Abstract
Combining single-cell methods and next-generation sequencing should provide a powerful means to understand single-cell biology and obviate the effects of sample heterogeneity. Here we report a single-cell identification method and seamless cancer gene profiling using semiconductor-based massively parallel sequencing. A549 cells (adenocarcinomic human alveolar basal epithelial cell line) were used as a model. Single-cell capture was performed using laser capture microdissection (LCM) with an Arcturus® XT system, and a captured single cell and a bulk population of A549 cells (≈ 106 cells) were subjected to whole genome amplification (WGA). For cell identification, a multiplex PCR method (AmpliSeq™ SNP HID panel) was used to enrich 136 highly discriminatory SNPs with a genotype concordance probability of 1031–35. For cancer gene profiling, we used mutation profiling that was performed in parallel using a hotspot panel for 50 cancer-related genes. Sequencing was performed using a semiconductor-based bench top sequencer. The distribution of sequence reads for both HID and Cancer panel amplicons was consistent across these samples. For the bulk population of cells, the percentages of sequence covered at coverage of more than 100 × were 99.04% for the HID panel and 98.83% for the Cancer panel, while for the single cell percentages of sequence covered at coverage of more than 100 × were 55.93% for the HID panel and 65.96% for the Cancer panel. Partial amplification failure or randomly distributed non-amplified regions across samples from single cells during the WGA procedures or random allele drop out probably caused these differences. However, comparative analyses showed that this method successfully discriminated a single A549 cancer cell from a bulk population of A549 cells. Thus, our approach provides a powerful means to overcome tumor sample heterogeneity when searching for somatic mutations.
Keywords: Single cell identification, Heterogeneity, Laser capture microdissection, Semiconductor-based sequencing
1. Introduction
Many areas of genomic research rely on pooled samples that include hundreds to millions of individual cells. When analyzing the genomic data of these samples, the results obtained are only average readouts. If these samples are mixtures or multi-clonal in nature, such as with tumor biopsies, then data interpretation may be hampered by low signal to noise ratios. Heterogeneity often limits data interpretation. Single-cell analysis has the potential to overcome this ambiguity in data interpretation. RNA sequencing to determine expression levels usually involves average values from bulk assays and single-cell analysis may obviate these heterogeneity issues. DNA sequence analysis also involves averaging (Shapiro et al., 2013).
Cancer research, in particular, would benefit from adopting single-cell analyses, as most tumor samples are mixtures of normal cells and cancer cells (Gerlinger et al., 2012). Recently, numerous next-generation sequencing (NGS) based studies have been conducted to provide a comprehensive molecular characterization of cancers to study tumor complexity, heterogeneity, and evolution (Shyr and Liu, 2013). Target enrichment methods for NGS are rapidly being developed and should be useful for cancer research by providing a powerful, cost effective method to study DNA and RNA in samples. Many PCR-based enrichment techniques are now available for this purpose (Mertes et al., 2011).
Currently, most cancer profiling still relies on average analyses, often because of methodological limitations. In these cases, genetic material is extracted from millions of cells. Despite the high sensitivity of modern NGS platforms, mutation frequencies of < 5% are difficult to detect even when using very high sequencing coverage (Harismendy et al., 2011). Thus, important somatic mutations may be missed due to the presence of contaminating wild-type cells or non-clonal contaminating cancer populations within the same sample (Swanton, 2012). However, research at the single-cell level enables unambiguous detection of rare variants and genetic characterization without this averaging effect of sample heterogeneity (Navin et al., 2011). Using this approach, cancer cells of different clonal origins, each containing a separate mutational profile, can be distinguished. However, single-cell level analysis carries an increased risk of contamination and analyte identification throughout the analysis is an important control step. Short tandem repeat (STR) analysis has been proposed as a means to overcome these limitations (Korzebor et al., 2013).
However, these methods are cumbersome and are not seamlessly integrated with functional analysis. Yet, this procedure can be applied to any routine NGS-based workflow. Combining single-cell methods and NGS would provide an effective means to understand single-cell biology and obviate the effects of sample heterogeneity. Here we report a single-cell identification method and seamless cancer gene profiling using semiconductor-based massively parallel sequencing.
2. Materials and methods
2.1. Cell culture and DNA extraction
A549 cells (adenocarcinomic human alveolar basal epithelial cells) were routinely maintained in RPMI 1640 medium with Glutamax-I supplemented with 10% fetal calf serum, penicillin (100 IU/ml), and streptomycin (100 ng/ml) (Life Technologies) with 5% CO2 in humidified air at 37 °C. Cell viability as estimated by trypan blue exclusion was > 95% prior to each experiment. For standard processing of a bulk cell population, DNA extraction and purification were performed using a PureLink™ genomic DNA kit (Life Technologies).
2.2. Single-cell capture
Single-cell capture was performed using laser capture microdissection (LCM) using an Arcturus® XT system (Life Technologies) (Pietersen et al., 2009) according to the manufacturer's instructions. A549 cells were cultured and adhered to a proton exchange membrane. A CapSure® LCM cap was placed over the target area. Laser pulsing through this cap caused a thermoplastic film to form a thin protrusion that bridged the membrane around a single A549 cell. The membrane around the A549 single cell was cut using a UV laser, and the cap was lifted to remove the target cell attached to it (Supplementary Fig. 1). A single captured cell and a blank sample, as a negative control, were subjected to whole genome amplification (WGA) using single-cell WGA kits (New England Bio Laboratories) (Zheng et al., 2011). The total amount of amplified DNA was 3.4 μg, as expected. After WGA, DNA from a single cell was purified using the PureLink™ PCR purification kit.
2.3. Library preparation
AmpliSeq technology is an ultra-high multiplex PCR method that utilizes up to 6144 PCR primer pairs in one tube (Yousem et al., 2013). Two primer pools were used for AmpliSeq target enrichment. For cell identification, the AmpliSeq™ SNP HID panel (Life Technologies) was used which interrogated 136 SNPs of high discriminatory power with a genotype concordance probability of 1031–35 (Pakstis et al., 2010, Sanchez et al., 2006). Although a 340 SNP panel was available for this technology, this panel provided sufficient discriminatory power and was cost effective.
For cancer gene profiling, we used AmpliSeq Cancer hotspot panel version 2 (Life Technologies), which included 207 primer pairs per tube to detect 50 cancer gene hotspots. DNA was extracted from a bulk population of A549 cells (≈ 106 cells), and 10 ng of DNA (≈ 3000 genome copies) was used as a PCR template. Amplicons were generated in a single PCR reaction tube with an endpoint thermal cycler. A total of 50 ng (single cell Library prep replicate #1) and 10 ng (single cell Library prep replicate #2) of WGA-amplified DNA from a single cell were subjected to PCR using the same conditions as above. The amplicons were partially digested and phosphorylated according to the manufacturer's instructions. Amplicons were ligated to adapters included in an Ion Xpress™ Barcode Adapters 1-16 kit (Life Technologies), nick-translated, and then subjected to another round of PCR to complete the linkage between adapters and amplicons. A BioAnalyzer High Sensitivity DNA kit (Agilent Technologies) was used to visualize the size range and determine the library concentration.
2.4. Semiconductor sequencing and data analysis
Individual and combined libraries were attached to Ion Sphere™ particles (ISPs) by emulsion PCR, and biotinylated ISPs were recovered from the emulsion using Dynabeads MyOne™ Streptavidin C1 beads (Life Technologies). Sequencing was performed using a semiconductor-based bench top sequencer (Ion PGM™, Life Technologies) (Rothberg et al., 2011). Four bar-coded samples were sequenced using an Ion PGM™ 200 Sequencing kit and an Ion 318™ Chip according to the manufacturer's instructions. Torrent Suite v3.2 software was used to parse bar-coded reads, to align reads to the reference genome, and to generate run metrics and total read counts and quality. Genetic variants were identified using Variant Caller v3.2 software.
2.5. Taqman® assay
A replication study was conducted using TaqMan® SNP genotyping assays with a step One Plus™ thermal cycler (Life technologies). To validate SNP HID sequencing results, allele-specific real-time PCR was used. Primers were used to identify any DNA sequence that contained a polymorphism. Allele discrimination could be determined when a fluorescent probe was hybridized in a complementary target region that should have been amplified.
3. Results
3.1. WGAs
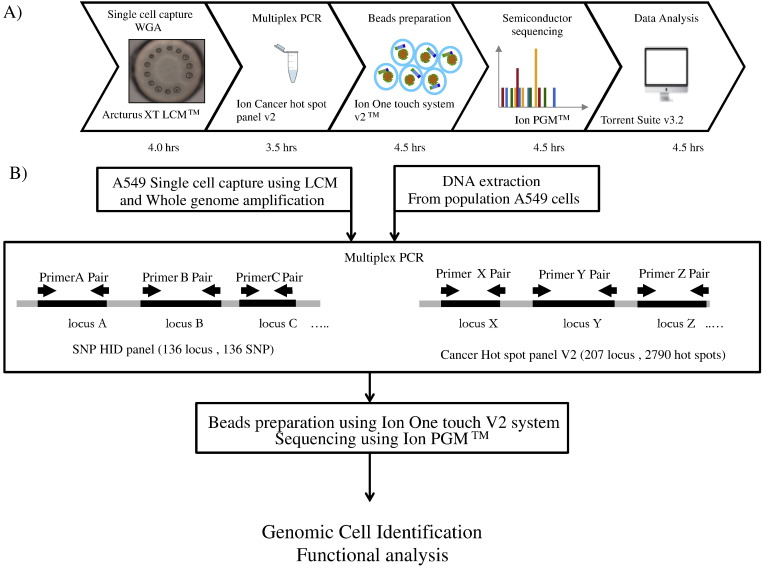
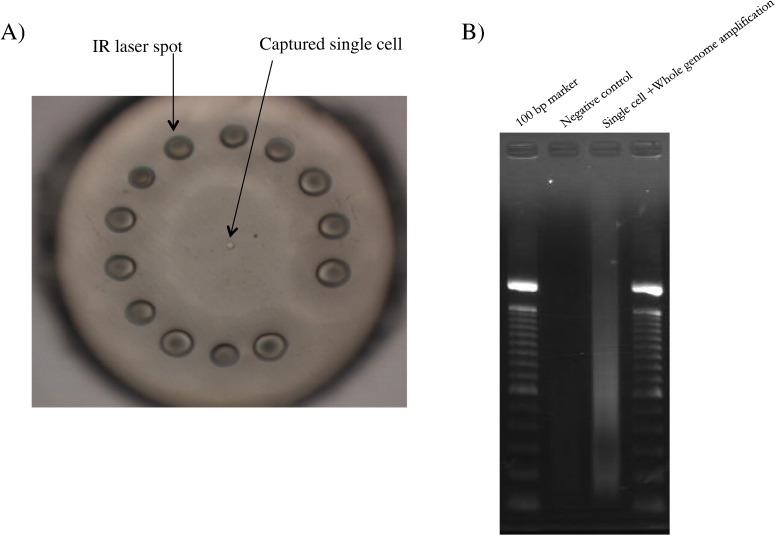
We used a semiconductor-based sequencing system in combination with a cancer hotspot panel for mutational profiling of a single cell. Single-cell capture was performed using LCM (Taylor et al., 2004), followed by WGA. The procedures used and the time required are shown in Fig. 1. The total time required for a single experiment was about 21 h. Successful amplification of the samples was confirmed by agarose gel electrophoresis (Fig. 2). Negative controls were included with each amplification batch. No amplification was observed for negative cell controls. This protocol utilized a highly multiplexed PCR amplification method (AmpliSeq™, Life Technologies) to enrich target sequence pools, a human identification pool, and a Cancer hotspot panel. Amplification from a bulk population of cells and a whole-genome amplified from a single cell from the same bulk population were compared.
Fig. 1.
Workflow used for this study.
A) Procedures used and the time required for an experiment. The total time required for a single experiment was approximately 21 h. B) Summary of single-cell identification and simultaneous functional sequence analysis with a semiconductor-based sequencer. Amplifications using a population of cells and a whole-genome amplified single cell from the same bulk population were compared.
Fig. 2.
Results for single-cell capture and WGA.
A) Image of a single cell. This cell was captured by LCM using an Arcturus® XT system (Life Technologies). B) Captured single cells and a blank sample included as a negative control were lysed and WGA was performed using single cell WGA kits (New England Bio Laboratories). Successful amplification of the samples was checked by agarose gel electrophoresis.
3.2. Sequencing analysis
Sequence coverage was assessed from the distribution of reads across target amplicons as shown in Table 1. After subtracting multiple-template reads and poor quality sequence reads, approximately 4.7 × 106 reads were obtained. An A549 bulk population of cells mapped approximately 1.5 × 106 sequence reads, while the A549 single cell Library prep replicate #1 derived sample mapped approximately 1.2 × 106 reads. The distribution of reads across both HID and Cancer panel amplicons was consistent across samples. The average coverage between samples ranged from 2591 to 4430, and was sufficient to evaluate normal samples.
Table 1.
Sequence data at SNP HID panel and Cancer hotspot panel v2.
| Basic reads information | ||
|---|---|---|
| Mapped reads (Cancer panel + HID panel) | Reads on targetb (Cancer panel + HID panel) | |
| A549 single cell Library prep replicate #1 | 1,129,189 | 90.06% |
| A549 single cell Library prep replicate #2 | ||
| Population cells | 1,562,883 | 96.22% |
| Read depth | 1 × coverage | 20 × coverage | 100 × coverage | Uniformity of coveragea | |
|---|---|---|---|---|---|
| SNP HID panel | |||||
| A549 single cell Library prep replicate #1 | 2851.93 | 69.65% | 62.31% | 55.93% | 45.57% |
| A549 single cell Library prep replicate #2 | |||||
| Population cells | 4430.34 | 100.00% | 99.16% | 99.04% | 73.32% |
| Cancer hotspot panel v2 | |||||
| A549 single cell Library prep replicate #1 | 2591.42 | 88.44% | 73.85% | 65.96% | 48.25% |
| A549 Single cell Library prep replicate #2 | |||||
| Population cells | 3740.26 | 98.84% | 98.84% | 98.83% | 95.73% |
Uniformity of coverage = percentage of bases covered at ≥ 20% of the mean coverage.
On-target reads = percentage of reads that mapped to target regions out of total mapped reads per run.
For the bulk population of cell,the percentages of sequence covered at coverage of more than 100 × were 99.04% for the HID panel and 98.83% for the Cancer panel, while the single cell percentages of sequence covered at coverage of more than 100 × were 55.93% for the HID panel and 65.96% for the Cancer panel. These differences were likely due to partial amplification failure or randomly distributed non-amplified regions across samples from single-cells during the WGA procedures or due to random allele drop out. Increased incidences of amplification failure and allele drop out have been previously reported (Garvin et al., 1998).
3.3. Comparative analysis between A549 single and bulk cells
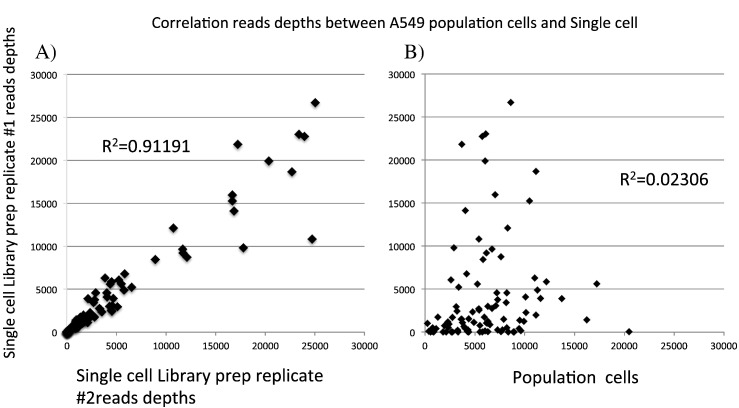
We made a comparative analysis between two A549 single-cell replicates (Library prep replicate #1 and Library prep replicate #2) and between an A549 single cell and an A549 population of cells. Correlations for read depths between two A549 single-cell replicates and between an A549 single cell and an A549 population of cells are shown in Fig. 3.
Fig. 3.
Correlations for read depths between two A549 single-cell replicates and between an A549 single cell and an A549 population of cells.
Comparative analyses were conducted for two A549 single-cell replicates and for an A549 single cell and an A549 population of cells. A) Correlation for read depths between single cell Library prep replicates #1 and #2 (#1 was from 50 ng of DNA templates and # 2 was from 10 ng of DNA templates). These results indicated a high correlation between these replicates. B) Correlation for read depths between A549 single cell Library prep replicate #1 and an A549 population of cells.
There was a high correlation between read depths for single cell Library prep replicates #1 and #2 (R2 = 0.91191). Single cell Library prep replicate #1 data were from 50 ng of DNA templates and single cell Library prep replicate # 2 data were from 10 ng of DNA templates. However, the correlation between the read depths of A549 single cell Library prep replicate #1 and an A549 population of cells was poor (R2 = 0.02306). This may also have been due to partial amplification failure or random non-amplified regions across samples from single cells during the WGA procedures or due to random allele drop out.
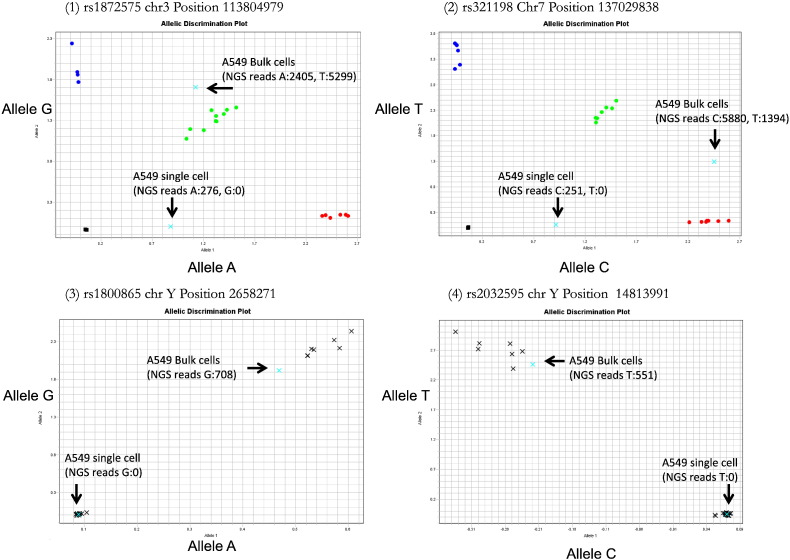
HID SNP typing showed high concordance rates between single cell Library prep replicate #1 and single cell Library prep replicate #2 and between an A549 single cell and an A549 population of cells. In particular, as for between single cell Library prep replicates #1 and #2, typing results were nearly the same. All 136 SNPs in the SNP HID panel were typed with the A549 population of cells, although some SNPs in the single-cell data set could not be detected. On autosomal chromosomes, 103 SNPs were typed, of which 86 SNPs were perfectly matched, 2 SNPs were partially matched, and 15 SNPs with autosomal chromosome locations had < 7 reads or had no coverage (Table 2). None of 33 SNP cells were detected on the Y chromosome with single-cell data. To validate the SNP HID sequencing results, allele-specific real-time PCR was performed using a Step One Plus™ thermal cycler with 4 primer pairs for selected non-perfectly matched SNPs (Fig. 4). This showed perfect matching between NGS typing and allele-specific real-time PCR typing results.
Table 2.
Comparison analysis at SNP HID panel on the autosomal chromosomes.
| Chromosome | Position | Target ID | A549 population cells |
A549 single cell Library prep replicate #1 |
A549 single cell Library prep replicate #2 |
Allele matching between population and single cell #1 | |
|---|---|---|---|---|---|---|---|
| Reads | Reads | Reads | |||||
| 1 | chr1 | 4367323 | rs1490413 | 783 | 506 | 887 | m |
| 2 | chr1 | 14155402 | rs7520386 | 8221 | 4566 | 4018 | m |
| 3 | chr1 | 160786670 | rs560681 | 2079 | 0 | 9 | n |
| 4 | chr1 | 238439308 | rs10495407 | 3626 | 1512 | 1923 | m |
| 5 | chr1 | 239881926 | rs891700 | 4255 | 91 | 147 | m |
| 6 | chr1 | 242806797 | rs1413212 | 3737 | 1078 | 2094 | m |
| 7 | chr2 | 114974 | rs876724 | 12185 | 5851 | 4528 | m |
| 8 | chr2 | 182413259 | rs12997453 | 3130 | 2359 | 4558 | m |
| 9 | chr3 | 961782 | rs1357617 | 4374 | 0 | 13 | n |
| 10 | chr3 | 59488340 | rs9866013 | 2409 | 951 | 1115 | m |
| 11 | chr3 | 113804979 | rs1872575 | 7710 | 276 | 311 | p |
| 12 | chr3 | 190806108 | rs1355366 | 8155 | 3452 | 2696 | m |
| 13 | chr3 | 193207380 | rs6444724 | 2755 | 1719 | 2107 | m |
| 14 | chr4 | 76425896 | rs13134862 | 1081 | 0 | 0 | n |
| 15 | chr4 | 169663615 | rs6811238 | 9403 | 405 | 0 | m |
| 16 | chr4 | 157489906 | rs1554472 | 511 | 0 | 325 | n |
| 17 | chr4 | 190318080 | rs1979255 | 7192 | 4561 | 2920 | m |
| 18 | chr5 | 2879395 | rs717302 | 16202 | 1432 | 934 | m |
| 19 | chr5 | 17374898 | rs159606 | 10053 | 4047 | 4056 | m |
| 20 | chr5 | 136633338 | rs13182883 | 3322 | 6 | 3 | m |
| 21 | chr5 | 159487953 | rs7704770 | 1111 | 434 | 917 | m |
| 22 | chr5 | 174778678 | rs251934 | 6320 | 2997 | 4315 | m |
| 23 | chr5 | 178690725 | rs338882 | 7017 | 15978 | 16675 | m |
| 24 | chr6 | 1135939 | rs1029047 | 372 | 105 | 350 | m |
| 25 | chr6 | 12059954 | rs13218440 | 6175 | 1066 | 1118 | m |
| 26 | chr6 | 55155704 | rs2811231 | 255 | 1019 | 1574 | m |
| 27 | chr6 | 120560694 | rs1478829 | 984 | 0 | 1 | n |
| 28 | chr6 | 123894978 | rs1358856 | 2631 | 0 | 0 | n |
| 29 | chr6 | 148761456 | rs2272998 | 4807 | 0 | 0 | n |
| 30 | chr6 | 152697706 | rs214955 | 3266 | 211 | 329 | m |
| 31 | chr6 | 165045334 | rs727811 | 8586 | 15 | 31 | m |
| 32 | chr7 | 4310365 | rs6955448 | 5403 | 2536 | 4468 | m |
| 33 | chr7 | 4457003 | rs917118 | 5497 | 792 | 716 | m |
| 34 | chr7 | 13894276 | rs1019029 | 6249 | 1272 | 1605 | m |
| 35 | chr7 | 137029838 | rs321198 | 7283 | 251 | 277 | p |
| 36 | chr7 | 155990813 | rs737681 | 13712 | 3881 | 2151 | m |
| 37 | chr8 | 28411072 | rs10092491 | 7523 | 67 | 121 | m |
| 38 | chr8 | 136839229 | rs4288409 | 4312 | 277 | 168 | m |
| 39 | chr8 | 139399116 | rs2056277 | 11214 | 4874 | 5782 | m |
| 40 | chr8 | 144656754 | rs4606077 | 1964 | 738 | 680 | m |
| 41 | chr9 | 14747133 | rs2270529 | 9489 | 1405 | 1911 | m |
| 42 | chr9 | 27985938 | rs7041158 | 3365 | 5217 | 6537 | m |
| 43 | chr9 | 126881448 | rs1463729 | 3678 | 21817 | 17240 | m |
| 44 | chr9 | 137417308 | rs10776839 | 7888 | 1506 | 1608 | m |
| 45 | chr10 | 3374178 | rs735155 | 2297 | 1198 | 1769 | m |
| 46 | chr10 | 17193346 | rs3780962 | 7251 | 3741 | 2789 | m |
| 47 | chr10 | 97172595 | rs1410059 | 5509 | 32 | 57 | m |
| 48 | chr10 | 118506899 | rs740598 | 9602 | 193 | 198 | m |
| 49 | chr10 | 132698419 | rs964681 | 10487 | 15260 | 16681 | m |
| 50 | chr11 | 5098714 | rs10768550 | 4277 | 69 | 101 | m |
| 51 | chr11 | 5099393 | rs10500617 | 8876 | 0 | 3 | n |
| 52 | chr11 | 5709028 | rs1498553 | 5400 | 10838 | 24725 | m |
| 53 | chr11 | 11096221 | rs901398 | 8284 | 12116 | 10728 | m |
| 54 | chr11 | 105912984 | rs6591147 | 3389 | 0 | 0 | n |
| 55 | chr11 | 122195989 | rs590162 | 1306 | 0 | 0 | n |
| 56 | chr12 | 888320 | rs2107612 | 868 | 16 | 49 | m |
| 57 | chr12 | 6909442 | rs2255301 | 6306 | 1009 | 933 | m |
| 58 | chr12 | 6945914 | rs2269355 | 6034 | 19889 | 20339 | m |
| 59 | chr12 | 106328254 | rs2111980 | 1820 | 14 | 12 | m |
| 60 | chr12 | 130761696 | rs10773760 | 20475 | 75 | 37 | m |
| 61 | chr13 | 22374700 | rs1886510 | 554 | 35 | 49 | m |
| 62 | chr13 | 84456735 | rs9546538 | 2879 | 9826 | 17784 | m |
| 63 | chr13 | 100038233 | rs1058083 | 2334 | 532 | 1110 | m |
| 64 | chr13 | 106938411 | rs354439 | 2635 | 2 | 4 | m |
| 65 | chr14 | 25850832 | rs1454361 | 9917 | 1251 | 915 | m |
| 66 | chr14 | 98845531 | rs873196 | 6715 | 2750 | 3275 | m |
| 67 | chr14 | 104769149 | rs4530059 | 6079 | 22987 | 23406 | m |
| 68 | chr15 | 39313402 | rs1821380 | 4909 | 1162 | 1481 | m |
| 69 | chr16 | 5606197 | rs729172 | 10102 | 2275 | 2360 | m |
| 70 | chr16 | 5868700 | rs2342747 | 2610 | 6048 | 5248 | m |
| 71 | chr16 | 78017051 | rs430046 | 6083 | 8464 | 11680 | m |
| 72 | chr16 | 80106361 | rs1382387 | 3983 | 443 | 444 | m |
| 73 | chr17 | 41286822 | rs2175957 | 7090 | 3093 | 4591 | m |
| 74 | chr17 | 41341984 | rs8070085 | 6172 | 9215 | 11726 | m |
| 75 | chr17 | 41691526 | rs1004357 | 5712 | 22482 | 23964 | m |
| 76 | chr17 | 80526139 | rs2291395 | 8351 | 19 | 9 | m |
| 77 | chr17 | 80531643 | rs4789798 | 6312 | 43 | 80 | m |
| 78 | chr17 | 80715702 | rs689512 | 6038 | 129 | 129 | m |
| 79 | chr17 | 80739859 | rs3744163 | 8121 | 480 | 652 | m |
| 80 | chr17 | 80765788 | rs2292972 | 7626 | 8748 | 12108 | m |
| 81 | chr18 | 1127986 | rs1493232 | 2756 | 18 | 34 | m |
| 82 | chr18 | 9749879 | rs9951171 | 6520 | 872 | 683 | m |
| 83 | chr18 | 22739001 | rs7229946 | 5395 | 2718 | 3322 | m |
| 84 | chr18 | 29311034 | rs985492 | 4040 | 14112 | 16841 | m |
| 85 | chr18 | 47371014 | rs521861 | 3959 | 0 | 0 | n |
| 86 | chr18 | 55225777 | rs1736442 | 5762 | 16 | 16 | m |
| 87 | chr18 | 75432386 | rs1024116 | 1300 | 1762 | 2807 | m |
| 88 | chr19 | 28463337 | rs719366 | 17160 | 5626 | 5487 | m |
| 89 | chr19 | 39559807 | rs576261 | 5800 | 8440 | 8923 | m |
| 90 | chr20 | 16241416 | rs12480506 | 11582 | 3938 | 4690 | m |
| 91 | chr20 | 23017082 | rs2567608 | 11131 | 18667 | 22692 | m |
| 92 | chr20 | 39487110 | rs1005533 | 4770 | 2345 | 3535 | m |
| 93 | chr20 | 51296162 | rs1523537 | 3113 | 2924 | 5102 | m |
| 94 | chr21 | 16685598 | rs722098 | 746 | 98 | 264 | m |
| 95 | chr21 | 28023370 | rs464663 | 5969 | 1750 | 1357 | m |
| 96 | chr21 | 33582722 | rs2833736 | 11000 | 6320 | 3864 | m |
| 97 | chr21 | 42415929 | rs914165 | 4174 | 6788 | 5841 | m |
| 98 | chr22 | 19920359 | rs9606186 | 8589 | 26670 | 25031 | m |
| 99 | chr22 | 23802171 | rs2073383 | 3889 | 659 | 798 | m |
| 100 | chr22 | 27816784 | rs733164 | 4370 | 1531 | 1524 | m |
| 101 | chr22 | 33559508 | rs987640 | 3691 | 1511 | 2156 | m |
| 102 | chr22 | 47836412 | rs2040411 | 11123 | 1985 | 1680 | m |
| 103 | chr22 | 48362290 | rs1028528 | 5252 | 5580 | 4370 | m |
m = match; p = partial match; n = no depth.
Fig. 4.
Allelic description plots as replication study using TaqMan® SNP genotyping assays.
To validate SNP HID sequencing results, allele-specific real-time PCR was performed. Four representative plots showing performance of four assays in analysis of A549 samples and reference samples. VIC signal (x-axis) is associated with the probe for allele A (graph (1), (3)) and allele C (graph (2), (4)), while FAM (y-axis) labels the allele G (graph (1), (3)) and allele T (graph (2), (4)) probes. Aqua blue × symbols indicate A549 bulk cells and a single cell with NGS reads data. Circles symbols and black × symbols indicate 20 Coriell gDNA samples as reference.
3.4. Cancer gene analysis
A Cancer gene panel was used for a functional analysis (Table 3). We again found high concordance rates between A549 single cell Library prep replicates #1 and #2 and between an A549 single cell and an A549 population of cells. A total of 11 variants were typed for both samples, of which 1 was partially matched and 5 SNPs were not detected because of low or no depth in the single cell Library prep replicates #1 and #2 data set. A total of 16 variants were detected in A549 single cell Library prep replicates #1 and #2 cell and 13 variants were detected in an A549 population of cells; 11 variant cells were perfectly consistent. Five SNPs were called as variants and some discrepancies were observed. No frameshifts or deletions were observed at 2790 hotspots.
Table 3.
Comparative analysis for the Cancer hotspot panel of 50 cancer-related genes.
| Chromosome | Position | Gene Sym | Hotspot ID | A549 population cells |
A549 single cell Library prep replicates #1 |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Zygosity | Ref | Variant | Var freq | Coverage | Ref cov | Var cov | Zygosity | Ref | Variant | Var freq | Coverage | Ref cov | Var cov | ||||
| Match pairs list | |||||||||||||||||
| chr4 | 1807894 | FGFR3 | – | Hom | G | A | 99.7 | 2003 | 6 | 1997 | Hom | G | A | 99.25 | 2135 | 14 | 2119 |
| chr4 | 55141055 | PDGFRA | – | Hom | A | G | 100 | 1605 | 0 | 1605 | Hom | A | G | 99.69 | 12002 | 21 | 11965 |
| chr5 | 149433597 | CSF1R | – | Hom | G | A | 97.6 | 1503 | 36 | 1467 | Hom | G | A | 96.18 | 12894 | 458 | 12402 |
| chr5 | 149433596 | CSF1R | – | Hom | T | G | 97.88 | 1464 | 1 | 1433 | Hom | T | G | 97.2 | 12285 | 33 | 11941 |
| chr7 | 55249063 | EGFR | – | Hom | G | A | 99.88 | 2456 | 2 | 2453 | Hom | G | A | 100 | 12 | 0 | 12 |
| chr10 | 43615633 | RET | – | Het | C | G | 66.46 | 3208 | 1075 | 2132 | Het | C | G | 64.45 | 422 | 149 | 272 |
| chr10 | 43613843 | RET | – | Hom | G | T | 99.85 | 6073 | 0 | 6064 | Hom | G | T | 99.56 | 1609 | 0 | 1602 |
| chr12 | 25398285 | KRAS | COSM517; | Hom | C | T | 99.62 | 4487 | 17 | 4470 | Hom | C | T | 100 | 24 | 0 | 24 |
| chr13 | 28610183 | FLT3 | – | Hom | A | G | 99.9 | 4910 | 4 | 4905 | Hom | A | G | 99.88 | 3342 | 4 | 3338 |
| chr17 | 7579472 | TP53 | Het | G | C | 91.03 | 2520 | 225 | 2294 | Het | G | C | 88.23 | 3865 | 446 | 3410 | |
| chr19 | 1207021 | STK11 | COSM12925; | Hom | C | T | 99.9 | 2909 | 3 | 2906 | Hom | C | T | 99.35 | 10927 | 47 | 10856 |
| Not mutch pairs list | |||||||||||||||||
| chr3 | 178917005 | PIK3CA | – | Hom | A | G | 99.57 | 1153 | 5 | 1148 | Not detected | ||||||
| chr4 | 55602749 | KIT | Not detected | Het | T | C | 46.36 | 4864 | 2581 | 2255 | |||||||
| chr4 | 55979623 | KDR | COSM32339 | Het | C | G | 48.72 | 2422 | 1243 | 1180 | Het | C | T | 73.68 | 19 | 5 | 14 |
| chr11 | 108155120 | ATM | Not detected | Het | G | T | 50 | 12 | 6 | 6 | |||||||
| chr11 | 108204661 | ATM | Not detected | Het | T | C | 70.54 | 258 | 76 | 182 | |||||||
| chr11 | 108204660 | ATM | Not detected | Hom | T | C | 91.89 | 259 | 21 | 238 | |||||||
4. Discussion
We have described a genomic single-cell identification method with simultaneous functional analysis using NGS. We used the A549 cell line to check for concordance rates between a single cell and ≈ 106 cells in a bulk population. Working with single cells requires careful monitoring, for which two approaches are primarily used: LCM and cell sorting.
Using these approaches, technical contamination should be ruled out. Sources of contamination can be unrelated genetic material that is inadvertently introduced into a sample. Simple and robust techniques to identify or confirm the genetic origin of a cellular material under investigation are a critical quality control step. With the application described here, we paired cell identification with cancer profiling.
HID SNP typing showed high concordance rates between an A549 single cell and an A549 population of cells. However, some SNPs on autosomal chromosomes and all SNP cells on the Y chromosome in a single-cell data set could not be detected. Depletion of the Y chromosome is often observed for transferred culture cells; thus, this may also have occurred with our preparations (Ono et al., 2001). There have been many reports regarding allele drop out and failed amplification rates after single cell WGA (Baslan et al., 2012, Spits et al., 2006, Handyside et al., 2004, Handyside et al., 2010, Konings et al., 2012).
Regarding the WGA methodology, some investigators have indicated that multiple displacement amplification (MDA), such as with QIAgen's REPLI-g technology, was more appropriate for microarray genotyping applications than PCR-based WGA, such as the NEB WGA kit used in this study (Treff et al., 2011). MDA-based WGA (Repli-G) may result in less allele dropout, which may suggest better results for the AmpliSeq protocol. We intend to compare amplification methodologies in future studies.
Although genomic instability or inefficient WGA may compromise analysis using single cells, we used 136 SNPs that were evenly distributed across the entire genome for discrimination purposes. Thus, despite the fact that some genome regions were missing in our single-cell data sets, the HID SNP set used here retained its discriminatory capability. To confirm the utility and robustness of our method, we intend to repeat our experiment using more single cell replicates and different cell-picking methods. The former should help to understand genomic instability or efficiency of WGA, the latter should help identify any background that results from using LCM. Although we plan to explore these issues in the future, in this report, we cannot deal with these issues because of the costs involved and the labor-intensive nature of the procedures used.
Regarding cancer gene analysis, 5 SNPs were called as variants and some discrepancies were found. Only 3 of 5 variants were detected for the ataxia telangiectasia mutated (ATM) gene. This was likely due to random non-amplified regions across samples of single cells during WGA.
Other possible applications for our method include forensics, transplantation medicine, regenerative medicine, and pre-natal testing using maternal blood (Fan et al., 2008). Forensic samples are often heterogeneous. In many cases, samples at crime scenes are mixtures from multiple subjects (e. g., offender, victim, or unrelated individual). Single-cell analysis should remove any ambiguity in data interpretation.
In conclusion, our method provides an easy to implement and effective method to investigate sample heterogeneity in various areas, such as tumor biology, forensics, regenerative medicine, and fetal DNA tracing in maternal blood samples.
The following are the supplementary data related to this article.
Workflow of living single-cell capture.
Completing interests
All authors work for Life Technologies Japan, Ltd.
Acknowledgment
The authors thank all members of the Life Technologies' Technical Department. We would like to thank Dr. Zhen Mahoney for the critical reading of the manuscript.
References
- Baslan T., Kendall J., Rodgers L., Cox H., Riggs M., Stepansky A., Troge J., Ravi K., Esposito D., Lakshmi B., Wigler M., Navin N., Hicks J. Genome-wide copy number analysis of single cells. Nat. Protoc. 2012;7:1024–1041. doi: 10.1038/nprot.2012.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan H.C., Blumenfeld Y.J., Chitkara U., Hudgins L., Quake S.R. Noninvasive diagnosis of fetal aneuploidy by shotgun sequencing DNA from maternal blood. Proc. Natl. Acad. Sci. U. S. A. 2008;105:16266–16271. doi: 10.1073/pnas.0808319105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garvin A.M., Holzgreve W., Hahn S. Highly accurate analysis of heterozygous loci by single cell PCR. Nucleic Acids Res. 1998;26:3468–3472. doi: 10.1093/nar/26.15.3468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlinger M., Rowan A.J., Horswell S., Larkin J., Endesfelder D., Gronroos E., Martinez P., Matthews N., Stewart A., Tarpey P., Varela I., Phillimore B., Begum S., McDonald N.Q., Butler A., Jones D., Raine K., Latimer C., Santos C.R., Nohadani M., Eklund A.C., Spencer-Dene B., Clark G., Pickering L., Stamp G., Gore M., Szallasi Z., Downward J., Futreal P.A., Swanton C. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handyside A.H., Robinson M.D., Simpson R.J., Omar M.B., Shaw M.A., Grudzinskas J.G., Rutherford A. Isothermal whole genome amplification from single and small numbers of cells: a new era for preimplantation genetic diagnosis of inherited disease. Mol. Hum. Reprod. 2004;10:767–772. doi: 10.1093/molehr/gah101. [DOI] [PubMed] [Google Scholar]
- Handyside A.H., Harton G.L., Mariani B., Thornhill A.R., Affara N., Shaw M.A., Griffin D.K. Karyomapping: a universal method for genome wide analysis of genetic disease based on mapping crossovers between parental haplotypes. J. Med. Genet. 2010;47:651–658. doi: 10.1136/jmg.2009.069971. [DOI] [PubMed] [Google Scholar]
- Harismendy O., Schwab R.B., Bao L., Olson J., Rozenzhak S., Kotsopoulos S.K., Pond S., Crain B., Chee M.S., Messer K., Link D.R., Frazer K.A. Detection of low prevalence somatic mutations in solid tumors with ultra-deep targeted sequencing. Genome Biol. 2011;12:R124. doi: 10.1186/gb-2011-12-12-r124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konings P., Vanneste E., Jackmaert S., Ampe M., Verbeke G., Moreau Y., Vermeesch J.R., Voet T. Microarray analysis of copy number variation in single cells. Nat. Protoc. 2012;7:281–310. doi: 10.1038/nprot.2011.426. [DOI] [PubMed] [Google Scholar]
- Korzebor A., Derakhshandeh-Peykar P., Meshkani M., Hoseini A., Rafati M., Purhoseini M., Ghaffari S.R. Heterozygosity assessment of five STR loci located at 5q13 region for preimplantation genetic diagnosis of spinal muscular atrophy. Mol. Biol. Rep. 2013;40:67–72. doi: 10.1007/s11033-012-2011-3. [DOI] [PubMed] [Google Scholar]
- Mertes F., Elsharawy A., Sauer S., van Helvoort J.M., van der Zaag P.J., Franke A., Nilsson M., Lehrach H., Brookes A.J. Targeted enrichment of genomic DNA regions for next-generation sequencing. Brief. Funct. Genomics. 2011;10:374–386. doi: 10.1093/bfgp/elr033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navin N., Kendall J., Troge J., Andrews P., Rodgers L., McIndoo J., Cook K., Stepansky A., Levy D., Esposito D., Muthuswamy L., Krasnitz A., McCombie W.R., Hicks J., Wigler M. Tumour evolution inferred by single-cell sequencing. Nature. 2011;472:90–94. doi: 10.1038/nature09807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono Y., Shimozawa N., Muguruma K., Kimoto S., Hioki K., Tachibana M., Shinkai Y., Ito M., Kono T. Production of cloned mice from embryonic stem cells arrested at metaphase. Reproduction. 2001;122:731–736. doi: 10.1530/rep.0.1220731. [DOI] [PubMed] [Google Scholar]
- Pakstis A.J., Speed W.C., Fang R., Hyland F.C., Furtado M.R., Kidd J.R., Kidd K.K. SNPs for a universal individual identification panel. Hum. Genet. 2010;127:315–324. doi: 10.1007/s00439-009-0771-1. [DOI] [PubMed] [Google Scholar]
- Pietersen C.Y., Lim M.P., Woo T.U. Obtaining high quality RNA from single cell populations in human postmortem brain tissue. J. Vis. Exp. Aug 2009;6(30):1–9. doi: 10.3791/1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothberg J.M., Hinz W., Rearick T.M., Schultz J., Mileski W., Davey M., Leamon J.H., Johnson K., Milgrew M.J., Edwards M., Hoon J., Simons J.F., Marran D., Myers J.W., Davidson J.F., Branting A., Nobile J.R., Puc B.P., Light D., Clark T.A., Huber M., Branciforte J.T., Stoner I.B., Cawley S.E., Lyons M., Fu Y., Homer N., Sedova M., Miao X., Reed B., Sabina J., Feierstein E., Schorn M., Alanjary M., Dimalanta E., Dressman D., Kasinskas R., Sokolsky T., Fidanza J.A., Namsaraev E., McKernan K.J., Williams A., Roth G.T., Bustillo J. An integrated semiconductor device enabling non-optical genome sequencing. Nature. 2011;475:348–352. doi: 10.1038/nature10242. [DOI] [PubMed] [Google Scholar]
- Sanchez J.J., Phillips C., Borsting C., Balogh K., Bogus M., Fondevila M., Harrison C.D., Musgrave-Brown E., Salas A., Syndercombe-Court D., Schneider P.M., Carracedo A., Morling N. A multiplex assay with 52 single nucleotide polymorphisms for human identification. Electrophoresis. 2006;27:1713–1724. doi: 10.1002/elps.200500671. [DOI] [PubMed] [Google Scholar]
- Shapiro E., Biezuner T., Linnarsson S. Single-cell sequencing-based technologies will revolutionize whole-organism science. Nat. Rev. Genet. 2013;14:618–630. doi: 10.1038/nrg3542. [DOI] [PubMed] [Google Scholar]
- Shyr D., Liu Q. Next generation sequencing in cancer research and clinical application. Biol. Proced. Online. 2013;15:4. doi: 10.1186/1480-9222-15-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spits C., Le Caignec C., De Rycke M., Van Haute L., Van Steirteghem A., Liebaers I., Sermon K. Optimization and evaluation of single-cell whole-genome multiple displacement amplification. Hum. Mutat. 2006;27:496–503. doi: 10.1002/humu.20324. [DOI] [PubMed] [Google Scholar]
- Swanton C. Intratumor heterogeneity: evolution through space and time. Cancer Res. 2012;72:4875–4882. doi: 10.1158/0008-5472.CAN-12-2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor T.B., Nambiar P.R., Raja R., Cheung E., Rosenberg D.W., Anderegg B. Microgenomics: identification of new expression profiles via small and single-cell sample analyses. Cytometry A. 2004;59:254–261. doi: 10.1002/cyto.a.20051. [DOI] [PubMed] [Google Scholar]
- Treff N.R., Su J., Tao X., Northrop L.E., Scott R.T., Jr. Single-cell whole-genome amplification technique impacts the accuracy of SNP microarray-based genotyping and copy number analyses. Mol. Hum. Reprod. 2011;17:335–343. doi: 10.1093/molehr/gaq103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yousem S.A., Dacic S., Nikiforov Y.E., Nikiforova M. Pulmonary Langerhans cell histiocytosis: profiling of multifocal tumors using next-generation sequencing identifies concordant occurrence of BRAF V600E mutations. Chest. 2013;143:1679–1684. doi: 10.1378/chest.12-1917. [DOI] [PubMed] [Google Scholar]
- Zheng Y.M., Wang N., Li L., Jin F. Whole genome amplification in preimplantation genetic diagnosis. J. Zhejiang Univ. Sci. B. 2011;12:1–11. doi: 10.1631/jzus.B1000196. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Workflow of living single-cell capture.