Abstract
Human longevity and diseases are likely influenced by multiple interacting genes within a few biologically conserved pathways. Using long-lived smokers as a phenotype (n = 90)—a group whose survival may signify innate resilience—we conducted a genome-wide association study comparing them to smokers at ages 52–69 (n = 730). These results were used to conduct a functional interaction network and pathway analysis, to identify single nucleotide polymorphisms that collectively related to smokers’ longevity. We identified a set of 215 single nucleotide polymorphisms (all of which had p <5×10−3 in the genome-wide association study) that were located within genes making-up a functional interaction network. These single nucleotide polymorphisms were then used to create a weighted polygenic risk score that, using an independent validation sample of nonsmokers (N = 6,447), was found to be significantly associated with a 22% increase in the likelihood of being aged 90–99 (n = 253) and an over threefold increase in the likelihood of being a centenarian (n = 4), compared with being at ages 52–79 (n = 4,900). Additionally, the polygenic risk score was also associated with an 11% reduction in cancer prevalence over up to 18 years (odds ratio: 0.89, p = .011). Overall, using a unique phenotype and incorporating prior knowledge of biological networks, this study identified a set of single nucleotide polymorphisms that together appear to be important for human aging, stress resistance, cancer, and longevity.
Keywords: Resilience, Polygenic risk score, Genetic network, Longevity, Cancer, HRS
Over time, nearly all biological organisms experience a progressive decline of cellular structure and function, resulting in a decreased ability for systems to adequately respond to environmental perturbations and maintain homeostasis. This process known as aging is the number one risk factor for mortality among humans, contributing to an individual’s susceptibility to a number of distinct conditions such as cardiovascular disease, cancer, diabetes, neurodegenerative diseases, sarcopenia, lung disease, vision/hearing impairment, and frailty (1,2). Accordingly, it has been suggested that slowing the aging process would not only increase life span but also postpone most major illnesses and disability (3).
In 1993, a paper by Schächter and coworkers discussed the enormous potential for identifying alleles that influence aging and longevity in humans (4). Since then, with the growing availability of sequencing data, the search for genes that regulate human aging and longevity has gained significant momentum (5–8). Using twin data, researchers have estimated that genetic differences account for 20%–30% of the variance in human life span, with the remainder being under the influence of environmental or stochastic factors (9,10). However, it has been suggested that the degree of genetic influence may also vary as a function of environment (11). Evidence from animal models suggests that genetic factors which influence longevity may be linked to innate stress resistance (12–15); if so, genetic endowment may contribute differentially to variability in life span within populations, as a function of environmental conditions. For instance, genes that promote somatic maintenance and repair may exhibit a larger effect among individuals who accumulate increased exposure to adverse environmental conditions. Exposure to damaging environmental stressors would likely result in significant reductions in life span for the majority of individuals; however, individuals genetically endowed with genes promoting somatic maintenance and repair may be able to better mitigate damage and thus would experience little to no reduction in life span. Conversely, the majority of those living under advantageous environmental conditions may have relatively long survival, due to minimal damage accumulation, and thus genetic differences would have less of an effect on life-span variability.
Smoking is one of the most consistent biological stressors among humans and has been shown to have drastic consequences for life span and disease progression, most notable heart disease and cancer (16,17). It is suggested that cigarette exposure may impact the risk of death and disease via its acceleration of the aging process (18,19). Yet, not all smokers experience earlier mortality—in fact, a small proportion manage to survive to extreme ages. For instance, centenarians have been shown to exhibit the same poor health behaviors as other members of their birth cohort (20). There is reason to believe that these long-lived smokers may represent a biologically distinct group, endowed with genetic variants allowing them to respond differentially to environmental stressors. In previous work, we showed that current heavy smokers who had survived to age 80 and beyond had mortality risks and inflammatory levels similar to nonsmoking individuals of the same age—suggesting that they may be innately equipped to offset the harmful effects of cigarette exposure (21).
In experimental studies, many of the genes associated with stress resistance and longevity in animal models have been found to be comprised within pathways, such as the insulin-like growth factor-I/insulin signaling pathway, that are evolutionarily conserved among yeast, drosophila, Caenorhabditis elegans, mice, and humans (14,22). However, most of these mutations occur so rarely in nature that it is unlikely that they would contribute to the variability of life span within the general population. Furthermore, most of these mutations were discovered in organisms with identical genetic backgrounds, which will certainly not be the case when identifying polymorphisms that influence life span in humans. For humans, it is likely that multiple polymorphisms may simultaneously influence life span. For this reason, we hypothesize that multiple genes, potentially within these conserved pathways, influence longevity in a polygenic manner. While work by Sebastiani and coworkers (8) successfully used Bayesian networks to quantify genetic signatures that were predictive of longevity, most genome-wide association studies (GWAS) of human longevity investigate the individual influences of single nucleotide polymorphisms (SNPs). Incorporating a priori information on networks may allow us to identify functionally related genes whose effects are too small to observe individually, yet jointly influence aging, longevity, and disease risks. Therefore, the current study aims to (a) investigate genes associated with the long-lived smoker phenotype, drawing on previous knowledge of functional interaction networks and pathways, in order to conceptualize GWAS results; (b) generate a polygenic risk score (PRS) based on GWAS and network-selected SNPs; (c) examine how the genetic score is related to age in the nonsmoking population of middle-aged and older adults; (d) examine how the genetic score is related to prevalence of disease within both the smoking and nonsmoking populations.
Methods
Discovery and Validation Samples
Participants were part of the 2006 and 2008 waves of the Health and Retirement Study (HRS), a nationally representative longitudinal study of health and aging in the United States (23). Our discovery sample was limited to white current smokers only. Cases (N = 90) were participants who reported that they currently smoked and who had survived to at least age 80 at the last wave they were interviewed, while controls (N = 730) were participants who reported that they currently smoked and who were less than 70 years of age at the last wave they were interviewed. It is well known that on average, smokers’ life expectancy is reduced by 10 years. Thus, one would expect that the mortality selection of smokers aged 80+ is similar to the mortality selection of nonsmokers aged 90+ (an age cutoff commonly used in longevity studies). Furthermore, we based our age cutoffs on our previous work, which provided evidence that heavy/current smokers who survived to age 80+ were a distinct group (21). We showed that a nationally representative group of 80+-year-old smokers did not have higher mortality rates (during up to 18 years of mortality follow-up) compared to 80+-year-old never smokers. They also had similar physiological functioning measures—inflammation, blood pressure, and immune function. On the other hand, smokers who were aged 50–69 had significantly higher mortality rates during follow-up and worse contemporaneous physiological functioning measures than never smokers of the same age. Finally, the mortality rates of the younger group suggested that the majority of 50- to 69-year-old current smokers will not survive to ages 80+. Overall, this suggests that smokers in their 80s and beyond likely represent a biologically resilient group (21).
Our validation sample (N = 6,447) was made up of HRS participants who self-reported as nonsmokers at the time of their last interview, were aged 52 and older, and who had complete genetic data from which to generate a PRS. Participants younger than 52 were excluded, given that HRS collects data on a nationally representative sample of older adults (aged 52 and older), and their spouses, and as a result, younger participants represent spouses of persons aged 52 and older and therefore may not be representative of the population their age. In the validation sample, 4,501 had missing genotype information for at least one of the SNPs used to create the PRS. When comparing excluded individuals (aged 50 and older) to our validation sample, we found that they did not significantly differ in age, sex, or smoking status (former vs never). However, our validation sample was made up of significantly more participants who self-reported their race as white (86%) than the excluded sample (83%).
Genotyping and Quality Control
Genotyping was performed for participants who provided saliva samples and signed consent forms in 2006 and 2008 and was carried out by the NIH Center for Inherited Disease Research (CIDR) using the Illumina Human Omni-2.5 Quad Beadchip, with coverage of approximately 2.5 million SNPs. Quality control filters were performed by CIDR and the Genetics Coordinating Center of the University of Washington (http://hrsonline.isr.umich.edu/sitedocs/genetics/HRS_QC_REPORT_MAR2012.pdf). These filters consisted of removal of: duplicate SNPs; missing call rates more than or equal to 2%; more than 4 discordant calls in 423 study duplicates; more than one Mendelian error; Hardy–Weinberg equilibrium p values less than 10−4 in European or African samples; sex differences in all allelic frequency more than or equal to 0.2; and sex differences in heterozygosity greater than 0.3. As a result, 2,201,371 SNPs remained. However, given our small sample of cases which could inflate p values for SNPs with small minor allele frequencies, we set our minor allele frequency cutoff at 0.05, which left us with a total of 1,224,285 SNPs for our analysis.
Principal components analysis was conducted by the HRS to account for population structure in accordance with the methods described by Patterson and coworkers (24). This analysis produced sample eigenvectors (EV). A screen plot generated by HRS showed that the 20 components produced by the principal components analysis only accounted for a small fraction of the overall genetic variance (<4%) for the full HRS genetic sample and that most of this was contained within the first two components (23). We used a logistic regression model to examine the relationship between the 20 EV and our phenotype and found that none of the EV were significantly associated with being a long-lived smoker. Nevertheless, we ultimately decided to adjust for the first four EV in all subsequent analyses. More information on QC checks and the principal components analysis is provided by HRS (25).
Functional Interaction Network
PLINK’s gene report command (26) was used to map SNPs with p less than 5×10−3 to Genes based on GRCh37/hg19 coordinates. Start and end genome positions from RefSeq genes were provided by the UCSC table browser (http://genome.ucsc.edu/cgi-bin/hgTables). For our analysis, we did not assign upstream or downstream SNPs to a gene; only those SNPs that fell within the designated GRCh37/hg19 coordinates of the gene were assigned to the gene—no border was added to the start and stop coordinates of each gene.
Cytoscape plugin Reactome FI (25) was used to identify and examine functional interaction networks based on results from the GWAS. The functional interaction networks in the Reactome FI plugin are based on data from the Reactome database (27,28), which contains information on manually curated human pathways—DNA replication, transcription, translation, the cell cycle, metabolism, and signaling cascades. “Reactions” encompass a number of biological processes, including binding, activation, translocation, degradation, and classical biochemical reactions. The information in the database comes from published research and is peer-reviewed and regularly updated by expert biologists. More information on the database is available through the Reactome website (http://www.reactome.org/).
Two gene sets—those whose assigned SNPs had p less than 5×10−3 in the GWAS and those whose assigned SNPs had p less than 5×10−4 in the GWAS—were selected for incorporation in the network analysis. These two thresholds were chosen to allow for more lenient significance criteria, which may address the problem of missing heritability (29), yet limiting the potential of overfitting which could weaken predictive ability in validation studies (30). After examining the network structures of both sets, the subset with p less than 5×10−3 was selected for further analysis and validation, given that the network comprised of genes from SNPs with p less than 5×10−4 significance had too few SNPs to generate large enough functional interaction networks to produce a meaningful PRS.
Using the genes which formed a functional interaction network, and whose assigned SNPs had p less than 5×10−3 in the GWAS, we ran pathway enrichment analysis using Reactome FI. This analysis examines whether the number of networked genes in a given pathway is significantly higher than what would be expected by chance alone. Probability and p value for the pathway enrichment analysis is determined by binomial test and false discovery rate based on 1,000 permutation test. The possible pathways are curated from a number of resources including Reactome, KEGG, CellMap, NCI PID, and BioCarta.
Polygenic Risk Score
PRS were developed as a means of examining the aggregate influence of multiple genetic markers (31). A PRS can be thought of as a measure of “genetic burden” (32) and has become increasingly used to facilitate understanding genetic associations with complex traits. To generate a PRS, the 215 genes in our final network were mapped back to the original SNPs. For those mapping to more than one SNP, the SNP with the lowest p value was selected to represent that gene. Next PRS were calculated for the discovery (smokers) and the validation samples (nonsmokers) from the HRS population.
The PRS assumes a dose–response effect, where for each SNP, persons who are homozygous for the negatively associated allele (major allele if the beta coefficient was positive, and minor allele if the beta coefficient was negative) are coded as 0, persons who are heterozygous are coded as 1, and persons who are homozygous for the positively associated allele are coded as 2. Finally, the allele counts for each SNP were weighted by the log of their odds ratio (OR) from the GWAS and summed across the 215 SNPs from our FI network, to generate the total and component PRS. Scores were then standardized to have a mean of 0 and a SD of 1.
Statistical Analysis
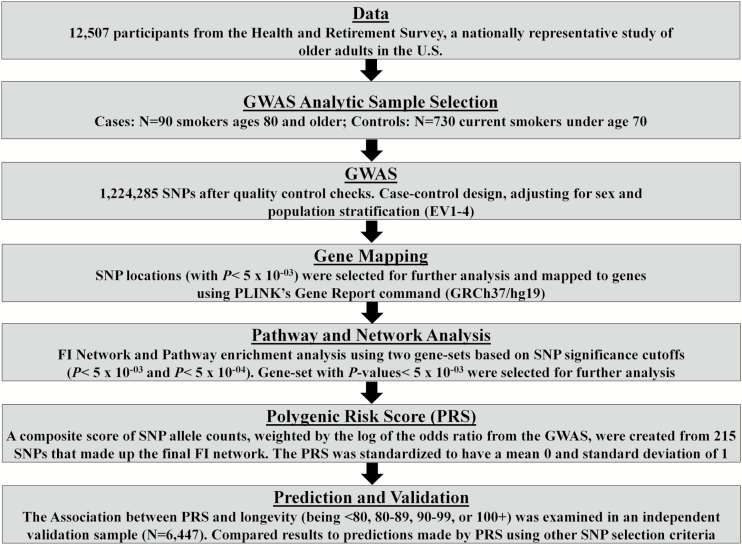
Study methodology is outlined in Figure 1. A case–control GWAS (long-lived vs normal lived smokers) was used to identify SNPs that are potentially associated with longevity and biological stress resistance. Moderately significant SNPs from the GWAS were then mapped to genes and used to build a genetic network based on a priori experimental proteomic evidence of identified genetic pathways and gene interactions. SNPs included within the gene network were used to calculate composite PRS for the entire HRS genetic sample. Using multinomial logistic regression, controlling for the first four EV and sex, we examined the association between PRS and longevity—operationalized as the probability of being in an older age group: ages 80–89, 90–99, or 100+, relative to being at ages 50–79 during the most recent wave interviewed—using a validation sample of nonsmokers from the HRS (n = 6,447). We then examined whether using the network-based approach to SNP selection for inclusion in the PRS improved predictive ability, we compared the association between being very old and our final PRS to the associations between being very old and four other PRS which utilized other SNP selection criteria—top hits, a random subset with p less than 5×10−3, a random subset of the 784 SNPs with p less than 5×10−3 that also mapped to genes, and the top hits of the 784 SNPs (p < 5×10−3 that also mapped to genes). To compare the PRS, logistic regression models controlling for the first four EV and sex were used, with participant aged 50–79 coded as 0 and participants aged 90 and older coded as 1. The cutoff age was increased to 90 versus 80, which was used in the initial GWAS, given that our validation sample was made up of nonsmokers for whom survival to age 80 is much more likely. Finally, we tested the association between PRS and disease prevalence for three major diseases of aging: heart disease, cancer (other than skin), and diabetes. Over 10 waves spanning from 1992 to 2010, participants were asked whether they had ever been diagnosed with each condition. Three logistic regression models incorporating the panel data and adjusting for repeated observations using random effects were run to assess the association between PRS and each of the three conditions. These models were run controlling for age, sex, the first four EV, self-reported race, education, smoking status at each wave, body mass index at each wave, and sample classification (discovery cases, discovery controls, or validation sample).
Figure 1.
Study approach. The study utilized data from the HRS to run a GWAS and network analysis using long-lived smokers as the phenotype of interest. SNPs identified through these processes were then used to create PRS. For validation and replication, we examined the association between the score and age or longevity among nonsmokers in the nationally representative population in HRS. GWAS = genome-wide association study; HRS = Health and Retirement Study; PRS = polygenetic risk score; SNP = single nucleotide polymorphism.
Results
Genome-Wide SNP Analysis
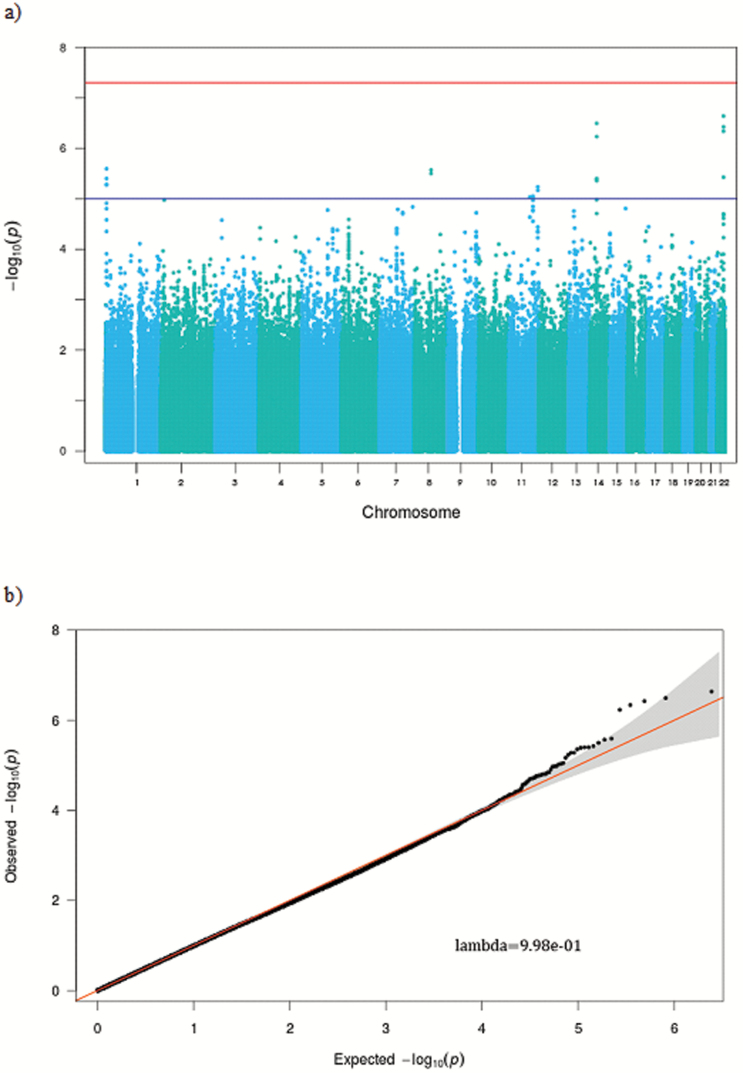
Our GWAS differentiating long-lived smokers from younger smokers was run controlling for sex and four EV which control for population stratification. Although no SNPs met the genome-wide significant threshold—which is not surprising given our small sample size—20 SNPs met the threshold for “suggestive” association (Figure 2a). Also, as shown in our Q-Q plot (Figure 2b), we observed a moderate departure from the null hypothesis of no association, beginning between p = 10−3 and 10−4. Studies of “missing heritability” have suggested that while for the most part, the SNPs meeting statistical significance cutoffs in GWAS only account for a relatively small proportion of the variance in a phenotype, there is evidence that the additional consideration of less significant loci may capture more of the association with phenotypic heterogeneity. Two p value thresholds were considered (p < 5×10−3 and p < 5×10−4) based on previous studies suggesting that the proportion of the variance explained for a phenotype increases when allowing p value threshold to relax to such levels (26), but that allowing for variables with higher p values than this reduced predictive power (33).
Figure 2.
GWAS results. We found that while no SNPs met the criteria for genome-wide significance, a number of SNPs had “suggestive” association with longevity among smokers (a). Additionally, our Q-Q plot shows that we had more SNPs that had p values <5×10−4 than might be expected by chance (b). GWAS = genome-wide association study; SNP = single nucleotide polymorphism.
Network and Pathway Analysis
SNP locations were mapped to Genes—for SNPs with p less than 5×10−3 and p less than 5×10−4 from the GWAS. Overall, there were 535 SNPs with p less than 5×10−4, which mapped to 115 genes, and 5,184 SNPs with p less than 5×10−3, which mapped to 784 unique genes. Cytoscape plugin Reactome FI was used to construct functional interaction networks and run subsequent pathway enrichment analyses. Reactome FI was designed to identify network patterns that relate to disease. The database covers more than 50% of human proteins which are used to build functional interaction networks based on a set of input genes.
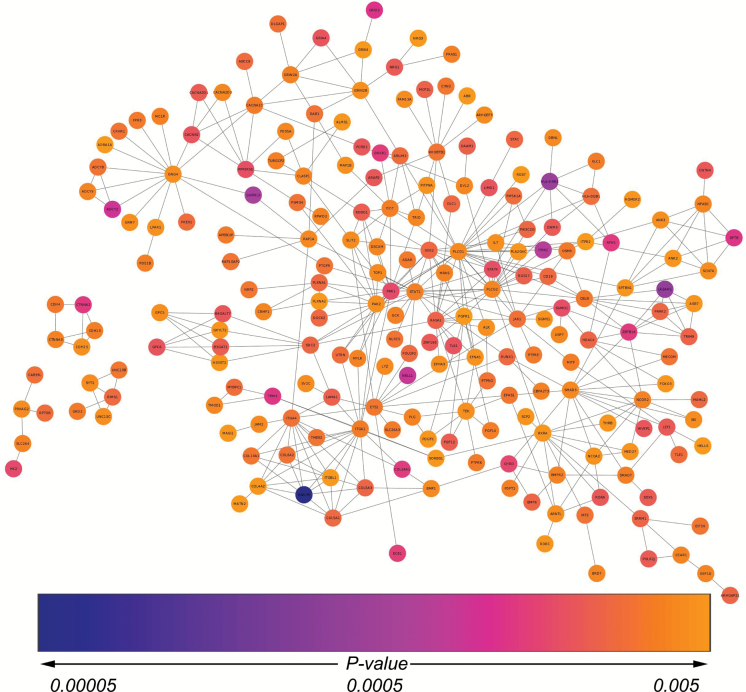
Using the 2013 FI network build, we found that 215 of our 784 genes (p < 5×10−3) made up functional networks that had five or more genes each, the largest of which was encompassed by 202 genes (Figure 3). The other 569 genes were either not functionally connected to any other genes that had SNPs with p less than 5×10−3 or formed networks of three or fewer genes. On the other hand, only three genes were comprised in the network that utilized a p value cutoff of p less than 5×10−4, therefore, the network with p less than 5×10−3 was selected for use in further investigation and validation.
Figure 3.
Functional interaction network. After mapping SNPs with p values <5×10−3 to genes, we found that 215 of them were comprised within functional interaction networks of five or more genes—200 genes were in a single network and 15 genes made up 3 networks of 5 genes each. Of the 215 SNPs (represented by their respective genes in the network), the majority had p values between 5×10−4 and 5×10−3, associations that would have been overlooked using a normal GWAS approach. GWAS = genome-wide association study; SNP = single nucleotide polymorphism.
Next, we ran Reactome FI’s pathway enrichment analysis for these 215 genes in the network using p less than 5×10−3 as the significance threshold and found 21 pathways that were enriched at false discovery rate less than 5×10−3. The 10 most highly enriched pathways, in order, included: P13K-Akt signaling, pathways in cancer, signaling by platelet-derived growth factor, glutamatergic synapse, Ras signaling pathway, Rap1 signaling pathway, L1CAM interactions, focal adhesion, Netrin-1 signaling, and Netrin-mediated signaling (Supplementary Table S1).
Validation Using a PRS
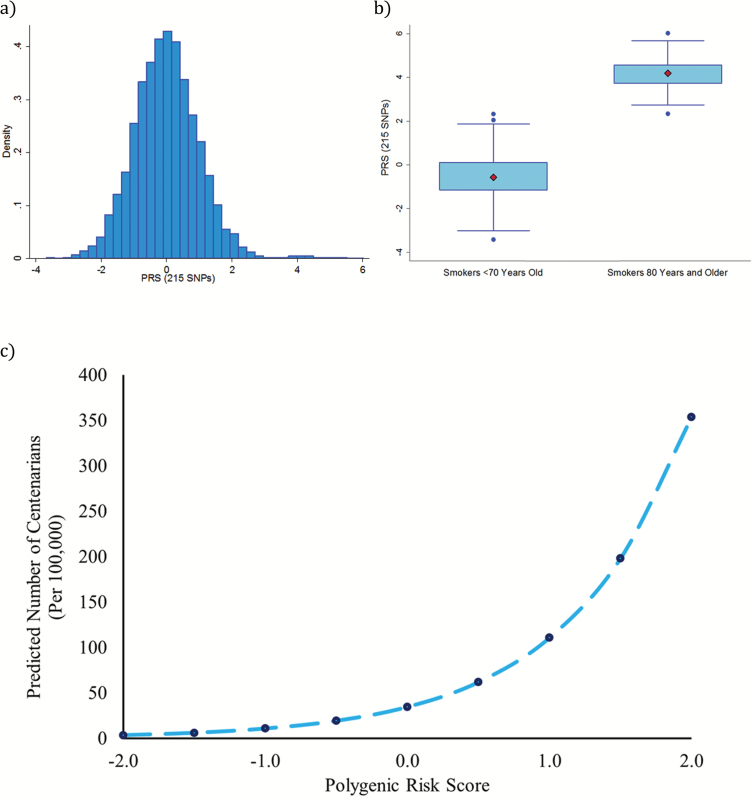
A standardized PRS was generated based on a weighted composite score of the 215 SNPs from the selected interaction network and was evenly distributed (Figure 4a), with a range from −3.68 to 6.02 in the overall HRS population (discovery and validation sample). Mean PRS were compared between our original cases and controls—smokers aged 80+ and smokers younger than 70, respectively—to determine how much of variation in the original phenotype was explained using a composite SNP score of only 215 SNPs from the original 1,224,285 SNPs. Results showed that the score completely accounted for group membership, with no overlap between the two groups (Figure 4b). Of our original 90 cases of long-lived smokers, 49 had complete data on all the SNPs needed to generate the overall PRS, and we found that for this group, PRS ranged from 2.34 to 6.02, with a mean of 4.17 and a SD of 0.78. Among the 730 controls, 422 had no missing genotype data for the 215 SNPs, and these participants had PRS ranging from −3.41 to 2.32, with a mean of −0.55 and a SD of 0.95. Not only were scores significantly higher for the long-lived group, but scores also appeared to be more homogeneous.
Figure 4.
Associations between PRS and longevity. Overall, the weighted PRS was found to be fairly evenly distributed and ranged between −3.68 and 6.02 (a). When comparing the scores of our GWAS cases and controls, we found that there was no overlap between the two groups (b)—cases all had scores of 2.34 or greater (with a mean of 4.17), while controls had scores ranging between −3.41 and 2.32 (with a mean of −0.55). A multinomial logistic regression model was used to examining the association between PRS and age in a validation sample (n = 6,447). Results were used to predict the proportion of centenarians in the population by PRS (c). We found that among individuals with a PRS of −2.0 (2 SDs below the mean), only 3.2 in 100,000 persons are predicted to be a centenarian. On the other hand, for individuals with a PRS of 2.0 (2 SDs above the mean), 340.3 in 100,000 persons are predicted to become centenarians. GWAS = genome-wide association study; PRS = polygenetic risk score; SNP = single nucleotide polymorphism.
Next using our validation sample, we performed a multinomial logistic regression, controlling for the four EV, sex, and race to determine if the PRS was associated with the probability of being in an older age group—aged 80–89 (n = 1,290), 90–99 (n = 253), and 100+ (n = 4)—relative to those aged 52–79 (n = 4,900). Our results showed (Table 1) that among these 6,447 participants, a higher PRS was associated with an increased likelihood of being at ages 90–99 or being a centenarian, relative to being in the youngest age group (50–79). Results showed that a one unit increase was associated with 20% greater likelihood of being at ages 90–99 compared to 29–79 (OR = 1.20, p = .007), and a 3.3-fold increases in the likelihood of being a centenarian (OR = 3.27, p = .027). Based on the parameter from Table 1, we estimated the predicted proportion of centenarians in the population of nonsmokers, by PRS (Figure 4c). We found that for individuals with a PRS that was 2 SDs below the mean (PRS = −2), only 3.2 in 100,000 were predicted to be centenarians. For individuals with a mean PRS (PRS = 0), 33.2 in 100,000 were predicted to be centenarians, and for individuals with a PRS that was 2 SDs above the mean (PRS = 2), 340.3 in 100,000 were predicted to be centenarians.
Table 1.
Odds Ratios for PRS From a Multinomial Regression Model for Longevity Using the Validation Sample (N = 6,447)
| Age Category | Odds Ratio | p Value |
|---|---|---|
| 80–89 | 1.04 | 0.204 |
| 90–99 | 1.20 | 0.007 |
| 100+ | 3.27 | 0.027 |
| <80 | (Reference category) | |
Notes: Model run adjusting for sex and the first four eigenvectors. PRS = polygenic risk score.
To provide evidence that using a network-based approach to select candidate SNPs improved our predictive measure, we compared the strength of the association between longevity and our measure to the associations between longevity and four other weighted PRSs from 215 SNPs selected via other means—top hits (the 215 most significant SNPs from the GWAS), a random subset of 215 SNPs with p less than 5×10−3 from the GWAS, a random subset of 215 SNPs that were part of the 784 SNPs that had p less than 5×10−3 from the GWAS and that also mapped to genes, and the 215 most significant SNPs out of the subset of 784 SNPs (those with p < 5×10−3 that also mapped to genes). To validate our approach, we used five separate logistic regression models—one per individual PRS—with the outcome being 1 if an individual was 90+ years old and 0 if he/she was aged 52–79. Models were run using our validation sample only and controlling for the four EV, sex, and self-reported race (Table 2). Of the five models, our original measure which utilized the functional interaction network to select SNPs for inclusion in the PRS was the only PRS variable found to be statistically significant (OR = 1.22, p = .005)—and it remained significant even after Bonferroni adjustment (p < .01). On the other hand, the four other PRSs were not significantly associated with being at ages 90 and older and had OR between 0.83 and 1.10 with p values ranging from .122 to .872. Lists of SNPs and corresponding gene sets used for the other PRS measures are available upon request.
Table 2.
Results From Logistic Regression Models (1 = 90+ years and 0 = 50–79 years) on the Validation Sample (N = 6,447) to Test the Association Between Longevity and Various PRS That Utilize Different SNP Selection Approaches
| PRS Method | Odds Ratio | p Value |
|---|---|---|
| Network-based PRSa | 1.21 | .005 |
| Top SNPsb | 0.95 | .649 |
| Random SNPsc | 0.83 | .122 |
| Random genesd | 0.98 | .872 |
| Top genese | 1.10 | .253 |
Notes: PRS = polygenic risk score; SNP = single nucleotide polymorphism.
aOriginal PRS generated from SNPs within the functional interaction network.
bPRS of the 215 top hits from the genome-wide association study.
cPRS from a random subset of 215 SNPs with p < 5×10−3.
dPRS from a random subset of 215 SNPs with p < 5×10−3 that also mapped to genes.
ePRS of the 215 most significant hits that also mapped to genes.
Given that the replication and discovery sets both came from the same population sample (HRS), we also examined the association between the PRS and longevity (90+) in a completely independent sample. Data for this analysis came from the English Longitudinal Study of Aging (ELSA). ELSA is a sister-study of the HRS and there has been an attempt to harmonize the sample design, the questions, and the genetic approach. Both are nationally representative panel studies of individuals aged 50 years and older. Our validation sample from ELSA consisted of 264 long-lived individuals (aged 90+) and 4,521 controls (aged 50–79). Approximately 11% of long-lived individuals were current smokers, whereas for controls, approximately 13% were current smokers. Genotyping was performed using the Illumina Omni 2.5-8 Beadchip and the same QC criteria that were used for HRS. The majority of the SNPs used in the original PRS were available for ELSA (n = 205). The 10 SNPs that were not genotyped in ELSA (and their corresponding genes) are listed in Supplementary Table S3. After standardizing the PRS, so that it had a mean of 0 and SD of 1, we used a logistic regression to test whether the PRS was associated with an increased probability of being at ages 90+ compared to ages 50–79. The model was run using all subjects, adjusting for population stratification (EV1–EV4). Results showed that a 1 SD increase in PRS was associated with a 7% increase in the likelihood of being at ages 90+, relative to ages 50–79 (OR = 1.07, p = .018). We were not able to test centenarian status given that ELSA top-codes mortality at 90, making it impossible to differentiate who survived to 100+. Subsequent models adjusting for smoking status (never, current, and former) were also run and did not appear to impact results (OR = 1.08, p = .014). Next, we examined whether the association between PRS and longevity was dependent on smoking status. We found that the interaction between PRS and smoking was not significant, and in stratified models, the effect size was similar for the never (OR = 1.06), former (OR = 1.08), and current smoking (OR = 1.08) groups.
Finally, using the HRS validation sample (nonsmokers), we examined the association between our original PRS and disease prevalence measures for heart disease, cancer, and diabetes. This analysis was restricted to participants who reached at least age 70 or older during the study, to ensure we were picking up aging-related disease. Over the 10 waves, 22.5% of participants self-reported having been diagnosed with heart disease, 15.3% self-reported having been diagnosed with diabetes, and 13.6% self-reported having been diagnosed with cancer other than skin cancer at some point during their lifetime. Logistic regression models were run on the validation sample. Repeated-measures data across all waves were pooled over time and nonindependence for repeated measures were accounted for using clustering. Our results showed that a higher PRS was significantly associated with lower prevalence of cancer (Table 3). For each 1 SD increase in the PRS, the likelihood that an individual had ever been diagnosed with cancer was reduced by nearly 11% (OR = 0.89; p = .011). When examining the association between PRS and either heart disease or diabetes, we found no statistically significant relationships (Table 3).
Table 3.
Random Effects Logistic Regression Models of the Association Between PRS and Disease Prevalence
| Cancer (nonskin)a | Heart Diseaseb | Diabetesc | ||||
|---|---|---|---|---|---|---|
| Odds Ratio | p Value | Odds Ratio | p Value | Odds Ratio | p Value | |
| PRS | 0.89 | .011 | 0.987 | .732 | 1.035 | .260 |
| Age (y) | 1.06 | <.001 | 1.073 | <.001 | 1.044 | <.001 |
| EV1 | 2.18E-06 | .026 | 4.22E-04 | .089 | 9.88E+10 | <.001 |
| EV2 | 2.96E-06 | .079 | 2.19E-09 | <.001 | 8.62E+04 | .008 |
| EV3 | 1.38E+03 | .355 | 7.509 | .687 | 9.48E+02 | .102 |
| EV4 | 73.44 | .303 | 80.830 | .225 | 9.16E-03 | .216 |
| BMI | ||||||
| Underweight (<18.5) | 1.242 | .344 | 0.942 | .776 | 1.156 | .711 |
| Overweight (25–29.9) | 1.000 | .998 | 1.144 | .043 | 1.813 | <.001 |
| Obese (30+) | 1.136 | .200 | 1.438 | <.001 | 4.045 | <.001 |
| Education | ||||||
| GED | 0.981 | .932 | 1.210 | .264 | 0.767 | .150 |
| High school | 1.324 | .026 | 0.855 | .113 | 0.795 | .036 |
| Some college | 1. 286 | .065 | 0.782 | .025 | 0.709 | .004 |
| College and above | 1.651 | <.001 | 0.631 | <.001 | 0.606 | <.001 |
| Sex (female = 1) | 0.945 | .523 | 0.543 | <.001 | 0.731 | <.001 |
| Ever smoked | 1.156 | .098 | 1.311 | <.001 | 1.096 | .260 |
| Constant | 0.001 | <.001 | 0.002 | <.001 | 0.006 | <.001 |
Notes: Disease prevalence run as three separate logistic regression models, adjusting for multiple observations over 10 waves by clustering. The reference categories for independent variables in each model were: “Normal weight (18.5–24.9)” for BMI and “No high school degree or equivalent” for Education. GED = general educational development degree; BMI = body mass index; EV = eigenvector; PRS = polygenic risk score.
aObservations = 49,891; log likelihood = -5,636.66.
bObservations = 49,931; log likelihood = -7,942.78.
cObservations = 49,920; log likelihood = -7,181.58.
N = 6,434.
Discussion
For most individuals, environment may play a major role in their probability of postponing disease and reaching old age. However, for those under chronic exposure to exogenous stressors, such as cigarette smoke, genetic variants may act as key factors in determining whether individuals are able to delay the age-related progressive decline in physiological functioning by offsetting damage though activation of somatic maintenance and repair mechanisms. As a result, survival among smokers may serve as a unique model for examining the genetics of stress resistance, aging, and longevity. Using long-lived smokers as our phenotype, we were able to identify a network of SNPs that, collectively, were strongly associated with extreme survival and lower cancer rates in an independent validation sample or nationally representative nonsmokers.
Our findings suggest that a 1 SD increase in the genetic load of the 215 SNPs we identified was associated with a 20% increase in the likelihood that an individual was aged 90–99 and an over threefold increase in the likelihood of being a centenarian. Additionally, our model predicted that approximately 340 in 100,000 individuals who had a PRS that was 2 SDs above the mean would be a centenarian, compared to only 33 and 3 in 100,000 who had a PRS at the mean or was 2 SDs below the mean, respectively. It has been reported in the literature that in 2010, there were just over 17 centenarians per 100,000 people in the United States (34), which appears similar to our estimate for mean PRS, taking into account that our validation sample is made up of participants aged 52 and older and only includes nonsmokers. However, additional studies utilizing different samples that include larger numbers of centenarians should be conducted to better understand the association between the PRS we generated in the current study and an individual’s likelihood of surviving to age 100 and beyond.
One of the major physiological risks of exogenous genotoxic exposure that accompanies smoking is the accumulation of DNA damage (35). However, it is likely that long-lived smokers possess variants which prevent genomic instability and allow them to survive to more extreme ages. Genomic instability also happens to be one of the hallmarks of cancer pathogenesis (36), thus the same genes that may promote survival among smokers may also be important for cancer prevention. This is consistent with our findings which showed that the genes we identified through our GWAS and network analysis on long-lived smokers were collectively associated with a nearly 11% lower cancer prevalence in the validation sample. Additionally, our functional interaction network of 215 genes was significantly enriched with pathways in cancer, as well as Ras signaling, Rap1 signaling pathways, and signaling by platelet-derived growth factor—all of which have implications for cancer pathogenesis (37–39).
Pathways which are believed to be potential regulators of the aging process were also enriched in our network. Overall, results showed that the PI3K/AKT signaling pathway had the highest enrichment score. This pathway has previously been shown to comprise genes related to stress resistance, DNA repair, cell death, protein turnover, and antioxidants (40). PI3K/AKT pathway is activated via insulin/insulin-like growth factor signaling. Insulin/insulin-like growth factor signaling is evolutionarily conserved has been shown to elicit a strong influence on life span in model organisms (14) and there is further evidence to suggest it may play an important role in human longevity (41). In worms, transcription factor Daf-16 (abnormal DAuer Formation-16) is a key regulator of insulin/insulin-like growth factor signaling and has been found to be fundamental for extreme life-span extension (42). The FOXO family of transcription factors are the human homolog for DAF-16, and FOXO3a has been shown to be one of the most consistently cited longevity genes in human populations (43). Our network analysis and PRS included SNP rs12203834 (Chr6:108975562), which is an intron variant in FOXO3a. While rs12203834 has not been previously cited, two SNPs in FOXO3 have been previously associated with extreme longevity in two distinct populations—American men of Japanese ancestry from Hawaii (rs13217795) and German men and women (rs9400239) (44,45). Furthermore, a SNP (rs10457180) in FOXO3 was one of only two markers to reach genome-wide significance in the longevity consortium (46). The other was an APOE marker, which unfortunately was not directly genotyped in our data.
Although there is evidence to suggest that FOXO3 is an important gene for aging and longevity, it is likely that additional genes may simultaneously be important for extreme survival, especially under adverse conditions. Previous studies have provided evidence that suggest life span is a polygenic trait (47), influenced by multiple alleles with individual small effects. Furthermore, Kirkwood and coworkers provide three explanations for why aging is likely polygenic: (a) aging is not programmed, (b) genes that influence life span are probably byproducts of selection for other traits, and (c) aging and life span are driven, for the most part, by stochastic damage accumulation (48). Using traditional single-SNP GWAS approaches, many alleles with small individual effects will go unnoticed due to the reliance on strict significance criteria, thus contributing to what is being termed the “missing heritability problem” (49). There is an urgent need for employing methods that both allow for the examination of cumulative associations across SNPs as well as reliable methods for selecting SNPs for inclusion in predictive measures. Our and others’ result illustrate the usefulness of polygenic measures (30,50); nevertheless best practices for SNP selection in the creation of these measures has been less concrete. Network-based analyses may be a useful tool for variant selection when creating polygenic scores (51). Given the evidence that phenotypes like longevity may be influenced by genes within specific pathways and networks (22), we believe that the use of prior knowledge, such as functional interaction network analysis, provides better inclusion criteria for composite scores than methods that only consider top GWAS hits. This is consistent with the present study which showed that PRSs composed of SNPs identified using other means—top hits—were not significantly associated with longevity, while the PRS made up of SNPs in a functional interaction network was found to be a significant predictor of whether an individual was 90 years or older. Furthermore, the strength of this association increased further when predicting whether a participant was a centenarian, remaining significant even with a very small sample size. This is consistent with previous studies reporting that genes may be more important for extreme longevity versus variations in life span within typical ranges (45).
There are limitations to the present study that should be noted. First, our discovery sample consisted of a very small number of cases, which could limit our ability to detect true associations that have small effect sizes. Second, smoking, age, and disease status were based on self-reports. Third, the network was based on curated information, which did not allow for the discovery of novel gene–gene interactions. Fourth, the p value threshold used to select SNPs for inclusion in the network analysis was based on the GWAS, which did not take into account gene–gene interactions. In moving forward, it will be important to incorporate network structure into the calculation of PRS and use statistical network-based methods to identify sets of functionally related genes.
Through our use of a unique phenotype, functional interaction networks to select SNPs, and methods allowing examination of associations with aging-related phenotypes using composite measures of multiple genetic variants, we developed a genetic risk score that was significantly associated with an individual’s likelihood of surviving to extreme old age and also found to predict lower cancer prevalence. Overall, our findings suggest that longevity may be under the regulation of complex genetic networks which influence stress resistance and genomic stability. In moving forward, it will be important to examine how functional variants associated with the SNPs in our score interact with one another to impact signaling within their respective pathways and how these alterations translate into differences in life span and cancer risk.
Supplementary Material
Please visit the article online at http://gerontologist.oxfordjournals.org/ to view supplementary material.
Funding
This research was supported by the National Institute on Aging (NIA): P30AG017265 and T32AG0037. The HRS is supported by NIA (U01AG009740) and the Social Security Administration.
Supplementary Material
Acknowledgment
Electronic resources: PLINK 1.9, Shaun Purcell: http://pngu.mgh.harvard.edu/purcell/plink/ and Cytoscape v3.1.0: http://www.cytoscape.org/
References
- 1. Harman D. The aging process: Major risk factor for disease and death. Proc Natl Acad Sci USA. 1991;88(12):5360–5363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Niccoli T, Partridge L. Ageing as a risk factor for disease. Curr Biol. 2012;22:R741–R752. doi:10.1016/j.cub.2012.07.024 [DOI] [PubMed] [Google Scholar]
- 3. Goldman DP, Cutler D, Rowe JW, et al. Substantial health and economic returns from delayed aging may warrant a new focus for medical research. Health Aff (Millwood). 2013;32(10):1698–1705. doi:10.1377/hlthaff.2013.0052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Schächter F, Cohen D, Kirkwood T. Prospects for the genetics of human longevity. Hum Genet. 1993;91:519–526. [DOI] [PubMed] [Google Scholar]
- 5. Barzilai N, Atzmon G, Schechter C, et al. Unique lipoprotein phenotype and genotype associated with exceptional longevity. JAMA. 2003;290:2030–2040. doi:10.1001/jama.290.15.2030 [DOI] [PubMed] [Google Scholar]
- 6. Beekman M, Blanché H, Perola M, et al. ; GEHA Consortium. Genome-wide linkage analysis for human longevity: Genetics of Healthy Aging Study. Aging Cell. 2013;12:184–193. doi:10.1111/acel.12039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Deelen J, Beekman M, Uh HW, et al. Genome-wide association meta-analysis of human longevity identifies a novel locus conferring survival beyond 90 years of age. Hum Mol Genet. 2014;23(16):4420–4432. doi:10.1093/hmg/ddu139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sebastiani P Solovieff N, Dewan AT, Walsh KM, Puca A et al. . Genetic signatures of exceptional longevity in humans. PloS One. 2012;7:e29848. doi:10.1371/journal.pone.0029848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Herskind AM, McGue M, Iachine IA, et al. Untangling genetic influences on smoking, body mass index and longevity: a multivariate study of 2464 Danish twins followed for 28 years. Hum Genet. 1996;98(4):467–475. [DOI] [PubMed] [Google Scholar]
- 10. Finch CE, Kirkwood TBL. Chance, Development and Aging. New York: Oxford University Press; 2000. [Google Scholar]
- 11. vB Hjelmborg J, Iachine I, Skytthe A, et al. Genetic influence on human lifespan and longevity. Hum Genet. 2006;119(3):312–321. doi:10.1007/s00439-006-0144-y [DOI] [PubMed] [Google Scholar]
- 12. Johnson TE, Henderson S, Murakami S, et al. Longevity genes in the nematode Caenorhabditis elegans also mediate increased resistance to stress and prevent disease. J Inherit Metab Dis. 2002;25(3):197–206. doi:10.1023/A:1015677828407 [DOI] [PubMed] [Google Scholar]
- 13. Finch CE, Morgan TE, Longo VD, de Magalhaes JP. Cell resilience in species life spans: A link to inflammation? Aging Cell. 2010;9(4):519–526. doi:10.1111/j.1474-9726.2010.00578.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gems D, Partridge L. Genetics of longevity in model organisms: Debates and paradigm shifts. Annu Rev Physiol. 2013;75:621–644. doi:10.1146/annurev-physiol-030212-183712 [DOI] [PubMed] [Google Scholar]
- 15. Vijg J, Suh Y. Genetics of longevity and aging. Annu Rev Med. 2005;56:193–212. doi:10.1146/annurev.med.56.082103.104617 [DOI] [PubMed] [Google Scholar]
- 16. Cancer Prevention Study II. The American Cancer Society Prospective Study. Stat Bull Metrop Insur Co. 1992;73(4):21–29. [PubMed] [Google Scholar]
- 17. Preston SH, Strokes A, Mehta NK, Cao B. Projecting the effects of changes in smoking and obesity on future life expectancy in the United States. Demography. 2013. doi:10.1007/s13524-013-0246-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Valdes AM, Andrew T, Gardner JP, et al. Obesity, cigarette smoking, and telomere length in women. Lancet. 2005;366(9486):662–664. doi:10.1016/S0140-6736(05)66630-5 [DOI] [PubMed] [Google Scholar]
- 19. Csiszar A, Podlutsky A, Wolin MS, Losonczy G, Pacher P, Ungvari Z. Oxidative stress and accelerated vascular aging: Implications for cigarette smoking. Front Biosci (Landmark Ed). 2009;14:3128–3144. doi:10.2741/4040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rajpathak SN, Liu Y, Ben-David O, et al. Lifestyle factors of people with exceptional longevity. J Am Geriatr Soc. 2011;59(8):1509–1512. doi:10.1111/j.1532-5415.2011.03498.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Levine M, Crimmins E. Not all smokers die young: A model for hidden heterogeneity within the human population. PLoS One. 2014;9(2):e87403. doi:10.1371/journal.pone.0087403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Longo VD, Finch CE. Evolutionary medicine: From dwarf model systems to healthy centenarians? Science. 2003;299(5611):1342–1346. doi:10.1126/science.1077991 [DOI] [PubMed] [Google Scholar]
- 23. Sonnega A, Faul JD, Ofstedal MB, Langa KM, Phillips JW, Weir DR. Cohort profile: The Health and Retirement Study (HRS). Int J Epidemiol. 2014;43(2):576–585. doi:10.1093/ije/dyu067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2(12):e190. doi:10.1371/journal.pgen.0020190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Health and Retirement Study. Quality Control Report for Genotypic Data University of Washington; 2012. http://hrsonline.isr.umich.edu/sitedocs/genetics/HRS_QC_REPORT_MAR2012.pdf. Accessed October 7, 2015. [Google Scholar]
- 26. Purcell S, Neale B, Todd-Brown K, et al. PLINK: A toolset for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi:10.1086/519795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Croft D, Mundo AF, Haw R, et al. The Reactome pathway knowledgebase. Nucleic Acids Res. 2014;42:D472–D477. doi:10.1093/nar/gkt1102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Milacic M, Haw R, Rothfels K, et al. Annotating cancer variants and anti-cancer therapeutics in reactome. Cancers (Basel). 2012;4(4):1180–1211. doi:10.3390/cancers4041180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhang G, Karns R, Sun G, et al. Finding missing heritability in less significant loci and allelic heterogeneity: Genetic variation in human height. PLoS One. 2012;7(12):e51211. doi:10.1371/journal.pone.0051211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lango Allen H, Estrada K, Lettre G, et al. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature. 2010;467:832–838. doi:10.1038/Nature09410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wray NR, Goddard ME, Visscher PM. Prediction of individual genetic risk to disease from genome-wide association studies. Genome Res. 2007;17(10):1520–1528. doi:10.1101/Gr.6665407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wray NR, Goddard ME, Visscher PM. Prediction of individual genetic risk of complex disease. Curr Opin Genet Dev. 2008;18(3):257–263. doi:10.1016/j.gde.2008.07.006 [DOI] [PubMed] [Google Scholar]
- 33. Shannon P, Markiel A, Ozier O, et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi:10.1101/Gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Meyer J. 2012. Centenarians: 2010. Washington, DC: 2010U.S. Census Bureau Special Report, No. C2010SR-03. [Google Scholar]
- 35. Soares JP, Cortinhas A, Bento T, et al. Aging and DNA damage in humans: A meta-analysis study. Aging. 2014;6(6):432–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Negrini S, Gorgoulis VG, Halazonetis TD. Genomic instability–an evolving hallmark of cancer. Nat Rev Mol Cell Biol. 2010;11(3):220–228. doi:10.1038/nrm2858 [DOI] [PubMed] [Google Scholar]
- 37. Shields JM, Pruitt K, McFall A, Shaub A, Der CJ. Understanding Ras: 'it ain't over 'til it's over'. Trends Cell Biol. 2000;10(4):147–154. [DOI] [PubMed] [Google Scholar]
- 38. Largaespada DA. A bad rap: Rap1 signaling and oncogenesis. Cancer Cell. 2003;4(1):3–4. [DOI] [PubMed] [Google Scholar]
- 39. Wang Z, Ahmad A, Li Y, et al. Emerging roles of PDGF-D signaling pathway in tumor development and progression. Biochim Biophys Acta. 2010;1806(1):122–130. doi:10.1016/j.bbcan.2010.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mercken EM, Crosby SD, Lamming DW, et al. Calorie restriction in humans inhibits the PI3K/AKT pathway and induces a younger transcription profile. Aging Cell. 2013;12(4):645–651. doi:10.1111/Acel.12088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Suh Y, Atzmon G, Cho MO, et al. Functionally significant insulin-like growth factor I receptor mutations in centenarians. Proc Natl Acad Sci USA. 2008;105(9):3438–3442. doi:10.1073/pnas.0705467105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kwon ES, Narasimhan SD, Yen K, Tissenbaum HA. A new DAF-16 isoform regulates longevity. Nature. 2010;466(7305):498–502. doi:10.1038/Nature09184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tazearslan C, Cho M, Suh Y. Discovery of functional gene variants associated with human longevity: Opportunities and challenges. J Gerontol A Biol Sci Med Sci. 2012;67(4):376–383. doi:10.1093/gerona/glr200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Willcox BJ, Donlon TA, He Q, et al. FOXO3A genotype is strongly associated with human longevity. Proc Natl Acad Sci USA. 2008;105:13987–13992. doi:10.1073/pnas.0801030105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Flachsbart F, Caliebe A, Kleindorp R, et al. Association of FOXO3A variation with human longevity confirmed in German centenarians. Proc Natl Acad Sci USA. 2009;106(8):2700–2705. doi:10.1073/pnas.0809594106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Broer L, Buchman AS, Deelen J, et al. GWAS of longevity in CHARGE consortium confirms APOE and FOXO3 candidacy. J Gerontol A Biol Sci Med Sci. 2015;70(1):110–118. doi:10.1093/gerona/glu166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yashin AI, Wu D, Arbeev KG, Ukraintseva SV. Joint influence of small-effect genetic variants on human longevity. Aging (Albany NY). 2010;2:612–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kirkwood TB, Cordell HJ, Finch CE. Speed-bumps ahead for the genetics of later-life diseases. Trends Genet. 2011;27(10):387–388. doi:10.1016/j.tig.2011.07.001 [DOI] [PubMed] [Google Scholar]
- 49. Dudridge F. Power and predictive accuracy of polygenic risk scores. PLoS Genet. 2013;9(3):e1003348. doi:10.1371/journal.pgen.1003348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Peterson RE, Maes HH, Holmans P, et al. Genetic risk sum score comprised of common polygenic variation is associated with body mass index. Hum Genet. 2011;129:221–230. doi:10.1007/s00439-010-0917-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Leiserson MD, Eldridge JV, Ramachandran S, Raphael BJ. Network analysis of GWAS data. Curr Opin Genet Dev. 2013;23(6):602–610. doi:10.1016/j.gde.2013.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.