be referred to as a nucleotide (although they are not exactly synonymous). Bases typically come in 5 versions: adenine (A), cytosine (C), guanine (G), thymine (T; typically found only in DNA), and uracil (U; typically found only in RNA as a substitute for T). The sequence of bases in a portion of a coding DNA molecule, called a gene, carries the instructions needed to assemble a protein.
Base pair—A single unit of the double helix of DNA, which has 2 complementary strands. Adenine in 1 strand pairs with thymine in the other strand, and cytosine pairs with guanine.
Bonferroni correction—A method by which to adjust for multiple statistical tests. In a genome-wide association study (GWAS), hundreds of thousands or even millions of association tests are performed, and the traditional threshold of P<0.05 for statistical significance is too permissive (ie, will result in too many false positives). The Bonferroni correction divides the P value threshold by the number of tests performed.
Carrier—A family member who carries 1 mutant allele but does not have disease, usually because the mutation is recessive.
Causal DNA variant—A DNA variant that directly contributes to a phenotype. Classically, causal DNA variants are located in the coding sequences of genes and modify the amino acid sequences of the protein products, thereby affecting protein function. DNA variants in non-coding DNA can be causal also, for example, by affecting binding sites of transcription factors in the promoters of genes.
Central dogma—An explanation of the flow of genetic information within a cell. Information is stored in the DNA of the genome, transcribed into RNA, and translated into protein. With a few exceptions, genetic information follows this path only in the forward direction.
Chip—A technology used to study many genes or polymorphisms at once. Also known as an array.
Chromosome—An organized package of DNA found in the nucleus of the cell. Different organisms have different numbers of chromosomes. Humans have 23 pairs of chromosomes: 22 pairs of numbered chromosomes, called autosomes, and 1 pair of sex chromosomes, X and Y.
Coding—Coding DNA sequences code for amino acids. These sequences are transcribed into mRNAs and then translated into the amino acid sequences of proteins.
Codominant—A quality found in the relationship between 2 alleles of a polymorphism or gene. If the alleles are different, codominant alleles will contribute equally to the phenotype. A normal allele and a mutated allele will combine to produce an intermediate phenotype (compared with 2 normal alleles or 2 mutated alleles), for example, a mild form of a disease.
Codon—A 3-base sequence of DNA or RNA that specifies a single amino acid on translation.
Common variant—An allele of a polymorphism that is found at high frequency (>5% of all alleles of the polymorphisms) in a population.
Complex disease—A disease that is influenced by >1 gene and, in many cases, environmental factors. Because of the involvement of multiple genes, the inheritance of the disease does not conform to the Mendel laws. Also known as polygenic disease.
Compound heterozygosity—Having 2 different disease-causing alleles (mutations) at a specific autosomal (or X chromosome in a woman) polymorphism or gene.
Conservative (conserved)—A type of substitution in which 1 amino acid in a protein is replaced with another amino acid that is structurally similar, presumably preserving the function of the protein.
Copy number variation (CNV)—A sequence of >1000 DNA base pairs that is repeated in such a way that the repeats lie adjacent to each other on the chromosome. The number of repeats varies from person to person, with the number typically ranging from zero copies up to a few copies.
Crossing over—The swapping of genetic material that occurs in the germline. During the formation of egg and sperm cells, also known as meiosis, paired chromosomes from each parent align so that similar DNA sequences from the paired chromosomes cross over one another. Crossing over results in a shuffling of genetic material and is an important cause of the genetic variation seen among offspring. This process is also known as meiotic recombination.
Deletion—A DNA alteration involving the loss of genetic material. It can be small, involving a single missing DNA base pair, or large, involving a piece of a chromosome.
Deoxyribonucleic acid (DNA)—The chemical name for the molecule that carries genetic instructions in all living things. The DNA molecule consists of 2 strands that wind around one another to form a shape known as a double helix. Each strand has a backbone made of alternating sugar (deoxyribose) and phosphate groups. Attached to each sugar is 1 of 4 bases: adenine (A), cytosine (C), guanine (G), and thymine (T). The 2 strands are held together by bonds between the bases: Adenine bonds with thymine, and cytosine bonds with guanine. The sequence of the bases along the backbones serves as instructions for assembling RNA and protein molecules.
DNA sequencing—A laboratory technique used to determine the exact sequence of bases (A, C, G, and T) in a DNA molecule. The DNA base sequence carries the information a cell needs to assemble RNA and protein molecules. DNA sequence information is important to scientists investigating the functions of genes. The technology of DNA sequencing was made faster and less expensive as a part of the Human Genome Project.
DNA variant—A site in the DNA sequence of the genome where there is variation among people in a population. Also known as a polymorphism.
Dominant—A quality found in the relationship between 2 alleles of a polymorphism or gene. If the alleles are different, the dominant allele will be expressed, whereas the effect of the other allele, called recessive, is masked. In the case of a dominant genetic disorder, an individual need only inherit 1 copy of the mutated allele for the disease to be present.
Double helix—A standard DNA molecule consists of 2 strands wound around each other in a helical configuration, with each strand held together by bonds between the bases.
Enzyme—A biological catalyst that speeds up the rate of a specific chemical reaction in the cell. Typically, enzymes are proteins, although some are made from RNA. The molecules produced by enzymes are called metabolites.
Exome—The regions of the genome (comprising ≈1% of the genome in humans) that code for proteins.
Exon—A region of the gene that codes for a protein.
Extranuclear inheritance—A nonmendelian pattern of inheritance characteristic of some genetic diseases. “Extranuclear” means that the gene in question is located outside the nucleus, typically in the mitochondria.
Frame (reading frame)—The grouping of DNA/RNA bases into codons. Because codons are 3-base sequences, there are 3 possible frames, only 1 of which typically codes for a functional protein (the other frames will result in amino acid sequences that do not make functional protein).
Frameshift variant—The addition or deletion of a number of DNA bases that is not a multiple of 3, thus causing a shift in the reading frame of the gene. This shift leads to a change in the reading frame of all parts of the gene that are downstream from the variant, often leading to a premature stop codon and ultimately to a truncated protein.
Gamete—A germ cell from a potential mother (egg cell) or father (sperm cell). Each gamete has a set of 23 unpaired chromosomes. Two human gametes (egg and sperm) combine to create a cell (zygote) that contains the full human genome of 23 paired chromosomes.
Genetic Information Nondiscrimination Act (GINA)—US federal legislation that makes it unlawful to discriminate against individuals on the basis of their genetic profiles in regard to health insurance and employment. These protections are intended to encourage Americans to take advantage of genetic testing as part of their medical care. President George W. Bush signed GINA into law on May 22, 2008.
Genetics—The study of the function of a single gene, including its interactions with other genes and environmental factors.
Genome—The entire set of genetic instructions, encoded in DNA, found in a cell. In humans, the genome consists of 23 pairs of chromosomes, found in the nucleus, and a small chromosome found in the mitochondria of the cells.
Genome-wide association study (GWAS)—An approach used in genetics research to associate specific genetic variations with particular diseases. The method involves scanning the genomes from many different people and looking for DNA variants that can be used to predict the presence of a disease. Once such DNA variants are identified, they can be used to understand how genes contribute to the disease and to develop better prevention and treatment strategies.
Genomics—The study of the functions and interactions of many genes in the genome, including their interactions with environmental factors.
Genotype—The set of 2 alleles inherited for a particular polymorphism or gene. “To genotype” means to determine the identity of the alleles.
Haplotype—A group of nearby alleles that are inherited together.
Heritability—The proportion of observable differences in a phenotype between individuals within a population that is attributable to genetic variation (rather than environmental factors or other nongenetic factors).
Heterozygosity—Having 2 different alleles at a specific autosomal (or X chromosome in a woman) polymorphism or gene. In the context of disease, this is usually taken to mean that there are 1 normal allele and 1 disease-causing allele. If the disease-causing allele is dominant, it will result in the person having disease. If the disease-causing allele is recessive, it will not result in disease on its own; instead, the person is considered a carrier.
Homozygosity—Having 2 identical alleles at a specific autosomal (or X chromosome in a woman) polymorphism or gene. In the context of disease, this is usually taken to mean that there are 2 copies of a disease-causing allele.
Imputation—The process of inferring the genotype of 1 polymorphism by directly genotyping another polymorphism, made possible by linkage disequilibrium (LD; a high degree of linkage) between the 2 polymorphisms.
Incomplete penetrance—Penetrance is the likelihood that a person carrying a particular mutation will have the disease. Incomplete penetrance means that the penetrance is <100%.
Indel—Contraction of insertion/deletion. A polymorphism involving variation of a DNA sequence in which it is either present (insertion) or absent (deletion) in the genome.
Intron—A region of a gene that does not code for a protein. Introns are considered noncoding DNA.
Isoform—Any of multiple different forms of the same protein. Different isoforms are often produced by alternative splicing.
Linkage (linked)—The close association of polymorphisms on the same chromosome. The closer 2 polymorphisms are to each other on the chromosome, the greater the probability is that their alleles will be inherited together.
Linkage disequilibrium (LD)—A state in which 2 polymorphisms on the same chromosome are in partial or complete linkage. This most often occurs when the 2 polymorphisms are in close proximity on the same chromosome and there are no recombination hotspots between them.
Linkage equilibrium—A state in which 2 polymorphisms have no linkage. The reason could be that the 2 polymorphisms are on different chromosomes or that they are far apart on the same chromosome, separated by numerous recombination hotspots.
Linkage study—An approach used in genetics research to map a disease-causing mutation in a family or a group of families. The method involves genotyping a large number of markers distributed across the genome and identifying the markers that are most closely linked to the disease. The degree of linkage can be calculated as a logarithm of the odds (LOD) score. Ideally, at least 1 marker will be identified to have a LOD score >3, which suggests that the marker is very close to the disease gene. Once such a marker is identified, nearby genes can be sequenced in an effort to identify the disease-causing mutation.
Locus—The specific physical location of a gene or other DNA sequence on a chromosome, like a genetic street address. In the context of GWASs, locus refers to a discrete chromosome region between 2 recombination hotspots.
Logarithm of the odds (LOD) score—In genetics, a statistical estimate of whether a polymorphism (also called a marker) and a disease gene are likely to be located near each other on a chromosome and are therefore likely to be inherited together. An LOD score of ≥3 is generally understood to mean that the marker and the disease gene are located close to each other on the chromosome.
Long noncoding RNA (lncRNA)—A transcribed non-coding RNA longer than 200 nucleotides in length. lncRNAs interact with genes or mRNAs and regulate their activity.
Low-frequency variant—An allele of a polymorphism that is found at low frequency (between 0.5% and 5% of all alleles of the polymorphism) in a population.
Major allele—The more common allele of a polymorphism that has 2 alleles in a population.
Marker—A polymorphism that is commonly used in linkage studies because it is easy to genotype and its location on a chromosome is well-defined. Typically microsatellites.
Maternal inheritance—A nonmendelian pattern of inheritance characteristic of some genetic diseases. “Maternal” means that the gene in question is located in the mitochondria, which are inherited solely from the mother. Accordingly, a disease can be transmitted to offspring only from the mother.
Meiosis—The formation of gametes (egg and sperm cells). In sexually reproducing organisms, body cells contain 2 sets of chromosomes (1 set from each parent). To maintain this state, the egg and sperm that unite during fertilization each contain a single set of chromosomes. During meiosis, diploid cells undergo DNA replication, followed by 2 rounds of cell division, producing 4 gametes, each of which has 1 set of chromosomes (for humans, 23 unpaired chromosomes). Recombination occurs during meiosis.
Mendelian disease—Same as monogenic disease. Named for the Austrian monk Gregor Mendel, who performed thousands of crosses with garden peas at his monastery during the middle of the 19th century. Mendel explained the results of his experiments by describing 2 laws of inheritance that introduced the idea of dominant and recessive genes.
Messenger RNA (mRNA)—A single-stranded RNA molecule that is complementary to 1 of the DNA strands of a gene. The mRNA is an RNA version of the gene that is processed (eg, splicing), leaves the cell nucleus, and moves to the cytoplasm where proteins are made.
Meta-analysis—In the context of genetics, a statistical technique that combines a number of smaller genetic studies into a single large genetic study.
Metabolite—A molecular intermediate or product of a chemical reaction carried out by an enzyme. In another sense, a product of metabolism.
Metabolomics—The study of a collection of many (if not all) metabolites in a cell, tissue, organ, or organism.
MicroRNA (miRNA)—A transcribed noncoding RNA ≈22 nucleotides in length. miRNAs bind to sequences in mRNAs and regulate their activity.
Microsatellites—Microsatellite sequences are repetitive DNA sequences usually several base pairs in length. They are a type of variable-number tandem repeats (VNTR).
Minor allele—The less common allele of a polymorphism that has 2 alleles in a population.
Minor allele frequency—The frequency of the less common allele of a polymorphism in a population (by definition, must be <50%).
Missense variant—Substitution of a single DNA base that results in a codon that specifies an alternative amino acid.
Monogenic disease—A disease caused by mutation of a single gene.
Mutation—A DNA variant (by some definitions, a DNA variant that occurs at low frequency or rarely in a population, distinguishing it from a polymorphism). Classically, mutations are responsible for monogenic (mendelian) diseases.
Natural selection—The process by which DNA variants become either more or less common in a population as a result of their effects on the survival and reproduction of the individuals carrying them. Can take the form of positive selection or negative selection.
Negative selection—A phenomenon in which a DNA variant decreases in frequency in a population because it adversely affects the survival and reproduction of the individuals who carry it relative to the rest of the population.
Next-generation DNA sequencing—A variety of DNA sequencing methodologies developed during the 2000s and 2010s, in the wake of the Human Genome Project, that have made whole-genome sequencing far cheaper and more efficient.
Noncoding—Noncoding DNA sequences do not code for amino acids. Most noncoding DNA lies between genes on the chromosome and has no known function. Other noncoding DNA, called introns, is found within genes. Some noncoding DNA plays a role in the regulation of gene expression.
Nonrecombinant—In the context of a linkage study, a gamete in which the mutation and a marker allele remain linked (either both are present or both are absent), that is, no meiotic recombination occurred between the 2, and their relationship in the gamete remains identical to their relationship in the parent's genome.
Nonsense variant—Substitution of a single DNA base that creates a stop codon, thus leading to premature truncation of a protein.
Nonsynonymous variant—A DNA variant that alters the coding sequence of a gene so that the amino acid sequence of the protein product is changed. Missense variants, nonsense variants, and (by some definitions) frameshift variants are considered nonsynonymous variants.
Pedigree—A genetic representation of a family tree that diagrams the inheritance of a trait or disease though several generations. The pedigree shows the relationships between family members and indicates which individuals express (affected) or silently carry (carrier) the trait in question.
Pharmacogenetics—A branch of pharmacology concerned with using DNA sequence data to correlate individual genetic variation with drug responses.
Phenotype—The clinical presentation or expression of a specific gene or genes, environmental factors, or both.
Polygenic disease—A disease that is influenced by >1 gene and, in many cases, environmental factors. Because of the involvement of multiple genes, the inheritance of the disease does not conform to the Mendel laws. Also known as complex disease.
Polymorphism—A DNA variant (by some definitions, a DNA variant for which the less frequent allele occurs at >1% frequency in a population, distinguishing it from a mutation).
Positive selection—A phenomenon in which a DNA variant increases in frequency in a population because it enhances the survival and reproduction of the individuals who carry it relative to the rest of the population.
Promoter—A sequence of DNA needed to turn a gene on or off. The process of transcription is initiated at the promoter. Usually found near the beginning of a gene, the promoter has a binding site for the enzymes used to make an mRNA molecule.
Protein—An important type of molecule found in all living cells. A protein is composed of ≥1 long chains of amino acids, the sequence of which corresponds to the DNA sequence of the gene that encodes it. Proteins play a variety of roles in the cell, including structural (cytoskeleton), mechanical (muscle), biochemical (enzymes), and cell signaling (hormones).
Proteomics—The study of a collection of many (if not all) proteins in a cell, tissue, organ, or organism.
Proxy—A polymorphism that reveals the alleles of another polymorphism without the need to directly genotype the second polymorphism as a result of the 2 polymorphisms being in LD (and therefore highly linked). See also tag SNP.
Rare variant—An allele of a polymorphism that is found at extremely low frequency (<0.5% of all alleles of the polymorphism) in a population. In some cases, a rare variant exists only in a single individual or family.
Recessive—A quality found in the relationship between 2 alleles of a polymorphism or gene. If the alleles are different, the dominant allele will be expressed, whereas the effect of the other allele, called recessive, is masked. In the case of a recessive genetic disorder, an individual must inherit 2 copies of the mutated allele for the disease to be present.
Recombinant—In the context of a linkage study, a gamete in which the mutation and a marker allele are no longer present on the same chromosome; that is, meiotic recombination occurred between the 2, and their relationship in the gamete (unlinked) is different from their relationship in the parent's genome (linked).
Recombination (meiotic recombination)—The swapping of genetic material that occurs in the germline. During the formation of egg and sperm cells, also known as meiosis, paired chromosomes from each parent align so that similar DNA sequences from the paired chromosomes recombine with one another. Recombination results in a shuffling of genetic material and is an important cause of the genetic variation seen among offspring. Also known as crossing over.
Recombination hotspot—A location on a chromosome where recombination occurs with high frequency compared with the surrounding regions of DNA.
Reduced-function variant—A DNA variant, usually in the coding sequence, that results in reduced function of the protein product.
Ribonucleic acid (RNA)—Unlike DNA, RNA is single stranded. An RNA strand has a backbone made of alternating sugar (ribose) and phosphate groups. Attached to each sugar is 1 of 4 bases: adenine (A), uracil (U), cytosine (C), or guanine (G).
Short tandem repeat (STR)—See definition of microsatellite.
Single-nucleotide polymorphism (SNP)—A type of polymorphism involving variation of a single base pair in the genome.
Single-sequence repeat (SSR)—See definition of microsatellite.
Splice site variant—Substitution of a single DNA base that, rather than changing a codon, leads to a change in the splicing of the mRNA transcribed from the gene.
Splicing factor—A protein that affects the splicing of an mRNA molecule. The presence versus absence of the protein may result in alternative splicing of the mRNA.
Stop (stop codon)—A codon that leads to the termination of a protein rather than to the addition of an amino acid. The 3 stop codons are UGA, UAA, and UAG.
Synonymous variant—A DNA variant that alters the coding sequence of a gene so that the amino acid sequence of the protein product is not changed.
Tag SNP—An SNP that reveals the alleles of another SNP without the need to directly genotype the second SNP, largely as a result of the 2 SNPs being in LD (and therefore highly linked). See also proxy.
Transcription (transcribed)—The process of making an RNA copy of a gene sequence. This copy, called an mRNA molecule, leaves the cell nucleus and enters the cytoplasm, where it directs the synthesis of the protein, which it encodes.
Transcription factor—A protein that affects the transcription of gene sequence, either increasing or decreasing the expression of a gene. Typically, but not always, the transcription factor will bind to a noncoding DNA sequence in the promoter of the gene.
Transcriptional enhancer—A type of transcription factor that increases expression of a gene.
Transcriptional repressor—A type of transcription factor that decreases expression of a gene.
Transcriptomics—The study of a collection of many (if not all) mRNAs (also known as RNA transcripts) in a cell, tissue, organ, or organism.
Translation (translated)—The process of “translating” the sequence of an mRNA molecule to a sequence of amino acids during protein synthesis. The genetic code describes the relationship between the sequence of base pairs in a gene and the corresponding amino acid sequence that it encodes. In the cell cytoplasm, the ribosome reads the sequence of the mRNA in groups of 3 bases (codons) to assemble the protein.
Unaffected—A family member who does not have disease.
Variable-number tandem repeats (VNTR) —A polymorphism in which a sequence of ≥2 DNA base pairs is repeated in such a way that the repeats lie adjacent to each other on the chromosome. The number of repeats varies from person to person. With a few exceptions, they are generally located in noncoding DNA.
Variant of unknown significance—A nonsynonymous DNA variant in coding DNA, typically discovered in a whole-exome or whole-genome sequencing experiment, the consequences of which on the function of the protein product are unknown. It might be entirely benign and have no clinical consequences; it might be deleterious and cause disease; or it may be somewhere in between.
Whole-exome sequencing—DNA sequencing of the entire exome.
Whole-genome sequencing—DNA sequencing of the entire genome.
X-linked dominant—A mendelian pattern of inheritance characteristic of some genetic diseases. “X-linked” means that the gene in question is located on the X chromosome. “Dominant” means that a single copy of the mutant gene is enough to cause the disease.
X-linked recessive—A mendelian pattern of inheritance characteristic of some genetic diseases. “X-linked” means that the gene in question is located on the X chromosome. “Recessive” means that either 2 copies of the mutant gene (in a woman, who is XX) are needed to cause the disease or 1 copy of the mutant gene (in a man, who is XY) causes the disease in the absence of a second, normal copy of the gene.
Zygote—A cell created by the fusion of 2 gametes (egg from the mother, sperm from the father) in the process of fertilization. A human zygote contains the full human genome of 23 paired chromosomes and, in favorable circumstances, gives rise to a human offspring.
Footnotes
The American Heart Association makes every effort to avoid any actual or potential conflicts of interest that may arise as a result of an outside relationship or a personal, professional, or business interest of a member of the writing panel. Specifically, all members of the writing group are required to complete and submit a Disclosure Questionnaire showing all such relationships that might be perceived as real or potential conflicts of interest.
This statement was approved by the American Heart Association Science Advisory and Coordinating Committee on October 30, 2014. A copy of the document is available at http://my.americanheart.org/statements by selecting either the “By Topic” link or the “By Publication Date” link. To purchase additional reprints, call 843-216-2533 or kelle.ramsay@wolterskluwer.com.
The American Heart Association requests that this document be cited as follows: Musunuru K, Hickey KT, Al-Khatib SM, Delles C, Fornage M, Fox CS, Frazier L, Gelb BD, Herrington DM, Lanfear DE, Rosand J; on behalf of the American Heart Association Council on Functional Genomics and Translational Biology, Council on Clinical Cardiology, Council on Cardiovascular Disease in the Young, Council on Cardiovascular and Stroke Nursing, Council on Epidemiology and Prevention, Council on Hypertension, Council on Lifestyle and Cardiometabolic Health, Council on Quality of Care and Outcomes Research, and Stroke Council. Basic concepts and potential applications of genetics and genomics for cardiovascular and stroke clinicians: a scientific statement from the American Heart Association. Circ Cardiovasc Genet. 2015;8:216–242.
Expert peer review of AHA Scientific Statements is conducted by the AHA Office of Science Operations. For more on AHA statements and guidelines development, visit http://my.americanheart.org/statements and select the “Policies and Development” link.
Permissions: Multiple copies, modification, alteration, enhancement, and/or distribution of this document are not permitted without the express permission of the American Heart Association. Instructions for obtaining permission are located at http://www.heart.org/HEARTORG/General/Copyright-Permission-Guidelines_UCM_300404_Article.jsp. A link to the “Copyright Permissions Request Form” appears on the right side of the page.
| Writing Group Disclosures | ||||||||
|---|---|---|---|---|---|---|---|---|
| Writing Group Member | Employment | Research Grant | Other Research Support | Speakers' Bureau/Honoraria | Expert Witness | Ownership Interest | Consultant/Advisory Board | Other |
| Kiran Musunuru | Harvard University | None | None | None | None | None | None | None |
| Kathleen T. Hickey | Columbia University | None | None | None | None | None | None | None |
| Sana M. Al-Khatib | Duke University Medical Center | None | None | None | None | None | None | None |
| Christian Delles | University of Glasgow | European Commission† | None | None | None | None | None | None |
| Myriam Fornage | University of Texas Health Science Center at Houston | None | None | None | None | None | None | None |
| Caroline Fox | NHLBI | None | None | None | None | None | None | None |
| Lorraine Frazier | University of Arkansas for Medical Sciences | None | None | None | None | None | None | None |
| Bruce D. Gelb | Icahn School of Medicine at Mount Sinai | Shire† | None | None | None | None | None | Royalties for PTPN11, SOS1, RAF1, and SHOC2 gene testing for Noonan syndrome from GeneDx* and Correlegan*; LabCorp*; Preventive Genetics*; Baylor College of Medicine*; Harvard Partners* |
| David M. Herrington | Wake Forest School of Medicine | NIH† | None | None | None | None | None | None |
| David E. Lanfear | Henry Ford Health System | None | None | None | None | None | None | None |
| Jonathan Rosand | Massachusetts General Hospital | NIH/NINDS† | None | None | None | None | None | None |
| Reviewer Disclosures | ||||||||
|---|---|---|---|---|---|---|---|---|
| Reviewer | Employment | Research Grant | Other Research Support | Speakers' Bureau/Honoraria | Expert Witness | Ownership Interest | Consultant/Advisory Board | Other |
| Taura Barr | West Virginia University | None | None | None | None | None | None | None |
| Jason Kovacic | Mount Sinai Medical Centre | None | None | None | None | None | None | None |
References
- 1.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K, Meldrim J, Mesirov JP, Miranda C, Morris W, Naylor J, Raymond C, Rosetti M, Santos R, Sheridan A, Sougnez C, Stange-Thomann N, Stojanovic N, Subramanian A, Wyman D, Rogers J, Sulston J, Ainscough R, Beck S, Bentley D, Burton J, Clee C, Carter N, Coulson A, Deadman R, Deloukas P, Dunham A, Dunham I, Durbin R, French L, Grafham D, Gregory S, Hubbard T, Humphray S, Hunt A, Jones M, Lloyd C, McMurray A, Matthews L, Mercer S, Milne S, Mullikin JC, Mungall A, Plumb R, Ross M, Shownkeen R, Sims S, Waterston RH, Wilson RK, Hillier LW, McPherson JD, Marra MA, Mardis ER, Fulton LA, Chinwalla AT, Pepin KH, Gish WR, Chissoe SL, Wendl MC, Delehaunty KD, Miner TL, Delehaunty A, Kramer JB, Cook LL, Fulton RS, Johnson DL, Minx PJ, Clifton SW, Hawkins T, Branscomb E, Predki P, Richardson P, Wenning S, Slezak T, Doggett N, Cheng JF, Olsen A, Lucas S, Elkin C, Uberbacher E, Frazier M, Gibbs RA, Muzny DM, Scherer SE, Bouck JB, Sodergren EJ, Worley KC, Rives CM, Gorrell JH, Metzker ML, Naylor SL, Kucherlapati RS, Nelson DL, Weinstock GM, Sakaki Y, Fujiyama A, Hattori M, Yada T, Toyoda A, Itoh T, Kawagoe C, Watanabe H, Totoki Y, Taylor T, Weissenbach J, Heilig R, Saurin W, Artiguenave F, Brottier P, Bruls T, Pelletier E, Robert C, Wincker P, Smith DR, Doucette-Stamm L, Rubenfield M, Weinstock K, Lee HM, Dubois J, Rosenthal A, Platzer M, Nyakatura G, Taudien S, Rump A, Yang H, Yu J, Wang J, Huang G, Gu J, Hood L, Rowen L, Madan A, Qin S, Davis RW, Federspiel NA, Abola AP, Proctor MJ, Myers RM, Schmutz J, Dickson M, Grimwood J, Cox DR, Olson MV, Kaul R, Raymond C, Shimizu N, Kawasaki K, Minoshima S, Evans GA, Athanasiou M, Schultz R, Roe BA, Chen F, Pan H, Ramser J, Lehrach H, Reinhardt R, McCombie WR, de la Bastide M, Dedhia N, Blöcker H, Hornischer K, Nordsiek G, Agarwala R, Aravind L, Bailey JA, Bateman A, Batzoglou S, Birney E, Bork P, Brown DG, Burge CB, Cerutti L, Chen HC, Church D, Clamp M, Copley RR, Doerks T, Eddy SR, Eichler EE, Furey TS, Galagan J, Gilbert JG, Harmon C, Hayashizaki Y, Haussler D, Hermjakob H, Hokamp K, Jang W, Johnson LS, Jones TA, Kasif S, Kaspryzk A, Kennedy S, Kent WJ, Kitts P, Koonin EV, Korf I, Kulp D, Lancet D, Lowe TM, McLysaght A, Mikkelsen T, Moran JV, Mulder N, Pollara VJ, Ponting CP, Schuler G, Schultz J, Slater G, Smit AF, Stupka E, Szustakowski J, Thierry-Mieg D, Thierry-Mieg J, Wagner L, Wallis J, Wheeler R, Williams A, Wolf YI, Wolfe KH, Yang SP, Yeh RF, Collins F, Guyer MS, Peterson J, Felsenfeld A, Wetterstrand KA, Patrinos A, Morgan MJ, de Jong P, Catanese JJ, Osoegawa K, Shizuya H, Choi S, Chen YJ, Szustakowki J, International Human Genome Sequencing Consortium Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 2.Ng SB, Turner EH, Robertson PD, Flygare SD, Bigham AW, Lee C, Shaffer T, Wong M, Bhattacharjee A, Eichler EE, Bamshad M, Nickerson DA, Shendure J. Targeted capture and massively parallel sequencing of 12 human exomes. Nature. 2009;461:272–276. doi: 10.1038/nature08250. doi: 10.1038/nature08250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Choi M, Scholl UI, Ji W, Liu T, Tikhonova IR, Zumbo P, Nayir A, Bakkaloglu A, Ozen S, Sanjad S, Nelson-Williams C, Farhi A, Mane S, Lifton RP. Genetic diagnosis by whole exome capture and massively parallel DNA sequencing. Proc Natl Acad Sci U S A. 2009;106:19096–19101. doi: 10.1073/pnas.0910672106. doi: 10.1073/pnas.0910672106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lupski JR, Reid JG, Gonzaga-Jauregui C, Rio Deiros D, Chen DC, Nazareth L, Bainbridge M, Dinh H, Jing C, Wheeler DA, McGuire AL, Zhang F, Stankiewicz P, Halperin JJ, Yang C, Gehman C, Guo D, Irikat RK, Tom W, Fantin NJ, Muzny DM, Gibbs RA. Whole-genome sequencing in a patient with Charcot-Marie-Tooth neuropathy. N Engl J Med. 2010;362:1181–1191. doi: 10.1056/NEJMoa0908094. doi: 10.1056/NEJMoa0908094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Musunuru K. Genome editing of human pluripotent stem cells to generate human cellular disease models. Dis Model Mech. 2013;6:896–904. doi: 10.1242/dmm.012054. doi: 10.1242/dmm.012054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hobbs HH, Brown MS, Goldstein JL. Molecular genetics of the LDL receptor gene in familial hypercholesterolemia. Hum Mutat. 1992;1:445–466. doi: 10.1002/humu.1380010602. doi: 10.1002/humu.1380010602. [DOI] [PubMed] [Google Scholar]
- 7.Soria LF, Ludwig EH, Clarke HR, Vega GL, Grundy SM, McCarthy BJ. Association between a specific apolipoprotein B mutation and familial defective apolipoprotein B-100. Proc Natl Acad Sci USA. 1989;86:587–591. doi: 10.1073/pnas.86.2.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abifadel M, Varret M, Rabès JP, Allard D, Ouguerram K, Devillers M, Cruaud C, Benjannet S, Wickham L, Erlich D, Derré A, Villéger L, Farnier M, Beucler I, Bruckert E, Chambaz J, Chanu B, Lecerf JM, Luc G, Moulin P, Weissenbach J, Prat A, Krempf M, Junien C, Seidah NG, Boileau C. Mutations in PCSK9 cause autosomal dominant hypercholesterolemia. Nat Genet. 2003;34:154–156. doi: 10.1038/ng1161. doi: 10.1038/ng1161. [DOI] [PubMed] [Google Scholar]
- 9.Garcia CK, Wilund K, Arca M, Zuliani G, Fellin R, Maioli M, Calandra S, Bertolini S, Cossu F, Grishin N, Barnes R, Cohen JC, Hobbs HH. Autosomal recessive hypercholesterolemia caused by mutations in a putative LDL receptor adaptor protein. Science. 2001;292:1394–1398. doi: 10.1126/science.1060458. doi: 10.1126/science.1060458. [DOI] [PubMed] [Google Scholar]
- 10.Hedley PL, Jørgensen P, Schlamowitz S, Wangari R, Moolman-Smook J, Brink PA, Kanters JK, Corfield VA, Christiansen M. The genetic basis of long QT and short QT syndromes: a mutation update. Hum Mutat. 2009;30:1486–1511. doi: 10.1002/humu.21106. doi: 10.1002/humu.21106. [DOI] [PubMed] [Google Scholar]
- 11.Brugada R, Campuzano O, Brugada P, Brugada J, Hong K. Brugada syndrome. In: Pagon RA, Adam MP, Bird TD, Dolan CR, Fong CT, Stephens K, editors. GeneReviews [Internet] University of Washington, Seattle; Seattle, WA: Apr 10, 2014. [November 26, 2014]. 1993–2014 http://www.ncbi.nlm.nih.gov/books/NBK1517/. [Google Scholar]
- 12.Cirino AL, Ho C. Familial hypertrophic cardiomyopathy overview. In: Pagon RA, Adam MP, Bird TD, Dolan CR, Fong CT, Stephens K, editors. GeneReviews [Internet] University of Washington, Seattle; Seattle (WA): Jan 16, 2014. [November 26, 2014]. 1993–2014 http://www.ncbi.nlm.nih.gov/books/NBK1768/. [Google Scholar]
- 13.McNally E, MacLeod H, Dellefave L. Arrhythmogenic right ventricular dysplasia/cardiomyopathy, autosomal dominant. In: Pagon RA, Adam MP, Bird TD, Dolan CR, Fong CT, Stephens K, editors. GeneReviews [Internet] University of Washington, Seattle; Seattle, WA: Jan 9, 2014. [November 26, 2014]. 1993–2014 http://www.ncbi.nlm.nih.gov/books/NBK1131/. [Google Scholar]
- 14.CARDIoGRAMplusC4D Consortium. Deloukas P, Kanoni S, Willenborg C, Farrall M, Assimes TL, Thompson JR, Ingelsson E, Saleheen D, Erdmann J, Goldstein BA, Stirrups K, König IR, Cazier JB, Johansson A, Hall AS, Lee JY, Willer CJ, Chambers JC, Esko T, Folkersen L, Goel A, Grundberg E, Havulinna AS, Ho WK, Hopewell JC, Eriksson N, Kleber ME, Kristiansson K, Lundmark P, Lyytikäinen LP, Rafelt S, Shungin D, Strawbridge RJ, Thorleifsson G, Tikkanen E, Van Zuydam N, Voight BF, Waite LL, Zhang W, Ziegler A, Absher D, Altshuler D, Balmforth AJ, Barroso I, Braund PS, Burgdorf C, Claudi-Boehm S, Cox D, Dimitriou M, Do R, DIAGRAM Consortium. CARDIOGENICS Consortium. Doney AS, El Mokhtari N, Eriksson P, Fischer K, Fontanillas P, Franco-Cereceda A, Gigante B, Groop L, Gustafsson S, Hager J, Hallmans G, Han BG, Hunt SE, Kang HM, Illig T, Kessler T, Knowles JW, Kolovou G, Kuusisto J, Langenberg C, Langford C, Leander K, Lokki ML, Lundmark A, McCarthy MI, Meisinger C, Melander O, Mihailov E, Maouche S, Morris AD, Müller-Nurasyid M, MuTHER Consortium. Nikus K, Peden JF, Rayner NW, Rasheed A, Rosinger S, Rubin D, Rumpf MP, Schäfer A, Sivananthan M, Song C, Stewart AF, Tan ST, Thorgeirsson G, van der Schoot CE, Wagner PJ, Wellcome Trust Case Control Consortium. Wells GA, Wild PS, Yang TP, Amouyel P, Arveiler D, Basart H, Boehnke M, Boerwinkle E, Brambilla P, Cambien F, Cupples AL, de Faire U, Dehghan A, Diemert P, Epstein SE, Evans A, Ferrario MM, Ferrières J, Gauguier D, Go AS, Goodall AH, Gudnason V, Hazen SL, Holm H, Iribarren C, Jang Y, Kähönen M, Kee F, Kim HS, Klopp N, Koenig W, Kratzer W, Kuulasmaa K, Laakso M, Laaksonen R, Lee JY, Lind L, Ouwehand WH, Parish S, Park JE, Pedersen NL, Peters A, Quertermous T, Rader DJ, Salomaa V, Schadt E, Shah SH, Sinisalo J, Stark K, Stefansson K, Trégouët DA, Virtamo J, Wallentin L, Wareham N, Zimmermann ME, Nieminen MS, Hengstenberg C, Sandhu MS, Pastinen T, Syvänen AC, Hovingh GK, Dedoussis G, Franks PW, Lehtimäki T, Metspalu A, Zalloua PA, Siegbahn A, Schreiber S, Ripatti S, Blankenberg SS, Perola M, Clarke R, Boehm BO, O'Donnell C, Reilly MP, März W, Collins R, Kathiresan S, Hamsten A, Kooner JS, Thorsteinsdottir U, Danesh J, Palmer CN, Roberts R, Watkins H, Schunkert H, Samani NJ. Large-scale association analysis identifies new risk loci for coronary artery disease. Nat Genet. 2013;45:25–33. doi: 10.1038/ng.2480. doi: 10.1038/ng.2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Traylor M, Farrall M, Holliday EG, Sudlow C, Hopewell JC, Cheng YC, Fornage M, Ikram MA, Malik R, Bevan S, Thorsteinsdottir U, Nalls MA, Longstreth W, Wiggins KL, Yadav S, Parati EA, Destefano AL, Worrall BB, Kittner SJ, Khan MS, Reiner AP, Helgadottir A, Achterberg S, Fernandez-Cadenas I, Abboud S, Schmidt R, Walters M, Chen WM, Ringelstein EB, O'Donnell M, Ho WK, Pera J, Lemmens R, Norrving B, Higgins P, Benn M, Sale M, Kuhlenbäumer G, Doney AS, Vicente AM, Delavaran H, Algra A, Davies G, Oliveira SA, Palmer CN, Deary I, Schmidt H, Pandolfo M, Montaner J, Carty C, de Bakker PI, Kostulas K, Ferro JM, van Zuydam NR, Valdimarsson E, Nordestgaard BG, Lindgren A, Thijs V, Slowik A, Saleheen D, Paré G, Berger K, Thorleifsson G, Hofman A, Mosley TH, Mitchell BD, Furie K, Clarke R, Levi C, Seshadri S, Gschwendtner A, Boncoraglio GB, Sharma P, Bis JC, Gretarsdottir S, Psaty BM, Rothwell PM, Rosand J, Meschia JF, Stefansson K, Dichgans M, Markus HS, Australian Stroke Genetics Collaborative. Wellcome Trust Case Control Consortium 2 (WTCCC2) International Stroke Genetics Consortium Genetic risk factors for ischaemic stroke and its subtypes (the METASTROKE collaboration): a meta-analysis of genome-wide association studies. Lancet Neurol. 2012;11:951–962. doi: 10.1016/S1474-4422(12)70234-X. doi: 10.1016/S1474-4422(12)70234-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ellinor PT, Lunetta KL, Albert CM, Glazer NL, Ritchie MD, Smith AV, Arking DE, Müller-Nurasyid M, Krijthe BP, Lubitz SA, Bis JC, Chung MK, Dörr M, Ozaki K, Roberts JD, Smith JG, Pfeufer A, Sinner MF, Lohman K, Ding J, Smith NL, Smith JD, Rienstra M, Rice KM, Van Wagoner DR, Magnani JW, Wakili R, Clauss S, Rotter JI, Steinbeck G, Launer LJ, Davies RW, Borkovich M, Harris TB, Lin H, Völker U, Völzke H, Milan DJ, Hofman A, Boerwinkle E, Chen LY, Soliman EZ, Voight BF, Li G, Chakravarti A, Kubo M, Tedrow UB, Rose LM, Ridker PM, Conen D, Tsunoda T, Furukawa T, Sotoodehnia N, Xu S, Kamatani N, Levy D, Nakamura Y, Parvez B, Mahida S, Furie KL, Rosand J, Muhammad R, Psaty BM, Meitinger T, Perz S, Wichmann HE, Witteman JC, Kao WH, Kathiresan S, Roden DM, Uitterlinden AG, Rivadeneira F, McKnight B, Sjögren M, Newman AB, Liu Y, Gollob MH, Melander O, Tanaka T, Stricker BH, Felix SB, Alonso A, Darbar D, Barnard J, Chasman DI, Heckbert SR, Benjamin EJ, Gudnason V, Kääb S. Meta-analysis identifies six new susceptibility loci for atrial fibrillation. Nat Genet. 2012;44:670–675. doi: 10.1038/ng.2261. doi: 10.1038/ng.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sotoodehnia N, Isaacs A, de Bakker PI, Dörr M, Newton-Cheh C, Nolte IM, van der Harst P, Müller M, Eijgelsheim M, Alonso A, Hicks AA, Padmanabhan S, Hayward C, Smith AV, Polasek O, Giovannone S, Fu J, Magnani JW, Marciante KD, Pfeufer A, Gharib SA, Teumer A, Li M, Bis JC, Rivadeneira F, Aspelund T, Köttgen A, Johnson T, Rice K, Sie MP, Wang YA, Klopp N, Fuchsberger C, Wild SH, Mateo Leach I, Estrada K, Völker U, Wright AF, Asselbergs FW, Qu J, Chakravarti A, Sinner MF, Kors JA, Petersmann A, Harris TB, Soliman EZ, Munroe PB, Psaty BM, Oostra BA, Cupples LA, Perz S, de Boer RA, Uitterlinden AG, Völzke H, Spector TD, Liu FY, Boerwinkle E, Dominiczak AF, Rotter JI, van Herpen G, Levy D, Wichmann HE, van Gilst WH, Witteman JC, Kroemer HK, Kao WH, Heckbert SR, Meitinger T, Hofman A, Campbell H, Folsom AR, van Veldhuisen DJ, Schwienbacher C, O'Donnell CJ, Volpato CB, Caulfield MJ, Connell JM, Launer L, Lu X, Franke L, Fehrmann RS, te Meerman G, Groen HJ, Weersma RK, van den Berg LH, Wijmenga C, Ophoff RA, Navis G, Rudan I, Snieder H, Wilson JF, Pramstaller PP, Siscovick DS, Wang TJ, Gudnason V, van Duijn CM, Felix SB, Fishman GI, Jamshidi Y, Stricker BH, Samani NJ, Kääb S, Arking DE. Common variants in 22 loci are associated with QRS duration and cardiac ventricular conduction. Nat Genet. 2010;42:1068–1076. doi: 10.1038/ng.716. doi: 10.1038/ng.716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.International Consortium for Blood Pressure Genome-Wide Association Studies. Ehret GB, Munroe PB, Rice KM, Bochud M, Johnson AD, Chasman DI, Smith AV, Tobin MD, Verwoert GC, Hwang SJ, Pihur V, Vollenweider P, O'Reilly PF, Amin N, Bragg-Gresham JL, Teumer A, Glazer NL, Launer L, Zhao JH, Aulchenko Y, Heath S, Sõber S, Parsa A, Luan J, Arora P, Dehghan A, Zhang F, Lucas G, Hicks AA, Jackson AU, Peden JF, Tanaka T, Wild SH, Rudan I, Igl W, Milaneschi Y, Parker AN, Fava C, Chambers JC, Fox ER, Kumari M, Go MJ, van der Harst P, Kao WH, Sjögren M, Vinay DG, Alexander M, Tabara Y, Shaw-Hawkins S, Whincup PH, Liu Y, Shi G, Kuusisto J, Tayo B, Seielstad M, Sim X, Nguyen KD, Lehtimäki T, Matullo G, Wu Y, Gaunt TR, Onland-Moret NC, Cooper MN, Platou CG, Org E, Hardy R, Dahgam S, Palmen J, Vitart V, Braund PS, Kuznetsova T, Uiterwaal CS, Adeyemo A, Palmas W, Campbell H, Ludwig B, Tomaszewski M, Tzoulaki I, Palmer ND, CARDIoGRAM CONSORTIUM. CKDGen Consortium. KidneyGen Consortium. EchoGen Consortium. CHARGE-HF Consortium. Aspelund T, Garcia M, Chang YP, O'Connell JR, Steinle NI, Grobbee DE, Arking DE, Kardia SL, Morrison AC, Hernandez D, Najjar S, McArdle WL, Hadley D, Brown MJ, Connell JM, Hingorani AD, Day IN, Lawlor DA, Beilby JP, Lawrence RW, Clarke R, Hopewell JC, Ongen H, Dreisbach AW, Li Y, Young JH, Bis JC, Kähönen M, Viikari J, Adair LS, Lee NR, Chen MH, Olden M, Pattaro C, Bolton JA, Köttgen A, Bergmann S, Mooser V, Chaturvedi N, Frayling TM, Islam M, Jafar TH, Erdmann J, Kulkarni SR, Bornstein SR, Grässler J, Groop L, Voight BF, Kettunen J, Howard P, Taylor A, Guarrera S, Ricceri F, Emilsson V, Plump A, Barroso I, Khaw KT, Weder AB, Hunt SC, Sun YV, Bergman RN, Collins FS, Bonnycastle LL, Scott LJ, Stringham HM, Peltonen L, Perola M, Vartiainen E, Brand SM, Staessen JA, Wang TJ, Burton PR, Soler Artigas M, Dong Y, Snieder H, Wang X, Zhu H, Lohman KK, Rudock ME, Heckbert SR, Smith NL, Wiggins KL, Doumatey A, Shriner D, Veldre G, Viigimaa M, Kinra S, Prabhakaran D, Tripathy V, Langefeld CD, Rosengren A, Thelle DS, Corsi AM, Singleton A, Forrester T, Hilton G, McKenzie CA, Salako T, Iwai N, Kita Y, Ogihara T, Ohkubo T, Okamura T, Ueshima H, Umemura S, Eyheramendy S, Meitinger T, Wichmann HE, Cho YS, Kim HL, Lee JY, Scott J, Sehmi JS, Zhang W, Hedblad B, Nilsson P, Smith GD, Wong A, Narisu N, Stančáková A, Raffel LJ, Yao J, Kathiresan S, O'Donnell CJ, Schwartz SM, Ikram MA, Longstreth WT, Jr, Mosley TH, Seshadri S, Shrine NR, Wain LV, Morken MA, Swift AJ, Laitinen J, Prokopenko I, Zitting P, Cooper JA, Humphries SE, Danesh J, Rasheed A, Goel A, Hamsten A, Watkins H, Bakker SJ, van Gilst WH, Janipalli CS, Mani KR, Yajnik CS, Hofman A, Mattace-Raso FU, Oostra BA, Demirkan A, Isaacs A, Rivadeneira F, Lakatta EG, Orru M, Scuteri A, Ala-Korpela M, Kangas AJ, Lyytikäinen LP, Soininen P, Tukiainen T, Würtz P, Ong RT, Dörr M, Kroemer HK, Völker U, Völzke H, Galan P, Hercberg S, Lathrop M, Zelenika D, Deloukas P, Mangino M, Spector TD, Zhai G, Meschia JF, Nalls MA, Sharma P, Terzic J, Kumar MV, Denniff M, Zukowska-Szczechowska E, Wagenknecht LE, Fowkes FG, Charchar FJ, Schwarz PE, Hayward C, Guo X, Rotimi C, Bots ML, Brand E, Samani NJ, Polasek O, Talmud PJ, Nyberg F, Kuh D, Laan M, Hveem K, Palmer LJ, van der Schouw YT, Casas JP, Mohlke KL, Vineis P, Raitakari O, Ganesh SK, Wong TY, Tai ES, Cooper RS, Laakso M, Rao DC, Harris TB, Morris RW, Dominiczak AF, Kivimaki M, Marmot MG, Miki T, Saleheen D, Chandak GR, Coresh J, Navis G, Salomaa V, Han BG, Zhu X, Kooner JS, Melander O, Ridker PM, Bandinelli S, Gyllensten UB, Wright AF, Wilson JF, Ferrucci L, Farrall M, Tuomilehto J, Pramstaller PP, Elosua R, Soranzo N, Sijbrands EJ, Altshuler D, Loos RJ, Shuldiner AR, Gieger C, Meneton P, Uitterlinden AG, Wareham NJ, Gudnason V, Rotter JI, Rettig R, Uda M, Strachan DP, Witteman JC, Hartikainen AL, Beckmann JS, Boerwinkle E, Vasan RS, Boehnke M, Larson MG, Järvelin MR, Psaty BM, Abecasis GR, Chakravarti A, Elliott P, van Duijn CM, Newton-Cheh C, Levy D, Caulfield MJ, Johnson T. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature. 2011;478:103–109. doi: 10.1038/nature10405. doi: 10.1038/nature10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Teslovich TM, Musunuru K, Smith AV, Edmondson AC, Stylianou IM, Koseki M, Pirruccello JP, Ripatti S, Chasman DI, Willer CJ, Johansen CT, Fouchier SW, Isaacs A, Peloso GM, Barbalic M, Ricketts SL, Bis JC, Aulchenko YS, Thorleifsson G, Feitosa MF, Chambers J, Orho-Melander M, Melander O, Johnson T, Li X, Guo X, Li M, Shin Cho Y, Jin Go M, Jin Kim Y, Lee JY, Park T, Kim K, Sim X, Twee-Hee Ong R, Croteau-Chonka DC, Lange LA, Smith JD, Song K, Hua Zhao J, Yuan X, Luan J, Lamina C, Ziegler A, Zhang W, Zee RY, Wright AF, Witteman JC, Wilson JF, Willemsen G, Wichmann HE, Whitfield JB, Waterworth DM, Wareham NJ, Waeber G, Vollenweider P, Voight BF, Vitart V, Uitterlinden AG, Uda M, Tuomilehto J, Thompson JR, Tanaka T, Surakka I, Stringham HM, Spector TD, Soranzo N, Smit JH, Sinisalo J, Silander K, Sijbrands EJ, Scuteri A, Scott J, Schlessinger D, Sanna S, Salomaa V, Saharinen J, Sabatti C, Ruokonen A, Rudan I, Rose LM, Roberts R, Rieder M, Psaty BM, Pramstaller PP, Pichler I, Perola M, Penninx BW, Pedersen NL, Pattaro C, Parker AN, Pare G, Oostra BA, O'Donnell CJ, Nieminen MS, Nickerson DA, Montgomery GW, Meitinger T, McPherson R, McCarthy MI, McArdle W, Masson D, Martin NG, Marroni F, Mangino M, Magnusson PK, Lucas G, Luben R, Loos RJ, Lokki ML, Lettre G, Langenberg C, Launer LJ, Lakatta EG, Laaksonen R, Kyvik KO, Kronenberg F, König IR, Khaw KT, Kaprio J, Kaplan LM, Johansson A, Jarvelin MR, Janssens AC, Ingelsson E, Igl W, Kees Hovingh G, Hottenga JJ, Hofman A, Hicks AA, Hengstenberg C, Heid IM, Hayward C, Havulinna AS, Hastie ND, Harris TB, Haritunians T, Hall AS, Gyllensten U, Guiducci C, Groop LC, Gonzalez E, Gieger C, Freimer NB, Ferrucci L, Erdmann J, Elliott P, Ejebe KG, Döring A, Dominiczak AF, Demissie S, Deloukas P, de Geus EJ, de Faire U, Crawford G, Collins FS, Chen YD, Caulfield MJ, Campbell H, Burtt NP, Bonnycastle LL, Boomsma DI, Boekholdt SM, Bergman RN, Barroso I, Bandinelli S, Ballantyne CM, Assimes TL, Quertermous T, Altshuler D, Seielstad M, Wong TY, Tai ES, Feranil AB, Kuzawa CW, Adair LS, Taylor HA, Jr, Borecki IB, Gabriel SB, Wilson JG, Holm H, Thorsteinsdottir U, Gudnason V, Krauss RM, Mohlke KL, Ordovas JM, Munroe PB, Kooner JS, Tall AR, Hegele RA, Kastelein JJ, Schadt EE, Rotter JI, Boerwinkle E, Strachan DP, Mooser V, Stefansson K, Reilly MP, Samani NJ, Schunkert H, Cupples LA, Sandhu MS, Ridker PM, Rader DJ, van Duijn CM, Peltonen L, Abecasis GR, Boehnke M, Kathiresan S. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. doi: 10.1038/nature09270. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McPherson R, Pertsemlidis A, Kavaslar N, Stewart A, Roberts R, Cox DR, Hinds DA, Pennacchio LA, Tybjaerg-Hansen A, Folsom AR, Boerwinkle E, Hobbs HH, Cohen JC. A common allele on chromosome 9 associated with coronary heart disease. Science. 2007;316:1488–1491. doi: 10.1126/science.1142447. doi: 10.1126/science.1142447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Helgadottir A, Thorleifsson G, Manolescu A, Gretarsdottir S, Blondal T, Jonasdottir A, Jonasdottir A, Sigurdsson A, Baker A, Palsson A, Masson G, Gudbjartsson DF, Magnusson KP, Andersen K, Levey AI, Backman VM, Matthiasdottir S, Jonsdottir T, Palsson S, Einarsdottir H, Gunnarsdottir S, Gylfason A, Vaccarino V, Hooper WC, Reilly MP, Granger CB, Austin H, Rader DJ, Shah SH, Quyyumi AA, Gulcher JR, Thorgeirsson G, Thorsteinsdottir U, Kong A, Stefansson K. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science. 2007;316:1491–1493. doi: 10.1126/science.1142842. doi: 10.1126/science.1142842. [DOI] [PubMed] [Google Scholar]
- 22.Samani NJ, Erdmann J, Hall AS, Hengstenberg C, Mangino M, Mayer B, Dixon RJ, Meitinger T, Braund P, Wichmann HE, Barrett JH, König IR, Stevens SE, Szymczak S, Tregouet DA, Iles MM, Pahlke F, Pollard H, Lieb W, Cambien F, Fischer M, Ouwehand W, Blankenberg S, Balmforth AJ, Baessler A, Ball SG, Strom TM, Braenne I, Gieger C, Deloukas P, Tobin MD, Ziegler A, Thompson JR, Schunkert H, WTCCC and the Cardiogenics Consortium Genomewide association analysis of coronary artery disease. N Engl J Med. 2007;357:443–453. doi: 10.1056/NEJMoa072366. doi: 10.1056/NEJMoa072366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Myocardial Infarction Genetics Consortium. Kathiresan S, Voight BF, Purcell S, Musunuru K, Ardissino D, Mannucci PM, Anand S, Engert JC, Samani NJ, Schunkert H, Erdmann J, Reilly MP, Rader DJ, Morgan T, Spertus JA, Stoll M, Girelli D, McKeown PP, Patterson CC, Siscovick DS, O'Donnell CJ, Elosua R, Peltonen L, Salomaa V, Schwartz SM, Melander O, Altshuler D, Ardissino D, Merlini PA, Berzuini C, Bernardinelli L, Peyvandi F, Tubaro M, Celli P, Ferrario M, Fetiveau R, Marziliano N, Casari G, Galli M, Ribichini F, Rossi M, Bernardi F, Zonzin P, Piazza A, Mannucci PM, Schwartz SM, Siscovick DS, Yee J, Friedlander Y, Elosua R, Marrugat J, Lucas G, Subirana I, Sala J, Ramos R, Kathiresan S, Meigs JB, Williams G, Nathan DM, MacRae CA, O'Donnell CJ, Salomaa V, Havulinna AS, Peltonen L, Melander O, Berglund G, Voight BF, Kathiresan S, Hirschhorn JN, Asselta R, Duga S, Spreafico M, Musunuru K, Daly MJ, Purcell S, Voight BF, Purcell S, Nemesh J, Korn JM, McCarroll SA, Schwartz SM, Yee J, Kathiresan S, Lucas G, Subirana I, Elosua R, Surti A, Guiducci C, Gianniny L, Mirel D, Parkin M, Burtt N, Gabriel SB, Samani NJ, Thompson JR, Braund PS, Wright BJ, Balmforth AJ, Ball SG, Hall A, Wellcome Trust Case Control Consortium. Schunkert H, Erdmann J, Linsel-Nitschke P, Lieb W, Ziegler A, König I, Hengstenberg C, Fischer M, Stark K, Grosshennig A, Preuss M, Wichmann HE, Schreiber S, Schunkert H, Samani NJ, Erdmann J, Ouwehand W, Hengstenberg C, Deloukas P, Scholz M, Cambien F, Reilly MP, Li M, Chen Z, Wilensky R, Matthai W, Qasim A, Hakonarson HH, Devaney J, Burnett MS, Pichard AD, Kent KM, Satler L, Lindsay JM, Waksman R, Knouff CW, Waterworth DM, Walker MC, Mooser V, Epstein SE, Rader DJ, Scheffold T, Berger K, Stoll M, Huge A, Girelli D, Martinelli N, Olivieri O, Corrocher R, Morgan T, Spertus JA, McKeown P, Patterson CC, Schunkert H, Erdmann E, Linsel-Nitschke P, Lieb W, Ziegler A, König IR, Hengstenberg C, Fischer M, Stark K, Grosshennig A, Preuss M, Wichmann HE, Schreiber S, Hólm H, Thorleifsson G, Thorsteinsdottir U, Stefansson K, Engert JC, Do R, Xie C, Anand S, Kathiresan S, Ardissino D, Mannucci PM, Siscovick D, O'Donnell CJ, Samani NJ, Melander O, Elosua R, Peltonen L, Salomaa V, Schwartz SM, Altshuler D. Genome-wide association of early-onset myocardial infarction with single nucleotide polymorphisms and copy number variants. Nat Genet. 2009;41:334–341. doi: 10.1038/ng.327. doi: 10.1038/ng.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Erdmann J, Grosshennig A, Braund PS, König IR, Hengstenberg C, Hall AS, Linsel-Nitschke P, Kathiresan S, Wright B, Trégouët DA, Cambien F, Bruse P, Aherrahrou Z, Wagner AK, Stark K, Schwartz SM, Salomaa V, Elosua R, Melander O, Voight BF, O'Donnell CJ, Peltonen L, Siscovick DS, Altshuler D, Merlini PA, Peyvandi F, Bernardinelli L, Ardissino D, Schillert A, Blankenberg S, Zeller T, Wild P, Schwarz DF, Tiret L, Perret C, Schreiber S, El Mokhtari NE, Schäfer A, März W, Renner W, Bugert P, Klüter H, Schrezenmeir J, Rubin D, Ball SG, Balmforth AJ, Wichmann HE, Meitinger T, Fischer M, Meisinger C, Baumert J, Peters A, Ouwehand WH, Deloukas P, Thompson JR, Ziegler A, Samani NJ, Schunkert H, Italian Atherosclerosis, Thrombosis, and Vascular Biology Working Group. Myocardial Infarction Genetics Consortium. Wellcome Trust Case Control Consortium. Cardiogenics Consortium New susceptibility locus for coronary artery disease on chromosome 3q22.3. Nat Genet. 2009;41:280–282. doi: 10.1038/ng.307. doi: 10.1038/ng.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gudbjartsson DF, Bjornsdottir US, Halapi E, Helgadottir A, Sulem P, Jonsdottir GM, Thorleifsson G, Helgadottir H, Steinthorsdottir V, Stefansson H, Williams C, Hui J, Beilby J, Warrington NM, James A, Palmer LJ, Koppelman GH, Heinzmann A, Krueger M, Boezen HM, Wheatley A, Altmuller J, Shin HD, Uh ST, Cheong HS, Jonsdottir B, Gislason D, Park CS, Rasmussen LM, Porsbjerg C, Hansen JW, Backer V, Werge T, Janson C, Jönsson UB, Ng MC, Chan J, So WY, Ma R, Shah SH, Granger CB, Quyyumi AA, Levey AI, Vaccarino V, Reilly MP, Rader DJ, Williams MJ, van Rij AM, Jones GT, Trabetti E, Malerba G, Pignatti PF, Boner A, Pescollderungg L, Girelli D, Olivieri O, Martinelli N, Ludviksson BR, Ludviksdottir D, Eyjolfsson GI, Arnar D, Thorgeirsson G, Deichmann K, Thompson PJ, Wjst M, Hall IP, Postma DS, Gislason T, Gulcher J, Kong A, Jonsdottir I, Thorsteinsdottir U, Stefansson K. Sequence variants affecting eosinophil numbers associate with asthma and myocardial infarction. Nat Genet. 2009;41:342–347. doi: 10.1038/ng.323. doi: 10.1038/ng.323. [DOI] [PubMed] [Google Scholar]
- 26.Clarke R, Peden JF, Hopewell JC, Kyriakou T, Goel A, Heath SC, Parish S, Barlera S, Franzosi MG, Rust S, Bennett D, Silveira A, Malarstig A, Green FR, Lathrop M, Gigante B, Leander K, de Faire U, Seedorf U, Hamsten A, Collins R, Watkins H, Farrall M, PROCARDIS Consortium Genetic variants associated with Lp(a) lipoprotein level and coronary disease. N Engl J Med. 2009;361:2518–2528. doi: 10.1056/NEJMoa0902604. doi: 10.1056/NEJMoa0902604. [DOI] [PubMed] [Google Scholar]
- 27.Schunkert H, König IR, Kathiresan S, Reilly MP, Assimes TL, Holm H, Preuss M, Stewart AF, Barbalic M, Gieger C, Absher D, Aherrahrou Z, Allayee H, Altshuler D, Anand SS, Andersen K, Anderson JL, Ardissino D, Ball SG, Balmforth AJ, Barnes TA, Becker DM, Becker LC, Berger K, Bis JC, Boekholdt SM, Boerwinkle E, Braund PS, Brown MJ, Burnett MS, Buysschaert I, Carlquist JF, Chen L, Cichon S, Codd V, Davies RW, Dedoussis G, Dehghan A, Demissie S, Devaney JM, Diemert P, Do R, Doering A, Eifert S, Mokhtari NE, Ellis SG, Elosua R, Engert JC, Epstein SE, de Faire U, Fischer M, Folsom AR, Freyer J, Gigante B, Girelli D, Gretarsdottir S, Gudnason V, Gulcher JR, Halperin E, Hammond N, Hazen SL, Hofman A, Horne BD, Illig T, Iribarren C, Jones GT, Jukema JW, Kaiser MA, Kaplan LM, Kastelein JJ, Khaw KT, Knowles JW, Kolovou G, Kong A, Laaksonen R, Lambrechts D, Leander K, Lettre G, Li M, Lieb W, Loley C, Lotery AJ, Mannucci PM, Maouche S, Martinelli N, McKeown PP, Meisinger C, Meitinger T, Melander O, Merlini PA, Mooser V, Morgan T, Mühleisen TW, Muhlestein JB, Münzel T, Musunuru K, Nahrstaedt J, Nelson CP, Nöthen MM, Olivieri O, Patel RS, Patterson CC, Peters A, Peyvandi F, Qu L, Quyyumi AA, Rader DJ, Rallidis LS, Rice C, Rosendaal FR, Rubin D, Salomaa V, Sampietro ML, Sandhu MS, Schadt E, Schäfer A, Schillert A, Schreiber S, Schrezenmeir J, Schwartz SM, Siscovick DS, Sivananthan M, Sivapalaratnam S, Smith A, Smith TB, Snoep JD, Soranzo N, Spertus JA, Stark K, Stirrups K, Stoll M, Tang WH, Tennstedt S, Thorgeirsson G, Thorleifsson G, Tomaszewski M, Uitterlinden AG, van Rij AM, Voight BF, Wareham NJ, Wells GA, Wichmann HE, Wild PS, Willenborg C, Witteman JC, Wright BJ, Ye S, Zeller T, Ziegler A, Cambien F, Goodall AH, Cupples LA, Quertermous T, März W, Hengstenberg C, Blankenberg S, Ouwehand WH, Hall AS, Deloukas P, Thompson JR, Stefansson K, Roberts R, Thorsteinsdottir U, O'Donnell CJ, McPherson R, Erdmann J, Samani NJ, Cardiogenics. CARDIoGRAM Consortium Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat Genet. 2011;43:333–338. doi: 10.1038/ng.784. doi: 10.1038/ng.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Coronary Artery Disease (C4D) Genetics Consortium A genome-wide association study in Europeans and South Asians identifies five new loci for coronary artery disease. Nat Genet. 2011;43:339–344. doi: 10.1038/ng.782. doi: 10.1038/ng.782. [DOI] [PubMed] [Google Scholar]
- 29.Kraft P, Hunter DJ. Genetic risk prediction: are we there yet? N Engl J Med. 2009;360:1701–1703. doi: 10.1056/NEJMp0810107. doi: 10.1056/NEJMp0810107. [DOI] [PubMed] [Google Scholar]
- 30.Priori SG, Schwartz PJ, Napolitano C, Bloise R, Ronchetti E, Grillo M, Vicentini A, Spazzolini C, Nastoli J, Bottelli G, Folli R, Cappelletti D. Risk stratification in the long-QT syndrome. N Engl J Med. 2003;348:1866–1874. doi: 10.1056/NEJMoa022147. doi: 10.1056/NEJMoa022147. [DOI] [PubMed] [Google Scholar]
- 31.Schwartz PJ, Priori SG, Spazzolini C, Moss AJ, Vincent GM, Napolitano C, Denjoy I, Guicheney P, Breithardt G, Keating MT, Towbin JA, Beggs AH, Brink P, Wilde AA, Toivonen L, Zareba W, Robinson JL, Timothy KW, Corfield V, Wattanasirichaigoon D, Corbett C, Haverkamp W, Schulze-Bahr E, Lehmann MH, Schwartz K, Coumel P, Bloise R. Genotype-phenotype correlation in the long-QT syndrome: gene-specific triggers for life-threatening arrhythmias. Circulation. 2001;103:89–95. doi: 10.1161/01.cir.103.1.89. doi: 10.1161/01.CIR.103.1.89. [DOI] [PubMed] [Google Scholar]
- 32.Ripatti S, Tikkanen E, Orho-Melander M, Havulinna AS, Silander K, Sharma A, Guiducci C, Perola M, Jula A, Sinisalo J, Lokki ML, Nieminen MS, Melander O, Salomaa V, Peltonen L, Kathiresan S. A multilocus genetic risk score for coronary heart disease: case-control and prospective cohort analyses. Lancet. 2010;376:1393–1400. doi: 10.1016/S0140-6736(10)61267-6. doi: 10.1016/S0140-6736(10)61267-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mega JL, Close SL, Wiviott SD, Shen L, Hockett RD, Brandt JT, Walker JR, Antman EM, Macias W, Braunwald E, Sabatine MS. Cytochrome p-450 polymorphisms and response to clopidogrel. N Engl J Med. 2009;360:354–362. doi: 10.1056/NEJMoa0809171. doi: 10.1056/NEJMoa0809171. [DOI] [PubMed] [Google Scholar]
- 34.Simon T, Verstuyft C, Mary-Krause M, Quteineh L, Drouet E, Méneveau N, Steg PG, Ferrières J, Danchin N, Becquemont L, French Registry of Acute ST-Elevation and Non-ST-Elevation Myocardial Infarction (FAST-MI) Investigators Genetic determinants of response to clopidogrel and cardiovascular events. N Engl J Med. 2009;360:363–375. doi: 10.1056/NEJMoa0808227. doi: 10.1056/NEJMoa0808227. [DOI] [PubMed] [Google Scholar]
- 35.Collet JP, Hulot JS, Pena A, Villard E, Esteve JB, Silvain J, Payot L, Brugier D, Cayla G, Beygui F, Bensimon G, Funck-Brentano C, Montalescot G. Cytochrome P450 2C19 polymorphism in young patients treated with clopidogrel after myocardial infarction: a cohort study. Lancet. 2009;373:309–317. doi: 10.1016/S0140-6736(08)61845-0. doi: 10.1016/S0140-6736(08)61845-0. [DOI] [PubMed] [Google Scholar]
- 36.Paré G, Mehta SR, Yusuf S, Anand SS, Connolly SJ, Hirsh J, Simonsen K, Bhatt DL, Fox KA, Eikelboom JW. Effects of CYP2C19 genotype on outcomes of clopidogrel treatment. N Engl J Med. 2010;363:1704–1714. doi: 10.1056/NEJMoa1008410. doi: 10.1056/NEJMoa1008410. [DOI] [PubMed] [Google Scholar]
- 37.Mega JL, Simon T, Collet JP, Anderson JL, Antman EM, Bliden K, Cannon CP, Danchin N, Giusti B, Gurbel P, Horne BD, Hulot JS, Kastrati A, Montalescot G, Neumann FJ, Shen L, Sibbing D, Steg PG, Trenk D, Wiviott SD, Sabatine MS. Reduced-function CYP2C19 genotype and risk of adverse clinical outcomes among patients treated with clopidogrel predominantly for PCI: a meta-analysis. JAMA. 2010;304:1821–1830. doi: 10.1001/jama.2010.1543. doi: 10.1001/jama.2010.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bauer T, Bouman HJ, van Werkum JW, Ford NF, ten Berg JM, Taubert D. Impact of CYP2C19 variant genotypes on clinical efficacy of antiplatelet treatment with clopidogrel: systematic review and meta-analysis. BMJ. 2011;343:d4588. doi: 10.1136/bmj.d4588. doi: 10.1136/bmj.d4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Holmes MV, Perel P, Shah T, Hingorani AD, Casas JP. CYP2C19 geno-type, clopidogrel metabolism, platelet function, and cardiovascular events: a systematic review and meta-analysis. JAMA. 2011;306:2704–2714. doi: 10.1001/jama.2011.1880. doi: 10.1001/jama.2011.1880. [DOI] [PubMed] [Google Scholar]
- 40.Bonello L, Armero S, Ait Mokhtar O, Mancini J, Aldebert P, Saut N, Bonello N, Barragan P, Arques S, Giacomoni MP, Bonello-Burignat C, Bartholomei MN, Dignat-George F, Camoin-Jau L, Paganelli F. Clopidogrel loading dose adjustment according to platelet reactivity monitoring in patients carrying the 2C19*2 loss of function polymorphism. J Am Coll Cardiol. 2010;56:1630–1636. doi: 10.1016/j.jacc.2010.07.004. doi: 10.1016/j.jacc.2010.07.004. [DOI] [PubMed] [Google Scholar]
- 41.Siller-Matula JM, Jilma B. Why have studies of tailored anti-platelet therapy failed so far? Thromb Haemost. 2013;110:628–631. doi: 10.1160/TH13-03-0250. doi: 10.1160/TH13-03-0250. [DOI] [PubMed] [Google Scholar]
- 42.Kimmel SE, French B, Kasner SE, Johnson JA, Anderson JL, Gage BF, Rosenberg YD, Eby CS, Madigan RA, McBane RB, Abdel-Rahman SZ, Stevens SM, Yale S, Mohler ER, 3rd, Fang MC, Shah V, Horenstein RB, Limdi NA, Muldowney JA, 3rd, Gujral J, Delafontaine P, Desnick RJ, Ortel TL, Billett HH, Pendleton RC, Geller NL, Halperin JL, Goldhaber SZ, Caldwell MD, Califf RM, Ellenberg JH, COAG Investigators A pharmacogenetic versus a clinical algorithm for warfarin dosing. N Engl J Med. 2013;369:2283–2293. doi: 10.1056/NEJMoa1310669. doi: 10.1056/NEJMoa1310669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Verhoef TI, Ragia G, de Boer A, Barallon R, Kolovou G, Kolovou V, Konstantinides S, Le Cessie S, Maltezos E, van der Meer FJ, Redekop WK, Remkes M, Rosendaal FR, van Schie RM, Tavridou A, Tziakas D, Wadelius M, Manolopoulos VG, Maitland-van der Zee AH, EU-PACT Group A randomized trial of genotype-guided dosing of acenocoumarol and phenprocoumon. N Engl J Med. 2013;369:2304–2312. doi: 10.1056/NEJMoa1311388. doi: 10.1056/NEJMoa1311388. [DOI] [PubMed] [Google Scholar]
- 44.Pirmohamed M, Burnside G, Eriksson N, Jorgensen AL, Toh CH, Nicholson T, Kesteven P, Christersson C, Wahlström B, Stafberg C, Zhang JE, Leathart JB, Kohnke H, Maitland-van der Zee AH, Williamson PR, Daly AK, Avery P, Kamali F, Wadelius M, EU-PACT Group A randomized trial of genotype-guided dosing of warfarin. N Engl J Med. 2013;369:2294–2303. doi: 10.1056/NEJMoa1311386. doi: 10.1056/NEJMoa1311386. [DOI] [PubMed] [Google Scholar]
- 45.Guttmacher AE, Collins FS, Drazen JM. Genomic Medicine: Articles from the New England Journal of Medicine. The Johns Hopkins University Press; Baltimore, MD: 2004. [Google Scholar]
- 46.National Human Genome Research Institute [June 16, 2014];Talking Glossary of Genetic Terms. http://www.genome.gov/glossary.cfm.