Abstract
The aim of the present study was to screen differentially co-expressed genes and the involved transcription factors (TFs) and microRNAs (miRNAs) in venous thromboembolism (VTE). Microarray data of GSE19151 were downloaded from Gene Expression Omnibus, including 70 patients with VTE and 63 healthy controls. Principal component analysis (PCA) was performed using R software. Differential co-expression analysis was performed using R, followed by screening of modules using Cytoscape. Functional annotation was performed using Database for Annotation, Visualization, and Integrated Discovery. Moreover, Fisher test was used to screen key TFs and miRNAs for the modules. PCA revealed the disease and healthy samples could not be distinguished at the gene expression level. A total of 4,796 upregulated differentially co-expressed genes (e.g. zinc finger protein 264, electron-transfer-flavoprotein, beta polypeptide and Janus kinase 2) and 3,629 downregulated differentially co-expressed genes (e.g. adenylate cyclase 7 and single-stranded DNA binding protein 2) were identified, which were further mined to obtain 17 and eight modules separately. Functional annotation revealed that the largest upregulated module was primarily associated with acetylation and the largest downregulated module was mainly involved in mitochondrion. Moreover, 48 TFs and 62 miRNA families were screened for the 17 upregulated modules, such as E2F transcription factor 4, miR-30 and miR-135 regulating the largest module. Conversely, 35 TFs and 18 miRNA families were identified for the 8 downregulated modules, including mitochondrial ribosomal protein S12 and miR-23 regulating the largest module. Differentially co-expressed genes regulated by TFs and miRNAs may jointly contribute to the abnormal acetylation and mitochondrion presentation in the progression of VTE.
Keywords: venous thromboembolism, differentially co-expressed gene pairs, modules, transcription factors, microRNAs
Introduction
Venous thromboembolism (VTE) is defined as deep vein thrombosis, pulmonary embolism or both, which affects an estimated 300,000–600,000 individuals in the U.S. each year (1). A population-based case-control study revealed that the 2-year cumulative incidence of VTE is between 0.8 and 8% (2). Recurrent VTE accounts for as many as 30% of patients after stopping a standard course of anticoagulant therapy (3). Cancer patients have seven-fold risk of developing VTE compared with patients without cancer (4). Inherited and acquired risk factors could increase individual's risk for developing VTE (5). The investigation of the molecular mechanisms underlying VTE could provide novel intervention methods for VTE.
Gene microarray is a powerful and cost-effective tool for large-scale analysis of gene expression features in disease (6). By using this tool, T cell-mediated immunity dysfunction has been reported to play an important role in the progression of VTE (7). The genes associated with immune cells, such as CD3, CD4 and CD8, exhibit dysregulation in VTE (8). Additionally, gene expression data of whole blood indicate the involvement of insulin-like growth-factor-1 receptor and peroxisome proliferator-activated receptor PPAR-δ in platelet aggregation in VTE (9). However, genes and their products cannot perform their functions in isolation, but are coordinate (10), and the dynamic switch of a gene from one community to another implies altered gene function (11). Thus, differential co-expression analysis could help to reveal molecular mechanisms of phenotypic changes through identifying subtle changes in gene expression coordination (12–14). Moreover, transcription factors, such as Kruppel-like factor 2, are involved in the pathogenesis of arterial thrombosis through exerting regulatory effect on gene expression (15). MicroRNAs (miRNAs) are also important for the development of thrombosis via regulating target genes (16). However, studies of transcription factors and miRNAs in VTE remain rare (17,18). The identification of the transcription factors and miRNAs involved in VTE may help to elucidate the gene expression alterations and provide novel therapeutic targets for VTE.
In the present study, based on the published gene expression profile data (GSE19151) of 70 VTE samples and 63 normal samples (9), the differentially co-expressed gene pairs in VTE samples were screened, followed by construction of densely connected network modules. Furthermore, functional analysis of modules was performed to reveal the underlying molecular mechanisms. Additionally, the key transcription factors and miRNAs of modules were screened to clarify the regulatory network of co-expressed genes in VTE.
Materials and methods
Data source and preprocessing
Gene expression profile data of GSE19151 (9) on platform GPL571 [HG-U133A_2] Affymetrix Human Genome U133A 2.0 Array were downloaded from Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/) at the National Center for Biotechnology Information (NCBI) online database (http://www.ncbi.nlm.nih.gov/pubmed/). These data included 70 blood samples from adult patients with VTE receiving warfarin and 63 blood samples from healthy controls. The raw expression profile data at the probe level were converted into the corresponding gene expression data based on the platform, followed by medium normalization using the preprocess Core package, version 1.28.0 (http://www.bioconductor.org/packages/3.0/bioc/html/preprocessCore.html). As a single gene may correspond to multiple probes, the expression level of the gene was calculated by taking the average of multiple probe expression values.
Principal component analysis (PCA)
PCA was performed to analyze the association between the gene expression profile and VTE using the plot PCA function provided by R software (http://www.bioconductor.org/). PCA is a multi-statistical analysis for detecting the correlation among multiple variables based on their variance, and is usually applied to identify the dominant PCA components from primary variables (19). Generally, the size of variance represents the information contained in the variable. The F1 with the largest variance in all linear combinations was considered to be the dominant PCA component. If the F1 could not include all the information of the variables, then F2 with the second largest variance was selected as the second PCA component. The information in F1 was not included in F2. Specifically, if Cover (F1, F2)=0, F2 was defined as the second dominant PCA component, and so on.
Identification of differentially co-expressed associations
The differential co-expression analysis was performed to investigate the differentially expressed gene interconnection at the expression level from a global perspective using the Derived Concentration Guidelines Level (DCGL) package in R (20). Briefly, Pearson correlation coefficient of paired genes (u,v) in normal samples and diseased samples were calculated based on the following formula (Formula 1):
In Formula 1, ‘ui’ represents the expression level of gene u in i status, and ‘vi’ represents the expression level of gene v in i status (i=1, diseased status; i=2, normal status).
We speculated that if the correlation coefficient of this gene pair in diseased samples obeyed the normal distribution: R1(u,v)~N(µ1, δ21 (µ, mean value of the random variables; and δ, variance of the random variables) and the correlation coefficient of this gene pair in normal samples obeyed the normal distribution: R1(u,v)~N(µ2, δ22), then the change of correlation coefficient of these paired genes would also obey the normal distribution: R1-R2~N(µ1-µ2, δ21+δ22). The differences between R1 and R2 for each gene pair were then treated with normalization and converted into corresponding T values, using the following formula (Formula 2):
In Formula 2, R represents the calculated Pearson correlation coefficient, µ represents the mean value of the random variables and δ represents the variance of the random variables (i=1, disease status; i=2, normal status). Subsequently, the T value for one pair of genes was calculated 100 times to ensure the precision of the T value. Then the results were set as a background to test the significance of T values. Gene pairs with P<0.05 were considered as differentially co-expressed gene pairs, of whose T-value was >0 were defined as an upregulated co-expressed gene pair, and T-value <0 was defined as a downregulated co-expressed gene pair.
Mining the network modules of differentially co-expressed genes
Here, the Molecular Complex Detection (MCODE) algorithm (21) from the plug-in clusterViz for Cytoscape software (22) was applied to mine the network modules of differentially co-expressed gene pairs. MCODE consisted of three stages: i) Vertex weighting; ii) complex prediction; and iii) an optional post-processing step. Briefly, the clustering coefficient of node i was defined as node weighting (Ci; Formula 3):
In Formula 3, Ki represented the number of nodes directly connected with node i, and n represented the number of edges among the Ki nodes. The node with highest weighting was set as seed of the module. Then, the node J with weighting ratio (Wj/Wseed) more than the threshold was selected and added into the module. Meanwhile, the weighting of nodes was rechecked and when no node meets the threshold, the module was successfully screened. The checked node was deleted from the network, so each node was not checked more than once. In addition, functional annotation analysis for genes in differentially co-expressed modules was performed using the Database for Annotation, Visualization and Integrated Discovery tool (23).
Selecting co-expressed transcription factors and miRNAs with target genes
To elucidate the potential co-expression associations between transcription factors and their target genes in VTE, an integrated transcription factor platform database was used, which possessed 4,105 putative transcription factors and 69,496 potential transcription factor-target pairs for humans (24). Furthermore, co-expression associations between miRNAs and target genes were downloaded from TargetScan database (http://www.targetscan.org/), which predicted miRNA-target genes basing on the base complementarity within the 3′UTR and the sequence conservativeness of target genes among species (25). Herein, the correlation between target genes of transcription factors and genes in differentially co-expressed network modules were calculated using Fisher's exact test by R software in Bioconductor (http://www.bioconductor.org/). The transcription factors with P<0.01 were screened out and considered to be key transcription factors. Similarly, using Fisher test, the miRNAs with P<0.01 were also screened out and considered to be key miRNAs.
Results
PCA
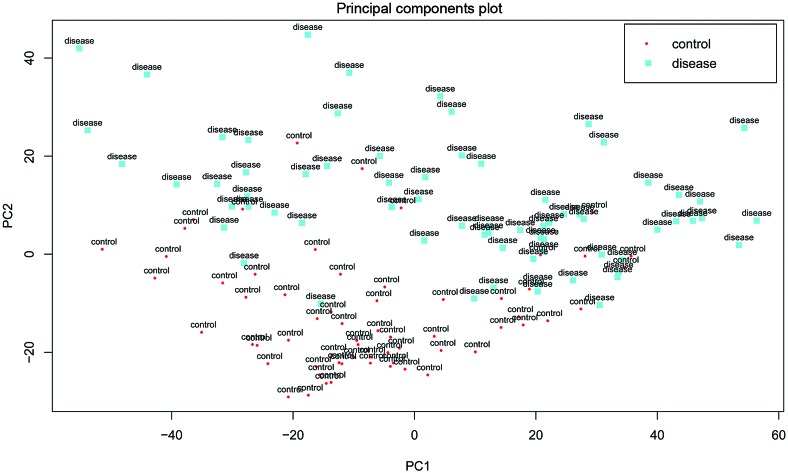
Following data preprocessing, a total of 13,035 genes were identified. Besides, based on the PCA analysis for gene expression profile data, it was found that the gene expression level in certain diseased samples was similar to that in normal samples, and the two groups were not distinguished absolutely at the gene expression level (Fig. 1).
Figure 1.
Principal component analysis plot of 63 normal samples and 70 venous thromboembolism samples.
Differentially co-expressed gene pairs
On the basis of the two formulas (1) and (2), a total of 27,913 gene pairs were identified in 4,796 differentially co-expressed genes with upregulation, such as zinc finger protein 264 (ZNF264), electron-transfer-flavoprotein beta polypeptide (ETFB) and Janus kinase 2 (JAK2) (Table I). Furthermore, 9,642 gene pairs were identified in 3,629 differentially co-expressed genes with downregulation, such as adenylate cyclase 7 (ADCY7) and single-stranded DNA binding protein 2 (SSBP2) (Table I).
Table I.
Top ten differentially co-expressed gene pairs.
| Gene 1 | Gene 2 | Normal group | Disease group | P-value |
|---|---|---|---|---|
| Upregulated | ||||
| ZNF264 | IARS2 | 0.673338 | −0.73511 | 3.89E-04 |
| SEC31B | GHITM | 0.701243 | −0.70213 | 1.41E-03 |
| ZNF264 | TSPYL1 | 0.584623 | −0.79107 | 1.62E-03 |
| GHITM | EIF6 | 0.658365 | −0.66949 | 1.67E-03 |
| ZNF264 | NNT | 0.527611 | −0.78110 | 2.55E-03 |
| MED4 | ETFB | 0.602538 | −0.69157 | 2.80E-03 |
| UBE2K | C19orf60 | 0.520002 | −0.77137 | 3.70E-03 |
| MFF | ETFB | 0.601631 | −0.68076 | 3.80E-03 |
| ZNF264 | PNMA1 | 0.539847 | −0.73907 | 6.30E-03 |
| SH3BGRL | SEC31B | 0.559038 | −0.71832 | 7.00E-03 |
| Downregulated | ||||
| ETFB | DUSP11 | 0.684080 | −0.61501 | 9.32E-05 |
| PLCG1 | ADCY7 | 0.686949 | −0.60522 | 9.62E-05 |
| RAB4A | C12orf44 | 0.766989 | −0.51036 | 1.00E-04 |
| CHD3 | ADCY7 | 0.654376 | −0.61336 | 1.10E-04 |
| MYL6 | IK | 0.666814 | −0.59576 | 1.90E-04 |
| C12orf44 | BCAS2 | 0.658958 | −0.59733 | 1.97E-04 |
| ZMIZ1 | TRAM1 | −0.746020 | 0.506433 | 1.97E-04 |
| OAT | C21orf45 | 0.666831 | −0.58034 | 2.00E-04 |
| UBE2K | ETFB | 0.672036 | −0.57243 | 2.10E-04 |
| TANK | PRRC1 | 0.631341 | −0.61247 | 3.19E-04 |
Differentially co-expressed gene pairs were ranked based on the P-value.
Modules of differentially co-expressed genes
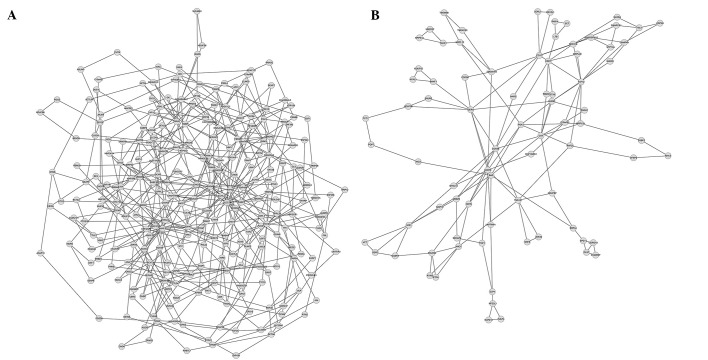
Using MCODE algorithm in the clusterViz plug-in for Cytoscape, 17 modules were mined in the co-expressed gene pairs with upregulation. The smallest module possessed three genes and three edges (lines; representing an interaction between two genes) while the largest module contained 263 genes and 482 edges (Fig. 2). By contrast, eight modules were mined in the differentially co-expressed gene pairs with downregulation. The smallest module had three genes and three edges while the largest module included 80 genes and 137 edges (Fig. 2). Functional annotation for the largest upregulated module was performed, and revealed that this module was primarily involved in functional term of acetylation (Table II). The functional annotation for the largest downregulated module revealed that it was mainly related to functional term of mitochondrion (Table II).
Figure 2.
Two largest modules of differentially co-expressed genes. (A) Upregulated module and (B) downregulated module.
Table II.
Top five most significantly enriched terms of differentially co-expressed genes in Module 1.
| Category | Term | Count | P-value |
|---|---|---|---|
| Upregulated genes | |||
| SP_PIR_KEYWORDS | Acetylation | 78 | 8.19E-12 |
| SP_PIR_KEYWORDS | Phosphoprotein | 148 | 1.62E-10 |
| GOTERM_CC_FAT | GO:0031981~nuclear lumen | 44 | 2.18E-07 |
| GOTERM_CC_FAT | GO:0005654~nucleoplasm | 32 | 4.58E-07 |
| GOTERM_CC_FAT | GO:0070013~intracellular organelle lumen | 48 | 1.50E-06 |
| Downregulated genes | |||
| GOTERM_CC_FAT | GO:0005739~mitochondrion | 20 | 1.43E-06 |
| SP_PIR_KEYWORDS | Mitochondrion | 15 | 6.54E-06 |
| SP_PIR_KEYWORDS | Transit peptide | 11 | 2.45E-05 |
| SP_PIR_KEYWORDS | Acetylation | 26 | 3.23E-05 |
| GOTERM_CC_FAT | GO:0044429~mitochondrial part | 13 | 4.54E-05 |
SP, splice patterns; PIR, protein information resource; GO, gene ontology; CC, cellular component.
Key transcription factors and miRNAs for differentially co-expressed modules
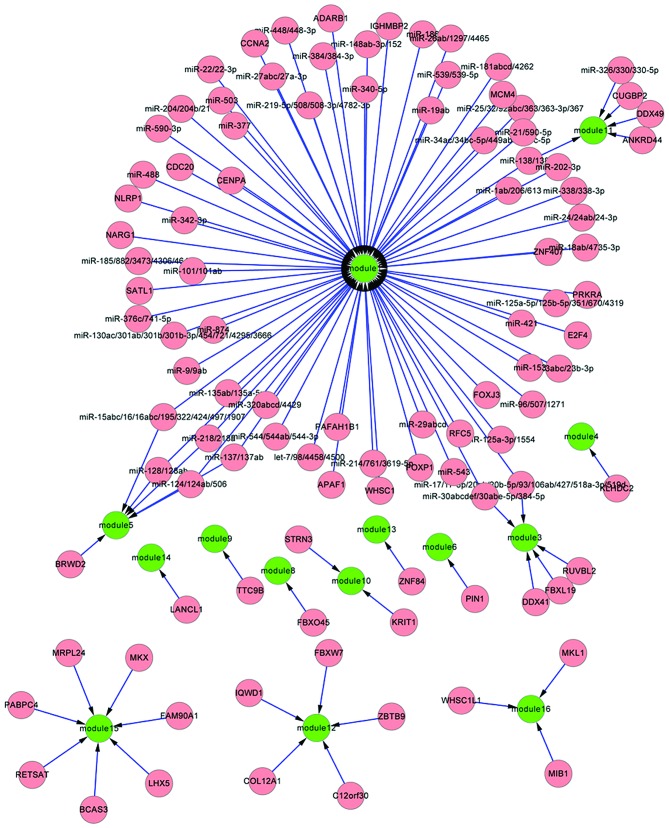
Using Fisher's test, 48 transcription factors were screened out for the 17 upregulated modules, such as JAK2 in module 1 regulated by E2F transcription factor 4 (E2F4), and low density lipoprotein receptor-related protein 1 (LRP1) in module 3 regulated by transcription factor F-box and leucine-rich repeat protein 19 (FBXL19) (Fig. 3). This suggested that these transcription factors could enhance the co-expression association between co-expressed gene pairs in modules under diseased status. Furthermore, 62 miRNA families were selected for 17 upregulated modules, including miR-30 and miR-135 regulating module 1 (Fig. 3).
Figure 3.
Regulatory networks between key transcription factors or micro (mi)RNAs with differentially co-expressed modules of upregulated genes. Green nodes represent differentially co-expressed modules, and pink nodes represent transcription factors or miRNA families.
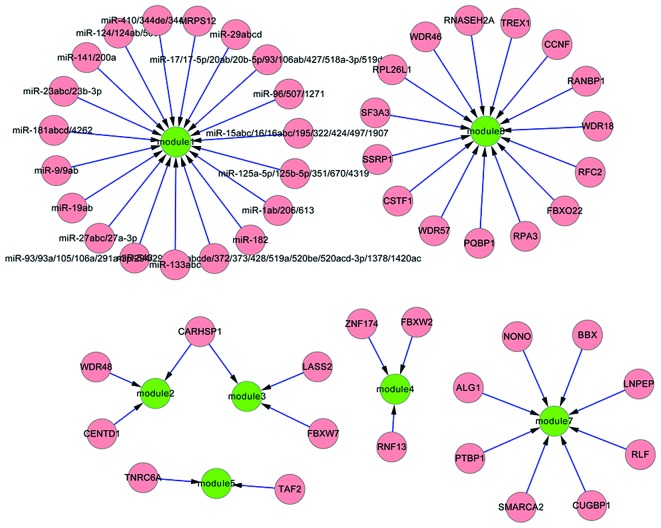
Furthermore, 35 transcription factors were identified for the eight downregulated modules, for instance, SSBP2 in module 1 regulated by transcription factor (mitochondrial ribosomal protein S12) MRPS12 (Fig. 4). This implied that these transcription factors could weaken the co-expressed associations between co-expressed gene pairs in modules under diseased status. Additionally, 18 miRNA families were selected for the eight downregulated modules, such as miR-23 targeting module 1.
Figure 4.
Regulatory association between key transcription factors or micro (mi)RNAs with differentially co-expressed modules of downregulated genes. Green nodes represent differentially co-expressed modules, and pink nodes represent transcription factors or miRNA families.
Discussion
By re-analyzing the gene expression profile data of 63 VTE samples and 70 normal samples, this study obtained a total of 27,913 pairs of 4,796 upregulated differentially co-expressed genes and 9,642 pairs of 3,629 downregulated differentially co-expressed genes. Altogether 17 upregulated and eight downregulated modules were screened based on the differentially co-expressed genes. Moreover, large numbers of transcription factors and miRNAs were identified to have a regulatory effect on the differentially co-expressed genes of modules.
Herein, co-expression analysis presented 4,796 upregulated differentially co-expressed genes in VTE, such as ZNF264 and ETFB. ZNF264 is reported to be associated with inflammation (26). ETFB has been identified as a candidate protein that participates in the mechanoregulation of fibroblast cell number in collagen gel culture (27). The upregulation of those differentially co-expressed genes may jointly contribute to abnormal inflammation-induced VTE. In addition, based on the differentially co-expressed genes, 17 modules were screened, among which the largest module was predominantly associated with the functional term of acetylation. A previous study reported that the abnormal acetylation of proteins may contribute to blood coagulation in the development of VTE (28). Thus, the genes involved in this module were speculated to induce abnormal acetylation, such as JAK2, whose mutation may be of help in early myeloproliferative neoplasms detection and clinical management of splanchnic venous thrombosis patients (29). Notably, transcription factor E2F4 was revealed to regulate the largest module. In previous studies, E2F4 has been identified as the most abundant E2F protein in numerous cell types (30) and the other family member E2F1 is related to histone acetylation (31), implying the involvement of E2F4 in acetylation. Furthermore, miR-30 and miR-135 family members were revealed to regulate the largest module. Previously, miR-30c has been shown to directly interact with PAI-1 and is suggested to be a potential biomarker for inflammatory and thrombotic disorders (32). MiR-135 has an important role in promoting portal vein tumor thrombus tumorigenesis (33). Those miRNAs may be involved in the progression of VTE via regulating acetylation. It may be therefore speculated that the upregulated differentially co-expressed genes may jointly contribute to abnormal acetylation in the progression of VTE. Transcription factors (e.g. E2F4) and miRNAs (e.g. miR-30 and miR-135) may contribute to the abnormal upregulation of the co-expressed genes.
In addition, 3,629 downregulated differentially co-expressed genes were revealed in VTE, including ADCY7, which may be the major form of adenylyl cyclase expressed in human platelets and is an important receptor-G protein-coupled effector involved in numerous neuronal functions in the central nervous system (34,35). Herein, on the basis of the downregulated differentially co-expressed genes, a total of eight modules were screened. The largest module was primarily related to the functional term mitochondrion. The activities of electron transport chain-associated enzymes are reduced in estrogen-receptor beta-deficiency mice, and mitochondrial energetic processes of platelets are associated with the production of circulating thrombogenic microvesicles, leading to thromboembolism (36). SSBP2 in the largest module is associated with normal protein C plasma level, which influences the risk of venous thrombosis (37), suggesting the involvement of SSBP2 in VTE. Notably, a number of transcription factors were revealed to exert a regulatory effect on the modules, such as mitochondrial ribosomal protein S12 (MRPS12) targeting the largest module. A previous study indicated that MRPS12 contains an array of four CCAAT boxes, and has been associated with mitochondrial stress (38). Furthermore, numerous miRNAs exert a regulatory effect on the modules. miR-23, targeting the largest module, is reported to be a novel therapeutic target in vascular disorders and ischemic heart disease (39). The transcription factors and miRNAs may be involved in the regulation of mitochondrion in the progression of VTE.
In conclusion, using bioinformatics a number of differentially co-expressed genes have been indicated to jointly contribute to the abnormal acetylation and mitochondrion presentation. Transcription factors (e.g. E2F4 and MRPS12) and miRNAs (e.g. miR-30, miR-135 and miR-23) may contribute to the dysregulation of the co-expressed genes. However, the present results require further validation. We anticipate that this study may provide novel insights into the mechanism and therapeutic targets associated with VTE, and further studies are required to investigate the function of these key transcription factors and miRNAs in VTE.
Acknowledgements
The present study was supported by the Science and Technology Program of Nantong, China (grant no. MS22015050).
Glossary
Abbreviations
- TFs
transcription factors miRNAs, microRNAs
- VTE
venous thromboembolism
- PCA
principal component analysis
- GEO
gene expression omnibus
- NCBI
national center for biotechnology information
- MCODE
molecular complex detection
- ZNF264
zinc finger protein 264
- ETFB
electron-transfer-flavoprotein beta polypeptide
- JAK2
janus kinase 2
- ADCY7
adenylate cyclase 7
- SSBP2
single-stranded DNA binding protein 2
- E2F4
E2F transcription factor 4
- LRP1
lipoprotein receptor-related protein 1
- FBXL19
F-box and leucine-rich repeat protein 19
References
- 1.Beckman MG, Hooper WC, Critchley SE, Ortel TL. Venous thromboembolism: A public health concern. Am J Prev Med. 2010;38(Suppl 4):S495–S501. doi: 10.1016/j.amepre.2009.12.017. [DOI] [PubMed] [Google Scholar]
- 2.Chew HK, Wun T, Harvey D, Zhou H, White RH. Incidence of venous thromboembolism and its effect on survival among patients with common cancers. Arch Intern Med. 2006;166:458–464. doi: 10.1001/archinte.166.4.458. [DOI] [PubMed] [Google Scholar]
- 3.Heit JA, Mohr DN, Silverstein MD, Petterson TM, O'Fallon WM, Melton LJ., III Predictors of recurrence after deep vein thrombosis and pulmonary embolism: A population-based cohort study. Arch Intern Med. 2000;160:761–768. doi: 10.1001/archinte.160.6.761. [DOI] [PubMed] [Google Scholar]
- 4.Mandalà M, Falanga A, Roila F. ESMO Guidelines Working Group: Management of venous thromboembolism (VTE) in cancer patients: ESMO clinical practice guidelines. Ann Oncol. 2011;22(Suppl 6):vi85–vi92. doi: 10.1093/annonc/mdr392. [DOI] [PubMed] [Google Scholar]
- 5.López JA, Kearon C, Lee AY. Deep venous thrombosis. ASH Education Program Book. 2004;2004:439–456. doi: 10.1182/asheducation-2004.1.439. [DOI] [PubMed] [Google Scholar]
- 6.Alavi-Majd H, Khodakarim S, Zayeri F, Rezaei-Tavirani M, Tabatabaei SM, Heydarpour-Meymeh M. Assessment of gene set analysis methods based on microarray data. Gene. 2014;534:383–389. doi: 10.1016/j.gene.2013.08.063. [DOI] [PubMed] [Google Scholar]
- 7.Song HM, Gong Z, Wang LM, Zhang XY. The expression of T cell immune-related gene mRNAs in peripheral blood mononuclear cells from patients with venous thromboembolism. Zhonghua Nei Ke Za Zhi. 2012;51:551–553. (In Chinese) [PubMed] [Google Scholar]
- 8.Duan Q, Gong Z, Song H, Wang L, Yang F, Lv W, Song Y. Symptomatic venous thromboembolism is a disease related to infection and immune dysfunction. Int J Med Sci. 2012;9:453–461. doi: 10.7150/ijms.4453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lewis DA, Stashenko GJ, Akay OM, Price LI, Owzar K, Ginsburg GS, Chi JT, Ortel TL. Whole blood gene expression analyses in patients with single versus recurrent venous thromboembolism. Thromb Res. 2011;128:536–540. doi: 10.1016/j.thromres.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rachlin J, Cohen DD, Cantor C, Kasif S. Biological context networks: A mosaic view of the interactome. Mol Syst Biol. 2006;2:66. doi: 10.1038/msb4100103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang Y, Li H, Hu H, Yan X, Waterman MS, Huang H, Zhou XJ. Systematic discovery of functional modules and context-specific functional annotation of human genome. Bioinformatics. 2007;23:i222–i229. doi: 10.1093/bioinformatics/btm222. [DOI] [PubMed] [Google Scholar]
- 12.Lee HK, Hsu AK, Sajdak J, Qin J, Pavlidis P. Coexpression analysis of human genes across many microarray data sets. Genome Res. 2004;14:1085–1094. doi: 10.1101/gr.1910904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Choi JK, Yu U, Yoo OJ, Kim S. Differential coexpression analysis using microarray data and its application to human cancer. Bioinformatics. 2005;21:4348–4355. doi: 10.1093/bioinformatics/bti722. [DOI] [PubMed] [Google Scholar]
- 14.Yu H, Liu BH, Ye ZQ, Li C, Li YX, Li YY. Link-based quantitative methods to identify differentially coexpressed genes and gene pairs. BMC Bioinformatics. 2011;12:315. doi: 10.1186/1471-2105-12-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nie XM, Su LX, Xu RX, Guo YL, Zhou YJ, Li JJ. Kruppel-like factor 2 might mediate the rapamycin-induced arterial thrombosis in vivo: Implications for stent thrombosis in patients. Chin Med J (Engl) 2013;126:2636–2640. [PubMed] [Google Scholar]
- 16.Blokhin IO, Lentz SR. Mechanisms of thrombosis in obesity. Curr Opin Hematol. 2013;20:437–444. doi: 10.1097/MOH.0b013e3283634443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kong M, Kim Y, Lee C. Functional investigation of a venous thromboembolism GWAS signal in a promoter region of coagulation factor XI gene. Mol Biol Rep. 2014;4:2015–2019. doi: 10.1007/s11033-014-3049-1. [DOI] [PubMed] [Google Scholar]
- 18.Qin J, Liang H, Shi D, Dai J, Xu Z, Chen D, Chen X, Jiang Q. A panel of microRNAs as a new biomarkers for the detection of deep vein thrombosis. J Thromb Thrombolysis. 2015;39:215–221. doi: 10.1007/s11239-014-1131-0. [DOI] [PubMed] [Google Scholar]
- 19.Khan J, Wei JS, Ringnér M, Saal LH, Ladanyi M, Westermann F, Berthold F, Schwab M, Antonescu CR, Peterson C, Meltzer PS. Classification and diagnostic prediction of cancers using gene expression profiling and artificial neural networks. Nat Med. 2001;7:673–679. doi: 10.1038/89044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu BH, Yu H, Tu K, Li C, Li YX, Li YY. DCGL: An R package for identifying differentially coexpressed genes and links from gene expression microarray data. Bioinformatics. 2010;26:2637–2638. doi: 10.1093/bioinformatics/btq471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Szalay-Beko M, Palotai R, Szappanos B, Kovács IA, Papp B, Csermely P. ModuLand plug-in for Cytoscape: Determination of hierarchical layers of overlapping network modules and community centrality. Bioinformatics. 2012;28:2202–2204. doi: 10.1093/bioinformatics/bts352. [DOI] [PubMed] [Google Scholar]
- 22.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:P3. doi: 10.1186/gb-2003-4-5-p3. [DOI] [PubMed] [Google Scholar]
- 24.Zheng G, Tu K, Yang Q, Xiong Y, Wei C, Xie L, Zhu Y, Li Y. ITFP: An integrated platform of mammalian transcription factors. Bioinformatics. 2008;24:2416–2417. doi: 10.1093/bioinformatics/btn439. [DOI] [PubMed] [Google Scholar]
- 25.Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: Determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Asimenos G, Sundquist A, Ganter B. Nucleotide-level variant analysis of next-generation sequencing data using a cloud-based data analysis pipeline. J Biomol Tech. 2011;22(Suppl):S31. [Google Scholar]
- 27.Hirokawa S, Shimanuki T, Kitajima H, Nishimori Y, Shimosaka M. Identification of ETFB as a candidate protein that participates in the mechanoregulation of fibroblast cell number in collagen gel culture. J Dermatol Sci. 2011;64:119–126. doi: 10.1016/j.jdermsci.2011.08.003. [DOI] [PubMed] [Google Scholar]
- 28.Undas A, Brummel-Ziedins K, Mann KG. Why does aspirin decrease the risk of venous thromboembolism? On old and novel antithrombotic effects of acetyl salicylic acid. J Thromb Haemost. 2014;12:1776–1787. doi: 10.1111/jth.12728. [DOI] [PubMed] [Google Scholar]
- 29.Colaizzo D, Amitrano L, Guardascione MA, Tiscia GL, D'Andrea G, Longo VA, Grandone E, Margaglione M. Outcome of patients with splanchnic venous thrombosis presenting without overt MPN: A role for the JAK2 V617F mutation re-evaluation. Thromb Res. 2013;132:e99–e104. doi: 10.1016/j.thromres.2013.07.014. [DOI] [PubMed] [Google Scholar]
- 30.Gaubatz S, Lees JA, Lindeman GJ, Livingston DM. E2F4 is exported from the nucleus in a CRM1-dependent manner. Mol Cell Biol. 2001;21:1384–1392. doi: 10.1128/MCB.21.4.1384-1392.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Taubert S, Gorrini C, Frank SR, Parisi T, Fuchs M, Chan HM, Livingston DM, Amati B. E2F-dependent histone acetylation and recruitment of the Tip60 acetyltransferase complex to chromatin in late G1. Mol Cell Biol. 2004;24:4546–4556. doi: 10.1128/MCB.24.10.4546-4556.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marchand A, Proust C, Morange PE, Lompré AM, Trégouët DA. MiR-421 and miR-30c inhibit SERPINE 1 gene expression in human endothelial cells. PloS One. 2012;7:e44532. doi: 10.1371/journal.pone.0044532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu S, Guo W, Shi J, Li N, Yu X, Xue J, Fu X, Chu K, Lu C, Zhao J, et al. MicroRNA-135a contributes to the development of portal vein tumor thrombus by promoting metastasis in hepatocellular carcinoma. J Hepatol. 2012;56:389–396. doi: 10.1016/j.jhep.2011.08.008. [DOI] [PubMed] [Google Scholar]
- 34.Hellevuo K, Welborn R, Menninger JA, Tabakoff B. Human adenylyl cyclase type 7 contains polymorphic repeats in the 3′untranslated region: Investigations of association with alcoholism. Am J Med Genet. 1997;74:95–98. doi: 10.1002/(SICI)1096-8628(19970221)74:1<95::AID-AJMG19>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 35.Sanabra C, Mengod G. Neuroanatomical distribution and neurochemical characterization of cells expressing adenylyl cyclase isoforms in mouse and rat brain. J Chem Neuroanat. 2011;41:43–54. doi: 10.1016/j.jchemneu.2010.11.001. [DOI] [PubMed] [Google Scholar]
- 36.Jayachandran M, Preston CC, Hunter LW, Jahangir A, Owen WG, Korach KS, Miller VM. Loss of estrogen receptor beta decreases mitochondrial energetic potential and increases thrombogenicity of platelets in aged female mice. Age (Dordr) 2010;32:109–121. doi: 10.1007/s11357-009-9119-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vossen CY, Koeleman BP, Hasstedt SJ, Nijman IJ, Renkens IJ, Callas PW, Rosendaal FR, Bovill EG. Genetic variants associated with protein C levels. J Thromb Haemost. 2013;11:715–723. doi: 10.1111/jth.12157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zanotto E, Lehtonen V, Jacobs HT. Modulation of Mrps12/Sarsm promoter activity in response to mitochondrial stress. Biochim Biophys Acta. 2008;1783:2352–2362. doi: 10.1016/j.bbamcr.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 39.Bang C, Fiedler J, Thum T. Cardiovascular importance of the microrna-23/27/24 family. Microcirculation. 2012;19:208–214. doi: 10.1111/j.1549-8719.2011.00153.x. [DOI] [PubMed] [Google Scholar]