TLR7 and TLR9 ligation induces M2-like macrophages
Keywords: activation, antigen presentation, macrophages, TLR
Abstract
The toll-like receptors (TLRs) are important innate receptors recognizing potentially pathogenic material. However, they also play a significant role in the development of Alzheimer’s disease, cancer, autoimmunity and the susceptibility to viral infections. Macrophages are essential for an effective immune response to foreign material and the resolution of inflammation. In these studies, we examined the impact of different TLR ligands on macrophage cell function. We demonstrate that stimulation of all TLRs tested increases the phagocytosis of apoptotic cells by macrophages. TLR7 and TLR9 ligation decreased the levels of the surface co-expression molecules CD86 and MHCII, which was associated with a concomitant reduction in antigen presentation and proliferation of T cells. This down-regulation in macrophage function was not due to an increase in cell death. In fact, exposure to TLR7 or TLR9 ligands promoted cell viability for up to 9 days, in contrast to TLR3 or TLR4. Additionally, macrophages exposed to TLR7/TLR9 ligands had a significantly lower ratio of Il-12/Il-10 mRNA expression compared with those treated with the TLR4 ligand, LPS. Taken together, these data demonstrate that TLR7/TLR9 ligands push the macrophage into a phagocytic long-lived cell, with a decreased capacity of antigen presentation and reminiscent of the M2 polarized state.
Introduction
An effective response to a microbial challenge depends upon the presence of different families of pathogen-recognition receptors (PRRs). The toll-like receptors (TLRs) are an important innate PRR family that recognizes specific pathogen-associated molecular patterns (PAMPs) (1). Following ligation with their respective ligand, these TLRs initiate a series of inflammatory events that depend upon the MyD88 and/or TRIF signaling pathways. They are known to influence the functions of many immune cells, but a comparative study on macrophages has yet to be completed.
Macrophages provide a first line of defense against invading microorganisms, sensing of which through PRRs triggers innate and adaptive immune responses (2). These cells are critical for the uptake and removal of potentially pathogenic material, antigen processing and T-lymphocyte presentation. However, the phagocytic processes involved can promote tissue destruction directly, through the generation of reactive oxygen species (ROS) and enzymes, and indirectly, by recruiting and activating other inflammatory cells. Macrophages also play a role in the resolution of inflammation and wound healing, through engulfing and clearing apoptotic cells and producing cytokines such as IL-1 receptor antagonist, IL-10 and TGF-β (3, 4). Therefore, dysregulation can have multiple effects on the system.
A hallmark of macrophages is their plasticity and ability for functional polarization following multiple environmental signals (5, 6). Although the polarization phenotypes can be diverse, two main macrophage polarization states, termed M1 and M2, have been defined in vitro. Proinflammatory M1 or classically activated macrophages are generated by priming with inflammatory cytokines such as IFN-γ in combination with LPS (5). These macrophages are characterized by the production of inflammatory cytokines, an IL-12hi/IL-10lo phenotype, and increased antigen-presentation capability. M2 macrophages can be induced by exposure to anti-inflammatory cytokines such as IL-4 and IL-13; by combined exposure to immune complexes (ICs) and TLR or IL-1R agonists; or by exposure to IL-10 (7). Although subtle functional differences exist between the M2 macrophages in terms of the stimuli used to generate them, they mainly share the common characteristic of an IL-12lo/IL-10hi phenotype and other features such as efficient scavenging ability and the promotion of immunoregulatory and tissue remodeling functions (5, 6, 8). Distinct macrophage subtypes perform markedly different functions based on their localization within the tissue (9). Additionally, the macrophage polarization state dictates the capacity of the macrophages to drive antigen-specific T-cell proliferation. For example, a subtype of M2 macrophages associated with the autoimmune disease SLE are generated by stimulation with LPS and ICs (7, 10). These cells express high levels of CD86 and MHCII and are efficient antigen-presenting cells (APCs) (8).
Macrophages can be activated by cytokines or potential pathogenic material. The bacterial cell wall component, LPS, is recognized by TLR4 that is located on the cell surface. TLR7 and TLR9 are located inside the cell, within endosomes, recognize ds-RNA and ss-RNA, respectively, and are dependent upon the MyD88 signaling pathway for an immune response. TLR3, activated by bacterial unmethylated ds-DNA, is TRIF dependent, whereas LPS stimulation of TLR4 may trigger both MyD88 and TRIF downstream signaling pathways.
In these studies, we evaluated the effects of multiple TLR ligands on macrophage functions important for pathogen recognition, immune activation and the resolution of inflammation. Surprisingly, we found that different TLR ligands induce distinct functional and biochemical properties in murine macrophages. In contrast to TLR4 stimulation by LPS, we determined that TLR7 and TLR9 ligation induced macrophages into a long-lived phagocytic state with a decreased capability of antigen presentation and T-cell proliferation. We speculate that this could be a protective mechanism that evolved to prevent presentation of self-antigens, such as self-nucleic acids, from dead cells ingested by macrophages.
Methods
Mice
Mice were bred at the Biological Resource Centre, A*STAR, in Singapore. Breeding pairs for C57BL6J (B6) and Rag1−/−/transgenic OT-II mice were originally purchased from the Jackson Laboratory (Bar Harbor, ME, USA). The care and use of laboratory animals conformed to the National Institutes of Health guidelines, and all experimental procedures conformed to an Institutional Animal Care and Use Committee approved animal protocol.
Reagents
TLR ligands CpG (type B, ODN 1826), R848, LPS-EB ultrapure and Poly (I:C) LMW were purchased from Invivogen (San Diego, CA, USA). Ovalbumin (OVA) and etoposide were from Sigma-Aldrich (St Louis, MO, USA). OVA peptide 323–339 (ISQAVHAAHAEINEAGR) was purchased from GenScript (Piscataway, NJ, USA). Reagents were re-suspended in endotoxin-free water and media was regularly checked for the absence of endotoxin, using a Limulus Amoebocyte Lysate test (Lonza, Walkersville, MD, USA). Unless otherwise indicated, cells were stimulated with 0.1 μM CpG, 1 μg ml–1 R848, 10ng ml–1 LPS and 1 μg ml–1 Poly I:C. Where indicated, cells were stimulated with IFN-γ (supernatants from X63 cell hybridoma transfected with IFN-γ) used at a 1:100 dilution, a gift from Dr Antonio Celada, University of Barcelona (Barcelona, Spain) (11).
Cell preparation and cell culture
Cells were cultured in complete RPMI 1640 (cRPMI), consisting of RPMI 1640 (Gibco, Life Technologies, Carlsbad, CA, USA) supplemented with 10 or 20% FBS (Hyclone, Thermo Fisher Scientific, Waltham, MA, USA), 15mM HEPES, 100 µg ml–1 streptomycin, 100U ml–1 penicillin, 200 µM l-glutamine, 10 µM non-essential amino acids, 100 µM sodium pyruvate (all from Gibco) and 45 µM 2-mercaptoethanol (Sigma-Aldrich, St Louis, MO, USA). Cells were maintained at 37°C in a 5% CO2 incubator. To obtain bone-marrow-derived macrophages (BMDMs), BM cells were isolated from femurs and tibia of 6- to 8-week-old female mice. They were then cultured on untreated Sterilin dishes (Thermo Fisher Scientific, Waltham, MA, USA) (12) for 7 days in cRPMI media containing 20% FBS supplemented with 30% L929-conditioned media (L929-cm). L929-cm was prepared in our laboratory as described previously (13). Macrophages were collected on day 7 by gentle scraping on ice and their phenotype was confirmed by the expression of CD11b, F4/80, CD64, CD16/CD32 and CD115 using flow cytometry (Supplementary Figure S1, available at International Immunology Online). BMDMs used for cell culture and stimulations were plated at 0.5×106 cells ml–1 and left to adhere for 3–4h in cRPMI before stimulation.
Bone-marrow-derived dendritic cells (BMDCs) were produced by culturing BM cells in cRPMI containing 10% FBS with 10ng ml–1 GM-CSF and 10ng ml–1 IL-4 (both from R&D Systems, Minneapolis, MN, USA) in T-75 vented flasks (Corning, NY, USA). On day 3, the supernatant and loosely adherent cells were collected, centrifuged and transferred into a new flask with fresh media and supplements. Immature BMDCs were then collected on day 7 and their phenotype confirmed by the expression of CD11c and CD11b using flow cytometry.
Peritoneal macrophages were obtained by lavage with ice-cold PBS immediately following euthanasia. They were seeded onto plates according to the number of live F4/80hiCD11bhi cells calculated by flow cytometry phenotyping and counting using a hemocytometer. Splenic DCs were sorted from collagenase-digested spleens using BD FACSAria III or V instruments. Briefly, spleens were crushed, re-suspended in digestion buffer, consisting of 1mg ml–1 collagenase IV (Sigma-Aldrich, St Louis, MO, USA) and DNase I (50 μg ml–1) in RPMI media, and incubated at 37°C for 30min. Cells were centrifuged and single-cell suspensions were obtained by passing through syringe needle. DCs were sorted as live CD11c+MHCII+ cells (F4/80−, CD64−, CD3−, CD19−, Gr-1−).
Flow cytometry
BMDMs, BMDCs, splenic or peritoneal cells were re-suspended in staining buffer, consisting of PBS/2% FBS and incubated with a combination of up to 12 directly conjugated antibodies (FITC, PE, PE-Texas-Red, PE-Cy5 or PerCP-Cy5.5, PE-Cy7, allophycocyanin, Alexa700, allophycocyanin-Cy7, Pacific Blue, Brilliant Violet 605/650). Antibodies were purchased from BD Biosciences (San Jose, CA, USA) or eBioscience (San Diego, CA, USA) with the exception of PE-Texas Red and Brilliant Violet 605/650 conjugates, which were from Invitrogen (Life Technologies, Carlsbad, CA, USA) and Biolegend (San Diego, CA, USA), respectively. DAPI or a live/dead fixable aqua dead-cell stain (Molecular probes, Life technologies, Carlsbad, CA, USA) were used to gate out dead cells. CFSE was evaluated in the FITC channel. Apoptosis was measured using antibodies to Annexin V-allophycocyanin and propidium iodide (PI). The staining was performed using the binding buffer according to the manufacturer’s instructions (eBioscience, San Diego, CA, USA). Where indicated, the absolute cell numbers were assessed by adding 25000 counting beads (SPHERO™ AccuCount blank, Spherotech, Lake Forest, IL, USA) to each 200 μl of sample. Samples were acquired on a BD Fortessa, BD FACSCanto II or BD LSRII flow cytometer and analyzed with FlowJo 7.6 for Windows (TreeStar).
Alamar blue cellular viability assay
BMDMs were plated in flat-bottomed 96-well plates at a density 5×104 cells per well in 100 µl cRPMI. They were stimulated overnight (20h), 6 days or 9 days and then 10 µl of Alamar Blue® reagent (Invitrogen, Life Technologies, Carlsbad, CA, USA) was added to each well, for 4h at 37°C. Absorbance of the reduced Alamar blue was measured on an EnVison plate reader (Perkin Elmer, Waltham, MA, USA) at 570nm with reference 600nm wavelengths. The percentage of the reduced Alamar blue was calculated as described previously (14).
RNA isolation and RT–PCR
RNA was isolated by TRIzol/chloroform extraction using a Qiagen RNeasy Mini purification kit according to the manufacturer’s instructions (Qiagen, Venlo, Netherlands). Concentration and purity was measured using a Nanodrop spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). cDNA was produced using TaqMan® Reverse Transcription Reagents (Applied Biosystems, Waltham, MA, USA). Real-time PCR was performed using the iQ SYBR Supermix on a iQ5 MyiQ Single Colour Detection System (Bio-Rad, Hercules, CA, USA) with the primers listed in Supplementary Table 1 (available at International Immunology Online).
Cytokine analysis by bead array
Supernatants harvested after overnight treatment (20h) were immediately frozen at −80°C and thawed on the day of the assay. The cytokines IL-6, TNF-α and IL-10 were analyzed with a FlowCytomix™ Multiplex Immunoassay (eBioscience, San Diego, CA, USA) according to the manufacturer’s protocol. The beads were acquired on a BD FACSCanto II flow cytometer and analyzed with the FlowCytomix Pro 3.0 software (eBioscience, San Diego, CA, USA).
Phagocytosis
Jurkat cells were washed in plain RPMI 1640 media and re-suspended at 2×107cells per ml. An equal volume of 2.5 µM CFSE (Molecular Probes, Life Technologies, Carlsbad, CA, USA) in plain RPMI was added to the cells for 10min in the dark. An equal volume of FBS was then added for 1min to stop CFSE labeling and cells were washed three times with cRPMI. CFSE-labeled Jurkats (CFSE-apo Jurkat) at a density of 2.5×106 cells per ml were incubated with 30 µM etoposide (Sigma-Aldrich, St Louis, MO, USA) for 18h to induce apoptosis. Cells were washed three times with cRPMI and an aliquot was used to assess the extent of apoptosis as previously described (15).
BMDMs were plated in 12-well plates at 0.5×106 cells per ml and stimulated with different TLR ligands. After overnight (18–22h) of incubation, supernatants were discarded and fresh cRPMI media containing CFSE-apo-Jurkat cells were added at a ratio 4:1 (apoptotic cells: macrophages). After 60min of incubation at 37°C, phagocytosis was stopped by placing the plates on ice. Supernatants were removed and cells were collected and stained on ice with specific antibodies. The percentage of CFSE-positive macrophages was used as a measure of phagocytosis.
Antigen presentation
Following overnight treatment with TLR ligands, cultured or purified macrophages, or DCs were incubated with the indicated concentrations of OVA or OVA peptide 323–339. After 4h, the media was replaced with a suspension of CFSE-stained splenocytes from Rag1−/−/transgenic OT-II mice. OT-II splenocytes were added at a ratio of 1:10 for macrophages or 1:20 for DCs. After 5 days of incubation, the proliferation of T cells was assessed by CFSE dilution using flow cytometry. When OVA peptide 323–339 was used, the T-cell proliferation was assessed after 3 days of incubation with CFSE-OTII-splenocytes.
Statistical analysis
Results are expressed as arithmetic means + SEM. Normality was tested using the Kolomogorov–Smirnov test and two groups were compared with an unpaired two-way Student’s t-test, whereas three or more groups were compared with a one-way ANOVA followed by Bonferroni’s multiple comparisons test. Data that did not pass the normality test were analyzed using a non-parametric Mann–Whitney U test or Kruskal–Wallis non-parametric ANOVA followed by Dunn’s multiple comparison test. For the Alamar blue and T-cell proliferation assays, results were first analyzed by two-way repeated measures ANOVA. Alamar blue was significant by time and by treatment and T-cell proliferation data were significant by OVA/peptide concentration and treatment (P values less than 0.05). Based on this, untreated–treated pairs were compared using a paired t-test followed by a multiple test correction. Statistical analysis was performed using GraphPad Prism 6.0 for Windows (GraphPad Software, San Diego, CA, USA). Two-way repeated measures ANOVA was carried out using the ez package in the R statistical language (version 2.15.2).
Results
Activation of BMDMs enhances phagocytosis of apoptotic cells
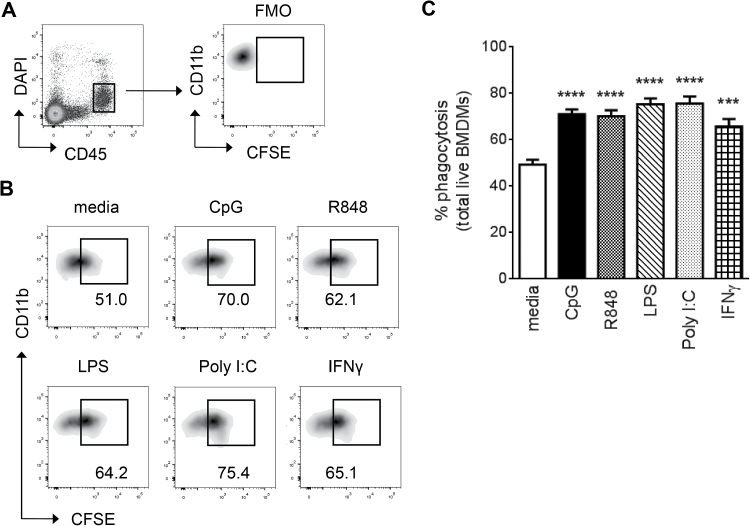
Phagocytosis of apoptotic cells and cellular debris constitutes a primary role of macrophages. To characterize the effects of TLR ligation on this principal macrophage function, we first compared Poly I:C (TLR3), LPS (TLR4), R848 (TLR7), CpG (TLR9) or IFN-γ on the ability of macrophages to phagocytose apoptotic cells in vitro. Following incubation with CFSE-labeled apoptotic Jurkat cells, increased phagocytosis was observed for all TLR-activated BMDMs, regardless of the stimulatory ligand used (Fig. 1). These results are consistent with an earlier report that examined the impact of TLRs 2, 3 and 9 on the phagocytic clearance of bacteria (16, 17). However, we show that this increase in macrophage function is unlikely a TLR-specific effect, since treatment with IFN-γ also resulted in enhanced phagocytosis (Fig. 1B and C), suggesting it is a common characteristic of macrophage activation.
Fig. 1.
Enhanced phagocytosis of activated BMDMs is not a TLR-specific effect. (A) TLR-stimulated BMDMs were incubated with CFSE-stained apoptotic Jurkat cells and gated on CD45+ live cells. An FMO control with unstained Jurkat cells was used to set the phagocytosis gate. Percentage of phagocytosis was assessed as percentage of CFSE+ CD11b+ BMDMs: (B) representative plots and (C) quantification of the data. Bars represent mean + SEM of data collected from 4 to 11 independent experiments with n = 8–15 per stimulation. P values were calculated by one-way ANOVA followed by a multiple comparisons test. ***P < 0.001, ****P < 0.0001 (compared to media). FMO, fluorescence minus one.
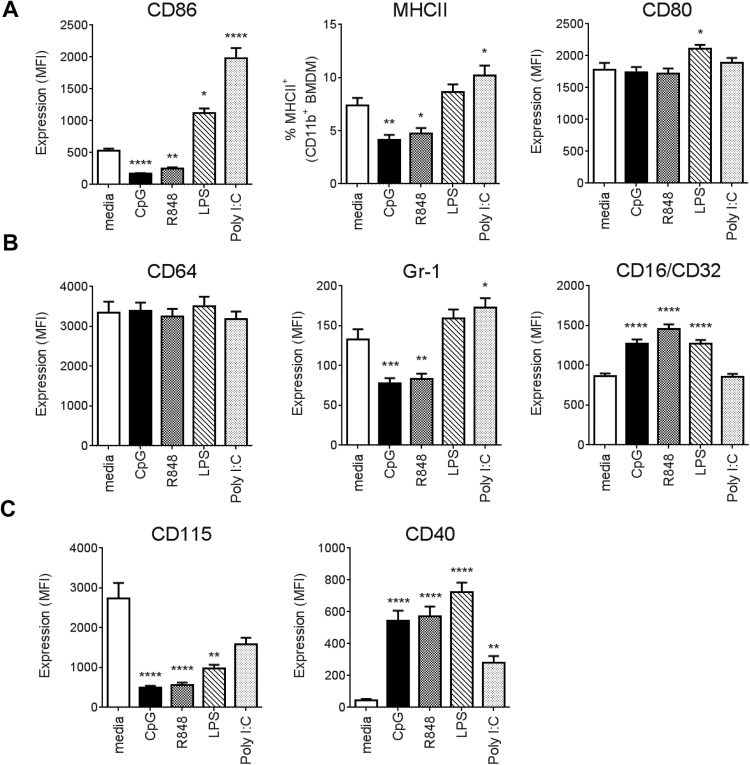
TLR7 and TLR9 ligands down-regulate co-stimulatory molecule expression on macrophages
TLR stimulation induces phenotypic maturation of myeloid cells, particularly DCs, characterized by the up-regulation of co-stimulatory molecules (e.g. MHCII, CD80, CD86) (18, 19). Therefore, we examined the expression of these cell-surface molecules on BMDMs following overnight exposure to the different TLR ligands. Interestingly, incubation with TLR7 and TLR9 ligands (R848 and CpG) resulted in a significant down-regulation of CD86 and MHCII expression (Fig. 2A). This was in contrast to TLR3 or TLR4 ligand exposure, which resulted in increased expression of both CD86 and MHCII or CD86, respectively. CD80 was unaffected by TLR3, 7 or 9 stimulation but was upregulated following treatment with LPS. As expected, overnight exposure of BMDCs from the same mice to each of the four TLR ligands resulted in an increase in these co-stimulatory molecules (Supplementary Figure S2, available at International Immunology Online).
Fig. 2.
TLR7/9 ligands down-regulate co-stimulatory molecules. BMDMs were collected on day 7 and stimulated overnight (18–22h) with TLR ligands. Surface expression of (A) CD86, MHCII, CD80, (B) CD64, Gr-1, CD16/CD32 and (C) CD115 and CD40 was measured by flow cytometry on CD45+, live, CD11b+CD64+ macrophages. Bars represent mean + SEM obtained from at least three independent experiments with minimum n = 10 per condition. P values were calculated by one-way ANOVA followed by a multiple comparisons test. *P < 0.05, **P< 0.01, ***P < 0.001, ****P < 0.0001 (compared to media).
The BMDMs produced by our culture conditions were characterized as Gr-1neg/lowCD64+, suggesting a mature phenotype (20) (Supplementary Figure S1, available at International Immunology Online). In addition, they expressed CD16/CD32. Stimulation with CpG and R848 further lowered the levels of Gr-1 while increasing CD16/CD32 expression (Fig. 2B). Treatment with LPS increased CD16/CD32 expression, whereas Poly I:C increased Gr-1 expression. . None of the tested ligands affected CD64 expression (Fig. 2B). Rapid CD115 down-regulation by LPS and bacterial/CpG DNA has been reported previously by Sester and colleagues (21). Our data confirmed this and demonstrated that the same effect can be mediated by TLR7 ligation (Fig. 2C). Additionally, all four TLR ligands increased cell-surface expression of CD40, a key co-stimulatory molecule that drives the adaptive immune response through interaction with antigen-activated CD154+ T cells (22).
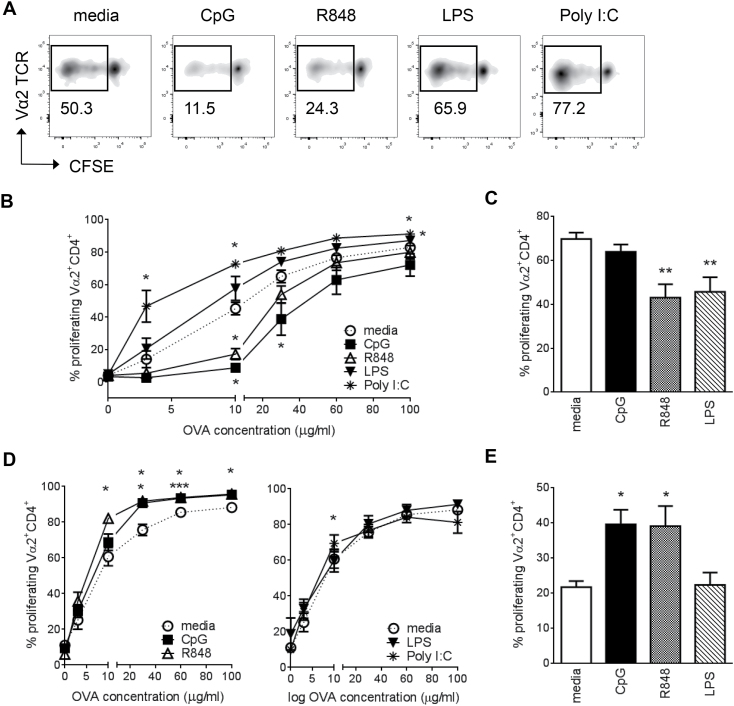
TLR7- and TLR9-activated macrophages regulate antigen presentation
Since CpG and R848 caused a distinctive down-regulation of CD86 and MHCII expression on BMDMs, we examined whether this would affect antigen-specific T-cell proliferation. BMDMs were treated with TLR ligands overnight and pulsed with the indicated concentrations of OVA. Proliferation of B6.OT-II derived, CD4+TCRVα2+ splenic T cells was measured by CFSE dilution using flow cytometry (Fig. 3A). Despite a low percentage of MHCII+ cells (Fig. 2B), BMDMs given antigen alone (media) were able to efficiently drive antigen-specific T-cell proliferation (Fig. 3A and B). When pre-treated with either CpG or R848, BMDMs induced significantly less T-cell proliferation, particularly at lower OVA concentrations (Fig. 3A and B). On the contrary, when BMDMs were pre-treated with TLR3 ligand Poly I:C, they induced significantly more T-cell proliferation compared with untreated cells across all the OVA concentrations tested. LPS significantly increased T-cell proliferation only at 100 μg ml–1 OVA (Fig. 3B). This effect is likely a direct consequence of altered expression of MHCII and CD86 (Fig. 2A) and not due to differential regulation of antigen processing (proteolysis) induced by TLR ligation. We tested the latter by pulsing BMDMs with the dominant immunogenic OVA peptide 323–339, which does not require proteolysis, and observed the same TLR-dependent modulation of antigen presentation as with full OVA (Supplementary Figure S3, available at International Immunology Online).
Fig. 3.
TLR7/9 ligands differentially regulate T-cell proliferation in macrophages and DCs. CFSE-stained OT-II splenocytes were co-cultured in the presence of OVA-pulsed APCs and T-cell proliferation was assessed by CFSE dilution displayed by live, CD45+CD4+TCRVα2+ T cells. (A) Representative flow cytometry plots of CD4+TCRVα2+ T-cell proliferation following co-culture with BMDMs pulsed with 10 µg ml–1 OVA. (B) Cumulative data of BMDMs pulsed with increasing concentrations of OVA. (C) Resting peritoneal macrophages pulsed with 30 μg ml–1 OVA. (D) BMDCs pulsed with increasing concentrations of OVA. (E) Splenic CD11c+MHCII+ DCs pulsed with 10 μg ml–1 OVA. Data shown for BMDMs and BMDCs are combined from two independent experiments, with total n = 5 for BMDMs and n = 7 for BMDCs. Data were first analyzed by two-way repeated measures ANOVA, followed by t-test for treated–untreated pairs. The P values were subsequently corrected for multiple tests and ranked using a Benjamini-Hockberg correction. Data shown in (C) and (E) are from three independent experiments with total n = 5–6 per condition. Significance was assessed by a two-way ANOVA followed by a multiple comparison test. *P < 0.05, **P < 0.01, ***P < 0.001 (compared to media).
We then examined the effects of TLR stimulation on antigen presentation by resident peritoneal macrophages. Overnight treatment of peritoneal macrophages with R848 resulted in decreased T-cell proliferation, which was consistent with our BMDM data (Fig. 3C). Additionally, the same effect was observed with LPS, but not with CpG (Fig. 3C). To show that the observed effects of TLR stimulation on T-cell proliferation were macrophage-specific, we performed the same experiments with BMDCs and sorted splenic DCs. As expected, R848 and CpG induced higher T-cell proliferation compared with untreated BMDCs (Fig. 3D) or splenic DCs (Fig. 3E). LPS and PolyI:C had minimal effects on enhancing T-cell proliferation (Fig. 3D, right panel, and 3E).
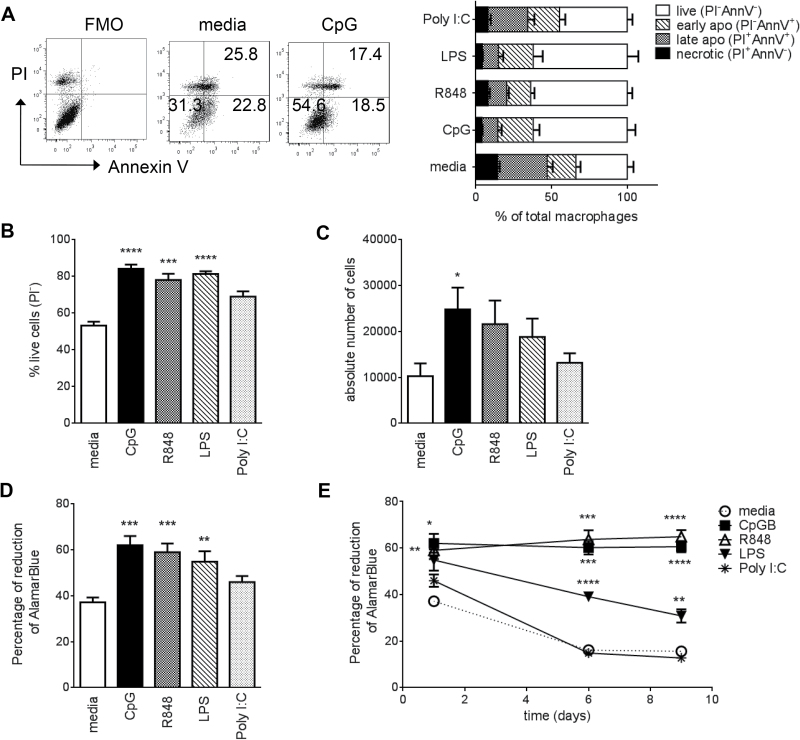
TLR7 and TLR9 ligands induce long-term survival of macrophages
To determine whether the reduction in cell-surface co-stimulation markers and antigen presentation were due to changes in cell viability, we assessed apoptosis in TLR ligand-treated macrophages for up to 9 days using two different methods. We initially used Annexin V (AnnV) and PI staining with flow cytometry (Fig. 4A). This is an established method that can define early (PI−AnnV+) and late (PI+AnnV+) apoptotic cells and necrosis (PI+AnnV−). We determined that overnight (20h) incubation of BMDM with TLR 4, 7 or 9 ligands significantly increased cell survival by reducing apoptosis (Fig. 4A and B). Additionally, CpG stimulation increased the total cell number (Fig. 4C). The increase in cell survival by TLR4, 7 and 9 ligands after overnight stimulation was confirmed using Alamar Blue (Fig. 4D). This is a non-toxic dye that is chemically reduced by the innate metabolic activity of cells into a colorimetric indicator. It is added directly to the cells in culture, avoiding cell scraping or other methods for detachment, which can cause severe damage to the cells. This method enabled assessment of macrophage viability in vitro for up to 9 days (Fig. 4E). Both CpG and R848 treatment resulted in statistically more viable and metabolically active macrophages during this time, comparable to day 1. While LPS reduced cell death, stimulation of TLR3 had no effect on cell survival (Fig. 4B–D). These results are supported by previous studies indicating that LPS or CpG can induce short-term (48h) survival of macrophages (21, 23, 24).
Fig. 4.
TLR7 and TLR9 ligands induce long-term macrophage survival. BMDMs were collected on day 7 and stimulated overnight (18–22h) with TLR ligands. (A) Macrophages were selected by gating on CD45+CD11b+ cells and then analyzed for PI and Annexin V staining. Left panel shows dot plots from a representative experiment. Right panel shows the collated data from two independent experiments (mean +SEM), total n = 6. (B) Percentage of live cells excluding PI (PI−) and (C) absolute cell numbers measured by flow cytometry of macrophages with added counting beads. (D) Cell viability assessed by Alamar blue reduction. For (B–D) data from three to four independent experiments are shown (mean + SEM) and P values were calculated by one-way ANOVA followed by a multiple comparison test. (E) Long-term effects of TLR stimulation (overnight, 6 days, 9 days) on macrophage viability assessed by Alamar blue reduction. Data from four independent experiments are shown (total n = 8). Data were first analyzed by a two-way repeated measures ANOVA, followed by t-test for treated–untreated pairs. P values underwent multiple test correction. *P < 0.05, **P< 0.01, ***P < 0.001, ****P< 0.0001 (compared to media). FMO, fluorescence minus one.
TLR9 and TLR4 differentially regulate late-phase gene expression in BMDMs
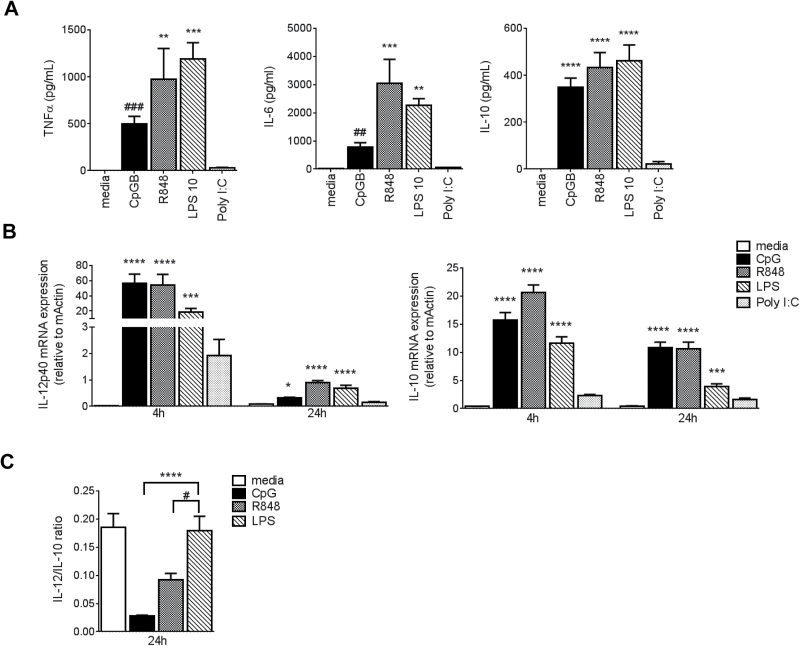
In light of the differential effect of TLR ligands on macrophage survival, we next analyzed BMDM cytokine production and their polarization. Macrophages produce several cytokines in response to microbial challenge, including TNF-α, IL-6 and IL-10. Our analyses of supernatants from overnight BMDM cultures showed that these were all produced in response to LPS, R848 and CpG (Fig. 5A). However, exposure to Poly I:C did not induce the release of any of these cytokines.
Fig. 5.
LPS and CpG/R848 differentially activate macrophages after prolonged exposure. BMDMs were collected on day 7 and incubated with TLR ligands. (A) Levels of TNF-α, IL-6 and IL-10 in 20h supernatants. (B) RNA was isolated after 4h and 24h of incubation with TLR ligands. Expression of polarization genes Il-12p40 and Il-10 was measured by real-time PCR and normalized to β-actin expression (×103). (C) Il-12/Il-10 mRNA ratios at 24h. Bars represent mean + SEM from one representative experiment (n = 5) for cytokines and from two independent experiments (total n = 9) for mRNA. P values were calculated by one-way ANOVA or Kruskal–Wallis test followed by a multiple correction test. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 (compared to media). # indicates P values calculated by an unpaired t-test or Mann–Whitney test between pairs of unstimulated (media) and CpG stimulated in (A) and between R848 and LPS stimulated in (C); # P < 0.05, ## P < 0.01, ### P < 0.001.
We then went on to evaluate whether the TLR ligands differentially affected the expression of macrophage polarization-related cytokine genes: Il-10 and Il-12p40 (7). RNA was isolated from BMDMs incubated with media, CpG, R848, LPS or Poly I:C for 4 and 24h and analyzed by RT–PCR. Short-term (4h) exposure to CpG, R848 and LPS resulted in a similar activation profile characterized by the induction of both M1/M2-related genes Il-12p40 and Il-10, while Poly I:C did not significantly affect their expression (Fig. 5B). In contrast, incubation for a longer period of time (24h) revealed TLR-specific differences in the IL-12/IL-10 gene expression profiles. Both CpG and R848 retained relatively high Il-10 transcription levels at 24h (Fig. 5B, right panel), resulting in a significantly lower ratio of Il-12/Il-10 mRNA compared with LPS (Fig. 5C). These data suggest that CpG and R848 may drive the macrophages to an M2-like state (IL-12lo/IL-10hi) in the later phase of activation.
Discussion
In these studies, we compare the different effects of commonly used ligands across several macrophage functions. We have created a reference point for future studies examining the innate mechanisms of macrophages. Furthermore, we have shown that in contrast to the effects of TLR4 and TLR3 ligands, TLR7 or TLR9 ligation results in a long-lived macrophage with heightened phagocytic ability and a decreased capability of antigen presentation (Table 1). This state may have evolved to protect the host from self-reactivity to nucleic acids from dead cells ingested by macrophages.
Table 1.
Summary of effects of TLR stimulation on BMDMs
| TLR7 | TLR9 | TLR4 | TLR3 | |
|---|---|---|---|---|
| Location | Endosomal | Endosomal | Surface | Endosomal |
| Signaling | MyD88 | MyD88 | MyD88/TRIF | TRIF |
| Effects on BMDMs | ||||
| Survival | ↑ | ↑ | ↑ | Unchanged |
| Phagocytosis | ↑ | ↑ | ↑ | ↑ |
| Co-stimulatory molecules | ↓MHCII, ↓CD86, ↑CD40 | ↓MHCII, ↓CD86, ↑CD40 | ↑CD86, ↑CD80, ↑CD40 | ↑CD86, ↑MHCII, ↑CD40 |
| Antigen presentation | ↓ | ↓ | ↑ | ↑ |
| Cytokine production | ↑TNFα, ↑IL-6, ↑IL-10 | ↑TNFα, ↑IL-6, ↑IL-10 | ↑TNFα, ↑IL-6, ↑IL-10 | Unchanged |
| IL-12/IL-10 ratio | ↓ | ↓ | Unchanged | ND |
↑, increased; ↓, decreased; ND, not defined.
The extended life span of macrophages for 9 days following TLR7/9 ligand treatment has not been described previously. However, it is supported by data showing that short-term (48h) exposure of macrophages to LPS or CpG prolongs cell survival (21, 23, 24). Interestingly, we show Poly I:C had no effect on the viability of these cells. This heightened longevity may have important consequences in light of macrophage ability to down-regulate antigen presentation. In contrast to DCs, which up-regulate co-stimulation molecules on TLR-induced maturation (18, 19, 25), macrophages are more plastic in their responses and the level of up-regulation of co-stimulatory molecules depends upon the type of activation. LPS stimulation results in an up-regulation of co-stimulatory molecule expression on BMDMs and RAW264.7 macrophages, including MHCII, CD80, CD86 and CD40 (26, 27). We have additionally shown that Poly I:C induced CD86, MHCII and CD40 on BMDMs. In contrast, TLR7 or TLR9 stimulation decreased CD86 and MHCII expression.
Although previous data have shown that IL-10 can down-regulate MHCII expression and antigen presentation (28), it is unlikely to be the sole contributor, since CpG, R848 and LPS all induced similar IL-10 levels following overnight incubation. Indeed, the decrease in co-stimulatory molecule expression that we observed following TLR7 or TLR9 ligation correlated with lower antigen presentation and T-cell proliferation in BMDMs. This mechanism might have a regulatory/protective role in autoimmunity, where enhanced antigen presentation can breach tolerance (29).
Long-term survival, decreased antigen presentation and an overall suppressive response are reminiscent of M2-like macrophages (30). Therefore, we ascertained if these effects were a reflection of the polarization state by assessing RNA levels of genes connected with macrophage profiling. We established that short-term (4h) exposure to CpG, R848 or LPS increased transcription of M1/M2-associated genes (Il-12p40 and Il-10). However, at 24h post-CpG or R848 stimulation, we detected a preferential skewing toward an M2-like state, defined by a lower ratio of Il-12/Il-10 compared with LPS. This distinctive profile and increase in long-term survival is consistent with a bimodal pattern of gene regulation observed after in vitro treatment and in vivo administration of CpG (31, 32). The initial peak (2–4h post-CpG) is characterized by a rapid up-regulation of genes associated with the activation of the immune system, such as NF-κB and TNF, followed by induction of the regulatory gene, Myc at 8h (31, 32). Subsequently, 5 days post-CpG administration, an up-regulation of genes associated with cell-cycle regulation and DNA damage responses are detected (32).
The macrophage polarization state dictates the ability to phagocytize pathogens. IFN-γ-induced M1 macrophages possess an increased ability to phagocytize Candida albicans (33), whereas IL-4-induced M2 macrophages have been shown to increase the uptake of parasites (34) while decreasing the phagocytosis of bacteria (35). In contrast to these findings, our studies do not support a difference in apoptotic cell uptake due to polarization state, but demonstrate simply that activation enhances phagocytosis.
In our studies, we chose to compare BM-derived cells to minimize the effects of unknown in vivo factors. The analysis of ex vivo naive peritoneal macrophages showed similar effects; however, in these cells, exposure to LPS also caused a reduction in T-cell proliferation. This is consistent with a previous study, where both CpG and LPS down-regulated MHCII expression in Con A-elicited peritoneal macrophages (36). It is possible that at different tissues or inflammatory sites, macrophages may respond to environmental signals in different ways. Two recent studies showed that thioglycollate-induced peritoneal macrophages are markedly different from naive resident macrophages and infection-elicited macrophages (37) and that tissue-resident macrophages respond differently to environmental challenges compared with monocyte-derived macrophages (38). This is consistent with emerging evidence showing that monocytes and macrophages have differences in their ontogeny and homeostasis [reviewed in (39)]. Tissue-resident macrophages play an important homeostatic role, including scavenging and the removal of cellular debris with intrinsically anti-inflammatory properties (40). Thus, they require protective mechanisms to prevent presentation of self-antigens from ingested dead cells. We suggest that TLR7/9-mediated stimulation with self-ligands may function as such a protective mechanism.
In summary, we have determined that TLR7/TLR9 ligands drive the macrophage into a phagocytic, long-lived state, with a decreased capacity for antigen presentation. Functionally, these cells resemble M2-like macrophages, which arise from immunosuppressive microenvironments. These observations have important consequences for homeostasis and the prevention of self-reactivity and tolerance.
Supplementary data
Supplementary data are available at International Immunology Online.
Funding
This work was supported by core funding from the Singapore Immunology Network (SIgN) to A.-M.F, J.E.C. and S.K.B.,by core funding from the Institute of Molecular and Cell Biology (IMCB) to J.C. at A*STAR, Singapore and by the NMRC/CBRG/0056/2014 grant to L.H.K.L.
Supplementary Material
Acknowledgements
We thank the flow cytometry core at SIgN, A*STAR, and Stephan Gasser, National University of Singapore (NUS), for critical review of the manuscript.
Conflict of interest statement: The authors declare no commercial or financial conflict of interest.
References
- 1. Medzhitov R. and Janeway C. A. Jr. 1997. Innate immunity: the virtues of a nonclonal system of recognition. Cell 91:295. [DOI] [PubMed] [Google Scholar]
- 2. Trinchieri G. and Sher A. 2007. Cooperation of toll-like receptor signals in innate immune defence. Nat. Rev. Immunol. 7:179. [DOI] [PubMed] [Google Scholar]
- 3. Huynh M. L. Fadok V. A. and Henson P. M. 2002. Phosphatidylserine-dependent ingestion of apoptotic cells promotes TGF-beta1 secretion and the resolution of inflammation. J. Clin. Invest. 109:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kinne R. W. Brauer R. Stuhlmuller B. Palombo-Kinne E. and Burmester G. R. 2000. Macrophages in rheumatoid arthritis. Arthritis Res. 2:189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Biswas S. K. and Mantovani A. 2010. Macrophage plasticity and interaction with lymphocyte subsets: cancer as a paradigm. Nat. Immunol. 11:889. [DOI] [PubMed] [Google Scholar]
- 6. Mosser D. M. and Edwards J. P. 2008. Exploring the full spectrum of macrophage activation. Nat. Rev. Immunol. 8:958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mantovani A. Sica A. Sozzani S. Allavena P. Vecchi A. and Locati M. 2004. The chemokine system in diverse forms of macrophage activation and polarization. Trends Immunol. 25:677. [DOI] [PubMed] [Google Scholar]
- 8. Edwards J. P. Zhang X. Frauwirth K. A. and Mosser D. M. 2006. Biochemical and functional characterization of three activated macrophage populations. J. Leukoc. Biol. 80:1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Davies L. C. Jenkins S. J. Allen J. E. and Taylor P. R. 2013. Tissue-resident macrophages. Nat. Immunol. 14:986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Orme J. and Mohan C. 2012. Macrophage subpopulations in systemic lupus erythematosus. Discov. Med. 13:151. [PubMed] [Google Scholar]
- 11. Llovera M., Carbo N., Lopez-Soriano J., et al. 1998. Different cytokines modulate ubiquitin gene expression in rat skeletal muscle. CancerLett. 133:83. [DOI] [PubMed] [Google Scholar]
- 12. Fleit S. A. Fleit H. B. and Zolla-Pazner S. 1984. Culture and recovery of macrophages and cell lines from tissue culture-treated and -untreated plastic dishes. J. Immunol. Methods 68:119. [DOI] [PubMed] [Google Scholar]
- 13. Weischenfeldt J. and Porse B. 2008. Bone marrow-derived macrophages (BMM): isolation and applications. CSH Protoc. 2008:pdb.prot5080. [DOI] [PubMed] [Google Scholar]
- 14. Larson E. M. Doughman D. J. Gregerson D. S. and Obritsch W. F. 1997. A new, simple, nonradioactive, nontoxic in vitro assay to monitor corneal endothelial cell viability. Invest. Ophthalmol. Vis. Sci. 38:1929. [PubMed] [Google Scholar]
- 15. Pereira-Lopes S. Celhar T. Sans-Fons G. Serra M. Fairhurst A. M. Lloberas J. and Celada A. 2013. The exonuclease Trex1 restrains macrophage proinflammatory activation. J. Immunol. 191:6128. [DOI] [PubMed] [Google Scholar]
- 16. Doyle S. E., O’Connell R. M., Miranda G. A., Vaidya S. A., Chow E. K., Liu P., T., Suzuki S., Suzuki N., Modlin R. L., Yeh W. C., Lane T. F., Cheng G. 2004. Toll-like receptors induce a phagocytic gene program through p38. J. Exp. Med. 199:81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang J. Huang W. L. and Liu R. Y. 2009. CpG-ODN enhances ingestion of apoptotic neutrophils by macrophages. Clin. Exp. Med. 9:37. [DOI] [PubMed] [Google Scholar]
- 18. Dearman R. J. Cumberbatch M. Maxwell G. Basketter D. A. and Kimber I. 2009. Toll-like receptor ligand activation of murine bone marrow-derived dendritic cells. Immunology 126:475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Steinman R. M. 2012. Decisions about dendritic cells: past, present, and future. Annu. Rev. Immunol. 30:1. [DOI] [PubMed] [Google Scholar]
- 20. Francke A. Herold J. Weinert S. Strasser R. H. and Braun-Dullaeus R. C. 2011. Generation of mature murine monocytes from heterogeneous bone marrow and description of their properties. J. Histochem. Cytochem. 59:813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sester D. P. Beasley S. J. Sweet M. J. Fowles L. F. Cronau S. L. Stacey K. J. and Hume D. A. 1999. Bacterial/CpG DNA down-modulates colony stimulating factor-1 receptor surface expression on murine bone marrow-derived macrophages with concomitant growth arrest and factor-independent survival. J. Immunol. 163:6541. [PubMed] [Google Scholar]
- 22. Mackey M. F. Barth R. J. Jr and Noelle R. J. 1998. The role of CD40/CD154 interactions in the priming, differentiation, and effector function of helper and cytotoxic T cells. J. Leukoc. Biol. 63:418. [DOI] [PubMed] [Google Scholar]
- 23. Sester D. P., Brion K., Trieu A., et al. 2006. CpG DNA activates survival in murine macrophages through TLR9 and the phosphatidylinositol 3-kinase-Akt pathway. J. Immunol. 177:4473. [DOI] [PubMed] [Google Scholar]
- 24. Lim E. J. Park D. W. Jeong T. W. Chin B. R. Bae Y. S. and Baek S. H. 2012. TRAIL is involved in CpG ODN-mediated anti-apoptotic signals. Oncol. Rep. 27:1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Askew D. Chu R. S. Krieg A. M. and Harding C. V. 2000. CpG DNA induces maturation of dendritic cells with distinct effects on nascent and recycling MHC-II antigen-processing mechanisms. J. Immunol. 165:6889. [DOI] [PubMed] [Google Scholar]
- 26. Saxena R. K. Vallyathan V. and Lewis D. M. 2003. Evidence for lipopolysaccharide-induced differentiation of RAW264.7 murine macrophage cell line into dendritic like cells. J. Biosci. 28:129. [DOI] [PubMed] [Google Scholar]
- 27. Marim F. M. Silveira T. N. Lima D. S. Jr and Zamboni D. S. 2010. A method for generation of bone marrow-derived macrophages from cryopreserved mouse bone marrow cells. PLoS One 5:e15263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. de Waal Malefyt R., Haanen J., Spits H., et al. 1991. Interleukin 10 (IL-10) and viral IL-10 strongly reduce antigen-specific human T cell proliferation by diminishing the antigen-presenting capacity of monocytes via downregulation of class II major histocompatibility complex expression. J. Exp. Med. 174:915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Garza K. M. Chan S. M. Suri R. Nguyen L. T. Odermatt B. Schoenberger S. P. and Ohashi P. S. 2000. Role of antigen-presenting cells in mediating tolerance and autoimmunity. J. Exp. Med. 191:2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mosser D. M. 2003. The many faces of macrophage activation. J. Leukoc. Biol. 73:209. [DOI] [PubMed] [Google Scholar]
- 31. Klaschik S. Gursel I. and Klinman D. M. 2007. CpG-mediated changes in gene expression in murine spleen cells identified by microarray analysis. Mol. Immunol. 44:1095. [DOI] [PubMed] [Google Scholar]
- 32. Klaschik S. Tross D. Shirota H. and Klinman D. M. 2010 Short- and long-term changes in gene expression mediated by the activation of TLR9. Mol. Immunol. 47:1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Maródi L. Schreiber S. Anderson D. C. MacDermott R. P. Korchak H. M. and Johnston R. B. Jr. 1993. Enhancement of macrophage candidacidal activity by interferon-gamma. Increased phagocytosis, killing, and calcium signal mediated by a decreased number of mannose receptors. J. Clin. Invest. 91:2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wirth J. J. Kierszenbaum F. and Zlotnik A. 1989. Effects of IL-4 on macrophage functions: increased uptake and killing of a protozoan parasite (Trypanosoma cruzi). Immunology 66:296. [PMC free article] [PubMed] [Google Scholar]
- 35. Varin A. Mukhopadhyay S. Herbein G. and Gordon S. 2010. Alternative activation of macrophages by IL-4 impairs phagocytosis of pathogens but potentiates microbial-induced signalling and cytokine secretion. Blood 115:353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chu R. S. Askew D. Noss E. H. Tobian A. Krieg A. M. and Harding C. V. 1999. CpG oligodeoxynucleotides down-regulate macrophage class II MHC antigen processing. J. Immunol. 163:1188. [PubMed] [Google Scholar]
- 37. Jenkins S. J., Ruckerl D., Cook P. C., et al. 2011. Local macrophage proliferation, rather than recruitment from the blood, is a signature of TH2 inflammation. Science 332:1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gundra U. M., Girgis N. M., Ruckerl D., et al. 2014. Alternatively activated macrophages derived from monocytes and tissue macrophages are phenotypically and functionally distinct. Blood 123:e110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ginhoux F. and Jung S. 2014. Monocytes and macrophages: developmental pathways and tissue homeostasis. Nat. Rev. Immunol. 14:392. [DOI] [PubMed] [Google Scholar]
- 40. Murray P. J. and Wynn T. A. 2011. Protective and pathogenic functions of macrophage subsets. Nat. Rev. Immunol. 11:723. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.