Abstract
The aim of the present study was to identify key genes and pathways in glioblastoma-associated stromal cells (GASCs) using bioinformatics. The expression profile of microarray GSE24100 was obtained from the Gene Expression Omnibus database, which included the expression profile of 4 GASC samples and 3 control stromal cell samples. Differentially expressed genes (DEGs) were identified using limma software in R language, and Gene Ontology and Kyoto Encyclopedia of Genes and Genomes pathway analysis of DEGs were performed using the Database for Annotation, Visualization and Integrated Discovery software. In addition, a protein-protein interaction (PPI) network was constructed. Subsequently, a sub-network was constructed to obtain additional information on genes identified in the PPI network using CFinder software. In total, 502 DEGs were identified in GASCs, including 331 upregulated genes and 171 downregulated genes. Cyclin-dependent kinase 1 (CDK1), cyclin A2, mitotic checkpoint serine/threonine kinase (BUB1), cell division cycle 20 (CDC20), polo-like kinase 1 (PLK1), and transcription factor breast cancer 1, early onset (BRCA1) were identified from the PPI network, and sub-networks revealed these genes as hub genes that were involved in significant pathways, including mitotic, cell cycle and p53 signaling pathways. In conclusion, CDK1, BUB1, CDC20, PLK1 and BRCA1 may be key genes that are involved in significant pathways associated with glioblastoma. This information may lead to the identification of the mechanism of glioblastoma tumorigenesis.
Keywords: glioblastoma-associated stromal cells, differentially expressed genes, pathways analysis, protein-protein interaction, bioinformatics analysis
Introduction
Glioblastoma is the most common and fatal malignant primary brain tumor in adults, with an incidence rate of 2.8 cases per 100,000 individuals per year and a perioperative mortality rate of 2.2% (1). It is estimated that 44,500 new cases of primary brain tumors were diagnosed in the USA in 2005, of which glioblastoma accounted for ~20% (2). The traditional treatment method is surgical resection combined with fractionated radiotherapy and adjuvant chemotherapy with temozolomide (3). However, despite advances in surgical techniques, postoperative supportive care, radiation and adjuvant systemic chemotherapy, the 5-year survival rate of glioblastoma remains at <10% (4). The disease generally recurs at the resection margin, and the median survival time is ~14 months; extremely few patients have a long-term survival, which highlights the importance of understanding the peripheral brain tumor region (5).
Glioblastoma cells are capable of infiltrating deep into the surrounding tissue, which allows these cells to migrate for long distances. This is typical behavior of neural stem cells, from which glioblastoma cells originate (6). Previous studies have demonstrated that malignant tumors may be affected by stromal cells, and that cancer cells may be controlled by the microenvironment; it has been reported that the non-neoplastic, stromal compartment of the majority of solid cancers is involved in tumor invasion, proliferation and metastasis (7–9).
In glioblastoma, a novel population of stromal cells that surround the tumor, termed glioblastoma-associated stromal cells (GASCs), has been isolated and analyzed. These cells have a different molecular expression profile compared with that of control stromal cells derived from non-glioblastoma peripheral brain tissues (7). GASCs have been revealed to have a phenotype and functional properties similar to that of cancer-associated fibroblasts located in the stroma of carcinomas, which are known to be important in the growth and progression of tumors (10). However, the genetic information concerning this novel cell population is relatively scarce.
The aim of the present study was to analyze the transcriptome and differentially expressed genes (DEGs) in GASCs. Bioinformatics analysis was performed using the microarray GSE24100, which is based on samples of GASCs and control stromal cells. In addition, functional and pathway enrichment analysis was performed and a protein-protein interaction (PPI) network was constructed. A sub-network was also constructed for additional analysis.
Materials and methods
Microarray data
Microarray data was obtained from the study by Clavreul et al (7), which is referenced in the Gene Expression Omnibus (GEO) database (www.ncbi.nlm.nih.gov/geo/) under accession number GSE24100. The microarray GSE24100 was detailed using Whole Human Genome Microarray 4×44K (catalog no., G4112F; design ID, 014850; Agilent Technologies, Santa Clara, CA, USA), and the data contains a total of 7 samples, consisting of 3 control stromal cell samples and 4 GASC samples.
Data preprocessing and DEG analysis
Using the limma model (11) on R/Bioconductor software version 2.15.1 (www.bioconductor.org/) and the microarray probe annotation profile from Brain Array Lab (brainarray.mbni.med.umich.edu/Brainarray/), the probe-level data was converted into expression measures, during which background correction, quantile normalization and probe summarization were performed. A t-test (12) was used to identify the significantly expressed DEGs in GASC samples, with a combination of P<0.05 and the |log2FC (fold change)| >1 used as the threshold. A heat map was generated using Z-score normalization of log2 expression values to illustrate the relative expression levels of DEGs in GASCs.
Gene ontology (GO) and pathway enrichment analysis of DEGs
GO is a commonly used approach for functional studies, and three independent ontologies (biological process, molecular function and cellular component) are accessible on the world-wide web (www.geneontology.org) (13). Kyoto Encyclopedia of Genes and Genomes (KEGG; www.genome.jp/kegg/) is a knowledge base for the systematic analysis of gene functions, which links genomic information with higher order functional information (14). In the present study, GO biological processes and KEGG pathway analysis were performed using the Database for Annotation, Visualization and Integrated Discovery; (http://david.abcc.ncifcrf.gov/home.jsp) (15) where P<0.05.
Functional annotation of DEGs
Functional annotation of DEGs was performed for the detection of transcription factors and tumor-associated genes. Two databases, Tumor Suppressor Gene Database version 2.0 (16) (bioinfo.mc.vanderbilt.edu/TSGene/)and Tumor Associated Gene database (last modified, 10/03/2014) (17) (www.binfo.ncku.edu.tw/TAG/GeneDoc.php) were used to screen tumor suppressor genes and oncogenes.
PPI network construction
The Search Tool for the Retrieval of Interacting Genes/Proteins (STRING; string-db.org/) database is a pre-computed global resource for the investigation and analysis of associations between proteins. The database reveals protein interactions, including experimental and predicted protein interaction information (18). In the present study, STRING was used to analyze the interactions between DEGs with the PPI required confidence (combined score, 0.9) and a PPI network was constructed. In addition, the degree of the nodes in the PPI network were calculated, and the nodes with a higher degree were deemed to be hub proteins compared with the other nodes in the PPI network.
Selection and pathway enrichment analysis of sub-network
To obtain additional information on the genes identified in the PPI network, a sub-network was constructed using CFinder (www.cfinder.org/) and Clique Percolation Method (k=3) (19). Four networks were obtained, but only one was associated with additional nodes and was additionally analyzed. GO and KEGG enrichment analysis were performed on the sub-network for the majority of nodes, and the interactions were selected using CFinder version 2.0.5 for the identification of significant pathways.
Results
DEG selection
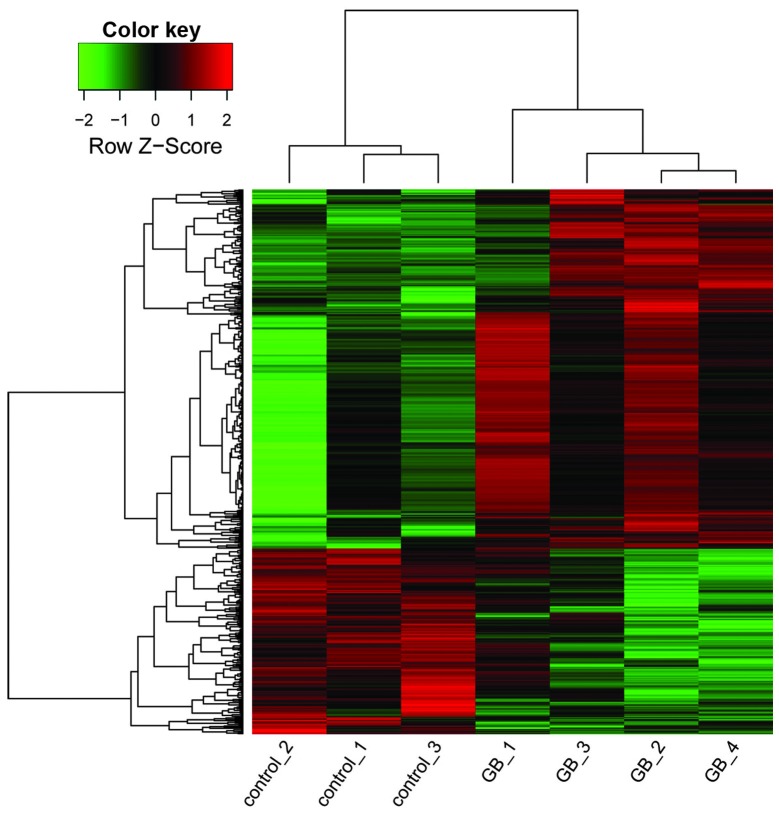
In total, 512 transcripts were observed to be expressed differentially, including 337 upregulated transcripts and 175 downregulated transcripts, corresponding to 331 upregulated genes and 171 downregulated genes. The heat map of DEGs in GASCs and control stromal cells is shown in Fig. 1.
Figure 1.
Heat map of gene expression in GB-associated stromal and control stromal cells. The rows indicate the expression levels of genes, and columns indicate different samples. Red, upregulated genes; green, downregulated genes. GB, glioblastoma.
GO categories and KEGG pathway enrichment analysis of DEGs
Pathways obtained by KEGG enrichment are presented in Table I. According to the results, upregulated genes were primarily enriched in pathways associated with the cell cycle, DNA replication, oocyte meiosis and p53 signaling (Table IA). Downregulated genes were primarily enriched in pathways associated with adipocytokine signaling, aldosterone-regulated sodium reabsorption and nucleotide oligomerization domain-like receptor signaling (Table IB).
Table I.
Enriched GO categories and KEGG pathway enrichment analysis of DEGs in glioblastoma-associated stromal cells.
| A, KEGG analysis of upregulated DEGs. | ||||
|---|---|---|---|---|
| Category | Term | Biological process | Count | P-value |
| KEGG | 4110 | Cell cycle | 124 | 0 |
| KEGG | 3030 | DNA replication | 36 | 3.47×10−13 |
| KEGG | 4114 | Oocyte meiosis | 112 | 5.49×10−10 |
| KEGG | 4914 | Progesterone-mediated oocyte maturation | 86 | 2.10×10−7 |
| KEGG | 4115 | p53 signaling pathway | 68 | 2.13×10−6 |
| KEGG | 3430 | Mismatch repair | 23 | 5.90×10−4 |
| KEGG | 240 | Pyrimidine metabolism | 99 | 1.57×10−3 |
| KEGG | 3410 | Base excision repair | 33 | 2.39×10−3 |
| KEGG | 3420 | Nucleotide excision repair | 44 | 6.85×10−3 |
| KEGG | 3440 | Homologous recombination | 28 | 1.22×10−2 |
| B, KEGG analysis of downregulated DEGs. | ||||
| Category | Term | Biological process | Count | P-value |
| KEGG | 4920 | Adipocytokine signaling pathway | 68 | 3.85×10−3 |
| KEGG | 4960 | Aldosterone-regulated sodium reabsorption | 42 | 7.26×10−3 |
| KEGG | 4621 | NOD-like receptor signaling pathway | 58 | 1.75×10−2 |
| KEGG | 4964 | Proximal tubule bicarbonate reclamation | 23 | 1.99×10−2 |
| KEGG | 640 | Propanoate metabolism | 32 | 3.69×10−2 |
| KEGG | 4060 | Cytokine-cytokine receptor interaction | 265 | 3.93×10−2 |
| C, GO analysis of upregulated DEGs. | ||||
| Category | Term | Biological process | Count | P-value |
| BP | GO:0000070 | Mitotic sister chromatid segregation | 53 | 0 |
| BP | GO:0000075 | Cell cycle checkpoint | 226 | 0 |
| BP | GO:0000226 | Microtubule cytoskeleton organization | 297 | 0 |
| BP | GO:0000278 | Mitotic cell cycle | 816 | 0 |
| BP | GO:0000280 | Nuclear division | 346 | 0 |
| BP | GO:0000819 | Sister chromatid segregation | 56 | 0 |
| BP | GO:0006259 | DNA metabolic process | 896 | 0 |
| BP | GO:0006260 | DNA replication | 277 | 0 |
| BP | GO:0006261 | DNA-dependent DNA replication | 100 | 0 |
| BP | GO:0006270 | DNA replication initiation | 29 | 0 |
| D, GO analysis of downregulated DEGs. | ||||
| Category | Term | Biological process | Count | P-value |
| BP | GO:0008217 | Reg. of blood pressure | 147 | 9.85×10−6 |
| BP | GO:0045776 | Negative regulation of blood pressure | 35 | 3.08×10−4 |
| BP | GO:0071260 | Cellular response to mechanical stimulus | 57 | 1.98×10−3 |
| BP | GO:0035094 | Response to nicotine | 31 | 2.98×10−3 |
| BP | GO:0016486 | Peptide hormone processing | 32 | 3.27×10−3 |
| BP | GO:0002864 | Reg. of acute inflammatory response to antigenic stimulus | 10 | 3.73×10−3 |
| BP | GO:0031272 | Reg. of pseudopodium assembly | 10 | 3.73×10−3 |
| BP | GO:0016485 | Protein processing | 160 | 3.95×10−3 |
| BP | GO:0051239 | Reg. of multicellular organismal processes | 1963 | 4.20×10−3 |
| BP | GO:0006952 | Defense response | 1372 | 4.25×10−3 |
GO, gene ontology; DEG, differentially expressed gene; BP, biological process; KEGG, Kyoto Encyclopedia of Genes and Genomes; Reg., regulation.
Several GO categories were enriched among DEGs and are shown in Table I. The upregulated genes were primarily enriched in categories associated with mitotic sister chromatid segregation, cell cycle checkpoint and DNA metabolic processes, which are all associated with cell mitosis and DNA replication (Table IC). Among downregulated genes, categories with increased transcripts included regulation of blood pressure and cellular response to mechanical stimulus (Table ID).
Functional annotation of DEGs
According to the annotation results (Table II), 11 transcriptional factors were upregulated, including breast cancer 1, early onset (BRCA1) and BRCA1 interacting protein C-terminal helicase 1, and 6 transcriptional factors were downregulated, including ary-hydrocarbon receptor nuclear translocator 2 and DNA damage inducible transcript 3 (DDIT3).
Table II.
Functional annotation of differentially expressed genes in glioblastoma-associated stromal cells.
| Category | n | Gene |
|---|---|---|
| Upregulated | ||
| TF | 11 | BRCA1, BRIP1, CDK2, HEYL, HMGB2, IRX5, MEF2C, MEIS2, MYBL2, RBL1, TBX2 |
| TAG oncogene | 9 | CCNA2, CCND2, CEP55, DUSP26, FGF5, HGF, MYBL2, NET1, PTTG1 |
| TAG tumor suppressor | 23 | AKAP12, BARD1, BLM, BMP2, BRCA1, BUB1B, CDH13, CHEK1, DAB2IP, E2F1, FANCD2, ID4, ITGB3, LIMD1, LIN9, MFSD2A, PCDH10, PTPN3, RBL1, STARD13, TFPI2, TMEFF2, ZFHX3 |
| Downregulated | ||
| TF | 6 | ARNT2, DDIT3, HES2, MITF, NFIA, NR3C2 |
| TAG oncogene | 3 | ARHGEF5, DDIT3, MRAS |
| TAG tumor suppressor | 10 | ATP8A2, BHLHE41, CABLES1, CDH4, DAB2, HRASLS2, LGI1, PLA2G16, RARRES3, RPS6KA2 |
TF, transcription factor; TAG, tumor-associated genes. BRCA1, breast cancer 1, early onset; BRIP1, BRCA1 interacting protein C-terminal helicase 1; CDK2, cyclin-dependent kinase 2; HEYL, hes-related family bHLH transcription factor with YRPW motif-like; HMGB2, high mobility group box 2; IRX5, iroquois homeobox 5; MEF2C, myocyte enhancer factor 2C; MEIS2, Meis homeobox 2; MYBL2, v-myb avian myeloblastosis viral oncogene homolog-like 2; RBL1, retinoblastoma-like 1; TBX2, T-box 2; CCNA2, cyclin A2; CCND2, cyclin D2; CEP55, centrosomal protein 55kDa; DUSP26, dual specificity phosphatase 26 (putative); FGF5, fibroblast growth factor 5; HGF, hepatocyte growth factor; NET1, neuroepithelial cell transforming 1; PTTG1, pituitary tumor-transforming 1; AKAP12, a kinase anchor protein; BARD1, BRCA1 associated RING domain 1; BLM, Bloom syndrome RecQ like helicase; BMP2, bone morphogenetic protein 2; BUB1B, BUB1 mitotic checkpoint serine/threonine kinase B; CDH13, cadherin 13; CHEK1, checkpoint kinase 1; DAB2IP, DAB2 interacting protein; E2F1, E2F transcription factor 1; FANCD2, Fanconi anemia complementation group D2; ID4, inhibitor of DNA binding 4, dominant negative helix-loop-helix protein; ITGB3, integrin subunit beta 3; LIMD1, LIM domains containing 1; LIN9, lin-9 DREAM MuvB core complex component; MFSD2A, major facilitator superfamily domain containing 2A; PCDH10, protocadherin 10; PTPN3, protein tyrosine phosphatase, non-receptor type 3; STARD13, StAR related lipid transfer domain containing 13; TFPI2, tissue factor pathway inhibitor 2; TMEFF2, transmembrane protein with EGF like and two follistatin like domains 2; ZFHX3, zinc finger homeobox 3; ARNT2, ary-hydrocarbon receptor nuclear translocator 2; DDIT3, DNA damage inducible transcript 3; HES2, hairy and enhancer of split 2; MITF, microphthalmia-associated transcription factor; NFIA, nuclear factor I/A; NR3C2, nuclear receptor subfamily 3 group C member 2; ARHGEF5, Rho guanine nucleotide exchange factor 5; MRAS, muscle RAS oncogene homolog; ATP8A2, ATPase, aminophospholipid transporter, class I, type 8A, member 2; BHLHE41, basic helix-loop-helix family member e41; CABLES1, Cdk5 and Abl enzyme substrate 1; CDH4, cadherin 4, type 1, R-cadherin; DAB2, Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila); HRASLS2, HRAS like suppressor 2; LGI1, leucine-rich, glioma inactivated 1; PLA2G16, phospholipase A2 group XVI; RARRES3, retinoic acid receptor responder (tazarotene induced) 3; RPS6KA2, ribosomal protein S6 kinase, 90kDa, polypeptide 2.
Additionally, among the upregulated genes, 9 oncogenes were identified [including cyclin A2 (CCNA2) and cyclin D2 (CCND2)] in addition to 23 tumor suppressor genes (including kinase anchoring protein 12 and BRCA1-associated RING domain 1). The downregulated genes included 3 oncogenes (such as Rho guanine nucleotide exchange factor 5 and DDIT3) and 10 tumor suppressor genes (such as cadherin 4, type 1, R-cadherin and ATPase, aminophospholipid transporter, class I, type 8A, member 2). The details are presented in Table II.
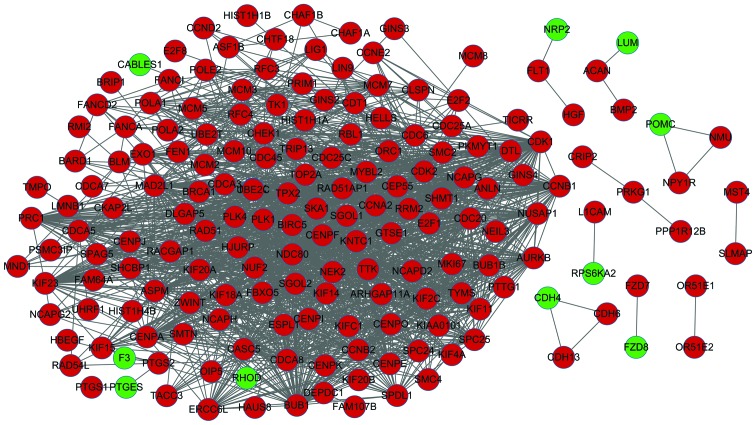
PPI network construction
The PPI network constructed for the DEGs is shown in Fig. 2, in which 181 nodes and 1,740 interactions were identified. In this network, 8 nodes with higher degrees were identified, including cyclin-dependent kinase 1 (CDK1), CCNA2, mitotic checkpoint serine/threonine kinase (BUB1), cell division cycle 20 (CDC20), kinetochore complex component 80 (NDC80), non-SMC condensing I complex, subunit G (NCAPG), cell division cycle associated-8 and polo-like kinase 1 (PLK1).
Figure 2.
Protein-protein interaction network of differentially expressed genes in glioblastoma-associated stromal cells. Red nodes, upregulated genes; green nodes, downregulated genes.
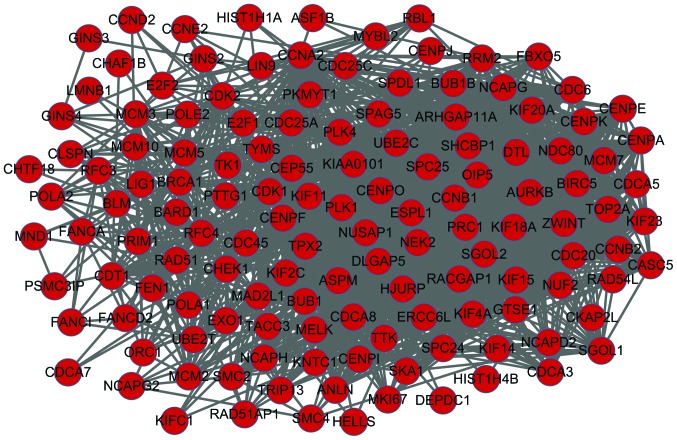
Analysis of sub-network
The sub-network obtained using CFinder is shown in Fig. 3, in which 135 nodes and 1,694 interactions were identified, and all nodes were upregulated genes. KEGG enrichment in the sub-network is presented in Table IIIA; cell cycle, p53 signaling pathway, oocyte meiosis and progesterone-mediated oocyte maturation were the predominant pathways enriched by these DEGs. GO enrichment analysis was also performed and is presented in Table IIIB; mitotic cell cycle, DNA metabolic process and nuclear division were the predominant categories.
Figure 3.
Sub-network constructed from the protein-protein interaction network. Red nodes, upregulated genes.
Table III.
GO terms and KEGG pathways of DEGs in the sub-network.
| A, Enriched pathways of DEGs in sub-network | ||||
|---|---|---|---|---|
| Category | Term | Biological Process | Count | P-value |
| KEGG | 3030 | DNA replication | 12 | 0 |
| KEGG | 4110 | Cell cycle | 29 | 0 |
| KEGG | 4114 | Oocyte meiosis | 15 | 1.55×10−14 |
| KEGG | 4914 | Progesterone-mediated oocyte maturation | 11 | 1.29×10−10 |
| KEGG | 4115 | p53 signaling pathway | 9 | 5.41×10−9 |
| KEGG | 240 | Pyrimidine metabolism | 7 | 2.25×10−5 |
| KEGG | 3430 | Mismatch repair | 4 | 4.28×10−5 |
| KEGG | 3420 | Nucleotide excision repair | 4 | 5.72×10−4 |
| KEGG | 3440 | Homologous recombination | 3 | 1.84×10−3 |
| KEGG | 3410 | Base excision repair | 3 | 2.97×10−3 |
| B, Enriched GO terms of DEGs in sub-network | ||||
| Category | Term | Biological Process | Count | P-value |
| BP | GO:0000070 | Mitotic sister chromatid segregation | 20 | 0 |
| BP | GO:0000075 | Cell cycle checkpoint | 27 | 0 |
| BP | GO:0000082 | G1/S transition of mitotic cell cycle | 24 | 0 |
| BP | GO:0000226 | Microtubule cytoskeleton organization | 36 | 0 |
| BP | GO:0000278 | Mitotic cell cycle | 101 | 0 |
| BP | GO:0000280 | Nuclear division | 62 | 0 |
| BP | GO:0000819 | Sister chromatid segregation | 21 | 0 |
| BP | GO:0006259 | DNA metabolic process | 72 | 0 |
| BP | GO:0006260 | DNA replication | 39 | 0 |
| BP | GO:0006261 | DNA-dependent DNA replication | 22 | 0 |
GO, gene ontology; DEG, differentially expressed gene; BP, biological process; KEGG, Kyoto Encyclopedia of Genes and Genomes.
Discussion
Glioblastoma is the most aggressive cerebral tumor in humans, and has a high annual mortality rate (20). GASCs represent a novel stromal cell population that express mesenchymal markers and exert tumor-promoting effects (7). In the present study, 3 samples of GASCs and 4 of control stromal cells were used to identify DEGs, and the functional categories associated with those DEGs, that are altered between GASCs and control stromal cells in glioblastoma. In total, 502 DEGs were identified, including 331 upregulated genes and 171 downregulated genes, including CDK1, BUB1, CDC20, CCNA2, NDC80, NCAPG and PLK1. These are hub genes and serve major roles in pathways of the cell cycle, p53 signaling, oocyte meiosis and progesterone-mediated oocyte maturation as determined from the results of KEGG pathway enrichment analysis. In addition, the upregulated gene BRCA1 was identified to be a transcription factor. The predominant pathway in which the majority of hub genes were enriched was the cell cycle, which is expected as glioblastoma cell invasion requires that cells have enhanced motility and the ability to degrade local tissue barriers (21).
CDK1 protein belongs to the CDK family, which controls the cell cycle by catalyzing the transfer of phosphate from ATP to specific protein substrates. CDKs have been established as master regulators of cell proliferation (22). As expected, in the present study, CDK1 was upregulated in GASCs and was primarily enriched in pathways involved in the cell cycle, mitotic cell cycle and DNA replication, all of which are closely associated with the mechanisms of tumor growth (23,24). In the cell cycle, CDK1 controls a widespread regulatory system, which involves phosphorylation of other regulatory molecules and phosphorylation of the molecular machinery that drives the cell-cycle (25). Furthermore, in the current study, CDK1 was observed to be enriched in the p53 signaling pathway, which is induced by a number of stress signals, including DNA damage, oxidative stress and activated oncogenes. The p53 signaling network is an integral tumor suppressor pathway in glioblastoma pathogenesis that affects cellular processes, including cell cycle control and cell death execution (26). In this pathway, the tumor suppressor p53 protein acts as a transcriptional activator of p53-regulated genes (27) and is primarily involved in control of numerous genes governing cell survival, cell proliferation, angiogenesis and metabolism (28). Stegh et al (26) reported that the p53 signaling pathway is inhibited in glioblastoma, which causes aberrant cell cycling and tumorigenesis. In the present study, several DEGs were enriched in the p53 signaling pathway, including CDK1, CDK2, CCNB1 and CCND2, which may be associated with the inhibition of p53 signaling (29). Therefore, according to the current study, upregulated CDK1 may increase the growth of glioblastoma by promoting cell cycle pathways and inhibiting the p53 signaling pathway.
BUB1 was identified to be upregulated in the present study, and was primarily enriched in biological processes associated with the mitotic cell cycle, including cell cycle chromatid segregation, G1/S transition of mitotic cells and DNA replication. The BUB family of genes encode proteins that are involved in a large multi-protein kinetochore complex, and are hypothesized to be key components of the checkpoint regulatory pathway (30). BUB1 encodes a serine/threonine-protein kinase that is critical in mitosis, and functions partly through the phosphorylation of members of the mitotic checkpoint complex and activation of the spindle checkpoint (31). BUB1 accumulates at unattached kinetochores where it mediates the recruitment of mitotic arrest deficient (Mad) dimers (32). Combined with Mad, BUB1 prevents the premature separation of sister chromatids until all the chromosomes are correctly attached to kinetochores, which leads to correct chromosome segregation (33). Therefore, BUB1 may promote the growth of cancer cells in glioblastoma primarily by regulating the mitotic cell cycle. In addition, it appears that the mutation of mitotic spindle checkpoint genes is associated with the evolution of certain human cancers, particularly those with aneuploidy (34). Glioblastoma exhibits a high degree of aneuploidy (35) and the upregulation of BUB1 in GASCs may increase the tumorigenesis of glioblastoma.
CDC20 appears to act as a regulatory protein interacting with several other proteins at multiple points in the cell cycle (36). In the present study, the CDC20 gene was upregulated and enriched in cell cycle and oocyte meiosis pathways. CDC20 is an activator protein that regulates the anaphase-promoting complex ubiquitin ligase, which is considered to be crucial in governing certain cellular processes (37), including the interaction with specific ubiquitin substrates for their subsequent degradation by the 26S proteasome at various points during cell cycle progression; this results in the forwards progression of the cell cycle in a unidirectional manner (38). Previous studies have demonstrated that CDC20 is highly expressed in various types of human tumors, including breast (39) and cervical cancer (40), where it functions as an oncoprotein. Marucci et al (41) reported that, in glioblastoma, CDC20 expression is upregulated, which is consistent with the present results. This implies that CDC20 may promote glioblastoma occurrence by regulating cellular processes. In addition, Bie et al (42) observed that the expression levels of mitotic spindle assembly checkpoint gene CDC20 is correlated with the grade of glioblastoma. The expression of CDC20 is regulated by BRCA1, a susceptibility gene that greatly increases the risk of breast and other types of cancer (43), and is expressed differently depending on the age of the patient (44). In the present study, BRCA1 and its target gene, CDC20, were upregulated. This leads to the hypothesis that BRCA1 acts on glioblastoma, and is expressed at various levels in patients of various ages, regulating the expression of target genes that are associated with tumor grade or age of the patient, including CDC20. Therefore, BRCA1 and its target genes are of significant value in clinical research, and BRCA1 may be used as an anti-cancer drug target.
According to the present study, PLK1 was upregulated and enriched in pathways associated with the cell cycle, oocyte meiosis and progesterone-mediated oocyte maturation. PLK1 is a serine/threonine kinase and is critical in centrosome maturation (45), mitotic entry (46), bipolar spindle formation (47,48), metaphase-to-anaphase transition (49) and cytokinesis (50) in the mitotic phase of the cell cycle. Foong et al (51) demonstrated that increased expression of PLK1 is an independent, negative prognostic factor in glioma and is associated with proliferative and mesenchymal molecular subclasses, which characterize highly recurrent and aggressive tumors (52). PLK1 has become a primary target in brain tumor treatment, and its inhibition has been shown to result in 80–90% growth suppression in a panel of pediatric cancer cells, including glioblastoma, following 72 h of treatment (52). Therefore, in GASCs, PLK1 upregulation may promote the cell cycle, leading to the growth of glioblastoma.
CCNA2 belongs to a highly conserved cyclin family and is expressed in almost all tissues of the human body (53). The encoded protein is crucial in the control of the cell cycle at G1/S and G2/M transition points, and this is essential in embryonic cells and the hematopoietic lineage (54). Overexpression of CCNA2 is involved in tumor transformation and progression in numerous types of cancer (55). Another member of the cyclin family, CCND2, is critical in cell cycle progression and tumorigenicity of glioblastoma stem cells (56). As expected, the present data revealed that CCNA2 was upregulated, which is in accordance with the function of CCNA2 in cancer. According to the pathway enrichment results, CCNA2 was enriched in cell cycle and progesterone-mediated oocyte maturation pathways, in which CDK1, BUB1 and PLK1 were also involved. The present results indicate that CCNA2 promotes the growth of glioblastoma by participating in the cell cycle. However, few studies have reported the association between oocyte maturation and glioblastoma, revealing that this may be a novel insight in glioblastoma.
In conclusion, the present study identified several significant genes in glioblastoma, including CDK1, BUB1, CDC20, CCNA2, PLK1 and BRCA1, which are all upregulated and may play various roles in the biological function of GASCs. These significant DEGs may promote the tumorigenesis of glioblastoma as they are involved in major biological pathways, including cell cycle, mitosis, p53 signaling and DNA replication. However, since the sample size used in this study is small and no experiments have been performed to confirm the conclusions, additional analyses of experimental studies are required to investigate the genes associated with glioblastoma.
References
- 1.McLendon RE, Rich JN. Glioblastoma stem cells: A Neuropathologist's View. J Oncol. 2011;2011:397195. doi: 10.1155/2011/397195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.States CBTRotU: CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2004–2006. 2010 doi: 10.1093/neuonc/not151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stupp R, Hegi ME, Mason WP, van den Bent MJ, Taphoorn MJ, Janzer RC, Ludwin SK, Allgeier A, Fisher B, Belanger K, et al. European Organisation for Research and Treatment of Cancer Brain Tumour and Radiation Oncology Groups; National Cancer Institute of Canada Clinical Trials Group: Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10:459–466. doi: 10.1016/S1470-2045(09)70025-7. [DOI] [PubMed] [Google Scholar]
- 4.Thumma SR, Fairbanks RK, Lamoreaux WT, Mackay AR, Demakas JJ, Cooke BS, Elaimy AL, Hanson PW, Lee CM. Effect of pretreatment clinical factors on overall survival in glioblastoma multiforme: A surveillance epidemiology and end results (SEER) population analysis. World J Surg Oncol. 2012;10:75. doi: 10.1186/1477-7819-10-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tseng YY, Liao JY, Chen WA, Kao YC, Liu SJ. Sustainable release of carmustine from biodegradable poly[(d,l)-lactide-co-glycolide] nanofibrous membranes in the cerebral cavity: In vitro and in vivo studies. Expert Opin Drug Deliv. 2013;10:879–888. doi: 10.1517/17425247.2013.758102. [DOI] [PubMed] [Google Scholar]
- 6.Gangemi RM, Griffero F, Marubbi D, Perera M, Capra MC, Malatesta P, Ravetti GL, Zona GL, Daga A, Corte G. SOX2 silencing in glioblastoma tumor-initiating cells causes stop of proliferation and loss of tumorigenicity. Stem Cells. 2009;27:40–48. doi: 10.1634/stemcells.2008-0493. [DOI] [PubMed] [Google Scholar]
- 7.Clavreul A, Etcheverry A, Chassevent A, Quillien V, Avril T, Jourdan ML, Michalak S, François P, Carré JL, Mosser J, et al. Isolation of a new cell population in the glioblastoma microenvironment. J Neurooncol. 2012;106:493–504. doi: 10.1007/s11060-011-0701-7. [DOI] [PubMed] [Google Scholar]
- 8.Mao Y, Keller ET, Garfield DH, Shen K, Wang J. Stromal cells in tumor microenvironment and breast cancer. Cancer Metastasis Rev. 2013;32:303–315. doi: 10.1007/s10555-012-9415-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Calon A, Espinet E, Palomo-Ponce S, Tauriello DV, Iglesias M, Céspedes MV, Sevillano M, Nadal C, Jung P, Zhang XH, et al. Dependency of colorectal cancer on a TGF-β-driven program in stromal cells for metastasis initiation. Cancer cell. 2012;22:571–584. doi: 10.1016/j.ccr.2012.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Östman A, Augsten M. Cancer-associated fibroblasts and tumor growth-bystanders turning into key players. Curr Opin Genet Dev. 2009;19:67–73. doi: 10.1016/j.gde.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 11.López-Romero P, González MA, Callejas S, Dopazo A, Irizarry RA. Processing of agilent microRNA array data. BMC Res Notes. 2010;3:18. doi: 10.1186/1756-0500-3-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hulsegge I, Kommadath A, Smits MA. Globaltest and GOEAST: Two different approaches for gene ontology analysis. BMC Proc. 2009;3(Suppl 4):S10. doi: 10.1186/1753-6561-3-s4-s10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. doi: 10.1186/gb-2003-4-5-p3. [DOI] [PubMed] [Google Scholar]
- 16.Zhao M, Sun J, Zhao Z. TSGene: A web resource for tumor suppressor genes. Nucleic Acids Res. 2013;41(Database issue):D970–D976. doi: 10.1093/nar/gks937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen JS, Hung WS, Chan HH, Tsai SJ, Sun HS. In silico identification of oncogenic potential of fyn-related kinase in hepatocellular carcinoma. Bioinformatics. 2013;29:420–427. doi: 10.1093/bioinformatics/bts715. [DOI] [PubMed] [Google Scholar]
- 18.Szklarczyk D, Franceschini A, Kuhn M, Simonovic M, Roth A, Minguez P, Doerks T, Stark M, Muller J, Bork P, et al. The STRING database in 2011: Functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011;39(Database issue):D561–D568. doi: 10.1093/nar/gkq973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adamcsek B, Palla G, Farkas IJ, Derényi I, Vicsek T. CFinder: Locating cliques and overlapping modules in biological networks. Bioinformatics. 2006;22:1021–1023. doi: 10.1093/bioinformatics/btl039. [DOI] [PubMed] [Google Scholar]
- 20.Ding T, Ma Y, Li W, Liu X, Ying G, Fu L, Gu F. Role of aquaporin-4 in the regulation of migration and invasion of human glioma cells. Int J Oncol. 2011;38:1521–1531. doi: 10.3892/ijo.2011.983. [DOI] [PubMed] [Google Scholar]
- 21.Baldwin RM, Barrett GM, Parolin DA, Gillies JK, Paget JA, Lavictoire SJ, Gray DA, Lorimer IA. Coordination of glioblastoma cell motility by PKCι. Mol Cancer. 2010;9:233. doi: 10.1186/1476-4598-9-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schwartz GK, Shah MA. Targeting the cell cycle: A new approach to cancer therapy. J Clin Oncol. 2005;23:9408–9421. doi: 10.1200/JCO.2005.01.5594. [DOI] [PubMed] [Google Scholar]
- 23.Mo W, Chen J, Patel A, Zhang L, Chau V, Li Y, Cho W, Lim K, Xu J, Lazar AJ, et al. CXCR4/CXCL12 mediate autocrine cell-cycle progression in NF1-associated malignant peripheral nerve sheath tumors. Cell. 2013;152:1077–1090. doi: 10.1016/j.cell.2013.01.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang TH, Huo L, Wang YN, Xia W, Wei Y, Chang SS, Chang WC, Fang YF, Chen CT, Lang JY, et al. EGFR potentiates MCM7-mediated DNA replication through tyrosine phosphorylation of Lyn kinase in human cancers. Cancer Cell. 2013;23:796–810. doi: 10.1016/j.ccr.2013.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ubersax JA, Woodbury EL, Quang PN, Paraz M, Blethrow JD, Shah K, Shokat KM, Morgan DO. Targets of the cyclin-dependent kinase Cdk1. Nature. 2003;425:859–864. doi: 10.1038/nature02062. [DOI] [PubMed] [Google Scholar]
- 26.Stegh AH, Brennan C, Mahoney JA, Forloney KL, Jenq HT, Luciano JP, Protopopov A, Chin L, Depinho RA. Glioma oncoprotein Bcl2L12 inhibits the p53 tumor suppressor. Genes Dev. 2010;24:2194–2204. doi: 10.1101/gad.1924710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao R, Gish K, Murphy M, Yin Y, Notterman D, Hoffman WH, Tom E, Mack DH, Levine AJ. Analysis of p53-regulated gene expression patterns using oligonucleotide arrays. Genes Dev. 2000;14:981–993. [PMC free article] [PubMed] [Google Scholar]
- 28.Levine A, Hu W, Feng Z. The P53 pathway: What questions remain to be explored? Cell Death Differ. 2006;13:1027–1036. doi: 10.1038/sj.cdd.4401910. [DOI] [PubMed] [Google Scholar]
- 29.Schwermer M, Lee S, Köster J, van Maerken T, Stephan H, Eggert A, Morik K, Schulte JH, Schramm A. Sensitivity to cdk1-inhibition is modulated by p53 status in preclinical models of embryonal tumors. Oncotarget. 2015;6:15425. doi: 10.18632/oncotarget.3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grabsch H, Takeno S, Parsons WJ, Pomjanski N, Boecking A, Gabbert HE, Mueller W. Overexpression of the mitotic checkpoint genes BUB1, BUBR1 and BUB3 in gastric cancer-association with tumour cell proliferation. J Pathol. 2003;200:16–22. doi: 10.1002/path.1324. [DOI] [PubMed] [Google Scholar]
- 31.Tang Z, Shu H, Qi W, Mahmood NA, Mumby MC, Yu H. PP2A is required for centromeric localization of Sgo1 and proper chromosome segregation. Dev Cell. 2006;10:575–585. doi: 10.1016/j.devcel.2006.03.010. [DOI] [PubMed] [Google Scholar]
- 32.Ricke RM, Jeganathan KB, van Deursen JM. Bub1 overexpression induces aneuploidy and tumor formation through Aurora B kinase hyperactivation. J Cell Biol. 2011;193:1049–1064. doi: 10.1083/jcb.201012035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kawashima SA, Yamagishi Y, Honda T, Ishiguro K, Watanabe Y. Phosphorylation of H2A by Bub1 prevents chromosomal instability through localizing shugoshin. Science. 2010;327:172–177. doi: 10.1126/science.1180189. [DOI] [PubMed] [Google Scholar]
- 34.Myrie KA, Percy MJ, Azim JN, Neeley CK, Petty EM. Mutation and expression analysis of human BUB1 and BUB1B in aneuploid breast cancer cell lines. Cancer Lett. 2000;152:193–199. doi: 10.1016/S0304-3835(00)00340-2. [DOI] [PubMed] [Google Scholar]
- 35.Telentschak S, Soliwoda M, Nohroudi K, Addicks K, Klinz FJ. Cytokinesis failure and successful multipolar mitoses drive aneuploidy in glioblastoma cells. Oncology Rep. 2015;33:2001–2008. doi: 10.3892/or.2015.3751. [DOI] [PubMed] [Google Scholar]
- 36.Hadjihannas MV, Bernkopf DB, Brückner M, Behrens J. Cell cycle control of Wnt/β-catenin signalling by conductin/axin2 through CDC20. EMBO Rep. 2012;13:347–354. doi: 10.1038/embor.2012.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Frescas D, Pagano M. Deregulated proteolysis by the F-box proteins SKP2 and β-TrCP: Tipping the scales of cancer. Nat Rev Cancer. 2008;8:438–449. doi: 10.1038/nrc2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Z, Wan L, Zhong J, Inuzuka H, Liu P, Sarkar FH, Wei W. Cdc20: A potential novel therapeutic target for cancer treatment. Curr Pharm Des. 2013;19:3210–3214. doi: 10.2174/1381612811319180005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jiang J, Jedinak A, Sliva D. Ganodermanontriol (GDNT) exerts its effect on growth and invasiveness of breast cancer cells through the down-regulation of CDC20 and uPA. Biochem Biophys Res Commun. 2011;415:325–329. doi: 10.1016/j.bbrc.2011.10.055. [DOI] [PubMed] [Google Scholar]
- 40.Rajkumar T, Sabitha K, Vijayalakshmi N, Shirley S, Bose MV, Gopal G, Selvaluxmy G. Identification and validation of genes involved in cervical tumourigenesis. BMC Cancer. 2011;11:80. doi: 10.1186/1471-2407-11-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marucci G, Morandi L, Magrini E, Farnedi A, Franceschi E, Miglio R, Calò D, Pession A, Foschini MP, Eusebi V. Gene expression profiling in glioblastoma and immunohistochemical evaluation of IGFBP-2 and CDC20. Virchows Archiv. 2008;453:599–609. doi: 10.1007/s00428-008-0685-7. [DOI] [PubMed] [Google Scholar]
- 42.Bie L, Zhao G, Cheng P, Rondeau G, Porwollik S, Ju Y, Xia XQ, McClelland M. The accuracy of survival time prediction for patients with glioma is improved by measuring mitotic spindle checkpoint gene expression. PloS one. 2011;6:e25631. doi: 10.1371/journal.pone.0025631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bae I, Rih JK, Kim HJ, Kang HJ, Haddad B, Kirilyuk A, Fan S, Avantaggiati ML, Rosen EM. BRCA1 regulates gene expression for orderly mitotic progression. Cell Cycle. 2005;4:1641–1666. doi: 10.4161/cc.4.11.2152. [DOI] [PubMed] [Google Scholar]
- 44.Bogdani M, Teugels E, De Grève J, Bourgain C, Neyns B, Pipeleers-Marichal M. Loss of nuclear BRCA1 localization in breast carcinoma is age dependent. Virchows Arch. 2002;440:274–279. doi: 10.1007/s004280100526. [DOI] [PubMed] [Google Scholar]
- 45.Lee C, Fotovati A, Triscott J, Chen J, Venugopal C, Singhal A, Dunham C, Kerr JM, Verreault M, Yip S, et al. Polo-like kinase 1 inhibition kills glioblastoma multiforme brain tumor cells in part through loss of SOX2 and delays tumor progression in mice. Stem Cells. 2012;30:1064–1075. doi: 10.1002/stem.1081. [DOI] [PubMed] [Google Scholar]
- 46.Roshak AK, Capper EA, Imburgia C, Fornwald J, Scott G, Marshall LA. The human polo-like kinase, PLK, regulates cdc2/cyclin B through phosphorylation and activation of the cdc25c phosphatas. Cell Signal. 2000;12:405–411. doi: 10.1016/S0898-6568(00)00080-2. [DOI] [PubMed] [Google Scholar]
- 47.Casenghi M, Barr FA, Nigg EA. Phosphorylation of Nlp by Plk1 negatively regulates its dynein-dynactin-dependent targeting to the centrosome. J Cell Sci. 2005;118:5101–5108. doi: 10.1242/jcs.02622. [DOI] [PubMed] [Google Scholar]
- 48.Feng Y, Yuan JH, Maloid SC, Fisher R, Copeland TD, Longo DL, Conrads TP, Veenstra TD, Ferris A, Hughes S, et al. Polo-like kinase 1-mediated phosphorylation of the GTP-binding protein Ran is important for bipolar spindle formation. Biochem Biophys Res Commun. 2006;349:144–152. doi: 10.1016/j.bbrc.2006.08.028. [DOI] [PubMed] [Google Scholar]
- 49.Hansen DV, Loktev AV, Ban KH, Jackson PK. Plk1 regulates activation of the anaphase promoting complex by phosphorylating and triggering SCFbetaTrCP-dependent destruction of the APC inhibitor Emi1. Mol Biol Cell. 2004;15:5623–5634. doi: 10.1091/mbc.E04-07-0598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Niiya F, Tatsumoto T, Lee KS, Miki T. Phosphorylation of the cytokinesis regulator ECT2 at G2/M phase stimulates association of the mitotic kinase Plk1 and accumulation of GTP-bound RhoA. Oncogene. 2006;25:827–837. doi: 10.1038/sj.onc.1209124. [DOI] [PubMed] [Google Scholar]
- 51.Foong CS, Sandanaraj E, Brooks HB, Campbell RM, Ang BT, Chong YK, Tang C. Glioma-propagating cells as an in vitro screening platform PLK1 as a case study. J Biomol Screen. 2012;17:1136–1150. doi: 10.1177/1087057112457820. [DOI] [PubMed] [Google Scholar]
- 52.Hu K, Lee C, Qiu D, Fotovati A, Davies A, Abu-Ali S, Wai D, Lawlor ER, Triche TJ, Pallen CJ, Dunn SE. Small interfering RNA library screen of human kinases and phosphatases identifies polo-like kinase 1 as a promising new target for the treatment of pediatric rhabdomyosarcomas. Mol Cancer Ther. 2009;8:3024–3035. doi: 10.1158/1535-7163.MCT-09-0365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ko E, Kim Y, Cho EY, Han J, Shim YM, Park J, Kim DH. Synergistic effect of Bcl-2 and Cyclin A2 on adverse recurrence-free survival in stage i non-small cell lung cancer. Ann Surg Oncol. 2013;20:1005–1012. doi: 10.1245/s10434-012-2727-2. [DOI] [PubMed] [Google Scholar]
- 54.Arsic N, Bendris N, Peter M, Begon-Pescia C, Rebouissou C, Gadéa G, Bouquier N, Bibeau F, Lemmers B, Blanchard JM. A novel function for Cyclin A2: Control of cell invasion via RhoA signaling. J Cell Biol. 2012;196:147–162. doi: 10.1083/jcb.201102085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Uhlen M, Oksvold P, Fagerberg L, Lundberg E, Jonasson K, Forsberg M, Zwahlen M, Kampf C, Wester K, Hober S, et al. Towards a knowledge-based human protein atlas. Nature Biotechnol. 2010;28:1248–1250. doi: 10.1038/nbt1210-1248. [DOI] [PubMed] [Google Scholar]
- 56.Koyama-Nasu R, Nasu-Nishimura Y, Todo T, Ino Y, Saito N, Aburatani H, Funato K, Echizen K, Sugano H, Haruta R, et al. The critical role of cyclin D2 in cell cycle progression and tumorigenicity of glioblastoma stem cells. Oncogene. 2013;32:3840–3845. doi: 10.1038/onc.2012.399. [DOI] [PubMed] [Google Scholar]