Abstract
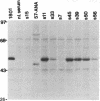
Overexpression of the p53 protein, resulting from gene mutations that increase protein stability, has been detected in greater than 25% of primary human breast cancers. In addition, approximately 10% of breast cancer patients have circulating antibodies to the p53 protein. In this study, the anti-p53 humoral response is correlated with the presence and type of mutant p53 protein expressed in the tumor. In a series of 60 breast cancer patients, 0 of 30 tumors with normal, low-level p53 expression induced anti-p53 antibodies, whereas 7 (23%) of 30 tumors with p53 overexpression elicited a specific anti-p53 antibody response. These 7 patients had anti-p53 antibodies that recognized wild-type p53 and a variety of mutant p53 proteins. A comparison of p53 mutations revealed that antibody-negative tumors had mutations exclusively in exons 7 and 8, whereas antibody-positive tumors had mutations primarily in exons 5 and 6. Moreover, all antibody-eliciting tumors contained complexes between p53 and a 70-kDa heat shock protein, whereas none of the antibody-negative tumors contained this complex. This study implicates a 70-kDa heat shock protein in the antigenic presentation of p53.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Band V., Sager R. Distinctive traits of normal and tumor-derived human mammary epithelial cells expressed in a medium that supports long-term growth of both cell types. Proc Natl Acad Sci U S A. 1989 Feb;86(4):1249–1253. doi: 10.1073/pnas.86.4.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banks L., Matlashewski G., Crawford L. Isolation of human-p53-specific monoclonal antibodies and their use in the studies of human p53 expression. Eur J Biochem. 1986 Sep 15;159(3):529–534. doi: 10.1111/j.1432-1033.1986.tb09919.x. [DOI] [PubMed] [Google Scholar]
- Bartek J., Iggo R., Gannon J., Lane D. P. Genetic and immunochemical analysis of mutant p53 in human breast cancer cell lines. Oncogene. 1990 Jun;5(6):893–899. [PubMed] [Google Scholar]
- Braithwaite A. W., Sturzbecher H. W., Addison C., Palmer C., Rudge K., Jenkins J. R. Mouse p53 inhibits SV40 origin-dependent DNA replication. Nature. 1987 Oct 1;329(6138):458–460. doi: 10.1038/329458a0. [DOI] [PubMed] [Google Scholar]
- Caron de Fromentel C., Nardeux P. C., Soussi T., Lavialle C., Estrade S., Carloni G., Chandrasekaran K., Cassingena R. Epithelial HBL-100 cell line derived from milk of an apparently healthy woman harbours SV40 genetic information. Exp Cell Res. 1985 Sep;160(1):83–94. doi: 10.1016/0014-4827(85)90238-1. [DOI] [PubMed] [Google Scholar]
- Chirico W. J., Waters M. G., Blobel G. 70K heat shock related proteins stimulate protein translocation into microsomes. Nature. 1988 Apr 28;332(6167):805–810. doi: 10.1038/332805a0. [DOI] [PubMed] [Google Scholar]
- Crawford L. V., Pim D. C., Bulbrook R. D. Detection of antibodies against the cellular protein p53 in sera from patients with breast cancer. Int J Cancer. 1982 Oct 15;30(4):403–408. doi: 10.1002/ijc.2910300404. [DOI] [PubMed] [Google Scholar]
- Davidoff A. M., Herndon J. E., 2nd, Glover N. S., Kerns B. J., Pence J. C., Iglehart J. D., Marks J. R. Relation between p53 overexpression and established prognostic factors in breast cancer. Surgery. 1991 Aug;110(2):259–264. [PubMed] [Google Scholar]
- Davidoff A. M., Humphrey P. A., Iglehart J. D., Marks J. R. Genetic basis for p53 overexpression in human breast cancer. Proc Natl Acad Sci U S A. 1991 Jun 1;88(11):5006–5010. doi: 10.1073/pnas.88.11.5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshaies R. J., Koch B. D., Werner-Washburne M., Craig E. A., Schekman R. A subfamily of stress proteins facilitates translocation of secretory and mitochondrial precursor polypeptides. Nature. 1988 Apr 28;332(6167):800–805. doi: 10.1038/332800a0. [DOI] [PubMed] [Google Scholar]
- Fields S., Jang S. K. Presence of a potent transcription activating sequence in the p53 protein. Science. 1990 Aug 31;249(4972):1046–1049. doi: 10.1126/science.2144363. [DOI] [PubMed] [Google Scholar]
- Finlay C. A., Hinds P. W., Tan T. H., Eliyahu D., Oren M., Levine A. J. Activating mutations for transformation by p53 produce a gene product that forms an hsc70-p53 complex with an altered half-life. Mol Cell Biol. 1988 Feb;8(2):531–539. doi: 10.1128/mcb.8.2.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halevy O., Michalovitz D., Oren M. Different tumor-derived p53 mutants exhibit distinct biological activities. Science. 1990 Oct 5;250(4977):113–116. doi: 10.1126/science.2218501. [DOI] [PubMed] [Google Scholar]
- Harris A. L. Mutant p53--the commonest genetic abnormality in human cancer? J Pathol. 1990 Sep;162(1):5–6. doi: 10.1002/path.1711620103. [DOI] [PubMed] [Google Scholar]
- Hinds P. W., Finlay C. A., Frey A. B., Levine A. J. Immunological evidence for the association of p53 with a heat shock protein, hsc70, in p53-plus-ras-transformed cell lines. Mol Cell Biol. 1987 Aug;7(8):2863–2869. doi: 10.1128/mcb.7.8.2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinds P. W., Finlay C. A., Quartin R. S., Baker S. J., Fearon E. R., Vogelstein B., Levine A. J. Mutant p53 DNA clones from human colon carcinomas cooperate with ras in transforming primary rat cells: a comparison of the "hot spot" mutant phenotypes. Cell Growth Differ. 1990 Dec;1(12):571–580. [PubMed] [Google Scholar]
- Hinds P., Finlay C., Levine A. J. Mutation is required to activate the p53 gene for cooperation with the ras oncogene and transformation. J Virol. 1989 Feb;63(2):739–746. doi: 10.1128/jvi.63.2.739-746.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollstein M., Sidransky D., Vogelstein B., Harris C. C. p53 mutations in human cancers. Science. 1991 Jul 5;253(5015):49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- Iggo R., Gatter K., Bartek J., Lane D., Harris A. L. Increased expression of mutant forms of p53 oncogene in primary lung cancer. Lancet. 1990 Mar 24;335(8691):675–679. doi: 10.1016/0140-6736(90)90801-b. [DOI] [PubMed] [Google Scholar]
- Kraiss S., Quaiser A., Oren M., Montenarh M. Oligomerization of oncoprotein p53. J Virol. 1988 Dec;62(12):4737–4744. doi: 10.1128/jvi.62.12.4737-4744.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine A. J., Momand J., Finlay C. A. The p53 tumour suppressor gene. Nature. 1991 Jun 6;351(6326):453–456. doi: 10.1038/351453a0. [DOI] [PubMed] [Google Scholar]
- Malkin D., Li F. P., Strong L. C., Fraumeni J. F., Jr, Nelson C. E., Kim D. H., Kassel J., Gryka M. A., Bischoff F. Z., Tainsky M. A. Germ line p53 mutations in a familial syndrome of breast cancer, sarcomas, and other neoplasms. Science. 1990 Nov 30;250(4985):1233–1238. doi: 10.1126/science.1978757. [DOI] [PubMed] [Google Scholar]
- Marks J. R., Davidoff A. M., Kerns B. J., Humphrey P. A., Pence J. C., Dodge R. K., Clarke-Pearson D. L., Iglehart J. D., Bast R. C., Jr, Berchuck A. Overexpression and mutation of p53 in epithelial ovarian cancer. Cancer Res. 1991 Jun 1;51(11):2979–2984. [PubMed] [Google Scholar]
- Milner J., Medcalf E. A. Cotranslation of activated mutant p53 with wild type drives the wild-type p53 protein into the mutant conformation. Cell. 1991 May 31;65(5):765–774. doi: 10.1016/0092-8674(91)90384-b. [DOI] [PubMed] [Google Scholar]
- Nigro J. M., Baker S. J., Preisinger A. C., Jessup J. M., Hostetter R., Cleary K., Bigner S. H., Davidson N., Baylin S., Devilee P. Mutations in the p53 gene occur in diverse human tumour types. Nature. 1989 Dec 7;342(6250):705–708. doi: 10.1038/342705a0. [DOI] [PubMed] [Google Scholar]
- Pinhasi-Kimhi O., Michalovitz D., Ben-Zeev A., Oren M. Specific interaction between the p53 cellular tumour antigen and major heat shock proteins. Nature. 1986 Mar 13;320(6058):182–184. doi: 10.1038/320182a0. [DOI] [PubMed] [Google Scholar]
- Raycroft L., Wu H. Y., Lozano G. Transcriptional activation by wild-type but not transforming mutants of the p53 anti-oncogene. Science. 1990 Aug 31;249(4972):1049–1051. doi: 10.1126/science.2144364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich N. C., Oren M., Levine A. J. Two distinct mechanisms regulate the levels of a cellular tumor antigen, p53. Mol Cell Biol. 1983 Dec;3(12):2143–2150. doi: 10.1128/mcb.3.12.2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues N. R., Rowan A., Smith M. E., Kerr I. B., Bodmer W. F., Gannon J. V., Lane D. P. p53 mutations in colorectal cancer. Proc Natl Acad Sci U S A. 1990 Oct;87(19):7555–7559. doi: 10.1073/pnas.87.19.7555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava S., Zou Z. Q., Pirollo K., Blattner W., Chang E. H. Germ-line transmission of a mutated p53 gene in a cancer-prone family with Li-Fraumeni syndrome. Nature. 1990 Dec 20;348(6303):747–749. doi: 10.1038/348747a0. [DOI] [PubMed] [Google Scholar]
- Stürzbecher H. W., Chumakov P., Welch W. J., Jenkins J. R. Mutant p53 proteins bind hsp 72/73 cellular heat shock-related proteins in SV40-transformed monkey cells. Oncogene. 1987 May;1(2):201–211. [PubMed] [Google Scholar]
- VanBuskirk A. M., DeNagel D. C., Guagliardi L. E., Brodsky F. M., Pierce S. K. Cellular and subcellular distribution of PBP72/74, a peptide-binding protein that plays a role in antigen processing. J Immunol. 1991 Jan 15;146(2):500–506. [PubMed] [Google Scholar]
- Wade-Evans A., Jenkins J. R. Precise epitope mapping of the murine transformation-associated protein, p53. EMBO J. 1985 Mar;4(3):699–706. doi: 10.1002/j.1460-2075.1985.tb03686.x. [DOI] [PMC free article] [PubMed] [Google Scholar]