Abstract
Overexpression of the components of the Janus kinase/signal transducer and activator of transcription (JAK/STAT) signaling pathway are key factors of the pathogenic mechanisms underlying systematic juvenile idiopathic arthritis (SJIA). The present study aimed to investigate the association between microRNA (miR)-19a, miR-21 and the JAK/STAT signaling pathway. A total of 20 patients with SJIA were included in the study, and peripheral blood mononuclear cells (PBMCs) from 20 normal controls were also collected. RNAiso was used to extract total RNA, and the RNA was then reverse transcribed into cDNA. Primers were designed to detect the mRNA of miR-19a and miR-21, and U6 was set as the internal parameter. In addition, the mRNA of STAT3, suppressor of cytokine signaling 3 (SOCS3), interleukin-6 (IL-6) and tumor necrosis factor-α (TNF-α) was detected, and β-actin was set as the internal parameter. Reverse transcription-quantitative polymerase chain reaction was performed to detect the expression levels of these proteins in patients with SJIA and control subjects, and non-parametric tests were used to analyze the statistical differences in 2−ΔΔCq between the two groups. The expression levels of miR-19a and miR-21 were significantly lower in the SJIA group compared with the control group (P<0.05). SOCS3, TNF-α and STAT3 were shown to be the target genes of miR-19a and miR-21, as determined by Targetscan. The expression levels of STAT3, SOCS3, TNF-α and IL-6 mRNA were significantly higher compared with those of the control group (P<0.05). In the PBMCs of sthe patients with SJIA, miR-19a and miR-21 expression levels were lower compared with those of the control group, and the JAK/STAT signaling pathway was activated, which indicated that miR-19a and miR-21 may participate in the activation of the JAK/STAT signaling pathway.
Keywords: systematic juvenile idiopathic arthritis, microRNA, Janus kinase/signal transducer and activator of transcription, signaling pathway
Introduction
Systematic juvenile idiopathic arthritis (SJIA) is a type of chronic arthritis that is characterized by systematic symptoms (high fever, rash, and enlargement of the liver, spleen and lymph nodes) and various increased inflammatory factors, such as C reactive proteins (CRP) and ferritin (FER) (1). SJIA is the most severe type of juvenile idiopathic arthritis (JIA), and can induce macrophage activation syndrome that results in mortality (2). Different from adult rheumatoid arthritis (RA), which presents as significant joint deformity, patients with JIA (specifically patients with SJIA) are diagnosed according to common symptoms and high levels inflammatory factors (3). However, there is a current requirement for more specific markers for the condition.
MicroRNA (miRNA) is a type of non-coding single stranded RNA that is widespread in the body and has a regulatory function (4). It binds specifically to the 3′-untranslated region of target mRNA in order to degrade or inhibit its translation, thereby participating in many basic bioprocesses, such as cell development, proliferation and differentiation, and the cell cycle (4). Therefore, miRNA has an important role in numerous diseases, including cancer, metabolic diseases, nerve disorders and infectious and autoimmune diseases (2). Recently, a number of studies suggested that miRNA serves an important role in regulating chronic inflammation, in which some miRNAs, such as miR-155 (5) and miR-146a (6), participated in the proliferation and differentiation of immune cells, such as T and B cells, and regulated their abnormal expression in autoimmune diseases. Increased expression levels of miR-146a partially contributes towards upregulated expression of TNF-α; however, the mechanism underlying this increase in expression and the site of action of miR-146a in signaling pathways remains unclear (7). Meanwhile, it could not explain JIA with normal TNF-α and bad efficacy of TNF-α antagonist to systematic arthritis, thus it was assumed that there was an interaction of a different signaling pathway in JIA. Despite recent random control studies suggesting that there are various expression levels of numerous types of miRNA, including miR-155, miR-19a, miR-203, miR-21 and miR-124a, in synovioblasts, PBMC and T cells from patients with RA (7–14), the studies did not investigate the target genes of miRNAs, which could demonstrate the immunological dysregulation, inflammation and differentiation of cells. Therefore, in the present study, miR-19a, miR-21 and their associated target genes involved in SJIA were used to investigate the roles of miRNA in SJIA.
Materials and methods
Inclusion criteria
A total of 20 patients with active SJIA (fever for >2 weeks; high fever type with or without articular pain; liver, spleen and lymph node enlargement; increased expression levels of FER; CRP ≥10 mg/l, or erythrocyte sedimentation rate ≥28 ram/1 h) were diagnosed in Guangzhou Women and Children's Medical Center (Guangdong, China) from January to March 2015, and 20 normal control patients were recruited from the Health Care Section of the hospital. Patients were diagnosed and classified according to the Task-force of the Pediatric Standing Committee of International League of Associations for Rheumatology discussion draft (15). Patients with recurrent bacterial, mycoplasma, viral, fungal and mycobacterial infections, an infection history, an active infection, or blood diseases detected by bone marrow cytology were excluded from the study. Baseline characteristics of the control subjects and patients with SJIA are presented in Table I. The study protocol was approved by the ethics committee of Guangzhou Women and Children's Medical Center, and written-informed consent was obtained from all subjects.
Table I.
Baseline data of the patients with systematic juvenile idiopathic arthritis and healthy controls.
| Clinical information | SJIA group (n=20) | Control group (n=20) |
|---|---|---|
| Age (years) | 7.36±3.64 | 8.90±3.69 |
| Male proportion (%) | 65 | 70 |
| Weight (kg) | 13.45±6.34 | 12.67±7.83 |
| DAS28 scores | 6.23±2.34 | – |
| Abnormal BMD or synovitis suggested by MRI | 16 (80%) | – |
| Anti-cytokeratin antibody | 1 (12.5%) | – |
| Anti-cyclic citrullinated peptide antibody (U/ml x±s) | 9.77±8.07 | – |
| RF-IgG (U/ml x±s) | 13.60±9.99 | – |
| Ferritin (ng/ml, x±s) | 1079.47±900.87 | – |
| CRP (mg/l, x±s) | 98.61±63.80 | – |
| ESR (mm/h, x±s) | 53.12±26.59 | – |
| AST median | 56.23 (6,74) | – |
| ALT median | 47.69 (11,16) | 24 (5,20) |
BMD, bone marrow density; MRI, magnetic resonance imaging; RF-IGg, rheumatoid factor immunoglobulin G; CRP, C-reactive protein; ESR, erythrocyte sedimentation rate; AST, aspartate aminotransferase; ALT, alanine transaminase; SJIA, systematic juvenile idiopathic arthritis; x±s, mean ± standard deviation.
Reagents
Ficoll Paque PLUS solution and phosphate-buffered saline (PBS) were purchased from GE Healthcare Life Sciences (Logan, UT, USA); the RNA extraction kit (9112; Takara Biotechnology Co., Ltd. (Dailan, China), reverse transcription kit, RNAiso (9753A), miR-X iRNA First Strand Synthesis kit (638315) and SYBR® Premix Ex Taq (Tli RNaseH Plus; RR420A) kits were purchased from Takara Biotechnology Co., Ltd.. The primers of miR-19a, miR-21 and U6 were synthesized by Takara Biotechnology Co., Ltd. The primer sequences were as follows: STAT3 forward, 5′-GCCAGAGAGCCAGGAGCA-3′, and reverse, 5′-ACACAGATAAACTTGGTCTTCAGGTATG-3′; SOCS3 forward, 5′-CAGCTCCAAGAGCGAGTACC-3′, and reverse, 5′-TGACGCTGAGCGTGAAGAAG-3′; TNF-α forward, 5′-ACCCTCACACTCAGATCATC-3′, and reverse, 5′-GAGTAGACAAGGTACAACCC-3′; IL-6 forward, 5′-AGCCAGAGTCCTTCAGAGAG-3′, and reverse, 5′-GATGGTCTTGGTCCTTAGCC-3′; β-actin forward, 5′-GAGCTACGAGCTGCCTGACG-3′, and reverse, 5′-GTAGTTTCGTGGATGCCACAG-3′. The primers were synthesized by Shenzen Huada Gene Technology Co., Ltd. (Shenzen, China) company.
Separation of PBMCs and extraction of total RNA
Fasting blood was drawn from the patients with SJIA and the control subjects, and placed in 4 ml ethylenediaminetetraacetic acid. Subsequently, 4 ml of PBS was added in order to dilute the sample. The same volume of Ficoll Paque PLUS solution and diluted blood was added into 15 ml centrifuge tubes and centrifuged at 160 × g for 20 min at 18–21°C. The cell pellet was then collected and added into pentaploid PBS, and centrifuged twice at 160 × g for 20 min. Waste liquid was discarded, and 1 ml RNAiso and 200–300 Ml chloroform (Sigma-Aldrich, St. Louis, MO, USA) was added. The samples were incubated for 5 min at 25–28°C and centrifuged at 12,000 × g for 15 min at 4°C. Approximately 400 µl supernatant was obtained, to which 1 ml cold isopropanol (Sigma-Aldrich) was added, mixed reversely and incubated at 4°C for 10 min, prior to being centrifuged at 12,000 × g for 10 min at 4°C. The supernatant was then discarded, and 10 ml 75% ethanol prepared with diethylpyrocarbonate (DEPC) (Sigma-Aldrich) water was added (ethanol:DEPC; 3:1), followed by centrifugation at 7,500 × g for 5 min at 4°C. Finally, 20 µl DEPC water was added to the samples, and the absorbance was measured using a spectrophotometer (Thermo NanoDrop 2000; Thermo Fisher Scientific, Inc., Waltham, MA, USA).
Reverse transcription-quantitative PCR (RT-qPCR)
A PrimeScript II 1st Strand cDNA Synthesis Kit (Takara Biotechnology Co., Ltd.) was used. RNAiso was used to extract total RNA. A total of 3.8 µl RNA was added into 0.2 ml PCR tubes, followed by the addition of 1.2 µl mRQ enzyme mix and 5 µl buffer. The samples were then incubated at 37°C for 1 h, and 95°C for 5 min. Following incubation, 90 µl ddH2O was added and the samples were stored at −20°C for the subsequent experiments. For qPCR, 9 µl dddH20, 12.5 µl SYBR Green, 0.5 µl upstream primers, 0.5 µl downstream primers, ROC 50×0.5 µl and 2 µl cDNA were used, and the samples were incubated at 95°C for 10 sec, 95°C for 5 sec, 60°C for 20 sec, then 40 cycles at 72°C for 5 min using a Roche LightCycler 480II (Roche Diagnostics, Basel, Switzerland). Each gene had three repeats. U6 was used to normalize the expression of miRNA, and β-actin was used to normalize the expression of STAT3, SOCS3, IL-6 and TNF-α. The ΔCq and ΔΔCq values were calculated, and the relative expression levels of mRNA, associated target genes and U6 were presented as the 2−ΔΔCq value (16).
Statistical analysis
Data were analyzed using SPSS version 13.0 software (SPSS, Inc., Chicago, IL, USA), and miR-19a and miR-21 were expressed as relative expression levels. A Kruskal-Wallis test was used for intergroup comparisons, and a Pearson's correlation coefficient was used to calculate the correlation between miR-19a, miR-21 and inflammatory factors. P<0.05 was considered to indicate a statistically significant difference.
Results
The expression levels of miR-21 and miR-19a are decreased in patients with SJIA compared with the controls
The expression levels of miR-21 and miR-19a were investigated in PBMCs from active state patients with SJIA and control subjects. The expression levels of miR-21 were lower in patients with SJIA compared with the control group (Fig. 1), and the relative expression levels of miR-21 in the control group were 7.7-fold higher compared with those of the patients with SJIA (Table II; P=0.036). In addition, the expression levels of miR-19a were lower in patients with SJIA compared with the control group (Fig. 2), and the relative expression levels of miR-19a in the control group were 11.3-fold higher compared with those of the patients with SJIA (Table III; P=0.014).
Figure 1.
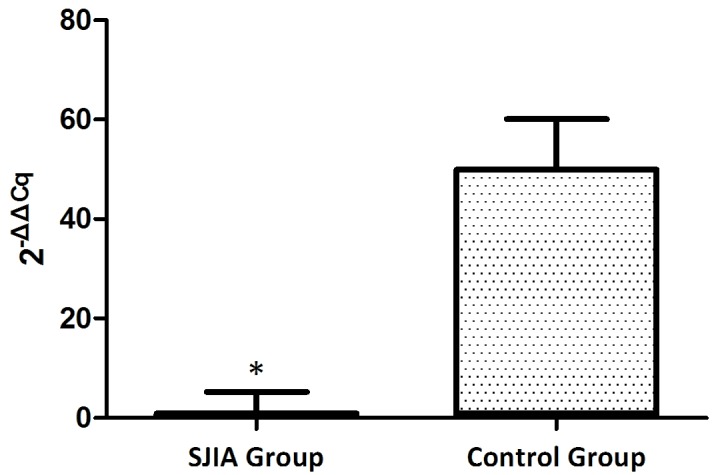
Expression levels of microRNA-21 in the SJIA and control group. *P<0.05 vs. the control group. SJIA, systemic juvenile idiopathic arthritis.
Table II.
Expression levels of miR-21 in PBMCs of patients with SJIA and the control group (2−ΔΔCq).
| Group | miR-21 Cq | U6 Cq | ΔCq | ΔΔCq | 2−ΔΔCq |
|---|---|---|---|---|---|
| SJIA groupa | 25.2±1.04 | 18.7±1.07 | 6.4±0.3 | 0.0±0.3 | 1.0 (0.8–1.2)b |
| Control groupa | 24.6±0.94 | 21.1±1.13 | 3.5±0.4 | −2.9±0.4 | 7.7 (7–8.5)b |
n=20
Z=−2.11; P=0.036. miR, microRNA, PBMC, peripheral blood mononuclear cell; SJIA, systematic juvenile idiopathic arthritis.
Figure 2.
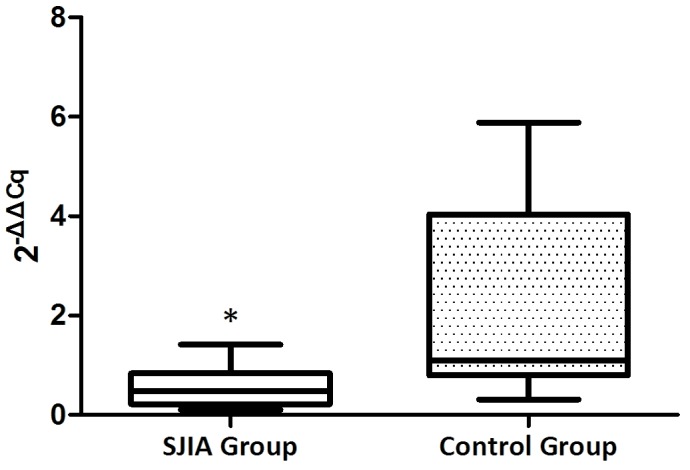
Expression levels of microRNA-19a in the SJIA and control group. *P<0.05 vs. the control group. SJIA, systemic juvenile idiopathic arthritis.
Table III.
Expression levels of miR-19a in PBMCs of patients with SJIA and the control group (2−ΔΔCq).
| Group | miR-19a Cq | U6 Cq | ΔCq | ΔΔCq | 2−ΔΔCq |
|---|---|---|---|---|---|
| SJIA groupa | 28.0±0.71 | 15.7±1.78 | 12.3±1.1 | 0.0±1.1 | 1.0 (0.9–1.3)b |
| Control groupa | 29.9±0.69 | 21.1±1.08 | 8.8±0.91 | −3.5±0.91 | 11.3 (10–12.1)b |
n=20
Z=−2.41, P=0.014. miR, microRNA, PBMC, peripheral blood mononuclear cell; SJIA, systematic juvenile idiopathic arthritis.
The mRNA expression levels of STAT3, TNF-α, IL-6 and SOCS3 are higher in patients with SJIA compared with the controls
The mRNA expression levels of STAT3, TNF-α, IL-6 and SOCS3 were expressed in the peripheral blood of patients with SJIA. The relative expression levels of STAT3 (Fig. 3), TNF-α (Fig. 4), IL-6 (Fig. 5) and SOCS3 (Fig. 6) mRNA were higher in patients with SJIA compared with the control group. In addition, the expression levels of STAT3 mRNA were negatively correlated with miR-21 2−ΔΔCq (Fig. 7; r=−0.5854; P=0.0067); this suggests that miR-21 may participate in the regulation of the expression of STAT3. TNF-α is the target gene of miR-21 (17); therefore, the expression of miR-21 may be related to TNF-α. However, no correlation was observed in the results of the present study (r=2.138; P=0.43). STAT3 can improve the expression of TNF-α mRNA, and this may restrict the negative regulation of miR-21. The expression levels of TNF-α (Fig. 8) and SOCS3 (Fig. 9) mRNA were negatively correlated with miR-19a 2−ΔΔCq (r=0.6642; P=0.0014 and r=−0.7903; P=0.001, respectively).
Figure 3.
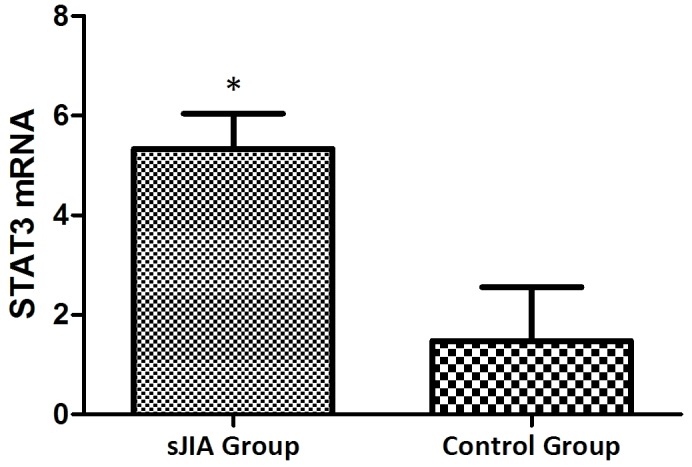
Expression levels of STAT3 in the SJIA and control group. *P<0.05 vs. the control group. STAT3, signal transducer and activator of transcription 3; SJIA, systemic juvenile idiopathic arthritis.
Figure 4.
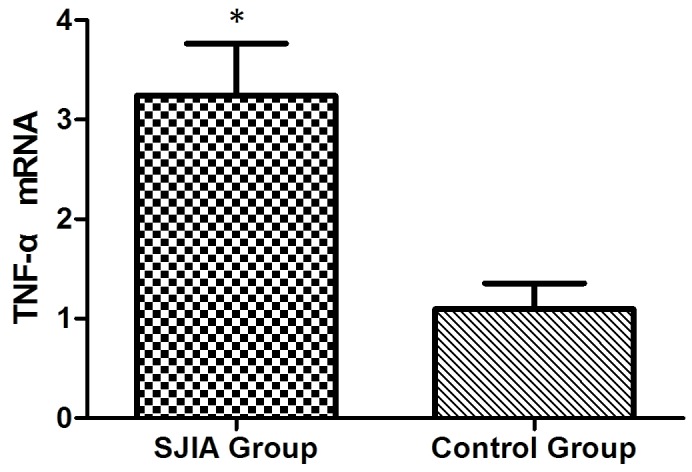
Expression levels of TNF-α in the SJIA and control group. *P<0.05 vs. the control group. TNF-α, tumor necrosis factor-α; SJIA, systemic juvenile idiopathic arthritis.
Figure 5.
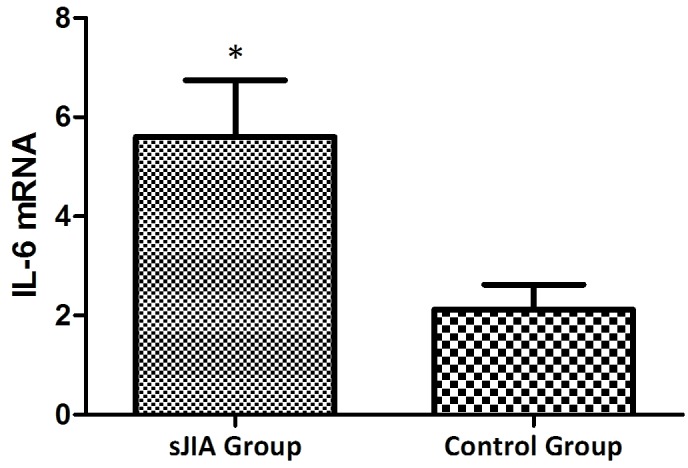
Expression levels of IL-6 in the SJIA and control group. *P<0.05 vs. the control group. IL-6, interleukin-6; SJIA, systemic juvenile idiopathic arthritis.
Figure 6.
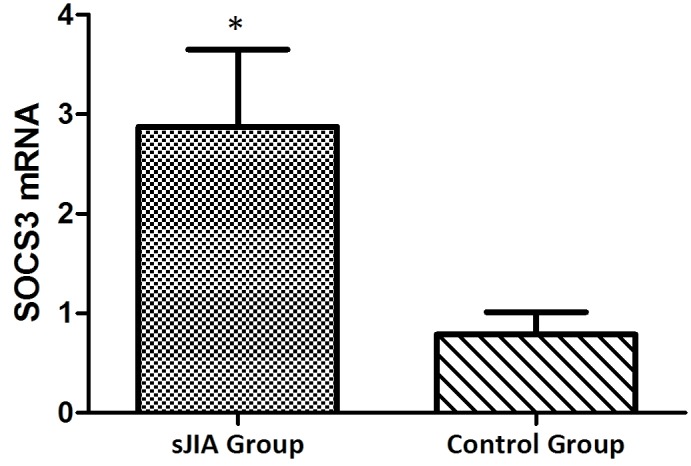
Expression levels of SOCS3 in the SJIA and control group. *P<0.05 vs. the control group. SOCS3, suppressor of cytokine signaling 3; SJIA, systemic juvenile idiopathic arthritis.
Figure 7.
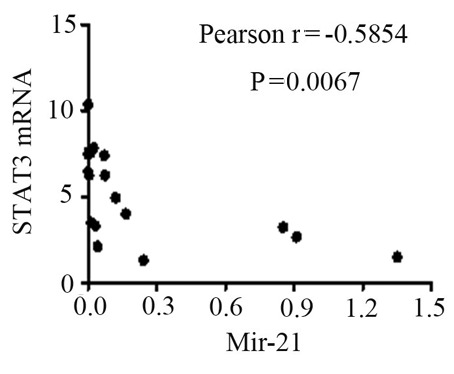
Correlation between STAT3 mRNA and the 2−ΔΔCq value of miR-21. STAT3, signal transducer and activator of transcription 3; miR-21, microRNA-21.
Figure 8.
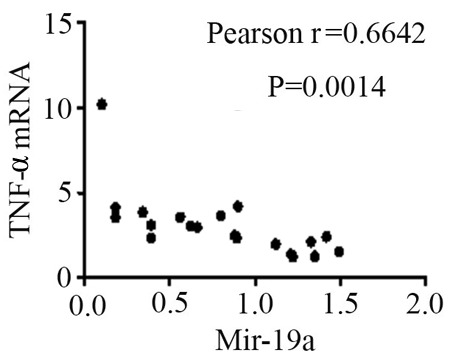
Correlation between TNF-α mRNA and the 2−ΔΔCq value of miR-21. TNF-α, tumor necrosis factor-α; miR-19a, microRNA-19a.
Figure 9.
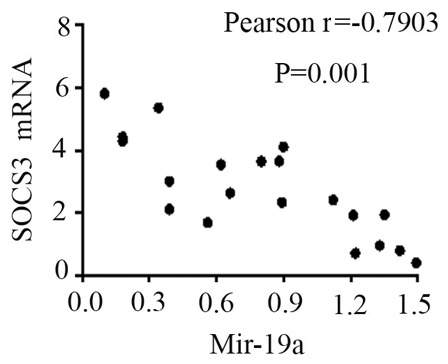
Correlation between SOCS3 mRNA and the 2−ΔΔCq value of miR-21. SOCS3, suppressor of cytokine signaling 3; miR-19a, microRNA-19a.
Discussion
Continuous or overexpression of the components of the JAK/STAT signaling pathway are the primary factors involved in the pathogenic mechanism underlying SJIA (18). IL-6 binds to the JAK receptor which results in the phosphorylation of STAT3, leading to the transcription of associated inflammatory genes, induced by downstream genes (18). Activated STAT3 is able to promote the differentiation of T-helper (Th)17 cells, induce high levels of receptor activator of nuclear factor-κB ligand protein expression in synovioblasts (19), and stimulate the production of inflammatory cytokines, including TNF-α, IL-1 and IL-6 (20). In addition, activated STAT3 is able to induce osteoclast formation and promote joint destruction (21).
The results from the present study suggest that STAT3 is expressed at high levels in patients with SJIA. miR-21 expression is upregulated in the majority of tumors, and may induce the transcription of STAT3 (22). Recently, numerous studies have suggested that miR21 participates in the development of inflammation, and is associated with the maturation and differentiation of T cells (23–26). miR-21 serves an important regulatory role in the interaction of Th1 and Th2 cells. A previous study demonstrated that the secretion of IL-12, IFN-α and IL-4 was decreased in murine Th cells lacking miR-21 following the induction of lipopolysaccharide, and may promote Th1 delayed hypersensitivity (23); the transcription repressor B cell lymphoma 6 (Bcl6) is an important regulator of Th cells, and could inhibit Th2-type inflammation (27). At present, it is thought that miR-21 is the target of Bcl16, and is able to downregulate Bcl16 expression and activate STAT3 (27). In addition, STAT3 is thought to upregulate miR-21 expression by binding to the specific miR-21 promoter site. It may therefore be hypothesized that miR-21 can promote the differentiation of Th2 cells (27).
Wang et al (27) demonstrated that in a transplanted tumor model, the knockdown of the miR-21 gene can inhibit the growth of a tumor, and inhibit the expression and phosphorylation of STAT3. In patients with RA, the expression levels of miR-21 gene are low, and those of STAT5/phosphorylated STAT5 proteins and forkhead box P3 mRNA were decreased following the expression and activation of STAT3, in which the negative feedback mechanism influenced the cell balance of Th17/Treg.
miR-19a may be involved in the development of SJIA by regulating the expression of SOCS3 and TNF-α. miR-19a is able to inhibit the expression of SOCS3, enhance the signal transduction of the JAK/STAT3 signaling pathway and promote the transcription of transcription factors (28). Collins et al (28) demonstrated using qPCR that miR-19a is able to regulate the JAK/STAT signaling pathway. The target genes of miR-19a, including the signaling factors of the JAK/STAT signaling pathway, were identified using bioinformatics to be SOCS1, SOCS3, SOCS5 and cullin 5 (28). A functional study demonstrated that miR-19a is able to downregulate the expression of SOCS3 mRNA and its proteins, whereas an antagonist of miR-19a is able to significantly reverse this inhibition (28). In addition, miR-19a decreases the expression levels of SOCS3, and enhances the transduction of IFN-α and IL-6/STAT3 (29). These results suggest that miR-19a positively regulates the JAK/STAT signaling pathway, and influences the incidence and development of inflammation.
Numerous studies demonstrated that miR-19a serves a negative regulatory role in inflammation. Philippe et al (30) reported that toll-like receptor 2 (TLR2) was highly expressed in the synovial cells of RA induced by LPS. In an miRNA chip, miR-19a/b was demonstrated to be downregulated in RA synovial cells, and miR-19a mimics were demonstrated to decrease the expression levels of TLR2, IL-6 and MMP3. Gantier et al (31) reported that miR-19a was able to inhibit A20/Tnfaip3, Rnf11, Fbxl11/Kdm2a, and Zbtb16, and promote the transduction of nuclear factor-κB, and that the downregulation of Rnf11 expression could decrease the expression levels of IL-6 and IL-8. In colitis tissue samples, miR-19a expression was downregulated, and the expression levels of TNF-α were increased (32). However, in the colitis mouse model, a luciferase vector demonstrated that TNF-α is the target gene of miR-19a (33).
The present study demonstrated that the relative expression levels of miR-19a in PBMCs of patients with SJIA were significantly lower compared with the control subjects. In addition, the mRNA of miR-19a target proteins, such as TNF-α and SOCS3, were highly expressed in patients with SJIA, and their expression was negatively correlated with miR-19a 2−ΔΔCq values (P<0.05). These results suggest that the low expression levels of miR-19a in patients with SJIA contribute towards the high expression levels of TNF-α, indirectly resulting in the increased expression levels of IL-6. Furthermore, SOCS3 was the negative feedback protein involved in the JAK/STAT signaling pathway, but high expression levels of SOCS3 did not downregulate the activation of STAT3; this may be associated with the functional disorder of SOCS3. These findings remain to be further studied.
An increasing number of miRNAs are being identified, and are thought to be closely associated with numerous diseases, resulting from their regulatory effect on signaling pathways (34). Investigation into the regulatory mechanism underlying the effect of miRNAs on the JAK/STAT signaling pathway may provide novel biomarkers for the diagnosis of SJIA. Further studies are required to examine the sites of action and effects of miR-21 and miR-19a on their target proteins, in order to identify their role in the immune mechanisms underlying SJIA.
Acknowledgements
The present study was funded by the Technology Project of Guangdong Province (grant no. 2014A020212010).
References
- 1.Beukelman T. Treatment advances in systemic juvenile idiopathic arthritis. F1000Prime Rep. 2014;6:21. doi: 10.12703/P6-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roderburg C, Luedde T. Circulating microRNAs as markers of liver inflammation, fibrosis and cancer. J Hepatol. 2014;61:1434–1437. doi: 10.1016/j.jhep.2014.07.017. [DOI] [PubMed] [Google Scholar]
- 3.Kemper AR, Van Mater HA, Coeytaux RR, Williams JW, Jr, Sanders GD. Systematic review of disease-modifying antirheumatic drugs for juvenile idiopathic arthritis. BMC Pediatr. 2012;12:29. doi: 10.1186/1471-2431-12-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tian TJ, Wang J, Zhou X. A review: microRNA detection methods. Org Biomol Chem. 2015;13:2226–2238. doi: 10.1039/C4OB02104E. [DOI] [PubMed] [Google Scholar]
- 5.Kurowska-Stolarska M, Alivernini S, Ballantine LE, Asquith DL, Millar NL, Gilchrist DS, Reilly J, Ierna M, Fraser AR, Stolarski B, et al. MicroRNA-155 as a proinflammatory regulator in clinical and experimental arthritis. Proc Natl Acad Sci USA. 2011;108:11193–11198. doi: 10.1073/pnas.1019536108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou Q, Haupt S, Kreuzer JT, Hammitzsch A, Proft F, Neumann C, Leipe J, Witt M, Schulze-Koops H, Skapenko A. Decreased expression of miR-146a and miR-155 contributes to an abnormal Treg phenotype in patients with rheumatoid arthritis. Ann Rheum Dis. 2015;74:1265–1274. doi: 10.1136/annrheumdis-2013-204377. [DOI] [PubMed] [Google Scholar]
- 7.Xie Q, Wang SC, Zhong J, Li J. MicroRNA-146a, a good biomarker and potential therapeutic target for rheumatoid arthritis. Genet Test Mol Biomarkers. 2013;17:91–92. doi: 10.1089/gtmb.2012.0333. [DOI] [PubMed] [Google Scholar]
- 8.Long L, Yu P, Liu Y, Wang S, Li R, Shi J, Zhang X, Li Y, Sun X, Zhou B, et al. Upregulated microRNA-155 expression in peripheral blood mononuclear cells and fibroblast-like synoviocytes in rheumatoid arthritis. Clin Dev Immunol. 2013;2013:296139. doi: 10.1155/2013/296139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen SY. MicroRNA-223: A double-edged sword in rheumatoid arthritis. Rheumatol Int. 2014;34:285–286. doi: 10.1007/s00296-013-2720-5. [DOI] [PubMed] [Google Scholar]
- 10.Dong L, Wang X, Tan J, Li H, Qian W, Chen J, Chen Q, Wang J, Xu W, Tao C, Wang S. Decreased expression of microRNA-21 correlates with the imbalance of Th17 and Treg cells in patients with rheumatoid arthritis. J Cell Mol Med. 2014;18:2213–2224. doi: 10.1111/jcmm.12353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shibuya H, Nakasa T, Adachi N, Nagata Y, Ishikawa M, Deie M, Suzuki O, Ochi M. Overexpression of microRNA-223 in rheumatoid arthritis synovium controls osteoclast differentiation. Mod Rheumatol. 2013;23:674–685. doi: 10.3109/s10165-012-0710-1. [DOI] [PubMed] [Google Scholar]
- 12.Feng ZT, Li J, Ren J, Lv Z. Expression of miR-146a and miR-16 in peripheral blood mononuclear cells of patients with rheumatoid arthritis and their correlation to the disease activity. Nan Fang Yi Ke Da Xue Xue Bao. 2011;31:320–323. (In Chinese) [PubMed] [Google Scholar]
- 13.Stanczyk J, Ospelt C, Karouzakis E, Filer A, Raza K, Kolling C, Gay R, Buckley CD, Tak PP, Gay S, Kyburz D. Altered expression of microRNA-203 in rheumatoid arthritis synovial fibroblasts and its role in fibroblast activation. Arthritis Rheum. 2011;63:373–381. doi: 10.1002/art.30115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu S, Pan W, Song X, Liu Y, Shao X, Tang Y, Liang D, He D, Wang H, Liu W, et al. The microRNA miR-23b suppresses IL-17-associated autoimmune inflammation by targeting TAB2, TAB3 and IKK-α. Nat Med. 2012;18:1077–1086. doi: 10.1038/nm.2815. [DOI] [PubMed] [Google Scholar]
- 15.ILAR 2001. Abstracts of the 20th Congress of the International League of Associations for Rheumatology. Edmonton, Alberta, Canada. August 26–30, 2001. J Rheumatol Suppll. 2001;63:1–120. [PubMed] [Google Scholar]
- 16.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 17.Zhao W, Dong Y, Wu C, Ma Y, Jin Y, Ji Y. MiR-21 overexpression improves osteoporosis by targeting RECK. Mol Cell Biochem. 2015;405:125–133. doi: 10.1007/s11010-015-2404-4. [DOI] [PubMed] [Google Scholar]
- 18.Sasi W, Sharma AK, Mokbel K. The role of suppressors of cytokine signalling in human neoplasms. Mol Biol Int. 2014;2014:630797. doi: 10.1155/2014/630797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ju JH, Heo YJ, Cho ML, Jhun JY, Park JS, Lee SY, Oh HJ, Moon SJ, Kwok SK, Park KS, et al. Modulation of STAT3 in rheumatoid synovial T cells suppresses Th17 differentiation and increases the proportion of Treg cells. Arthritis Rheum. 2012;64:3543–3552. doi: 10.1002/art.34601. [DOI] [PubMed] [Google Scholar]
- 20.Garbers C, Aparicio-Siegmund S, Rose-John S. The IL-6/gp130/STAT3 signaling axis: Recent advances towards specific inhibition. Curr Opin Immunol. 2015;34:75–82. doi: 10.1016/j.coi.2015.02.008. [DOI] [PubMed] [Google Scholar]
- 21.Kotake S, Udagawa N, Takahashi N, Matsuzaki K, Itoh K, Ishiyama S, Saito S, Inoue K, Kamatani N, Gillespie MT, et al. IL-17 in synovial fluids from patients with rheumatoid arthritis is a potent stimulator of osteoclastogenesis. J Clin Invest. 1999;103:1345–1352. doi: 10.1172/JCI5703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sawant DV, Wu H, Kaplan MH, Dent AL. The Bcl6 target gene microRNA-21 promotes Th2 differentiation by a T cell intrinsic pathway. Mol Immunol. 2013;54:435–442. doi: 10.1016/j.molimm.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu TX, Hartner J, Lim EJ, Fabry V, Mingler MK, Cole ET, Orkin SH, Aronow BJ, Rothenberg ME. MicroRNA-21 limits in vivo immune response-mediated activation of the IL-12/IFN-gamma pathway, Th1 polarization and the severity of delayed-type hypersensitivity. J Immunol. 2011;187:3362–3373. doi: 10.4049/jimmunol.1101235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iliopoulos D, Jaeger SA, Hirsch HA, Bulyk ML, Struhl K. STAT3 activation of miR-21 and miR-181b-1 via PTEN and CYLD are part of the epigenetic switch linking inflammation to cancer. Mol Cell. 2010;39:493–506. doi: 10.1016/j.molcel.2010.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park HK, Jo W, Choi HJ, Jang S, Ryu JE, Lee HJ, Lee H, Kim H, Yu ES, Son WC. Time-course changes in the expression levels of miR-122, −155, and −21 as markers of liver cell damage, inflammation, and regeneration in acetaminophen-induced liver injury in rats. J Vet Sci. 2015;4:64–71. doi: 10.4142/jvs.2016.17.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peacock O, Lee AC, Cameron F, Tarbox R, Vafadar-Isfahani N, Tufarelli C, Lund JN. Inflammation and MiR-21 pathways functionally interact to downregulate PDCD4 in colorectal cancer. PLoS One. 2014;9:e110267. doi: 10.1371/journal.pone.0110267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang YY, Sun G, Luo H, Wang XF, Lan FM, Yue X, Fu LS, Pu PY, Kang CS, Liu N, You YP. MiR-21 modulates hTERT through a STAT3-dependent manner on glioblastoma cell growth. CNS Neurosci Ther. 2012;18:722–728. doi: 10.1111/j.1755-5949.2012.00349.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Collins AS, McCoy CE, Lloyd AT, O'Farrelly C, Stevenson NJ. miR-19a: An effective regulator of SOCS3 and enhancer of JAK-STAT signalling. PLoS One. 2013;8:e69090. doi: 10.1371/journal.pone.0069090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qin S, Ai F, Ji WF, Rao W, Zhang HC, Yao WJ. miR-19a promotes cell growth and tumorigenesis through targeting SOCS1 in gastric cancer. Asian Pac J Cancer Prev. 2013;14:835–840. doi: 10.7314/APJCP.2013.14.2.835. [DOI] [PubMed] [Google Scholar]
- 30.Philippe L, Alsaleh G, Suffert G, Meyer A, Georgel P, Sibilia J, Wachsmann D, Pfeffer S. TLR2 expression is regulated by MicroRNA miR-19 in rheumatoid fibroblast-like synoviocytes. J Immunol. 2012;188:454–461. doi: 10.4049/jimmunol.1102348. [DOI] [PubMed] [Google Scholar]
- 31.Gantier MP, Stunden HJ, McCoy CE, Behlke MA, Wang D, Kaparakis-Liaskos M, Sarvestani ST, Yang YH, Xu D, Corr SC, et al. A miR-19 regulon that controls NF-kB signaling. Nucleic Acids Res. 2012;40:8048–8058. doi: 10.1093/nar/gks521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen B, She S, Li D, Liu Z, Yang X, Zeng Z, Liu F. Role of miR-19a targeting TNF-α in mediating ulcerative colitis. Scand J Gastroenterol. 2013;48:815–824. doi: 10.3109/00365521.2013.800991. [DOI] [PubMed] [Google Scholar]
- 33.Zhou P, Chen B, Hu P, Sun Y. Role of miR-19a in ulcerative colitis in mice. Nan Fang Yi Ke Da Xue Xue Bao. 2013;33:1325–1328. (In Chinese) [PubMed] [Google Scholar]
- 34.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Notebook J, O'Brian KC, et al. Circulating microRNAs as stableblood-based markers for cancer detection. Proc Natl Acad Sci USA. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]


