Abstract
The field of medical genomics involves translating high throughput genetic methods to the clinic, in order to improve diagnostic efficiency and treatment decision making. Technical questions related to sample enrichment, sequencing methodologies and variant identification and calling algorithms, still need careful investigation in order to validate the analytical step of next generation sequencing techniques for clinical applications. However, the main foreseeable challenge will be interpreting the clinical significance of the variants observed in a given patient, as well as their significance for family members and for other patients.
Every step in the variant interpretation process has limitations and difficulties, and its quote of contribution to false positive and false negative results. There is no single piece of evidence enough on its own to make firm conclusions on the pathogenicity and disease causality of a given variant.
A plethora of automated analysis software tools is being developed that will enhance efficiency and accuracy. However a risk of misinterpretation could derive from biased biorepository content, facilitated by annotation of variant functional consequences using previous datasets stored in the same or linked repositories. In order to improve variant interpretation and avoid an exponential accumulation of confounding noise in the medical literature, the use of terms in a standard way should be sought and requested when reporting genetic variants and their consequences. Generally, stepwise and linear interpretation processes are likely to overrate some pieces of evidence while underscoring others. Algorithms are needed that allow a multidimensional, parallel analysis of diverse lines of evidence to be carried out by expert teams for specific genes, cellular pathways or disorders.
Keywords: Genetic variant, Interpretation challenge, Pathogenicity assessment, Clinical significance, Informatics pipeline, Family co-segregation
1. Introduction
Next generation sequencing (NGS) technologies are rapidly becoming a routine tool in the diagnostic workup of patients with diverse conditions, including tumor profiling. Medical genomics refers to the ability to simultaneously query the diagnostically relevant gene set of a given person for clinical decisions. Sequencing of the complete set of protein coding exons of an individual — whole exome sequencing (WES) — has enhanced the identification of genetic defect of rare diseases (Wan et al., 2012, Keller et al., 2013). These technologies can also be applied to decipher more common syndromes (Cirulli and Goldstein, 2010, Kiezun et al., 2012). Decision-making in oncology can now be based on the singular molecular signature of the tumor with implications in pathology and response to treatment or individual prognosis (Normanno et al., 2013). Another approach to the diagnosis of genetically heterogeneous disorders is the simultaneous sequence of a panel of genes associated with a given syndrome. NGS also harbors potential to delineate an individual's pharmacogenetic profile (Patrinos et al., 2013). The use of high throughput molecular analysis for clinical decision making is often referred to as personalized medicine or personal genomics, although warnings have also been raised about myths and inflated expectations that may come along with these somewhat blurry terms (Salari et al., 2012).
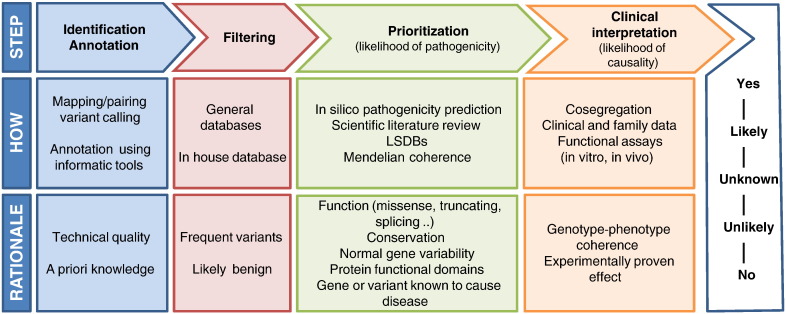
How far are we still from being able to interpret all genetic variations accurately in a clinical context? Many challenges lie ahead before NGS can be integrated as part of routine medical care. The process to know which one among the thousands of genetic variants harbored within an individual's genome is clinically relevant generally involving a number of steps summarized in Fig. 1. In the following sections we review some of the challenges and limitations encountered along this path, as well as potential sources of errors that must be taken into account for an adequate clinical interpretation of genetic variants.
Fig. 1.
Stepwise evidence pipeline for clinical interpretation genetic variants. After identification and automatic annotation, likely benign variants are filtered out and the remaining variants are prioritized. The weight of different lines of evidence leads to final clinical interpretation.
2. Need for accurate use of terms on genetic variations and their consequences
A first source of difficulty comes from the imprecise use of vocabulary referred to genetic variations and their consequences. The terms polymorphism and mutation do not bear implications on their functional consequences, however they are often used with that meaning. A polymorphism is a genetic variant present in ≥ 1% of the population, whereas a mutation is any change in the DNA sequence compared to the previous state or wild type. Neither concepts imply whether they are or are not disease-causing. Just because a polymorphism is not so rare, it does not necessarily mean that it is benign (not associated with a disorder) or neutral (without functional consequences). Because of the potential for misinterpretation of polymorphism and mutation, the term genetic variant is currently favored, as defined by the presence of a particular allele — at a nucleotide position, gene or locus — that is not the most commonly encountered allele in the general population. Thus, the term genetic variant does not imply any a priori assumption on the frequency of the variant allele or its potential effect on the health of the individual carrying it. Also, terms such as neutral, benign, functional, pathogenic, deleterious, damaging, disease-associated and causal, when referring to a genetic variant, are often used in ill-defined manner throughout the medical literature. For instance, pathogenic is often equaled to disease-causing, which is not necessarily always the case. While functionality, deleteriousness, pathogenicity and disease causality may be strongly related terms, they are not interchangeable. As for the term phenotype, it must be specified whether the authors mean abnormalities detectable at a cellular/organ level, to biochemical alterations that can be measured, or to abnormal clinical traits that can be observed in an individual, animal model or cellular construct. A phenotype can be made up of several endophenotypes that may provide useful clinical measures (Mann et al., 2009).
Another issue is the system level at which the consequence of a genetic variant is being described. For example, the variant may be deleterious at a cellular level (causes a loss of function in a given cellular process), but not necessarily deleterious for the organ or individual. When discussing the potential effects of a given genetic variant on disease, there is a tendency to classify the variant in a simple three or five-tiered scheme (pathogenic, likely pathogenic, unlikely pathogenic, non pathogenic, unknown). This scheme, however, ignores the complexity of biological processes that can imply other types of relationships between a variant and a clinical manifestation (predisposing, triggering, modifying, protective, etc.), as well as digenic or polygenic disorders. We call for using terminology – and requesting its use in scientific publications – with more precision when describing the consequences of genetic variants, such as done in the recent paper by MacArthur et al. (2014). While a consensus is developed by the genetics community on the definition of these terms and how they should be used, it would be a good practice that curators of genetic databases define their intended meaning.
3. Variant identification and annotation
The first step towards genetic variant interpretation is the ability to correctly determine the presence of, and subsequently annotate, the alleles at each position of the target sequence. Obviously, variants that have not been identified and annotated will not be subject to further scrutiny. However, the problem of non called or incorrectly annotated variants is not limited to the possibility of missing the actual causing mutation. It also increases the likelihood of overinterpreting the variants that were identified and annotated. The ability to correctly identify all the genetic variants through different NGS platforms and informatics tools still needs to be studied in more depth. The sensitivity and specificity for variant detection, false positive and false negative rates, depend on the genomic region, mutation type, sample source and quality, and clinical problem, among other factors. Parameters such as coverage and quality, that can be influenced by enrichment and sequencing methods (Mamanova et al., 2010), affect efficiency of variant identification, and there is a trade-off between sensitivity and specificity. Variant calling programs appear to have a low concordance rate (O'Rawe et al., 2013). There is a long way to go before quality control of the sequencing platforms is standardized. In order to apply NGS to diagnosis, Coonrod et al. (2013) suggested that it will be critical to empirically determine read coverage requirements for a given platform to achieve accurate variant calling. Variant calling can also be affected by the presence of structural variations such as indels, repetitive and homopolymorphic regions that can be differently interpreted by different callers, even if coverage and sequence quality were high (Linderman et al., 2014). Another technical issue is that highly homologous sequences such as pseudogenes or gene families can be co-captured with the target gene of interest, potentially generating both false positive and false negative results. The GC content can also bias variant identification, since regions with high or low GC content are captured less efficiently (Clark et al., 2011).
The identified variants are then annotated, i.e., described in a standard way through comparison with a reference sequence. Variant annotation, usually carried out with automatic software tools such as ANNOVAR (Wang et al., 2010), is performed to indicate its position and functional classification. One of the challenges for variant annotation is the existence of diverse reference sequences and versions of genomic regions, genes or transcripts as new knowledge is acquired (Pruitt et al., 2014). At least theoretically, the functional consequences of a given variant can differ depending on the reference sequence used. For instance, a missense variant in a given transcript sequence may be intronic if annotated on a different transcript where that particular exon was not present. Thus, when interpreting NGS variants totally different conclusions may be reached depending on what transcript isoform was used for annotation. Since it is often unknown which of the alternative transcripts of a gene is relevant for a given organ, symptom, disorder or disease stage, available informatics tools often annotate variants according to all known transcripts. Sometimes the largest transcript is used, other times, the canonical transcript or the one most relevant by consensus. Other pipelines only show the annotations that have a priori higher functional impact, e.g. the predicted missense effect of certain variant on a given transcript would be shown, but the intronic consequence of the same variant on another transcript would not be shown in the resulting file. This may lead to a high rate of false positive predictions, as well as the dismissal of a deleterious variant that might actually cause disease through the functional consequence of its presence in an intron (on splicing, on a micro RNA, etc.). The Locus Reference Genomic (LRG) project was proposed with the aim of establishing a fixed annotation layer for each gene with clinical implications, containing essential transcripts and a stable exon numbering system (Dalgleish et al., 2010). The magnitude of this project makes its widespread adoption difficult to achieve at this time.
4. Literature and database search
Another usual step in genetic variant interpretation consists of scrutinizing two types of variant databases.
-
1)
Collections of genetic variants observed in the general population (such as the 1000 Genomes Project or the NHLBI Exome Sequencing Project).
In-house databases of variants observed in the local population are also available in many laboratories. The rationale is that a sequence variant present in random individuals above a given frequency threshold is likely to be benign or, at least, not a high penetrant, disease-causing variant. While this premise is generally correct, caution is advisable. For instance, interpreting recessive variants can be more difficult in inbred populations, where a relatively high carrier frequency of non neutral variants may be encountered (Azmanov et al., 2013). Moreover, we should expect to find potentially pathogenic mutations in the general population just by chance, leading us to wrongly conclude that they are harmless (false negative). The contrary is also true: a neutral variant may not be detected in control individuals because it is rare, and this might lead us to over-interpret its clinical significance (false positive). In fact, most DNA variants in the human genome occur with a frequency below 1% (Mitchell et al., 2005). Thus, it is likely that a benign variant is not found when screening the general population. Furthermore, a harmless and relatively frequent variant in a given population might actually have deleterious consequences in a different population within other haplotype, epigenetic or environmental influences. An example of this is the c.35delG and c.101 T > C variants in GJB2, a gene causing congenital non syndromic sensorineural hearing impairment. There has been some controversy on the pathogenicity of these variants that were reported as either disease-causing low penetrance or non pathogenic alleles in different ethnicities (Hall et al., 2012). Kenna et al. (2013) questioned the pathogenicity of 51 variants associated with amyotrophic lateral sclerosis, based on their population frequency. While this is true in case of a full-penetrant variant with homogeneous clinical consequences (i.e. all carriers have the same manifestations), their argument would not hold up under a model of recessive inheritance, incomplete or late penetrance, di- or multi-genic disease and/or variable clinical expressivity of that variant.
-
2)
Information on genes and gene variants with reported association to disease.
In a broad sense, this includes gene-disease or variant-disease relationships described in any repository of biomedical information: published scientific papers (PubMed), online genetics resources (OMIM, GeneReviews, ClinVar) and mutation databases (HGMD®, public mutation databases). Other sources of information that may be useful for variant interpretation are repositories with data on cellular pathways and gene expression, protein domain structure and modeling, animal models and others. The ultimate goal of this search is to weigh the available evidence in favor of a likely role of the genetic variants encountered in a patient (variant present in other patients or related medical literature, gene expressed in the affected tissue, protein–protein interactions in relevant biological pathway, etc.) against lack thereof (patient clinical characteristics not reminiscent of what was previously known to be caused by that gene, variant present in a significant percentage of the general population, variant located in a functionally little relevant protein domain, etc.).
The importance of promoting a collective and open effort to build locus specific databases (LSDBs) has been highlighted by the Human Variome Project (HVP) (Cotton et al., 2009). There are manifold obstacles to developing high quality LSDBs for all human genes, including the rates at which genetic variants are identified with NGS. Also the fast accumulation of biological and medical data relevant to any given gene makes it difficult to carry out comprehensive and high quality curation of each variant. One downside of the open and non peer-reviewed information as currently published in LSDBs is the lack of quality control of the data. Because the accuracy of published research is critical both for scientists, physicians and patients who rely on these results, curation of LSDBs should not be a task left to personnel in training, but a commitment of experts in the particular gene or disorder. LSDB publishers have ethical obligations with regard to the content of the LSDB they curate (Povey et al., 2010), especially since non expert readers will also be able to access the information. All professionals involved in the production of both traditional journals and genetic databases should ensure good quality data, since incorrect information might cause misinterpretation of a genetic finding and lead to wrong medical decisions with consequences for the patients. The HVP is working on developing database quality assessment criteria.
5. Functional predictions
The functional annotation of genetic variants generally refers to predicting their probable consequences at the protein level (missense, splicing effect, truncating, etc.). However, a variant's potential pathogenicity cannot be inferred based on its functional classification only. Missense changes can be benign, whereas some synonymous variants can be deleterious. Although frameshift and truncating mutations are generally considered deleterious, truncating variants have been found in the X chromosome that is apparently inconsequential on the carrier individual's health (Raymond et al., 2009). Also, a truncating variant could be deleterious in a recessive disease (loss-of function model) but irrelevant in a dominant disease caused by a gain of abnormal function. In fact, truncating loss of function variants can be evolutionary favorable (Xue et al., 2006, Behe, 2010). Furthermore, the tendency to acquire beneficial null mutations may be variable for different functional gene classes (Hottes et al., 2013).This may imply that not only the functional type of mutation, but also the function of the encoded protein has to be considered when evaluating the likelihood of pathogenicity.
Given the complexity of the variant-function relationship, it is not surprising that in silico predictors of the theoretical consequences of genetic variants are imperfect. Many prediction tools have been developed to estimate whether a given variant is likely to be deleterious for the encoded protein, like SIFT, PolyPhen, GERP, SNAP, SNPs&Go, PhyloP, and MutationTaster(reviewed in Frousios et al., 2013). Some prediction programs are based on the nucleotide sequence, while others are based on the protein sequence, and may include analysis such as conservation among species, biochemical properties of the encoded amino acids, splicing predictions and three-dimensional calculations of the effect on protein structure, among other things. One obvious limitation is that these estimates are based on a priori assumptions and general knowledge of biological processes. For instance, it is assumed that a variant that changes the predicted codon nucleotide composition will lead to an amino acid substitution at the protein level, which may not apply for a given protein domain, gene, organ, or disease. Also, while the level of conservation among species is generally high for exons, the “no conservation = not functionally important” rule does not necessarily hold true for splicing regions, UTRs, promoters and other regulatory elements, since regulation of gene expression can be highly variable between species and even tissues. Our current ability to interpret the functional consequences of sequence variations outside coding regions is highly limited.
Another limitation is that these informatics pathogenicity prediction tools are tested with different datasets, which may lead to variable interpretations (Vihinen, 2012). It should also be emphasized that in silico predictions are based on previous knowledge and information already annotated in databases, which could have a snowball effect on accumulation of errors. The positive and negative prediction values of most in silico functional prediction tools have not been evaluated for specific genes or disorders. To overcome the limitations of individual prediction tools, scoring systems have recently been proposed that integrate the output of diverse prediction tools into a unified classification (González-Pérez and López-Bigas, 2011, Capriotti et al., 2013, Kircher et al., 2014).
6. Experimental evidence of pathogenicity
When available, patient samples are used to check for evidence of abnormal gene expression, either at RNA or protein level. The lack of correlation between in silico predictions and gene expression measured by real time PCR in colon tissue underlines the need to verify predictions experimentally (Penney et al., 2013). Since a sample from appropriate tissue cannot always be obtained, in vitro splicing assays have been developed, which have shown that in silico calculations of a splicing effect do not necessarily imply that the predicted abnormal splicing actually happens in the cell. Antagonistic splicing factors and other elements could affect the fine balance of exon identity in a disease context. Even if inferences of in silico and in vitro assays on splicing were always correct, this does not necessarily imply that such variation in the splicing process is relevant in a given individual and tissue, let alone that it is disease-causing. Such thinking ignores the natural functions of alternative splicing and how little we still know about ethnic, gender, age and tissue variability of splicing patterns (Shargunov et al., 2014).
The ultimate proof of pathogenicity is often claimed to come from showing that, when introduced to cultured cells or laboratory animals, the suspect variant causes alterations reminiscent of the phenotype, and these abnormalities are rescued by methods that recover the wild type function. Experimental studies, however, also have limitations and their results cannot be simply extrapolated to human disease. Discordant data from in vitro and in vivo assays may lead to question the role of a gene in a given disorder. For instance, the p.G191V variant in ZFYVE27, the gene encoding protrudin, was first identified in a small German family with hereditary spastic paraplegia (HSP), assigned to the SPG33 locus (Mannan et al., 2006). Using a yeast two-hybrid assay, ZFYVE27 was proposed to interact with SPAST — the most common gene causing autosomal dominant HSP. Discordant in vivo results, together with the high frequency of the p.G191V variant in some populations (7.2% in African Americans) prompted Martignoni et al. (2008) to question the pathogenic role of ZFYVE27. The controversy was nurtured by more recent functional studies suggesting that protrudin is functionally related to other endoplasmic reticulum proteins causing HSP (Pantakani et al., 2011, Chang et al., 2013). However, definitive evidence that the p.G191V variant causes HSP is still lacking, additional mutations in ZFYVE27 causing HSP have not been reported, and some authors do not recommend including this gene in the routine genetic diagnosis of this group of disorders (Finsterer et al., 2012).
7. Family co-segregation
The importance of clinical history and examination, as well as family data for variant interpretation cannot be overemphasized. The causal role of a genetic variant in a given patient is less plausible if the variant is not present in all affected family members and/or it is carried by unaffected individuals. However, the weight of clinical and genealogical evidence on the estimated likelihood of causality must also be taken cautiously. First, establishing affectation status is not always easy for late-onset diseases that may show mild or variable symptoms in different family members. On the other hand, similar symptoms can be due to a different — genetic or environmental — cause in some family members (phenocopies). In a revision of 160 families with Parkinson's disease, Klein et al. (2011) found that up to 1.3% of all relatives with PD were phenocopies. This highlights the complexity of interpreting familial co-segregation in disorders where the genetic disease can be indistinguishable from idiopathic forms.
Another challenge comes from assumptions on inheritance models and genetics rules. For instance, that there is just one causal variant at play in a family with a dominant disease, that the disease causing variant will be homozygous in a consanguineous family or that a given variant or gene always acts as either dominant or recessive. Establishing the inheritance pattern in most families is far from straightforward. In the clinical diagnostic setting genetic variants are typically evaluated one by one, and thus the possibility that other variants present in a particular patient could influence clinical presentation is generally overlooked. In a patient with both myopathic and neuropathic signs,Ardissone et al. (2014) observed two previously known mutations in CLCN1, a gene causing autosomal recessive congenital myopathy. The patient also had a duplication in the PMP22 gene causing Charcot–Marie–Tooth disease, an autosomal dominant hereditary neuropathy, which he had inherited from his father. In the asymptomatic parents, neurophysiological studies showed a demyelinating polyneuropathy in the father, and mild myotonia in the mother. This case exemplifies several facts that can further complicate clinical interpretation of genetic findings. First of all, the difficulty to accurately establish family history, clinical status and inheritance pattern. In many disorders, clinical manifestations may be variable, including subclinical signs that go unnoticed. Secondly, disease-causing variants in two or more genes may be contributing to the particular combination of clinical features in a given patient. In HSP families with atlastin mutations,Varga et al. (2013) also warn on this issue, discussing the challenge of assessing family history correctly and suggest that some cases with as yet unidentified genetic basis might be due to misinterpretation of the inheritance pattern.
Upon familial co-segregation analysis for variant pathogenicity assessment, another issue that deserves more attention is the interpretation of de novo mutations. Genetic variants that arose de novo in a patient — not present in any of the parents — are generally considered more likely deleterious. Although de novo mutations might underlie many rare developmental disorders, most of an individual's de novo variants are expected to be clinically irrelevant (Veltman and Brunner, 2012). The type of mutations and polymorphisms, mutation rate in different genetic regions, as well as other factors that may influence de novo mutation rates in different individuals or populations should be taken into account before drawing conclusions on the pathogenic role of a de novo variant. Around 74 de novo single-nucleotide mutations per genome is expected, a frequency that is influenced by paternal age (Kong et al., 2012). It must also be kept in mind that the possibility of sequencing artifacts is higher among apparently de novo mutations.
8. Weighing the evidence: likelihood of pathogenicity scoring and clinical interpretation
Eventually, all the lines of evidence described above, as well as other sources and types of related knowledge need to be evaluated in order to achieve the most accurate insight on the potential role of a given variant in a given disease or symptom, in a given patient. For this purpose, pathogenicity scoring systems have been developed by researchers or expert consortia, such as those proposed to classify breast cancer or mismatch repair gene variants (Plon et al., 2008, Thompson et al., 2014). Classification schemes generally involve multifactorial likelihood analysis of quantitative and qualitative data (Goldgar et al., 2008). A 5-tiered system is commonly used, which recognizes variants as benign, probably benign, of uncertain significance, probably pathogenic, and pathogenic. However, this scoring system is clearly over-simplistic and for the most part applicable to Mendelian disorders only. If all genetic and non genetic factors are considered, such as in complex human diseases, other categories also exist to define the relationship between a genetic variant and the disease (susceptibility variant, prognostic factor, and modifier of drug-response, among others).
Some LSDBs and other databases summarize useful information about evidence of pathogenicity for each variant. ClinVar is a public repository providing access to clinically relevant variants with supporting evidence (Landrum et al., 2014). Assessment of significance is provided by the submitter, and conflicts in interpretation by different submissions are tagged in the aggregate records. A distinction is also made between variants classified by single or multiple submitters or not classified by any submitter. A collective, wiki-like open system to sum up the evidence of pathogenicity from different sources could also be envisioned, with potential advantages and disadvantages.
Increasingly, informatics packages and online portals are being developed that exploit data from multiple sources, taking into account different levels of evidence in a combined way (Hermida et al., 2013). Such machine learning systems are becoming more powerful to compile and mine biomedical information. They can carry out iterative operations to contrast hypothesis, identify relationship patterns (variant-disorder, gene-symptom, protein-organ, variant-treatment), and generate meta-data to facilitate biomedical interpretation. However, these in silico knowledge building processes are still in early development stages for clinical applications. Machine-learning installations can use semantic similarity searches and phenotype ontologies to extract information from the millions of articles in the medical literature (Köhler et al., 2009). To name just one challenge, there is good and bad scientific literature, and also an immense amount of biomedical information uploaded in the internet that would not pass minimum quality standard to even be called scientific literature (i.e., conclusions not drawn with scientific methods). Thus, automatic data integrators would have to give different weights to reliable and non reliable sources of data. With the increasing amount of non peer-reviewed publications, there is a risk that mere information is treated as if it was knowledge. What will happen if machine knowledge acquisition is based on thousands of data retrieved from non peer reviewed biomedical literature, full of errors? Even peer-reviewed literature of course contains mistakes, and the chance that this happens is likely to grow exponentially, as the capacity to acquire and analyze biomedical data speeds up. This is reflected in the fact that NGS technologies are uncovering a growing number of published genes and genetic variants whose supposed to be associated to a given disease is now being questioned (Kenna et al., 2013). If genetic variants whose link to certain disease was not well validated are later identified in patients with different symptoms, this may lead to an “expansion of the phenotype spectrum” caused by that gene. The new genotype–phenotype relationship will in turn populate general repositories such as PubMed, OMIM, and OrphaNet, potentially contributing to bias our interpretation of genetic findings.
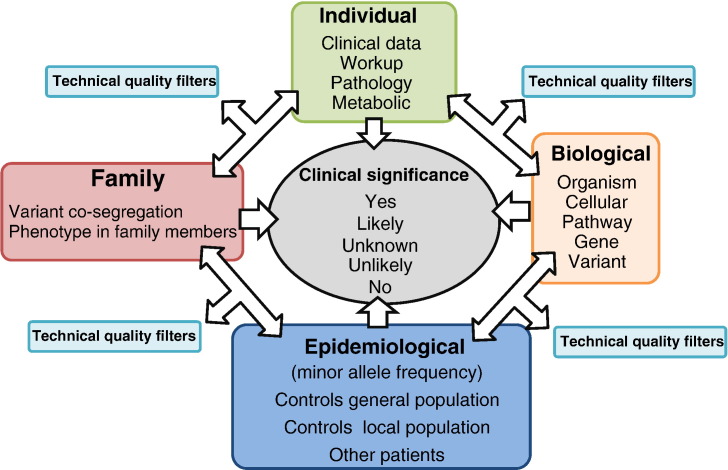
In summary, interpreting the clinical significance of genetic variants is a complex process that goes beyond functional studies and does not even end with pathogenicity assessment. Demonstrating that the variant has a deleterious consequence on the function of the encoded protein does not automatically translate into clinical relevance. Even if assessment of pathogenicity — defined as a negative impact on cell processes — could be robustly determined for a given variant, its relationship with the patient's symptoms, or lack thereof, and its implications in medical decision making need to be unequivocally established for it to be useful in clinical practice. Table 1 summarizes some of the difficulties in this process that starts with variant identification and entails scoring genetic and non genetic evidence. There is no single piece of evidence enough on its own for clinical variant interpretation. Rather than stepwise criteria that apply generally for any variant in any gene, specific pathogenicity scoring algorithms should be developed by experts on each gene, gene family, biological pathway or disorder. They should involve multidimensional analyses, where information in one domain contributes to set the significance threshold and weigh the evidence from other domains (Fig. 2). For instance, the minor allele frequency threshold to filter out likely benign variants depends on the inheritance pattern, as well as on the clinical features (a not very rare variant may cause a phenotype that is mild and/or late onset, however it would unlikely be the cause of an early-onset, severe phenotype). The presence of characteristic clinical or biochemical signatures would modify the a priori likelihood of causality of variants in a given gene. Also, the quality criteria set in the variant evaluation pipeline to filter out likely sequencing artifacts can be less strict to minimize false negative results in a priori candidate genes.
Table 1.
Difficulties for interpretation of the clinical significance of genetic variants.
| Step/methods | Example challenges |
|---|---|
| Variant identification | -Variable performance of sequencing platforms and strategy -Bioinformatic tools with different mapping and variant calling parameters -Sensitivity/specificity may depend on genetic region, variant type and clinical question. -Coverage and quality standards and thresholds not well established - Failure to identify relevant variants may lead to overrate variants that are observed. |
| Variant annotation | - Not universally adopted nomenclature - Gene-centric and exon-centric annotation system - Variable and evolving reference sequences - Incomplete knowledge of alternative transcripts and regulatory elements |
| Search scientific literature | - Exponential number of gene-disease associations, many are not validated - Publication bias towards positive results - Publication bias towards certain ethnic groups/populations |
| Search general databases | - Increasing content of rare, potentially pathogenic variants - There is no absolute frequency threshold to prove that a variant is likely benign or likely pathogenic. It depends on disease model, clinical characteristics, etc. - Many populations are not represented. - No information on phenotype |
| Search LSDBs | - Not available for most genes - Contradictory information in different databases - May not be updated, not peer-reviewed - Biased with data obtained from patients |
| Search in house database | - Overrepresentation of technical errors - Regions of repeatedly deficient coverage - Neutral variants may be rare and non neutral may be frequent in specific populations |
| A priori biological knowledge | - Assumption that some variant types are always more likely to cause disease than others. E.g. truncating/frameshift more than synonymous. - Assumption of compliance with Mendelian rules - Equal interpretation of a given genetic variant in different patients, disorders, populations, disregarding other genetic and/or environmental factors |
| In silico missense predictions | - Based on general biological principles that may not always apply - May vary with the amount of input sequence, transcript isoform and the information available in biorepositories - The complexity of splicing regulation is poorly understood |
| In vitro splicing analysis and expression studies | - Technically demanding and susceptible to be influenced by experimental conditions - Expensive to set up in a routine pathogenicity assessment pipeline - Observations may not reflect what happens in the cell, organ and disease state |
| Animal experiments | - Technically demanding, expensive - Results do not necessarily parallel to what happens in humans, in disease state |
| Family co-segregation | - Inheritance pattern is not always clear - Family members are not always near, alive, or willing to be studied - Biological causes of apparent lack of co-segregation. E.g. incomplete penetrance, anticipation, variable phenotype, phenocopies. - Ethical and legal implications: need for genetic counseling, follow-up of family members may appropriate according to results (time consuming, expensive) |
Fig. 2.
Multidimensional analysis for clinical interpretation of the evidence of causality. The weight of results from one domain in the estimation of a variant's clinical significance depends on information from other domains. Technical and quality variant filtering criteria are not universally fixed, but are influenced by the a priori evidence.
Finally, the ethical implications of premature delivery of incorrect/incomplete variant interpretation include patient misdiagnosis, and its impact on the patient as well as potentially on family members, investment of resources in the study of a gene that may be irrelevant, and challenges interpretation of further variants. While seeking ways to make clinically useful genetic data openly and promptly available worldwide, we should not ignore the basic rules to produce high quality scientific knowledge. As it starts to show, the literature is abundant in reports of misclassified genetic variants. The immediate clinical use of new information on genes and genetic variants from research studies, without further validation, may lead to premature conclusions and misinterpretation, and might cause irreparable harm.
Acknowledgments
We are mostly indebted to the patients and families who participate in our research and teach us every day. We are grateful to Dr. Patricia Blanco for fruitful discussions on variant interpretation and clinical reporting, as well as to Dr. Jorge Amigo for bioinformatics support. We acknowledge useful input from many members of the Human Variome Project (HVP), especially from the HVP pathogenicity assessment committee. This work was partly supported by PN I+D+I 2008-2011 and the Fondo de Investigaciones Sanitarias (FIS PS09/01830). MJS and BQ are recipients of research contracts by the Institute of Health Carlos III and Servicio Galego de Saúde, Spain. All this grants has been cofinanced with FEDER funds (2007-2013).
References
- Ardissone A., Brugnoni R., Gandioli C., Milani M., Ciano C., Uziel G., Moroni I. Double-trouble in pediatric neurology: myotonia congenita combined with Charcot–Marie–Tooth disease type 1a. Muscle Nerve. 2014 doi: 10.1002/mus.24205. (Feb 11) [DOI] [PubMed] [Google Scholar]
- Azmanov D.N., Chamova T., Tankard R., Gelev V., Bynevelt M., Florez L., Tzoneva D., Zlatareva D., Guergueltcheva V., Bahlo M., Tournev I., Kalaydjieva L. Challenges of diagnostic exome sequencing in an inbred founder population. Mol. Genet. Genomic Med. 2013;1:71–76. doi: 10.1002/mgg3.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behe M.J. Experimental evolution, loss-of-function mutations, and “the first rule of adaptive evolution”. Q. Rev. Biol. 2010;85:419–445. doi: 10.1086/656902. [DOI] [PubMed] [Google Scholar]
- Capriotti E., Altman R.B., Bromberg Y. Collective judgment predicts disease-associated single nucleotide variants. BMC Genomics. 2013;14(Suppl. 3):S2. doi: 10.1186/1471-2164-14-S3-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J., Lee S., Blackstone C. Protrudin binds atlastins and endoplasmic reticulum-shaping proteins and regulates network formation. Proc. Natl. Acad. Sci. U. S. A. 2013;110:14954–14959. doi: 10.1073/pnas.1307391110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirulli E.T., Goldstein D.B. Uncovering the roles of rare variants in common disease through whole-genome sequencing. Nat. Rev. Genet. 2010;11:415–425. doi: 10.1038/nrg2779. [DOI] [PubMed] [Google Scholar]
- Clark M.J., Chen R., Lam H.Y., Karczewski K.J., Chen R., Euskirchen G., Butte A.J., Snyder M. Performance comparison of exome DNA sequencing technologies. Nat. Biotechnol. 2011;29:908–914. doi: 10.1038/nbt.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coonrod E.M., Durtschi J.D., Margraf R.L., Voelkerding K.V. Developing genome and exome sequencing for candidate gene identification in inherited disorders: an integrated technical and bioinformatics approach. Arch. Pathol. Lab. Med. 2013;137:415–433. doi: 10.5858/arpa.2012-0107-RA. [DOI] [PubMed] [Google Scholar]
- Cotton R.G., Al Aqeel A.I., Al-Mulla F., Carrera P., Claustres M., Ekong R., Hyland V.J., Macrae F.A., Marafie M.J., Paalman M.H., Patrinos G.P., Qi M., Ramesar R.S., Scott R.J., Sijmons R.H., Sobrido M.J., Vihinen M., Members of the Human Variome Project Data Collection from Clinics, Data Collection from Laboratories and Publication, Credit and Incentives Working Groups Capturing all disease-causing mutations for clinical and research use: toward an effortless system for the Human Variome Project. Genet Med. 2009;11:843–849. doi: 10.1097/GIM.0b013e3181c371c5. [DOI] [PubMed] [Google Scholar]
- Dalgleish R., Flicek P., Cunningham F., Astashyn A., Tully R.E., Proctor G., Chen Y., McLaren W.M., Larsson P., Vaughan B.W., Béroud C., Dobson G., Lehväslaiho H., Taschner P.E., den Dunnen J.T., Devereau A., Birney E., Brookes A.J., Maglott D.R. Locus reference genomic sequences: an improved basis for describing human DNA variants. Genome Med. 2010;2:24. doi: 10.1186/gm145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finsterer J., Löscher W., Quasthoff S., Wanschitz J., Auer-Grumbach M., Stevanin G. Hereditary spastic paraplegias with autosomal dominant, recessive, x-linked, or maternal trait of inheritance. J. Neurol. Sci. 2012;318:1–18. doi: 10.1016/j.jns.2012.03.025. [DOI] [PubMed] [Google Scholar]
- Frousios K., Iliopoulos C.S., Schlitt T., Simpson M.A. Predicting the functional consequences of non-synonymous DNA sequence variants—evaluation of bioinformatics tools and development of a consensus strategy. Genomics. 2013;102:223–228. doi: 10.1016/j.ygeno.2013.06.005. [DOI] [PubMed] [Google Scholar]
- Goldgar D.E., Easton D.F., Byrnes G.B., Spurdle A.B., Iversen E.S. Greenblatt MS; IARC Unclassified Genetic Variants Working Group. Genetic evidence and integration of various data sources for classifying uncertain variants into a single model. Hum. Mutat. 2008;29:1265–1272. doi: 10.1002/humu.20897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Pérez A., López-Bigas N. Improving the assessment of the outcome of nonsynonymous SNVs with a consensus deleteriousness score, Condel. Am. J. Hum. Genet. 2011;88:440–449. doi: 10.1016/j.ajhg.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall A., Pembrey M., Lutman M., Steer C., Bitner-Glindzicz M. Prevalence and audiological features in carriers of GJB2 mutations, c.35delG and c.101 T > C (p.M34T), in a UK population study. BMJ Open. 2012;2 doi: 10.1136/bmjopen-2012-001238. (pii: e001238) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermida L., Poussin C., Stadler M.B., Gubian S., Sewer A., Gaidatzis D., Hotz H.R., Martin F., Belcastro V., Cano S., Peitsch M.C., Hoeng J. Confero: an integrated contrast data and gene set platform for computational analysis and biological interpretation of omics data. BMC Genomics. 2013;14:514. doi: 10.1186/1471-2164-14-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hottes A.K., Freddolino P.L., Khare A., Donnell Z.N., Liu J.C., Tavazoie S. Bacterial adaptation through loss of function. PLoS Genet. 2013;9:e1003617. doi: 10.1371/journal.pgen.1003617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller A., Westenberger A., Sobrido M.J., García-Murias M., Domingo A., Sears R.L., Lemos R.R., Ordoñez-Ugalde A., Nicolas G., da Cunha J.E., Rushing E.J., Hugelshofer M., Wurnig M.C., Kaech A., Reimann R., Lohmann K., Dobričić V., Carracedo A., Petrović I., Miyasaki J.M., Abakumova I., Mäe M.A., Raschperger E., Zatz M., Zschiedrich K., Klepper J., Spiteri E., Prieto J.M., Navas I., Preuss M., Dering C., Janković M., Paucar M., Svenningsson P., Saliminejad K., Khorshid H.R., Novaković I., Aguzzi A., Boss A., Le Ber I., Defer G., Hannequin D., Kostić V.S., Campion D., Geschwind D.H., Coppola G., Betsholtz C., Klein C., Oliveira J.R. Mutations in the gene encoding PDGF-B cause brain calcifications in humans and mice. Nat. Genet. 2013;45:1077–1082. doi: 10.1038/ng.2723. [DOI] [PubMed] [Google Scholar]
- Kenna K.P., McLaughlin R.L., Hardiman O., Bradley D.G. Using reference databases of genetic variation to evaluate the potential pathogenicity of candidate disease variants. Hum. Mutat. 2013;34:836–841. doi: 10.1002/humu.22303. [DOI] [PubMed] [Google Scholar]
- Kiezun A., Garimella K., Do R., Stitziel N.O., Neale B.M., McLaren P.J., Gupta N., Sklar P., Sullivan P.F., Moran J.L., Hultman C.M., Lichtenstein P., Magnusson P., Lehner T., Shugart Y.Y., Price A.L., de Bakker P.I., Purcell S.M., Sunyaev S.R. Exome sequencing and the genetic basis of complex traits. Nat. Genet. 2012;44:623–630. doi: 10.1038/ng.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher M., Witten D.M., Jain P., O'Roak B.J., Cooper G.M., Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein C., Chuang R., Marras C., Lang A.E. The curious case of phenocopies in families with genetic Parkinson's disease. Mov. Disord. 2011;26:1793–1802. doi: 10.1002/mds.23853. [DOI] [PubMed] [Google Scholar]
- Köhler S., Schulz M.H., Krawitz P., Bauer S., Dölken S., Ott C.E., Mundlos C., Horn D., Mundlos S., Robinson P.N. Clinical diagnostics in human genetics with semantic similarity searches in ontologies. Am. J. Hum. Genet. 2009;85:457–464. doi: 10.1016/j.ajhg.2009.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong A., Frigge M.L., Masson G., Besenbacher S., Sulem P., Magnusson G., Gudjonsson S.A., Sigurdsson A., Jonasdottir A., Jonasdottir A., Wong W.S., Sigurdsson G., Walters G.B., Steinberg S., Helgason H., Thorleifsson G., Gudbjartsson D.F., Helgason A., Magnusson O.T., Thorsteinsdottir U., Stefansson K. Rate of de novo mutations and the importance of father's age to disease risk. Nature. 2012;488:471–475. doi: 10.1038/nature11396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landrum M.J., Lee J.M., Riley G.R., Jang W., Rubinstein W.S., Church D.M., Maglott D.R. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 2014;42(Database issue):D980–D985. doi: 10.1093/nar/gkt1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linderman M.D., Brandt T., Edelmann L., Jabado O., Kasai Y., Kornreich R., Mahajan M., Shah H., Kasarskis A., Schadt E.E. Analytical validation of whole exome and whole genome sequencing for clinical applications. BMC Med. Genomics. 2014;7:20. doi: 10.1186/1755-8794-7-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacArthur D.G., Manolio T.A., Dimmock D.P., Rehm H.L., Shendure J., Abecasis G.R., Adams D.R., Altman R.B., Antonarakis S.E., Ashley E.A., Barrett J.C., Biesecker L.G., Conrad D.F., Cooper G.M., Cox N.J., Daly M.J., Gerstein M.B., Goldstein D.B., Hirschhorn J.N., Leal S.M., Pennacchio L.A., Stamatoyannopoulos J.A., Sunyaev S.R., Valle D., Voight B.F., Winckler W., Gunter C. Guidelines for investigating causality of sequence variants in human disease. Nature. 2014;508:469–476. doi: 10.1038/nature13127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamanova L., Coffey A.J., Scott C.E., Kozarewa I., Turner E.H., Kumar A., Howard E., Shendure J., Turner D.J. Target-enrichment strategies for next-generation sequencing. Nat. Methods. 2010;7:111–118. doi: 10.1038/nmeth.1419. [DOI] [PubMed] [Google Scholar]
- Mann J.J., Arango V.A., Avenevoli S., Brent D.A., Champagne F.A., Clayton P., Currier D., Dougherty D.M., Haghighi F., Hodge S.E., Kleinman J., Lehner T., McMahon F., Mościcki E.K., Oquendo M.A., Pandey G.N., Pearson J., Stanley B., Terwilliger J., Wenzel A. Candidate endophenotypes for genetic studies of suicidal behavior. Biol. Psychiatry. 2009;65:556–563. doi: 10.1016/j.biopsych.2008.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannan A.U., Krawen P., Sauter S.M., Boehm J., Chronowska A., Paulus W., Neesen J., Engel W. ZFYVE27 (SPG33), a novel spastin-binding protein, is mutated in hereditary spastic paraplegia. Am. J. Hum. Genet. 2006;79:351–357. doi: 10.1086/504927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martignoni M., Riano E., Rugarli E. The role of ZFYVE27/protudin in hereditary spastic paraplegia. Am. J. Hum. Genet. 2008;83:127–128. doi: 10.1016/j.ajhg.2008.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell A.A., Chakravarti A., Cutler D.J. On the probability that a novel variant is a disease-causing mutation. Genome Res. 2005;15:960–966. doi: 10.1101/gr.3761405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normanno N., Rachiglio A.M., Roma C., Fenizia F., Esposito C., Pasquale R., La Porta M.L., Iannaccone A., Micheli F., Santangelo M., Bergantino F., Costantini S., De Luca A. Molecular diagnostics and personalized medicine in oncology: challenges and opportunities. J. Cell. Biochem. 2013;114:514–524. doi: 10.1002/jcb.24401. [DOI] [PubMed] [Google Scholar]
- O'Rawe J., Jiang T., Sun G., Wu Y., Wang W., Hu J., Bodily P., Tian L., Hakonarson H., Johnson W.E., Wei Z., Wang K., Lyon G.J. Low concordance of multiple variant-calling pipelines: practical implications for exome and genome sequencing. Genome Med. 2013;5:28. doi: 10.1186/gm432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantakani D.V., Czyzewska M.M., Sikorska A., Bodda C., Mannan A.U. Oligomerization of ZFYVE27 (Protrudin) is necessary to promote neurite extension. PLoS One. 2011;6:e29584. doi: 10.1371/journal.pone.0029584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrinos G.P., Baker D.J., Al-Mulla F., Vasiliou V., Cooper D.N. Genetic tests obtainable through pharmacies: the good, the bad, and the ugly. Hum. Genomics. 2013;7:17. doi: 10.1186/1479-7364-7-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penney R.B., Lundgreen A., Yao-Borengasser A., Koroth-Edavana V., Williams S., Wolff R., Slattery M.L., Kadlubar S. Lack of correlation between in silico projection of function and quantitative real-time PCR-determined gene expression levels in colon tissue. Pharmacogenomics Pers. Med. 2013;6:99–103. doi: 10.2147/PGPM.S49199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plon S.E., Eccles D.M., Easton D., Foulkes W.D., Genuardi M., Greenblatt M.S., Hogervorst F.B., Hoogerbrugge N., Spurdle A.B. Tavtigian SV; IARC Unclassified Genetic Variants Working Group. Sequence variant classification and reporting: recommendations for improving the interpretation of cancer susceptibility genetic test results. Hum. Mutat. 2008;29:1282–1291. doi: 10.1002/humu.20880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Povey S., Al Aqeel A.I., Cambon-Thomsen A., Dalgleish R., den Dunnen J.T., Firth H.V., Greenblatt M.S., Barash C.I., Parker M., Patrinos G.P., Savige J., Sobrido M.J., Winship I., Cotton R.G., Ethics Committee of the Human Genome Organization (HUGO) Practical guidelines addressing ethical issues pertaining to the curation of human locus-specific variation databases (LSDBs) Hum. Mutat. 2010;31:1179–1184. doi: 10.1002/humu.21339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt K.D., Brown G.R., Hiatt S.M., Thibaud-Nissen F., Astashyn A., Ermolaeva O., Farrell C.M., Hart J., Landrum M.J., McGarvey K.M., Murphy M.R., O'Leary N.A., Pujar S., Rajput B., Rangwala S.H., Riddick L.D., Shkeda A., Sun H., Tamez P., Tully R.E., Wallin C., Webb D., Weber J., Wu W., DiCuccio M., Kitts P., Maglott D.R., Murphy T.D., Ostell J.M. RefSeq: an update on mammalian reference sequences. Nucleic Acids Res. 2014;42(Database issue):D756–D763. doi: 10.1093/nar/gkt1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond F.L., Whibley A., Stratton M.R., Gecz J. Lessons learnt from large-scale exon re-sequencing of the X chromosome. Hum. Mol. Genet. 2009;18:R60–R64. doi: 10.1093/hmg/ddp071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salari K., Watkins H., Ashley E.A. Personalized medicine: hope or hype? Eur. Heart J. 2012;33:1564–1570. doi: 10.1093/eurheartj/ehs112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shargunov A.V., Krasnov G.S., Ponomarenko E.A., Lisitsa A.V., Shurdov M.A., Zverev V.V., Archakov A.I., Blinov V.M. Tissue-specific alternative splicing analysis reveals the diversity of chromosome 18 transcriptome. J. Proteome Res. 2014;13:173–182. doi: 10.1021/pr400808u. [DOI] [PubMed] [Google Scholar]
- Thompson B.A., Spurdle A.B., Plazzer J.P., Greenblatt M.S., Akagi K., Al-Mulla F., Bapat B., Bernstein I., Capellá G., den Dunnen J.T., du Sart D., Fabre A., Farrell M.P., Farrington S.M., Frayling I.M., Frebourg T., Goldgar D.E., Heinen C.D., Holinski-Feder E., Kohonen-Corish M., Robinson K.L., Leung S.Y., Martins A., Moller P., Morak M., Nystrom M., Peltomaki P., Pineda M., Qi M., Ramesar R., Rasmussen L.J., Royer-Pokora B., Scott R.J., Sijmons R., Tavtigian S.V., Tops C.M., Weber T., Wijnen J., Woods M.O., Macrae F., Genuardi M., InSiGHT Application of a 5-tiered scheme for standardized classification of 2,360 unique mismatch repair gene variants in the InSiGHT locus-specific database. Nat. Genet. 2014;46:107–115. doi: 10.1038/ng.2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga R.E., Schüle R., Fadel H., Valenzuela I., Speziani F., Gonzalez M., Rudenskaia G., Nürnberg G., Thiele H., Altmüller J., Alvarez V., Gamez J., Garbern J.Y., Nürnberg P., Zuchner S., Beetz C. Do not trust the pedigree: reduced and sex-dependent penetrance at a novel mutation hotspot in ATL1 blurs autosomal dominant inheritance of spastic paraplegia. Hum. Mutat. 2013;34:860–863. doi: 10.1002/humu.22309. [DOI] [PubMed] [Google Scholar]
- Veltman J.A., Brunner H.G. De novo mutations in human genetic disease. Nat. Rev. Genet. 2012;13:565–575. doi: 10.1038/nrg3241. [DOI] [PubMed] [Google Scholar]
- Vihinen M. How to evaluate performance of prediction methods? Measures and their interpretation in variation effect analysis. BMC Genomics. 2012;13(Suppl. 4):S2. doi: 10.1186/1471-2164-13-S4-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan J., Yourshaw M., Mamsa H., Rudnik-Schöneborn S., Menezes M.P., Hong J.E., Leong D.W., Senderek J., Salman M.S., Chitayat D., Seeman P., von Moers A., Graul-Neumann L., Kornberg A.J., Castro-Gago M., Sobrido M.J., Sanefuji M., Shieh P.B., Salamon N., Kim R.C., Vinters H.V., Chen Z., Zerres K., Ryan M.M., Nelson S.F., Jen J.C. Mutations in the RNA exosome component gene EXOSC3 cause pontocerebellar hypoplasia and spinal motor neuron degeneration. Nat. Genet. 2012;44:704–708. doi: 10.1038/ng.2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Li M., Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y., Daly A., Yngvadottir B., Liu M., Coop G., Kim Y., Sabeti P., Chen Y., Stalker J., Huckle E., Burton J., Leonard S., Rogers J., Tyler-Smith C. Spread of an inactive form of caspase-12 in humans is due to recent positive selection. Am. J. Hum. Genet. 2006;78:659–670. doi: 10.1086/503116. [DOI] [PMC free article] [PubMed] [Google Scholar]