TLR7 senses guanosine and its analogs synergistically with oligoribonucleotides
Key words: 8-OHdG, 8-OHG, dG, G, TLR7
Abstract
Toll-like receptor (TLR) 7and 8 were considered to recognize single-strand RNA (ssRNA) from viruses. Although these receptors also respond to synthetic small chemical ligands, such as CL075 and R848, it remains to be determined whether these receptors sense natural small molecules or not. In the structure of human TLR8 (huTLR8) with ssRNA, there are two ligand-binding sites: one binds a uridine and the other binds an oligoribonucleotide (ORN). This finding demonstrates that huTLR8 recognizes degradation products of ssRNA, suggesting the presence of natural small ligands. We here show that TLR7 works as the sensor for guanosine (G)/2′-deoxyguanosine (dG) in the presence of ORN where ORN strengthens TLR7 interaction with G/dG. In addition, modified nucleosides such as 7-methylguanosine, 8-hydroxyguanosine (8-OHG) and 8-hydroxydeoxyguanosine (8-OHdG) activated TLR7 with ORNs. Importantly, 8-OHdG—a well-known oxidative DNA damage marker with unknown function—induced strong cytokine production comparable to G and dG both in mouse and human immune cells. Although 8-OHdG bound TLR7/ORN with lower affinity than dG did in isothermal titration calorimetry, administered 8-OHdG was metabolically more stable than dG in the serum, indicating that 8-OHdG acts on TLR7 as an endogenous ligand in vivo. To address a role of G analogs in the disease state, we also examined macrophages from Unc93b1 D34A/D34A mice, which suffer from TLR7-dependent systemic inflammation, and found that Unc93b1 D34A/D34A macrophages showed significantly enhanced response to G alone or 8-OHdG with ORN. In conclusion, our results provide evidence that G, dG, 8-OHG and 8-OHdG are novel endogenous ligands for TLR7.
Introduction
Toll-like receptors (TLRs) recognize a variety of microbial products and mount immune responses (1, 2). Nucleic acid (NA) is one of the principal TLR ligands. TLR7 and TLR8 respond to single-strand RNA (ssRNA) derived from viruses and bacteria in humans. In contrast to human TLR8 (huTLR8), mouse TLR8 does not function as an ssRNA sensor. Over-expression of mouse TLR7 and huTLR8 in mice causes diseases such as lupus nephritis or arthritis (3, 4), suggesting that TLR7 and TLR8 have a risk of responding to endogenous ligands in the disease state. Consistent with this, TLR7 and TLR8 are shown to respond to RNA-associated autoantigens (5–7).
In addition to ssRNA, synthetic small molecules are able to activate TLR7 and TLR8 and used to treat genital warts and skin cancers (8, 9). The structure of huTLR8 complexed with the small chemical ligand shows the ligand-binding site located in the dimer interface (10). Moreover, the structure of huTLR8 complexed with ssRNA shows that degradation products of ssRNA bind to two different sites (11). The site for synthetic small chemical ligands was occupied by uridine (U), whereas an oligoribonucleotide (ORN) binds another site and strengthened huTLR8 interaction with U, leading to synergistic huTLR8 activation. These results demonstrate that huTLR8 primarily responds to U and ORN. Considering that TLR7 is a paralogue of huTLR8, TLR7 is likely to interact with nucleosides and those ligands may contribute to autoimmune diseases caused by TLR7.
RNA is modified by methylation or hydroxylation more commonly in higher eukaryotes than prokaryotes. TLR7 and TLR8 responses to modified RNAs are much weaker than to unmodified RNAs (12), suggesting that self-pathogen discrimination by TLR7 and TLR8 depends on RNA modification. In this regard, it is interesting to study TLR7 and TLR8 responses to modified nucleosides. DNAs are modified by reactive oxygen species (ROS). Formation of 8-hydroxydeoxyguanosine (8-OHdG) is the most common and well-studied modification (13, 14). DNA containing 8-OHdG turns resistant to degradation by DNase III, and accumulated DNA activates cytoplasmic DNA sensors, leading to type I interferon (IFN) production in systemic lupus erythematosus (15). These results demonstrate that 8-hydroxylation of guanosine (G) in DNAs influences DNA degradation, but not DNA recognition by cytoplasmic DNA sensors. Despite the resistance against DNase III, modified DNA is eventually degraded into 8-OHdG, which is detectable in the circulation or urine and used as a marker for DNA damages caused by oxidative stresses such as UV irradiation and inflammatory responses (13). Modified nucleosides such as 8-OHdG may have an impact on innate immune responses by directly acting on TLR7 or TLR8 when innate immune cells are exposed to oxidative stresses.
The present study focused on nucleoside ligands of TLR7 and TLR8. HuTLR8 strongly responded to U in the presence of ORN. In sharp contrast, mouse and human TLR7 was synergistically activated by G in the presence of ORN, demonstrating that G is an endogenous TLR7 ligand. Although 8-OHdG showed lower affinity to TLR7 than 2′-deoxyguanosine (dG) did, it comparably activated TLR7 and induced cytokine production in vitro and in vivo. Exogenous addition of dG did not alter the serum level of dG, probably because of rapid turnover of dG in vivo. In contrast, injection of 8-OHdG led to a rapid and long-lasting increase in serum 8-OHdG. 8-hydroxylation is likely to change the metabolism of dG. These results demonstrate that TLR7 responds to G, dG and 8-OHdG in the presence of ORN. Considering that these nucleosides are metabolites present in the steady state and disease state, TLR7 responses to these nucleosides may have a role in autoimmune diseases, as well as in infectious diseases.
Methods
Mice
C57BL/6N mice were purchased from Japan SLC Inc. (Shizuoka, Japan). C57BL/6 background Tlr7−/− mice were kindly provided by Prof. S. Akira (Osaka University, Japan). Mice were kept under specific pathogen-free conditions in the animal center at the Institute of Medical Science, The University of Tokyo (IMSUT). All the mouse experiments were approved by the Institutional Animal Care and Use Committee in the IMSUT.
Reagents
Guanosine (G) and uridine (U) were purchased from MP Biomedicals (Santa Ana, CA, USA), Sigma-Aldrich Japan (Tokyo, Japan) and Wako Pure Chemical Industries (Osaka, Japan). G and U from multiple manufacturers showed similar activity in immune response via TLR7 and TLR8, respectively. Adenosine (A), inosine (I), cytidine (C), thymidine (T), uridine-5′-monophosphate disodium dihydrate (UMP), uridine-5′-diphosphate disodium salt (UDP), uridine-5′-triphosphate trisodium salt (UTP), uracil, guanosine-5′-monophosphate (GMP), guanosine-5′-diphosphate disodium salt (GDP), guanosine-5′-triphosphate trisodium salt (GTP), guanine and pseudouridine were purchased from MP Biomedicals (Santa Ana, CA, USA). 5-Methyluridine was purchased from Wako Pure Chemical Industries (Osaka, Japan). 2′-Deoxyuridine hydrate (dU) and 7-methylguanosine were purchased from Sigma-Aldrich Japan (Tokyo, Japan). 2′-Deoxyguanosine hydrate (dG) and 8-hydroxy-2′-deoxyguanosine (8-OHdG, 8-Oxo-dG) were obtained from Jena Bioscience (Jena, Germany). 8-Hydroxyguanosine (8-OHG) was obtained from Enzo Life Sciences. CL075 was purchased from Invivogen (San Diego, CA, USA). 8-Mercaptoguanosine (8-SGuo) was purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA, USA).
RNA9.2s (UsGsUsCsCsUsUsCsAsAsUsGsUsCsC sUsUsCsAsA), ssRNA40 (GsCsCsCsGsUsCsUsGsUsUsGsU sGsUsGsAsCsUsC), ssRNA41 (GsCsCsCsGsUsCsUsGsUs UsGsUsGsUsGsAsCsUsC), ORN06 (UsUsGsUsUsGsUsUsG sUsUsGsUsUsGsUsUsGsUsU), polyA (19mer, AsAsAsAsAsAsAsAsAsAsAsAsAsAsAsAsAsAsA), polyG (19mer, GsGsGsGsGsGsGsGsGsGsGsGsGsGsGsGsGsGsG), polyC (19mer, CsCsCsCsCsCsCsCsCsCsCsCsCsCsCsCsCsCsC), polyU (19mer, UsUsUsUsUsUsUsUsUsUsUsUsUsUsUsUsUsUsU) and Sa12 (GsAsCsGsGsAsAsAsGsAsCsC), in which ‘s’ depicts a phosphothioate linkage, were synthesized by FASMAC (Kanagawa, Japan).
PE anti-rat CD2 (OX-34) used to check the expression level of TLR7 and TLR8 in HEK293T cells was purchased from Biolegend (San Diego, CA, USA).
NF-κB-dependent luciferase reporter assay
HEK293T cells were cultured in DMEM (Gibco, Paisley, UK) with 10% FBS, 2mM l-glutamine (Gibco, Paisley, UK) and 50 μM 2-ME. To check the activity of human TLR8 (huTLR8), HEK293T cells were seeded in collagen-coated 6-well plates at a density of 1×106 cells per well and transiently transfected with wild-type (WT) or mutant huTLR8 cDNAs in pMX-puro-IRES-rat CD2 (kindly supplied by Prof. Kitamura, University of Tokyo, Japan) (1 μg), together with a pELAM1-luc reporter plasmid (5ng) (16), using PEI (Polyethylenimine ‘Max’, MW40000; Polysciences, Inc., Warrington, PA, USA) at 30h before stimulation. To check the activity of mouse or human TLR7, HEK293T cells were plated on collagen-coated 10-cm dishes at a density of 6×106 cells per well and transiently transfected with WT mouse or human TLR7 cDNAs in pMX-puro-IRES-rat CD2 (6 μg), together with WT mouse or human Unc93B1 cDNA in pMX-puro (6 μg) and a pELAM1-luc reporter plasmid (30ng), using PEI at 30h before stimulation. Twenty-four hours after transfection, cells were reseeded in collagen-coated, flat 96-well plates (Corning, Oneonta, NY, USA) at a density of 1×105 cells per well. After pre-culture for 6h, attached cells were stimulated with various ligands for 6h. Then stimulated cells were lysed by 50 μl of Cell Culture Lysis Reagent (Promega, Madison, WI, USA) and 6 μl of lysate was subjected to a luciferase assay using the Luciferase Assay System (Promega, Madison, WI, USA). The relative light unit (RLU) of chemiluminescence was measured by GloMax 96 Microplate Luminometer (Promega, Madison, WI, USA). When cells were stimulated by ssRNA alone (Figs 1B, 2C and D) or U analogs with or without ssRNA (Fig. 1A and C–E), 30 μl ml–1 N-[1-(2,3-Dioleoyloxy)propyl]-N, N, N-trimethylammonium methyl-sulfate (DOTAP) transfection reagent (Roche Diagnostics, Indianapolis, IN, USA) was used. Transfection efficiency of cDNAs in pMX-puro-IRES-rat CD2 was evaluated by checking the cell-surface rat CD2 expression level using a FACSCalibur flow cytometer (BD Biosciences, San Jose, CA, USA).
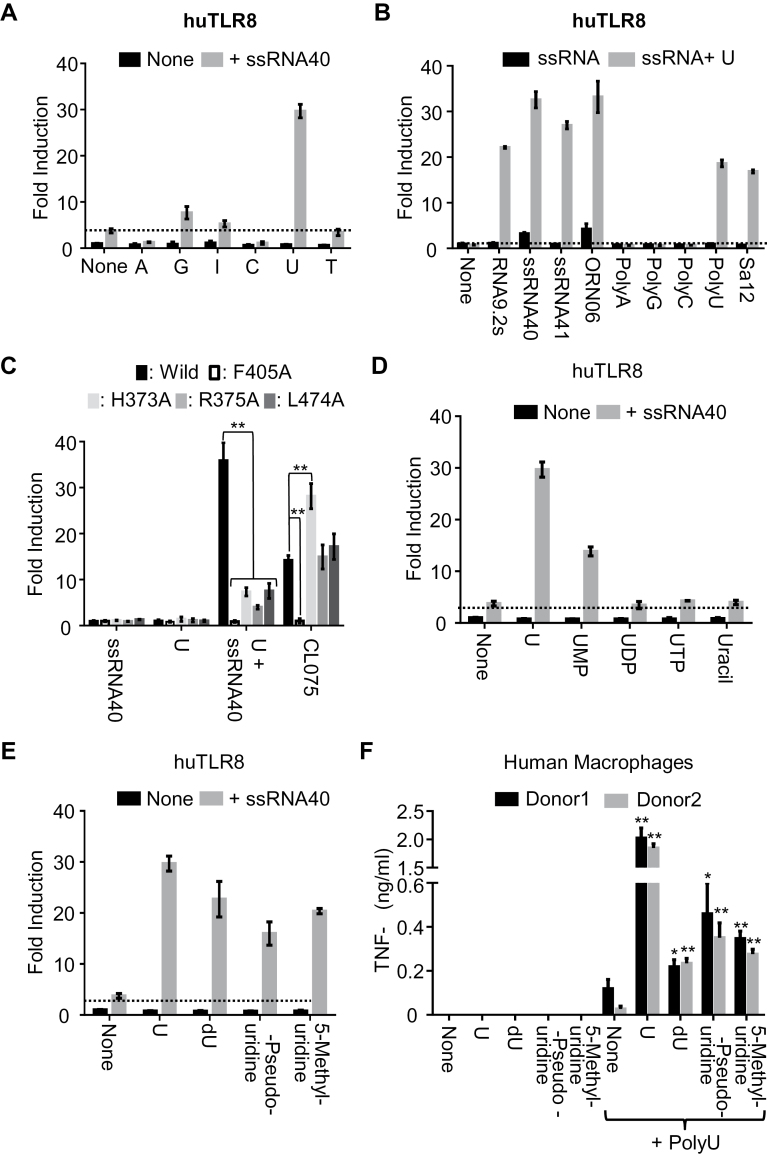
Fig. 1.
Human TLR8 is synergistically activated by U analogs and ssRNA. Human TLR8 (huTLR8) response was analyzed by NF-κB-dependent luciferase reporter assay using HEK293T cells expressing huTLR8. Cells were stimulated with (A) a variety of 1mM nucleosides with or without 20 μg ml–1 ssRNA40; (B) a variety of 20 μg ml–1 ssRNA with or without 1mM U; (D) 1mM U, U nucleotides or uracil with or without 20 μg ml–1 ssRNA40; (E) 1mM U, deoxyuridine (dU), β-pseudouridine or 5-methyluridine with or without 20 μg ml–1 ssRNA40, in the presence of DOTAP. (C) The NF-κB activation by WT huTLR8, first-site mutants (F405A) and second-site mutants (H373A, R375A, L474A) of huTLR8 in response to 20 μg ml–1 ssRNA40, 1mM U, 1mM U + 20 μg ml–1 ssRNA40 or 5 μg ml–1 CL075 with DOTAP. Data represent the mean fold induction of NF-κB activity (n = 3, ±SD), calculated as the RLU of stimulated cells divided by the RLU of non-stimulated cells, and two-tailed Student’s t-test was used to evaluate the statistical significance between WT and mutant huTLR8 (**P < 0.01). (F) Human macrophages derived from two healthy donors were stimulated by indicated U derivatives in the presence or absence of ssRNA40 (20 μg ml–1) with DOTAP. TNF-α production was determined by ELISA. Two-tailed Student’s t-test was used to evaluate the statistical significance between polyU only and U derivatives with polyU (**P < 0.01; *P < 0.05). Data shown are representative of more than three independent experiments.
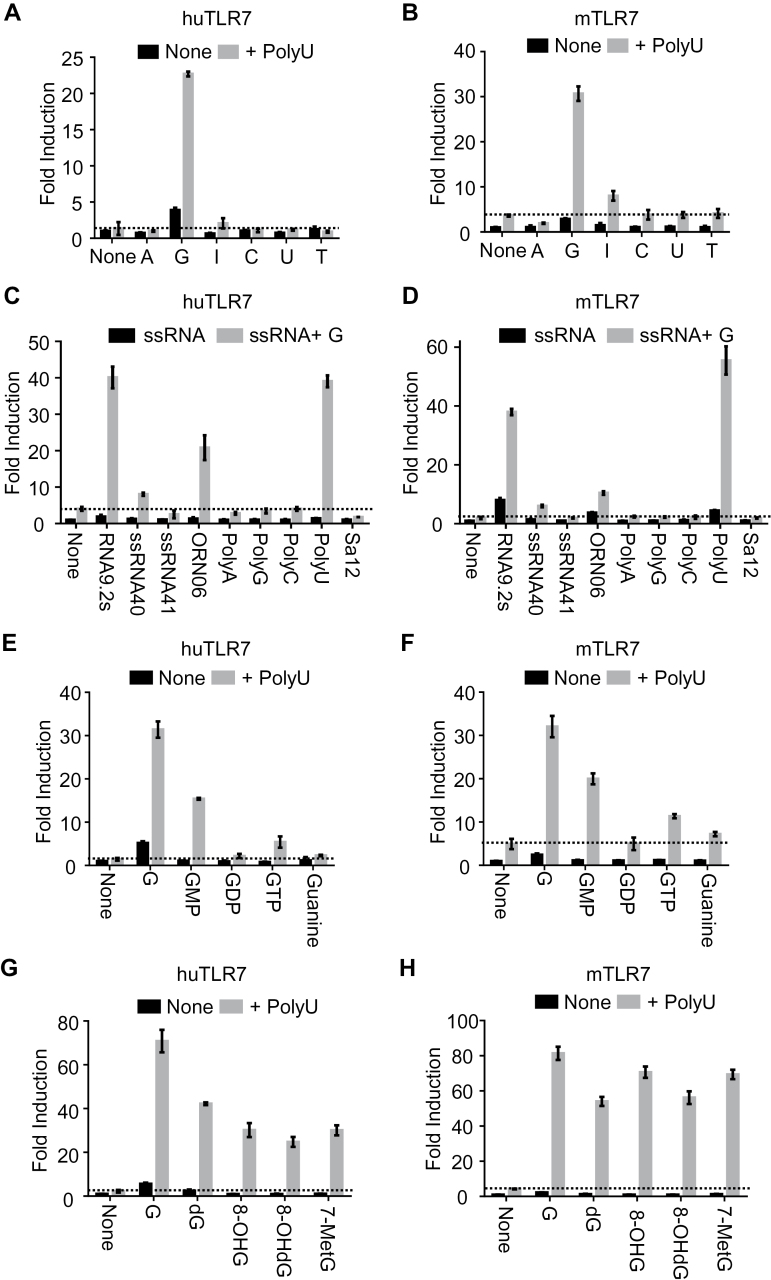
Fig. 2.
Human and mouse TLR7 are activated by G or modified Gs in the presence of ssRNA. Human (hu) and mouse (m) TLR7 responses were analyzed by NF-κB-dependent luciferase reporter assay using HEK293T cells co-expressing huTLR7 and huUnc93B1 (A, C, E and G), or mTLR7 and mUnc93B1 (B, D, F and H). Cells were stimulated with (A and B) a variety of 1mM nucleosides with or without 20 μg ml–1 polyU; (C and D) a variety of 20 μg ml–1 ssRNA with or without 1mM guanosine (G); (E and F) 1mM G, G nucleotides, or guanine with or without 20 μg ml–1 PolyU; (G and H) 1mM G, deoxyguanosine (dG), 8-hydroxyguanosine (8-OHG) or 8-hydroxy-2′-deoxyguanosine (8-OHdG) with or without 20 μg ml–1 PolyU, in the absence of DOTAP. Data represent the mean fold induction of NF-κB activity (n = 3, ±SD), calculated as the RLU of stimulated cells divided by the RLU of non-stimulated cells. Data shown are representative of more than three independent experiments.
Induction of bone-marrow-derived macrophages, cDCs and pDCs
Bone marrow (BM) cells were collected from WT or TLR7 tibiae and femora. For preparation of BM-derived macrophages (BM-MCs), BM cells were plated at a density of 7×106 cells per one non-tissue culture polystyrene 94-mm petri dish (Greiner Bio-One, Frickenhausen, Germany) and cultured in 10ml DMEM medium (Gibco, Paisley, UK) supplemented with 10% FBS, Penicillin-Streptomycin-Glutamine (Gibco, Paisley, UK), 50 μM 2-ME and 100ng ml–1 recombinant murine macrophage colony-stimulating factor (M-CSF, PeproTech Inc., Rocky Hill, NJ, USA) for 6 or 7 days. For BM-derived conventional DCs (BM-cDCs), BM cells were plated at a density of 1×106 cells per well in 24-well plates and cultured in 1ml RPMI 1640 medium (Gibco, Paisley, UK) supplemented with 10% FBS, Penicillin-Streptomycin-Glutamine, 50 μM 2-ME and 10ng ml–1 of recombinant murine granulocyte–macrophage colony-stimulating factor (GM-CSF, PeproTech Inc., Rocky Hill, NJ, USA) for 7 days. Half volume of medium was changed every other day. For BM-derived plasmacytoid DCs (BM-pDCs), BM cells in 10-cm cell-culture dishes (Greiner Bio-One, Frickenhausen, Germany) with a density of 2.5×107 cells per dish were cultured in 10ml RPMI 1640 medium (Gibco, Paisley, UK) supplemented with 10% FBS, Penicillin-Streptomycin-Glutamine, 50 μM 2-ME and 100ng ml–1 of recombinant murine fms-like tyrosine kinase-3 ligand (Flt3L, PeproTech Inc., Rocky Hill, NJ, USA) for 7 days. Before use as BM-pDCs, the CD11c+B220+ population was sorted by FACSAria flow cytometers (BD Biosciences, San Jose, CA, USA). Cell staining for sorting was performed in 1× PBS-based staining buffer complemented with 10% FBS, 10mM HEPES and 1mM sodium pyruvate.
Human macrophages and pDCs
Human macrophages were induced from human peripheral blood mononuclear cells (hPBMCs), purchased from Allcells (Alameda, CA, USA) and Cellular Technology Limited (Shaker Height, OH, USA). hPBMCs were plated in 10-cm cell-culture dishes with a density of 0.75–1.0×107 cells per dish and cultured in 10ml RPMI 1640 medium (Gibco, Paisley, UK) supplemented with 10% FBS, Penicillin-Streptomycin-Glutamine (Gibco, Paisley, UK), 50 μM 2-ME, 50ng ml–1 of recombinant human M-CSF (PeproTech Inc., Rocky Hill, NJ, USA) and 20ng of recombinant human IL-4 (PeproTech Inc., Rocky Hill, NJ, USA) for 7 days. Human pDCs were isolated from PBMCs from adult healthy donors, as described previously (17). BDCA4+/CD11c-/lineage-/CD4+ pDCs were isolated by FACS Aria (BD Biosciences, San Jose, CA, USA) to reach greater than 99% purity according to BDCA2 staining. This study was approved by the Institutional Review Board of Kansai Medical University (IRB No. 1414).
Cytokine measurement
Human macrophages or mouse BM-MCs and BM-cDCs were cultured in flat-bottomed 96-well plates (BD Falcon, Durham, NC, USA) at 1×104 per well or 1×105 per well, respectively. Mouse BM-pDCs were cultured in round-bottomed 96-well plates (BD Falcon, Durham, NC, USA) at 5×104 per well. Purified human pDCs were cultured in flat-bottomed 96-well plates at 5×104 cells. All types of immune cells in 200 μl of medium were stimulated with indicated ligands for 20h and supernatants were collected to measure cytokines by ELISA. The concentration of human TNF-α from human macrophages and mouse IL-12p40 in supernatant was measured with Ready-Set-Go! ELISA kits (eBioscience, San Diego, CA, USA). The concentration of human TNF-α and IL-6 from human pDCs was detected by Quantikine ELISA Kit (R&D, Minneapolis, MN, USA). The concentrations of mouse and human IFN-α in supernatant were measured by IFN-α ELISA Kit (PBL Assay Science, Piscataway, NJ, USA).
Preparation of the TLR7 ectodomain
The DNA encoding the extracellular domain of TLR7 (residues 26–838) from Macaca mulatta with a C-terminal thrombin cleavage sequence (LVPRGS) followed by protein A tag was inserted into the expression vector pMT/BiP/V5-His (Drosophila Expression System; ThermoFisher Scientific). Three mutations (N166Q, N388Q and N487Q) for reducing glycosylation sites and a replacement of residues 439–444 by thrombin cleavage sequence to cleave the loop region between LRR14 and LRR15 artificially were introduced. Drosophila S2 cells were co-transfected with the TLR7 and pCoHygro vectors. Stably transfected cells were selected in Sf-900 II SFM medium (Life Technologies, Gaithersburg, MD, USA) containing 300 μg ml–1 hygromycin. Protein secreted to the supernatant was purified by IgG Sepharose 6 Fast Flow (Life Technologies, Gaithersburg, MD, USA) affinity chromatography, the loop and protein A tag cleavage by thrombin, followed by Superdex 200 (Life Technologies, Gaithersburg, MD, USA) gel filtration chromatography.
Isothermal titration calorimetry
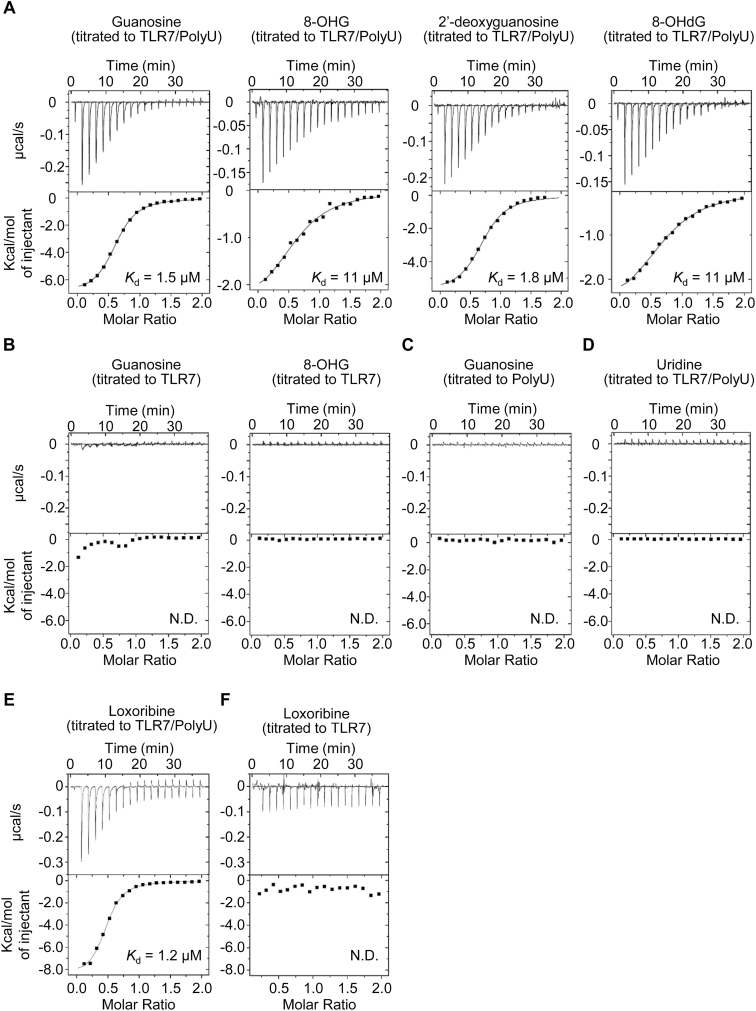
Isothermal titration calorimetry (ITC) experiments were performed at 298K in a buffer composed of 10mM 2-(N-morpholino)ethanesulfonic acid buffer pH 5.5 and 150mM NaCl using a MicroCal iTC200 (GE Healthcare, Piscataway, NJ, USA). The titration sequence included a single 0.4 μl injection followed by 19 injections, 2 μl each, with a spacing of 120s between the injections. The titration conditions were as follows: 300 μM G into 30 μM TLR7/polyU; 500 μM 8-OHG into 50 μM TLR7/polyU; 300 μM dG into 30 μM TLR7/polyU; 500 μM 8-OHdG into 50 μM TLR7/polyU; 300 μM G into 30 μM TLR7; 500 μM 8-OHG into 50 μM TLR7; 300 μM G into 30 μM polyU; or 3mM U into 30 μM TLR7/polyU. PolyU (19-mer) with normal phosphodiester linkage was used for ITC analyses. OrigineLab software (GE Healthcare, Piscataway, NJ, USA) was used to analyze the raw ITC data. Thermodynamic parameters were extracted from curve fitting analysis with a single-site binding model.
Cytokine production in vivo
Two micromoles of guanosine analogs (G, dG, 8-OHG or 8-OHdG) with or without 10 μg polyU dissolved in 100 μl 1× PBS containing 10% mouse serum per mouse were injected into mice using 1ml Terumo Myjector 29G insulin syringes (Myjector 29G; Terumo corporation, Tokyo, Japan) by intravenous route. At 3h after injection, blood was collected and serum IL-12p40 level was measured using Ready-Set-Go! ELISA kits (eBioscience, San Diego, CA, USA).
Determination of the serum levels of dG and 8-OHdG
Two micromoles of dG or 8-OHdG dissolved in 100 μl 1× PBS per mouse was injected into mice using Myjector 29G by the intravenous route. At 5, 15, 30 and 60min after injection, blood was collected and serum samples mixed with an antioxidant stock solution were subjected to analysis of dG and 8-OHdG concentration by TAS Project Co. Ltd. (Fukuoka, Japan). In brief, 8-OHdG in samples was quantified by an electrochemical detector system (HITEC; EICOM, Kyoto, Japan). After measuring 8-OHdG, dG in the same sample was serially measured with an ultraviolet absorptivity (254nm).
Statistical analysis
Data from triplicate (Fig. 1C and F, Fig. 4A) or seven (Fig. 5A) samples were used for statistical analysis. Statistical significance was calculated by the two-tailed Student’s t-test. A P value of less than 0.05 was considered to be significant.
Fig. 4.
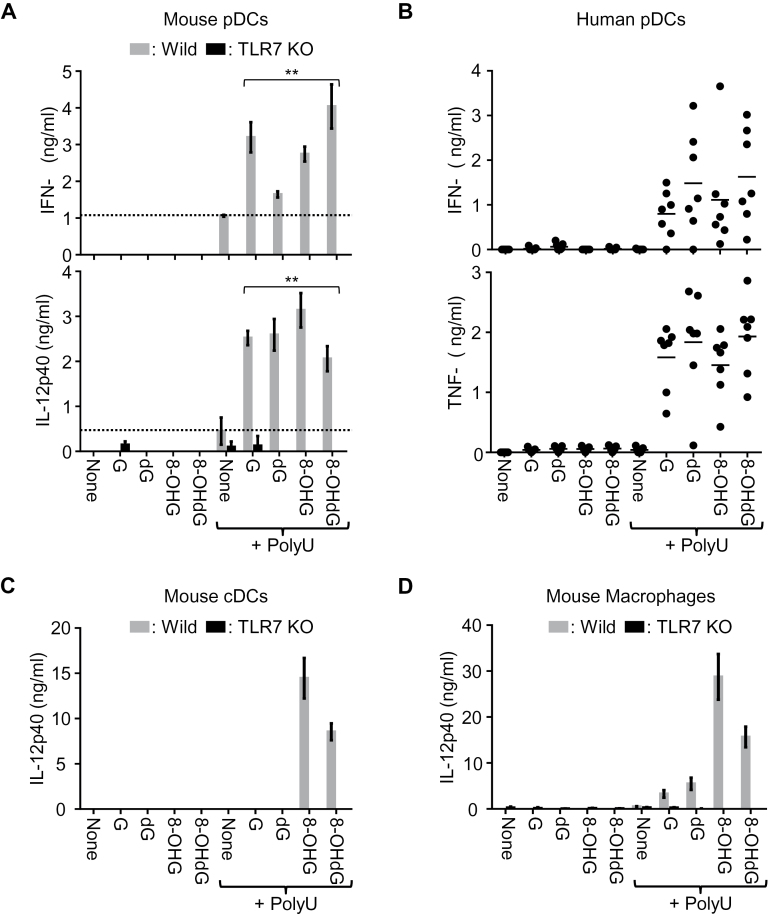
8-OHG and 8-OHdG are comparable to G and dG in activating TLR7 in vitro. Cytokine production from various immune cells was determined by ELISA. (A) Mouse BM-pDCs from WT and Tlr7 −/− mice were stimulated with 100 μM G analogs with or without poly U (20 μg ml–1). Two-tailed Student’s t-test was used to evaluate the statistical significance between polyU only and guanosine derivatives with polyU (**P < 0.01). (B) Human pDCs from seven healthy donors were analyzed in the same way as in (A). (C and D) BM-cDCs (C) and BM-MCs (D) from WT or Tlr7 −/− mice were analyzed as in (A). Data represent the mean ± SD of triplicate samples. All the results except (B) are representative of more than three independent experiments.
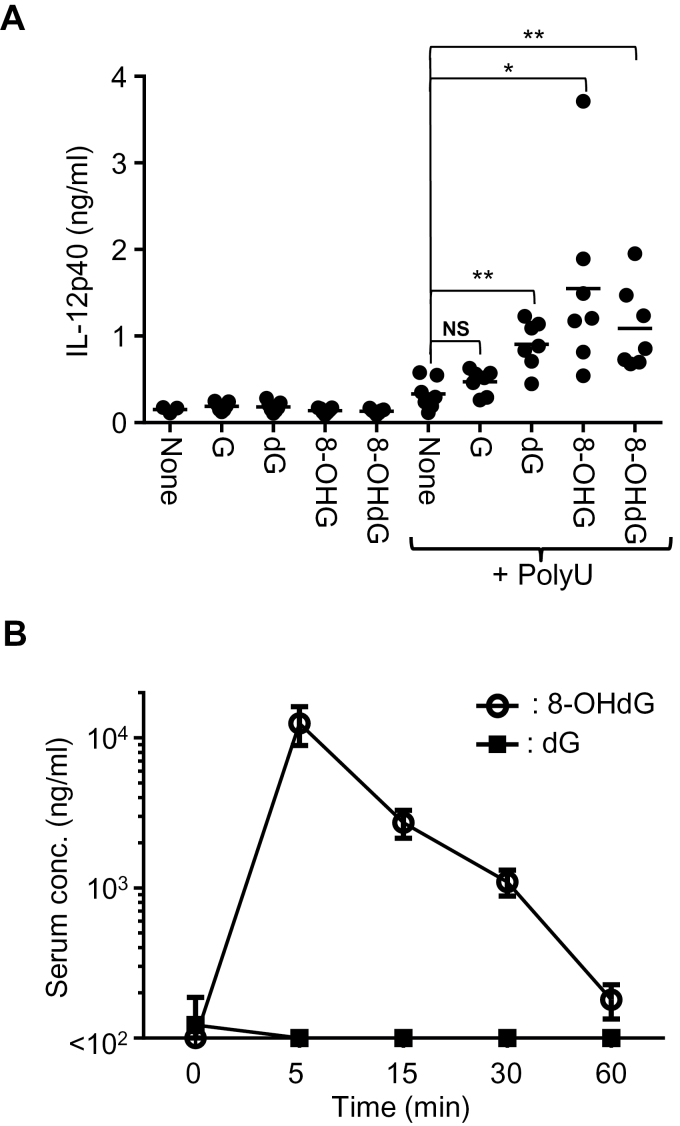
Fig. 5.
The cytokine-inducing activity and serum level of 8-OHdG after in vivo injection. (A) Mice (n = 7) were administered with G and modified G with or without polyU. After 3h, blood was collected and serum IL-12 p40 was determined by ELISA. Two-tailed Student’s t-test was used to evaluate the statistical significance between polyU only and G derivatives with polyU (**P < 0.01; *P < 0.05; NS, not significant). (B) Mice (n = 6) were administered with dG or 8-OHdG and blood was collected at each time point. Serum dG and 8-OHdG were determined.
Results
Uridine and ORN synergistically activate huTLR8
To study the specificity of TLR8 for nucleosides, NF-κB luciferase reporter assay using HEK293 cells was conducted. As reported previously (11), huTLR8 was activated by U and ssRNA40. Other nucleosides such as A, G, I, C and T failed to activate huTLR8 (Fig. 1A). A variety of ssRNAs were next used to stimulate huTLR8. HuTLR8 was synergistically activated by U with a variety of ssRNAs including RNA9.2s, ssRNA40 and ORN06 (Fig. 1B). Unexpectedly, huTLR8 was co-stimulated even by non-agonistic ORNs including ssRNA41, a non-agonistic control for ssRNA40, TLR7 ligand PolyU and mouse TLR13 ligand Sa12, 12 mer ORNs from S. aureus 23S ribosomal RNA (18, 19) (Fig. 1B), while polyA, polyG or polyC failed to activate huTLR8. This finding demonstrates that human huTLR8 responds to a broad range of ORNs in the presence of U. Considering that TLR13 does not function in humans, huTLR8 is likely to have a role in sensing bacterial 23S ribosomal RNA in humans.
U and ORN bind to huTLR8 at the dimerization interface and the concave surface of the huTLR8 horseshoe structure, respectively (11). To clarify the role of these binding sites in synergistic activation of huTLR8 by U and ssRNA40, we performed the mutational analysis on huTLR8. Whereas the mutation at the U-binding site (F405A) completely abolished TLR8 responses to both a synthetic small chemical ligand CL075 and U with ssRNA40, the three mutations at the ORN-binding site (H373A, R375A and L474A) did not change CL075 response but significantly impaired huTLR8 response to U with ssRNA40 (Fig. 1C). These results indicate that the U-binding site is primarily responsible for huTLR8 activation and that the ORN-binding site strengthens huTLR8 activation by U.
In addition to U, there are various type of modified Us in vivo. To study the activity of these U-related molecules on huTLR8 response, NF-κB luciferase reporter assay was also conducted. Compared to U, 5′-phosphorylation of U (UMP, UDP, UTP) clearly attenuated huTLR8 response in synergy with ssRNA40. HuTLR8 was not activated by uracil nucleobase (Fig. 1D). In contrast, modified Us such as deoxyuridine (dU), β-pseudouridine and 5-methyluridine were able to activate huTLR8 in the presence of ssRNA40 (Fig. 1E). To further confirm these synergistic response via huTLR8 in primary immune cells, human macrophages were stimulated by U or modified Us with ssRNA40. As a result, U with ssRNA40 robustly induced TNF-α production (Fig. 1F). Weak but significant production of TNF-α was also induced by stimulation with dU, β-pseudouridine or 5-methyluridine in the presence of ssRNA40. These results demonstrate that huTLR8 responds to U and modified Us under stimulation with ssRNA.
Guanosine and ssRNA synergistically activate TLR7
TLR7 is similar to human TLR8 in responding to both synthetic small chemical ligand and ssRNA. To examine whether TLR7 responds to nucleosides, human or mouse TLR7 was treated by various nucleosides under stimulation with polyU in the NF-κB reporter assay. G synergistically activated human and mouse TLR7 with polyU (Fig. 2A and B). It is to be noted that TLR7 was activated by G and polyU without DOTAP, whereas U and ssRNA40 required DOTAP to activate huTLR8. This finding indicates that TLR7 is able to detect extracellular G. In the presence of G, TLR7 was activated by a variety of ssRNAs such as RNA9.2s, ORN06 and polyU, although the TLR7 response to ORN06 was weaker than to RNA9.2s and polyU (Fig. 2C and D). This finding indicates that the ORN specificity of TLR7 is much more restricted than that of huTLR8.
Additionally, we studied the activity of various types of modified Gs on TLR7 response, using NF-κB luciferase reporter assay. As with modified Us on huTLR8 response, 5′-phosphorylation of G (GMP, GDP, GTP) attenuated TLR7 response in synergy with polyU and guanine nucleobase did not activate TLR7 at all (Fig. 2E and F). On the other hand, human and mouse TLR7 was appreciably activated by dG, 8-OHG and 8-OHdG in synergy with polyU (Fig. 2G and H). Since 8-OHdG is a well-known marker for oxidative damage to genomic DNA, our finding that 8-OHdG is a ligand for TLR7 suggests the involvement of TLR7 in oxidative stress response. We also found that human and mouse TLR7 synergistically responded to 7-methylguanosine, which could be released by mRNA decapping enzyme, with polyU. These results demonstrate that not only G and dG but also various modified Gs are ligands for human and mouse TLR7.
ssRNA enhances the binding of G, dG, 8-OHG and 8-OHdG to TLR7
HuTLR8 is shown to bind to U with Kd of 55 μM, which is enhanced up to 1.0 μM in the presence of ssRNA (11). To determine the affinity of TLR7 to G, ITC was performed with the purified simian TLR7 ectodomain. The affinity of G to TLR7+polyU was determined to be 1.5 μM (Fig. 3A). Modified Gs such as dG, 8-OHG and 8-OHdG bound to TLR7 + polyU with Kd of 1.8, 11 and 11 μM, respectively. In the absence of polyU, the heat released or absorbed upon TLR7-ligand interaction was too small to determine the affinity constants (Fig. 3B). The interaction of G with polyU alone was not detected in ITC analyses (Fig. 3C). Consistent with functional analyses in Fig. 2(A and B), U also did not release or absorb heat in the ITC experiment (Fig. 3D). Finally, we studied a synthetic TLR7 ligand loxoribine. Loxoribine binding to TLR7 + polyU but not to TLR7 alone was detected (Fig. 3E and F). These results demonstrate that TLR7 directly interacts with G and modified Gs only in the presence of polyU. The affinities of 8-OHG and 8-OHdG for simian TLR7 were weaker than those of G and dG.
Fig. 3.
TLR7 interacts with G in the presence of ssRNA. The binding affinities of G, dG, 8-OHG and 8-OHdG for simian TLR7 ectodomain (sTLR7) with or without PolyU were determined by ITC. (A, B and D–F) ITC analysis of guanosine analogs (G, dG, 8-OHG, 8-OHdG), loxoribine and U binding to sTLR7 with (A, D and E) or without (B and F) PolyU. (C) ITC analysis of the binding of G to PolyU. Calculated Kd are also shown. N.D. means ‘not determined’.
8-OHdG shows comparable or even stronger co-stimulatory activity than G/dG in vitro
Next, we examined the responses of innate immune cells to nucleosides and ORNs. BM cells were allowed to differentiate into BM-pDCs, BM-cDCs and BM-MCs. Maturation and purity of these innate immune cells were confirmed by FACS analyses and no alteration was seen in Tlr7 −/− mice (Supplementary Figure 1, available at International Immunology Online). In mouse BM-pDCs, 8-OHG and 8-OHdG were as potent as G and dG to induce IFN-α and IL-12 p40 production in synergy with polyU (Fig. 4A). Consistent with mouse BM-pDCs, human pDCs robustly produced IFN-α and TNF-α in response to G, dG, 8-OHG, or 8-OHdG in the presence of poly U (Fig. 4B). In sharp contrast with pDCs, when BM-cDCs or BM-MCs were stimulated with nucleosides and polyU, G and dG showed much weaker IL-12 p40 production than 8-OHG and 8-OHdG (Fig. 4C and D). In all immune cell types, nucleoside alone failed to induce cytokine production. 8-mercaptoguanosine (8-SGuo) is reported to activate TLR7 in B cells (20). 8-SGuo alone, however, failed to activate BM-MCs. Only in the presence of polyU, did 8-SGuo induce IL-12p40, IL-6 and TNF-α production in WT but not Tlr7 −/− BM-MCs (Supplementary Figure 2, available at International Immunology Online). These results demonstrate that modified G such as 8-OHG and 8-OHdG were comparable to G or dG as TLR7 ligands in vitro, despite the finding that 8-OHG and 8-OHdG showed lower affinities to TLR7 than G or dG in ITC experiments.
8-OHdG is more stable than dG in vivo
Finally, the synergistic effect of G derivatives and ORNs was studied in vivo. When dG, 8-OHG or 8-OHdG was intravenously administered together with polyU, the serum IL-12p40 level was significantly increased compared with polyU only (Fig. 5A), demonstrating that dG, 8-OHG and 8-OHdG could work as TLR7 ligands in vivo. dG is reported to be rapidly metabolized in the presence of rat liver homogenates, whereas 8-OHdG is stable (21). We also studied in vivo turnover of dG and 8-OHdG by evaluating the serum concentration of dG and 8-OHdG after intravenous administration of 2 μmol dG or 8-OHdG. The serum dG level was about 100ng ml–1 at steady state and the administration of dG did not alter the serum level of dG (Fig. 5B). In contrast, 8-OHdG was undetectable at steady state and the administration of 8-OHdG clearly increased the serum 8-OHdG level to 10 μg ml–1 at 5min after administration (Fig. 5B). 8-OHdG appeared to be metabolically stable in vivo, whereas serum dG is strictly controlled.
Unc93b1D34A/D34A mice show hyper-responsiveness to 8-OHG and 8-OHdG in the presence of polyU
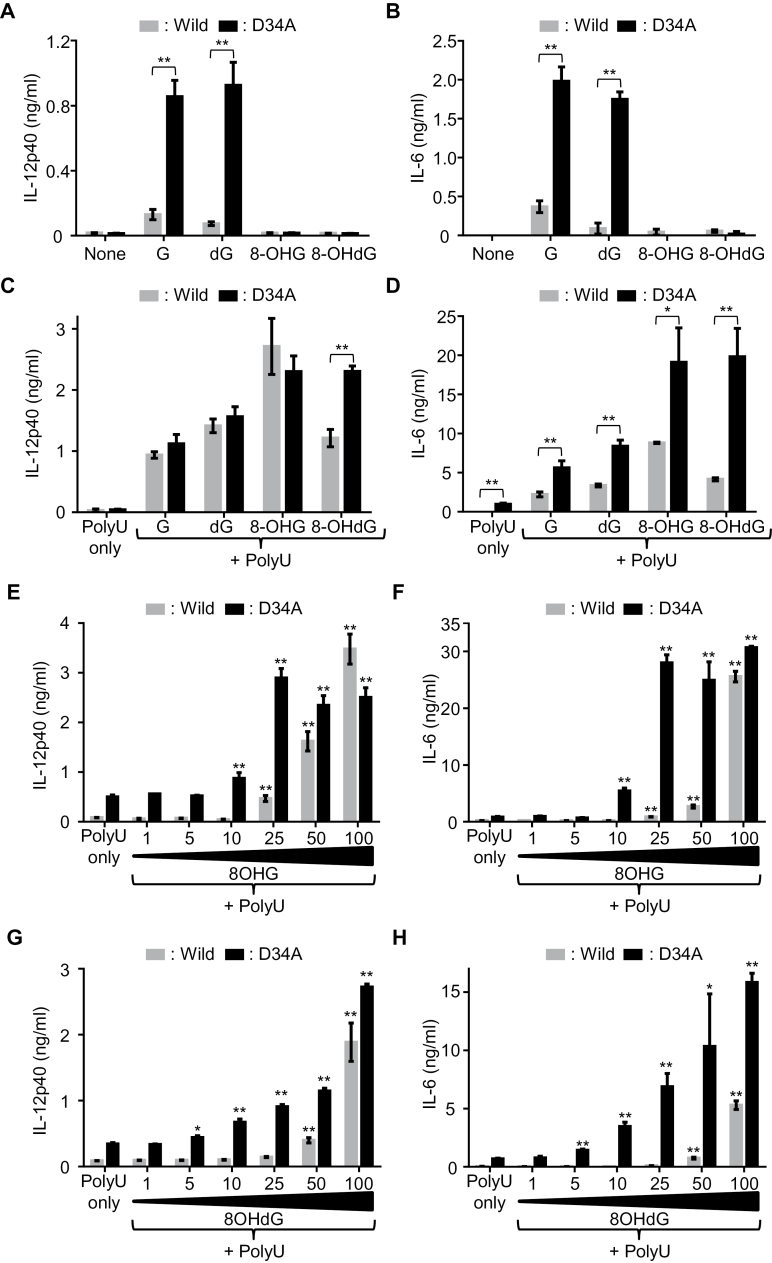
Finally, to ask whether TLR7 responses to G analogs have a pathogenic role in inflammatory disease, we studied Unc93b1 D34A/D34A mice, which suffer from systemic inflammatory diseases due to TLR7 hyper-responsiveness (22). BM-MCs from WT or Unc93b1 D34A/D34A mice were stimulated with G, dG, 8-OHG or 8-OHdG with or without polyU, and the production of IL-12p40 and IL-6 was measured. In sharp contrast with WT BM-MCs, Unc93b1 D34A/D34A BM-MCs robustly responded to G or dG alone (Fig. 6A and B). Interestingly, 8-OHG or 8-OHdG alone failed to activate Unc93b1 D34A/D34A BM-MCs. In the presence of polyU, Unc93b1 D34A/D34A BM-MCs showed significantly higher production of IL-12p40 and IL-6 than WT BM-MCs in response to 8-OHG or 8-OHdG (Fig. 6C and D). Since oxidized G analogs induced stronger cytokine production in Unc93b1 D34A/D34A BM-MCs, we studied lower concentrations of 8-OHG/8-OHdG than 100 μM. 8-OHG and 8-OHdG induced IL-12p40 production in WT BM-MCs at 50 or 100 μM (Fig. 6B). Unc93b1 D34A/D34A BM-MCs responded to the modified Gs at lower concentrations of 5–10 μM and produced a larger amount of IL-12p40 and IL-6. These findings identified G/dG only or 8-OHG/8-OHdG with ssRNA as candidates for endogenous pathogenic TLR7 ligands in Unc93b1 D34A/D34A mice.
Fig. 6.
Macrophages from Unc93b1 D34A/D34A mice are more sensitive to G analogs. Mouse BM-MCs from WT and Unc93b1 D34A/D34A mice were stimulated with indicated ligands. The production of IL-12p40 (A, C, E and G) and IL-6 (B, D, F and H) in culture supernatant was determined by ELISA. (A–D) BM-MCs were stimulated with 200 μM G analogs without polyU (A and B), or 100 μM G analogs with polyU (10 μg ml–1) (C and D). Two-tailed Student’s t-test was used to evaluate the statistical significance between WT and Unc93b1 D34A/D34A (*P < 0.05; **P < 0.01). (E–H) In the presence of polyU (10 μg ml–1), BM-MCs were stimulated with titrated concentrations of 8-OHG (1–100 μM) (E and F) or 8-OHdG (1–100 μM) (G and H). Two-tailed Student’s t-test was used to evaluate the statistical significance of each data compared with polyU only both in WT and Unc93b1 D34A/D34A (*P < 0.05; **P < 0.01). Data represent the mean ± SD of triplicate samples. All the results are representative of three independent experiments.
Discussion
Although TLR7 and TLR8 have been thought to sense ssRNA, synthetic small chemical ligands are able to activate TLR7 and TLR8, raising a question about how two structurally and chemically distinct ligands activate TLR7 and TLR8. Our previous study clarifies that huTLR8 binds U and ORNs, degradation products of ssRNA (11), indicating that huTLR8 is a U sensor. In the present study, we show that U and ssRNA actually induces cytokine production from human macrophages.
In addition, we provide evidence that TLR7 is a G sensor. The interaction between TLR7 and G was clearly enhanced by ssRNA, indicating that G and ssRNA independently bind to TLR7. Although our ITC assay could detect the heat released by the interaction between TLR7 and its ligand in the presence of polyU, we failed to detect any heat released or absorbed in the absence of polyU. We interpreted these results as enhancement of the binding affinity of ligand for TLR7 in the presence of polyU, although we are aware that molecular interactions sometimes give low ITC signals and the possibility of ligand binding to TLR7 alone still remains. Moreover, since native full-length TLR7 in the membrane might have stronger binding affinity than the soluble TLR7 ectodomain, it would be possible that G analogs alone directly bind to TLR7 in the cells.
TLR7 also responded to GMP and dG, as well as to G. As UMP does not interact with huTLR8 (11), GMP would not bind to TLR7. GMP conversion into G would enable TLR7 activation. In contrast to GMP, dG directly bound to TLR7 in ITC analyses. Considering that dG is generated by DNA degradation, TLR7 might be able to sense DNA degradation in the presence of ssRNA.
It is important to understand the situation where TLR7 and TLR8 are activated by nucleosides and ssRNAs. RNA is known to be degraded by endonucleases and exonucleases during viral infection. By accelerating host mRNA decay, many viruses facilitate their own mRNA translation and limit host-defense responses. Decapping enzymes and endonucleases for mRNAs are encoded by a variety of viruses including herpesviruses, influenza viruses, coronaviruses and vaccinia viruses (23). On the other hand, RNA digestion by cellular nucleases is known to have a role in restricting viral infection (23, 24). First, the nonsense-mediated decay (NMD) pathway has a primary role in surveying mRNA integrity and eliminating atypical mRNAs. Interestingly, NMD restricts positive-strand RNA viruses. Second, type I IFN induces the zinc-finger antiviral protein, which binds to viral RNA and recruits it to the exosome, cellular RNA decay machinery. TLR7 and TLR8 are likely to sense RNA degradation by both viral and host nucleases.
The present study also shows that TLR7 and TLR8 respond to modified nucleosides. Previously, modification of RNAs by pseudouridine and/or methylated nucleosides was shown to attenuate TLR7 and TLR8 responses (12). It is possible that ssRNA interaction with TLR7 is impaired by RNA modification. Alternatively, RNA modification may alter RNA degradation, as DNA degradation by DNase III is impaired by 8-hydroxylation (15). Altered RNA degradation may impair TLR7 and TLR8 activation. In contrast to modified RNA, modified nucleosides were able to activate TLR7 and TLR8. Despite the discrimination between self and pathogen RNA based on RNA modification, TLR7 and TLR8 are known to be activated in autoimmune diseases (4, 22, 25). Considering BM-MCs from autoimmune prone Unc93b1 D34A/D34A mice showed enhanced response to 8-OHG/8-OHdG in the presence of ssRNA, modified nucleosides may have a role as endogenous ligands in a variety of autoimmune diseases.
We stress again that TLR7 responded to 8-OHG and 8-OHdG with ssRNA. 8-hydroxylation of guanine bases on DNA and RNA is the most common modification upon oxidative stress conditions induced by inflammation, ultraviolet light irradiation, and radiation. For example, ultraviolet sunlight causes the generation and the accumulation of 8-OHdG in the skin. Oxidative damage to genomic DNA induces the removal and repair of oxidative DNA modification mediated by repair enzyme systems, resulting in the excretion of 8-OHdG into circulation and urine (26). 8-OHdG is therefore used for a marker of oxidative stress in vivo. Patients with lupus erythematosus often develop skin lesions after UV exposure. 8-OHdG is known to accumulate in skin lesions but not in UV-exposed skin of healthy control (15), suggesting higher sensitivity to oxidative stress and/or impaired clearance of modified genomic DNA in autoimmune diseases. 8-hydroxilation also occurs to phagocytosed microbial DNA by exposure to ROS in lysosomes. In inflammatory lesions, the oxidative burst results in 8-OHdG generation in neutrophil extracellular traps (15). Subsequent degradation of modified DNAs/RNAs would release G, dG, 8-OHG and 8-OHdG, as well as ssRNA fragments. Interestingly, although TLR7 had weaker affinity for 8-OHdG than that for dG, 8-OHdG, but not G and dG, induced synergistic production of IL-12 p40 with polyU in mouse BM-cDCs. Since 8-OHdG is reported to be more stable than dG in treatment with liver homogenate (21), 8-OHdG may be resistant to degradation by immune cells. Consistent with this possibility, we also found that 8-OHdG was more stable than dG in the serum, suggesting that 8-OHdG can probably escape from the metabolic absorption and the degradation pathway for dG in vivo. These findings uncover the important role of 8-OHdG as an inducer of inflammation via TLR7 under oxidative stress. In addition, G is metabolized into guanine by an enzyme called purine nucleoside phosphorylase (27). TLR7 responses to G or dG with polyU in BM-cDCs and BM-MCs were much weaker than those in BM-pDCs. Conversion from G into guanine may be much faster in BM-cDCs and BM-MCs than in BM-pDCs. It may be reasonable that cells with phagocytosis activity have a robust system metabolizing NAs.
Unc93b1 D34A/D34A mice show systemic inflammation such as splenomegaly and hepatitis due to myeloid cell infiltration (22). These phenotypes are caused by TLR7 hyper-responsiveness. Unc93B1 controls TLR7 distribution and D34A mutation increases TLR7 distribution in lysosomes (22). Unc93b1 D34A/D34A BM-MCs responded to G or dG alone, whereas WT BM-MCs did not. Further, Unc93b1 D34A/D34A BM-MCs showed stronger responses to synergistic stimulation by 8-OHG or 8-OHdG with polyU. The D34A mutation of Unc93B1 may increase TLR7 exposure to RNA metabolites such as G, modified Gs and short RNA fragments. TLR7 in WT BM-MCs is likely to be kept away from RNA metabolites as a mechanism preventing autoimmune responses. This might be the reason why TLR7 responses to G or modified Gs with polyU is detectable only at high concentrations of those G analogs. A large amount of G or modified G would be required to reach TLR7. When massive cell deaths occur, the RNA-ribonucleoprotein complex is released and endocytosed by immune cells. The RNA-protein complex may induce alterations in the distribution of RNA metabolites or TLR7. Such alteration might induce TLR7 exposure to RNA/DNA metabolites and cause TLR7 activation.
Intraperitoneal injection of hydrocarbon oils such as pristane induces lupus-like disease in mice (28). TLR7 promotes disease progression in pristane-induced lupus. However, it has never been clarified how TLR7 is activated in this model. Massive cell death induced by pristane are likely to activate TLR7. Interestingly, oxidative damage is reported to contribute to this disease model (29). In this model, TLR7 may be activated by 8-OHdG and ssRNA derived from damaged or dead cells.
This is the first report to identify endogenous small-molecule ligands for TLR7. TLR7 recognizes various nucleosides such as G, dG, 7-MetG, 8-OHG and 8-OHdG in the presence of ssRNA to induce innate immune response. Because oxidative damage generates metabolically stable 8-OHdG, TLR7 may have an important role in inducing inflammation under oxidative stress condition.
Supplementary data
Supplementary data are available at International Immunology Online.
Funding
This work was supported in part by Grant-in-Aid from the Ministry of Education, Culture, Sports, Science and Technology for Scientific Research A (25253032) and C (24591472); and Grant-in Aid for Exploratory Research (26670235 and 15K15148).
Supplementary Material
Acknowledgements
We thank Prof. Shizuo Akira (Osaka Univ.) for providing the Tlr7 −/− mice.
Conflict of interest statement: The authors declare no competing financial results.
References
- 1. Miyake K. 2007. Innate immune sensing of pathogens and danger signals by cell surface Toll-like receptors. Semin. Immunol. 19:3. [DOI] [PubMed] [Google Scholar]
- 2. Kaisho T. and Akira S. 2006. Toll-like receptor function and signaling. J. Allergy Clin. Immunol. 117:979. [DOI] [PubMed] [Google Scholar]
- 3. Deane J. A., Pisitkun P., Barrett R. S., et al. 2007. Control of toll-like receptor 7 expression is essential to restrict autoimmunity and dendritic cell proliferation. Immunity 27:801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Guiducci C., Gong M., Cepika A. M., et al. 2013. RNA recognition by human TLR8 can lead to autoimmune inflammation. J. Exp. Med. 210:2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lau C. M., Broughton C., Tabor A. S., et al. 2005. RNA-associated autoantigens activate B cells by combined B cell antigen receptor/Toll-like receptor 7 engagement. J. Exp. Med. 202:1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Barrat F. J., Meeker T., Gregorio J., et al. 2005. Nucleic acids of mammalian origin can act as endogenous ligands for Toll-like receptors and may promote systemic lupus erythematosus. J. Exp. Med. 202:1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Vollmer J., Tluk S., Schmitz C., et al. 2005. Immune stimulation mediated by autoantigen binding sites within small nuclear RNAs involves Toll-like receptors 7 and 8. J. Exp. Med. 202:1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ahn C. S. and Huang W. W. 2014. Imiquimod in the treatment of cutaneous warts: an evidence-based review. Am. J. Clin. Dermatol. 15:387. [DOI] [PubMed] [Google Scholar]
- 9. Micali G. Lacarrubba F. Dinotta F. Massimino D. and Nasca M. R. 2010. Treating skin cancer with topical cream. Expert Opin. Pharmacother. 11:1515. [DOI] [PubMed] [Google Scholar]
- 10. Tanji H. Ohto U. Shibata T. Miyake K. and Shimizu T. 2013. Structural reorganization of the Toll-like receptor 8 dimer induced by agonistic ligands. Science 339:1426. [DOI] [PubMed] [Google Scholar]
- 11. Tanji H., Ohto U., Shibata T., et al. 2015. Toll-like receptor 8 senses degradation products of single-stranded RNA. Nat. Struct. Mol. Biol. 22:109. [DOI] [PubMed] [Google Scholar]
- 12. Karikó K. Buckstein M. Ni H. and Weissman D. 2005. Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity 23:165. [DOI] [PubMed] [Google Scholar]
- 13. Kroese L. J. and Scheffer P. G. 2014. 8-hydroxy-2’-deoxyguanosine and cardiovascular disease: a systematic review. Curr. Atheroscler. Rep. 16:452. [DOI] [PubMed] [Google Scholar]
- 14. Wiseman H. and Halliwell B. 1996. Damage to DNA by reactive oxygen and nitrogen species: role in inflammatory disease and progression to cancer. Biochem. J. 313(Pt 1):17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gehrke N., Mertens C., Zillinger T., et al. 2013. Oxidative damage of DNA confers resistance to cytosolic nuclease TREX1 degradation and potentiates STING-dependent immune sensing. Immunity 39:482. [DOI] [PubMed] [Google Scholar]
- 16. Muta T. and Takeshige K. 2001. Essential roles of CD14 and lipopolysaccharide-binding protein for activation of toll-like receptor (TLR)2 as well as TLR4 Reconstitution of TLR2- and TLR4-activation by distinguishable ligands in LPS preparations. Eur. J. Biochem. 268:4580. [DOI] [PubMed] [Google Scholar]
- 17.Ito, T., Kanzler, H., Duramad, O., Cao, W. and Liu, YJ. 2006. Specialization, kinetics, and repertoire of type 1 interferon responses by human plasmacytoid predendritic cells. Blood 107:6. [DOI] [PubMed] [Google Scholar]
- 18. Li X. D. and Chen Z. J. 2012. Sequence specific detection of bacterial 23S ribosomal RNA by TLR13. Elife 1:e00102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Oldenburg M., Krüger A., Ferstl R., et al. 2012. TLR13 recognizes bacterial 23S rRNA devoid of erythromycin resistance-forming modification. Science 337:1111. [DOI] [PubMed] [Google Scholar]
- 20. Tsukamoto Y., Nagai Y., Kariyone A., et al. 2009. Toll-like receptor 7 cooperates with IL-4 in activated B cells through antigen receptor or CD38 and induces class switch recombination and IgG1 production. Mol. Immunol. 46:1278. [DOI] [PubMed] [Google Scholar]
- 21. Shi M. Takeshita H. Komatsu M. Xu B. Aoyama K. and Takeuchi T. 2005. Generation of 8-hydroxydeoxyguanosine from DNA using rat liver homogenates. Cancer Sci. 96:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fukui R., Saitoh S., Kanno A., et al. 2011. Unc93B1 restricts systemic lethal inflammation by orchestrating Toll-like receptor 7 and 9 trafficking. Immunity 35:69. [DOI] [PubMed] [Google Scholar]
- 23. Abernathy E. and Glaunsinger B. 2015 Emerging roles for RNA degradation in viral replication and antiviral defense. Virology 479–480:600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rigby R. E. and Rehwinkel J. 2015. RNA degradation in antiviral immunity and autoimmunity. Trends Immunol. 36:179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Christensen S. R. Shupe J. Nickerson K. Kashgarian M. Flavell R. A. and Shlomchik M. J. 2006. Toll-like receptor 7 and TLR9 dictate autoantibody specificity and have opposing inflammatory and regulatory roles in a murine model of lupus. Immunity 25:417. [DOI] [PubMed] [Google Scholar]
- 26. Wu L. L. Chiou C. C. Chang P. Y. and Wu J. T. 2004. Urinary 8-OHdG: a marker of oxidative stress to DNA and a risk factor for cancer, atherosclerosis and diabetics. Clin. Chim. Acta 339:1. [DOI] [PubMed] [Google Scholar]
- 27. Nyhan W. L. 2005. Disorders of purine and pyrimidine metabolism. Mol. Genet. Metab. 86:25. [DOI] [PubMed] [Google Scholar]
- 28. Lee P. Y., Kumagai Y., Li Y., et al. 2008. TLR7-dependent and FcgammaR-independent production of type I interferon in experimental mouse lupus. J. Exp. Med. 205:2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Minhas U. Das P. and Bhatnagar A. 2011. Role of reactive intermediates in the immunopathogenesis of the pristane-induced Balb/c model of lupus. Lupus 20:1421. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.