Abstract
Several recent studies link parental environments to phenotypes in subsequent generations. Here, we investigate the mechanism by which paternal diet affects offspring metabolism. Protein restriction in mice affects small RNA levels in mature sperm, with decreased let-7 levels and increased levels of 5’ fragments of glycine tRNAs. tRNA fragments are scarce in testicular sperm, but are gained as sperm mature in the epididymis. Epididymosomes – vesicles that fuse with sperm during epididymal transit – carry RNA payloads matching those of mature sperm, and deliver RNAs to immature sperm in vitro. Functionally, tRNA-Gly-GCC fragments repress genes associated with the endogenous retroelement MERVL, both in ES cells and embryos. Our results shed light on small RNA biogenesis, and its dietary regulation, during post-testicular sperm maturation, and link tRNA fragments to regulation of endogenous retroelements active in the preimplantation embryo.
RESULTS
There is accumulating evidence that parental environments can affect the health of offspring. For example, paternal nutrition influences offspring metabolism in mammals(1). Our prior published work showed that male mice consuming Low Protein diet fathered offspring exhibiting altered hepatic cholesterol biosynthesis, relative to offspring of Control males(2). The mechanisms by which paternal conditions reprogram offspring phenotype remain elusive, as males can influence offspring via the sperm epigenome, microbiome transfer, seminal fluid signaling, or maternal judgement of mate quality(3,4). We therefore first tested whether metabolic gene expression was altered in offspring generated via in vitro fertilization (IVF) using sperm obtained from animals consuming Control or Low Protein (19% or 10% protein, respectively) diet. Despite the potential for IVF and embryo culture to obscure paternal effects on offspring metabolism, we nonetheless found that IVF-derived offspring of Low Protein males exhibited significant hepatic upregulation of the gene encoding the cholesterol biosynthesis enzyme squalene epoxidase, compared to Control IVF offspring (2)(Fig.S1), demonstrating that paternal diet can affect offspring metabolism via information located in sperm.
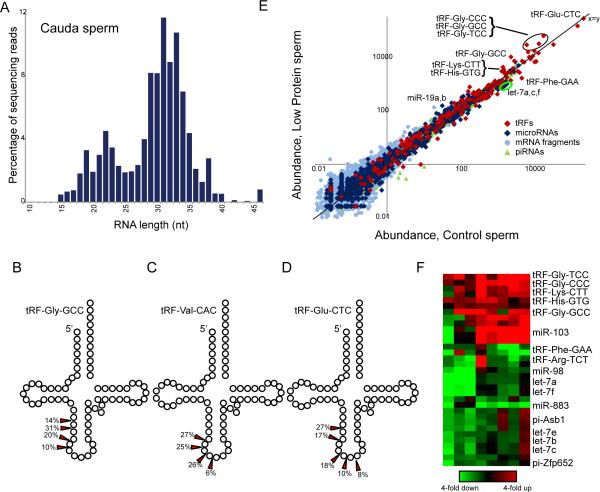
As small RNAs are central to a broad range of epigenetic phenomena(5), we isolated cauda sperm from males consuming Control or Low Protein diet, and purified small (<40nt) RNAs for analysis by deep sequencing. Resulting sequencing libraries reveal remarkably abundant (~80% of small RNAs) ~28-34nt tRNA fragments (tRFs), predominantly derived from the 5’ ends of tRNAs (Fig.1A-D,Tables S1-S2)(6). 5’ tRFs are also abundant in cauda sperm from the bull B. taurus (Table S3), suggesting that tRNA cleavage in gametes is conserved among mammals, and perhaps more broadly(7). Given the low RNA content of sperm relative to oocytes, we focus our analyses on highly abundant small RNAs in sperm. Low Protein diet affected levels of multiple small RNAs, including highly abundant tRNA fragments, across eight pairs of sperm samples (Fig.1E-F). Most notably, 5’ fragments of tRNA-Gly-CCC, TCC, and GCC exhibited a ~2-3-fold increase in Low Protein sperm, and tRF-Lys-CTT and tRF-His-GTG were similarly upregulated. In addition to tRFs, other RNA species differ in abundance between sperm samples, with several let-7 species being downregulated in Low Protein sperm.
Fig.1. Dietary effects on small RNAs in sperm.
(A) Size distribution of sequencing reads for cauda sperm small RNAs.
(B-D) 5’ tRNA fragments are shown schematically, with arrows indicating dominant 3’ ends.
(E) Dietary effects on sperm small RNA content. Scatterplot shows RNA abundance (ppm) for sperm isolated from Control (x axis, log10) compared to Low Protein sperm (y axis), with various RNA classes indicated. Multiple points for tRFs result from sequence differences between genes encoding a given tRNA isoacceptor.
(F) Heatmap showing RNAs responding to diet across eight paired sperm samples.
We next assayed levels of intact tRNAs in testis, finding no correlation between dietary effects on testicular tRNA levels and tRF changes in cauda sperm (Fig.S2). This argues against the hypothesis that tRFs in mature sperm result simply from random degradation of tRNAs utilized during spermatogenesis. Moreover, deep sequencing and Northern blot analyses (Figs.2A,C,S3,Tables S1-S2) revealed very low levels of tRNA fragments in testes or in various purified testicular spermatocyte/spermatid populations, raising the question of when sperm gain tRFs during maturation. After exiting the testis, sperm continue to mature for several days in the epididymis, and we find robust tRNA cleavage throughout this tissue (Figs.2B,D,S4). Not only do overall tRF levels increase distally in the male reproductive system, but the spectrum of specific tRFs differs between testis, proximal caput epididymis, and distal cauda epididymis (Fig.2D, Table S2).
Fig.2. tRNA cleavage predominantly occurs in the epididymis.
(A) Sperm RNA payload diverges dramatically from testicular RNA. Scatterplot shows small RNAs in testis vs. sperm, as in Fig.1E.
(B) Schematic of epididymis. Sperm exiting the testis enter the proximal (caput) epididymis, then proceed distally to corpus and cauda epididymis, and exit via the vas deferens.
(C) tRFs are primarily generated in epididymis. Northern blots for 5’ ends of tRNA-Gly-GCC or tRNA-Val-CAC on RNA extracted from testis, caput, and cauda epididymis. Arrow indicates ~30-34 nt 5’ tRFs. 5S RNA serves as loading control.
(D) Pie charts showing percentage of small RNAs mapping to the indicated features.
(E) Scatterplot of small RNA abundance for sperm vs. epididymosomes. Sperm-enriched RNAs include piRNAs and fragments of mRNAs involved in spermatogenesis (eg Prm1), and represent RNAs synthesized during testicular spermatogenesis.
Since our data suggest that small RNAs in mature sperm could have originated at multiple locations throughout the reproductive tract, we assessed the effect of paternal diet on small RNAs in testis (n=9 pairs), caput epididymis (n=6), and cauda epididymis (n=5) (Fig.S5). Intriguingly, two prominent dietary effects on the cauda sperm RNA repertoire – increased abundance of glycine tRFs, decreased abundance of let-7 – were recapitulated in the testis and epididymis, but not in liver, muscle, or blood (Table S1). Thus, tissues throughout the male reproductive tract – including mature sperm – exhibit consistent changes in glycine tRFs and let-7 in response to Low Protein diet, suggesting that similar diet-responsive pathways are present throughout the tract, and providing technical replication of the fundamental epigenomic changes wrought by Low Protein diet.
The finding of robust tRNA cleavage in the epididymis, but not testis, raises the possibility that the abundant tRFs in cauda sperm might be trafficked to sperm from the epididymal epithelium, rather than arising during testicular spermatogenesis. During transit through the epididymis, sperm fuse with small extracellular vesicles known as epididymosomes(8-11). To test the hypothesis that epididymosomes deliver small RNAs(12,13) to sperm, we purified epididymosomes (Fig.S6) and characterized their small RNA payload by deep sequencing. Epididymosomes carry high levels (~87% of reads) of 5’ tRFs such as tRF-Glu-CTC and tRF-Gly-GCC, and small RNAs found in purified epididymosomes closely mirror (r = 0.96) those in cauda sperm (Figs.2E,S6). Epididymosomal RNAs were resistant to RNAse treatment, and were found in epididymosomes from spermless Tdrd1-/- mice, ensuring that vesicles purified from the epididymis are not generated from maturing sperm (Fig.S6G).
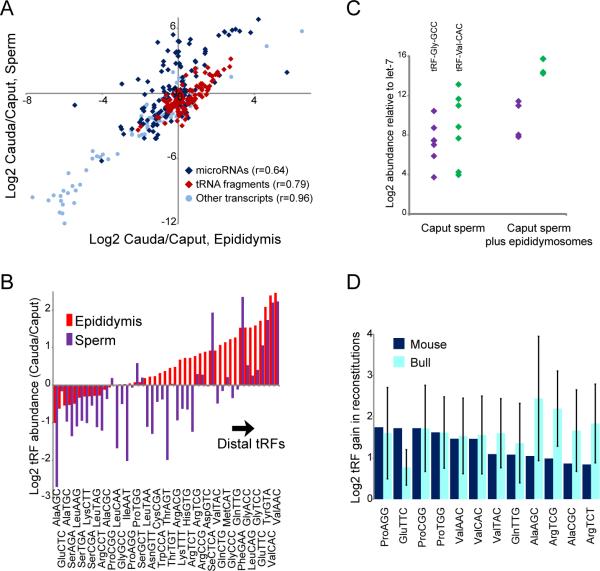
To further test the hypothesis that epididymosomes are responsible for shaping the RNA payload of sperm, we characterized small RNAs in sperm isolated from the proximal caput epididymis, finding that the RNA payload of caput sperm differs substantially from that of distal cauda sperm (Figs.3,S7)(14). Proximal-distal biases for specific tRFs along the epididymis were reflected in maturing sperm, showing a dramatic ~10-fold enrichment of tRF-Val-CAC, for example, in cauda relative to caput samples. To directly test whether epididymosomes can deliver their RNAs to caput sperm, we purified caput sperm and incubated them with cauda epididymosomes, then pelleted and washed resulting “reconstituted” sperm. Epididymosomal fusion with caput sperm was sufficient to deliver tRF-Val-CAC and other cauda-enriched tRFs to caput sperm in both mouse and bull (Fig.3C-D)(15). Taken together, these results are most consistent with a mechanism of RNA biogenesis in mammalian sperm in which tRFs generated in the epididymis are trafficked to sperm in epididymosomes, although they do not rule out the alternative hypothesis that intact tRNAs in immature sperm (Fig.S7F) are cleaved as sperm mature in the epididymis.
Fig.3. Changes in sperm tRF payload during epididymal transit.
(A) Proximal-distal biases observed for RNAs in epididymis are recapitulated in sperm samples.
(B) Proximal-distal biases for tRFs, aggregated by anticodon, in epididymis and sperm.
(C) TaqMan of the indicated tRFs in caput sperm and reconstituted sperm, showing gain of tRFs relative to let-7 (t-test p=0.05 for Gly-GCC, 0.004 for Val-CAC).
(D) Deep sequencing of reconstituted sperm. tRFs are aggregated by codon, and normalized to levels of tRF-Glu-CTC. Caput vs. cauda differences were broadly recapitulated in reconstitutions, with tRFs such as tRF-Val-CAC being delivered to caput sperm via fusion with cauda epididymosomes.
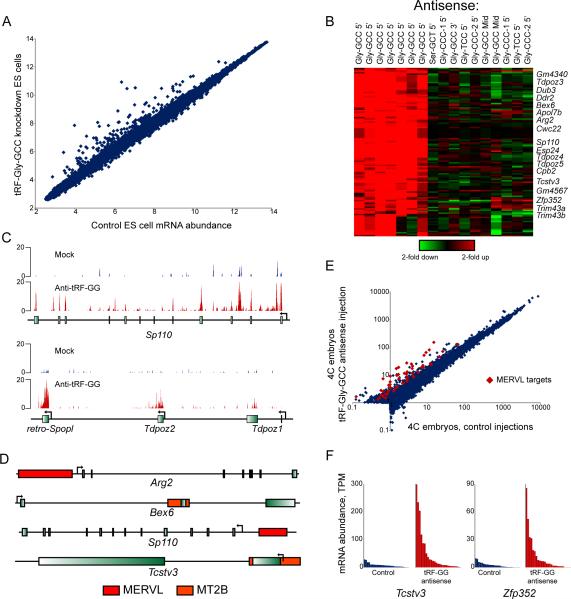
We next turned to potential functions of diet-regulated tRFs, using an embryonic stem (ES) cell system amenable to mechanistic analysis. Here, we interfered with the function of specific tRFs using antisense LNA-containing oligonucleotides, and assayed mRNA abundance as a readout of tRF inhibition. Most antisense oligos had no effect on mRNA abundance (Table S4), suggesting that the targeted tRFs are not functional in ES cells, or exert regulatory effects that are not assayed by mRNA abundance(16). In contrast, interfering with tRF-Gly-GCC resulted in dramatic upregulation of ~70 genes (Fig.4A-C). These genes were unaffected by antisense oligos against other tRNA-Gly isoacceptors or against the middle or 3’ end of tRNA-Gly-GCC (Fig.4B). Curiously, the genes derepressed upon tRF-Gly-GCC inhibition are highly expressed in preimplantation embryos, and are regulated by the long terminal repeat (LTR) of the endogenous retroelement MERVL(17) (Fig.4D). tRF-Gly-GCC regulation of MERVL LTR-driven transcription could be recapitulated in LTR reporter cell lines, and is not secondary to translational effects of tRF-Gly-GCC inhibition (Figs.S8,S9,Table S5). Microinjection of antisense oligos targeting tRF-Gly-GCC into zygotes also resulted in derepression of MERVL targets later in development (Fig.4E-F,Table S6), indicating that tRF-Gly-GCC also regulates MERVL in a more physiological context.
Fig.4. tRF-Gly-GCC regulates MERVL-driven transcripts.
(A) Scatterplot shows mRNA abundance in ESCs transfected with GFP siRNA vs. LNA antisense to 5’ end of tRF-Gly-GCC.
(B) Effect of tRF-Gly-GCC inhibition on MERVL-regulated genes is isoacceptor-specific. Affymetrix data for ESCs transfected with LNA antisense oligos, relative to matched GFP controls, showing genes changing 2-fold in 2 or more samples.
(C) RNA-Seq data for ESCs transfected with GFP siRNA, or anti-tRF-Gly-GCC.
(D) Genomic context of tRF-Gly-GCC target genes, showing nearby MERVL LTRs.
(E) tRF-Gly-GCC inhibition affects MERVL targets in embryos. Averaged single embryo RNA-Seq data for control (n=28) or tRF-inhibited (n=27) 4-cell stage embryos. Among 2-fold upregulated genes, known MERVL targets(17) are indicated.
(F) Single embryo data for two MERVL targets.
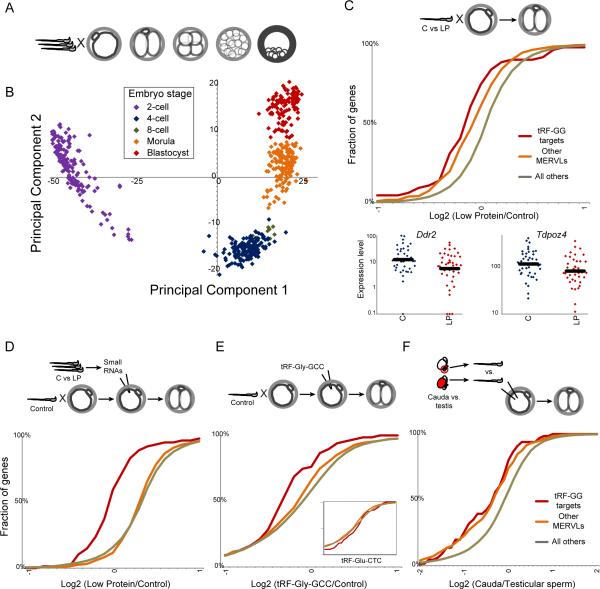
Given the robust connection between a diet-regulated small RNA and a highly specific set of target genes, we asked whether tRF-Gly-GCC targets are affected in preimplantation embryos generated using sperm from animals consuming Control or Low Protein diet. Single-embryo RNA-Seq(18,19) of individual mouse embryos cultured to various stages of development robustly clustered embryos by developmental stage (Figs.5A-B,S10,Table S7), with the first two principal components of the dataset representing oocyte-derived transcripts (PC1), and the products of embryonic genome activation (PC2).
Fig.5. Paternal dietary effects on preimplantation development.
(A) Embryos generated by IVF were cultured for varying times, then subject to single embryo RNA-Seq.
(B) Single-embryo data for preimplantation embryos represented via PCA: first two principal components explain 74% of dataset variance.
(C) mRNA abundance in 2-cell embryos generated via IVF using Control vs. Low Protein sperm (n=41 C and 39 LP). Cumulative distribution plots for tRF-Gly-GCC targets (p=4.5×10-7, KS test), other MERVL targets (17) (p=2.5×10-13), and all remaining genes, showing percentage of genes with the average Log2(LP/C) indicated on the x axis. Low Protein embryos exhibit a significant shift to lower expression of MERVL targets. Bottom panels show individual embryo data for two targets.
(D) Small RNAs isolated from Control or Low Protein cauda sperm were microinjected into control zygotes. RNA-Seq (n=42 C and 46 LP embryos) reveals downregulation of tRF-Gly-GCC targets (p=4.8×10-14) driven by Low Protein RNAs.
(E) Effects of synthetic tRF-Gly-GCC on 2-cell gene regulation, showing significant (p=0.0001) downregulation of target genes in embryos injected with tRF-Gly-GCC (n=26) vs. GFP controls (n=11). Inset shows effects of tRF-Glu-CTC (n=6).
(F) Effects of epididymal passage on embryonic gene regulation. Intact sperm isolated from rete testis (n=12), or cauda epididymis (n=9), were injected into control oocytes, and mRNA abundance was analyzed as above.
As single embryo RNA-Seq data are not suitable for identification of modest changes in individual mRNAs, we searched for consistent changes in larger genesets: the subset of MERVL targets that respond to tRF-Gly-GCC inhibition (Fig.4), and remaining MERVL targets(17). At the 2-cell stage both tRF-Gly-GCC targets and remaining MERVL targets were downregulated in Low Protein embryos relative to Control (Fig.5C), consistent with the hypothesis that tRF-Gly-GCC in sperm affects expression of MERVL targets in early embryos. We carried out several independent tests of this hypothesis. First, we injected <40nt RNA populations purified from Control and Low Protein sperm into control zygotes, finding that Low Protein RNAs could inhibit tRF-Gly-GCC targets in 2-cell embryos (Figs.5D,S11A), indicating that paternal diet can affect preimplantation gene regulation via RNAs in sperm. Second, further defining the relevant RNA from Low Protein sperm, microinjection of a synthetic tRF-Gly-GCC oligo resulted in repression of MERVL target genes in 2-cell embryos (Figs.5E,S11B). Finally, as tRFs in sperm are gained during epididymal transit (Fig.S3), we generated embryos via intracytoplasmic sperm injection (ICSI) using testicular spermatozoa or cauda sperm. Consistent with the higher levels of tRF-Gly-GCC in cauda sperm, embryos generated using cauda sperm expressed MERVL targets at lower levels than embryos generated using testicular sperm (Figs.5F,S12). Together, these findings all support the hypothesis that tRF-Gly-GCC in sperm is capable of delaying or repressing MERVL target expression in 2-cell embryos.
Lastly, we note that tRF-Gly-GCC is one of several abundant RNAs regulated by Low Protein diet, and MERVL-driven genes are not the only diet-responsive genes in preimplantation embryos. Most notably, ribosomal protein genes (RPGs) were downregulated in Low Protein embryos, and, correspondingly, Low Protein embryos develop slower than Controls (Fig.S13)(20). It remains to be determined whether altered preimplantation growth kinetics, or regulation of MERVL targets, could be responsible for eventual metabolic consequences in offspring. However, as the MERVL program is linked to totipotency(17), we speculate that tRF-Gly-GCC repression of MERVL-regulated genes could affect placental size or function, causing downstream effects on metabolism secondary to altered placentation(1).
Taken together, our data identify a role for paternal diet in regulating the sperm epigenome in mammals. We show 1) that paternal diet can influence offspring phenotype via information in sperm, 2) that diet alters the level of small RNAs, including tRNA fragments, throughout the male reproductive tract and in mature sperm, and 3) that tRNA fragments can regulate expression of transcripts driven by endogenous retroelements. Our data also illuminate temporal dynamics of small RNA biogenesis during post-testicular sperm maturation, and suggest that epididymosomes traffic RNAs from somatic cells of the epididymis to maturing gametes.
Supplementary Material
One sentence summary.
Abundant tRNA fragments in mammalian sperm are gained during post-testicular maturation in the epididymis, and regulate expression of endogenous retroelements upon delivery to the embryo.
“In Science” summary.
Paternal dietary conditions have been shown to influence metabolic phenotypes in offspring in mammals. Sharma et al show that paternal diet affects levels of small RNAs in mammalian sperm. Small RNAs such as tRNA fragments are gained as sperm exit the testis and transit the epididymis, suggesting a role for soma to germline communication in small RNA biogenesis in mammals. Functionally, a key diet-regulated tRNA fragment, tRF-Gly-GCC, was shown to repress genes linked to an endogenous retroelement active in the preimplantation embryo. These results illuminate surprising aspects of small RNA biogenesis and function in mammalian reproduction.
ACKNOWLEDGEMENTS
We thank H.Florman, P.Visconti, R.Sadeh, N.Friedman for reading the manuscript, P.Visconti, M.Jungnickel for training in caput sperm purification, K.Sutton for training in ICSI, A.Shalek, A.Regev for training in single cell RNA-Seq, T.Fazzio, S.Hainer, B.Chen for assistance with ES studies, and J.Lawrence, K.Creamer for help with microscopy. Research was supported by NIH grants R01HD080224, DP1ES025458, March of Dimes grant FY13-1268. CCC was supported by a Helen Hay Whitney Postdoctoral Fellowship, AB by a Human Frontiers Science Program Fellowship. Data can be accessed at GEO, accessions GSE74537,GSE75613,GSE75622.
Footnotes
SUPPLEMENTARY MATERIALS
Materials and Methods
Figs.S1 to S13
Datasets S1 to S8
References (21) to (35)
References
- 1.Rando OJ, Simmons RA. I'm Eating for Two: Parental Dietary Effects on Offspring Metabolism. Cell. 2015;161:93–105. doi: 10.1016/j.cell.2015.02.021. published online EpubMar 26 (10.1016/j.cell.2015.02.021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carone BR, Fauquier L, Habib N, Shea JM, Hart CE, Li R, Bock C, Li C, Gu H, Zamore PD, Meissner A, Weng Z, Hofmann HA, Friedman N, Rando OJ. Paternally induced transgenerational environmental reprogramming of metabolic gene expression in mammals. Cell. 2010;143:1084–1096. doi: 10.1016/j.cell.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Daxinger L, Whitelaw E. Understanding transgenerational epigenetic inheritance via the gametes in mammals. Nature reviews. Genetics. 2012;13:153–162. doi: 10.1038/nrg3188. [DOI] [PubMed] [Google Scholar]
- 4.Rando OJ. Daddy issues: paternal effects on phenotype. Cell. 2012;151:702–708. doi: 10.1016/j.cell.2012.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heard E, Martienssen RA. Transgenerational epigenetic inheritance: myths and mechanisms. Cell. 2014;157:95–109. doi: 10.1016/j.cell.2014.02.045. published online EpubMar 27 (10.1016/j.cell.2014.02.045) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peng H, Shi J, Zhang Y, Zhang H, Liao S, Li W, Lei L, Han C, Ning L, Cao Y, Zhou Q, Chen Q, Duan E. A novel class of tRNA-derived small RNAs extremely enriched in mature mouse sperm. Cell Res. 2012;22:1609–1612. doi: 10.1038/cr.2012.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Couvillion MT, Sachidanandam R, Collins K. A growth-essential Tetrahymena Piwi protein carries tRNA fragment cargo. Genes & development. 2010;24:2742–2747. doi: 10.1101/gad.1996210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dacheux JL, Dacheux F. New insights into epididymal function in relation to sperm maturation. Reproduction. 2013 doi: 10.1530/REP-13-0420. [DOI] [PubMed] [Google Scholar]
- 9.Sullivan R, Frenette G, Girouard J. Epididymosomes are involved in the acquisition of new sperm proteins during epididymal transit. Asian journal of andrology. 2007;9:483–491. doi: 10.1111/j.1745-7262.2007.00281.x. [DOI] [PubMed] [Google Scholar]
- 10.Sullivan R, Saez F. Epididymosomes, prostasomes, and liposomes: their roles in mammalian male reproductive physiology. Reproduction. 2013;146:R21–35. doi: 10.1530/REP-13-0058. [DOI] [PubMed] [Google Scholar]
- 11.Krapf D, Ruan YC, Wertheimer EV, Battistone MA, Pawlak JB, Sanjay A, Pilder SH, Cuasnicu P, Breton S, Visconti PE. cSrc is necessary for epididymal development and is incorporated into sperm during epididymal transit. Developmental biology. 2012;369:43–53. doi: 10.1016/j.ydbio.2012.06.017. published online EpubSep 1 (10.1016/j.ydbio.2012.06.017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nature cell biology. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 13.Regev-Rudzki N, Wilson DW, Carvalho TG, Sisquella X, Coleman BM, Rug M, Bursac D, Angrisano F, Gee M, Hill AF, Baum J, Cowman AF. Cell-cell communication between malaria-infected red blood cells via exosome-like vesicles. Cell. 2013;153:1120–1133. doi: 10.1016/j.cell.2013.04.029. [DOI] [PubMed] [Google Scholar]
- 14.Nixon B, Stanger SJ, Mihalas BP, Reilly JN, Anderson AL, Tyagi S, Holt JE, McLaughlin EA. The MicroRNA Signature of Mouse Spermatozoa Is Substantially Modified During Epididymal Maturation. Biology of reproduction. 2015 doi: 10.1095/biolreprod.115.132209. published online EpubSep 2 (10.1095/biolreprod.115.132209) [DOI] [PubMed] [Google Scholar]
- 15.Caballero JN, Frenette G, Belleannee C, Sullivan R. CD9-positive microvesicles mediate the transfer of molecules to Bovine Spermatozoa during epididymal maturation. PloS one. 2013;8:e65364. doi: 10.1371/journal.pone.0065364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sobala A, Hutvagner G. Transfer RNA-derived fragments: origins, processing, and functions. Wiley Interdiscip Rev RNA. 2011;2:853–862. doi: 10.1002/wrna.96. [DOI] [PubMed] [Google Scholar]
- 17.Macfarlan TS, Gifford WD, Driscoll S, Lettieri K, Rowe HM, Bonanomi D, Firth A, Singer O, Trono D, Pfaff SL. Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature. 2012;487:57–63. doi: 10.1038/nature11244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ramskold D, Luo S, Wang YC, Li R, Deng Q, Faridani OR, Daniels GA, Khrebtukova I, Loring JF, Laurent LC, Schroth GP, Sandberg R. Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nature biotechnology. 2012;30:777–782. doi: 10.1038/nbt.2282. published online EpubAug (10.1038/nbt.2282) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shalek AK, Satija R, Adiconis X, Gertner RS, Gaublomme JT, Raychowdhury R, Schwartz S, Yosef N, Malboeuf C, Lu D, Trombetta JJ, Gennert D, Gnirke A, Goren A, Hacohen N, Levin JZ, Park H, Regev A. Single-cell transcriptomics reveals bimodality in expression and splicing in immune cells. Nature. 2013;498:236–240. doi: 10.1038/nature12172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mitchell M, Bakos HW, Lane M. Paternal diet-induced obesity impairs embryo development and implantation in the mouse. Fertility and sterility. 2011;95:1349–1353. doi: 10.1016/j.fertnstert.2010.09.038. published online EpubMar 15 (10.1016/j.fertnstert.2010.09.038) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.