Abstract
GT factors are trihelix transcription factors that specifically regulate plant development and stress responses. Recently, several GT factors have been characterized in different plant species; however, little is known about the role of GT factors in wheat. Here, we show that TaGT2L1A, TaGT2L1B, and TaGT2L1D are highly homologous in hexaploid wheat, and are localized to wheat chromosomes 2A, 2B, and 2D, respectively. These TaGT2L1 genes encode proteins containing two SANT domains and one central helix. All three homologs were ubiquitously expressed during wheat development and were responsive to osmotic stress. Functional analyses demonstrated that TaGT2L1D acts as a transcriptional repressor; it was able to suppress the expression of AtSDD1 in Arabidopsis by binding directly to the GT3 box in its promoter that negatively regulates drought tolerance. TaGT2L1D overexpression markedly increased the number of stomata and reduced drought tolerance in gtl1-3 plants. Notably, ectopic expression of TaGT2L1D also affected floral organ development and overall plant growth. These results demonstrate that TaGT2L1 is an ortholog of AtGTL1, and that it plays an evolutionarily conserved role in drought resistance by fine tuning stomatal density in wheat. Our data also highlight the role of TaGT2L1 in plant growth and development.
Drought is a major environmental factor limiting crop productivity and quality. The identification and application of genetic resources associated with dehydration responses and drought resistance are essential for screening and breeding new crops with increased quality and stress tolerance1. In recent decades, the signaling networks involved in abiotic stress responses and tolerance have been thoroughly defined and characterized. Transcription factor families, including WRKY, NAC, AP2/ERF, bHLH and MYB, play significant roles in regulating the expression of functional genes2,3,4,5,6,7. Thus, many studies have investigated the application of transcription factors to plant genetic engineering. In Arabidopsis, the NAC transcription factor ATAF1 influences abiotic and biotic stress responses, probably through a signaling pathway involving reactive oxygen species8; it also affects DREB2A-regulated water stress-responsive gene expression to modulate plant drought tolerance9. In crops, genetic engineering using various transcription factors has yielded plants with an increased ability to adapt to various stresses. For example, the overexpression of TaPIMP1 (a MYB gene) significantly improved the drought tolerance capacity of transgenic wheat through the regulation of stress-responsive genes belonging to a hormone signaling pathway10. In rice, the drought-induced gene OsWRKY47 has been shown to enhance drought tolerance, probably through the regulation of CaBP and CRRSP11.
Notably, GT factors, which belong to the trihelix transcription factor family, have also been shown to be crucial for plant abiotic stress responses12. GT factors contain a conserved trihelix DNA-binding domain, called a SANT (Swi 3 [switching-defective protein 3], Ada2 [adaptor 2], N-Cor [nuclear receptor co-repressor], TFIIB [transcription factor IIIB]) domain13. This domain binds specifically to the GT element located in a target gene’s promoter14,15,16. A genome-wide analysis revealed 319 putative trihelix genes in various plant species, including 30 members in Arabidopsis12,17, and functional analyses have revealed that GT factors are involved in multiple biological processes, including seed scattering, embryo development, floral organ morphogenesis, and most importantly, biotic and abiotic stress resistance in various plants18,19,20,21,22,23,24,25,26,27.
Stomata play a central role in plant drought responses by modulating transpiration and the uptake of carbon dioxide28. The plant hormone abscisic acid (ABA) initiates signaling pathways that control the drought response of guard cells29,30. Under drought conditions, the ABA content increases and triggers the activation of OPEN STOMATA 1 (SnRK2.6), a core regulatory component of the ABA signaling pathway; it phosphorylates and activates SLOW ANION CHANNEL ASSOCIATED 1, resulting in stomatal closure and plant tolerance to drought31,32. In addition to stomatal movement, stomatal size and density dramatically affect transpiration and carbon dioxide uptake in plants. It has been demonstrated that stomatal development is precisely controlled by a gene regulatory network33. In Arabidopsis, SPEECHLESS (SPCH), MUTE, and FAMA promote stomatal differentiation and development from meristemoid mother cells to guard cells, while the TOO MANY MOUTHS-ERECTA (TMM-ER) complex acts as a negative regulator34,35,36,37. The TMM-ER complex is activated by STOMATAL DENSITY AND DISTRIBUTION1 (SDD1) encoding a subtilisin-like serine protease38,39, which causes the activation of a mitogen-activated protein kinase cascade and the phosphorylation of BREAKING OF ASYMMETRY IN THE STOMATAL LINEAGE and SPCH; this ultimately attenuates the basal signaling pathway underlying stomatal development40,41,42.
It is worth nothing that among the GT transcription factor family, GT-2 LIKE 1 (GTL1) has been identified as an important regulator of stomatal development in Arabidopsis27. Loss-of-function gtl1 mutations result in a reduced number of stomata and lower stomatal density; as a result, gtl1 mutant plants exhibit reduced stomatal conductance and leaf transpiration, leading to enhanced drought tolerance and water use efficiency27. Further analysis has shown that GTL1 binds directly to the GT3 box in the SDD1 promoter, repressing its transcription; this in turn affects a key regulator of stomatal development, the TMM-ER complex, and, ultimately, modulates stomatal density27. Recently, it was demonstrated that the AtGTL1 ortholog in poplar, PtaGTL1, has the same biological functions as AtGTL1; PtaGTL1 expression triggered by the AtGTL1 promoter completely restored all of the defects observed in gtl1 mutant (e.g., trichome size, stomatal development, water use efficiency, and drought tolerance). Importantly, PtaGTL1 binds not only to the AtSDD1 promoter to regulate AtSDD1 expression, it also binds to the promoter of the putative PtaSDD1 gene, which encodes a protein with high identity to AtSDD143. These results suggest that GTL1-mediated stomatal regulation and plant responses to water status represent a conserved mechanism in dicots.
Despite of our understanding of the role of GT factors in stomatal development in dicots, much less is known about the functions of GT factors in monocots. In rice, a total of 30 trihelix members have been identified12,17, and the downstream key regulators OsSPCH2, OsFAMA, and OsMUTE have been confirmed to have the same regulatory functions in stomatal development in monocots as they do in dicots44. These results suggest that GT factor-mediated mechanisms and downstream stomatal development might be conserved in monocots. This also led us to wonder whether GT factors function similarly to AtGTL1 in regulating stomatal development and drought tolerance in bread wheat. In this report, we demonstrate that TaGT2L1 is an ortholog of AtGTL1. TaGT2L1 contains two SANT domains and is a member of the typical GT-2 superfamily. Functional characterization of TaGT2L1D revealed similar functions to AtGTL1 in stomatal density and drought tolerance. TaGT2L1D was found to be responsive to osmotic stress and acted as a transcriptional repressor of AtSDD1 expression by binding to the GT3 box located in the AtSDD1 promoter. Notably, TaGT2L1 genes were ubiquitously expressed in multiple organs of wheat during plant development, with the highest level in floral organs during reproductive development; moreover, ectopic expression of TaGT2L1D resulted in abnormal plant growth and floral organ development. These results reveal the evolutionarily conserved role of GT-2 LIKE genes in planta, and they provide insight into the novel functions of trihelix transcription factors in bread wheat.
Results
TaGT2L1 proteins are GT-2 family members in bread wheat
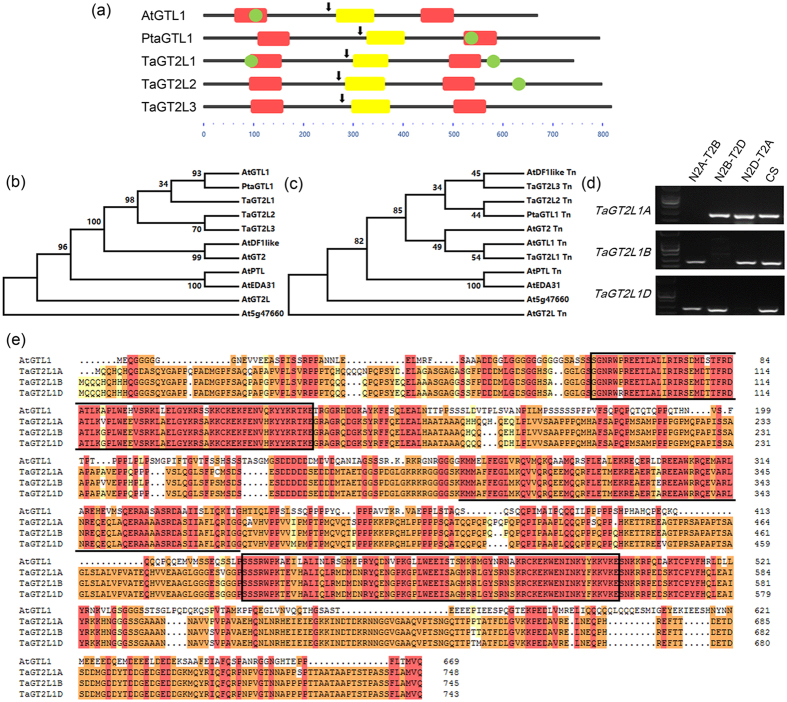
To identify potential GT-2 LIKE genes, we performed a TBLASTN search of the Triticum aestivum transcription factor database and Ensembl Plants database using the sequences of AtGTL1 and PtaGTL1 as bait. Three predicted proteins containing two typical SANT domains were identified; they were named TaGT2L1, TaGT2L2, and TaGT2L3, respectively. To investigate whether these predicted proteins are encoded by functional genes in wheat, we isolated the full-length coding sequences of TaGT2L1, TaGT2L2, and TaGT2L3 from hexaploid wheat and analysed their corresponding proteins. Similar to AtGTL1 and PtaGTL1, all three predicted proteins were found to contain an N-terminal SANT domain, a C-terminal SANT domain, a central helix, and a predicted nuclear localization signal; in addition, TaGT2L1 and TaGT2L2 were found to contain a predicted calmodulin (CaM)-binding site (Fig. 1a). It has been demonstrated that the SANT domain confers DNA-binding activity15, while the CaM-binding site enhances or reduces transcription factor activity45,46,47, and the nuclear localization signal directs proteins to the nucleus. Thus, it was deemed highly likely that TaGT2L1, TaGT2L2, and TaGT2L3 are nuclear proteins that function as transcription factors in wheat. A phylogenetic analysis of the full-length or N-terminus of the three homologs revealed that TaGT2L1 shares the highest identity with AtGTL1 (Fig. 1b,c). The results indicate that TaGT2L1 is a potential ortholog of AtGTL1. Thereafter, we focused our research on the biochemical and biological functions of TaGT2L1.
Figure 1. Identification of a bread wheat GT-2 family member, TaGT2L1.
(a) Schematic diagrams of AtGTL1, PtaGTL1, TaGT2L1, TaGT2L2, and TaGT2L3. Red boxes highlight the predicted SANT domains; yellow boxes represent the predicted central helices; and green ovals indicate the positions of the predicted CaM-binding sites. Arrows indicate the positions of the predicted nuclear localization sequences. (b,c) Phylogenetic analysis of the A. thaliana GT-2 superfamily transcription factors, PtaGTL1, and TaGT2Ls. Tn indicates the N-terminal SANT domain. (d) PCR-basis analysis for chromosomal localization of the TaGT2L1s. N indicates nullisomic; T indicates tetrasomic. The letters A, B, and D in the line name indicate the three genomes of hexaploid wheat. CS indicates wild type Chinese Spring as a positive control. (e) Alignment of the TaGT2L1s with AtGTL1. The SANT domains are framed in black boxes; the conserved central helix is underlined.
The identification of three copies of wheat TaGTL2L1
Hexaploid wheat contains six sets of chromosomes, and each gene is usually present in three copies. To further characterize the functions of TaGT2L1, we first identified three homologous sequences from hexaploid wheat (cv. Shimai15). Based on the nucleotide sequence of TaGT2L1, we performed a BLAST search in the Ensembl Plants database. Three homologous sequences were located in wheat chromosomes 2A, 2B and 2D, respectively. Thus, the corresponding genes were named TaGT2L1A, TaGT2L1B, and TaGT2L1D. Specific primers based on the sequence information were designed and PCR cloning was performed to get the three homologous mixture. As these three homologous sequences are too similar in sequence to be separated using different primers, the PCR products were cloned into T-vector and sequenced to separate individual homolog (see Supplementary Fig. S1).
Next, we attempted to determine their locations on chromosomes A, B, and D using Chinese Spring nulli-tetrasomic (NT) lines. As shown in Fig. 1d, no PCR product was detected when PCR was performed using 2A nullisomic wheat genome DNA and TaGT2L1A specific primer as template and primer. However, TaGT2L1B and TaGT2L1D PCR products were detected using same template and corresponding primers. These results indicated that TaGT2L1A is located in the wheat chromosome 2A. Similar results also indicated that TaGT2L1B and TaGT2L1D is located in the wheat chromosome 2B and 2D, respectively. TaGT2L1A, TaGT2L1B, and TaGT2L1D were found to be highly conserved and to share a high level of identity, ranging from 93.89 to 97.08% (Fig. 1e). Notably, the N-terminal SANT domains of these TaGT2L1s were found to be highly identical to each other, and to be 85.51–86.96% identical to that of AtGTL1, suggesting that they exhibit similar DNA-binding activity and functionality.
The TaGT2L1s were expressed in multiple organs and were responsive to osmotic stress
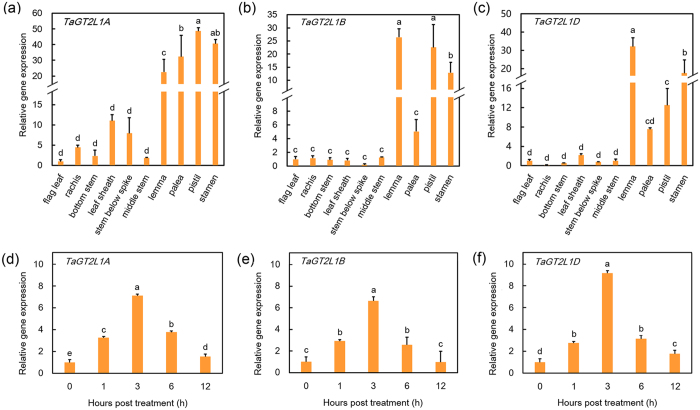
To investigate whether TaGT2L1A, TaGT2L1B, and TaGT2L1D are involved in wheat development and are responsive to osmotic stress, we analysed their expression patterns using real-time quantitative PCR (qPCR). The TaGT2L1s showed universal expression in multiple organs at the anthesis stage in wheat (cv. Shimai15), indicating a universal function in wheat development (Fig. 2a–c). Interestingly, the expression levels of the TaGT2L1s in both pistil and stamen tissues were much higher than those in leaves and stems, although the exact expression levels of the three TaGT2L1s were different. These results suggest that TaGT2L1s play a major role in floral organ development in wheat plants.
Figure 2. TaGT2L1 expression patterns.
Expression patterns of TaGT2L1A (a), TaGT2L1B (b), and TaGT2L1D (c) in different tissues were analysed at the anthesis stage. Changes in the expression patterns of TaGT2L1A (d), TaGT2L1B (e), and TaGT2L1D (f) in response to exposure to 20% (w/v) PEG6000 were analysed using one-week-old wheat seedling leaves. Total RNA was isolated and reverse-transcribed (n = 6). The qPCR was performed using TaACTIN gene as an internal reference. All values are the means ± standard deviations of three independent experiments. Different letters indicate significant differences (Student-Newman-Keuls test, P < 0.05).
Because AtGTL1 and PtaGTL1 mediate plant drought tolerance27,43, we examined whether TaGT2L1A, TaGT2L1B, and TaGT2L1D are responsive to osmotic stress. To this end, we performed qPCR and analysed the expression patterns of the TaGT2L1s in one-week-old wheat seedlings under osmotic stress. In the presence of 20% (w/v) PEG6000, all three homologs were sharply up-regulated and reached a peak within 3 h after treatment. Following the peak, all three homologs were down-regulated (Fig. 2d–f). The observation of similar expression patterns for TaGT2L1A, TaGT2L1B, and TaGT2L1D indicates that these homologs may play a similar role in the response of plants to osmotic stress.
It has been reported that AtGTL1 was suppressed by drought stress27. The qPCR analysis showed that three weeks drought treatment obviously suppressed TaGT2L1s expression in wheat seedlings, while expression level of TaGT2L1D showed greatest reduction (see Supplementary Fig. S2), indicating its greater impact on regulating plant drought response. Thus, TaGT2L1D gene was selected for further functional analysis.
TaGT2L1D is located in the nucleus
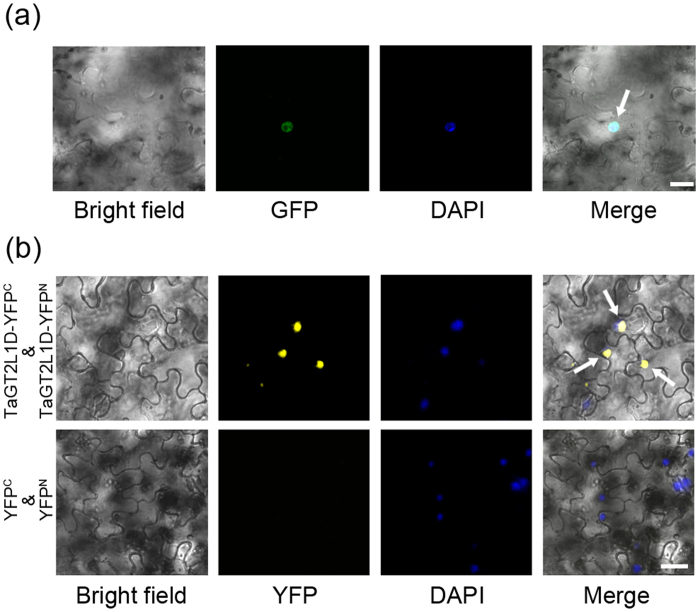
Because TaGT2L1D has a putative nuclear localization signal and DNA-binding domains, we predicted that it should be localized to the nucleus. To confirm this prediction, we generated a construct harboring the full-length coding sequence of TaGT2L1D fused with the green fluorescent protein (GFP) gene under the control of the cauliflower mosaic virus (CaMV) 35S promoter and determined the subcellular localization of TaGT2L1D by transiently expressing the GFP-TaGT2L1D fusion protein in Nicotiana benthamiana leaf cells. As shown in Fig. 3a, strong GFP fluorescence was detected exclusively in the nucleus of N. benthamiana leaf cells; the nuclear localization of the fusion protein was confirmed by 4′,6-diamidino-2-phenylindole (DAPI) staining, which was used as a nuclear indicator.
Figure 3. Subcellular localization of TaGT2L1D.
(a) The construct harboring GFP-TaGT2L1D was transiently transformed to N. benthamiana leaves. (b) The construct harboring TaGT2L1D-YFPC and TaGT2L1D-YFPN were transiently co-transformed to N. benthamiana leaves. Constructs harboring YFPC and YFPN were used as negative control. All transformed leaves were stained with DAPI and then photographed by confocal microscopy. Bright field images show the complete epidermal cell profile in bright-field view, GFP/YFP or DAPI images show fluorescent signals in dark view, and Merge images show the overlay of three signals including bright field, GFP/YFP fluorescence and DAPI fluorescence. White arrows indicate the GFP or YFP fluorescence merged to the DAPI fluorescence. Scale bar = 50 μm.
It has been shown that the central helix facilitates the formation of a coiled coil48, which is associated with protein dimerization12. The structural similarities between TaGT2L1D, AtGTL1, and PtaGTL1 prompted us to test whether dimerization also occurs between TaGT2L1D proteins. To do so, we examined whether TaGT2L1D can interact with itself in plant cells via a bimolecular fluorescence complementation (BiFC) assay. Yellow fluorescent protein (YFP) signal should be detected in corresponding location when protein-protein interaction does exist. As expected, obvious YFP signal in the nucleus of N. benthamiana leaf cells was observed when TaGT2L1D-YFPN with TaGT2L1D-YFPC were co-expressed (Fig. 3b), which indicate that TaGT2L1D is located in the nucleus probably in a homodimer manner. These results demonstrate that TaGT2L1D may function as a homodimer in the nucleus.
TaGT2L1D overexpression restores the trichome phenotype of gtl1-3
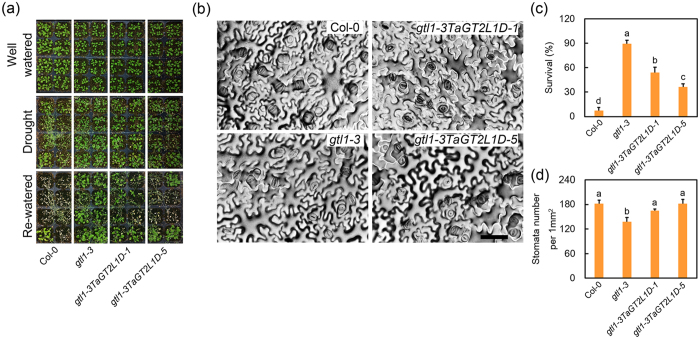
It has been shown that AtGTL1 loss-of-function mutant gtl1-3 reveals an increase in trichome size19. To analyse whether TaGT2L1D is a functional AtGTL1 ortholog in wheat, we expressed a GFP-TaGT2L1D fusion protein under the control of the CaMV 35S promoter in gtl1-3 mutant plants using the Agrobacterium tumefaciens-mediated floral-dip method. Five independent kanamycin-resistant T1 lines displayed a similar trichome size and whole plant size with wild-type Columbia-0 (Col-0); among the progeny of these plants, two T3 homozygous lines gtl1-3TaGT2L1D-1 and gtl1-3TaGT2L1D-5 were selected for further analysis. As shown in Fig. 4a, four-week-old gtl1-3TaGT2L1D-1 and gtl1-3TaGT2L1D-5 plants showed a similar phenotype in terms of rosette size to Col-0 and gtl1-3 plants. The trichome size in gtl1-3TaGT2L1D-1 and gtl1-3TaGT2L1D-5 was completely restored to normal (Fig. 4b). RT-PCR analysis showed that TaGT2L1D transcripts were detected in the leaves of the transgenic lines, but not in Col-0 or gtl1-3 plants (see Supplementary Fig. S3a).
Figure 4. TaGT2L1D expression suppressed the leaf trichome phenotype of gtl1-3.
(a) Photographs of rosette leaves in four-week-old Col-0, gtl1-3, gtl1-3TaGT2L1D-1, and gtl1-3TaGT2L1D-5 plants were taken using a digital camera. Scale bar = 2 cm. (b) Trichomes on the adaxial surface of fully expanded rosette leaves from four-week-old plants. Photographs were taken using a dissecting microscope. Scale bar = 0.5 cm.
Since AtGTL1 can suppress the expression of CELL CYCLE SWITCH 52 A1 (CCS52A1), leading to the termination of cell growth49, we questioned whether TaGT2L1D regulates trichome size in Arabidopsis through CCS52A1. To answer this question, we performed qPCR and examined the expression of CCS52A1 in gtl1-3TaGT2L1D-1 and gtl1-3TaGT2L1D-5 lines. As expected, CCS52A1 expression was significantly decreased in the transgenic lines compared with the high level of CCS52A1 in the gtl1-3 mutant (see Supplementary Fig. S3b), suggesting that TaGT2L1D functions similarly to AtGTL1 in modulating trichome development.
TaGT2L1D negatively regulates plant drought tolerance
As mentioned, AtGTL1 and PtaGTL1 are involved in plant drought tolerance27,43. The fact that TaGT2L1D restored the trichome phenotype of gtl1-3 suggested a similar role for TaGT2L1D in plant drought tolerance. To test this possibility, we performed a phenotypic analysis of gtl1-3TaGT2L1D-1 and gtl1-3TaGT2L1D-5 to determine whether TaGT2L1D expression could complement the drought-responsive phenotype of gtl1-3. The Col-0, gtl1-3, gtl1-3TaGT2L1D-1, and gtl1-3TaGT2L1D-5 plants were watered thoroughly and uniformly for two weeks and then grown without watering for the next two weeks. When the leaves of almost all of the plants were no longer turgid, the plants were re-watered and the survival rate was measured after four days of re-watering (Fig. 5a). The survival rate of Col-0 was 12.5%, while that of gtl1-3 was 90.6%, indicating that the loss of function of GTL1 produced a better drought resistance phenotype in accordance with a previous report27. By contrast, the gtl1-3TaGT2L1D-1 and gtl1-3TaGT2L1D-5 lines exhibited substantially reduced survival rates (50.0 and 34.4%, respectively; Fig. 5c). TaGT2L1D expression partially restored the drought resistance of the gtl1-3 mutant, suggesting a role for TaGT2L1D in plant drought responses.
Figure 5. TaGT2L1D is involved in plant drought tolerance.
(a) The plant water-deficit tolerance of two-week-old gtl1-3TaGT2L1D-1, and gtl1-3TaGT2L1D-5 plants (16 h/8 h photoperiod) was evaluated. When the leaves of almost all of the plants were no longer turgid, the plants were re-watered. (b) An abaxial epidermal peel in the middle of the leaves from Col-0, gtl1-3, gtl1-3TaGT2L1D-1, and gtl1-3TaGT2L1D-5 plants was printed and photographed to show the stomatal density. Scale bar = 50 μm. (c) The survival rate was measured after four days of re-watering. (d) The number of stomata per mm2 in the abaxial epidermis of fully expanded rosette leaves (four-week-old plants, 16 h/8 h photoperiod). All values are the means ± standard deviations of three independent experiments. Different letters indicate significant differences (Student-Newman-Keuls test, P < 0.05).
Since GTL1 regulates plant drought tolerance by modulating stomatal density27, the stomatal density in the rosette leaves of four-week-old Col-0, gtl1-3, gtl1-3TaGT2L1D-1, and gtl1-3TaGT2L1D-5 plants were measured. The abaxial stomatal density in gtl1-3 (138 ± 10 per mm2) was decreased by 24% compared with that in Col-0 (181 ± 9 per mm2), while the gtl1-3TaGT2L1D-1 and gtl1-3TaGT2L1D-5 lines showed significantly increased abaxial stomatal densities (165 ± 4 and 181 ± 10 per mm2, respectively) compared with gtl1-3; moreover, their stomatal densities were identical to that in wild-type (Fig. 5b,d).
It has been reported that GTL1 does not regulate stomatal aperture and ABA-induced stomatal closure27. Similarly, the stomatal aperture in the gtl1-3TaGT2L1D-1 and gtl1-3TaGT2L1D-5 lines under water-sufficient conditions was comparable to that in Col-0 and gtl1-3 (see Supplementary Fig. S4a,b). Moreover, ABA treatment caused equivalent stomatal closure (see Supplementary Fig. S4a). These results indicate that TaGT2L1D is not involved in stomatal aperture or the response of plants to ABA, consistent with the role of AtGTL127. Together, these data indicate that TaGT2L1D modulates plant drought tolerance by influencing stomatal density.
TaGT2L1D trans-repressed SDD1 expression in Arabidopsis
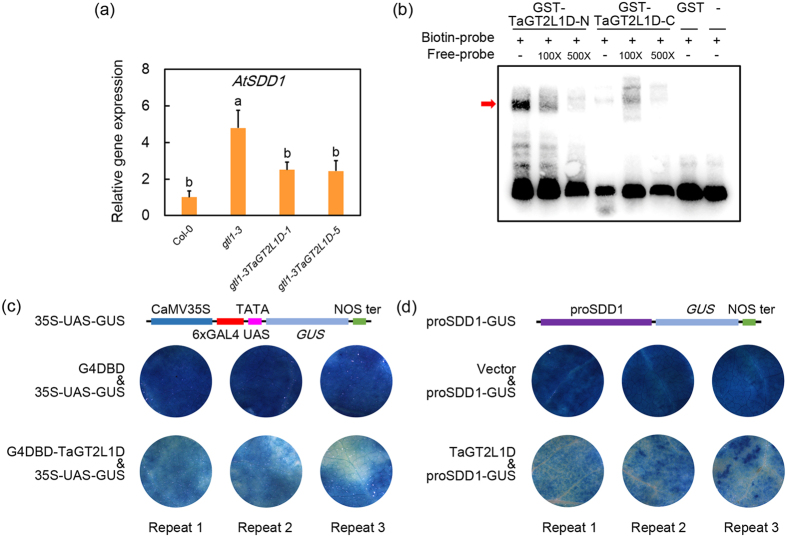
Since AtGTL1 and PtaGTL1 regulate stomatal development by trans-repressing the expression of AtSDD127,43, we performed qPCR to analyse the effect of TaGT2L1D expression on AtSDD1 expression in fully expanded leaves of gtl1-3TaGT2L1D-1 and gtl1-3TaGT2L1D-5 plants. Our results show that the up-regulation of AtSDD1 in gtl1-3 plants was markedly repressed by TaGT2L1D expression (Fig. 6a), indicating the negative regulatory effect of TaGT2L1D on AtSDD1. To verify that TaGT2L1D acts as a repressor, we performed a GAL4/UAS-based system assay for transcriptional activity50. TaGT2L1D fused with the GAL4 DNA-binding domain (G4DBD) was generated and co-transformed with a 35S-UAS-GUS reporter into N. benthamiana leaves. GUS (β glucuronidase) expression was repressed when TaGT2L1D was fused with the G4DBD (Fig. 6c), indicating that TaGT2L1D is a transcriptional repressor.
Figure 6. TaGT2L1D trans-represses SDD1 expression in Arabidopsis.
(a) AtSDD1 expression in fully expanded rosette leaves from six-week-old Col-0, gtl1-3, gtl1-3TaGT2L1D-1, and gtl1-3TaGT2L1D-5 plants (16 h/8 h photoperiod) was examined. Total RNA was isolated and reverse-transcribed (n = 6). The qPCR was performed using AtACTIN gene as an internal reference. All values are the means ± standard deviations of three independent experiments. Different letters indicate significant differences (Student-Newman-Keuls test, P < 0.05). (b) EMSAs were conducted using biotin-labeled DNA probes corresponding to a fragment of the AtSDD1 promoter harboring the GT3 box (5′-GGTAAA-3′). The arrow indicates the interaction between the TaGT2L1D N-terminal fragment and the GT3 box probe. GST protein was used as a negative control. Three biological replications were performed. (c) The trans-repression activity of TaGT2L1D was tested in N. benthamiana leaves using a GAL4/UAS-based system. The G4DBD or G4DBD-TaGT2L1D was transiently co-expressed with the 35S-UAS-GUS reporter in N. benthamiana leaves, and the GUS staining was imaged. Three biological replications were performed. (d) TaGT2L1D represses the activity of the SDD1 promoter in N. benthamiana leaves. The proSDD1-GUS reporter and GFP-TaGT2L1D were transiently co-expressed in N. benthamiana leaves and the GUS staining was imaged. GFP (Vector) was used as a negative control. Three biological replications were performed.
To determine whether TaGT2L1D trans-repressed AtSDD1 expression directly, we generated a reporter construct harboring proSDD1-GUS and co-transformed it with 35S-GFP-TaGT2L1D into N. benthamiana leaves. The GUS gene was well expressed when the proSDD1-GUS reporter was co-transformed with 35S-GFP; by sharp contrast, GUS expression was repressed when 35S-GFP-TaGT2L1D was co-transformed with proSDD1-GUS (Fig. 6d). AtGTL1 and PtaGTL1 repress AtSDD1 expression by binding directly to the GT3 box (5′-GGTAAA-3′) located in the promoter of AtSDD127,43. The fact that the N-terminal SANT domain of TaGT2L1D showed high identity with that of AtGTL1 suggested that TaGT2L1D regulates AtSDD1 expression in the same way. To verify this, we purified the N and C-terminal SANT domain fragments of TaGT2L1D fused with a GST (glutathione S-transferase) tag (GST-TaGT2L1D-N and GST-TaGT2L1D-C, see Supplementary Fig. S5) to perform an electrophoretic mobility shift assay (EMSA) with a 28-nt AtSDD1 promoter fragment that included the GT3 box. Mobility shift should be detected when protein-DNA interaction does exist. Our results show that TaGT2L1D-N, but not TaGT2L1D-C, caused a mobility shift in the biotin-labeled probe, and that unlabeled probe could eliminate the shift (Fig. 6b). These data indicates that TaGT2L1D modulates stomatal density and drought tolerance through the transcriptional repression of AtSDD1 in Arabidopsis.
TaGT2L1D is involved in plant development
In addition to its conserved roles in trichome and stomatal development, we observed that ectopic TaGT2L1D expression in gtl1-3 mutant plants caused developmental defects in the floral organs of the transgenic lines. The filaments of gtl1-3TaGT2L1D-1 and gtl1-3TaGT2L1D-5 lines were obviously shortened (Fig. 7a), leading to reduced fertility compared with Col-0 and gtl1-3 plants. When TaGT2L1D was overexpressed in Col-0 under the control of the CaMV 35S promoter (see Supplementary Fig. S6), floral organ development in two overexpression lines, TaGT2L1DOE-7 and TaGT2L1DOE-23, was visibly suppressed (Fig. 7a). This observation suggests that TaGT2L1D plays a role in floral organ development, which is favored by the high expression level of TaGT2L1 in wheat floral organs.
Figure 7. TaGT2L1D is involved in plant development.
(a) Mature flowers from Col-0, gtl1-3, gtl1-3TaGT2L1D-1, gtl1-3TaGT2L1D-5, TaGT2L1DOE-7, and TaGT2L1DOE-23 plants. Scale bar = 2 mm. (b) The size of rosette in four-week-old Col-0, TaGT2L1DOE-7, and TaGT2L1DOE-23 plants (16 h/8 h photoperiod). Scale bar = 2 cm. (c) All rosette leaves were taken from four-week-old Col-0, TaGT2L1DOE-7, and TaGT2L1DOE-23 plants (16 h/8 h photoperiod). Scale bar = 2 cm.
Notably, lines TaGT2L1DOE-7 and TaGT2L1DOE-23 exhibited small rosettes compared with Col-0 (Fig. 7b) in accordance with the phenotype caused by the ectopic expression of AtGTL149. Specifically, a significant decrease in rosette leaf size and slightly early senescence were observed (Fig. 7b,c). Moreover, lines TaGT2L1DOE-7 and TaGT2L1DOE-23 exhibited slightly early bolting (Fig. 7b). These results demonstrate that TaGT2L1D is involved in whole-plant development.
Discussion
In recent years, the functions of GT factors in abiotic stress responses have been identified in various plant species. However, no functional analysis has been published regarding GT factors in bread wheat. Here, we characterized a drought-responsive gene, TaGT2L1, encoding a GT transcription factor in bread wheat. Our results demonstrate that TaGT2L1 is an ortholog of AtGTL1 and that it plays an evolutionarily conserved roles in stomatal development and drought tolerance. Our data also indicate roles for TaGT2L1 in plant growth and development, implying that this GT factor performs novel functions in bread wheat.
AtGTL1 is a transcription factor that belongs to the GT-2 superfamily of trihelix family proteins. It is a nuclear protein with DNA-binding activity that contains two SANT domains and one central helix12,17. AtGTL1 regulates stomatal development in Arabidopsis by repressing the expression of AtSDD1 through binding to its promoter27. In this study, we identified orthologs of AtGTL1 (TaGT2L1s) in bread wheat; several pieces of evidence support this conclusion. First, the TaGT2L1s are structurally similar to AtGTL1, and they are evolutionarily close to AtGTL1. As a diploid plant, there is a single copy of the GTL1 gene in Arabidopsis; in comparison, bread wheat is hexaploid and carries three AtGTL1 homologs, TaGT2L1A, TaGT2L1B, and TaGT2L1D on chromosomes 2A, 2B, and 2D, respectively (Fig. 1d). Importantly, TaGT2L1 proteins contain two SANT domains and one central helix; further, the N-terminal SANT domain is highly conserved compared with that of AtGTL1 (Fig. 1e).
Second, we used TaGT2L1D to confirm that TaGT2L1 proteins are indeed localized to the nucleus (Fig. 3a). Although there is no classical nuclear localization signal in TaGT2L1D, its sequence does contain a predicted nuclear localization signal, 284KRKRGGGGSK293, which may mediate the nuclear localization of the protein. The predicted nuclear localization signal of TaGT2L1D is close to the central helix, which is believed to be associated with protein dimerization12. Notably, TaGT2L1D is able to interact with itself, which suggest a dimerization. It is likely that TaGT2L1D dimerization through the predicted nuclear localization signal in each monomer ensures the nuclear localization of the protein. Further analysis is needed to confirm the predicted dimerization of TaGT2L1s and to elucidate the role of this dimerization in the functions of these proteins.
Third, we found that TaGT2L1D has DNA binding activity and trans-repression activity, similar to AtGTL1 (Fig. 6b,c). Interestingly, a potential ethylene-responsive element binding factor-associated amphiphilic repression (EAR) motif, 460GLSLAL465, was identified in TaGT2L1D. It has been suggested that EAR motif-containing trans-repressors suppress the expression of their target genes through histone deacetylation at regulatory regions by physically interacting with co-repressors51,52,53. It is conceivable that TaGT2L1s repress the transcription of their target genes through a similar mechanism. The identification of these co-repressors and an analysis of chromatin modification (especially the histone acetylation status of candidate target genes) will reveal the molecular mechanism underlying the repression of gene expression by TaGT2L1s. Unexpectedly, there is no typical EAR motif in AtGTL1, indicating that AtGTL1 may utilize a different regulatory mechanism to repress the transcription of its target genes. To our knowledge, this is the third report to show that a GT factor can function as a trans-repressor (after GTL1 and ASR3). GTL1 is a trans-repressor that can suppress the expression of SDD1 and CCS52A127,49, while ASR3 is a trans-repressor that can suppress the gene expression via its ERF-associated amphiphilic repression binding activity22. The demonstrated functional similarity in transcriptional regulation supports the notion that TaGT2L1s are orthologs of AtGTL1.
The most important piece of evidence supporting the idea that TaGT2L1s are AtGTL1 orthologs is that TaGT2L1 proteins have similar biological functions to AtGTL1. The previous study has shown that AtGTL1 was a drought-responsive gene and was suppressed by dehydration stress27. In this study, we found that TaGT2L1s were also down-regulated in leaves when wheat seedlings were exposed to drought stress (See Supplementary Fig. S2). The similar expression patterns of TaGT2L1s to AtGTL1 in response to water stress indicate that these genes might play similar roles in drought tolerance of wheat. Further functional analysis results favor our hypothesis. Overexpression of TaGT2L1D complemented both the trichome size and stomatal density phenotypes of gtl1-3 (Figs 4b and 5b). As a result, the stress tolerance of the gtl1-3 mutant plants was largely restored by ectopic TaGT2L1D expression, confirming that TaGT2L1D modulates plant responses to water stress through the same regulatory mechanism as AtGTL1.
In addition, GTL1 binds to the GT3 box in the SDD1 promoter and represses its expression, thereby inactivating the downstream TMM-ER complex and mitogen-activated protein kinase pathway to promote stomatal development54. In the present study, we found that TaGT2L1D was able to bind directly to the GT3 box in the AtSDD1 promoter and suppress AtSDD1 promoter activity (Fig. 6b,d). Like AtGTL1, TaGT2L1D appears to regulate plant drought resistance by modulating stomatal density. However, TaGT2L1D overexpression did not completely restore the drought tolerance of gtl1-3 mutant plants (Fig. 5a). This may be explained by the functional discrepancy suggested by the structural differences between AtGTL1 and TaGT2L1D, including the EAR motif and amino acid sequences outside of the conserved SANT domains. These results imply that TaGT2L1 proteins have both a different regulatory mode, and novel biological roles in bread wheat.
Our results also confirm that TaGT2L1 mediates plant growth and development in Arabidopsis. In addition to complementing of the trichome and stomata phenotypes of gtl1-3, TaGT2L1D overexpression suppressed filament elongation in the mutant (Fig. 7a), resulting in short siliques and reduced fertility in our transgenic lines. Taking into account the finding that the three TaGT2L1 genes were expressed at high levels in wheat stamens during reproductive growth (Fig. 2a–c), it is possible that TaGT2L1s play crucial roles in filament elongation and reproduction in wheat. Identification of the targets of TaGT2L1s that modulate filament development will enhance our understanding of TaGT2L1s-mediated filament development and reproductive development in wheat. AtGTL1 expression driven by the ATML1 promoter was previously shown to cause a strong reduction in plant size49.It is supposed that GTL1 may control earlier progression of endocycles in non-trichome cells potentially by modulating the CCS52A1 expression49. In this study, TaGT2L1D overexpression also suppressed the expression of CCS52A1 and development of the whole plant in wild-type Arabidopsis, including floral organs (Fig. 7), indicating the function of TaGT2L1D in terminating cell growth. The universal expression of TaGT2L1 in wheat supports its multiple biological functions in various developmental processes through trans-repression.
In summary, in this study we identified GT factors and orthologs of AtGTL1 in bread wheat. In addition to their roles in stomatal density regulation and plant drought tolerance, these TaGT2L1s modulate plant growth and development. Our findings provide novel insight into the functions of GT factors in drought tolerance and development in bread wheat. Further study of these TaGT2L1s and their underlying regulatory network will help us elucidate the genetic basis for development and drought tolerance in wheat, and it will promote the development of crop plants with increased stress tolerance.
Materials and Methods
Cloning and sequence analyses
Sequence information of TaGT2L1s, TaGT2L2, and TaGT2L3 were obtained via a TBLASTN search of the plant transcription factors database (http://planttfdb.cbi.pku.edu.cn/) and Ensembl Plants database (http://plants.ensembl.org/index.html) using the sequences of AtGTL1 and PtaGTL1 as bait. Gene-specific primers were designed based on sequences from the plant transcription factors database and Ensembl Plants database and all primer sequences are listed in the Supplementary Table S1.
Total RNA was extracted from leaves of bread wheat (cv. Shimai15) using Trizol reagent (Invitrogen, Corp., Carlsbad, CA, USA) and treated with DNase I (Promega, Madison, WI, USA). A total of 5 μg total RNA was used for reverse transcription with a FastQuant RT Kit (Tiangen Biotech [Beijing] Co. Ltd., Beijing, China) according to the manufacturer’s protocol. The cDNA was used as template for cloning full-length coding sequences of TaGT2L1s, TaGT2L2, and TaGT2L3. PCR was performed using KOD FX DNA Polymerase (Toyobo, Tokyo, Japan) according to the manufacturer’s protocol. Positive PCR products were cloned into pMD19-T vector and independent clones were sequenced to identify individual sequence.
The amino acid sequences of TaGT2L1s, TaGT2L2, and TaGT2L3 were determined using the ExPASy Translate tool (http://web.expasy.org/translate/). The SANT domains were predicted through an NCBI conserved domain search (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi). The central helices were predicted using Jpred 4 (http://www.compbio.dundee.ac.uk/jpred4/index_up.html). The CaM-binding sites were predicted using a CaM-binding site search (http://calcium.uhnres.utoronto.ca/ctdb/ctdb/sequence.html). The position of the potential nuclear localization signal was predicted using NLStradamus (http://www.moseslab.csb.utoronto.ca/NLStradamus/). Sequence alignments were performed using Clustal Omega 1.2.1 (http://www.ebi.ac.uk/Tools/msa/clustalo/). Full-length amino acid sequences and N-terminal SANT domain sequences (according to Kaplan’s definition12, from four upstream of the first conserved tryptophan, to 12 downstream of the third conserved phenylalanine) were used to construct a phylogenetic tree by the neighbor-joining method with MEGA 6.05 and 1,000 bootstrap replicates.
Chromosomal locations of the three TaGT2L1 homologs
Three Chinese Spring NT lines, N2A-T2B (2A chromosome nullisomic line), N2B-T2D (2B chromosome nullisomic line), and N2D-T2A (2D chromosome nullisomic line) were used for the PCR-basis analysis of TaGT2L1 chromosomal locations. Gene-specific primers for the three TaGT2L1 homologs were designed based on the differences in their 3′-untranslated regions. PCR was performed to determine the chromosomal locations of the genes using wheat NT line genomic DNA as the template. The PCR conditions were as follows: 95 °C for 5 min; 35 cycles of 95 °C for 30 s, 61 °C for 30 s, and 72 °C for 30 s; and 72 °C for 10 min. The PCR products were separated by 2% agarose gel electrophoresis. All primer sequences are listed in the Supplementary Table S1.
Gene expression analysis
Total RNA extraction and reverse transcription were performed as described previously. For tissue expression pattern analysis of TaGT2L1s, wheat plants (cv. Shimai15) were cultivated in the field under natural conditions with well irrigation. A variety of tissues, including leaf, stem, lemma, palea, pistil, stamen, and rachis, were sampled at the anthesis stage for the qPCR analysis. For examining TaGT2L1s expression pattern response to PEG treatment, wheat seedlings (cv. Shimai15) were grown in a controlled chamber with a 16 h/8 h photoperiod at 22 °C. One week after germination, the plants were exposed to 20% (w/v) PEG6000 solution for 1, 3, 6, and 12 h, after which their leaves were sampled for the qPCR analysis. For examining TaGT2L1s expression pattern in response to drought treatment, one-week-old wheat seedlings (cv. Shimai15) were grown in pots (10 cm in length, 10 cm in width, and 10 cm in height) in a controlled chamber with a 16 h/8 h photoperiod at 22 °C with or without watering for three weeks. Leaf samples were harvest at the same time for the qPCR analysis. For the gene expression analysis in Arabidopsis, plants were grown in pots (8 cm in length, 8 cm in width, and 8 cm in height) in a controlled chamber environment under a 16 h/8 h photoperiod at 22 °C and leaf samples were harvest for qPCR analysis.
The qPCR was performed in a Bio-Rad CFX Connect™ Real-Time PCR Detection System (Hercules, CA, USA) using a SYBR Green SuperReal PreMix Plus (Tiangen Biotech [Beijing] Co. Ltd.). The PCR parameters were: 95 °C for 15 min, followed by 40 cycles of 95 °C for 10 s and 61 °C for 30 s. Three biological replicates were used with three technical replicates for each biological replicate. Wheat ACTIN gene (GenBank accession number KC775780) and Arabidopsis ACTIN gene (GenBank accession number NM_179953) were used as internal references. Relative gene expression levels were detected using the 2−ΔΔCT method55. All primer sequences are listed in the Supplementary Table S1.
Subcellular localization and protein-protein interaction analysis
To verify the subcellular localization of TaGT2L1D, full-length coding sequence of TaGT2L1D were amplified by PCR and then cloned into pTF101-GFP vector with XmaI to create a 35S-GFP-TaGT2L1D fusion. The plasmid was introduced into A. tumefaciens (strain EHA105). The infiltration of N. benthamiana was performed as described previously56. Briefly, transformed A. tumefaciens cells were grown at 28 °C in Luria-Bertani liquid medium and then incubated at room temperature for 3 h, followed by the infiltration into the abaxial air spaces of N. benthamiana leaves. Epidermal cell layers were stained with DAPI and assayed for fluorescence 3 d after infiltration. GFP and DAPI fluorescence were observed with a confocal microscope (Leica TCS-SP8; Leica Microsystems, Wetzlar, Germany).
To verify the potential interaction between TaGT2L1D itself in plant cells, a widely used BiFC assay was performed as described previously57. The full-length coding sequence of TaGT2L1D was cloned into pDONR207 using BP Clonase Enzyme (Invitrogen Corp.) and then recombined in the Gateway-compatible destination vectors pEarleyGate201-YN (N-terminal YFP) and pEarleyGate202-YC (C-terminal YFP) using LR Clonase Enzyme (Invitrogen Corp.) according to the manufacturer’s protocol. The resulting plasmids were transiently co-transformed into N. benthamiana leaves using A. tumefaciens (strain EHA105) as described previously. The reconstituted YFP fluorescence was observed with a confocal microscope (Leica TCS-SP8; Leica Microsystems). Empty vectors were used as a negative control.
These experiments were performed with three biological replications. All primer sequences are listed in the Supplementary Table S1.
Generation of TaGT2L1D transgenic Arabidopsis plants
Arabidopsis thaliana ecotype Col-0 and GTL1 loss-of-function mutant gtl1-3 (SALK_005966) plants were used to generate TaGT2L1D transgenic plants. The plants were germinated on Murashige and Skoog (MS; pH 5.7) medium containing 1% (w/v) sucrose. One-week-old seedlings were transferred to pots and grown in a controlled chamber environment under a 16 h/8 h photoperiod at 22 °C. The full-length coding sequence of TaGT2L1D was amplified by PCR and cloned into the EcoRI/XbaI sites in pEZR(K)-LC vector to put gene under the control of the CaMV 35S promoter. The resulting plasmid was introduced into A. tumefaciens (strain EHA105), and transgenic Arabidopsis plants were generated using the A. tumefaciens-mediated floral dip method. Homozygous T3 plants were screened on plates containing MS medium with 75 mg/L of kanamycin. TaGT2L1D expression in transgenic Arabidopsis plants was examined by Semi-quantitative RT-PCR, which was conducted as follows: 95 °C for 5 min; 30 cycles of 95 °C for 30 s, 57 °C for 30 s, and 72 °C for 30 s; and 72 °C for 10 min. The sequences of the primers used are listed in the Supplementary Table S1.
Drought treatment, stomatal density and stomatal aperture measurement
Col-0, gtl1-3, gtl1-3TaGT2L1D-1 and gtl1-3TaGT2L1D-5 plants were cultivated in pots (50 cm in length, 25 cm in width, and 5 cm in height) under controlled chamber environment with a 16 h/8 h photoperiod at 22 °C. For drought stress treatment, plants were grown with sufficient watering for two weeks after germination. Water was withheld for the next two weeks. When the leaves of almost all of the plants were no longer turgid, the plants were re-watered and the survival rate was measured after four days of re-watering.
For stomatal density measurement, fully expanded four-week-old leaves from Col-0, gtl1-3, gtl1-3TaGT2L1D-1 and gtl1-3TaGT2L1D-5 lines were used. The number of stomata was determined in a 0.144 mm2 area using an abaxial epidermal peel taken from the middle of each leaf. Four areas per leaf were measured, and six leaves from each different individuals were analysed. The epidermis was printed using Super Glue (Ontario, CA) and the imprint on the glass slide was observed with a confocal microscope (Leica TCS-SP8; Leica Microsystems).
The stomatal aperture was analysed using abaxial epidermal peels of fully expanded leaves according to a previous report58. For ABA treatment, fully expanded four-week-old leaves from Col-0, gtl1-3, gtl1-3TaGT2L1D-1 and gtl1-3TaGT2L1D-5 plants were incubated in a buffer solution (20 mM KCl, 5 mM MES-KOH, and 1 mM CaCl2, pH 6.15) for 2 h to open stomata, and then incubated with 20 μM ABA for 1 h. The aperture width of each stomatal pore was determined.
Transcriptional activity analysis of TaGT2L1D
An effective GAL4/UAS-based system was performed as described previously59 to assess the transcriptional activity of TaGT2L1D in plant. The coding sequence of TaGT2L1D was cloned into EcoRI/SalI of pYF503 vector to produce a G4DBD-TaGT2L1D fusion. The G4DBD and G4DBD-TaGT2L1D fragments were cloned into pDONR207 and then recombined in the Gateway-compatible destination vector pGWB2 to generate the effector constructs. The 35S-UAS-GUS reporter plasmid and effector constructs were then transformed into A. tumefaciens (strain EHA105). The two effectors were then transiently co-transformed with the reporter into N. benthamiana leaves. After 24 h of incubation in the dark and 72 h of incubation in the light, the leaves were subjected to histochemical GUS staining as described previously60. Leaf samples were incubated in freshly prepared buffer containing 5-bromo-4-chloro-3-indolyl-β-D-glucuronic acid (X-gluc) overnight, followed by the cleanup with 75% ethanol.
For the repression of AtSDD1 promoter activity by TaGT2L1D, The promoter of AtSDD1 (1987-bp upstream of translation initiation site) was cloned into PstI/BamHI of pCambia3301 vector to generate a proSDD1-GUS reporter construct. pTF101-GFP-TaGT2L1D was used as a positive effector, while pTF101-GFP was used as a negative control effector. A transient co-expression assay was performed in N. benthamiana leaves, followed by histochemical GUS staining as described previously.
These experiments were performed with three biological replications. All primer sequences are listed in the Supplementary Table S1.
EMSAs
To determine DNA binding activity and the specificity of TaGT2L1D in vitro, EMSAs were performed using a LightShift Chemiluminescent EMSA Kit (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s protocol. Protein samples were designed to be fused with GST tag for purification process. For the GST-TaGT2L1D-N and GST-TaGT2L1D-C fusion proteins, the 5′ end 513-bp fragment and 3′ end 831-bp fragment were amplified and cloned using EcoRI and SalI into pGEX-4T-1, respectively. The recombinant proteins were expressed in Escherichia coli and purified using amylose resin (New England Biolabs, Ipswich, MA, USA) according to the manufacturer’s protocol. For the nucleotide probes, an oligonucleotide containing the GT3 box (5′-GGTAAA-3′) from the AtSDD1 promoter was synthesized and labeled with biotin at its 5′ end (Invitrogen Corp.). After 30 min of incubation at room temperature, the protein-probe mixture was separated on a 6% polyacrylamide gel and transplanted to a Biodyne B nylon membrane (Pall Corp., Port Washington, NY, USA). Migration of the biotin-labeled probes was detected by chemiluminescence according to the manufacturer’s protocol. This experiment was performed three times. All primer and probe sequences are listed in the Supplementary Table S1.
Statistics
Statistical analysis was performed using the SPSS 21.0 software. Unpaired two-tailed t test was used for comparison between drought treatment group and control treatment group in the TaGT2L1s expression analysis. One-way analysis of variance (one-way ANOVA) followed by Student-Newman-Keuls test (as a post hoc test) was used for the rest of the statistical comparisons. All data are presented as mean ± standard deviations. P < 0.05 was considered as significant in the statistical analysis.
Additional Information
Accession codes: Sequences of the genes in this paper may be found in the GenBank database library under the following accession numbers: KU559336 (TaGT2L1), AK336250 (TaGT2L2), AK333756 (TaGT2L3), KC775780 (TaACTIN), NM_179953 (AtACTIN), NM_100292 (SDD1), AY099585 (CCS52A1).
How to cite this article: Zheng, X. et al. The Wheat GT Factor TaGT2L1D Negatively Regulates Drought Tolerance and Plant Development. Sci. Rep. 6, 27042; doi: 10.1038/srep27042 (2016).
Supplementary Material
Acknowledgments
We thank Dr. Genji Qin (Peking University) and Dr. Xigang Liu (The Center for Agricultural Resources Research, Institute of Genetics and Developmental Biology of CAS) for kindly providing the 35S-UAS-GUS reporter construct and strain EHA105, respectively. We are grateful to Dr. Mengjun Li (Shijiazhuang Academy of Agriculture and Forestry Sciences) and Dr. Junming Li (The Center for Agricultural Resources Research, Institute of Genetics and Developmental Biology of CAS) for technical assistance and some valuable suggestions. This work was supported by the National Transgenic Key Project of MOA (2014ZX08002-002 to X.L.), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDA08010209), the key project of National Science and Technology Pillar Program of China (2013BAD05B02), and the National Natural Science Foundation of China (31571612 and 31100191).
Footnotes
Author Contributions X.Z. performed all of the experiments, analysed the data, and prepared the manuscript. H.L. prepared the manuscript. H.J. and Y.W. contributed to the writing of the manuscript. B.D. and Y.Q. analysed the data. X.L. and M.L. designed and supervised the project, and modified the manuscript. All authors have reviewed the manuscript.
References
- Valliyodan B. & Nguyen H. T. Understanding regulatory networks and engineering for enhanced drought tolerance in plants. Curr Opin Plant Biol 9, 189–195 (2006). [DOI] [PubMed] [Google Scholar]
- Baldoni E., Genga A. & Cominelli E. Plant MYB transcription factors: their role in drought response mechanisms. Int J Mol Sci 16, 15811–15851 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castilhos G., Lazzarotto F., Spagnolo-Fonini L., Bodanese-Zanettini M. H. & Margis-Pinheiro M. Possible roles of basic helix-loop-helix transcription factors in adaptation to drought. Plant Sci 223, 1–7 (2014). [DOI] [PubMed] [Google Scholar]
- Chen L. et al. The role of WRKY transcription factors in plant abiotic stresses. Biochim Biophys Acta 1819, 120–128 (2012). [DOI] [PubMed] [Google Scholar]
- Mizoi J., Shinozaki K. & Yamaguchi-Shinozaki K. AP2/ERF family transcription factors in plant abiotic stress responses. Biochim Biophys Acta 1819, 86–96 (2012). [DOI] [PubMed] [Google Scholar]
- Nakashima K., Takasaki H., Mizoi J., Shinozaki K. & Yamaguchi-Shinozaki K. NAC transcription factors in plant abiotic stress responses. Biochim Biophys Acta 1819, 97–103 (2012). [DOI] [PubMed] [Google Scholar]
- Nakashima K., Yamaguchi-Shinozaki K. & Shinozaki K. The transcriptional regulatory network in the drought response and its crosstalk in abiotic stress responses including drought, cold, and heat. Front Plant Sci 5, 170 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y. et al. Dual function of Arabidopsis ATAF1 in abiotic and biotic stress responses. Cell Res 19, 1279–1290 (2009). [DOI] [PubMed] [Google Scholar]
- Sakuma Y. et al. Dual function of an Arabidopsis transcription factor DREB2A in water-stress-responsive and heat-stress-responsive gene expression. Proc Natl Acad Sci USA 103, 18822–18827 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z. et al. An R2R3 MYB transcription factor in wheat, TaPIMP1, mediates host resistance to Bipolaris sorokiniana and drought stresses through regulation of defense- and stress-related genes. New phytol 196, 1155–1170 (2012). [DOI] [PubMed] [Google Scholar]
- Raineri J., Wang S., Peleg Z., Blumwald E. & Chan R. L. The rice transcription factor OsWRKY47 is a positive regulator of the response to water deficit stress. Plant Mol Biol 88, 401–413 (2015). [DOI] [PubMed] [Google Scholar]
- Kaplan-Levy R. N., Brewer P. B., Quon T. & Smyth D. R. The trihelix family of transcription factors–light, stress and development. Trends Plant Sci 17, 163–171 (2012). [DOI] [PubMed] [Google Scholar]
- Boyer L. A., Latek R. R. & Peterson C. L. The SANT domain: a unique histone-tail-binding module? Nat Rev Mol Cell Biol 5, 158–163 (2004). [DOI] [PubMed] [Google Scholar]
- Nagano Y. Several features of the GT-factor trihelix domain resemble those of the Myb DNA-binding domain. Plant physiol 124, 491–494 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano Y., Inaba T., Furuhashi H. & Sasaki Y. Trihelix DNA-binding protein with specificities for two distinct cis-elements: both important for light down-regulated and dark-inducible gene expression in higher plants. J Biol Chem 276, 22238–22243 (2001). [DOI] [PubMed] [Google Scholar]
- Ni M., Dehesh K., Tepperman J. M. & Quail P. H. GT-2: in vivo transcriptional activation activity and definition of novel twin DNA binding domains with reciprocal target sequence selectivity. Plant Cell 8, 1041–1059 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Y. et al. Evolutionary history of trihelix family and their functional diversification. DNA Res 21, 499–510 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr M. S., Willmann M. R. & Jenik P. D. Is there a role for trihelix transcription factors in embryo maturation? Plant Signal Behav 7, 205–209 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breuer C. et al. The trihelix transcription factor GTL1 regulates ploidy-dependent cell growth in the Arabidopsis trichome. Plant cell 21, 2307–2322 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan-Levy R. N., Quon T., O’Brien M., Sappl P. G. & Smyth D. R. Functional domains of the PETAL LOSS protein, a trihelix transcription factor that represses regional growth in Arabidopsis thaliana. Plant J 79, 477–491 (2014). [DOI] [PubMed] [Google Scholar]
- Lampugnani E. R., Kilinc A. & Smyth D. R. PETAL LOSS is a boundary gene that inhibits growth between developing sepals in Arabidopsis thaliana. Plant J 71, 724–735 (2012). [DOI] [PubMed] [Google Scholar]
- Li B. et al. Phosphorylation of trihelix transcriptional repressor ASR3 by MAP KINASE4 negatively regulates Arabidopsis immunity. Plant Cell 27, 839–856 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Zhou A. & Sang T. Rice domestication by reducing shattering. Science 311, 1936–1939 (2006). [DOI] [PubMed] [Google Scholar]
- Tzafrir I. et al. Identification of genes required for embryo development in Arabidopsis. Plant Physiol 135, 1206–1220 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi J., Qiu Y., Du L. & Poovaiah B. W. Plant-specific trihelix transcription factor AtGT2L interacts with calcium/calmodulin and responds to cold and salt stresses. Plant Sci 185–186, 274–280 (2012). [DOI] [PubMed] [Google Scholar]
- Xie Z. M. et al. Soybean Trihelix transcription factors GmGT-2A and GmGT-2B improve plant tolerance to abiotic stresses in transgenic Arabidopsis. PLoS One 4, e6898 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Yoo C. Y. et al. The Arabidopsis GTL1 transcription factor regulates water use efficiency and drought tolerance by modulating stomatal density via transrepression of SDD1. Plant Cell 22, 4128–4141 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson T. & Blatt M. R. Stomatal size, speed, and responsiveness impact on photosynthesis and water use efficiency. Plant Physiol 164, 1556–1570 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaerle L., Saibo N. & Van Der Straeten D. Tuning the pores: towards engineering plants for improved water use efficiency. Trends Biotechnol 23, 308–315 (2005). [DOI] [PubMed] [Google Scholar]
- Wasilewska A. et al. An update on abscisic acid signaling in plants and more. Mol Plant 1, 198–217 (2008). [DOI] [PubMed] [Google Scholar]
- Geiger D. et al. Activity of guard cell anion channel SLAC1 is controlled by drought-stress signaling kinase-phosphatase pair. Proc Natl Acad Sci USA 106, 21425–21430 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustilli A. C., Merlot S., Vavasseur A., Fenzi F. & Giraudat J. Arabidopsis OST1 protein kinase mediates the regulation of stomatal aperture by abscisic acid and acts upstream of reactive oxygen species production. Plant Cell 14, 3089–3099 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann D. C. & Sack F. D. Stomatal development. Annu Rev Plant Biol 58, 163–181 (2007). [DOI] [PubMed] [Google Scholar]
- Dong J., MacAlister C. A. & Bergmann D. C. BASL controls asymmetric cell division in Arabidopsis. Cell 137, 1320–1330 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacAlister C. A., Ohashi-Ito K. & Bergmann D. C. Transcription factor control of asymmetric cell divisions that establish the stomatal lineage. Nature 445, 537–540 (2007). [DOI] [PubMed] [Google Scholar]
- Ohashi-Ito K. & Bergmann D. C. Arabidopsis FAMA controls the final proliferation/differentiation switch during stomatal development. Plant Cell 18, 2493–2505 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillitteri L. J., Sloan D. B., Bogenschutz N. L. & Torii K. U. Termination of asymmetric cell division and differentiation of stomata. Nature 445, 501–505 (2007). [DOI] [PubMed] [Google Scholar]
- Berger D. & Altmann T. A subtilisin-like serine protease involved in the regulation of stomatal density and distribution in Arabidopsis thaliana. Genes Dev 14, 1119–1131 (2000). [PMC free article] [PubMed] [Google Scholar]
- Von Groll U., Berger D. & Altmann T. The subtilisin-like serine protease SDD1 mediates cell-to-cell signaling during Arabidopsis stomatal development. Plant Cell 14, 1527–1539 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann D. C., Lukowitz W. & Somerville C. R. Stomatal development and pattern controlled by a MAPKK kinase. Science 304, 1494–1497 (2004). [DOI] [PubMed] [Google Scholar]
- Lampard G. R., Macalister C. A. & Bergmann D. C. Arabidopsis stomatal initiation is controlled by MAPK-mediated regulation of the bHLH SPEECHLESS. Science 322, 1113–1116 (2008). [DOI] [PubMed] [Google Scholar]
- Wang H., Ngwenyama N., Liu Y., Walker J. C. & Zhang S. Stomatal development and patterning are regulated by environmentally responsive mitogen-activated protein kinases in Arabidopsis. Plant Cell 19, 63–73 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng H., Yoo C. Y., Gosney M. J., Hasegawa P. M. & Mickelbart M. V. Poplar GTL1 is a Ca2+/calmodulin-binding transcription factor that functions in plant water use efficiency and drought tolerance. PLoS One 7, e32925 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T., Ohashi-Ito K. & Bergmann D. C. Orthologs of Arabidopsis thaliana stomatal bHLH genes and regulation of stomatal development in grasses. Development 136, 2265–2276 (2009). [DOI] [PubMed] [Google Scholar]
- Choi M. S. et al. Isolation of a calmodulin-binding transcription factor from rice (Oryza sativa L.). J Biol Chem 280, 40820–40831 (2005). [DOI] [PubMed] [Google Scholar]
- Kim H. S. et al. Identification of a calmodulin-binding NAC protein as a transcriptional repressor in Arabidopsis. J Biol Chem 282, 36292–36302 (2007). [DOI] [PubMed] [Google Scholar]
- Kim M. C., Chung W. S., Yun D. J. & Cho M. J. Calcium and calmodulin-mediated regulation of gene expression in plants. Mol Plant 2, 13–21 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn R. M., Caspar T., Dehesh K. & Quail P. H. DNA binding factor GT-2 from Arabidopsis. Plant Mol Biol 23, 337–348 (1993). [DOI] [PubMed] [Google Scholar]
- Breuer C. et al. Transcriptional repression of the APC/C activator CCS52A1 promotes active termination of cell growth. EMBO J 31, 4488–4501 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Q. et al. The TIE1 transcriptional repressor links TCP transcription factors with TOPLESS/TOPLESS-RELATED corepressors and modulates leaf development in Arabidopsis. Plant Cell 25, 421–437 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagale S., Links M. G. & Rozwadowski K. Genome-wide analysis of ethylene-responsive element binding factor-associated amphiphilic repression motif-containing transcriptional regulators in Arabidopsis. Plant Physiol 152, 1109–1134 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagale S. & Rozwadowski K. Small yet effective: the ethylene responsive element binding factor-associated amphiphilic repression (EAR) motif. Plant Signal Behav 5, 691–694 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagale S. & Rozwadowski K. EAR motif-mediated transcriptional repression in plants: an underlying mechanism for epigenetic regulation of gene expression. Epigenetics 6, 141–146 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo C. Y., Hasegawa P. M. & Mickelbart M. V. Regulation of stomatal density by the GTL1 transcription factor for improving water use efficiency. Plant Signal Behav 6, 1069–1071 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J. & Schmittgen T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408 (2001). [DOI] [PubMed] [Google Scholar]
- Hu Y. et al. Arabidopsis transcription factor WRKY8 functions antagonistically with its interacting partner VQ9 to modulate salinity stress tolerance. Plant J. 74, 730–745 (2013). [DOI] [PubMed] [Google Scholar]
- Liu X. et al. HDA6 directly interacts with DNA methyltransferase MET1 and maintains transposable element silencing in Arabidopsis. Plant Physiol. 158, 119–129 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Assmann S. M. & Albert R. Predicting essential components of signal transduction networks: a dynamic model of guard cell abscisic acid signaling. PLoS Biol 4, e312 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. N. et al. Soybean miR172c targets the repressive AP2 transcription factor NNC1 to activate ENOD40 expression and regulate nodule initiation. Plant Cell 26, 4782–4801 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y. K., Wang T., Zhang W. S. & Li X. SOS3 mediates lateral root development under low salt stress through regulation of auxin redistribution and maxima in Arabidopsis. New Phytol 189, 1122–1134 (2011). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.