Abstract
Objectives:
Malaria has been a major global health problem in recent times with increasing mortality. Current treatment methods include parasiticidal drugs and vaccinations. However, resistance among malarial parasites to the existing drugs has emerged as a significant area of concern in anti-malarial drug design. Researchers are now desperately looking for new targets to develop anti-malarials drug which is more target specific. Malarial parasites harbor a plastid-like organelle known as the ‘apicoplast’, which is thought to provide an exciting new outlook for the development of drugs to be used against the parasite. This review elaborates on the current state of development of novel compounds targeted againstemerging malaria parasites.
Methods:
The apicoplast, originates by an endosymbiotic process, contains a range of metabolic pathways and housekeeping processes that differ from the host body and thereby presents ideal strategies for anti-malarial drug therapy. Drugs are designed by targeting the unique mechanism of the apicoplasts genetic machinery. Several anabolic and catabolic processes, like fatty acid, isopenetyl diphosphate and heme synthess in this organelle, have also been targeted by drugs.
Results:
Apicoplasts offer exciting opportunities for the development of malarial treatment specific drugs have been found to act by disrupting this organelle’s function, which wouldimpede the survival of the parasite.
Conclusion:
Recent advanced drugs, their modes of action, and their advantages in the treatment of malaria by using apicoplasts as a target are discussed in this review which thought to be very useful in desigining anti-malarial drugs. Targetting the genetic machinery of apicoplast shows a great advantange regarding anti-malarial drug design. Critical knowledge of these new drugs would give a healthier understanding for deciphering the mechanism of action of anti-malarial drugs when targeting apicoplasts to overcome drug resistance.
Keywords: anti-malarial drugs, apicoplast, drug resistance, malaria
1. Introduction
Malaria, a profound human health problem that has gained importance and continues to stimulate research, affecting the major tropical and subtropical regions of the world, including parts of America, Asia, and Africa. Each year approximately 515 million cases of malaria are reported. According to a recent report, 214 million malaria cases were reported worlwide in 2015. That year saw an estimated 438,000 malaria deaths worldwide, and 90% of those deaths occurred in Africa followed by 7% in Southeast Asia and 2% in the Eastern Mediterranean region (2%) [1]. Strategies such as vaccination, anti-malarial drug formulation, and vector control have been undertaken to combat this parasitic disease. Among these, parasitical drugs are the main line of defense. Unfortunately, the emergence of malarial parasites with resistance to existing drugs is a burning crisis [2]. Therefore, the design of a new line of anti-malarial drugs would represent a major breakthrough that needs to be explored.
The recent identification of a chloroplast-like organelle called the apicoplast in malarial parasites has great implications for overcoming their resistance to anti-malarial drugs. The apicomplexan plastid, which is called a apicoplast, arises from the endosymbiotic process, just as with other plastids, and is integrated into the parasite’s function [3]. The nuclear gene of the parasite is post-translationally targeted to the organelle via the secretory pathway via the endoplasmic reticulum. Vesicles bearing toxoplasma apicoplast membrane proteins persist following loss of the relict plastid or golgi body disruption [4]. On the other hand, the apicoplast appears to serve its host with multiple functions, though it apparently lacks the capacity to photosynthesize; however, it retains the typical plastid functions, such as fatty acid, isoprenoid, and heme syntheses, among others. It also bears all cellular processes such as deoxyribonucleic acid (DNA) replication, transcription, translation, and post-translational modification. The products of the metabolic pathways might be exported from the apicoplast for the use of the parasite itself, and some components of the products are essential for host cell invasion [5, 6]. Each of these processes is a potential drug target because they differ from the processes in the host, which are fundamentally eukaryotic. In this review, we will examine the apicoplast’s genome organization and structural peculiarities, as well as anti-malarial drugs designed to target the apicoplast.
2. Present status of anti-malarial drugs
Malaria is caused by the protozoan parasite Plasmodium. Only four types of the Plasmodium parasite can infect humans. People get malaria because of their having been bitten by an infective female Anopheles mosquito that carries the apicomplexan parasite, Plasmodium. The prophylactic drug treatments against malaria are often too expensive for most people who live in endemic areas. Most adults from endemic areas possess partial immunity in terms of resistance to those drugs and become susceptible to severe malaria [2].
The emergence of resistance to existing drugs among malarial parasites has been a deepening crisis. Resistance to the most effective classes of anti-malarial drugs has emerged, and this has been responsible for the recent increase in malaria-related mortality, particularly in Africa. The genetic events that confer anti-malarial drug resistance (while retaining parasite viability) are thought to be autonomous and independent of the drug used. The causes of drug resistance are generally mutations or changes in the copy number of genes relating to the drug’s parasite target. However, influx/efflux pumps, which alter the intraparasitic concentrations of the drug, also help to develop the resistance. For example, chloroquine resistance in Palsomdium falciparum (P. falciparum) may be multigenic and is initially conferred by mutations in a gene encoding transporter Palsomdium falciparum chloroquine-resistance transporter (PfCRT). Resistance to atovaquone results from point mutations in the gene cytochrome b (cytB), coding for cytB [7]. Resistance to mefloquine (MQ) and other structurally-related arylaminoalcohols in P. falciparum results from duplications in Palsomdium falciparum multi-drug resistance gene (Pfmdr), which encodes the p-glycoprotein pump (Pgh) [8]. For P. falciparum and Plasmodium vivax (P. vivax), resistance to pyrimethamine and cycloguanil results from the sequential acquisition of mutations in dihydrofolate reductase (DHFR) [9, 10]. The resistance to such anti-malarial compounds in malarial parasites is thought to appear relatively frequently, and recent reports have suggested the emergence of resistance toone of the most widely used anti-malarial drugs, artemisinin [11].
Current reports have suggested that patients with severe malaria who were treated with quinine did not recover [12]. Resistance to chloroquinine is associated with a resistance transporter PfCRT, and the mutated form of the PfCRT gene. Moreover, additional mutations on the Pfmdr1 are also linked with the resistance [13]. On the other hand, recent evidence has shown resistance to MQ to be due to a mutation in Pfmdr-1 [14]. Piperaquine (PPQ) resistance was recently reported and was thoughtto be due to a variation on chromosome 5 that includes Pfmdr-1 [15]. Point mutations in the Palsomdium falciparum dihydrofolate reductase (PfDHFR) and the Palsomdium falciparum dihydropteroate synthetase (PfDHPS) genes confer resistance to sulfadoxine-pyrimethamine (SP) [16]. Point mutations and copy number variations in Pfmdr-1 have also been implicated in resistance to artemisinin. However, the exact mechanism is unclear, and clinical trials were unable to identify the role of Pfmdr-1 in artemisinin resistance [17, 18, 19]. These findings suggest that prevention of drug resistance will be very problematic; thus, control efforts would be better directed at limiting the subsequent spread of resistance (Table 1). For this reason, new lines of drugs must be explored before existing drugs lose their efficacy.
Table. 1. Anti-malarial drugs and their mechanisms of resistance.
| Name of anti-malarial drug | Mechanism of resistance |
|---|---|
| Chloroquinine | Mutations in a gene encoding a transporter, PfCRT and mutations on the Pfmdr-1 [13, 17] |
| Atovaquone | Point mutations in the gene of cytochrome b [7] |
| Mefloquine | Duplications in Pfmdr, which encodes the Pgh [14] |
| Arylaminoalcohols | Duplications in Pfmdr, which encodes the Pgh [8] |
| Pyrimethamine | Sequential acquisition of mutations in the DHFR gene [16] |
| Cycloguanil | Sequential acquisition of mutations in the DHFR gene [7, 9] |
| Artemisinin | Not clearly understood; probably by mutation in C580y or point mutations and copy number variations in Pfmdr-1 [17, 18, 19] |
PfCRT, Plasmodium falciparum chloroquine resistance transporter; Pfmdr-1, Plasmodium falciparum multidrug resistance gene-1; Pgh, p-glycoprotein pump , DHFR, dihydrofolate reductase.
3. Emergence of the apicoplast
The recent identification of the apicoplast in malarial parasites has insightful implications for drug therapies to overcome the problem of resistance. To assess the potential of the apicoplast as a drug target, we need to understand how this vital organelle arises. Consensus phylogenetic analyses have divided eukaryotes into six supergroups. Apicomplexa in the stramenophile, alveolate and rhizaria supergroups is distinct from its animal hosts. Apicomplexa started their evolutionary journey to being a parasite from aphotosynthetic entity; a path that has recently been established by the discovery of fully photosynthetic apicomplexan-like algae. Phylogenetic studies reveal that these two groups and the dinoflagellate algae arose from a common photosynthetic ancestor. Evolutionary data suggest that Apicomplexa are often confused with ‘animal cells with a plastid’ or ‘parasitic plants’. However, that is not so. Rather, they have many unique biological characteristics with a unique evolutionary history [10]. The apicoplast retains the basic features of a plastid, which makes this organelle a promising drug target [20, 21, 22]. However, that the apicoplasts differ markedly from plant plastids is increasing apparent. This divergence of the apicoplast was shown in a recent phylogenetic analysis of plastid origins, in which the apicoplast was placed in a completely evolutionarily distinct group from that of the plastids of green plants and Archaeplastida algae. Evolutionary divergence and the large number of unknown genes within this novel apicomplexan-related plastid lineagepresent considerable opportunities in targeting the apicoplast with anti-malarials.
4. General structure and genome organization of an apicoplast
All apicomplexan parasites, except Cryptosporidium sp., possess three active components for replication/transcription/ translation: the nucleus, the apicoplast, and the mitochondrion. As Cryptosporidium sp. bears mitochondria that do not contain DNA, noorganelle of an endosymbiotic origin containing DNA, such as an apicoplast, could have evolved in the genomic machinery of that organism [23]. On the other hand, although an organism like Chromera velia (C. velia) contains a photosynthetic plastid, the majority of apicomplexan parasites contain either a non-photosynthetic plastid or no plastid at all. This indicates that the apicomplexan malarial parasites might have evolved from a significant and essential organelle like an apicoplast [24]. As C. velia contains the original apicoplast, it has been used as a model target for anti-malarial drug development.
The nucleus contains a 23-Mb nuclear genome composed of 14 linear chromosomes with approximately 5,300 genes whereas specialized organelles each contain a circular genome: a 35-kb apicoplast genome with around 50 genes and a 3-kb mitochondrial genome encoding only 3 protein genes and ribosomal RNA (rRNA) genes. The apicoplast is a non-photosynthetic plastid-like organelle of protozoan parasites of Apicomplexan-including pathogens like Plasmodium, Toxoplasma and Cryptosporidum. This organelle might have originated as a process of secondary endosymbiosis. However, a hypothesis is that apicomplexans were once able to use apicoplasts to synthesize energy by photosynthesis. Therfore, the ancient apicomplexans and their related progenitors might have had a symbiotic relationship with their surrounding coral reef. To achieve that, those early organisms would have possessed a working chloroplast. However, over the course of time, that autotrophic ability was lost, and apicomplexans slowly evolved to become parasitic species dependent on their hosts for survival.
The apicoplast is ovoid and intimately associated with mitochondria. It shows three distinct membrane-like structures: an inner envelope membrane (IEM), an outer envelope membrane (OEM) and an outer membrane system (OMS). In Plasmodium sp. and Toxoplasma sp., the apicoplast seems to be surrounded by a basic set of membranes. Most of the genes appearing in an apicoplast are involved in organelle transcription and translation [6, 10, 20].
5. Apicoplast as a drug target
Creating food from carbon dioxide and sunlight is not the only task that apicoplasts perform for their host; they are also major sites of a number of anabolic processes whose products are supplied to the surrounding host cell. These metabolic services make plastids indispensable to plant and alga, even to non-photosynthetic ones like apicomplexan parasites. The Plasmodium apicoplast was revealed to be a stripped-down version of a chloroplast, in which about 500 nucleus-encoded and apicoplast-targeted proteins lie and are combined with 35 proteins encoded within the apicoplast by its circular genome.
The first portrait of apicoplast metabolic pathways showed the apicoplast as carrying four identifiable metabolic pathways, isoprenoid precursor synthesis, fatty acid synthesis, heme synthesis and ironsulfur cluster biogenesis, and as having the functions of genome replication, transcription, translation, post-translational modification and protein turnover [6, 20]. Among all of these apicoplast functions, fatty acid synthesis, isoprenoid precursor synthesis and “housekeeping” functions are clearly bacterial in nature and are, therefore, potential targets for existing anti-biotic drugs because they differ from the processes in the host, which are fundamentally eukaryotic. Many such anti-biotics have been successfully tested on Plasmodium in culture [19, 20].
As a drug target, the apicoplast can be considered to be an important organelle in which all cellular processes, such as DNA replication, transcription, translation, post-translational modification, catabolism and anabolism, occur. Each of these processes is bacterial in nature and potentially a drug target.
5.1. Drugs targeting apicoplast DNA replication
Replication of apicoplast DNA occurs in a rolling circular mechanism initiated at sites outside the inverted repeat. However, the inverted repeat is not coded by the apicoplast genome itself. That nuclear gyrase B (GyrB) encodes a functional subunit of the P. falciparum gyrase involved in apicoplast DNA replication has been clear for some time.
Fluro quinolene ciprofloxacine is a selective and target- specific inhibitor of prokaryote-type DNA gyrase, which stabilizes DNA complexes that are nicked. The coding sequences for two recognized apicoplast-targeted proteins, gyrase A (GyrA) and GyrB, have been identified from genomic information on P. falciparum [25] and suggest that the apicoplast utilizes these molecules for its DNA replication. The GyrA subunit contains DNA cleavage and wrapping domains whereas the GyrB subunit contains adenosine triphosphase (ATPase) and DNA binding/GyrA interaction domains, exhibiting strong homology to their Escherichia coli (E. coli) counterparts.
The aminocoumarin drug novobiocin, which inhibits the ATPase activity of GyrB in E. coli, was shown to specifically and competitively inhibit the apicoplast’s ATPase activity and the DNA replication in P. falciparum, leading to a lag in trophozoite-to-schizont conversion [26]. Ciprofloxacin has been shown to inhibit apicoplast DNA replication in P. falciparum, sparing the nuclear replication of the parasite [27]. Novobiocin also inhibits PfGyrB by disrupting the activity of its ATPase, which causes parasite death in cultures. On the other hand, it also reduces the apicoplast/nuclear DNA ratio, which indicates that the drug targets apicoplast DNA replication [28]. Several derivatives of quinolones and fluoro quinolones have also been shown to have anti- parasiticidal activity against Toxoplasma gondii, further supporting apicoplast DNA replication as the drug target of choice in the parasite [29].
Recent studies have revealed that apicoplasts are abnormal in the progeny of doxycycline-treated parasites, as evidenced by a block in apicoplast genome replication and a failure to elongate and segregate during schizogeny. Clindamycin and functionally-related compounds also inhibit plastid replication upon drug application, as revealed by the work reported in Refs. [30, 31].
5.2. Drugs targeting apicoplast transcription
Transcription in plastids utilizes an RNA polymerase homologous to that of cyanobacteria and other eubacteria. Like a bacterial genome, the polymerase of an apicoplast recognises − 10, − 35 promoters and transcribes polycistronic RNAs from operons. The apicoplast genome also encodes the β and the β' subunits of the polymerase [RNA polymerase gene (Rpo) RpoB, RpoC1, and RpoC2 genes], which strongly suggests that it uses a transcription system similar to that of bacteria [32].
The eubacterial form of transcription is extremely sensitive to the anti-biotic rifampin, showing anti-malarial activity both in vitro and in vivo. Furthermore, rifampin has been shown to be selective in targeting transcripts of apicoplast- encoded genes. The α2, β polymerase of an apicoplast is highly sensitive to rifampin, and the anti-malarial activity of rifampin suggests that this drug blocks apicoplast transcription.
A phage-like RNA polymerase was recently identified in P. falciparum [33]. However, it is believed to be the mitochondrial polymerase. No drugs that are specific to phagetype polymerase are known to us. The apicoplast genome encodes the RpoB, RpoC1 and RpoC2 subunits of an RNA polymerase. Indirect transciption inhibition by blocking translation of these RNA polymerase subunits seemed promising to several researchers. In that approach, the natural cyclic oligopeptide anti-biotic thiostrepton was found to reduce the expressions of RpoB and RpoC [34]. Similarly, anti-biotic doxycycline also appeared to reduce the expressions of those two subunits and to inhibit the synthesis of nucleotides and deoxynucleotides [35].
The anti-biotic rifampin (or rifadin) also reduces the expression of RpoB and RpoC transcripts without the delayed- death effect [36]. However, rifampin does not target translation of RNA polymerase subunits, but rather transcription by binding to RNA polymerase. Then, why does targeting different elements of transcription/translation result in different death phenotypes? The answer may lie in the off-apicoplast targets of some of the drugs like thiostrepton. However, another explanation might lie in the extra-apicoplast utilization of apicoplast-borne RNA.
5.3. Drugs targeting apicoplast translation machinery
At present, no direct evidence for translation in the apicoplast exists. However, ribosome-like particles, which have sizes similar to those of bacteria and polysomes containing plastid rRNA and messenger RNA (mRNA), can be obtained from the erythrocyte stages of parasites. This intervention provides indirect evidence that apicoplast genes are certainly translated [37]. Recently, a single putative organellar Palsomdium falciparfum initiation factor (PfIF)-1 was found to be targeted at the apicoplast. This typical translation initiation factor interacts with ribosome and exhibits nucleic-acid binding activity, which mediates transcription anti-termination. This suggests a prominent functional role for PfIF-1 in the apicoplast genome, indicating that PfIF-1 may be a good target for a drug [38].
Numerous drugs have been developed to target the protein synthesis pathways of apicoplasts. In P. falciparum, most anti-biotics instigate delayed death by inhibiting translation and specifically interacting with the apicoplast’s ribosome. Their mode of action is associated with the prevention of nascent peptide elongation, mostly interfering with the peptide exit tunnel formed within ribosomal subunits [39, 40, 41].
A complete review of nuclear, apicoplastic, and mitochondrial translation, which summarizes the current efforts in understanding protein translation in Plasmodium, has recently been published [42]. Lincosamides and macrolides inhibit protein synthesis by binding to the peptidyl transferase domain of the prokaryote-type 23s synthesis after binding to the peptidyl transferase domain of the prokaryote-type 23s RNA. Two thiopetides, such as thiostrepton, micrococcinetc., also bind to 23s rRNAs, specially with the GTPase domain, where they interact with key nucleotides. Thus, P. falciparum has shown high sensitivity to thiostrepton and micrococcin.
Studies examining the effect of drugs that target apicoplast ‘housekeeping’ processes, like replication, transcription and translation, have revealed an unfamiliar aspect of apicomplexan biology, which is called ‘delayed death’ [30, 42], where daughter cells find a new infection but fail to grow and produce further progeny. When growing in human cells, Plasmodium sp. undergoes a complex division process to produce multiple daughter cells within a single parasitophorous vacuole. These daughter cells are released by falling out of the vacuole and invading new cells. When treated with compounds inhibiting apicoplast genome replication and transcription, protein translation, post-translation modification or protein turnover, the parasites continue to grow, divide and release daughter cells that are still capable of invading new host cells [43, 44]. The effect is identical to the growth pattern of apicoplast segregation mutants lacking an apicoplast. This suggests that ‘delayed death’ relates to the function of the apicoplast and not simply to accessibility of the drug to the parasites. In P. falciparum, thecase of delayed death is less clear. Chloramphenicol does not have a delayed effect in Plasmodium as compared with Toxoplasma gondii. However, thiostreptom is reported to be a slow-acting drug.
5.4. Drugs that target fatty acid biosynthesis
The presence of a distinct and prokaryotic-like pathway for the biosynthesis of fatty acids in parasites offers tremendous potential for parasite-selective drugs. Bacteria and chloroplasts synthesize fatty acids by using a type-II fatty acid synthase (FAS)-II. This utilizes several dissimilar enzymes (proteins) for the steps in the FAS biosynthetic pathway. Emerging evidence indicates that the apicoplast harbors a type-II FAS machinery. This anabolic pathway may be involved in the synthesis of the lipids required by the apicoplast. A suggestion is that P. falciparum synthesizes fatty acids to form a parasitophorous vacuole (PV) during invasion of host erythrocytes, and the requirement for apicoplast activity for successful infection is consistent with this postulate.
The first breakthrough in the identification of a fatty acid biosynthesis pathway was the identification of genes encoding apicoplasts that were homologues of several bacterial type-II fatty acid biosynthesis sub-units. One of these sub-units, β-ketoacyl-acyl carrier protein (ACP) synthase (FabH), is targeted by the anti-biotic thiolactomycin, showing in vitro inhibition of Plasmodium. Another type- II FAS sub-unit, enoyl-ACP reductase (FabI), has also been identified in the P. falciparum apicoplast [45]. Thiolactomycin is drug that inhibits type-II fatty acid synthetase, and several analogs of this have shown almost five-fold greater efficacy.
Cerulenin targets β-ketoacyl-ACP sythetase I and II and inhibits Plasmodium [46]. Furthermore, triclosan is another drug that is selective by targeting FabI and that has shown efficacy as an anti-malarial in a mouse model. Recently, triclosan was shown to inhibit purified P. falciparum enoyl-ACP reductase, as well as parasite growth, in vitro and in vivo. However, similarities between the FabI proteins of E. coli and P. falciparum suggest that a similar mutation might result in triclosan resistance in P. falciparum, thereby compromising the usefulness of triclosan as an anti-malarial [47]. Recent studies have shown that FASII is not a suitable target for drug design because the drugs targeting FASII were found to be effective only in in vitro, not in in vitro, due to different patterns of essentiality [42, 45, 48, 49].
5.5. Drugs that target isopenetyl diphosphate (IPP) synthesis
Isopentenyl diphosphate (IPP) is the precursor for a structurally-diverse isoprenoid class of compounds. In animals and plants, IPP is synthesized via the classical mevalonate pathway. An alternate pathway proceeding via 1-deoxy-D-xylulose 5-phosphate (DOXP) has recently been elucidated in chloroplasts and bacteria, and two enzymes from this pathway have been identified in P. falciparum [50]. One of those enzymes, DOXP reductoisomerase, is the target of the anti-biotic fosmidomycin in recombinant P. falciparum. DOXP reductoisomerase is inhibited by fosmidomycin. It also inhibits the growth of P. falciparum in and is known to cure malaria in a mouse model [51, 52]. Assays to test potential inhibitors of DOXP enzymes in bacterial systems and in plant test systems have already exposed new inhibitors of DOXP synthase (the first step of IPP synthesis). Remaining to be seen is whether or not these compounds act against P. falciparum DOXP synthase, and if they do, whether they inhibit parasite growth.
On the other hand, during the blood stages, parasites lacking an apicoplast can grow in the presence of IPP, indicating that it is the only metabolite produced in the apicoplast that is needed outside the organelle. One of the isoprenoid biosynthesis enzymes is predicted to dependon iron-sulfur (FeS) cluster cofactors [53, 54]. Recently, reaserchers have targeted this cofactor as a potential drug target. Anti-malarial drugs like primaquine have been used to kill the gametocyte forms of P. falciparum and the hepatic forms of P. vivax. However, the mechanism of action for this drug is poorly understood. Though its mechanism of action is still poorly understood, recent studies have indicated that the drug might targets Fe-S cluster proteins and, thus, show anti-malarial efficacy [55, 56].
5.6. Drugs that target heme synthesis
Animals are known to synthesize heme from glycine and succinyl CoA via the Shemin pathway. However, plastids use an unusual pathway starting with glutamate ligated to transfer RNA (tRNA)-Glu. The Shemin pathway apparently occurs in P. falciparum, presumably in the mitochondrion and cytosol, but a cyanobacterial-type heme pathway may also exist in the apicoplast. For instance, a likely apicoplast targeted dehydratase (a key enzyme of heme synthesis) has been identified in P. falciparum, and it has groups with predominantly Mg2+ binding plastid dehydratases rather than the Zn2+ binding mitochondrial equivalent [57, 58]. However, another heme synthesis enzyme that bears an apparent mitochondrial-targeting signal has also been identified. Moreover, a recent study has also speculated that P. falciparum uses an additional dehydratase imported from the erythrocyte cytosol.
Localization of the enzymes recognized to date and characterization of the remaining unidentified enzymes of the heme synthesis pathway should help clarify this currently confusing issue. Heme synthesis is an established target for herbicides both in the blood and the liver stages of the parasites, and given that P. falciparum possesses at least one plant-like heme synthesis enzyme, the elucidation of the full pathway should further illuminate potential drug targets [59, 60, 61, 62].
5.7. Recent advances and application of bioinformatics on apicoplast targeted drug designing
The structures of the inhibitors were extracted from the Protein Data Bank (PDB) files of a large subunit of the 70S ribosome of an apicoplast co-complexed with respective inhibitors such as chloramphenicol, clindamycin, erythromycin, azithromycin and thiostrepton. Molecular docking was performed, by using Autodock ver. 4, on ribosome models with ribosome as a receptor to dock the inhibitors of interest [63]. Kollman charges were assigned with 40 × 40 × 40 grid points of 0.375 Å spacing. One hundred Autodock runs were performed for each inhibitor. To validate the docking program, we used Autodock ver. 4 to dock the inhibitor co-complexed with a template in which the limit of Autodock to read the maximum number of atoms in the macromolecules was kept constant at the default value; therefore, 35 Å around the ligands was considered only after superimposing the modelled structure on the E. coli ribosome structure [64].
On the other hand, apicoplast-targeted tRNA guanine transglycosylase and its potential inhibitors were also designed by using docking and simulation studies [65]. In this field, computational models have also been generated, and on the basis of which apicoplast-targeted proteins were identified, target-specific drugs were designed [66].
6. Future aspects
Anti-malarial drug design targeting apicoplasts, like organelles, has enetered a new dimension in the development of parasitical drugs. Genetic evidence confirmed that apicoplast FAS is not essential in vivo. Thus, a clearer understanding of how human malaria parasites respond to loss of FAS in the liver stage is needed before this pathway can be enlisted as a viable drug target. The heme biosynthesis pathway is biologically fascinating as it shows a curious nature in apicomplexan parasites. During their evolution, apicomplexans have combined the Shemin pathway. This pathway involves all the enzymatic reactions in the mitochondrion and cytosol with the plastid localized pathway found in plants to finally form a chimeric pathway where synthesis begins in the mitochondrion, proceeds through the cytosol and apicoplasts, and is eventually completed in the mitochondrion. Whether apicoplast-deficient parasites remain viable in their sexual or liver stages has not yet been determioned. Therefore, interpreing the role of heme synthesis in these life stages is difficult, but again, heme synthesis may play a vital a role in gametes and liver-stage parasites. Considering this, one can appreciably state that heme synthesis may be an important drug target for transmission blocking and prophylaxis.
The malaria parasite can be killed both by inhibitors like fosmidomycin, which is known to have a direct action on isoprenoid precursor biosynthesis, and by many other common anti-biotics, which act indirectly by disrupting the viability of the apicoplast itself. Beside this, dual targeting drugs have been designed and applied. A dual targeted aminoacyl-tRNA synthase in P. falciparum changes both the cytosolic and the apicoplast tRNACys and inhibits the growth of the parasite [67, 68]. At present, research on apicoplasts has established a detailed prediction of an apicoplast metabolism in Plasmodium sp based on a bioinformatics approach [63-66]. Furthermore, metabolic maps, the functional significance of the apicoplast, and the importance of the organelle to the parasite have been analyzed extensively. Researchers have identified the malaria plastid as having a broad range of potential new chemotherapeutic target options in combating malaria. Already, a large number of anti-malarial compounds with assumed targets in the apicoplast have been identified. If these target options are successful in trials, that would be a huge advancement in the field of drug development. Progress has been rapid due to the availability of genome data for the apicoplast [21, 22, 42, 69].
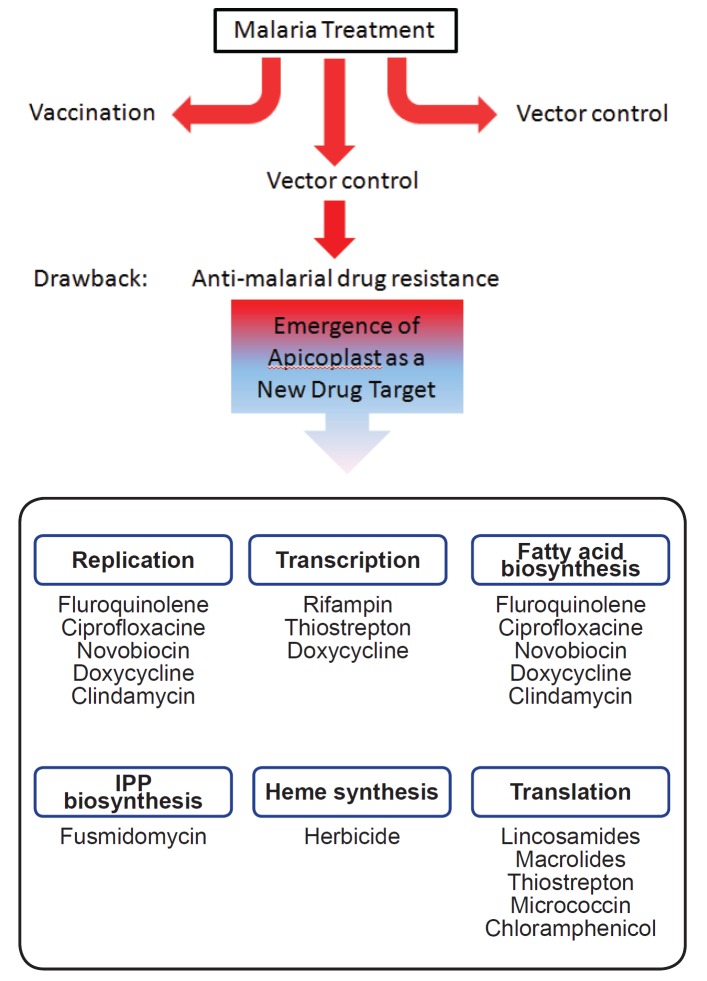
In conclusion, the mode of action and the potential mechanisms of resistance to anti-malarial drugs, such as chloroquine, atovaquone, mefloquine, pyrimethamine, cycloguanil, artemisinin etc. [7, 8, 9, 11], have been deciphered. However, the development of new anti-malarial drugs targeting apicoplasts may represent a significant breakthrough in drug development, provided future clinical trials are successful (Fig. 1). Now, we are in a sphere of activities where a new system of drug development using apicoplasts as a targets in Plasmodium can be seen on the horizon. This review has discussed the immense opportunities that are at hand to neutralize the curse of malaria on human lives, but further trials are needed if the unstinted benefits of these new drugs are to be harvested.
Fig. 1. Schematic representation of apicoplast-targeted anti-malarial drugs and their modes of action.
IPP, Isopentenyl diphosphate.
Footnotes
Conflict of interest The authors declare that there are no conflict of interest.
References
- 1.World Health Organization. World Malaria Report 2015. World Health Organization; India: 2015. [[Cited 2015]]. Available from: http://www.who.int/wer/2015/wer9045.pdf. [Google Scholar]
- 2.Hayton K, Su XZ. Drug resistance and genetic mapping in Plasmodium falciparum. Curr Genet. 2008;54(5):223–239. doi: 10.1007/s00294-008-0214-x. [DOI] [PubMed] [Google Scholar]
- 3.Gleeson MT. The plastid in Apicomplexa: what use is it? Int J Parasitol. 2000;30(10):1053–1070. doi: 10.1016/S0020-7519(00)00100-4. [DOI] [PubMed] [Google Scholar]
- 4.Bouchut A, Geiger JA, Derocher AE, Parsons M. Vesicles bearing Toxoplasma apicoplast membrane proteins persist following loss of the relict plastid or Golgi body disruption. PLoS One. 2014;9(11):e112096. doi: 10.1371/journal.pone.0112096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kalanon M, McFadden GI. Malaria, Plasmodium falciparum and its apicoplast. Biochem Soc Trans. 2010;38(3):775–782. doi: 10.1042/BST0380775. [DOI] [PubMed] [Google Scholar]
- 6.Ralph SA, van Dooren, GG, Waller RF, Crawford MJ, Fraunholz MJ, Foth BJ et al. Tropical infectious diseases: metabolic maps and functions of the Plasmodium falciparum apicoplast. Nat Rev Microbiol. 2004;2(3):203–216. doi: 10.1038/nrmicro843. [DOI] [PubMed] [Google Scholar]
- 7.White NJ. Antimalarial drug resistance. J Clin Invest. 2004;113(8):1084–1092. doi: 10.1172/JCI21682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cowman AF, Galatis D, Thompson JK. Selection for mefloquine resistance in Plasmodium falciparum is linked to amplification of the pfmdr1 gene and cross-resistance to halofantrine and quinine. Proc Natl Acad Sci USA. 2004;91(3):1143–1147. doi: 10.1073/pnas.91.3.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Plowe CV. Monitoring antimalarial drug resistance: making the most of the tools at hand. J Exp Biol. 2003;206(21):3745–3752. doi: 10.1242/jeb.00658. [DOI] [PubMed] [Google Scholar]
- 10.Arisue N, Hashimoto T. Phylogeny and evolution of apicoplasts and apicomplexan parasites. Parasitol Int. 2015;64(3):254–259. doi: 10.1016/j.parint.2014.10.005. [DOI] [PubMed] [Google Scholar]
- 11.Miotto O, Amato R, Ashley EA, MacInnis B, Almagro- Garcia, J, Amaratunga C et al. Genetic architecture of artemisinin-resistant Plasmodium falciparum. Nat Genet. 2015;47(3):226–234. doi: 10.1038/ng.3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chandey M, Mannan R, Bhasin TS, Manjari M. Quinine- resistant severe falciparum malaria in north India: documentation. Indian Journal of Medical Specialities. 2013;4(1):99–102. doi: 10.7713/ijms.2012.0063. [DOI] [Google Scholar]
- 13.Awab GR, Pukrittayakamee S, Imwong M, Dondorp AM, Woodrow CJ, Lee SJ et al. Dihydroartemisinin-piperaquine versus chloroquine to treat vivax malaria in Afghanistan: an open randomized, non-inferiority, trial. Malar J. 2010;9:105. doi: 10.1186/1475-2875-9-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Farooq U, Mahajan RC. Drug resistance in malaria. J Vector Borne Dis. 2004;41(3-4):45–53. [PubMed] [Google Scholar]
- 15.Eastman RT, Dharia NV, Winzeler EA, Fidock DA. Piperaquine resistance is associated with a copy number variation on chromosome 5 in drug-pressured Plasmodium falciparum parasites. Antimicrob Agents Chemother. 2011;55(8):3908–3916. doi: 10.1128/AAC.01793-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barnes KI, Little F, Smith PJ, Evans A, Watkins WM, White NJ. Sulfadoxine-pyrimethamine pharmacokinetics in malaria: pediatric dosing implications. Clin Pharmacol Ther. 2006;80(6):582–596. doi: 10.1016/j.clpt.2006.08.016. [DOI] [PubMed] [Google Scholar]
- 17.Dondorp AM, Nosten F, Yi P, Das D, Phyo AP, Tarning J et al. Artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med. 2009;361(5):455–467. doi: 10.1056/NEJMoa0808859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Imwong M, Dondorp AM, Nosten F, Yi P, Mungthin M, Hanchana S et al. Exploring the contribution of candidate genes to artemisinin resistance in Plasmodium falciparum. Antimicrob Agents Chemother. 2010;54(7):2886–2892. doi: 10.1128/AAC.00032-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heuchert A, Abduselam N, Zeynudin A, Eshetu T, Löscher T, Wieser A et al. Molecular markers of anti- malarial drug resistance in southwest Ethiopia over time: regional surveillance from 2006 to 2013. Malar J. 2015;14:208. doi: 10.1186/s12936-015-0723-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liting L, Geoffrey IM. The evolution, metabolism and functions of the apicoplast. Philos Trans R Soc Lond B Biol Sci. 2010;365(1541):749–763. doi: 10.1098/rstb.2009.0273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Macrae JI, Maréchal E, Biot C, Botté CY. The apicoplast: a key target to cure malaria. Curr Pharm Des. 2012;18(24):3490–3504. doi: 10.2174/138161212801327275. [DOI] [PubMed] [Google Scholar]
- 22.Stuart A. Ralph, Marthe C. D’Ombrain, Geoffrey I. Mc- Fadden. The apicoplast as an antimalarial drug target. Drug Resistance Updates. 2001;4(3):145–151. doi: 10.1054/drup.2001.0205. [DOI] [PubMed] [Google Scholar]
- 23.Abrahamsen MS, Templeton TJ, Enomoto S, Abrahante JE, Zhu G, Lancto CA et al. Complete genome sequence of the apicomplexan, Cryptosporidium parvum. Science. 2004;304(5669):441–445. doi: 10.1126/science.1094786. [DOI] [PubMed] [Google Scholar]
- 24.Moore RB, Oborník M, Janouskovec J, Chrudimský T, Vancová M, Green DH et al. A photosynthetic alveolate closely related to apicomplexan parasites. Nature. 2008;451(7181):959–963. doi: 10.1038/nature06635. [DOI] [PubMed] [Google Scholar]
- 25.Dar MA, Sharma A, Mondal N, Dhar SK. Molecular cloning of apicoplast-targeted Plasmodium falciparum DNA gyrase genes: unique intrinsic ATPase activity and ATP-independent dimerization of PfGyrB subunit. Eukaryot Cell. 2007;6(3):398–412. doi: 10.1128/EC.00357-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Raghu Ram, EV, Kumar A, Biswas S, Kumar A, Chaubey S, Siddiqi MI et al. Nuclear gyrB encodes a functional subunit of the Plasmodium falciparum gyrase that is involved in apicoplast DNA replication. Mol Biochem Parasitol. 2007;154(1):30–39. doi: 10.1016/j.molbiopara.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 27.Goodman CD, Su V, McFadden GI. The effects of anti- bacterials on the malaria parasite Plasmodium falciparum. Mol Biochem Parasitol. 2007;152(2):181–191. doi: 10.1016/j.molbiopara.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 28.Singh D, Kumar A, Raghu Ram, EV, Habib S. Multiple replication origins within the inverted repeat region of the Plasmodium falciparum apicoplast genome are differentially activated. Mol Biochem Parasitol. 2005;139(1):99–106. doi: 10.1016/j.molbiopara.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 29.Onodera Y, Tanaka M, Sato K. Inhibitory activity of quinolones against DNA gyrase of Mycobacterium tuberculosis. J Antimicrob Chemother. 2001;47(4):447–450. doi: 10.1093/jac/47.4.447. [DOI] [PubMed] [Google Scholar]
- 30.Dahl EL, Rosenthal PJ. Multiple antibiotics exert delayed effects against the Plasmodium falciparum apicoplast. Antimicrob Agents Chemother. 2007;51(10):3485–3490. doi: 10.1128/AAC.00527-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goodman CD, McFadden GI. Ycf93 (Orf105), a small apicoplast-encoded membrane protein in the relict plastid of the malaria parasite Plasmodium falciparum that is conserved in Apicomplexa. PLoS One. 2014;9(4):e91178. doi: 10.1371/journal.pone.0091178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gray MW, Lang BF. Transcription in chloroplasts and mitochondria: a tale of two polymerases. Trends Microbiol. 1998;6(1):1–3. doi: 10.1016/S0966-842X(97)01182-7. [DOI] [PubMed] [Google Scholar]
- 33.Li J, Maga JA, Cermakian N, Cedergren R, Feagin JE. Identification and characterization of a Plasmodium falciparum RNA polymerase gene with similarity to mitochondrial RNA polymerases. Mol Biochem Parasitol. 2001;113(2):261–269. doi: 10.1016/S0166-6851(01)00223-7. [DOI] [PubMed] [Google Scholar]
- 34.McConkey GA, Rogers MJ, McCutchan TF. Inhibition of Plasmodium falciparum protein synthesis. targeting the plastid-like organelle with thiostrepton. J Biol Chem. 1997;272(4):2046–2049. doi: 10.1074/jbc.272.4.2046. [DOI] [PubMed] [Google Scholar]
- 35.Dahl EL, Shock JL, Shenai BR, Gut J, DeRisi JL, Rosenthal PJ. Tetracyclines specifically target the apicoplast of the malaria parasite Plasmodium falciparum. Antimicrob Agents Chemother. 2006;50(9):3124–3131. doi: 10.1128/AAC.00394-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pradel G, Schlitzer M. Antibiotics in malaria therapy and their effect on the parasite apicoplast. Curr Mol Med. 2010;10(3):335–349. doi: 10.2174/156652410791065273. [DOI] [PubMed] [Google Scholar]
- 37.Chaubey S, Kumar A, Singh D, Habib S. The apicoplast of Plasmodium falciparum is translationally active. Mol Microbiol. 2005;56(1):81–89. doi: 10.1111/j.1365-2958.2005.04538.x. [DOI] [PubMed] [Google Scholar]
- 38.Haider A, Allen SM, Jackson KE, Ralph SA, Habib S. Targeting and function of proteins mediating translation initiation in organelles of Plasmodium falciparum. Mol Microbiol. 2015;96(4):796–814. doi: 10.1111/mmi.12972. [DOI] [PubMed] [Google Scholar]
- 39.Gupta A, Mir SS, Jackson KE, Lim EE, Shah P, Sinha A et al. Recycling factors for ribosome disassembly in the apicoplast and mitochondrion of Plasmodium falciparum. Mol Microbiol. 2013;88(5):891–905. doi: 10.1111/mmi.12230. [DOI] [PubMed] [Google Scholar]
- 40.Budimulja AS, Syafruddin, Tapchaisri P, Wilairat P, Marzuki S. The sensitivity of Plasmodium protein synthesis to prokaryotic ribosomal inhibitors. Mol Biochem Parasitol. 1997;84(1):137–141. doi: 10.1016/S0166-6851(96)02781-8. [DOI] [PubMed] [Google Scholar]
- 41.Lemgruber L, Kudryashev M, Dekiwadia C, Riglar DT, Baum J, Stahlberg H et al. Cryo-electron tomography reveals four-membrane architecture of the Plasmodium apicoplast. Malar J. 2013;12:25. doi: 10.1186/1475-2875-12-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Botté CY, Dubar F, McFadden GI, Maréchal E, Biot C. Plasmodium falciparum apicoplast drugs: targets or off-targets? Chem Rev. 2012;112(3):1269–1283. doi: 10.1021/cr200258w. [DOI] [PubMed] [Google Scholar]
- 43.Lindner J, Meissner KA, Schettert I, Wrenger C. Trafficked proteins-druggable in Plasmodium falciparum? Int J Cell Biol. 2013;2013:e435981. doi: 10.1155/2013/435981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sahu R, Walker LA, Tekwani BL. In vitro and in vivo anti-malarial activity of tigecycline, a glycylcycline antibiotic, in combination with chloroquine. Malar J. 2014;13:414. doi: 10.1186/1475-2875-13-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shears MJ, Botté CY, McFadden GI. Fatty acid metabolism in the Plasmodium apicoplast: Drugs, doubts and knockouts. Mol Biochem Parasitol. 2015;199(1-2):34–50. doi: 10.1016/j.molbiopara.2015.03.004. [DOI] [PubMed] [Google Scholar]
- 46.Waller RF, Keeling PJ, Donald RGK, Striepen B, Handman E, Lang-Unnasch N et al. Nuclear-encoded proteins target to the plastid in Toxoplasma gondii and Plasmodium falciparum. Proc Natl Acad Sci USA. 1998;95(21):12352–12357. doi: 10.1073/pnas.95.21.12352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McLeod R, Muench SP, Rafferty JB, Kyle DE, Mui EJ, Kirisits MJ et al. Triclosan inhibits the growth of Plasmodium falciparum and Toxoplasma gondii by inhibition of apicomplexan Fab I. Int J Parasitol. 2001;31(2):109–113. doi: 10.1016/S0020-7519(01)00111-4. [DOI] [PubMed] [Google Scholar]
- 48.Haussig JM, Matuschewski K, Kooij TW. Inactivation of a Plasmodium apicoplast protein attenuates formation of liver merozoites. Mol Microbiol. 2011;81(6):1511–1525. doi: 10.1111/j.1365-2958.2011.07787.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Qidwai T, Priya A, Khan NA, Tripathi H, Khan F, Darokar MP et al. Antimalarial drug targets and drugs targeting dolichol metabolic pathway of Plasmodium falciparum. Curr Drug Targets. 2014;15(4):374–409. doi: 10.2174/13894501113149990169. [DOI] [PubMed] [Google Scholar]
- 50.Lichtenthaler HK, Schwender J, Disch A, Rohmer M. Biosynthesis of isoprenoids in higher plant chloroplasts proceeds via a mevalonate-independent pathway. FEBS Letters. 1997;400(3):271–274. doi: 10.1016/S0014-5793(96)01404-4. [DOI] [PubMed] [Google Scholar]
- 51.Baumeister S, Wiesner J, Reichenberg A, Hintz M, Bietz S, Harb OS et al. Fosmidomycin uptake into Plasmodium and Babesia-infected erythrocytes is facilitated by parasite-induced new permeability pathways. PLoS One. 2011;6(5):e19334. doi: 10.1371/journal.pone.0019334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jomaa H, Wiesner J, Sanderbrand S, Altincicek B, Weidemeyer C, Hintz M et al. Inhibitors of the nonmevalonate pathway of isoprenoid biosynthesis as antimalarial drugs. Science. 1999;285(5433):1573–1576. doi: 10.1126/science.285.5433.1573. [DOI] [PubMed] [Google Scholar]
- 53.Gisselberg JE, Dellibovi-Ragheb TA, Matthews KA, Bosch G, Prigge ST. The suf iron-sulfur cluster synthesis pathway is required for apicoplast maintenance in malaria parasites. PLoS Pathog. 2013;9(9):e1003655. doi: 10.1371/journal.ppat.1003655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dellibovi-Ragheb TA, Gisselberg JE, Prigge ST. Parasites FeS up: iron-sulfur cluster biogenesis in eukaryotic pathogens. PLoS Pathog. 2013;9(4):e1003227. doi: 10.1371/journal.ppat.1003227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Seliverstov AV, Zverkov OA, Istomina SN, Pirogov SA, Kitsis PS. Comparative analysis of apicoplast-targeted protein extension lengths in apicomplexan parasites. Biomed Res Int. 2015;2015:e452958. doi: 10.1155/2015/452958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lalève A, Vallières C, Golinelli-Cohen MP, Bouton C, Song Z, Pawlik G et al. The antimalarial drug primaquine targets Fe-S cluster proteins and yeast respiratory growth. Redox Biol. 2016;7:21–29. doi: 10.1016/j.redox.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wilson CM, Smith AB, Baylon RV. Characterization of the delta-aminolevulinate synthase gene homologue in P. falciparum. Mol Biochem Parasitol. 1996;75(2):271–276. doi: 10.1016/0166-6851(95)02531-6. [DOI] [PubMed] [Google Scholar]
- 58.Surolia N, Pasmanaban G. de novo biosynthesis of heme offers a new chemotherapeutic target in the human malarial parasite. Biochem Biophys Res Commun. 1992;187(2):744–750. doi: 10.1016/0006-291x(92)91258-r. [DOI] [PubMed] [Google Scholar]
- 59.Bonday ZQ, Dhanasekaran S, Rangarajan PN, Padmanaban G. Import of host delta-aminolevulinate dehydratase into the malarial parasite: identification of a new drug target. Nat Med. 2000;6(8):898–903. doi: 10.1038/78659. [DOI] [PubMed] [Google Scholar]
- 60.Ke H, Sigala PA, Miura K, Morrisey JM, Mather MW, Crowley JR et al. The heme biosynthesis pathway is essential for Plasmodium falciparum development in mosquito stage but not in blood stages. J Biol Chem. 2014;289(50):34827–34837. doi: 10.1074/jbc.M114.615831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nagaraj VA, Sundaram B, Varadarajan NM, Subramani PA, Kalappa DM, Ghosh SK et al. Malaria parasite-synthesized heme is essential in the mosquito and liver stages and complements host heme in the blood stages of infection. PLoS Pathog. 2013;9(8):e1003522. doi: 10.1371/journal.ppat.1003522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yeh E, DeRisi JL. Chemical rescue of malaria parasites lacking an apicoplast defines organelle function in blood-stage Plasmodium falciparum. PLoS Biol. 2011;9(8):e1001138. doi: 10.1371/journal.pbio.1001138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS et al. AutoDock4 and AutoDock- Tools4: automated docking with selective receptor flexibility. J Comput Chem. 2009;30(16):2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gupta A, Shah P, Haider A, Gupta K, Siddiqi MI, Ralph SA et al. Reduced ribosomes of the apicoplast and mitochondrion of Plasmodium spp. and predicted interactions with antibiotics. Open Biol. 2014;4(5):e140045. doi: 10.1098/rsob.140045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sawhney B, Chopra K, Misra R, Ranjan A. Identification of Plasmodium falciparum apicoplast-targeted tRNA-guanine transglycosylase and its potential inhibitors using comparative genomics, molecular modelling, docking and simulation studies. J Biomol Struct Dyn. 2015;33(11):2404–2420. doi: 10.1080/07391102.2015.1040074. [DOI] [PubMed] [Google Scholar]
- 66.Cilingir G, Broschat SL, Lau AO. ApicoAP: the first computational model for identifying apicoplast-targeted proteins in multiple species of Apicomplexa. PLoS One. 2012;7(5):e36598. doi: 10.1371/journal.pone.0036598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pham JS, Sakaguchi R, Yeoh LM, De Silva, NS, McFadden GI, Hou YM et al. A dual-targeted aminoacyl-tRNA synthetase in Plasmodium falciparum charges cytosolic and apicoplast tRNACys. Biochem J. 2014;458(3):513–523. doi: 10.1042/BJ20131451. [DOI] [PubMed] [Google Scholar]
- 68.Jackson KE, Pham JS, Kwek M, De Silva, NS, Allen SM, Goodman CD et al. Dual targeting of aminoacyl-tRNA synthetases to the apicoplast and cytosol in Plasmodium falciparum. Int J Parasitol. 2012;42(2):177–186. doi: 10.1016/j.ijpara.2011.11.008. [DOI] [PubMed] [Google Scholar]
- 69.McFadden GI, Roos DS. Apicomplexan plastids as drug targets. Trends Microbiol. 1999;7(8):328–333. doi: 10.1016/S0966-842X(99)01547-4. [DOI] [PubMed] [Google Scholar]