Abstract
SNARE proteins are the core machinery to drive fusion of a vesicle with its target membrane. Inspired by the tethering proteins that bridge the membranes and thus prepare SNAREs for docking and fusion, we developed a lipid conjugated ssDNA mimic that is capable of regulating SNARE function, in situ. The DNA-lipid tethers consist of a 21 base pairs binding segment at the membrane distal end that can bridge two liposomes via specific base-pair hybridization. A linker at the membrane proximal end is used to control the separation distance between the liposomes. In the presence of these artificial tethers, SNARE-mediated lipid mixing is significantly accelerated, and the maximum fusion rate is obtained with the linker shorter than 40 nucleotides. As a programmable tool orthogonal to any native proteins, the DNA-lipid tethers can be further applied to regulate other biological processes where capturing and bridging of two membranes are the prerequisites for the subsequent protein function.
Keywords: membrane proteins, liposomes, membrane fusion, ssDNA-lipid conjugation, mimicking protein function
Specific and programmable DNA hybridization has broadened the applications of DNA beyond biological genetic information carrier. Membrane incorporation of DNA-lipid hybrid molecules, rendered by functional modification of DNA using terminal cholesterol,[1] dialkyllipid,[2] or biotin [3] allows DNA controlled assembly of soft membrane structures, such as liposome to liposome, and liposomes to solid supported membrane. These technologies have been applied to generate arrays or clusters of liposomes on supported-lipid-bilayers, to drive liposome-liposome fusion, and to serve as a distance ruler based on the defined length of dsDNA. Herein we introduce a new application of DNA-lipid hybrid molecules to directly, and in situ, regulate a biological process, the membrane fusion reaction mediated by SNARE (soluble N-ethylmaleimide-sensitive factor attachment protein receptor) protein.
Purified t- and v-SNAREs, when reconstituted into distinct vesicles are capable of driving vesicle fusion, albeit at a rate in minutes. [4,5] This slow rate, together with the observation that the rate-limiting step, N-terminal assembly, requires overcoming high energy barrier, suggest that under physiological conditions, additional protein factors are required to facilitate SNARE-complex formation.[5,6] Among such proteins, are the tethering factors that, in most cases, belong to the family of Rab effectors. Prior to SNARE-driven membrane fusion, tethering factors, residing on the target membrane, link vesicles to the active zone (on target membrane) by interacting with the GTP bound Rab proteins on vesicles, [4, 7] and thus get SNAREs ready to function.
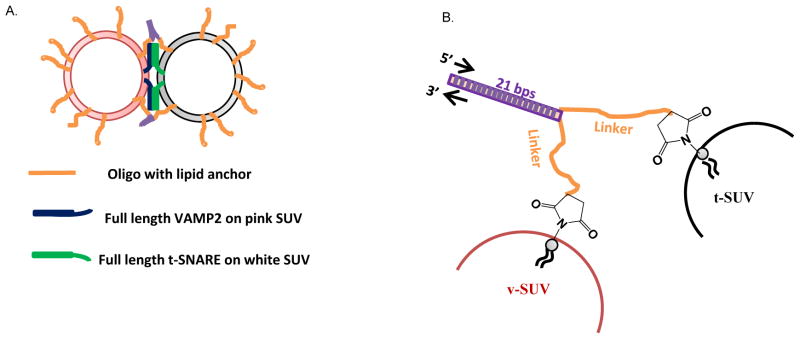
Here, we developed a non-fusogenic DNA-lipid mimic of tethering factor (Figure 1a). Without directly interacting with SNAREs, it regulates SNARE function by capturing and keeping two apposed membranes within controlled distance, and we were able to control the distance between two membranes, within which, SNARE-driven membrane fusion reaches its maximum rate.
Figure 1.
Schematic design of the DNA-lipid tether system. (A) A pair of complementary DNA-lipid tethers were incorporated into two populations of liposomes, t- and v-liposomes, respectively, at a loaded lipid:oligo molar ratio of 200:1 (100 per liposome), unless otherwise specified. The t-liposome (mole%, 85% POPC, and 15% DOPS) contains co-expressed t-SNARE (syntaxin 1/SNAP25, Cys free) with loaded lipid:protein molar ratio of 200:1 (100 per liposome), and the fluorescent v-liposome (mole%, 82% POPC, 15% DOPS, 1.5% NBD-DPPE, and 1.5% Rho-DPPE) contains v-SNARE (VAMP2, Cys free) with loaded lipid:protein molar ratio of 400:1 (50 per liposome). (B) Structure of DNA-lipid tethers. From the membrane distal end, a DNA-lipid tether hybrid molecule contains a 21 bp hybridization region (purple), a linker region containing 5 to 63 consecutive nucleotides, Thymidine (T) (orange), and a thiol-maleimide linked membrane anchor.
The non-fusogenic DNA-lipid hybrid molecules, serving as artificial tethers in this study, were constructed with three consecutive structural segments from the membrane-distal to membrane-proximal ends (Figure 1): (i) a 21-bp hybridization region, responsible for physically bridging two membranes via parallel hybridization; (ii) a linker region containing 5 to 63 consecutive Thymidine (T) nucleotides (Table S1), that is inserted to regulate liposome distance and minimize possible nonspecific fusion caused by the tethers alone; and (iii) the lipid anchor residing in the lipid bilayer, that is cross-linked to the DNA sequence via a thiol-maleimide linkage.
To directly monitor the in situ regulation effect of tethers on SNARE-driven membrane fusion, we reconstituted a pair of complementary DNA-lipid tethers (noted as “t-tethers” and “v-tethers”) together with either t- or v-SNARE into two populations of proteo-liposomes (See SI. Materials and Methods section for detailed procedure), hereinafter called t-liposomes (with t-tethers and t-SNAREs) and v-liposomes (with v-tethers and v-SNAREs), by detergent-dialysis procedure (Figure S3) (See Table S1 for oligonucleotide sequences and nomenclature). The presence of both DNA-lipid tethers and SNARE proteins was confirmed by SDS-PAGE. Transmission Electron Microscopy (TEM) showed that the diameter of the final liposomes was 54.5 ± 16.3 nm (±SD).
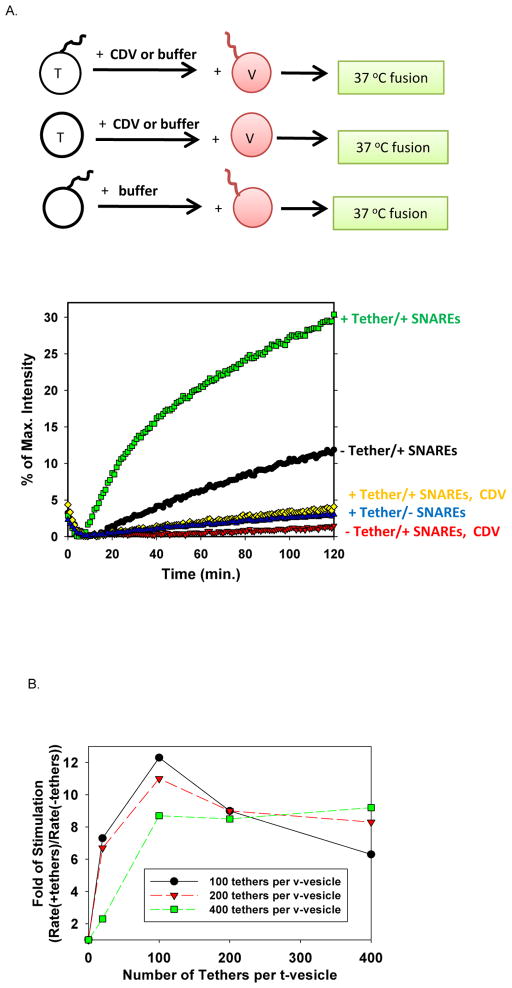
We evaluated the tethering effects on SNARE function using liposome-liposome fusion assay.[8] We mixed the t-liposome (with or without t-tethers) containing no fluorescent probes and the v-liposome (with or without v-tethers) containing fluorescently quenched lipid probes, NBD-DOPE and Rho-DOPE, that de-quench during membrane fusion as a result of the dilution of lipid upon lipid mixing. Hence, fusion was measured by following the fluorescence intensity increase of NBD-DOPE over time (Figure 2).
Figure 2.
DNA-lipid tether significantly increased the initial rate of SNARE-mediated membrane fusion. (A) In the SNARE-driven liposome-liposome fusion assay, the initial reaction rate was increased 10-fold by about 50 externally facing i-42T DNA-lipid tethers on each side of the t- and v-liposomes. (B) DNA-lipid tethers stimulated SNARE-driven lipid mixing in a dose-dependent manner (using i-42T tether pair).
As shown in (Figure 2a) and (Figure S1), DNA-lipid tethers stimulated SNARE-dependent membrane fusion. Under our assay conditions, liposomes containing DNA-lipid tethers alone only contributed to the background level of membrane fusion. Proteo-liposomes (t-SNARE, loaded at 100 per liposome; v-SNARE, loaded at 50 per liposome) alone showed 12% of the maximum fluorescence intensity increase after 2 hr. However, when DNA-lipid and SNARE were both present in liposome (same loaded density as controls), 30% of intensity increase was observed. Moreover, membrane fusion could be blocked by soluble cytoplasmic domain of VAMP2 (CDV) protein (the counterpart of t-SNARE on white liposome), indicating that all the fusion observed here was specifically SNARE-dependent. Noticeably, the initial reaction rate (within the first 20 min), measured by the fluorescence intensity change in unit time, increased by 10 fold for about 50 externally facing i-42T DNA-lipid tethers on each side of the t- and v-liposomes.
We also observed that DNA-lipid tethers stimulated SNARE-driven membrane fusion in a dose-dependent way (Figure 2b). Below 100–200 tethers per vesicle (loaded, with about half of the tethers on the outer leaflet of each vesicle), increasing the tether density on liposome enhanced the stimulation effect. However, this stimulation effect plateaued and even decreased when the density of tethers was over 200 per vesicle on either t- or v-liposome (loaded, i.e. over about 100 of the tethers on the outer leaflet of each vesicle). ssDNA (total of 63 nt in each i-42T tether) is generally considered to behave as polymer chains. The configuration of polymer chains that are adsorbed or grafted to a surface depends on their density. At low surface density, in which the neighboring polymer chains do not overlap with each other laterally, they display the “mushroom” configuration. In contrast, at high density, the neighboring chains are close enough to overlap, and the resulting layer adopts the “brush” structure.[9][10] Under our assay conditions, at the density of 100 tethers on the outer leaflet of one vesicle, the conformation of ssDNA changes from “mushroom” to extended “brush” (See SI. Calculations), which makes the t- and v-SNAREs harder to overlap and assemble after two vesicles are bridged. Therefore, the stimulation effect of tethers is compromised beyond this density.
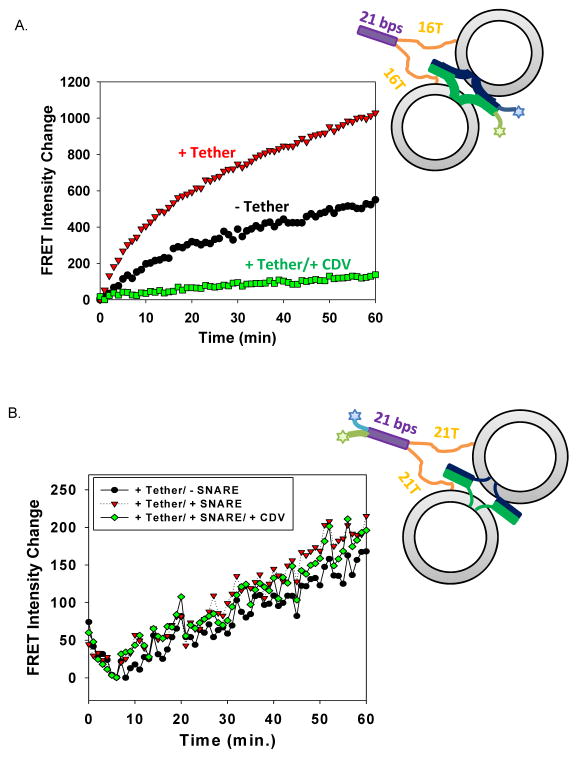
To evaluate how DNA-lipid tethers affect the assembly of t- and v- SNAREs, we employed Fluorescence Resonance Energy Transfer (FRET) experiment with an Alexa488-Texas Red donor-acceptor pair. In this assay, residue 20 on SNAP25 was labeled with Alexa 488 and residue 28 on VAMP2 was labeled with Texas Red. Since the residues are located at the N-termini of t- and v-SNAREs, respectively, the change of FRET intensity over time directly reflects the kinetics of N-terminal assembly of SNAREs during membrane fusion.
As shown in Figure 3a, the assembly rate of N-termini of SNAREs was significantly increased by DNA-lipid tethers within the first 20 min of the fusion reaction. In the later stage of membrane fusion, the two curves (with and without DNA-lipid tethers) gradually increased in a more parallel manner, indicating that the stimulation effect of DNA-lipid tethers became less significant in the later stage of reaction. When Texas Red labeled VAMP-4X mutant (VAMP2 L70D, A74R, A81D, L84D), a mutant of VAMP2 that forms stable complex with t-SNARE without triggering fusion,[11] was used in the fusion assay and the FRET assay (Figure S2), in the presence of DNA-lipid tethers, SNARE-driven lipid mixing was completely blocked; however, the acceleration of the N-terminal assembly of SNAREs in the initial stage of fusion reaction was unaffected. These observations further confirmed that the designed DNA-lipid tethers did not drive membrane fusion by themselves, and that their only function was to promote the formation of SNARE complexes, probably during the initial stage of fusion reaction.
Figure 3.
DNA-lipid tether increased the rate of N-terminal assembly of t- and v-SNAREs. (A) Rate of SNARE N-terminal assembly during liposome-liposome fusion. The change of FRET intensity between the full-length t-SNAREs, SNAP25 Q20C labeled with Alexa 488, and the full-length v-SNARE, S28C labeled with Texas Red (right before SNARE domain) (using i-16T tethering pairs) was followed during fusion. (B) Rate of DNA-lipid assembly during liposome-liposome fusion. The change of FRET intensity between the tether, i-21T, labeled with Alexa 488 and its complementary tether, anti i-21T, labeled with Texas Red was followed during fusion.
To compare the assembly rate of DNA-lipid tethers with that of t- and v-SNARE, we put the same donor-acceptor pair at the distal ends of DNA-lipid tethers and used unlabeled proteins and unlabeled liposomes in the FRET assay. As shown in Figure 3b, in contrast to the kinetics of SNARE assembly during fusion, DNA-lipid assembled gradually with a constant rate, and this rate was not affected by the presence of SNAREs or CDV. This observation suggested that hybridization of just a small number of DNA-lipid tethers was sufficient to promote the assembly of t- and v-SNAREs (drastic increase of SNARE assembly) and, therefore, significantly accelerate the resulting fast membrane fusion (drastic increase in lipid mixing).
The dsDNA segments at the membrane distal end of the DNA-lipid tethers were designed to hybridize in a parallel manner. Further zippering of this hybridization segment was prevented by the ssDNA linker segment. Assuming the dsDNA are fully hybridized (in an orientation away from the linkage between two membranes), the distance spanning the bridged membranes is determined by the length of the ssDNA linker segment. Therefore, by varying the number of nucleotides in the linker segment, we are able to control the separation distance between the membranes. However, ssDNA is flexible and non-structural under our assay conditions. To our knowledge, it is impossible to experimentally determine the precise end-to-end length of ssDNA under our conditions, in which random pulling forces are applied by colliding liposomes to the tethers. So our best estimation of the actual separation distance is based on the contour length of ssDNA (L= # of bases × 0.5 nm per base),[12] which gives the maximum length of the linker segment. Under low forces (up to a few pN and ~ 70% extension), the length of the tethers linearly increases with the load. It is also worth mentioning that ssDNA only reaches its maximal length under extremely high stretching forces, which do not exist in our fusion system. Hence, the actual distance between tethered liposomes will fluctuate by Brownian motion but will remain lower than this estimated maximum length. In addition, multiple tethers, hybridizing in parallel between opposing liposomes, self-exclude and thus certainly bring the liposomes much closer than the separation distance estimated based on the contour length.
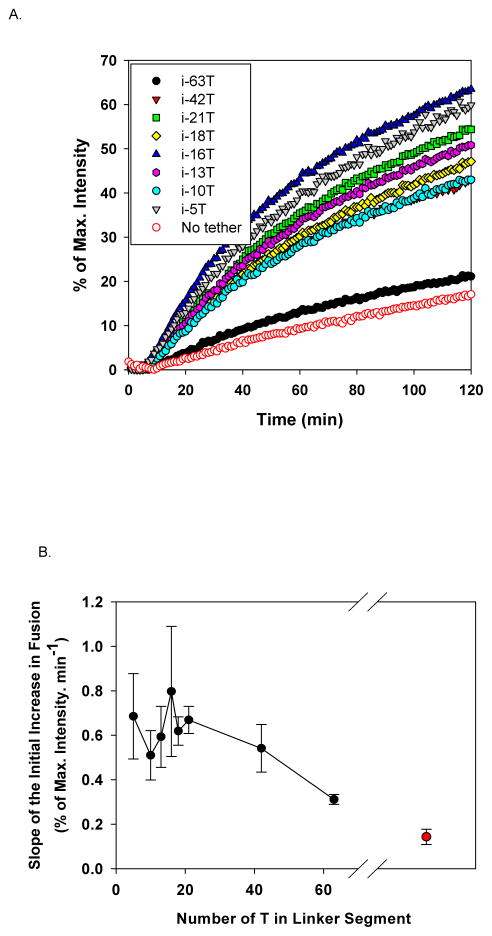
In Figure 4a, a series of representative kinetic curves for fusion reactions between two populations of liposomes were shown, each of which contains DNA-lipid tethers with specified number of oligonucleotides (nt), T, in the linker segment. The fusion reactions in the presence of the longest tether, i-63T (Table S1), was slightly faster, but not distinctly different from those in the absence of tethers (SNARE alone). This suggests that when two liposomes are 63 nm (maximal) apart, the rate of SNARE complex formation between two bridged liposomes is close to that between two freely diffusing liposomes, analogous to using infinitely long tethers. Note that, at our lipid concentration (1 mM), the average distance between two liposomes is ~ 300 nm (See SI. Calculation). When the linker length ranged from 5 to 42 nt, we observed that all the fusion reactions were stimulated by tethers and they followed very similar kinetics with fluctuations within a small range. We have done similar experiments at higher lipid concentration and lower v-SNARE density and the acceleration with 42 nt linker length is conserved (Figure S4). All these results indicate that when two liposomes are kept within this distance range (maximal 40 nm), the SNARE complex formation was promoted by the tethers at rates relatively independent of the linker length.
Figure 4.
Length of the linker region affects the stimulation effect of DNA-lipid tethers. (A) Representative fusion curves in the absence and in the presence of DNA-lipid tethers bearing different length of linkers. (B) The initial fusion rate (first 10 min) as a function of the length of linker. Under assay conditions, when the linker region contains 40 or less nts, tethers stimulated SNARE driven-membrane fusion most efficiently.
In Figure 4b, we plotted the initial fusion rate (the first 10 min after the reaction started) of SNARE-driven membrane fusion, measured by the percentage of NBD fluorescence intensity change in unit time, against the length of the linker segment. This plot demonstrated that SNARE activity inversely correlates with the maximum distance between two bridged membranes. Although the exact maximal distance could not be determined and the actual distance fluctuates in time because of Brownian motion, we qualitatively showed that when two membranes are kept closer to each other, membrane fusion driven by SNAREs occurs with increasing rate, until reaching a maximal distance of at most 40 nm, within which the reaction rate plateaued. This distance-dependent activity of SNAREs can be regulated and controlled using our non-fusogenic DNA-lipid tethers, which are designed to implement the bridging function of tethering factors.
Zippering of t- and v-SNARE into a four-helix bundle generates sufficient force to trigger membrane fusion. The energy landscape of neuronal and other SNAREs,[5d, 13] determined by high-resolution dual-trap optical tweezer experiments, reveals a sequential three-stage model for SNARE assembly. It includes a slow intrinsic assembly of N-terminal domain (NTD), an intrinsic pause near the ionic layer, and unobstructed, fast zippering of the C-terminal domain (CTD) and the linker domain (LD) to open the fusion pore. According to this proposed model, SNARE function can be regulated at any stage of its assembly.
Examples of regulatory proteins, such as Munc18 that stimulates membrane fusion by stabilizing SNARE complexes,[14] and Complexin that inhibits membrane fusion by clamping further zippering after NTD, [11, 15] have been investigated in details. Examples of conformational switches, such as C- or N-terminal segment of the soluble v-SNARE (termed as Vc- or Vn- peptides), that can activate membrane fusion by directly binding to, and thereby pre-arranging t-SNARE conformation into its fusion-competent conformation, have also been reported.[6,16] Different from the regulatory proteins and Vc/ Vn peptides, DNA-lipid tethers regulate SNARE assembly by bridging two membranes close to each other without directly interacting with SNARE proteins. Consequently, local concentrations of cognate SNARE proteins are increased, and thereby, the encounter probability of t- and v-SNAREs also significantly increased.
DNA-lipid tethers provide an in situ tool to control the separation distance between two membranes. Previous investigations on the membrane-embedded neuronal SNARE proteins, measured by surface forces apparatus (SFA),[5c] demonstrated that during the approaching of two SNARE bilayers, no force was detected until the membranes reach a separation distance of 20 nm, where t- and v-SNAREs begin overlapping (not binding yet), and then, SNAREpins start assembling when the separation distance reaches 8 nm. In fact, using tethers containing shorter and shorter linkers mimics the procedure of approaching two facing membranes in a controlled manner in solution. Consistent with the general trend of SFA results, we observed similar correlation between SNARE activity and the distance between two apposed membranes, that is DNA-lipid tether does not stimulate SNARE function efficiently until the linker is shorter than 40 nt and the stimulation effect stays at plateau level with even shorter tethers.
In general, DNA-lipid tethers provide an independent tool (other than native proteins) to regulate biological processes. It can be further applied, in situ, to facilitate function of proteins that require pre-assembly of two membranes within certain distance, and to mimic or bypass functions of membrane binding proteins. In addition, replacement of DNA with its enzyme resistant analogue, peptide nucleotide acid (PNA), will allow incorporation of such artificial tethers into cells to build synthetic and controllable biological system in vivo.
Supplementary Material
Acknowledgments
We would like to thank Dr. S. Krishnakumar and Dr. F. Li for providing some protein plasmids and labelled protein and for discussion on the results. We would like to thank Dr. C. Lin for discussion on designing the DNA sequences. This work was supported by US National Institutes of Health (NIH) grant DK027044-39 and Agence Nationale de la Recherche (ANR) ANR-14-1CHN-0022-01 grant to J.E.R..
Contributor Information
James E. Rothman, Email: james.rothman@yale.edu.
Frederic Pincet, Email: pincet@lps.ens.fr.
References
- 1.a) Pfeiffer I, Hook F. J Am Chem Soc. 2004;126:10224–10225. doi: 10.1021/ja048514b. [DOI] [PubMed] [Google Scholar]; b) Beales PA, Vanderlick TK. J Phys Chem A. 2007;111:12372–12380. doi: 10.1021/jp075792z. [DOI] [PubMed] [Google Scholar]; c) Gunnarsson A, Jonsson P, Marie R, Tegenfeldt JO, Hook F. Nano Lett. 2008;8:183–188. doi: 10.1021/nl072401j. [DOI] [PubMed] [Google Scholar]
- 2.a) Yoshina-Ishii C, Boxer SG. J Am Chem Soc. 2003;125:3696–3697. doi: 10.1021/ja029783+. [DOI] [PubMed] [Google Scholar]; b) Jakobsen U, Simonsen AC, Vogel S. J Am Chem Soc. 2008;130:10462. doi: 10.1021/ja8030054. [DOI] [PubMed] [Google Scholar]; c) Chan YH, van Lengerich B, Boxer SG. Proc Natl Acad Sci U S A. 2009;106:979–984. doi: 10.1073/pnas.0812356106. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Lin CC, Seikowski J, Perez-Lara A, Jahn R, Hobartner C, Walla PJ. Nat Commun. 2014;5:5859. doi: 10.1038/ncomms6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Graneli A, Yeykal CC, Prasad TK, Greene EC. Langmuir. 2006;22:292–299. doi: 10.1021/la051944a. [DOI] [PubMed] [Google Scholar]
- 4.Jahn R, Lang T, Sudhof TC. Cell. 2003;112:519–533. doi: 10.1016/s0092-8674(03)00112-0. [DOI] [PubMed] [Google Scholar]
- 5.a) Weber T, Zemelman BV, McNew JA, Westermann B, Gmachl M, Parlati F, Sollner TH, Rothman JE. Cell. 1998;92:759–772. doi: 10.1016/s0092-8674(00)81404-x. [DOI] [PubMed] [Google Scholar]; b) Sudhof TC, Rothman JE. Science. 2009;323:474–477. doi: 10.1126/science.1161748. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Li F, Pincet F, Perez E, Eng WS, Melia TJ, Rothman JE, Tareste D. Nat Struct Mol Biol. 2007;14:890–896. doi: 10.1038/nsmb1310. [DOI] [PubMed] [Google Scholar]; d) Gao Y, Zorman S, Gundersen G, Xi Z, Ma L, Sirinakis G, Rothman JE, Zhang Y. Science. 2012;337:1340–1343. doi: 10.1126/science.1224492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li F, Kummel D, Coleman J, Reinisch KM, Rothman JE, Pincet F. J Am Chem Soc. 2014;136:3456–3464. doi: 10.1021/ja410690m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.a) Pfeffer SR. Nat Cell Biol. 1999;1:E17–22. doi: 10.1038/8967. [DOI] [PubMed] [Google Scholar]; b) Sudhof TC. Annu Rev Neurosci. 2004;27:509–547. doi: 10.1146/annurev.neuro.26.041002.131412. [DOI] [PubMed] [Google Scholar]; c) Hutagalung AH, Novick PJ. Physiol Rev. 2011;91:119–149. doi: 10.1152/physrev.00059.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scott BL, Van Komen JS, Liu S, Weber T, Melia TJ, McNew JA. Methods Enzymol. 2003;372:274–300. doi: 10.1016/S0076-6879(03)72016-3. [DOI] [PubMed] [Google Scholar]
- 9.Milner ST. Science. 1991;251:905–914. doi: 10.1126/science.251.4996.905. [DOI] [PubMed] [Google Scholar]
- 10.de Gennes PG. Conformation of polymers attached to an interface. 1980;13:1069–1075. [Google Scholar]
- 11.Krishnakumar SS, Radoff DT, Kummel D, Giraudo CG, Li F, Khandan L, Baguley SW, Coleman J, Reinisch KM, Pincet F, Rothman JE. Nat Struct Mol Biol. 2011;18:934–940. doi: 10.1038/nsmb.2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smith SB, Cui Y, Bustamante C. Science. 1996;271:795–799. doi: 10.1126/science.271.5250.795. [DOI] [PubMed] [Google Scholar]
- 13.Zorman S, Rebane AA, Ma L, Yang G, Molski MA, Coleman J, Pincet F, Rothman JE, Zhang Y. eLife. 2014;3:e03348. doi: 10.7554/eLife.03348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.a) Shen J, Tareste DC, Paumet F, Rothman JE, Melia TJ. Cell. 2007;128:183–195. doi: 10.1016/j.cell.2006.12.016. [DOI] [PubMed] [Google Scholar]; b) Parisotto D, Malsam J, Scheutzow A, Krause JM, Sollner TH. J Biol Chem. 2012;287:31041–31049. doi: 10.1074/jbc.M112.386805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.a) Malsam J, Parisotto D, Bharat TA, Scheutzow A, Krause JM, Briggs JA, Sollner TH. EMBO J. 2012;31:3270–3281. doi: 10.1038/emboj.2012.164. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Giraudo CG, Eng WS, Melia TJ, Rothman JE. Science. 2006;313:676–680. doi: 10.1126/science.1129450. [DOI] [PubMed] [Google Scholar]
- 16.a) Melia TJ, Weber T, McNew JA, Fisher LE, Johnston RJ, Parlati F, Mahal LK, Sollner TH, Rothman JE. J Cell Biol. 2002;158:929–940. doi: 10.1083/jcb.200112081. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Pobbati AV, Stein A, Fasshauer D. Science. 2006;313:673–676. doi: 10.1126/science.1129486. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.