Abstract
PURPOSE
We hypothesized that adverse prognostic associations of specific tumor molecular factors vary by patient age at colorectal cancer (CRC) diagnosis.
EXPERIMENTAL DESIGN
We examined the prognostic associations and interactions by age at CRC diagnosis (<60 vs. 60–74 vs. ≥75 years old) of key molecular factors – CpG island methylator phenotype (CIMP), microsatellite instability (MSI), KRAS, BRAF, and PIK3CA mutations, and nuclear CTNNB1 expression status – on CRC-specific survival and overall survival, utilizing 1280 incident CRC cases (median age 69 years, range 38–91 years) within the Nurses’ Health Study (NHS) and Health Professionals Follow-up Study (HPFS) cohorts.
RESULTS
MSI-high was associated with better survival while BRAF mutation was associated with worse survival, but these associations did not appreciably differ by age group. Status of CIMP, KRAS mutation, or PIK3CA mutation was not associated with prognosis regardless of age. Nuclear CTNNB1 expression was associated with a trend toward worse prognosis among older adults (age ≥75) [multivariate hazard ratio (HR), 1.67; 95% confidence interval (CI) 0.89 to 3.13 (for CRC-specific survival); multivariate HR 1.44; 95% CI 0.93 to 2.24 (for overall survival)] but not among younger patients, and there was a statistically significant interaction by age (p-interaction=0.03 for CRC-specific survival; p-interaction=0.007 for overall survival).
CONCLUSIONS
Tumor nuclear CTNNB1 expression may be associated with higher mortality among older CRC patients but not among younger patients. Our findings need to be confirmed in independent datasets. Detailed exploration of tumor molecular signatures in older CRC patients in large populations is warranted.
INTRODUCTION
Colorectal cancer (CRC) is the 4th leading cancer diagnosis and 2nd cause of cancer death in the United States, with an estimated 140,000 diagnoses and 50,000 deaths annually.(1) Despite 36% of CRC patients being age 75 or older at diagnosis, guidance on how to approach the treatment of this population is limited. Among those selected to receive chemotherapy, survival outcomes are similar to younger patients in most,(2–4) but not all studies.(5) Yet, only one-third of older adults receive indicated chemotherapy.(6) There is a paucity of research concerning which factors should determine patient selection for treatment and the resulting survival outcomes. Most of this limited research focuses on age alone as a predictor of treatment and survival outcomes.
Advancing age has long been recognized as a potent risk factor for the development of cancer, further underscored by the fact that nearly 60% of CRC is diagnosed in those age ≥65. Several investigators have postulated mechanisms by which aging impacts CRC carcinogenesis, including accumulation of somatic mutations over time and epigenetic silencing.(7) Baseline rates of detectable somatic mosaicism in the general population are low, but generally higher in older adults (~2% in people with age ≥75) than in younger adults (< 0.5% in people with age <50).(7, 8) However, the degree to which CRC carcinogenesis differs by age at diagnosis, as driven by somatic mutations and epigenetic changes, is not well known. We hypothesized that adverse prognostic associations of key molecular factors would be disproportionately higher in older adults than younger adults at diagnosis of CRC.
To test this hypothesis, we examined the prognostic associations of CpG island methylator phenotype (CIMP), microsatellite instability (MSI), KRAS, BRAF, and PIK3CA mutations, and CTNNB1 (ß-catenin) nuclear expression status according to age group (at CRC diagnosis) among 1280 cases of CRC within two large prospective longitudinal cohorts. We tested the statistical interaction of age at CRC diagnosis with each molecular factor on CRC-specific survival and overall survival. The combined prospective cohorts used for analysis provide the unique advantage of a large age distribution of incident, previously untreated CRC cases with well-annotated tumor molecular data to address the hypothesis.
MATERIALS AND METHODS
Study Population
Initiated in 1976, the Nurses’ Health Study (NHS)(9) is a prospective U.S. nationwide cohort of 121,700 female registered nurses age 30 to 55 years at the time of enrollment, who responded to a mailed questionnaire regarding cancer and cardiovascular risk. The Health Professionals Follow-up Study (HPFS) subsequently enrolled 51,529 male health professionals, age 40 to 75 years beginning in 1986. Both cohorts continue to complete biennial follow-up questionnaires updating information on medical history and potential risk factors. The studies were approved by the Human Subjects Committees at Harvard T.H. Chan School of Public Health and Brigham and Women’s Hospital (both Boston, MA). All participants signed informed consent permitting questionnaire, blood and tumor data to be used in research studies.
The study population consists of NHS and HPFS subjects with pathologically confirmed colon or rectal carcinoma diagnosed up to June 1, 2010 for NHS and January 1, 2010 for HPFS with available CRC tumor specimen for analysis (Figure 1). Subjects with other cancer occurring within 3 years before colorectal cancer diagnosis (except non-melanoma skin cancer) were excluded from the analysis (n=33). The final cohort includes 1280 subjects including 690 from NHS and 590 from HPFS.
Figure 1.
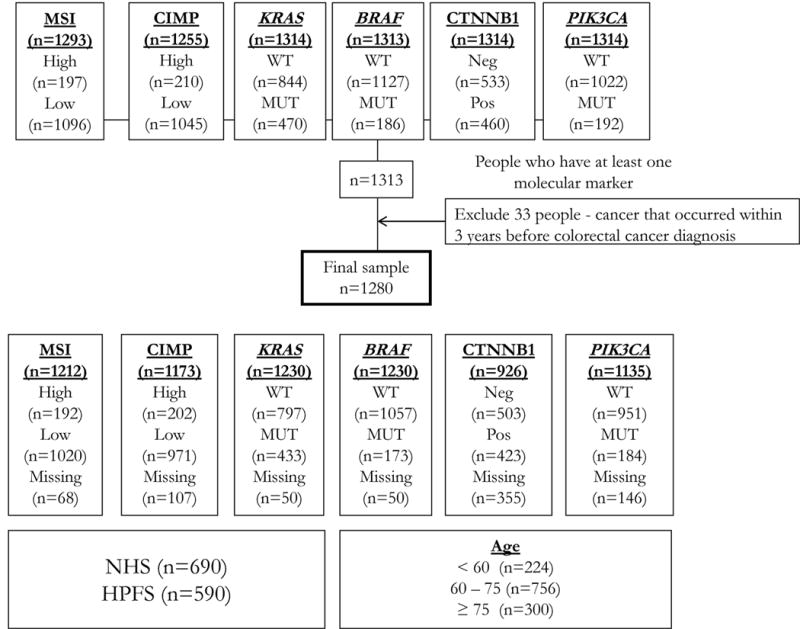
Nurses’ Health Study (NHS) and Health Professionals Follow-up Study (HPFS) incident colorectal case cohort
Identification of Colorectal Cancer
For respondents reporting a diagnosis of CRC within the prior 2 years, we requested permission to review all hospital and pathology records pertaining to CRC. Once obtained, study physicians blinded to patient outcomes extracted information on American Joint Committee on Cancer (AJCC) stage, histology, tumor location and date of diagnosis. Cause and date of death were obtained from the National Death Index (NDI) for non-respondents.(10) Nearly 96% of all incident CRC cases were identified by either of these two methods.(11) For deceased participants with known or suspected cancer for which we have not been able to obtain medical records, we contacted the state tumor registry to confirm and classify the cancer. CRC treatment data are not available in these databases.
Analysis of Tumor Molecular Factors
The term molecular factors is used in this study to denote the accumulated somatic mutations associated with promotion of CRC. Archival CRC tumor specimens were collected from the hospitals at which subjects underwent resection or biopsy of CRC. All genomic DNA extraction from paraffin-embedded tissue and whole genome amplification by polymerase chain reaction (PCR) were performed as previously described.(9) All CRC tumor block specimens and hematoxylin and eosin (H&E)-stained tissue sections were reviewed by a pathologist (S.O.) with established quality control measures consistent with the strategy used in prior NHS/HPFS analyses.(9) We use HUGO (Human Genome Organisation)-approved official symbols for genes and gene products – including BRAF, CTNNB1 [catenin (cadherin-associated protein), beta 1, 88kDa; so-called β-catenin], KRAS, and PIK3CA.(12)
Analysis of PIK3CA, KRAS, BRAF and CIMP
PCR and pyrosequencing of PIK3CA (exons 9 and 20),(11, 13) KRAS (codons 12, 13, 61 and 146),(14, 15) and BRAF (codon 600) were performed as previously described.(16) Using MethyLight assay,(17) DNA methylation was quantified in 8 CIMP-specific promoters [CACNA1G, CDKN2A (p16), CRABP1, IGF2, MLH1, NEUROG1, RUNX3, and SOCS1].(18, 19) Using these 8 markers, CIMP-high was defined as ≥6 methylated markers, CIMP-negative as 0 methylated markers, and the remainder as CIMP-low, as the previously established criteria.(20)
Analysis of MSI
MSI status was quantified using a 10-marker panel using D2S123, D5S346, D17S250, BAT25, BAT26, BAT40, D18S55, D18S56, D18S67, and D18S487.(21) MSI-high was defined as presence of instability in ≥30% of the markers, microsatellite instability low (MSI-low as 1–29% unstable markers, and microsatellite stability (MSS) as no unstable marker. Given no difference in prognosis between MSI-low and MSS tumors in prior analysis,(9) they were combined in the present study.
Analysis of CTNNB1 (β-catenin)
Immunohistochemistry for CTNNB1 nuclear expression was performed as previously described, and interpreted as negative (weak or no expression) or positive (moderate or strong expression) by a pathologist (T.M.).(22) A subset of cases (n=292) were independently interpreted by a second pathologist (S.O.); agreement between the two pathologists was 0.90 for CTNNB1 nuclear expression (κ=0.80; p<0.0001), indicating good to substantial agreement.
Definition of Age
The age at diagnosis was classified into three age groups (<60, 60–74 and ≥75). Older adults are defined as those age ≥75 years at diagnosis. Sensitivity analyses were conducted to examine other definitions of age to allow comparisons consistent with prior studies in the epigenetic,(9) CRC and geriatric oncology(5, 23) literature.
Outcome Measurement
Patients are observed until death and censored at last questionnaire prior to data analysis as of January 1, 2011. The two primary outcomes are CRC-specific survival and overall survival. For NHS/HPFS, follow-up began from date of CRC diagnosis. CRC-specific survival is defined as the time from CRC diagnosis to CRC-specific death; deaths from other causes are censored at the time of death. Overall survival is defined as the time from CRC diagnosis to death due to any cause. Date of death was obtained by report from family, postal authority or confirmation via the NDI.(10) Cause of death was assigned by study physicians blinded to questionnaire responses. Nearly 98% of deaths were confirmed by these methods.(9)
Covariate Assessment
Known and potential prognostic factors affecting CRC-specific and overall survival were extracted from the hospital and pathology records, including AJCC stage, grade of tumor differentiation, histology, date and age of diagnosis. Body mass index (BMI), prediagnosis activity level and diagnosis of type 2 diabetes mellitus (DM), myocardial infarction (MI)/congestive heart failure (CHF) and cerebrovascular accident (CVA) were taken from the biennial questionnaire preceding date of diagnosis as previously reported.(22, 24–28)
Statistical Analysis
Cox proportional hazards models were used to calculate hazard ratios (HRs) of death or death resulting from CRC according to molecular factors, adjusted using the stepwise variable selection method including BMI (<30, 30+, missing), prediagnosis activity (<18, 18+, missing), tumor location, tumor differentiation, and other markers (MSI, CIMP, KRAS, BRAF, CTNNB1 and PIK3CA). Gender, regular aspirin use and comorbidity (DM, MI/CHF, CVA) were forced in the model. Regular aspirin use is defined as at least 2 tablets/week in NHS and at least 2 times/week in HPFS. Disease stage was used as a stratifying variable. The analysis results are from Cox regression models where all patient data is censored at 5 years or 10 years for the respective CRC-specific and overall survival. We tested interaction impact of age on the association between each molecular factor and survival by including the cross-product of age as a continuous variable and each molecular factor in the model. We evaluated the distribution of molecular tumor factors using the X2 test (categorical variables) and analysis of variance (ANOVA, continuous variables) across patient and disease factors. We considered multiple hypothesis testing adjusting the p for significance level to p=0.01 given 5 molecular factors evaluated. All analyses used SAS software, version 9.3 (SAS Institute, Inc, Cary, NC). Smoothing splines of log hazard were used to visualize the relationship between CRC-specific and overall survival at 5 years by molecular factor status.
RESULTS
Patient characteristics
The incident CRC cases with tumor sample availability in the combined NHS/HPFS cohort included 1280 patients (n=224 age <60, 756 age 60–74 and 300 age ≥75). We assessed patient and disease characteristics according to age at diagnosis (Table 1). Older patients were more likely to be male (65% vs. 30% age <60, 43% age 60–74), have a lower body mass index (93% BMI <30 vs. 86% age <60, 83% age 60–74), present with lower rate of stage IV disease at diagnosis (8% vs. 16% age <60, 14% age 60–74) and have tumors in the proximal colon (52% vs. 36% age <60, 49% age 60–74) (all p<0.001). Of those with reporting presence of comorbid medical conditions, older adults had the highest rate of prior cerebrovascular accident (56% vs. 1% age <60, 43% age 60–74) but second highest rate of DM (35% vs. 7% age <60, 57% age 60–74) and MI or CHF (45% vs. 2% age <60, 54% age 60–74) (Table 1). We adjusted all analyses for the presence of cerebrovascular accident, diabetes mellitus and myocardial infarction/congestive heart failure. There were no appreciable differences in race, year of diagnosis, tumor differentiation, number of lymph nodes examined, and number of lymph nodes positive.
Table 1.
Baseline characteristics of colorectal cancer (CRC) cases by age group
| Characteristic | Class | Age at CRC diagnosis | |||
|---|---|---|---|---|---|
| Overall N | <60 years | 60–74 years | 75+ years | ||
| Age | N | 1280 | 224 | 756 | 300 |
| Mean | 54.8 | 68.1 | 79.4 | ||
| SD | 4.1 | 4.1 | 3.5 | ||
| Gender | Female | 690 | 156 (70%) | 430 (57%) | 104 (35%) |
| Male | 590 | 68 (30%) | 326 (43%) | 196 (65%) | |
| Race | White | 1214 | 218 (97%) | 715 (95%) | 281 (94%) |
| Black | 20 | 2 (0.9%) | 13 (2%) | 5 (2%) | |
| Asian | 9 | 2 (0.9%) | 5 (0.7%) | 2 (0.7%) | |
| Year of diagnosis | Before 2002 | 986 | 214 (96%) | 610 (81%) | 162 (54%) |
| 2002 or later | 294 | 10 (5%) | 146 (19%) | 138 (46%) | |
| BMI (kg/m2) | <30 | 1098 | 192 (86%) | 626 (83%) | 280 (93%) |
| 30+ | 154 | 29 (13%) | 108 (14%) | 17 (6%) | |
| Tumor location | Proximal colon | 603 | 80 (36%) | 367 (49%) | 156 (52%) |
| Distal colon | 389 | 76 (34%) | 242 (32%) | 71 (24%) | |
| Rectum | 282 | 66 (30%) | 147 (19%) | 69 (23%) | |
| Disease stage | I | 297 | 48 (21%) | 173 (23%) | 76 (25%) |
| II | 375 | 52 (23%) | 238 (32%) | 85 (28%) | |
| III | 329 | 70 (31%) | 185 (25%) | 74 (25%) | |
| IV | 161 | 35 (16%) | 103 (14%) | 23 (8%) | |
| Tumor differentiation | Well/moderate | 1144 | 201 (90%) | 667 (88%) | 276 (92%) |
| Poor | 125 | 21 (9%) | 81 (11%) | 23 (8%) | |
| No. of lymph node examined | <12 | 556 | 103 (46%) | 345 (46%) | 108 (36%) |
| 12+ | 596 | 101 (45%) | 338 (45%) | 157 (52%) | |
| No. of lymph node positive for metastasis | 0 | 713 | 119 (53%) | 429 (57%) | 165 (55%) |
| 1–3 | 248 | 47 (21%) | 150 (20%) | 51 (17%) | |
| 4+ | 249 | 53 (24%) | 136 (18%) | 60 (20%) | |
| Comorbid conditions | Diabetes mellitus | 122 | 9 (4%) | 70 (9%) | 43 (14%) |
| Myocardial Infarction (MI) | 83 | 1 (0.4%) | 47 (6%) | 35 (12%) | |
| CVA | 90 | 1 (0.4%) | 39 (5%) | 50 (17%) | |
| COPD | 57 | 3 (1%) | 35 (5%) | 19 (6%) | |
| CHF | 82 | 2 (0.9%) | 39 (5%) | 41 (14%) | |
| MI/CHF | 125 | 2 (0.9%) | 67 (9%) | 56 (19%) | |
Abbreviations: BMI, body mass index; No., number; CHF, congestive heart failure; COPD, chronic obstructive pulmonary disease; CRC, colorectal cancer; CVA, cerebrovascular accident; MI, myocardial infarction; SD, standard deviation.
Prevalence of molecular factors by age
We examined the distribution of potentially prognostic and/or predictive molecular changes in CRC tumor samples by age (Table 2). Fifteen percent of the overall cohort had MSI-high tumors; there was a statistically significant difference in the rate of MSI-high across age groups (8% age < 60, 17% age 60–74, 16% age ≥75; p=0.006) (Table 2). Similarly, 16% of the overall cohort has CIMP-high tumors and the distribution of CIMP-high was statistically significantly increased in older adults (5% age < 60; 18% age 60–74, and 18% age ≥75; p<0.0001). There were no differences in rates of KRAS mutation (p=0.53), BRAF mutation (p=0.14), PIK3CA mutation (p=0.50), and CTNNB1 nuclear expression positive (p=0.69) among the three age groups.
Table 2.
Distribution of tumor molecular features according to age group at colorectal cancer (CRC) diagnosis
| Characteristic | Class | Age Group
|
Overall N=1280 |
P-value | ||
|---|---|---|---|---|---|---|
| <60 years (n=224) |
60–74 years (n=756) |
75+ years (n=300) |
||||
|
| ||||||
| MSI | 0.006 | |||||
|
| ||||||
| Low/MSS | 192 (86%) | 590 (78%) | 238 (79%) | 1020 (80%) | ||
| High | 18 (8%) | 126 (17%) | 48 (16%) | 192 (15%) | ||
| Missing | 14 (6%) | 40 (5%) | 14 (5%) | 68 (5%) | ||
|
| ||||||
| CIMP | <.0001 | |||||
|
| ||||||
| Low/negative | 202 (90%) | 566 (75%) | 203 (68%) | 971 (76%) | ||
| High | 11 (5%) | 136 (18%) | 55 (18%) | 202 (16%) | ||
| Missing | 11 (5%) | 54 (7%) | 42 (14%) | 107 (8%) | ||
|
| ||||||
| KRAS | 0.53 | |||||
|
| ||||||
| Wild-type | 133 (59%) | 446 (59%) | 168 (56%) | 747 (58%) | ||
| Mutant | 81 (36%) | 280 (37%) | 122 (41%) | 483 (38%) | ||
| Missing | 10 (5%) | 30 (4%) | 10 (3%) | 50 (4%) | ||
|
| ||||||
| BRAF | 0.14 | |||||
|
| ||||||
| Wild-type | 194 (87%) | 615 (81%) | 248 (83%) | 1057 (83%) | ||
| Mutant | 21 (9%) | 108 (14%) | 44 (15%) | 173 (14%) | ||
| Missing | 9 (4%) | 33 (4%) | 8 (3%) | 50 (4%) | ||
|
| ||||||
| CTNNB1 (nuclear expression) | 0.69 | |||||
|
| ||||||
| Negative | 109 (49%) | 308 (41%) | 86 (29%) | 503 (39%) | ||
| Positive | 86 (38%) | 256 (34%) | 81 (27%) | 423 (33%) | ||
| Missing | 29 (13%) | 192 (25%) | 133 (44%) | 354 (28%) | ||
|
| ||||||
| PIK3CA | 0.50 | |||||
|
| ||||||
| Wild-type | 160 (71%) | 563 (75%) | 228 (76%) | 951 (74%) | ||
| Mutant | 27 (12%) | 106 (14%) | 51 (17%) | 184 (14%) | ||
| Missing | 37 (17%) | 87 (12%) | 21 (7%) | 145 (11%) | ||
Abbreviations: CIMP, CpG island methylator phenotype; MSI, microsatellite instability; MSS, microsatellite stable.
Prognostic utility of molecular factors by age
At 5 years following diagnosis, 297 (23%) patients died of CRC (22% <60; 23% age 60–74, 24% ≥75; p=0.90) and 372 (29%) patients died of CRC or other causes (23% <60; 28% age 60–74, 35% ≥75; p=0.008). Similar differences in CRC-specific and overall survival were noted at 10 years (p=0.98 and <0.001, respectively). Despite the observed similar rates of events at each time point, older age was associated with inferior CRC-specific and overall survival at 5 (CRC-specific survival p=0.003, overall survival p<0.0001) and 10 years (CRC-specific survival p=0.0002, overall survival p<0.0001), adjusting for gender, regular aspirin use, comorbid medical conditions (DM, MI/CHF, CVA), BMI, prediagnosis physical activity, tumor location, and tumor differentiation, stratifying by disease stage (Table 3), possibly reflecting difference in treatment receipt and tolerance.
Table 3.
Colorectal cancer -specific and overall survival by age group
| Age group
|
p-within age strata | ||||||
|---|---|---|---|---|---|---|---|
| Survival Characteristic | Model | <60 years (n=224) |
60–74 years (n=756) |
75+ years (n=300) |
|||
|
| |||||||
| CRC-specific survival | 5yr | Event, N (%) | 50 (22%) | 175 (23%) | 72 (24%) | ||
|
| |||||||
| Unadjusted, HR (95% CI) | 1 (Referent) | 1.05 (0.76 to 1.43) | 1.14 (0.79 to 1.63) | 0.57 | |||
| Adjusted, HR (95% CI) | 1 (Referent) | 1.46 (1.03 to 2.09) | 2.13 (1.39 to 3.27) | 0.003 | |||
|
| |||||||
| 10yr | Event, N (%) | 60 (27%) | 207 (27%) | 81 (27%) | |||
|
| |||||||
| Unadjusted, HR (95% CI) | 1 (Referent) | 1.06 (0.80 to 1.42) | 1.15 (0.82 to 1.60) | 0.38 | |||
| Adjusted, HR (95% CI) | 1 (Referent) | 1.45 (1.06 to 1.99) | 2.15 (1.46 to 3.17) | 0.0002 | |||
|
| |||||||
| Overall survival | 5yr | Event, N (%) | 52 (23%) | 214 (28%) | 106 (35%) | ||
|
| |||||||
| Unadjusted, HR (95% CI) | 1 (Referent) | 1.24 (0.91 to 1.67) | 1.63 (1.17 to 2.27) | 0.002 | |||
| Adjusted, HR (95% CI) | 1 (Referent) | 1.67 (1.19 to 2.34) | 2.68 (1.81 to 3.96) | <.0001 | |||
|
| |||||||
| 10yr | Event, N (%) | 63 (28%) | 304 (40%) | 152 (51%) | |||
|
| |||||||
| Unadjusted, HR (95% CI) | 1 (Referent) | 1.56 (1.19 to 2.05) | 2.32 (1.73 to 3.12) | <.0001 | |||
| Adjusted, HR (95% CI) | 1 (Referent) | 1.91 (1.42 to 2.57) | 3.20 (2.29 to 4.49) | <.0001 | |||
Abbreviations: HR, hazard ratio; CI, confidence interval
The associations of molecular factors on CRC-specific and overall survival by age are depicted in Table 4 and Table 5. Adjusting for the afore-mentioned covariates as well as other molecular factors (e.g. for analysis of MSI, adjusting for CIMP, KRAS, BRAF, PIK3CA and CTNNB1), we examined the CRC-specific and overall survival at 10 years among the three age groups by each molecular factor. For the overall cohort, MSI-high was associated with improved CRC-specific and overall survival (data not shown) but there was no statistically significant interaction by age (p=0.17 for CRC-specific survival and p=0.94 for overall survival). In contrast, CIMP-high, KRAS mutation and PIK3CA was not associated with CRC-specific and overall survival (data not shown) and not associated with a statistically significant interaction by age (CRC-specific survival: p=0.92, 0.89, and 0.24, respectively; overall survival: p=0.53, 0.57, and 0.09, respectively). In contrast, BRAF mutation was associated with inferior CSS and OS within age group 60–74 years (CRC-specific survival: p=0.002; overall survival: p=0.02) but not in the other age groups [(age <60 – CRC-specific survival: p=0.65; overall survival: p=0.73), (age ≥75 – CRC-specific survival: p=0.83; overall survival: p=0.94)]. There was no statistically significant interaction of BRAF and CRC-specific and overall survival by age (p=0.25, 0.71 respectively). Although statistical power was limited in subgroup analyses, among those patients whose tumors are both MSI-high and CIMP-high, BRAF mutation might be prognostic of inferior survival [HR 1.48 (95% CI 0.97 to 2.24) for CRC-specific survival at 10 years; HR 1.28 (95% CI 0.91 to 1.80) for overall survival at 10 years]. However, there was no statistically significant interaction of age and BRAF mutation, among MSI-high/CIMP-high (p-within age strata = 0.07 for CRC-specific survival, p-within age strata = 0.15 for overall survival).
Table 4.
Colorectal cancer (CRC)-specific survival at 10 years by age and molecular factor
| Molecular Factor | Model | Age group | Class | P-within age strata | P-interaction | |
|---|---|---|---|---|---|---|
|
| ||||||
| MSI | Low/MSS | High | ||||
|
| ||||||
| Event/N | <60 years | 56/192 | 2/18 | |||
| 60–74 years | 179/590 | 17/126 | ||||
| 75+ years | 75/238 | 3/48 | ||||
| Unadjusted, HR (95% CI) | <60 years | 1 (Referent) | 0.36 (0.09 to 1.49) | 0.16 | 0.11 | |
| 60–74 years | 1 (Referent) | 0.42 (0.26 to 0.69) | 0.0007 | |||
| 75+ years | 1 (Referent) | 0.16 (0.05 to 0.52) | 0.002 | |||
| Adjusted, HR (95% CI) | <60 years | 1 (Referent) | 0.47 (0.11 to 1.97) | 0.30 | 0.17 | |
| 60–74 years | 1 (Referent) | 0.42 (0.24 to 0.72) | 0.002 | |||
| 75+ years | 1 (Referent) | 0.18 (0.06 to 0.58) | 0.004 | |||
|
| ||||||
| CIMP | Low/negative | High | ||||
|
| ||||||
| Event/N | <60 years | 55/202 | 2/11 | |||
| 60–74 years | 164/566 | 29/136 | ||||
| 75+ years | 64/203 | 10/55 | ||||
| Unadjusted, HR (95% CI) | <60 years | 1 (Referent) | 0.67 (0.16 to 2.74) | 0.58 | 0.21 | |
| 60–74 years | 1 (Referent) | 0.75 (0.50 to 1.11) | 0.15 | |||
| 75+ years | 1 (Referent) | 0.52 (0.27 to 1.01) | 0.06 | |||
| Adjusted, HR (95% CI) | <60 years | 1 (Referent) | 0.37 (0.09 to 1.59) | 0.18 | 0.92 | |
| 60–74 years | 1 (Referent) | 0.89 (0.53 to 1.48) | 0.65 | |||
| 75+ years | 1 (Referent) | 0.68 (0.32 to 1.44) | 0.32 | |||
|
| ||||||
| KRAS | Wild-type | Mutant | ||||
|
| ||||||
| Event/N | <60 years | 34/133 | 24/81 | |||
| 60–74 years | 109/446 | 88/280 | ||||
| 75+ years | 41/168 | 39/122 | ||||
| Unadjusted, HR (95% CI) | <60 years | 1 (Referent) | 1.21 (0.72 to 2.04) | 0.47 | 0.80 | |
| 60–74 years | 1 (Referent) | 1.30 (0.98 to 1.73) | 0.07 | |||
| 75+ years | 1 (Referent) | 1.36 (0.88 to 2.11) | 0.17 | |||
| Adjusted, HR (95% CI) | <60 years | 1 (Referent) | 1.12 (0.64 to 1.94) | 0.70 | 0.89 | |
| 60–74 years | 1 (Referent) | 0.96 (0.71 to 1.29) | 0.78 | |||
| 75+ years | 1 (Referent) | 1.02 (0.64 to 1.61) | 0.94 | |||
|
| ||||||
| BRAF | Wild-type | Mutant | ||||
|
| ||||||
| Event/N | <60 years | 53/194 | 5/21 | |||
| 60–74 years | 162/615 | 34/108 | ||||
| 75+ years | 72/248 | 8/44 | ||||
| Unadjusted, HR (95% CI) | <60 years | 1 (Referent) | 0.92 (0.37 to 2.30) | 0.86 | 0.52 | |
| 60–74 years | 1 (Referent) | 1.36 (0.94 to 1.97) | 0.11 | |||
| 75+ years | 1 (Referent) | 0.58 (0.28 to 1.19) | 0.14 | |||
| Adjusted, HR (95% CI) | <60 years | 1 (Referent) | 0.80 (0.31 to 2.07) | 0.65 | 0.25 | |
| 60–74 years | 1 (Referent) | 2.06 (1.31 to 3.23) | 0.002 | |||
| 75+ years | 1 (Referent) | 1.09 (0.49 to 2.43) | 0.83 | |||
|
| ||||||
| CTNNB1 (nuclear expression) | Negative | Positive | ||||
|
| ||||||
| Event/N | <60 years | 33/109 | 17/86 | |||
| 60–74 years | 95/308 | 60/256 | ||||
| 75+ years | 18/ 86 | 25/81 | ||||
| Unadjusted, HR (95% CI) | <60 years | 1 (Referent) | 0.59 (0.33 to 1.06) | 0.08 | 0.03 | |
| 60–74 years | 1 (Referent) | 0.72 (0.52 to 0.99) | 0.04 | |||
| 75+ years | 1 (Referent) | 1.55 (0.85 to 2.85) | 0.15 | |||
| Adjusted, HR (95% CI) | <60 years | 1 (Referent) | 0.64 (0.35 to 1.16) | 0.14 | 0.03 | |
| 60–74 years | 1 (Referent) | 0.69 (0.49 to 0.96) | 0.03 | |||
| 75+ years | 1 (Referent) | 1.67 (0.89 to 3.13) | 0.11 | |||
|
| ||||||
| PIK3CA | Wild-type | Mutant | ||||
|
| ||||||
| Event/N | <60 years | 44/160 | 9/27 | |||
| 60–74 years | 152/563 | 28/106 | ||||
| 75+ years | 63/228 | 12/51 | ||||
| Unadjusted, HR (95% CI) | <60 years | 1 (Referent) | 1.33 (0.65 to 2.71) | 0.44 | 0.25 | |
| 60–74 years | 1 (Referent) | 0.98 (0.65 to 1.46) | 0.92 | |||
| 75+ years | 1 (Referent) | 0.80 (0.43 to 1.48) | 0.48 | |||
| Adjusted, HR (95% CI) | <60 years | 1 (Referent) | 1.42 (0.68 to 2.95) | 0.35 | 0.24 | |
| 60–74 years | 1 (Referent) | 0.88 (0.58 to 1.33) | 0.54 | |||
| 75+ years | 1 (Referent) | 0.72 (0.38 to 1.36) | 0.31 | |||
Abbreviations: HR, hazard ratio; CI, confidence interval; CIMP, CpG island methylator phenotype; MSI, microsatellite instability; MSS, microsatellite stable.
Table 5.
Overall survival at 10 years by age and molecular factor
| Molecular Factor | Model | Age group | Class | P- within age strata | P-interaction | |
|---|---|---|---|---|---|---|
|
| ||||||
| MSI | Low | High | ||||
|
| ||||||
| Event/N | <60 years | 57/192 | 3/18 | |||
| 60–74 years | 251/590 | 38/126 | ||||
| 75+ years | 129/238 | 17/48 | ||||
| Unadjusted, HR (95% CI) | <60 years | 1 (Referent) | 0.53 (0.16 to 1.68) | 0.28 | 0.57 | |
| 60–74 years | 1 (Referent) | 0.66 (0.47 to 0.93) | 0.02 | |||
| 75+ years | 1 (Referent) | 0.53 (0.32 to 0.88) | 0.01 | |||
| Adjusted, HR (95% CI) | <60 years | 1 (Referent) | 0.53 (0.16 to 1.72) | 0.29 | 0.94 | |
| 60–74 years | 1 (Referent) | 0.62 (0.40 to 0.95) | 0.03 | |||
| 75+ years | 1 (Referent) | 0.53 (0.30 to 0.93) | 0.03 | |||
|
| ||||||
| CIMP | Low | High | ||||
|
| ||||||
| Event/N | <60 years | 58/202 | 2/11 | |||
| 60–74 years | 235/566 | 51/136 | ||||
| 75+ years | 110/203 | 27/55 | ||||
| Unadjusted, HR (95% CI) | <60 years | 1 (Referent) | 0.61 (0.15 to 2.50) | 0.49 | 0.84 | |
| 60–74 years | 1 (Referent) | 0.90 (0.67 to 1.22) | 0.51 | |||
| 75+ years | 1 (Referent) | 0.83 (0.54 to 1.26) | 0.38 | |||
| Adjusted, HR (95% CI) | <60 years | 1 (Referent) | 0.53 (0.13 to 2.20) | 0.38 | 0.53 | |
| 60–74 years | 1 (Referent) | 1.11 (0.76 to 1.64) | 0.59 | |||
| 75+ years | 1 (Referent) | 1.17 (0.73 to 1.87) | 0.52 | |||
|
| ||||||
| KRAS | Wild-type | Mutant | ||||
|
| ||||||
| Event/N | <60 years | 36/133 | 24/81 | |||
| 60–74 years | 171/446 | 119/280 | ||||
| 75+ years | 82/168 | 67/122 | ||||
| Unadjusted, HR (95% CI) | <60 years | 1 (Referent) | 1.16 (0.69 to 1.94) | 0.58 | 0.87 | |
| 60–74 years | 1 (Referent) | 1.13 (0.89 to 1.42) | 0.32 | |||
| 75+ years | 1 (Referent) | 1.17 (0.85 to 1.62) | 0.33 | |||
| Adjusted, HR (95% CI) | <60 years | 1 (Referent) | 1.07 (0.63 to 1.81) | 0.81 | 0.57 | |
| 60–74 years | 1 (Referent) | 0.88 (0.69 to 1.12) | 0.29 | |||
| 75+ years | 1 (Referent) | 0.87 (0.62 to 1.21) | 0.405 | |||
|
| ||||||
| BRAF | Wild-type | Mutant | ||||
|
| ||||||
| Event/N | <60 years | 55/194 | 5/21 | |||
| 60–74 years | 241/615 | 48/108 | ||||
| 75+ years | 132/248 | 18/44 | ||||
| Unadjusted, HR (95% CI) | <60 years | 1 (Referent) | 0.86 (0.34 to 2.15) | 0.75 | 0.38 | |
| 60–74 years | 1 (Referent) | 1.29 (0.95 to 1.76) | 0.10 | |||
| 75+ years | 1 (Referent) | 0.69 (0.42 to 1.13) | 0.14 | |||
| Adjusted, HR (95% CI) | <60 years | 1 (Referent) | 0.84 (0.33 to 2.16) | 0.73 | 0.71 | |
| 60–74 years | 1 (Referent) | 1.56 (1.07 to 2.27) | 0.02 | |||
| 75+ years | 1 (Referent) | 0.98 (0.56 to 1.71) | 0.94 | |||
|
| ||||||
| CTNNB1 (nuclear expression) | Negative | Positive | ||||
|
| ||||||
| Event/N | <60 years | 34/109 | 19/86 | |||
| 60–74 years | 135/308 | 96/256 | ||||
| 75+ years | 39/ 86 | 47/81 | ||||
| Unadjusted, HR (95% CI) | <60 years | 1 (Referent) | 0.63 (0.36 to 1.10) | 0.11 | 0.007 | |
| 60–74 years | 1 (Referent) | 0.80 (0.61 to 1.03) | 0.09 | |||
| 75+ years | 1 (Referent) | 1.37 (0.89 to 2.09) | 0.15 | |||
| Adjusted, HR (95% CI) | <60 years | 1 (Referent) | 0.66 (0.38 to 1.17) | 0.16 | 0.007 | |
| 60–74 years | 1 (Referent) | 0.78 (0.60 to 1.03) | 0.08 | |||
| 75+ years | 1 (Referent) | 1.44 (0.93 to 2.24) | 0.10 | |||
|
| ||||||
| PIK3CA | Wild-type | Mutant | ||||
|
| ||||||
| Event/N | <60 years | 44/160 | 10/27 | |||
| 60–74 years | 228/563 | 37/106 | ||||
| 75+ years | 115/228 | 24/51 | ||||
| Unadjusted, HR (95% CI) | <60 years | 1 (Referent) | 1.51 (0.76 to 3.00) | 0.24 | 0.17 | |
| 60–74 years | 1 (Referent) | 0.85 (0.60 to 1.20) | 0.35 | |||
| 75+ years | 1 (Referent) | 0.87 (0.56 to 1.36) | 0.55 | |||
| Adjusted, HR (95% CI) | <60 years | 1 (Referent) | 1.52 (0.76 to 3.05) | 0.23 | 0.09 | |
| 60–74 years | 1 (Referent) | 0.77 (0.54 to 1.10) | 0.15 | |||
| 75+ years | 1 (Referent) | 0.69 (0.44 to 1.09) | 0.11 | |||
Abbreviations: HR, hazard ratio; CI, confidence interval; CIMP, CpG island methylator phenotype; MSI, microsatellite instability; MSS, microsatellite stable.
Positive CTNNB1 nuclear expression was associated with a trend toward inferior survival at 10 years and the adverse prognostic impact of positive CTNNB1 nuclear expression was significantly greater among older patients (p-interaction=0.03 for CRC-specific survival and 0.007 for overall survival). As depicted in Supplemental Figure 1, splines show the association of CTNNB1 nuclear expression status with CRC-specific and overall survival. The inflections within the splines for hazard ratios greater than 1 were observed at older age, as suggested in the trend toward inferior survival noted in Cox proportional hazards analyses. This association was not consistently modified by tumor location or presence of comorbid medical conditions. Older age was associated with nuclear CTNNB1 expression in the proximal colon (8% age <60, 15% age 60–75, 22% age ≥75; p=0.03). However, CTTNB1 expression in the distal colon or rectum was not appreciably different by age (data not shown; p=0.66 and p=0.30, respectively). Further, there was not a statistically significant difference in positive nuclear CTNNB1 expression by presence of diabetes mellitus (3% age <75 vs. 7% age ≥75; p=0.67), myocardial infarction/congestive heart failure (3% age <75 vs. 10% age ≥75; p=0.39) or cerebrovascular accident (2% age <75 vs. 7% age ≥75; p=0.07).
The observed association of examined molecular factors on CRC-specific and overall survival was not altered when alternative modeling of age is used (data not shown). For example, we examined age as a continuous variable as well as divided into two categories (age <70, ≥70) consistent with prior oncologic and geriatric literature,(2, 5, 29–32) noting no change in associations of CTNNB1 on CRC-specific and overall survival.
DISCUSSION
In this prospective cohort of men and women diagnosed with colorectal cancer (CRC), we hypothesized that adverse prognostic associations of key molecular factors would be disproportionately higher in older adults than younger adults at diagnosis of CRC. We noted a higher prevalence of MSI-high and CIMP-high as well as a similar prevalence of KRAS mutation, BRAF mutation, positive CTNNB1 (β-catenin) nuclear expression and PIK3CA mutation in older adults compared to younger counterparts. Regardless of age, MSI-high was associated with better prognosis and BRAF mutation was associated with worse survival (consistent with earlier analysis(9, 33)), whereas neither CIMP, KRAS, nor PIK3CA status was associated with prognosis. Positive CTNNB1 nuclear expression in CRC tumors was associated with a trend toward worse prognosis among older adults with a statistically significant interaction by age, making CTNNB1 an interesting molecular factor of interest for older adults diagnosed with CRC.
Analyses of tumor molecular features of CRC have become important in clinical practice and research.(34–38) Prognostic associations of tumor molecular features according to age at diagnosis of CRC have not been adequately studied. Given a recent trend of increasing age at CRC diagnosis, our prospective cohort studies could provide us with an unique opportunity to address this critical unmet need.
CTNNB1 (the β-catenin gene) is a mediator of the canonical WNT signaling pathway regulating key genes, including those involved in CRC carcinogenesis and tumor progression.(39) CTNNB1 nuclear expression has been associated inversely with CIMP-high, independent of MSI status.(40) Obesity and physical activity have been associated with higher risk of CTNNB1 nuclear-negative CRC,(28) and with higher CRC mortality in CTNNB1 nuclear-negative subtype.(22) There is no known age-specific data in cancer but it has been associated with other aging-related disease. WNT pathway activation triggers accelerated cellular senescence in klotho mouse model of accelerated aging,(41) failure of vascular cell proliferation necessary for vascular repair,(42) Alzheimer’s disease,(39) and has been implicated in the pathogenesis of osteoarthritis.(39, 43) In CRC, the tumor suppressor gene APC is implicated in hereditary CRC, Familial Adenomatous Polyposis, and most of sporadic CRCs have somatic APC mutations. APC is a known negative regulator of the WNT pathway.
Inhibitors of the WNT pathway have yet to be tested in CRC or specifically in older adults with cancer; however, evidence suggests that NSAIDs, such as aspirin, may act as modifiers of CTNNB1-associated CRC carcinogenesis and progression via modification of the WNT/CTNNB1 pathway.(44) Evaluation of the impact of regular aspirin use on the observed age-related difference in CTNNB1 and CRC-specific and overall survival among older adults with CRC and unaffected counterparts is needed to understand how aspirin use may modify survival among CRC tumors lacking CTNNB1 nuclear expression. Finally, additional research is needed to determine the correlation of MSI/CIMP status with rates of somatic mutations that may have downstream implications for prognosis, response to therapy, and potentially for treatment resistance. Investigators of The Cancer Genome Atlas Network performed exome capture DNA sequencing on 224 tumor and normal pairs of human CRC cases.(45) Age-specific analysis within this cohort is forthcoming with validation within the NHS/HPFS cohort, linked to Medicare claims data on chemotherapy treatment, to determine the impact of explored somatic mutations on CRC-specific and overall survival. Such analysis will provide additional insights because the current analysis lacks information on treatment or bench-marking against normal cases of older adults without CRC, particularly given the association of CTNNB1 with other age-related conditions.
The combined NHS/HPFS database provided the unique advantage of integrative molecular pathological epidemiology(46) data within a large age distribution of incident previously untreated CRC cases. Nonetheless, there are limitations to this analysis. We lack available treatment data for the cohort. While the yield and quality of CRC tumor specimens in NHS/HPFS were high, there were some incident CRC cases for which tumor specimens were not available. However, there were no substantial differences in patient or tumor characteristics between incident CRC patients with and without available tumor specimens.(9) In addition, residual confounding might be an issue in any observational study; however, one of the advantages of the NHS/HPFS cohorts is availability of data on potential confounders including comorbidities and detailed clinical and tumor characteristics. Lastly, the majority of the younger cohort age <60 were diagnosed with CRC before 2002. This is a consequence of the age at which patients were enrolled in the NHS and HPFS cohorts, potentially negatively impacting the overall cancer specific survival within this age group.
In conclusion, our findings suggest an age-specific pattern of molecular factors associated with CRC-specific and overall survival among older adults diagnosed with CRC. Specifically, we observed trend toward an inferior survival among older adults by tumor CTNNB1 nuclear expression status compared to younger counterparts. Given the call to integrate molecular, histopathologic and physiologic factors in the study of aging and cancer,(7) subsequent planned investigation includes determination of the molecular characterization of CRC by age and evaluation of interaction with chemotherapy treatment among older adults. Determination of the mechanisms underlying observed differences in survival and treatment response for older adults diagnosed with CRC may ultimately be translated from the laboratory to patient care to inform subsequent development of prevention strategies, targeted therapies and treatment selection for this population.
Supplementary Material
STATEMENT OF TRANSLATIONAL RELEVANCE.
It is not known whether the prognostic associations of colorectal cancer (CRC) molecular factors vary by patient age at diagnosis. Advancing age has long been recognized as a potent risk factor for the development of cancer. Yet, whether and how CRC progression differs by age at diagnosis, influenced by tumor molecular features, remain poorly understood. We hypothesized that prognostic associations of key molecular factors would be disproportionately pronounced in older adults than younger adults at diagnosis of CRC. We found that positive CTNNB1 nuclear expression appeared to confer a greater adverse association among older patients. Evaluation of inhibitors of the WNT signaling pathway, such as aspirin, on the observed age-related association of CTNNB1 expression with CRC-specific and overall survival is warranted.
Acknowledgments
We would like to thank the participants and staff of the Nurses Health Study and Health Professionals Follow-up Study for their valuable contributions as well as the following state cancer registries for their help: AL, AZ, AR, CA, CO, CT, DE, FL, GA, ID, IL, IN, IA, KY, LA, ME, MD, MA, MI, NE, NH, NJ, NY, NC, ND, OH, OK, OR, PA, RI, SC, TN, TX, VA, WA, WY. The authors assume full responsibility for analyses and interpretation of these data.
This project was supported in part by the National Comprehensive Cancer Network Young Investigator Award and Dana-Farber Cancer Institute Gloria Spivak Faculty Advancement Fund (to N.J.M), the Dana-Farber Cancer Institute / Harvard Cancer Center SPORE (NCI grant P50 CA127003 to C.S.F.), NCI grants R01 CA151993 (to S.O.), R35 CA197735 (to S.O.), K07 CA190673 (to R.N.), UM1 CA186107 (to M.J. Stampfer), P01 CA87969 (to S.E. Hankinson), R01 CA49449 (to S.E. Hankinson), P01 CA55075 (to W.C. Willett), and UM1 CA167552 (to W.C. Willett); and supports from the Paula and Russell Agrusa Fund for Colorectal Cancer Research (to C.S.F.), and the Friends of the Dana-Farber Cancer Institute (to S.O.).
Footnotes
Disclosure of potential conflicts of interest: Consultant for Amgen, Eli Lilly, Sanofi, Pfizer, Takeda Pharmaceuticals, Acceleron, Momenta Pharmaceuticals, Bayer Oncology, Genentech, Roche, Medimmune, Pharmacyclics, Vertex Pharmaceuticals, Pozen, Celgene, Merck, Gilead, Macrogenics (for C.S.F).
References
- 1.Society AC. Cancer Facts & Figures 2014. Atlanta: 2014. [Google Scholar]
- 2.McCleary NJ, Meyerhardt JA, Green E, Yothers G, de Gramont A, Van Cutsem E, et al. Impact of age on the efficacy of newer adjuvant therapies in patients with stage II/III colon cancer: findings from the ACCENT database. J Clin Oncol. 2013;31:2600–6. doi: 10.1200/JCO.2013.49.6638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yothers G, O’Connell MJ, Allegra CJ, Kuebler JP, Colangelo LH, Petrelli NJ, et al. Oxaliplatin as adjuvant therapy for colon cancer: updated results of NSABP C-07 trial, including survival and subset analyses. J Clin Oncol. 2011;29:3768–74. doi: 10.1200/JCO.2011.36.4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tournigand C, Andre T, Bonnetain F, Chibaudel B, Lledo G, Hickish T, et al. Adjuvant Therapy With Fluorouracil and Oxaliplatin in Stage II and Elderly Patients (between ages 70 and 75 years) With Colon Cancer: Subgroup Analyses of the Multicenter International Study of Oxaliplatin, Fluorouracil, and Leucovorin in the Adjuvant Treatment of Colon Cancer Trial. J Clin Oncol. 2012 doi: 10.1200/JCO.2012.42.5645. [DOI] [PubMed] [Google Scholar]
- 5.Sanoff HK, Carpenter WR, Sturmer T, Goldberg RM, Martin CF, Fine JP, et al. Effect of Adjuvant Chemotherapy on Survival of Patients With Stage III Colon Cancer Diagnosed After Age 75 Years. J Clin Oncol. 2012 doi: 10.1200/JCO.2011.41.1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schrag D, Cramer LD, Bach PB, Begg CB. Age and adjuvant chemotherapy use after surgery for stage III colon cancer. J Natl Cancer Inst. 2001;93:850–7. doi: 10.1093/jnci/93.11.850. [DOI] [PubMed] [Google Scholar]
- 7.de Magalhaes JP. How ageing processes influence cancer. Nat Rev Cancer. 2013;13:357–65. doi: 10.1038/nrc3497. [DOI] [PubMed] [Google Scholar]
- 8.Laurie CC, Laurie CA, Rice K, Doheny KF, Zelnick LR, McHugh CP, et al. Detectable clonal mosaicism from birth to old age and its relationship to cancer. Nat Genet. 2012;44:642–50. doi: 10.1038/ng.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ogino S, Nosho K, Kirkner GJ, Kawasaki T, Meyerhardt JA, Loda M, et al. CpG island methylator phenotype, microsatellite instability, BRAF mutation and clinical outcome in colon cancer. Gut. 2009;58:90–96. doi: 10.1136/gut.2008.155473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sathiakumar N, Delzell E, Abdalla O. Using the National Death Index to obtain underlying cause of death codes. J Occup Environ Med. 1998;40:808–13. doi: 10.1097/00043764-199809000-00010. [DOI] [PubMed] [Google Scholar]
- 11.Liao X, Lochhead P, Nishihara R, Morikawa T, Kuchiba A, Yamauchi M, et al. Aspirin use, tumor PIK3CA mutation, and colorectal-cancer survival. N Engl J Med. 2012;367:1596–606. doi: 10.1056/NEJMoa1207756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gray KAYB, Seal RL, Wright MW, Bruford EA. genenames.org: the HUGO Gene Nomenclature Committee resources in 2015. Nucleic acids research. 2015 Jan;43:D1079–85. doi: 10.1093/nar/gku1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nosho K, Kawasaki T, Ohnishi M, Suemoto Y, Kirkner GJ, Zepf D, et al. PIK3CA mutation in colorectal cancer: relationship with genetic and epigenetic alterations. Neoplasia. 2008;10:534–41. doi: 10.1593/neo.08336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ogino S, Kawasaki T, Brahmandam M, Yan L, Cantor M, Namgyal C, et al. Sensitive sequencing method for KRAS mutation detection by Pyrosequencing. J Mol Diagn. 2005;7:413–21. doi: 10.1016/S1525-1578(10)60571-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Imamura Y, Lochhead P, Yamauchi M, Kuchiba A, Qian ZR, Liao X, et al. Analyses of clinicopathological, molecular, and prognostic associations of KRAS codon 61 and codon 146 mutations in colorectal cancer: cohort study and literature review. Mol Cancer. 2014;13:135. doi: 10.1186/1476-4598-13-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ogino S, Kawasaki T, Kirkner GJ, Loda M, Fuchs CS. CpG island methylator phenotype-low (CIMP-low) in colorectal cancer: possible associations with male sex and KRAS mutations. J Mol Diagn. 2006;8:582–8. doi: 10.2353/jmoldx.2006.060082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ogino S, Kawasaki T, Brahmandam M, Cantor M, Kirkner GJ, Spiegelman D, et al. Precision and performance characteristics of bisulfite conversion and real-time PCR (MethyLight) for quantitative DNA methylation analysis. J Mol Diagn. 2006;8:209–17. doi: 10.2353/jmoldx.2006.050135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nosho K, Irahara N, Shima K, Kure S, Kirkner GJ, Schernhammer ES, et al. Comprehensive biostatistical analysis of CpG island methylator phenotype in colorectal cancer using a large population-based sample. PLoS ONE. 2008;3:e3698. doi: 10.1371/journal.pone.0003698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hinoue T, Weisenberger DJ, Lange CP, Shen H, Byun HM, Van Den Berg D, et al. Genome-scale analysis of aberrant DNA methylation in colorectal cancer. Genome research. 2012;22:271–82. doi: 10.1101/gr.117523.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ogino S, Kawasaki T, Kirkner GJ, Kraft P, Loda M, Fuchs CS. Evaluation of markers for CpG island methylator phenotype (CIMP) in colorectal cancer by a large population-based sample. J Mol Diagn. 2007;9:305–14. doi: 10.2353/jmoldx.2007.060170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lochhead P, Kuchiba A, Imamura Y, Liao X, Yamauchi M, Nishihara R, et al. Microsatellite instability and BRAF mutation testing in colorectal cancer prognostication. J Natl Cancer Inst. 2013;105:1151–56. doi: 10.1093/jnci/djt173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morikawa T, Kuchiba A, Yamauchi M, Meyerhardt JA, Shima K, Nosho K, et al. Association of CTNNB1 (beta-catenin) alterations, body mass index, and physical activity with survival in patients with colorectal cancer. JAMA. 2011;305:1685–94. doi: 10.1001/jama.2011.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jackson McCleary N, Meyerhardt J, Green E, Yothers G, de Gramont A, Van Cutsem E, et al. Impact of older age on the efficacy of newer adjuvant therapies in >12,500 patients (pts) with stage II/III colon cancer: Findings from the ACCENT Database. Journal of Clinical Oncology. 2009;27:4010. doi: 10.1200/JCO.2013.49.6638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meyerhardt JA, Giovannucci EL, Holmes MD, Chan AT, Chan JA, Colditz GA, et al. Physical activity and survival after colorectal cancer diagnosis. J Clin Oncol. 2006;24:3527–34. doi: 10.1200/JCO.2006.06.0855. [DOI] [PubMed] [Google Scholar]
- 25.Meyerhardt JA, Giovannucci EL, Ogino S, Kirkner GJ, Chan AT, Willett W, et al. Physical activity and male colorectal cancer survival. Arch Intern Med. 2009;169:2102–8. doi: 10.1001/archinternmed.2009.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meyerhardt JA, Heseltine D, Niedzwiecki D, Hollis D, Saltz LB, Mayer RJ, et al. Impact of physical activity on cancer recurrence and survival in patients with stage III colon cancer: findings from CALGB 89803. J Clin Oncol. 2006;24:3535–41. doi: 10.1200/JCO.2006.06.0863. [DOI] [PubMed] [Google Scholar]
- 27.Meyerhardt JA, Ogino S, Kirkner GJ, Chan AT, Wolpin B, Ng K, et al. Interaction of molecular markers and physical activity on mortality in patients with colon cancer. Clin Cancer Res. 2009;15:5931–36. doi: 10.1158/1078-0432.CCR-09-0496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morikawa T, Kuchiba A, Lochhead P, Nishihara R, Yamauchi M, Imamura Y, et al. Prospective analysis of body mass index, physical activity, and colorectal cancer risk associated with beta-catenin (CTNNB1) status. Cancer Res. 2013;73:1600–10. doi: 10.1158/0008-5472.CAN-12-2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Andre T, Boni C, Navarro M, Tabernero J, Hickish T, Topham C, et al. Improved overall survival with oxaliplatin, fluorouracil, and leucovorin as adjuvant treatment in stage II or III colon cancer in the MOSAIC trial. J Clin Oncol. 2009;27:3109–16. doi: 10.1200/JCO.2008.20.6771. [DOI] [PubMed] [Google Scholar]
- 30.Tournigand A, Bachet, Teixeira, Boni, Clingan, Hickish, Tabernero, de Gramont, for the Multicenter International Study of Oxaliplatin/5-Fluorouracil/Leucovorin in the Adjuvant Treatment of Colon Cancer (MOSAIC) Investigators and GERCOR FOLFOX4 as adjuvant therapy in elderly patients with colon cancer: Subgroup analysis of the MOSAIC trial. Journal of Clinical Oncology. 2010;28(supplement):3522. abstract. [Google Scholar]
- 31.Goldberg RM, Tabah-Fisch I, Bleiberg H, de Gramont A, Tournigand C, Andre T, et al. Pooled analysis of safety and efficacy of oxaliplatin plus fluorouracil/leucovorin administered bimonthly in elderly patients with colorectal cancer. J Clin Oncol. 2006;24:4085–91. doi: 10.1200/JCO.2006.06.9039. [DOI] [PubMed] [Google Scholar]
- 32.Sargent DJ, Goldberg RM, Jacobson SD, Macdonald JS, Labianca R, Haller DG, et al. A pooled analysis of adjuvant chemotherapy for resected colon cancer in elderly patients. N Engl J Med. 2001;345:1091–7. doi: 10.1056/NEJMoa010957. [DOI] [PubMed] [Google Scholar]
- 33.Ogino S, Shima K, Meyerhardt JA, McCleary NJ, Ng K, Hollis D, et al. Predictive and prognostic roles of BRAF mutation in stage III colon cancer: results from intergroup trial CALGB 89803. Clin Cancer Res. 2012;18:890–900. doi: 10.1158/1078-0432.CCR-11-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Colussi D, Brandi G, Bazzoli F, Ricciardiello L. Molecular pathways involved in colorectal cancer: implications for disease behavior and prevention. International journal of molecular sciences. 2013;14:16365–85. doi: 10.3390/ijms140816365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Phipps AI, Limburg PJ, Baron JA, Burnett-Hartman AN, Weisenberger DJ, Laird PW, et al. Association between molecular subtypes of colorectal cancer and patient survival. Gastroenterology. 2015;148:77–87 e2. doi: 10.1053/j.gastro.2014.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bishehsari F, Mahdavinia M, Vacca M, Malekzadeh R, Mariani-Costantini R. Epidemiological transition of colorectal cancer in developing countries: environmental factors, molecular pathways, and opportunities for prevention. World journal of gastroenterology: WJG. 2014;20:6055–72. doi: 10.3748/wjg.v20.i20.6055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seppala TT, Bohm JP, Friman M, Lahtinen L, Vayrynen VM, Liipo TK, et al. Combination of microsatellite instability and BRAF mutation status for subtyping colorectal cancer. Br J Cancer. 2015;112:1966–75. doi: 10.1038/bjc.2015.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yoon HH, Tougeron D, Shi Q, Alberts SR, Mahoney MR, Nelson GD, et al. KRAS codon 12 and 13 mutations in relation to disease-free survival in BRAF-wild-type stage III colon cancers from an adjuvant chemotherapy trial (N0147 alliance) Clin Cancer Res. 2014;20:3033–43. doi: 10.1158/1078-0432.CCR-13-3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Klaus A, Birchmeier W. Wnt signalling and its impact on development and cancer. Nat Rev Cancer. 2008;8:387–98. doi: 10.1038/nrc2389. [DOI] [PubMed] [Google Scholar]
- 40.Kawasaki T, Nosho K, Ohnishi M, Suemoto Y, Kirkner GJ, Dehari R, et al. Correlation of beta-catenin localization with cyclooxygenase-2 expression and CpG island methylator phenotype (CIMP) in colorectal cancer. Neoplasia. 2007;9:569–77. doi: 10.1593/neo.07334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu H, Fergusson MM, Castilho RM, Liu J, Cao L, Chen J, et al. Augmented Wnt signaling in a mammalian model of accelerated aging. Science. 2007;317:803–6. doi: 10.1126/science.1143578. [DOI] [PubMed] [Google Scholar]
- 42.Marchand A, Atassi F, Gaaya A, Leprince P, Le Feuvre C, Soubrier F, et al. The Wnt/beta-catenin pathway is activated during advanced arterial aging in humans. Aging cell. 2011;10:220–32. doi: 10.1111/j.1474-9726.2010.00661.x. [DOI] [PubMed] [Google Scholar]
- 43.Corr M. Wnt-beta-catenin signaling in the pathogenesis of osteoarthritis. Nature clinical practice Rheumatology. 2008;4:550–6. doi: 10.1038/ncprheum0904. [DOI] [PubMed] [Google Scholar]
- 44.Nan H, Morikawa T, Suuriniemi M, Imamura Y, Werner L, Kuchiba A, et al. Aspirin use, 8q24 single nucleotide polymorphism rs6983267, and colorectal cancer according to CTNNB1 alterations. J Natl Cancer Inst. 2013;105:1852–61. doi: 10.1093/jnci/djt331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Network TCGA. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–37. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ogino S, Chan AT, Fuchs CS, Giovannucci E. Molecular pathological epidemiology of colorectal neoplasia: an emerging transdisciplinary and interdisciplinary field. Gut. 2011;60:397–411. doi: 10.1136/gut.2010.217182. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


