Abstract
microRNA (miR)-153 has been shown to play a role in several solid malignancies; however, its expression and function in laryngeal squamous cell carcinoma (LSCC) have not been fully explored. In the present study, reverse transcription-quantitative polymerase chain reaction analysis was performed in order to detect the expression levels of miR-153 in mucosal specimens isolated from patients undergoing total laryngectomy. In addition, in vitro experiments were performed to analyze the cellular proliferation and invasion abilities of human epithelial type 2 (HEp-2) cells transfected with miR-153 mimics or miR-153 antisense oligonucleotide (ASO). It was found that miR-153 was downregulated in LSCC tissues. Furthermore, while miR-153 mimics inhibited cell proliferation and invasion, miR-153 ASO promoted HEp-2 cell growth and invasion. At a molecular level, it was predicted by bioinformatics that kruppel-like factor 5 (KLF5) has a miR-153 binding site. Luciferase and protein expression analyses revealed that miR-153 inhibited the protein expression of KLF5. These results suggest that miR-153 may act as a tumor suppressor during LSCC progression via the suppression of KLF5, and could potentially serve as a therapeutic target for LSCC.
Keywords: laryngeal squamous cell carcinoma, microRNA, miR-153, kruppel-like factor 5
Introduction
Laryngeal squamous cell carcinomas (LSCCs) account for >85% of all malignant tumors occurring in the larynx (1), and despite the great progress that has been made in the treatment of the disease, including surgery or radiotherapy (2,3), the molecular basis of LSCC remains unclear.
Numerous studies have shown that microRNAs (miRNAs), a class of small, single-stranded, non-coding RNAs, play a critical role in the regulation of tumorigenesis (4,5). The aberrant expression of certain miRNAs has been shown to regulate cell proliferation, apoptosis, invasion and angiogenesis in human malignancies, either through the induction of mRNA degradation, or through translational inhibition (5,6).
microRNA-153 (miR-153) is located in intron 19 of genes encoding two of the major type 1 diabetes autoantigens, islet-associated protein (IA)-2 and IA-2β (7). Kim et al found that miR-153 was downregulated and correlated with advanced clinical stage in ovarian epithelial tumors (8). In addition, the downregulation of miR-153 has been found to contribute to epithelial-mesenchymal transition and tumor metastasis in human epithelial cancer (9). Furthermore, the levels of miR-153 have been found to be decreased in LSCC tissues compared with the adjacent tissues (10). In cultured lung cancer cells, the overexpression of miR-153 has been shown to significantly inhibit cell proliferation and migration, and promote apoptosis in through the suppression of protein kinase B (AKT1) (10); however, miR-153 has been observed to be upregulated in human colorectal cancer (CC) cells, thereby supporting CC progression and chemotherapeutic resistance (11). Based on the aforementioned results, the role of miR-153 in human cancers may be cell- or tissue-specific.
In the present study, the expression of miR-153 in LSCC tissues was evaluated, and the effect of miR-153 transfection on human epithelial cells was evaulated in vitro, with the aim of elucidating the role and mechanisms of miR-153 in laryngeal tumorigenesis.
Materials and methods
Cinical tissue samples
LSCC tissue and adjacent control mucosa specimens were collected from 38 patients (age, 22–54 years; males, 22; females, 16) who had undergone total or partial laryngectomy between May 2011 and December 2013. Informed consent was obtained from each patient prior to sample collection. The present study was reviewed and approved by the Ethics Committee of Zhangjiagang First People's Hospital (Suzhou, China).
Cell culture and transfection
Human epithelial type 2 (HEp-2) cells were purchased from American Type Culture Collection (Rockville, MD, USA). Cells were cultured in RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc., Shanghai, China) supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.). The cultured cells were incubated at 37°C with 5% CO2 in a humidified incubator. Human miR-153 mimics (miR-153-1), antisense oligonucleotide (ASO) and negative controls (NCs) were obtained from Shanghai GenePharma Co., Ltd. (Shanghai, China). For transfection, cells were cultured in a 6-well plate and transiently transfected at 70–80% confluence using Lipofectamine 2000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.) following the manufacturer's protocol.
Cell viability assay
The viability of the cells was determined by assaying the reduction of 3-(4,5-dimethylthiazol-2-yl)-2,5-di-phenyltetrazolium bromide (MTT; Beyotime Institute of Biotechnology Co., Shanghai, China) to formazan, following the manufacturer's protocol.
Cell proliferation, cell cycle, and invasion assays
Cell proliferation was evaluated by analyzing the incorporation of bromodeoxyuridine (BrdU) into cellular DNA during DNA synthesis using an enzyme-linked immunosorbent assay (BrdU kit; Beyotime Institute of Biotechnology Co., Shanghai, China), following the manufacturer's instructions. All experiments were performed in triplicate. Absorbance was measured using the Spectra Max 190 ELISA reader (Molecular Devices, Sunnyvale, CA, USA) at 450 nm. For cell-cycle analysis, cells were labeled for 20 min with propidium iodide (Beyotime Institute of Biotechnology Co.) and immediately analyzed using Gallios™ flow cytometry (Beckman Coulter, Inc., Brea, CA, USA). A total of 10,000 cells were acquired and analyzed for DNA content. All data were collected, stored, and analyzed using ModFit 3.2 software (Verity Software House Company, Topsham, ME, USA). Histograms represent the percentage of cells in each phase of the cell cycle (G0/G1, S and G2/M).
Cell invasion assays were performed using extracellular matrix-coated invasion chambers (ECM550; EMD Millipore, Temecula, CA, USA), according to the manufacturer's instructions. Briefly, cells were harvested and resuspended in serum-free medium following pretreatment for 24 h with miR-153 mimic/miR-153 antisense or negative control. A total of 2×105 cells were plated into the top well of an extracellular matrix-coated invasion chamber, whereas the bottom well of the chamber contained 500 µl Dulbecco's Modified Eagle's Medium supplemented with 10% FBS. After 24-h incubation, the noninvading cells were removed using a cotton swab and the invading cells on the underside of the membrane were stained with Giemsa cell stain solution (Millipore) for 10 min. Following washing three times with water, 100 µl extraction buffer (EMD Millipore) was used to remove the stain from each membrane and quantified using a colorimetric microplate reader at 570 nm (Bio-Rad Laboratories, Shanghai, China), according to the manufacturer's instructions.
RNA isolation and reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA from fresh tissues and cultured cells was harvested using an mirVana miRNA Isolation kit (Ambion, Austin, TX, USA) according to the manufacturer's protocol. A NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA) was used to determine the RNA concentration. RNA was treated with DNase (PureLink DNase Set; 12185010; Thermo Fisher Scientific, Inc.) prior to analysis. RT-qPCR was performed using TaqMan Universal PCR Master mix (Applied Biosystems; Thermo Fisher Scientific, Inc.) and an ABI Prism 7900HT Real-Time PCR System (Thermo Fisher Scientific, Inc.). The primer for miR-153 was GATCACTTTTGTGACTATGCAACT. Small nuclear U6 snRNA (Applied Biosystems) was used as an internal control for the expression of miR-153.
To analyze the relative mRNA levels of cyclin B1 and cyclin D1, total RNA was isolated using TRIzol reagent according to the manufacturer's protocol (Invitrogen; Thermo Fisher Scientific, Inc.). First-strand complementary DNA (cDNA) synthesis was conducted for each RNA sample using the Promega Reverse Transcription System (Promega Corporation, Madison, WI, USA). Oligo dT was employed to prime cDNA synthesis. qPCR was performed using Takara SYBR Master mix (Takara Bio., Inc., Shiga, Japan) and a LightCycler 480 instrument (Roche Diagnostics, Basel, Switzerland). PCR conditions included an initial holding period at 95°C for 5 sec and 60°C for 30 sec for 45 cycles. The primer sequences were: Cyclin B1, forward: 5′-AATAAGGCGAAGATCAACATGGC-3′, and reverse: 5′-TTTGTTACCAATGTCCC CAAGAG-3′; cyclin D1, forward: 5′-TGGAGCCCGTGAAAAAGAGC-3′, and reverse: 5′-TCTCCTTCATCTTAGAGGCCAC-3′). Differences in gene expression were calculated using the 2−ΔΔCq method (12) and expressed as a fold-change. Each experiment was repeated ≥3 times.
Western blot analysis
Cells or tissues were harvested and lysed with lysis buffer (150 mm NaCl, 50 mm Tris-HCl, 1% NP-40, pH 7.5; Beyotime Institute of Biotechnology Co.). Proteins were quantified and separated in 8% sodium dodecyl sulfate-agarose gel and then transferred to nitrocellulose membranes (Amersham Biosciences, Little Chalfont, Buckinghamshire, UK). Following blocking with 10% nonfat milk in phosphate-buffered saline, membranes were immunoblotted with primary antibodies as listed below. The membranes were incubated with primary antibodies at 4°C overnight and washed three times in 0.1% Tris-buffered saline and Tween 20 (TBST; Beyotime Institute of Biotechnology Co.). They were then incubated with mouse anti-rabbit (1:1,500; sc-2491; Santa Cruz Biotechnology, Inc., Dallas, TX, USA) and rabbit anti-mouse IgG (1:2,000; ab193651; Abcam, Cambridge, MA, USA) secondary antibodies for 2 h at 25°C and washed three times in 0.1% TBST. They were then incubated with secondary antibodies (Santa Cruz Company, California, USA) for 2 hr at 25℃ and washed three times each in 0.1 %TBST. The signals were detected using SuperSignal West Pico Chemiluminescent Substrate kit (Pierce Biotechnology, Inc., Rockford, IL, USA) according to the manufacturer's instructions. The primary antibodies anti-cyclin B1 (mouse monoclonal antibody; ab72; 1:3,000), anti-cyclin D1 (rabbit polyclonal antibody; ab16663; 1:2,000), anti-kruppel-like factor 5 (KLF5; rabbit polyclonal antibody; ab24331; 1:2,000), anti-AKT1 (rabbit polyclonal antibody; ab32505; 1:1,000), anti-forkhead box O3 (FOXO3a; rabbit polyclonal antibody; ab12162; 1:1,000) and anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH; rabbit polyclonal antibody; ab181602, 1:1,000) were purchased from Abcam. Protein levels were normalized to total GAPDH expression.
Luciferase reporter assay
Potential target genes for miR-153 were searched for using bioinformatics software (miRWalk) (13). The 3′-untranslated region (3′-UTR) of the human gene KLF5 was predicted to interact with miR-153. The 3′-UTR of KLF5 was synthesized and inserted into the luciferase reporter construct pMir-Report (Ambion; Thermo Fisher Scientific, Inc.), yielding pMir-Report-KLF5. Mutations within potential miR-153 binding sites were generated by nucleotide replacement of the wild type sequence to inhibit miR-153 binding. HEp-2 cells were transfected with either miR-153 mimics or NC, together with pMir-Report vectors containing wild-type or mutant KLF5 3′-UTR variants for 36 h. The pRL-SV40 vector (Promega Corporation) carrying the Renilla luciferase gene was used as an internal control to normalize the transfection efficiency, and the Dual-Luciferase Reporter Assay System (Promega Corporation) was used to determine the luciferase values.
Statistical analysis
Data were collected from at least four separate experiments and are expressed as the mean ± standard error. The differences between groups were analyzed using the Student's t-test or one-way analysis of variance. P<0.05 was considered to indicate a statistically significant difference.
Results
Downregulation of miR-153 in LSCC tissues
RT-qPCR was performed to determine the expression levels of miR-153 in LSCC tissues and control mucosa specimens. miR-153 was found to be significantly downregulated in the LSCC tissues compared with the control (P<0.001; Fig. 1).
Figure 1.
Expression levels of miR-153 in LSCC tissues. miR-153 expression was determined using reverse transcription-quantitative polymerase chain reaction in human LSCC and adjacent normal tissues (n=38). ***P<0.001 vs. the normal group. miR-153, microRNA-153; LSCC, laryngeal squamous cell carcinoma.
Effects of miR-153 on HEp-2 cell proliferation and invasion
In order to evaluate the effect of miR-153 on cell growth, HEp-2 cells were transiently transfected with miR-155 mimics or NCs (Fig. 2A). The forced expression of miR-153 mimics markedly inhibited cell viability and proliferation, as shown by the results of MTT and BrdU incorporation assays (P<0.01; Fig. 2B and C). In addition, cell cycle analysis revealed that the percentage of miR-153 mimic-transfected cells in the G1/G0 phase was notably higher than the percentage in the S phase (Fig. 2D). Furthermore, the invasive abilities of the HEp-2 cells were also found to be reduced by miR-153 mimics (Fig. 2E).
Figure 2.
miR-153 mimics inhibit cell proliferation and invasion in laryngeal squamous cell carcinoma. (A) Expression levels of miR-153 following transfection of miR-153 mimics or NC in HEp-2 cells for 24 h. (B) Cell viability and (C) proliferative potential determined in HEp-2 cells transfected with miR-153 mimics or NC. (D) Cell-cycle phases of HEp-2 cells transfected with miR-153 mimics or NC determined using flow cytometry. (E) Cell invasion assay results for HEp-2 cells transfected with miR-153 mimics or NC. *P<0.05, **P<0.01 and ***P<0.001 vs. the NC group. miR-153, microRNA-153; NC, negative control; HEp-2, human epithelial type 2; MTT, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide; BrdU, bromodeoxyuridine.
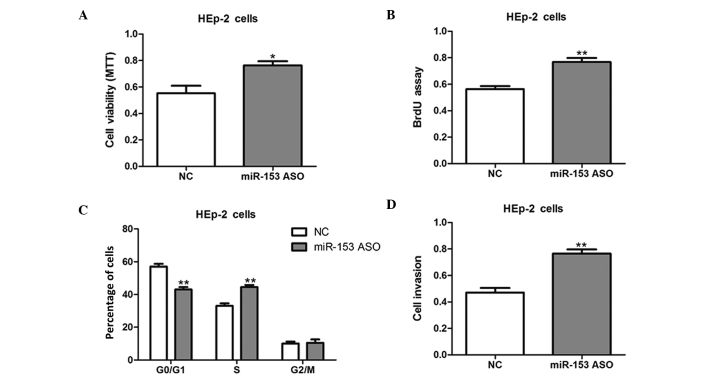
HEp-2 cells were then transfected with miR-153 ASO to knock down miR-153 expression. The inhibition of miR-153 promoted the cell growth and invasion of HEp-2 cells, as compared with NC-transfected cells (Fig. 3); therefore, the present results suggest that miR-153 inhibits cell proliferation and invasion in vitro.
Figure 3.
miR-153 ASO promotes laryngeal squamous cell carcinoma cell growth. (A) Cell viability and (B) proliferative potential assay results for HEp-2 cells transfected with miR-153 ASO or NC for 24 h. (C) Cell-cycle phases of HEp-2 cells transfected with miR-153 ASO or NC as determined using flow cytometry. (D) Cell invasion assay results for HEp-2 cells transfected with miR-153 ASO or NC. *P<0.05 and **P<0.01 vs. the NC group. miR-153, microRNA-153; HEp-2, human epithelial type 2; NC, negative control; ASO, antisense oligonucleotide; MTT, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide; BrdU, bromodeoxyuridine.
miR-153 suppresses KLF5 in LSCC cells
AKT1 and FOXO3a have been shown to be negatively regulated by miR-153 in human cancers (10,11); however, no changes in the protein levels of AKT1 and FOXO3a among the HEp-2 cells transfected with miR-153 mimics, ASO or NC were detected in the present study (Fig. 4A and B). Potential target genes for miR-153 were therefore searched for using bioinformatics software. It was found that KLF5 harbors a potential miR-153 binding site in its 3′-UTR (Fig. 4C). Subsequently, the 3′-UTR of the KLF5 gene (wild type and mutant) was cloned and inserted into a luciferase reporter construct. As shown in Fig. 4D, the overexpression of miR-153 mimics led to a reduction in luciferase activity when the luciferase reporter construct contained the wild-type KLF5 3′-UTRs; however, mutation of the potential miR-153 binding site abolished the reduction in luciferase expression (Fig. 4D). Furthermore, the transfection of miR-153 mimics in HEp-2 cells resulted in a reduction in the level of KLF5 protein expression (Fig. 4E). Consistent with these findings, a marked upregulation of KLF5 was observed in the HEp-2 cells with miR-153 inhibition (Fig. 4F). The results of the present study therefore suggest that KLF5 may be a target of miR-153 in LSCC cells.
Figure 4.
miR-153 negatively regulates KLF5 expression in LSCC. (A and B) Western blot analysis of AKT1 and FOXO3a expression in HEp-2 cells transfected with miR-153 mimics, ASO or NC. (C) Informatics prediction of miR-153 binding sites in the 3′-UTRs of KLF5. (D) Luciferase reporter assays in HEp-2 cells. Cells were transfected with wild-type 3′-UTR-reporter or mutant constructs together with miR-153 mimics or NC. Representative western blot analysis of KLF5 expression in HEp-2 cells transfected with (E) miR-153 mimics or NC and (F) miR-153 antisense or NC. **P<0.01 between the two groups. KLF5, anti-kruppel-like factor 5; miR-153, microRNA-153; HEp-2, human epithelial type 2; 3′-UTR, 3′-untranslated region; NC, negative control; ASO, antisense oligonucleotide; LSCC, laryngeal squamous cell carcinoma; AKT1, protein kinase B; FOXO3a, forkhead box O3.
Regulation of cyclins B1 and D1 by miR-153
Previous studies have shown that KLF5 may promote cell proliferation through the upregulation of cell-cycle regulators, such as cyclins B1 and D1 (14,15). In the present study, it was found that the transfection of HEp-2 cells with miR-153 mimics reduced the mRNA and protein expression levels of cyclin B1 and cyclin D1 (Fig. 5A and B), and the expression levels of these cyclins in HEp-2 cells were upregulated by miR-153 ASO (Fig. 5C and D).
Figure 5.
Modulation of cell-cycle regulators by miR-153. (A) mRNA and (B) protein levels of cyclins B1 and D1 were determined using reverse transcription-quantitative polymerase chain reaction and western blot analysis in HEp-2 cells transfected with miR-153 mimics or NC. (C) mRNA and (D) protein levels of cyclins B1 and D1 in HEp-2 cells transfected with miR-153 ASO or NC. **P<0.01, ***P<0.001 between the two groups. miR-153, microRNA-153; NC, negative control; HEp-2, human epithelial type 2; ASO, antisense oligonucleotide; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Upregulation of KLF5 in LSCC tissues
The protein levels of KLF5 in the LSCC tissues and control mucosa specimens were determined. Consistent with the downregulation of miR-153, KLF5 was markedly upregulated in the LSCC tissues (Fig. 6).
Figure 6.
Upregulation of KLF5 in LSCC tissues. Representative protein levels of KLF5 as determined using western blot analysis in LSCC or normal tissues from two patients. LSCC, laryngeal squamous cell carcinoma; KLF5, anti-kruppel-like factor 5.
Discussion
In the present study, miR-153 expression was found to be downregulated in LSCC tissues. In vitro experiments further indicated that miR-153 may inhibit LSCC cell proliferation and progression through the suppression of KLF5; however, the exact mechanism of the inhibition of miR-153 expression in LSCC remains unclear. Notably, a previous study has found that cytosine methylation in miR-153 gene promoters could modulate its expression in human kidney cells (16); therefore, it may be speculated that epigenetic regulation might be involved in the downregulation of miR-153 in LSCC tissues.
Previous studies have identified AKT1 and FOXO3a as translational targets for miR-153 (10,11); however, the present study showed that the abundance of these proteins was not affected in HEp-2 cells transfected with miR-153 mimics or ASO. Although the reason for this inconsistency is not known, the role of miR-153 in human cancers may be tissue- or cell-specific. Furthermore, KLF5, a new direct and functional target of miR-153, was identified in LSCC. miR-153 was shown to be able to bind to the 3′-UTR of KLF5 to suppress its protein contents. Studies have shown that KLF5 acts as an oncogene in various types of human cancers (17,18). The upregulation of KLF5 has been observed in breast, colon, gastric and prostate cancers (17–21). In addition, KLF5 promotes cell survival and inhibits apoptosis in cancer cells via multiple mechanisms (22,23). For instance, KLF5 has been shown to interact with tumor suppressor p53 in regulating the expression of the inhibitor-of-apoptosis protein survivin, which may play a role in the pathological process of cancer (24). KLF5 has also been found to promote lung tumorigenesis through the upregulation of the Sox4 gene (25). Despite the aforementioned findings, KLF5 remains a poor candidate as a drug target, due to the lack of a ligand-binding domain and broad transcriptional signature; therefore, a better understanding of the regulatory pathways for the manipulation of its expression could provide impetus to the identification of pharmacological approaches to treat human cancers, including LSCC.
In conclusion, a downregulation of miR-153 was observed in LSCC tissues, and it was demonstrated that miR-153 may act as a tumor suppressor in LSCC initiation and/or progression. It was further found that miR-153 possesses the potency to suppress LSCC growth and invasion through the suppression of KLF5, suggesting that miR-153 could potentially serve as a therapeutic target for LSCC.
References
- 1.Nadal A, Cardesa A. Molecular biology of laryngeal squamous cell carcinoma. Virchows Arch. 2003;442:1–7. doi: 10.1007/s00428-002-0726-6. [DOI] [PubMed] [Google Scholar]
- 2.Lionello M, Staffieri A, Marioni G. Potential prognostic and therapeutic role for angiogenesis markers in laryngeal carcinoma. Acta Otolaryngol. 2012;132:574–582. doi: 10.3109/00016489.2011.652308. [DOI] [PubMed] [Google Scholar]
- 3.Caicedo-Granados E, Beswick DM, Christopoulos A, Cunningham DE, Razfar A, Ohr JP, Heron DE, Ferris RL. Oncologic and functional outcomes of partial laryngeal surgery for intermediate-stage laryngeal cancer. Otolaryngol Head Neck Surg. 2013;148:235–242. doi: 10.1177/0194599812466367. [DOI] [PubMed] [Google Scholar]
- 4.Schwarzenbach H, Nishida N, Calin GA, Pantel K. Clinical relevance of circulating cell-free microRNAs in cancer. Nat Rev Clin Oncol. 2014;11:145–156. doi: 10.1038/nrclinonc.2014.5. [DOI] [PubMed] [Google Scholar]
- 5.Kasinski AL, Slack FJ. Epigenetics and genetics. MicroRNAs en route to the clinic: Progress in validating and targeting microRNAs for cancer therapy. Nat Rev Cancer. 2011;11:849–864. doi: 10.1038/nrc3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ryan BM, Robles AI, Harris CC. Genetic variation in microRNA networks: The implications for cancer research. Nat Rev Cancer. 2010;10:389–402. doi: 10.1038/nrc2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mandemakers W, Abuhatzira L, Xu H, Caromile LA, Hébert SS, Snellinx A, Morais VA, Matta S, Cai T, Notkins AL, De Strooper B. Co-regulation of intragenic microRNA miR-153 and its host gene Ia-2 β: Identification of miR-153 target genes with functions related to IA-2β in pancreas and brain. Diabetologia. 2013;56:1547–1556. doi: 10.1007/s00125-013-2901-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim TH, Kim YK, Kwon Y, Heo JH, Kang H, Kim G, An HJ. Deregulation of miR-519a, 153 and 485-5p and its clinicopathological relevance in ovarian epithelial tumours. Histopathology. 2010;57:734–743. doi: 10.1111/j.1365-2559.2010.03686.x. [DOI] [PubMed] [Google Scholar]
- 9.Xu Q, Sun Q, Zhang J, Yu J, Chen W, Zhang Z. Downregulation of miR-153 contributes to epithelial-mesenchymal transition and tumor metastasis in human epithelial cancer. Carcinogenesis. 2013;34:539–549. doi: 10.1093/carcin/bgs374. [DOI] [PubMed] [Google Scholar]
- 10.Yuan Y, Du W, Wang Y, Xu C, Wang J, Zhang Y, Wang H, Ju J, Zhao L, Wang Z, et al. Suppression of AKT expression by miR-153 produced anti-tumor activity in lung cancer. Int J Cancer. 2015;136:1333–1340. doi: 10.1002/ijc.29103. [DOI] [PubMed] [Google Scholar]
- 11.Zhang L, Pickard K, Jenei V, Bullock MD, Bruce A, Mitter R, Kelly G, Paraskeva C, Strefford J, Primrose J, et al. MiR-153 supports colorectal cancer progression via pleiotropic effects that enhance invasion and chemotherapeutic resistance. Cancer Res. 2013;73:6435–6447. doi: 10.1158/0008-5472.CAN-12-3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Erickson HS, Albert PS, Gillespie JW, Wallis BS, Rodriguez-Canales J, Linehan WM, Gonzalez S, Velasco A, Chuaqui RF, Emmert-Buck MR. Assessment of normalization strategies for quantitative RT-PCR using microdissected tissue samples. Lab Invest. 2007;87:951–962. doi: 10.1038/labinvest.3700659. [DOI] [PubMed] [Google Scholar]
- 13.Dweep H, Gretz N, Sticht C. MiRWalk database for miRNA-target interactions. Methods Mol Biol. 2014;1182:289–305. doi: 10.1007/978-1-4939-1062-5_25. [DOI] [PubMed] [Google Scholar]
- 14.Nandan MO, Chanchevalap S, Dalton WB, Yang VW. Kruppel-like factor 5 promotes mitosis by activating the cyclin B1/Cdc2 complex during oncogenic Ras-mediated transformation. FEBS Lett. 2005;579:4757–4762. doi: 10.1016/j.febslet.2005.07.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Y, Wen JK, Dong LH, Zheng B, Han M. Kruppel-like factor (KLF) 5 mediates cyclin D1 expression and cell proliferation via interaction with c-Jun in Ang II-induced VSMCs. Acta Pharmacol Sin. 2010;31:10–18. doi: 10.1038/aps.2009.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bao B, Rodriguez-Melendez R, Zempleni J. Cytosine methylation in miR-153 gene promoters increases the expression of holocarboxylase synthetase, thereby increasing the abundance of histone H4 biotinylation marks in HEK-293 human kidney cells. J Nutr Biochem. 2012;23:635–639. doi: 10.1016/j.jnutbio.2011.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ghaleb AM, Nandan MO, Chanchevalap S, Dalton WB, Hisamuddin IM, Yang VW. Kruppel-like factors 4 and 5: The yin and yang regulators of cellular proliferation. Cell Res. 2005;15:92–96. doi: 10.1038/sj.cr.7290271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dong JT, Chen C. Essential role of KLF5 transcription factor in cell proliferation and differentiation and its implications for human diseases. Cell Mol Life Sci. 2009;66:2691–2706. doi: 10.1007/s00018-009-0045-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu R, Dong JT, Chen C. Role of KLF5 in hormonal signaling and breast cancer development. Vitam Horm. 2013;93:213–225. doi: 10.1016/B978-0-12-416673-8.00002-2. [DOI] [PubMed] [Google Scholar]
- 20.Bialkowska AB, Liu Y, Nandan MO, Yang VW. A colon cancer-derived mutant of Kruppel-like factor 5 (KLF5) is resistant to degradation by glycogen synthase kinase 3β (GSK3β) and the E3 ubiquitin ligase F-box and WD repeat domain-containing 7α (FBW7 α) J Biol Chem. 2014;289:5997–6005. doi: 10.1074/jbc.M113.508549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frigo DE, Sherk AB, Wittmann BM, Norris JD, Wang Q, Joseph JD, Toner AP, Brown M, McDonnell DP. Induction of Kruppel-like factor 5 expression by androgens results in increased CXCR4-dependent migration of prostate cancer cells in vitro. Mol Endocrinol. 2009;23:1385–1396. doi: 10.1210/me.2009-0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li X, Liu X, Xu Y, Liu J, Xie M, Ni W, Chen S. KLF5 promotes hypoxia-induced survival and inhibits apoptosis in non-small cell lung cancer cells via HIF-1α. Int J Oncol. 2014;45:1507–1514. doi: 10.3892/ijo.2014.2544. [DOI] [PubMed] [Google Scholar]
- 23.Nakaya T, Ogawa S, Manabe I, Tanaka M, Sanada M, Sato T, Taketo MM, Nakao K, Clevers H, Fukayama M, et al. KLF5 regulates the integrity and oncogenicity of intestinal stem cells. Cancer Res. 2014;74:2882–2891. doi: 10.1158/0008-5472.CAN-13-2574. [DOI] [PubMed] [Google Scholar]
- 24.Zhu N, Gu L, Findley HW, Chen C, Dong JT, Yang L, Zhou M. KLF5 Interacts with p53 in regulating survivin expression in acute lymphoblastic leukemia. J Biol Chem. 2006;281:14711–14718. doi: 10.1074/jbc.M513810200. [DOI] [PubMed] [Google Scholar]
- 25.Li Q, Dong Z, Zhou F, Cai X, Gao Y, Wang LW. Kruppel-like factor 5 promotes lung tumorigenesis through upregulation of Sox4. Cell Physiol Biochem. 2014;33:1–10. doi: 10.1159/000356645. [DOI] [PubMed] [Google Scholar]