Abstract
Previous studies have indicated that miR-200c is a promising cancer biomarker. However, different studies have presented conflicting results. Therefore, the aim of the present study was to perform a meta-analysis of miR-200c based on 34 relevant studies. The Materials and methods sections of papers were carefully identified using the databases PubMed, Web of Science and Embase for publications up to December 4, 2015. Pooled hazard ratios (HRs) and 95% confidence intervals (95% CIs) were systematically calculated to investigate the association between the expression of miR-200c and cancer prognosis. The results demonstrated that elevated expression levels of miR-200c indicated significantly worse overall survival rates (HR=1.37, 95% CI: 1.01, 1.85), and a high level of miR-200c was considered an indicator of an unfavorable prognosis in patients from Europe and America (HR=1.85, 95% CI: 1.27, 2.69). Furthermore, overexpression of miR-200c was significantly associated with progression of the disease in the subgroups of tissue and blood samples (HR=0.68 and 2.45, respectively), and inferior overall survival rates for the blood subgroup were revealed (HR=2.21, 95% CI: 1.04, 4.72). In addition, miR-200c was of prognostic value in several disease subgroups. Taken together, high expression levels of miR-200c are of significant prognostic value in various human malignancies.
Keywords: cancer, miR-200c, prognosis, hazard ratio, meta-analysis
Introduction
Cancer has become the primary cause of mortality in the majority of countries and regions worldwide, and the incidence of cancer has increased substantially in recent years (1). In 2012, 14.1 million new cancer cases, 8.2 million cancer mortalities and 32.6 million individuals living with cancer (within 5 years of diagnosis) were reported worldwide. Specifically, 57% (8 million) of new cancer cases, 65% (5.3 million) of cancer mortalities and 48% (15.6 million) of the 5-year prevalent cancer cases occurred in less developed regions (2). Due to the difficulty of early diagnosis and the low survival rate of multiple cancer types, reliable biomarkers that are associated with the diagnosis or prognosis of cancer are urgently required.
MicroRNAs are conserved small non-coding RNAs with a length of ~18–25 nucleotides, which regulate the expression of target genes and exert vital roles in various biological processes (3,4). They were first identified in 1993 (5). Thereafter, an understanding of their roles in the cell cycle, apoptosis, proliferation and differentiation has greatly advanced (6,7). Furthermore, several microRNAs were identified that function as either oncogenes or tumor suppressors, and the expression levels of certain microRNAs were associated with the degree of malignancy (8–10). Due to their good stability and unique expression profiles in human malignancies, microRNAs hold great promise as conceivable biomarkers for cancer diagnosis and prognosis (11,12).
MicroRNA-200c (miR-200c), the most representative member of the microRNA-200 family, has been widely investigated during the last few years. miR-200c was revealed to exert a critical role in the regulation of epithelial-to-mesenchymal transition (EMT) and mesenchymal-to-epithelial transition (MET) (13,14). In addition, there have been numerous studies demonstrating the association between an aberrant expression level of miR-200c and the prognosis of various human malignancies, including endometrial cancer (8,9), gastric cancer (13,15–17), ovarian cancer (18–21), clear cell renal cell carcinoma (ccRCC) (22–24), breast cancer (25–28), colorectal cancer (14,29), non-small cell lung cancer (NSCLC) (30–35), prostate cancer (36), esophageal cancer (37–39), diffuse large B-cell lymphoma (40), bladder cancer (41,42) and pancreatic cancer (43). Approximately half of these studies verified the anti-oncogenic function of miR-200c in certain cancer types, indicating the potential correlation of elevated expression levels of miR-200c and superior prognosis (13–15,18,20–22,28,29,31,35,41–43). However, other studies have provided opposing evidence, suggesting that miR-200c serves as an oncogene (23–27,36–39). Therefore, miR-200c is a noteworthy biomarker for cancer prognosis, and a meta-analysis of its precise role is required. To clarify the value of miR-200c as a prognostic biomarker, data from studies of miR-200c in various cancer types were systematically collected and evaluated.
Materials and methods
Search strategy
Relevant studies were identified by carefully searching the online databases PubMed, Web of Science and Embase up to December 4th, 2015. The following combination of keywords was simultaneously applied for the literature search: ‘microRNA-200c’ or ‘microrna-200c’ or ‘miRNA-200c’ or ‘miR-200c’ and ‘tumor’ or ‘cancer’ or ‘carcinoma’ or ‘neoplasm’ or ‘malignancies’. In addition, the following criteria for the study characteristics were used to improve the search further: i) English language publications; ii) studies that concentrated on patients with malignancies; and iii) studies that demonstrated the association of miR-200c expression with cancer prognosis. This comprehensive online search was independently performed by two authors (Jia-Yi Zhang and Ya-Min Wang).
Inclusion and exclusion criteria
The present meta-analysis was performed strictly following the guidelines of the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) statement (44). Articles were considered eligible if they met the following criteria: i) the expression level of miR-200c had been assessed in tissue or peripheral blood samples from cancer patients; ii) dichotomous categorization of expression levels of miR-200c had been investigated according to a cut-off value; and iii) an investigation had been made of the association of expression levels of miR-200c with survival rates or recurrence, together with a corresponding hazard ratio (HR) or survival curve. If more than one article had been published on the identical study cohort, only the most comprehensive study was selected for the present meta-analysis. In addition, letters, review articles and experiments on animals were excluded. A flow diagram of the study selection process with further details is shown in Fig. 1.
Figure 1.
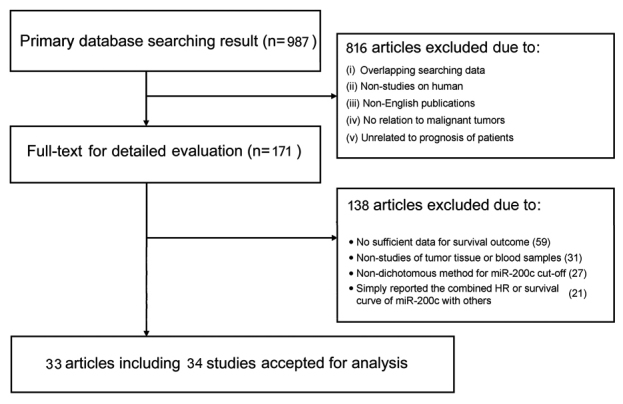
Flow diagram with details of the study selection process.
Data extraction
All eligible studies were identified by Ya-Min Wang and Le-Bin Song, and uncertain data were reassessed by Ning-Hong Song. The data extraction included the following elements: i) the first author and publication year; ii) characteristics of the studied population, including patient nationality, number, mean or median age, disease type and stage, and sample examined; iii) study design, assay method and cut-off definition; iv) HRs of elevated expression levels of miR-200c for cancer-specific survival (CSS), overall survival (OS), recurrence-free survival (RFS), progression-free survival (PFS) and disease-free survival (DFS); and v) mean or median follow-up duration. If HRs were not directly reported in the studies, then the data were extracted from Kaplan-Meier survival plots using Engauge Digitizer v.5.1 (license type: GPL; developed by Mark Mitch; Category: C:Science/CAD) to calculate HRs with 95% confidence intervals (95% CIs) using methods that are previously described (45). Furthermore, if both the univariate and multivariate results were reported, then only the latter was selected, since these results were adjusted for confounding factors. All the above-mentioned data are comprehensively shown in Tables I and II.
Table I.
The predominant characteristics of the included studies.
| Authors, year | Patient's nationality | Patient's number | Mean or median age | Study design | Malignant disease | Disease stage | Detected sample | Mean or median follow-up time | Refs. |
|---|---|---|---|---|---|---|---|---|---|
| Antolín et al, 2015 | Spain | 57 | 55.4 | R | Breast cancer | I–IV | Blood | 264.6OS/235.3PFS weeks | (27) |
| Butz et al, 2015 | Canada | 425 | NM | R | ccRCC | I–IV | Tissue | NM | (24) |
| Song et al, 2015 | China | 134 | 50.0 | R | Breast cancer | I–IV | Tissue | NM | (28) |
| Zhao et al, 2015 | China | 78 | 61.4 | P | NSCLC | IIB-IIIB | Tissue | NM | (35) |
| Zhou et al, 2015 | China | 63 | 65.0 | R | Gastric cancer | IIB-IV | Tissue | NM | (13) |
| Gao et al, 2015 | China | 93 | NM | R | Ovarian cancer | I–IV | Blood | NM | (18) |
| Vergho et al, 2014 | Germany | 37 | 66.8 | R | ccRCC | I–IV | Tissue | 45.6 months | (22) |
| Tuomarila et al, 2014 | Finland | 172 | 60.4 | P | Breast cancer | I–IV | Tissue | 9.7 years | (25) |
| Toiyama et al, 2014 | Japan | 156T/182S | 68.0 | R | Colorectal cancer | I–IV | Both | 20 months | (14) |
| Tejero et al, 2014 | Spain | 155 | 65.0 | R | NSCLC | I–III | Tissue | 43 months | (30) |
| Song et al, 2014 | China | 373 | 60.5 | R | Gastric cancer | I–IV | Tissue | 35 months | (17) |
| Lin et al, 2014 | Australia | 97 | 68.0 | P | Prostate cancer | III–IV | Blood | 12 months | (36) |
| Li et al, 2014 | China | 150 | 59.0 | R | NSCLC | IIIB, IV | Tissue | 16.7 months | (31) |
| Kim et al, 2014 | South Korea | 72 | 64.0 | R | NSCLC | I–IV | Tissue | 31 months | (32) |
| Diaz et al, 2014 | Spain | 127 | 67.4 | R | Colorectal cancer | I–III | Tissue | 113 months | (29) |
| Cao et al, 2014 | China | 100 | 58.0 | R | Ovarian cancer | I–IV | Tissue | 36.8 months | (19) |
| Berghmans et al, 2013 | Belgium | 38 | 59.0 | P | NSCLC | NM | Tissue | NM | (33) |
| Yu et al, 2013 | China | 157 | 60.0 | R | Esophageal cancer | III, IV | Blood | 20 months | (37) |
| Wotschofsky et al, 2013 | Germany | 111 | >60.0 | R | ccRCC | NM | Tissue | NM | (23) |
| Tang et al, 2013 | China | 167 | 60.0 | R | Gastric cancer | I–IV | Tissue | NM | (15) |
| Tanaka et al, 2013 | Japan | 64 | 67.5 | R | Esophageal cancer | II–IV | Blood | NM | (38) |
| Berglund et al, 2013 | Sweden | 61 | >60.0 | R | DLBCL | I–IV | Tissue | NM | (40) |
| Madhavan et al, 2012 | Germany | 193 | NM | R | Breast cancer | III–IV | Blood | NM | (26) |
| Valladares-Ayerbes et al, 2012 | Spain | 52 | 65.3 | P | Gastric cancer | I–IV | Blood | 24 months | (16) |
| Torres et al, 2013 | Poland | 108 | 62.8 | P | Endometrial cancer | I–IV | Tissue | NM | (9) |
| Liu et al, 2012 | China | 70 | 60.0 | P | NSCLC | I–IV | Tissue | NM | (34) |
| Karaayvaz et al, 2012 | USA | 34 | NM | R | Endometrial cancer | I–IV | Tissue | NM | (8) |
| Wszolek et al, 2011 | USA | 57 | >60.0 | R | Bladder cancer | NM | Tissue | 92 months | (41) |
| Marchini et al, 2011 | Italy | 89 | 52.0 | R | Ovarian cancer | I | Tissue | 110 months | (20) |
| Marchini et al, 2011 | Italy | 55 | 57.0 | R | Ovarian cancer | I | Tissue | 108 months | (20) |
| Hamano et al, 2011 | Japan | 98 | >60.0 | R | Esophageal cancer | I–IV | Tissue | 28.8 months | (39) |
| Wiklund et al, 2011 | Denmark | 100 | NM | R | Bladder cancer | I | Tissue | NM | (42) |
| Yu et al, 2010 | Japan | 99 | 65.7 | R | Pancreatic cancer | I–IV | Tissue | NM | (43) |
| Leskelä et al, 2010 | Spain | 72 | 57.0 | R | Ovarian cancer | I–IV | Tissue | NM | (21) |
The study design is described as prospective (P) or retrospective (R) T, tissue sample of patients was assayed; S, serum sample of patients was assayed; ccRCC, clear cell renal cell carcinoma; NSCLC, non-small cell lung cancer; DLBCL, diffuse large B-cell lymphoma; NM, not mentioned; OS, overall survival; PFS, progression-free survival.
Table II.
HRs of included studies.
| HRs | ||||||
|---|---|---|---|---|---|---|
| Authors, year | Main assay of miR200c | Cut-off | Resource of HR | OS/CSS | RFS/PFS/DFS | Refs. |
| Antolín et al, 2015 | RT-qPCR | Mean | Reported | 2.79 | 3.33 | (27) |
| Butz et al, 2015 | RT-qPCR | Mean | Reported | 2.73 | 3.57 | (24) |
| Song et al, 2015 | ISH | Mean | a | 0.18 | 0.19 | (28) |
| Zhao et al, 2015 | RT-qPCR | Median | Reported | – | 0.35 | (35) |
| Zhou et al, 2015 | RT-qPCR | Median | a | – | 0.49 | (13) |
| Gao et al, 2015 | RT-qPCR | Mean | Reported | 0.32 | – | (18) |
| Vergho et al, 2014 | RT-qPCR | 2.73 | Reported | 0.95 | – | (22) |
| Tuomarila et al, 2014 | RT-qPCR | Median | a | 2.78 | 3.16 | (25) |
| Toiyama et al, 2014 | RT-qPCR | Median | Reported | 0.56T/2.67S | 4.51S | (14) |
| Tejero et al, 2014 | RT-qPCR | Mean | a | 1.95 | – | (30) |
| Song et al, 2014 | RT-qPCR | Lowest quartile | Reported | 1.32 | 1.06 | (17) |
| Lin et al, 2014 | RT-qPCR | Median | Reported | 2.30 | – | (36) |
| Li et al, 2014 | RT-qPCR | 0.01385 | Reported | 0.57 | 0.55 | (31) |
| Kim et al, 2014 | RT-qPCR | Mean | Reported | 3.67 | – | (32) |
| Diaz et al, 2014 | RT-qPCR | NM | a | 0.51 | 0.55 | (29) |
| Cao et al, 2014 | RT-qPCR | 3.84 | Reported | 16.22 | – | (19) |
| Berghmans et al, 2013 | RT-qPCR | Median | Reported | 1.51 | – | (33) |
| Yu et al, 2013 | RT-qPCR | Median | Reported | 1.67 | – | (37) |
| Wotschofsky et al, 2013 | RT-qPCR | Median | Reported | – | 1.40 | (23) |
| Tang et al, 2013 | RT-qPCR/ISH | Mean | Reported | 0.40 | 0.51 | (15) |
| Tanaka et al, 2013 | RT-qPCR | Median | Reported | – | 2.79 | (38) |
| Berglund et al, 2013 | RT-qPCR | Mean | a | 2.68 | – | (40) |
| Madhavan et al, 2012 | RT-qPCR | Lower quartile | a | 15.27 | 2.20 | (26) |
| Valladares-Ayerbes et al, 2012 | RT-qPCR | 62.4 | Reported | 2.24 | 2.27 | (16) |
| Torres et al, 2013 | RT-qPCR | Median | Reported | 2.72 | – | (9) |
| Liu et al, 2012 | RT-qPCR | 2.00 | Reported | 6.02 | – | (34) |
| Karaayvaz et al, 2012 | RT-qPCR | 35.5 | a | 1.28 | – | (8) |
| Wszolek et al, 2011 | RT-qPCR | Mean | a | 0.09 | – | (41) |
| Marchini et al, 2011 | RT-qPCR | Median | Reported | 0.24 | 0.42 | (20) |
| Marchini et al, 2011 | RT-qPCR | Median | Reported | 0.09 | 0.04 | (20) |
| Hamano et al, 2011 | RT-qPCR | Median | a | 1.71 | – | (39) |
| Wiklund et al, 2011 | ISH | NM | a | 0.52 | – | (42) |
| Yu et al, 2010 | RT-qPCR | 0.64 | Reported | 0.45 | – | (43) |
| Leskelä et al, 2010 | RT-qPCR | Median | Reported | – | 0.85 | (21) |
The study design is described as prospective (P) or retrospective (R).
Data extracted from the survival curve. In the OS/CSS and RFS/PFS/DFS columns, ‘T’ denotes the HR of miR-200c overexpression in tumor tissue, and ‘S’ denotes the HR of miR-200c overexpression in serum sample; HR, hazard ratio; OS, overall survival; CSS, cancer specific survival; RFS, relapse-free survival; PFS, progression-free survival; DFS, disease-free survival; RT-qPCR, quantitative reverse transcription-polymerase chain reaction; ISH, in situ hybridization.
Statistical analysis
In the present meta-analysis, HRs and corresponding 95% CIs were combined to estimate the value of high expression levels of miR-200c for cancer prognosis. An individual or pooled HR of >1.0 indicated poorer prognosis in patients with miR-200c overexpression, and an HR of <1.0 represented an improved prognosis. Furthermore, a fixed-effects model using the Mantel-Haenszel method or a random-effects model using the DerSimonian-Laird method was applied for the meta-analysis, according to the heterogeneity between the pooled studies (46). Statistical heterogeneity was evaluated by performing the Chi-square test (assessing the P-value) and by calculating the Higgins I2 statistic. If significant heterogeneity was observed (P<0.10 or I2>50%), the random-effects model was applied; otherwise, the fixed-effects model was used. Subgroup analyses were further performed to investigate the source of the identified heterogeneity. In addition, sensitivity analyses were implemented to avoid biases in the results due to certain low-quality studies, and the publication bias was estimated using Begg's and Egger's tests. All P-values were two-sided, and P<0.05 was considered to indicate a statistically significant value. All statistical analyses were conducted using Stata v.12.0 (StataCorp, College Station, Texas, USA), and Microsoft Excel (v.2010, Microsoft Corporation, Redmond, Washington, USA).
Results
Summary of the included studies
In total, 987 articles were initially collected from a primary retrieval using the databases PubMed, Web of Science and Embase. Of these articles, 33 articles that included 34 studies [the article of Marchini et al (20) included independent studies of two different cohorts, tissue collections A and B (20)] were ultimately considered eligible by screening the titles, abstracts and full texts (Fig. 1). Of these studies of the association between expression levels of miR-200c and the survival rate or disease recurrence in human malignancies, 27 were retrospective, and seven were prospective. In total, 3,940 patients from China, Germany, Finland, Japan, Spain, Australia, South Korea, Belgium, Sweden, Poland, USA, Italy or Denmark were included; these patients were diagnosed with a variety of cancer types, including endometrial cancer (8,9), gastric cancer (13,15–17), ovarian cancer (18–21), ccRCC (22–24), breast cancer (25–28), colorectal cancer (14,29), NSCLC (30–35), prostate cancer (36), esophageal cancer (37–39), diffuse large B-cell lymphoma (40), bladder cancer (41,42) and pancreatic cancer (43). Tissue samples were predominantly used to determine expression levels of miR-200c, although six studies detected expression levels of miR-200c in serum or plasma, and one study used tissue and blood samples (14). To assess miRNA-200c expression, quantitative real-time polymerase chain reaction (RT-qPCR) had predominantly been used in 32 studies, and in situ hybridization (ISH) was performed in three studies. The characteristics of primary studies are systematically summarized in Table I.
Association of CSS/OS with miR-200c overexpression
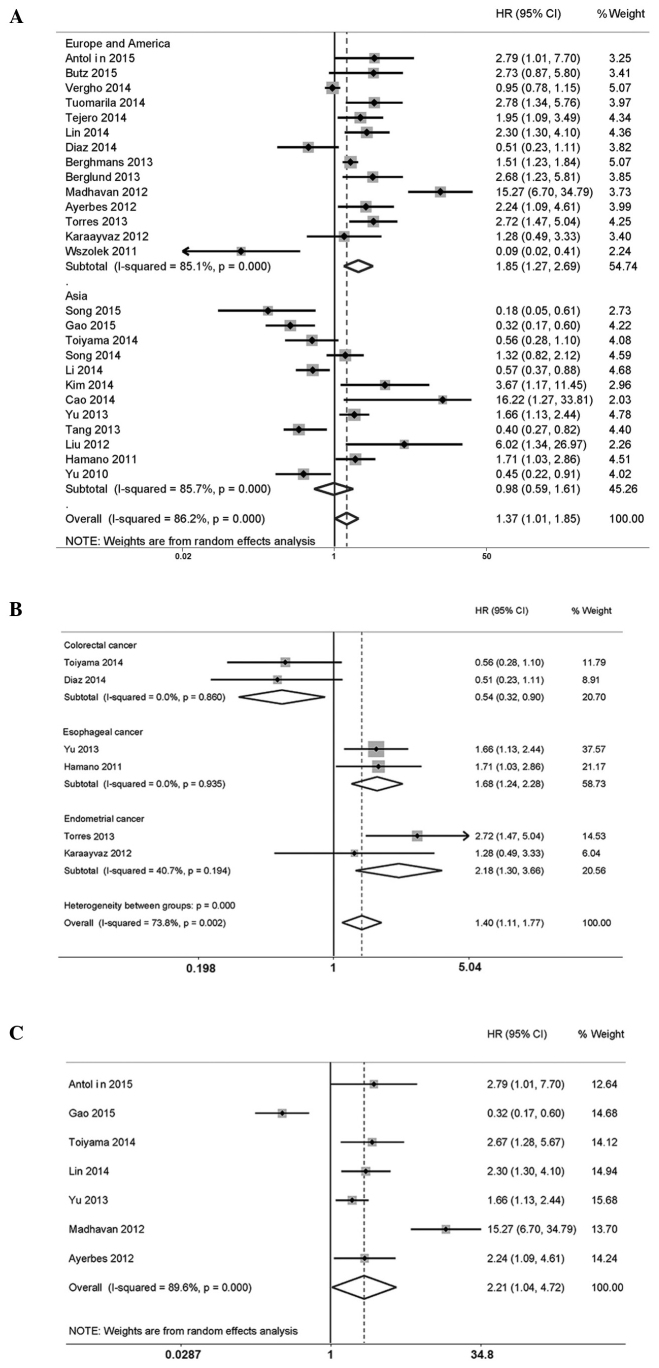
In total, 26 studies were included in the meta-analysis of the association between miR-200c overexpression and CSS/OS, and a random-effects model was applied due to the high level of heterogeneity (P<0.001, I2=86.2%). Three studies were excluded, since they enrolled cancer patients of only stage I (20,42). The pooled value of HRs from individual studies was 1.37 (95% CI: 1.01, 1.85), along with a P-value of 0.040 (Fig. 2A). Therefore, this result indicated a significant correlation of CSS/OS with high expression levels of miR-200c.
Figure 2.
Forest plots of the combined analyses of the association of CSS/OS and expression levels of miR-200c. (A) Forest plots of the pooled analysis of CSS/OS. Squares and horizontal lines correspond to study-specific HRs and 95% CIs, respectively. The area of the squares correlates with the weight, and the diamonds represent the pooled HRs and 95% CIs. (B) Forest plots of the pooled analysis of CSS/OS in different disease type subgroups. (C) Forest plots of the pooled analysis of CSS/OS in blood sample subgroup. CSS, cancer-specific survival; OS, overall survival; HR, hazard ratio; CI, confidence interval.
Furthermore, subgroup analyses were performed on specific study characteristics, including region, disease type and sample detection. All the pooled HRs with 95% CIs of the subgroups are shown in Fig. 2B and C. First, in the subgroup analysis of patients from Europe and America, the pooled outcome demonstrated that a high expression level of miR-200c was significantly associated with worse OS (HR=1.85, 95% CI: 1.27, 2.69). Secondly, the subgroups of esophageal cancer and endometrial cancer exhibited an identical association, with HR values of 1.68 and 2.18, respectively. However, the colorectal cancer subgroup demonstrated the opposite result (HR=0.54, 95% CI: 0.32, 0.90). Thirdly, the result for the blood sample subgroup significantly revealed that miR-200c overexpression was associated with worse OS, with an HR value of 2.21 (95% CI: 1.04–4.72). No significant results were identified in the other subgroups.
Association of disease progress with miR-200c overexpression
A total of 16 studies were included in the present meta-analysis of the association of miR-200c overexpression and RFS/PFS/DFS; the random-effects model was used due to the high level of heterogeneity (P<0.001, I2=87.1%). Two studies from the literature were excluded, since they enrolled cancer patients at only stage I (20). The pooled HR from individual studies was 1.03, along with a P-value of 0.880, indicating a lack of statistical significance (Fig. 3A). Therefore, subgroup analyses were performed to reduce the confounding influence of the apparent heterogeneity (Fig. 3B).
Figure 3.
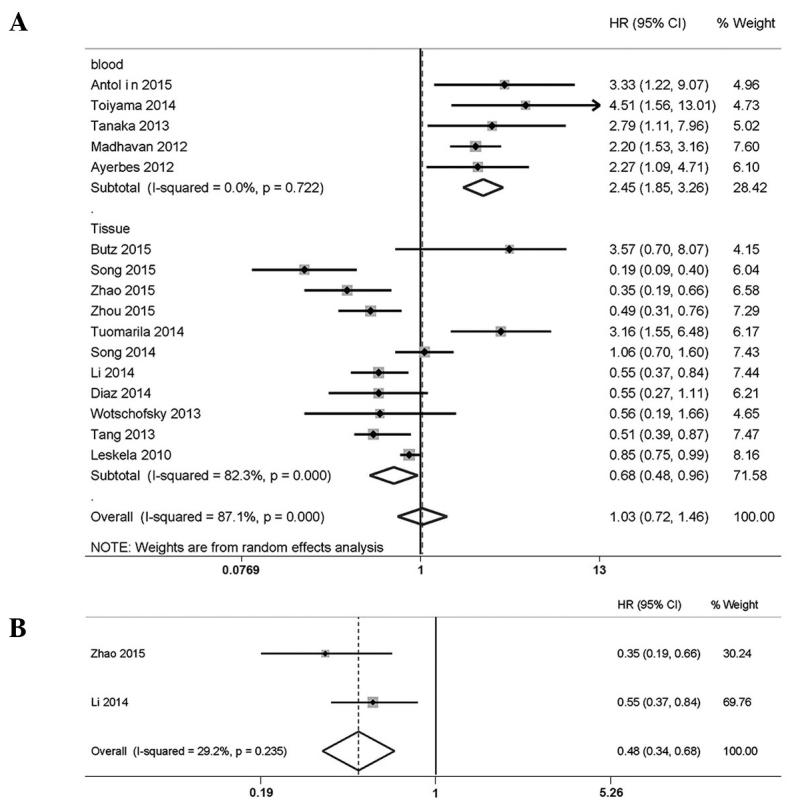
Forest plots. (A) Forest plots of the pooled analysis of the association of RFS/PFS/DFS and miR-200c expression in different sample subgroups. (B) Forest plots of the pooled analysis of RFS/PFS/DFS in the in the non-small cell lung cancer subgroup. RFS, recurrence-free survival; PFS, progression-free survival; DFS, disease-free survival; HR, hazard ratio; CI, confidence interval.
First, high expression levels of miR-200c were shown to correlate significantly with improved disease progression in patients with NSCLC (HR=0.48, 95% CI: 0.34, 0.68; fixed-effects model: P=0.235, I2=29.2% for the heterogeneity test). Secondly, the pooled HR of the tissue subgroup was 0.68 (95% CI: 0.48, 0.96, random-effects model; P<0.001, I2=82.3% for the heterogeneity test), suggesting an association between miR-200c overexpression and favorable patient prognosis. However, the blood subgroup revealed a significant correlation of expression levels of miR-200c with unfavorable patient prognosis (HR=2.45, 95% CI: 1.85, 3.26; fixed-effects model: P=0.722, I2=0.0% for heterogeneity test).
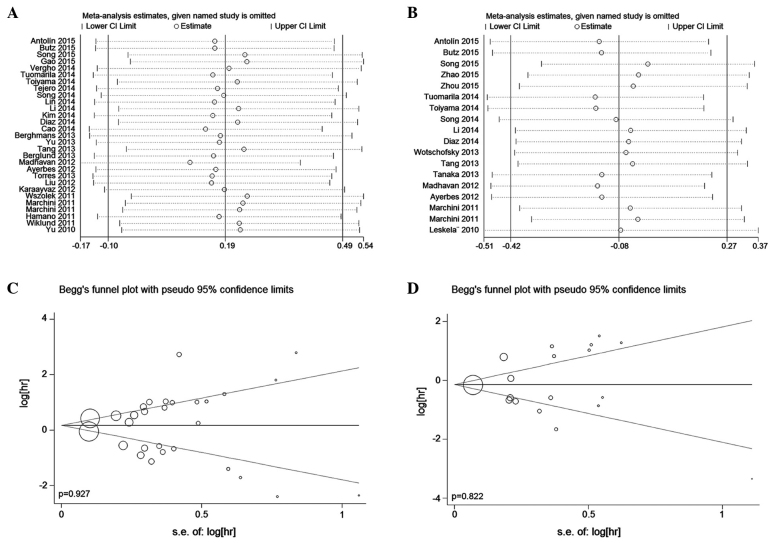
Sensitivity analysis
In the studies of CSS/OS and RFS/PFS/DFS, our sensitivity analyses did not reveal any alterations in the results due to the inclusion of any individual study (Fig. 4A and B), indicating that no single study significantly influenced the pooled HRs and 95% CIs.
Figure 4.
Sensitivity analyses and Begg's funnel plots. (A) Sensitivity analysis of the effect of individual studies on the CSS/OS results. (B) Sensitivity analysis of the effect of individual studies on the RFS/PFS/DFS results. (C) Begg's funnel plots to test for the publication bias in the overall analysis of CSS/OS. Each point represents a separate study. (D) Begg's funnel plots to test for publication bias in the overall analysis of RFS/PFS/DFS. RFS, recurrence-free survival; PFS, progression-free survival; DFS, disease-free survival; HR, hazard ratio; CI, confidence interval.
Publication bias
Egger's test and Begg's funnel plot were used for the analysis of publication bias. The funnel plots of the CSS/OS and RFS/PFS/DFS analyses were almost symmetrical, and all P-values from the Egger's test were >0.05 (Fig. 4C and D). Therefore, no significant publication bias was observed in the present meta-analysis.
Discussion
The EMT is a well-established mechanism that includes intercellular contact disruption and enhanced cell motility (47). Additionally, a burgeoning body of evidence has clearly demonstrated the involvement of EMT in the invasion and migration of tumor cells (14,32,43). Intracellular and extracellular factors are known to be capable of promoting or inhibiting EMT progression. In particular, the miR-200 family has been proposed to suppress EMT by directly targeting the transcriptional repressors of E-cadherin, zinc finger E-box binding homeobox 1 (ZEB1) and zinc finger E-box binding homeobox 2 (ZEB2), thus inducing E-cadherin upregulation (48). Conversely, inhibition of the miR-200 family would induce mesenchymal-like spindle morphology, which promotes cancer metastasis.
As the most representative microRNA among the miR-200 family, miR-200c fulfills important roles in EMT inhibition and in MET promotion. For instance, Marchini et al (20) demonstrated that several downstream targets of miR-200c, including vascular endothelial growth factor A (VEGFA) and tubulin, beta 3 class III (TUBB3), were significantly upregulated in patients with ovarian cancer who relapsed (20). Leskelä et al (21) suggested an inverse correlation between miR-200c and TUBB3 expression in advanced ovarian cancer. Furthermore, a marked inverse correlation of the expression levels of miR-200c and the mRNA levels of VEGFA was demonstrated in two independent cohorts of ccRCC and normal tissues (49). Thus, miR-200c has been considered a tumor suppressor.
However, a potential oncogenic role of miR-200c in human malignancies has also been reported. Tuomarila et al (25) demonstrated that progesterone receptor (PR)-negative cases with local or distant recurrence had higher expression levels of miR-200c compared with those without recurrence, suggesting that a high expression level of miR-200c is an independent factor for predicting poor survival rates in PR-negative breast cancer. Tejero et al (30) reported that high expression levels of miR-200c were associated with shorter OS of patients with NSCLC due to MET and angiogenesis (30). In addition, Hamano et al (39) indicated that the miR-200c-induced chemoresistance of esophageal cancer was mediated by the Akt pathway, showing that miR-200c overexpression was significantly correlated with a shortened OS (39).
In the present meta-analysis, a significant association of the expression of miR-200c with outcome was observed for pooled CSS/OS (Fig. 2A). However, there was heterogeneity in both groups of outcomes (P<0.001, I2=86.2% for CSS/OS; P<0.001, I2=87.1% for RFS/PFS/DFS). Heterogeneity may be caused by the different characteristics of the patients, including race, disease type and clinical stage, as well as the selected cut-off value of the expression level of miR-200c. To minimize the influence of these confounding factors, subgroup analyses that focused on region, disease type and sample detection were performed. On the basis of the subgroup analyses of the included studies, heterogeneity in several subgroups was greatly reduced, and valuable results were obtained (Fig. 2B and C). These results indicate that miR-200c may be used as a prognostic biomarker; however, several details require further refinement. First, different cut-off values were used in the studies of miR-200c. The majority of the available studies established a median or mean expression cut-off value, although certain studies used a lower quartile value. Furthermore, several studies used a ternary method, separating the expression levels of miR-200c into high, intermediate and low categories (50). Due to the lack of standardized miR-200c expression data, evaluating its prognostic role in malignancies would produce inaccurate results. Secondly, whether miR-200c functions as an independent tumor biomarker or, more likely, as a component of a predictive microRNA signature, has yet to be firmly established. For instance, Yeh et al (51) demonstrated that the downregulation of miR-141 and miR-200c serves as an independent predictor of DFS in hepatocellular carcinoma. In the multivariate analysis performed by Blanco-Calvo et al (52), the combination of high levels of growth differentiation factor 15, matrix metalloproteinase 7 and miR-200c was considered an independent predictor of mortality in gastric cancer. In addition, using a linear combination of the microRNA cycle threshold values and Cox regression coefficients as weights, Berghmans et al (33) revealed that a certain microRNA signature (miR-200c, miR-124, miR-29c and miR-424) is of prognostic value for OS in patients with NSCLC (33). Thirdly, although significant results were identified for several subgroups based on the subgroup analyses, the results for other subgroups were not decisive. Empirically, a prognostic factor is considered decisive when the HR is >2.0 or < 0.5 (53). Finally, several HRs were extracted from the survival curves, which unavoidably resulted in several slight statistical errors.
In conclusion, we have demonstrated that high expression levels of miR-200c are of significant prognostic value in various human malignancies. In addition, expression levels of miR-200c in tumor tissue and blood samples were considered to serve as a reliable predictive biomarker for disease progression in cancer patients. Due to the complex role of miR-200c in tumor progression and metastasis, further investigations at a larger scale are required to establish the usefulness of miR-200c as a prognostic biomarker.
Acknowledgements
We would like to thank the authors of the primary studies.
References
- 1.Bray F, Ren JS, Masuyer E, Ferlay J. Global estimates of cancer prevalence for 27 sites in the adult population in 2008. Int J Cancer. 2013;132:1133–1145. doi: 10.1002/ijc.27711. [DOI] [PubMed] [Google Scholar]
- 2.http://globocan.iarc.fr/Pages/fact_sheets_cancer.aspx Estimated Cancer Incidence, Mortality and Prevalence Worldwide in 2012. 2012 Globocan. [Google Scholar]
- 3.Hannon GJ, Rossi JJ. Unlocking the potential of the human genome with RNA interference. Nature. 2004;431:371–378. doi: 10.1038/nature02870. [DOI] [PubMed] [Google Scholar]
- 4.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 5.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-Y. [DOI] [PubMed] [Google Scholar]
- 6.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 7.Yang W, Lee DY, Ben-David Y. The roles of microRNAs in tumorigenesis and angiogenesis. Int J Physiol Pathophysiol Pharmacol. 2011;3:140–155. [PMC free article] [PubMed] [Google Scholar]
- 8.Karaayvaz M, Zhang C, Liang S, Shroyer KR, Ju J. Prognostic significance of miR-205 in endometrial cancer. PloS One. 2012;7:e35158. doi: 10.1371/journal.pone.0035158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Torres A, Torres K, Pesci A, Ceccaroni M, Paszkowski T, Cassandrini P, Zamboni G, Maciejewski R. Diagnostic and prognostic significance of miRNA signatures in tissues and plasma of endometrioid endometrial carcinoma patients. Int J Cancer. 2013;132:1633–1645. doi: 10.1002/ijc.27840. [DOI] [PubMed] [Google Scholar]
- 10.Yu CC, Tsai LL, Wang ML, Yu CH, Lo WL, Chang YC, Chiou GY, Chou MY, Chiou SH. mir145 targets the SOX9/ADAM17 axis to inhibit tumor-initiating cells and IL-6-mediated paracrine effects in head and neck cancer. Cancer Res. 2013;73:3425–3440. doi: 10.1158/0008-5472.CAN-12-3840. [DOI] [PubMed] [Google Scholar]
- 11.Nana-Sinkam P, Croce CM. MicroRNAs in diagnosis and prognosis in cancer: What does the future hold? Pharmacogenomics. 2010;11:667–669. doi: 10.2217/pgs.10.57. [DOI] [PubMed] [Google Scholar]
- 12.Ferracin M, Veronese A, Negrini M. Micromarkers: miRNAs in cancer diagnosis and prognosis. Expert Rev Mol Diagn. 2010;10:297–308. doi: 10.1586/erm.10.11. [DOI] [PubMed] [Google Scholar]
- 13.Zhou X, Wang Y, Shan B, Han J, Zhu H, Lv Y, Fan X, Sang M, Liu XD, Liu W. The downregulation of miR-200c/141 promotes ZEB1/2 expression and gastric cancer progression. Med Oncol. 2015;32:428. doi: 10.1007/s12032-014-0428-3. [DOI] [PubMed] [Google Scholar]
- 14.Toiyama Y, Hur K, Tanaka K, Inoue Y, Kusunoki M, Boland CR, Goel A. Serum miR-200c is a novel prognostic and metastasis-predictive biomarker in patients with colorectal cancer. Ann Surg. 2014;259:735–743. doi: 10.1097/SLA.0b013e3182a6909d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tang H, Deng M, Tang Y, Xie X, Guo J, Kong Y, Ye F, Su Q, Xie X. miR-200b and miR-200c as prognostic factors and mediators of gastric cancer cell progression. Clin Cancer Res. 2013;19:5602–5612. doi: 10.1158/1078-0432.CCR-13-1326. [DOI] [PubMed] [Google Scholar]
- 16.Valladares-Ayerbes M, Reboredo M, Medina-Villaamil V, Iglesias-Díaz P, Lorenzo-Patiño MJ, Haz M, Santamarina I, Blanco M, Fernández-Tajes J, Quindós M, et al. Circulating miR-200c as a diagnostic and prognostic biomarker for gastric cancer. J Transl Med. 2012;10:186. doi: 10.1186/1479-5876-10-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Song F, Yang D, Liu B, Guo Y, Zheng H, Li L, Wang T, Yu J, Zhao Y, Niu R, et al. Integrated microRNA network analyses identify a poor-prognosis subtype of gastric cancer characterized by the miR-200 family. Clin Cancer Res. 2014;20:878–889. doi: 10.1158/1078-0432.CCR-13-1844. [DOI] [PubMed] [Google Scholar]
- 18.Gao YC, Wu J. MicroRNA-200c and microRNA-141 as potential diagnostic and prognostic biomarkers for ovarian cancer. Tumour Biol. 2015;36:4843–4850. doi: 10.1007/s13277-015-3138-3. [DOI] [PubMed] [Google Scholar]
- 19.Cao Q, Lu K, Dai S, Hu Y, Fan W. Clinicopathological and prognostic implications of the miR-200 family in patients with epithelial ovarian cancer. Int J Clin Exp Pathol. 2014;7:2392–2401. [PMC free article] [PubMed] [Google Scholar]
- 20.Marchini S, Cavalieri D, Fruscio R, Calura E, Garavaglia D, Fuso Nerini IF, Mangioni C, Cattoretti G, Clivio L, Beltrame L, et al. Association between miR-200c and the survival of patients with stage I epithelial ovarian cancer: A retrospective study of two independent tumour tissue collections. Lancet Oncol. 2011;12:273–285. doi: 10.1016/S1470-2045(11)70012-2. [DOI] [PubMed] [Google Scholar]
- 21.Leskelä S, Leandro-García LJ, Mendiola M, Barriuso J, Inglada-Pérez L, Muñoz I, Martínez-Delgado B, Redondo A, de Santiago J, Robledo M, et al. The miR-200 family controls beta-tubulin III expression and is associated with paclitaxel-based treatment response and progression-free survival in ovarian cancer patients. Endocr Relat Cancer. 2010;18:85–95. doi: 10.1677/ERC-10-0148. [DOI] [PubMed] [Google Scholar]
- 22.Vergho DC, Kneitz S, Kalogirou C, Burger M, Krebs M, Rosenwald A, Spahn M, Löser A, Kocot A, Riedmiller H, Kneitz B. Impact of miR-21, miR-126 and miR-221 as prognostic factors of clear cell renal cell carcinoma with tumor thrombus of the inferior vena cava. PloS One. 2014;9:e109877. doi: 10.1371/journal.pone.0109877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wotschofsky Z, Busch J, Jung M, Kempkensteffen C, Weikert S, Schaser KD, Melcher I, Kilic E, Miller K, Kristiansen G, et al. Diagnostic and prognostic potential of differentially expressed miRNAs between metastatic and non-metastatic renal cell carcinoma at the time of nephrectomy. Clin Chim Acta. 2013;416:5–10. doi: 10.1016/j.cca.2012.11.010. [DOI] [PubMed] [Google Scholar]
- 24.Butz H, Szabó PM, Khella HW, Nofech-Mozes R, Patocs A, Yousef GM. miRNA-target network reveals miR-124 as a key miRNA contributing to clear cell renal cell carcinoma aggressive behaviour by targeting CAV1 and FLOT1. Oncotarget. 2015;6:12543–12557. doi: 10.18632/oncotarget.3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tuomarila M, Luostari K, Soini Y, Kataja V, Kosma VM, Mannermaa A. Overexpression of microRNA-200c predicts poor outcome in patients with PR-negative breast cancer. PloS One. 2014;9:e109508. doi: 10.1371/journal.pone.0109508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Madhavan D, Zucknick M, Wallwiener M, Cuk K, Modugno C, Scharpff M, Schott S, Heil J, Turchinovich A, Yang R, et al. Circulating miRNAs as surrogate markers for circulating tumor cells and prognostic markers in metastatic breast cancer. Clin Cancer Res. 2012;18:5972–5982. doi: 10.1158/1078-0432.CCR-12-1407. [DOI] [PubMed] [Google Scholar]
- 27.Antolín S, Calvo L, Blanco-Calvo M, Santiago MP, Lorenzo-Patiño MJ, Haz-Conde M, Santamarina I, Figueroa A, Antón-Aparicio LM, Valladares-Ayerbes M. Circulating miR-200c and miR-141 and outcomes in patients with breast cancer. BMC Cancer. 2015;15:297. doi: 10.1186/s12885-015-1238-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Song C, Liu LZ, Pei XQ, Liu X, Yang L, Ye F, Xie X, Chen J, Tang H, Xie X. miR-200c inhibits breast cancer proliferation by targeting KRAS. Oncotarget. 2015;6:34968–34978. doi: 10.18632/oncotarget.5198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Diaz T, Tejero R, Moreno I, Ferrer G, Cordeiro A, Artells R, Navarro A, Hernandez R, Tapia G, Monzo M. Role of miR-200 family members in survival of colorectal cancer patients treated with fluoropyrimidines. J Surg Oncol. 2014;109:676–683. doi: 10.1002/jso.23572. [DOI] [PubMed] [Google Scholar]
- 30.Tejero R, Navarro A, Campayo M, Viñolas N, Marrades RM, Cordeiro A, Ruíz-Martínez M, Santasusagna S, Molins L, Ramirez J, Monzó M. miR-141 and miR-200c as markers of overall survival in early stage non-small cell lung cancer adenocarcinoma. PloS One. 2014;9:e101899. doi: 10.1371/journal.pone.0101899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li J, Li X, Ren S, Chen X, Zhang Y, Zhou F, Zhao M, Zhao C, Chen X, Cheng N, et al. miR-200c overexpression is associated with better efficacy of EGFR-TKIs in non-small cell lung cancer patients with EGFR wild-type. Oncotarget. 2014;5:7902–7916. doi: 10.18632/oncotarget.2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim MK, Jung SB, Kim JS, Roh MS, Lee JH, Lee EH, Lee HW. Expression of microRNA miR-126 and miR-200c is associated with prognosis in patients with non-small cell lung cancer. Virchows Arch. 2014;465:463–471. doi: 10.1007/s00428-014-1640-4. [DOI] [PubMed] [Google Scholar]
- 33.Berghmans T, Ameye L, Willems L, Paesmans M, Mascaux C, Lafitte JJ, Meert AP, Scherpereel A, Cortot AB, Cstoth I, et al. Identification of microRNA-based signatures for response and survival for non-small cell lung cancer treated with cisplatin-vinorelbine A ELCWP prospective study. Lung Cancer. 2013;82:340–345. doi: 10.1016/j.lungcan.2013.07.020. [DOI] [PubMed] [Google Scholar]
- 34.Liu XG, Zhu WY, Huang YY, Ma LN, Zhou SQ, Wang YK, Zeng F, Zhou JH, Zhang YK. High expression of serum miR-21 and tumor miR-200c associated with poor prognosis in patients with lung cancer. Med Oncol. 2012;29:618–626. doi: 10.1007/s12032-011-9923-y. [DOI] [PubMed] [Google Scholar]
- 35.Zhao J, Zhao Y, Wang Z, Xuan Y, Luo Y, Jiao W. Loss expression of micro ribonucleic acid (miRNA)-200c induces adverse post-surgical prognosis of advanced stage non-small cell lung carcinoma and its potential relationship with ETAR messenger RNA. Thorac Cancer. 2015;6:421–426. doi: 10.1111/1759-7714.12193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin HM, Castillo L, Mahon KL, Chiam K, Lee BY, Nguyen Q, Boyer MJ, Stockler MR, Pavlakis N, Marx G, et al. Circulating microRNAs are associated with docetaxel chemotherapy outcome in castration-resistant prostate cancer. Br J Cancer. 2014;110:2462–2471. doi: 10.1038/bjc.2014.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu H, Duan B, Jiang L, Lin M, Sheng H, Huang J, Gao H. Serum miR-200c and clinical outcome of patients with advanced esophageal squamous cancer receiving platinum-based chemotherapy. Am J Transl Res. 2013;6:71–77. [PMC free article] [PubMed] [Google Scholar]
- 38.Tanaka K, Miyata H, Yamasaki M, Sugimura K, Takahashi T, Kurokawa Y, Nakajima K, Takiguchi S, Mori M, Doki Y. Circulating miR-200c levels significantly predict response to chemotherapy and prognosis of patients undergoing neoadjuvant chemotherapy for esophageal cancer. Ann Surg Oncol. 2013;20(Suppl 3):S607–S615. doi: 10.1245/s10434-013-3093-4. [DOI] [PubMed] [Google Scholar]
- 39.Hamano R, Miyata H, Yamasaki M, Kurokawa Y, Hara J, Moon JH, Nakajima K, Takiguchi S, Fujiwara Y, Mori M, Doki Y. Overexpression of miR-200c induces chemoresistance in esophageal cancers mediated through activation of the Akt signaling pathway. Clin Cancer Res. 2011;17:3029–3038. doi: 10.1158/1078-0432.CCR-10-2532. [DOI] [PubMed] [Google Scholar]
- 40.Berglund M, Hedström G, Amini RM, Enblad G, Thunberg U. High expression of microRNA-200c predicts poor clinical outcome in diffuse large B-cell lymphoma. Oncol Rep. 2013;29:720–724. doi: 10.3892/or.2012.2173. [DOI] [PubMed] [Google Scholar]
- 41.Wszolek MF, Rieger-Christ KM, Kenney PA, Gould JJ, Silva Neto B, Lavoie AK, Logvinenko T, Libertino JA, Summerhayes IC. A MicroRNA expression profile defining the invasive bladder tumor phenotype. Urol Oncol. 2011;29:794–801. doi: 10.1016/j.urolonc.2009.08.024. e1. [DOI] [PubMed] [Google Scholar]
- 42.Wiklund ED, Bramsen JB, Hulf T, Dyrskjøt L, Ramanathan R, Hansen TB, Villadsen SB, Gao S, Ostenfeld MS, Borre M, et al. Coordinated epigenetic repression of the miR-200 family and miR-205 in invasive bladder cancer. Int J Cancer. 2011;128:1327–1334. doi: 10.1002/ijc.25461. [DOI] [PubMed] [Google Scholar]
- 43.Yu J, Ohuchida K, Mizumoto K, Sato N, Kayashima T, Fujita H, Nakata K, Tanaka M. MicroRNA, hsa-miR-200c, is an independent prognostic factor in pancreatic cancer and its upregulation inhibits pancreatic cancer invasion but increases cell proliferation. Mol Cancer. 2010;9:169. doi: 10.1186/1476-4598-9-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moher D, Liberati A, Tetzlaff J, Altman DG. PRISMA Group. Preferred reporting items for systematic reviews and meta-analyses: The PRISMA statement. J Clin Epidemiol. 2009;62:1006–1012. doi: 10.1016/j.jclinepi.2009.06.005. [DOI] [PubMed] [Google Scholar]
- 45.Tierney JF, Stewart LA, Ghersi D, Burdett S, Sydes MR. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials. 2007;8:16. doi: 10.1186/1745-6215-8-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 47.Guarino M, Rubino B, Ballabio G. The role of epithelial-mesenchymal transition in cancer pathology. Pathology. 2007;39:305–318. doi: 10.1080/00313020701329914. [DOI] [PubMed] [Google Scholar]
- 48.Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 49.Liu H, Brannon AR, Reddy AR, Alexe G, Seiler MW, Arreola A, Oza JH, Yao M, Juan D, Liou LS, et al. Identifying mRNA targets of microRNA dysregulated in cancer: With application to clear cell renal cell carcinoma. BMC Syst Biol. 2010;4:51. doi: 10.1186/1752-0509-4-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vilming Elgaaen B, Olstad OK, Haug KB, Brusletto B, Sandvik L, Staff AC, Gautvik KM, Davidson B. Global miRNA expression analysis of serous and clear cell ovarian carcinomas identifies differentially expressed miRNAs including miR-200c-3p as a prognostic marker. BMC Cancer. 2014;14:80. doi: 10.1186/1471-2407-14-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yeh TS, Wang F, Chen TC, Yeh CN, Yu MC, Jan YY, Chen MF. Expression profile of microRNA-200 family in hepatocellular carcinoma with bile duct tumor thrombus. Ann Surg. 2014;259:346–354. doi: 10.1097/SLA.0000000000000223. [DOI] [PubMed] [Google Scholar]
- 52.Blanco-Calvo M, Tarrío N, Reboredo M, Haz-Conde M, García J, Quindós M, Figueroa A, Antón-Aparicio L, Calvo L, Valladares-Ayerbes M. Circulating levels of GDF15, MMP7 and miR-200c as a poor prognostic signature in gastric cancer. Future Oncol. 2014;10:1187–1202. doi: 10.2217/fon.13.263. [DOI] [PubMed] [Google Scholar]
- 53.Hayes DF, Isaacs C, Stearns V. Prognostic factors in breast cancer: Current and new predictors of metastasis. J Mammary Gland Biol Neoplasia. 2001;6:375–392. doi: 10.1023/A:1014778713034. [DOI] [PubMed] [Google Scholar]