Abstract
We analyzed more than 600 red deer (Cervus elaphus) from large parts of its European distribution range at 13 microsatellite loci, presenting the first continent-wide study of this species using nuclear markers. Populations were clearly differentiated (overall F ST = 0.166, Jost’s D est = 0.385), and the BAPS clustering algorithm yielded mainly geographically limited and adjacent genetic units. When forced into only 3 genetic clusters our data set produced a very similar geographic pattern as previously found in mtDNA phylogeographic studies: a western group from Iberia to central and parts of Eastern Europe, an eastern group from the Balkans to Eastern Europe, and a third group including the threatened relict populations from Sardinia and Mesola in Italy. This result was also confirmed by a multivariate approach to analyzing our data set, a discriminant analysis of principal components. Calculations of genetic diversity and effective population sizes (linkage disequilibrium approach) yielded the lowest results for Italian (Sardinia, Mesola; N e between 2 and 8) and Scandinavian red deer, in line with known bottlenecks in these populations. Our study is the first to present comparative nuclear genetic data in red deer across Europe and may serve as a baseline for future analyses of genetic diversity and structuring in this widespread ungulate.
Keywords: Cervus elaphus, effective population size, Europe, microsatellites, phylogeography, Red deer
Introduction
The present genetic structure of large mammals in Europe is mainly due to: 1) signatures of glacial-interglacial cycles with (in temperate species) southern refugia during glacials and subsequent recolonization of northern regions, particularly after the Last Glacial Maximum (LGM) (Hewitt 2000); 2) anthropogenic influences over the past centuries, e. g. selective hunting, habitat fragmentation and translocations (Hartl et al. 2003; Frantz et al. 2006). Genetic consequences of human interference are thus grafted onto natural phylogeographic patterns, often blurring the intraspecific structuring of pre-human eras.
The red deer (Cervus elaphus) is arguably the most important European game species and, consequently, has been impacted by humans for centuries or even millennia (Hartl et al. 2003). A multitude of studies on its genetic structure in Europe have been carried out, both on a local, regional and continental scale (Zachos and Hartl 2011; Carden et al. 2012; Niedziałkowska et al. 2012; Fernández-García et al. 2014; Karaiskou et al. 2014, Krojerová-Prokesová et al. 2015). Studies on mitochondrial DNA (for a review see Zachos and Hartl 2011) have uncovered 3 main phylogeographic lineages in Europe: a western haplogroup (designated A) distributed from Iberia through France and northern Central Europe to the British Isles, Scandinavia and parts of Eastern Europe; an eastern haplogroup (designated C) in the Balkans and parts of Eastern and Central Europe; and an isolated lineage B restricted to the Tyrrhenian red deer (C. e. corsicanus) on Sardinia and Corsica and the North African Barbary red deer (C. e. barbarus). The suture zone between the lineages A and C appears to run from Austria through Poland and Belarus to the Baltic States (Niedziałkowska et al. 2011; Fickel et al. 2012; Krojerová-Prokesová et al. 2015).
While the overall continental pattern does not seem to have been blurred by human interference (there are only few geographic outliers with respect to the 3 lineages, Nussey et al. 2006), the areas where geographic lineages meet may or may not show the natural distribution patterns. In red deer, this is a general issue, even more so at regional and local scales, where it is often not clear if and to what extent populations are “pure”, i.e. free from artificial introductions. Translocations of farmed red deer have occurred countless times, and what is known about them is still only the tip of the iceberg (Linnell and Zachos 2011; Apollonio et al. 2014, especially Table 3.1, and references therein). Even if, as the mtDNA phylogeographic studies suggest, translocations have mainly been carried out within the main lineages rather than between them, these translocations have often covered large geographic distances (Linnell and Zachos 2011). Of course, between-lineage translocations of stags would not leave a signature in mitochondrial patterns; however, there does not seem to be a male bias in translocations, and available documentation confirms that most of the times females were translocated as well (Niethammer 1963). Local or regional red deer stocks have also been intensively studied from a population genetic point of view, often taking into account human impacts (Kuehn et al. 2003, 2004; Zachos et al. 2007; Frantz et al. 2008; Haanes et al. 2010a, 2010b; Niedziałkowska et al. 2012; Fernández-García et al. 2014).
In this study, we present the first microsatellite data set covering most of the European range of the red deer to infer its nuclear genetic structuring. Contrary to the roe deer and wild boar, the other 2 widespread European ungulate species (Randi et al. 2004; Scandura et al. 2008), no such analysis exists for red deer to date. The present study aims at closing this gap, for the first time allowing for nuclear genetic comparisons across the whole continent. In particular, our study aims are:
(1) to uncover the large-scale nuclear genetic structure of the red deer in Europe and compare it to the known mtDNA phylogeography that is believed to bear signatures of the Quaternary climatic cycles, especially those of refugia during the LGM and subsequent recolonization events;
(2) to use our microsatellite data set to calculate genetic diversity and effective population sizes (N e) at a continent-wide comparative level to get further insights into the distribution of genetic variability in this game species and to produce important and directly comparable diversity parameters for the endangered Tyrrhenian (Corsica, Sardinia) and Mesola (NE Italy) subspecies.
Material and Methods
Sample Collection and Laboratory Work
The present study was based on 638 red deer tissue samples from 27 locations throughout the continent (see Table 1; Figure 1). We also included 30 samples from a French deer farm (Boisgervilly) in an attempt to understand the origin of these individuals and to test the potential to identify farmed individuals in the European data set. The samples included both new material and individuals already analyzed in previous studies (Kuehn et al. 2003; 2004, Feulner et al. 2004; Hmwe et al. 2006a, 2006b; Zachos et al. 2007, Skog et al. 2009; Dellicour et al. 2011; Niedziałkowska et al. 2012; Pérez-González et al. 2012). DNA was extracted from new samples using a chloroform-based extraction method (Doyle and Doyle 1990). All samples (old and new) were genotyped at 13 microsatellite loci (BM1818, Cer14, CSPS115, CSSM14, CSSM16, CSSM19, CSSM22, CSSM66, ETH225, Haut14, ILSTS06, INRA35, and MM12; for references see Kuehn et al. 2003) in 3 multiplex polymerase chain reactions (PCR) using the Qiagen Multiplex kit (Qiagen, Hilden, Germany). Detailed information on the PCR composition and reaction times can be found in Dellicour et al. (2011). Reactions were performed using a Verity thermocycler (Applied Biosystems, Warrington, UK). PCR products were separated using an ABI 3100 automated DNA sequencer (Applied Biosystems), and the data were analyzed using GeneMapper version 3.7 (Applied Biosystems). All individuals were genotyped at 11 loci or more, with 629 of the 668 sampled having a complete 13-locus profile.
Table 1.
Geographic distribution of the European samples analyzed in this study and summary of genetic diversity measures. A R: allelic richness; H o: observed heterozygosity; He u: unbiased expected heterozygosity
| Country | Region within country | n | Microsatellite diversity | ||
|---|---|---|---|---|---|
| A R | Ho | He u | |||
| Belgium | Wallonia NE | 20 | 5.31 | 0.68 | 0.69 |
| Belgium | Wallonia central | 20 | 5.08 | 0.67 | 0.67 |
| Belgium | Wallonia West | 20 | 5.39 | 0.69 | 0.69 |
| Croatia | E Croatia | 53 | 4.82 | 0.63 | 0.68 |
| Croatia/Slovenia | NW Croatia/S Slovenia | 49 | 5.36 | 0.70 | 0.68 |
| France | Central France (Châteauroux) | 23 | 4.51 | 0.60 | 0.58 |
| France | E France (Meurthe) | 27 | 4.70 | 0.56 | 0.67 |
| France | NW France (Hardouinais, Brittany) | 22 | 4.20 | 0.61 | 0.65 |
| Germany | E Germany (Saxony) | 15 | 6.09 | 0.60 | 0.69 |
| Germany | N Germany (Schleswig-Holstein) | 19 | 5.53 | 0.53 | 0.60 |
| Germany | NE Bavaria (Fichtelberg/Goldkronach) | 31 | 5.27 | 0.64 | 0.68 |
| Germany | NE Germany (Mecklenburg) | 10 | 5.15 | 0.56 | 0.61 |
| Germany | SE Germany (Berchtesgaden) | 29 | 5.65 | 0.70 | 0.73 |
| Italy | N Italy (Southern Tyrol/Vinschgau) | 26 | 5.71 | 0.61 | 0.71 |
| Italy | NE Italy (Mesola) | 22 | 2.76 | 0.43 | 0.45 |
| Italy | Sardinia | 16 | 2.69 | 0.39 | 0.49 |
| Liechtenstein | 29 | 5.85 | 0.68 | 0.72 | |
| Norway | W Norway (Sogn og Fjordane) | 31 | 2.85 | 0.65 | 0.65 |
| Poland | E Poland (Białowieża) | 21 | 6.32 | 0.66 | 0.66 |
| Poland | NE Poland (Warmia-Masuria Province) | 23 | 5.84 | 0.69 | 0.70 |
| Poland | SE Poland (N Carpathians) | 25 | 6.10 | 0.29 | 0.33 |
| Romania | SE Romania (Carpathians) | 17 | 5.36 | 0.42 | 0.46 |
| Scotland | 25 | 5.71 | 0.67 | 0.69 | |
| Serbia | NE Serbia (Bachka) | 19 | 4.55 | 0.66 | 0.69 |
| Spain | SE Spain (Andalucía) | 15 | 4.50 | 0.63 | 0.62 |
| Spain | W Spain (Extremadura) | 15 | 5.19 | 0.63 | 0.68 |
| Sweden | S Sweden (Skåne) | 16 | 2.52 | 0.64 | 0.67 |
| Deer Farm | France, Brittany (Boisgervilly) | 30 | 5.59 | 0.63 | 0.69 |
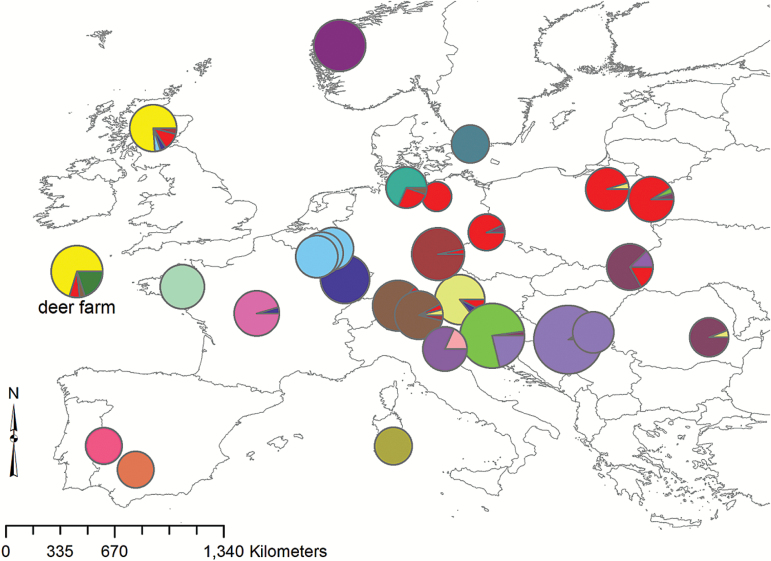
Figure 1.
Location of the genetic populations inferred using the individual-based BAPS algorithm. The size of the pie charts indicates the number of samples collected from a locality, while the pattern of the pie chart indicates the identity of the genetic clusters. The 4 deer that had been sampled in Croatia/Slovenia, Norway, SE Germany, and SE Poland and that formed single-individual partitions were omitted from the plot. For the pattern based on only 3 genetic clusters, see Supplementary Figure S3 in the Appendix.
Data Analysis
We tested for the significance of heterozygote deficiency or excess (i.e. deviation from Hardy–Weinberg equilibrium) in the 26 European sampling locations with N ≥ 15 (Supplementary Table S1, excluding the deer farm) with the Markov-chain method in GENEPOP 3.4 (Raymond and Rousset 1995), with 10 000 dememorization steps, 500 batches and 10 000 subsequent iterations. The populations were tested for pairwise linkage disequilibrium (LD) between loci using an exact test based on a Markov-chain method as implemented in GENEPOP 3.4. The false discovery rate technique was used to eliminate false assignment of significance by chance (Verhoeven et al. 2005). Mean allelic richness per locus for each pre-defined European population was calculated with FSTAT v. 2.9.3 (Goudet 1995) to standardize measures for a population size of 10 diploid individuals. Observed (Ho) and unbiased expected (He u) heterozygosities (Nei 1978) for the same populations were estimated using GENETIX 4.05.2 (Belkhir 2004).
We used STRUCTURE v2.3.1 (Pritchard et al. 2000) to estimate the number of subpopulations (K). Ten independent runs of K = 1–10 were carried out with 106 Markov chain Monte Carlo (MCMC) iterations after a burn-in period of 105 to 106 iterations, using the model with correlated allele frequencies and assuming admixture. ALPHA, the Dirichlet parameter for the degree of admixture, was allowed to vary between populations. After deciding on the most probable number of subpopulations based on the log-likelihood values (and their convergence) associated with each K, we calculated each individual’s percentage of membership (q), averaging q over 10 runs. Bar plots of assignments were generated using DISTRUCT 1.1 (Rosenberg 2004). We also used BAPS v5.4 (Corander et al. 2004) to perform a population mixture analysis based on clustering individuals. This algorithm partitions the data into populations with non-identical allele frequencies. The program was run for K = 2 to 30 with 10 replications for each K.
A discriminant analysis of principal components (DAPC) was performed using the R-package adegenet (Jombart 2008; Jombart et al. 2010) for R v. 2.12. (R Development Core Team 2011). This method, which is not based on any assumptions regarding the population genetic model, first extracts information by applying a principal component analysis (PCA). In a second calculation step, a discriminant analysis (DA) maximizes the between-group component of the genetic variation. The result of the DAPC can be visualized by using RGB color coding; the similarity of the dot color represents the genetic similarity of the populations (Jombart 2008; Jombart et al. 2010). In the first step of this procedure, 50 principal components of PCA were retained in order to explain approximately 90% of the total variation of the data set analyzed in this study. We carried out the DAPC at the population level as we did not have coordinates for a large number of single deer specimens.
To quantify overall genetic differentiation within our data set, we calculated the overall F ST value (indicating which portion of the overall variance was due to differentiation among populations) with Arlequin 3.5 (Excoffier and Lischer 2010) and an estimator of Jost’s D (D est) with GenAlEx 6.502 (Peakall and Smouse 2012). These calculations were carried out over all populations listed in Table 1 (excluding the deer farm). GenAlEx was also used to identify private alleles and their frequencies.
We estimated effective population sizes (N e) using a bias-corrected version of the LD method by Waples and Do (2008) as implemented in the NEEstimator v2 software (Do et al. 2014). This approach is based on the rationale that in small populations with few parent individuals genetic drift will create non-random combinations of alleles of different loci, i.e. LD. In general this approach is reliable if effective population sizes are not much larger than ca. 200 and the data set is based on 10 or more loci and population sample sizes of 25 or more. These conditions are not met for all our populations, so results should be viewed with due caution in these cases. Since rare alleles (which occur frequently in highly polymorphic markers like microsatellites) may have a disproportionately high impact on the linkage values, the software offers different critical threshold values (we chose the default values of 0.05, 0.02, and 0.01) below which alleles are not considered. We were particularly interested in the values for the endangered subspecies from Sardinia and Mesola which have undergone serious bottlenecks and for which no values of N e derived from genetic data have ever been published. The present data set, the largest nuclear genetic on European red deer so far, offers a good opportunity to estimate effective population sizes of these deer and put them into a comparative perspective. We calculated N e values for pre-defined populations, not for the clusters retrieved by BAPS because 1) differences between the 2 were often small and 2) BAPS uses marker independence as one of the clustering parameters, so LD values might be affected by this clustering approach. Additionally, we only chose those populations which had a sample size of n ≥ 15 and which were not obviously part of a much larger continuous population (which is why we did not include the red deer from the Carpathians).
RESULTS
After correcting for multiple tests, we observed 15 instances of a locus deviating from HWE in one of the 26 pre-defined populations with n ≥ 15 (Supplementary Table S1). Four loci (BM1818, CSSM19, CSSM66, ETH225) significantly deviated from HWE in more than one population, 16 populations showed no deviations from HWE at all. We concluded that no locus systematically deviated from HWE, but that the genetic characteristics of some populations (e.g. Wahlund effect, immigrants, non-random sampling) led to the majority of the significant deviations from HWE. All loci were therefore retained in subsequent analyses. No pairs of loci were characterized by systematic LD. Diversity values (allelic richness, observed and expected heterozygosities) are given in Table 1. Across all 3 diversity parameters, Sardinia and Mesola showed the lowest genetic diversity (as expected), but other populations showed similarly low values for allelic richness (Norway, Sweden) or heterozygosity (Polish and Romanian Carpathians). As expected given our comprehensive geographical sampling we found private alleles in several populations. Most populations only had a single private allele, but Berchtesgaden in Germany had 5 (all at low frequencies of less than 3.5%). Sardinia, arguably the evolutionarily most divergent population in our data set, only had a single private allele which, however, had a frequency of 40.6%!
Genetic Structuring Across Europe
The results of the STRUCTURE analysis showed that the independent runs did not converge on an optimal solution. Log-likelihood values gradually increased, without reaching a higher value of K where they converge reasonably well, and started to decline again after K = 12 (Supplementary Figure S1). The best convergence of log-likelihoods was obtained for K = 2, K = 4, and K = 7. However, even at these 3 values of K, assignments of individuals differed fairly widely between runs of the same K. For example, at K = 2, we obtained 6 different clustering solutions (Supplementary Figure S2).
The individual-based modal population mixture analysis in BAPS inferred the presence of 26 genetic populations. The majority of the sampling locations formed distinct groups (Figure 1). The samples from eastern Poland formed a genetic cluster with the samples from northern and eastern Germany. Seven clusters consisted of 6 individuals or less. Four deer, which had been sampled in Croatia/Slovenia, Norway, SE Germany, and SE Poland, respectively, formed single-individual partitions (not shown in Figure 1). The SE Polish and NE Italian (Mesola) populations, as well as the deer farm, each contained a few individuals that formed distinct clusters (Figure 1). The remaining deer farm individuals were grouped with the Scottish and Eastern European clusters and it was not possible to unequivocally identify farmed individuals in the rest of the data set.
When forced to assign all European individuals to only 3 clusters (K = 3), BAPS produced a geographical pattern very much like that known from the 3 mtDNA lineages (Supplementary Figure S3): there is a clear separation of western from eastern red deer with an overlap of these 2 groups in eastern Central Europe and Poland. Sardinia, together with Mesola (NE Italy) and Norway, constitutes the third cluster (at K = 4, Norway is separated from Sardinia/Mesola). We checked for the occurrence of otherwise rare alleles in these 3 populations as a possible explanation for their clustering. However, while we did find rare alleles at several loci shared by Mesola and Sardinia, we did not find any that were shared also by Norway.
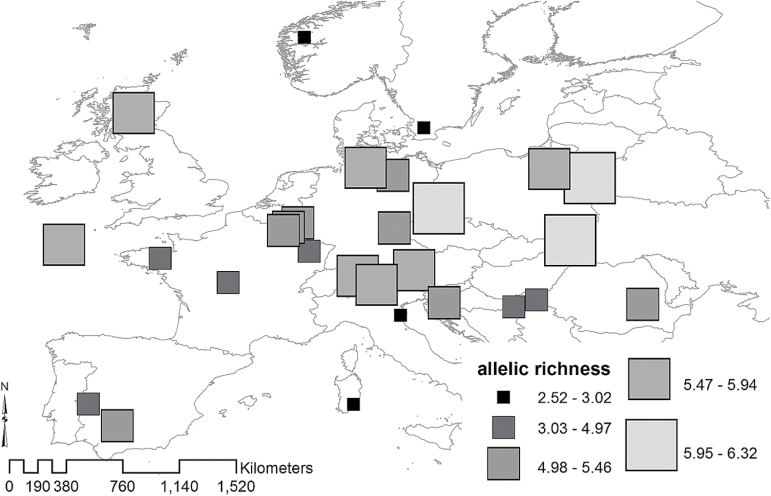
The DAPC yielded results in accordance with those from BAPS (Figure 2). Sardinia, Mesola, Norway, and Sweden were most divergent genetically, masking differentiation among the remaining populations. But again, when removing these 4 outliers from the analysis, we found a west–east dichotomy of red deer populations (Figure 2, right map). The Belgian red deer were somewhat genetically differentiated from its surrounding populations. The allelic richness values characterizing the populations from Sardinia, Mesola, Norway, and Sweden were the lowest in the data set (Figure 3; Table 1). Furthermore, populations in France, eastern Croatia, and Iberia had low levels of allelic richness, while deer in central Europe and Poland had the highest. The overall F ST value was 0.166 (P < 0.00001), indicating that 16.6% of all genetic variance was due to differences among populations as opposed to variability within populations (83.4%). Jost’s D est was 0.385 (significant at P = 0.001).
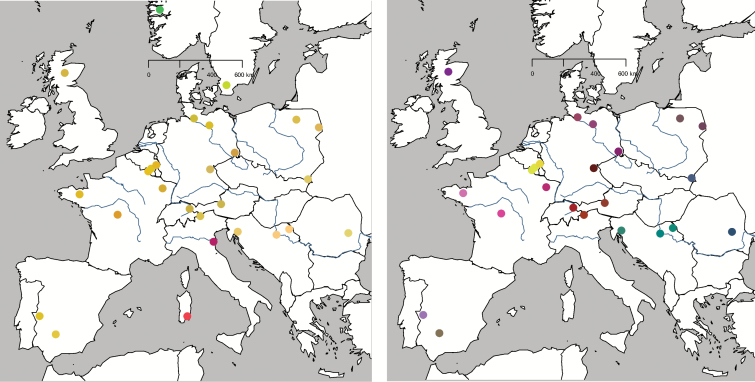
Figure 2.
DAPC of European red deer populations. Similar RGB colour codes signify genetic similarity. Sardinia, Mesola, Norway, and Sweden are genetically very different from the rest of Europe, effectively veiling differentiation among the latter (left). When running the analysis without these 4 outliers (right), the European pattern largely shows a dichotomy between a western group (red-purple colors) that ranges from Iberia through western Europe and the British Isles to eastern central Europe, and an eastern group (green-blue colors) in the Balkans and southern central Europe. In Central and Eastern Europe, these groups admix (brownish colors). This is in accordance with both mtDNA phylogeography and the BAPS results for K = 3. The Belgian red deer are the only outliers in this pattern (yellow dots).
Figure 3.
Microsatellite-based allelic richness measures for 27 pre-defined European red deer populations and the deer farm. The estimate of allelic richness is based on a sample of 10 diploid individuals and 13 microsatellite markers. Sardinia, Mesola, Norway, and Sweden show the lowest values (little black squares).
Effective Population Sizes of European Red Deer
N e values as calculated with the LD method are given in Table 2. Values above 200 and for samples much smaller than 25 should be viewed with due caution (see above). In line with the diversity parameters (Table 1) values for Sardinia and Mesola were the lowest (between 2.0 and 8.2), and no other populations show similarly low effective population sizes, although some do show values that are below 50 (e.g. Sweden, Norway and Schleswig-Holstein in northern Germany), which is often viewed as a threshold below which inbreeding depression is likely to occur. The comparison of different threshold values also shows that rare alleles sometimes have a large impact on the result for a given population but do not change the overall picture.
Table 2.
Effective population sizes (N e) as calculated with N e Estimator based on the LD approach. For each population, N e values are given for 3 different thresholds for the lowest allele frequency used. The values in parentheses are the 95% confidence intervals based on jackknifing on loci. n: sample size. “infinite” values of N e refer to cases where there is no evidence of variation of the genetic characteristic due to finite numbers of parental individuals, i.e. all can be explained by sampling error (Do et al. 2014). Only those populations are included for which evidence was present that they were not just artificially designated sample sites and for which n was ≥ 15. The 2 Spanish populations, after mostly yielding infinite values separately, were pooled.
| Population | n | Effective population size (N e) | ||
|---|---|---|---|---|
| Frequency threshold: 0.05 | 0.02 | 0.01 | ||
| Sardinia | 16 | 4.3 (2.3–10.4) | 8.2 (3.3–17.1) | 8.2 (3.3–17.1) |
| Mesola | 22 | 2.0 (1.4–3.0) | 2.6 (1.8–5.3) | 2.6 (1.8–5.3) |
| Sweden | 16 | Infinite (9.8–infinite) | 20.4 (7.8–549.8) | 20.4 (7.8–549.8) |
| Norway | 31 | 40.6 (16.9–532.1) | 30.2 (15.5–87.8) | 10.3 (3.9–21.6) |
| Schleswig-Holstein | 19 | 19.2 (13.7–29.0) | 26.2 (18.6–40.5) | 26.2 (18.6–40.5) |
| Saxony | 15 | Infinite (122.2–infinite) | 283.5 (69.8–infinite) | 283.5 (69.8–infinite) |
| Serbia | 19 | 303.7 (41.1–infinite) | 131.3 (37.1–infinite) | 131.3 (37.1–infinite) |
| E Croatia | 53 | 480.6 (128.7–infinite) | 1384.1 (206.6–infinite) | 1127.7 (217.6–infinite) |
| NW Croatia/S Slovenia | 49 | 84.5 (52.5–177.2) | 155.4 (93.1–394.9) | 139.5 (80.6–397.4) |
| Berchtesgaden | 29 | 46.2 (32.1–75.4) | 51.7 (35.2–89.1) | 41.4 (28.0–71.2) |
| Fichtelgebirge | 31 | 28.8 (20.2–44.7) | 38.3 (27.7–57.7) | 41.1 (30.4–59.9) |
| Liechtenstein | 29 | 139.6 (60.3–infinite) | 166.0 (75.3–infinite) | 156.3 (74.0–infinite) |
| Vinschgau (Italy) | 26 | 101.0 (45.6–infinite) | 119.5 (54.9–infinite) | 149.5 (62.3–infinite) |
| E France (Meurthe) | 27 | 42.0 (22.1–144.8) | 57.6 (30.3–229.9) | 79.2 (40.8–424.2) |
| NW France (Hardouinais) | 22 | 77.7 (26.6–infinite) | 176.4 (45.1–infinite) | 176.4 (45.1–infinite) |
| C France (Châteauroux) | 23 | 62.1 (27.0–infinite) | 85.1 (40.6–1797.9) | 85.1 (40.6–1797.9) |
| Spain (pooled) | 30 | 38.0 (24.9–68.8) | 42.1 (28.8–70.4) | 68.0 (45.3–124.8) |
| NE Wallonia | 20 | Infinite (118–infinite) | Infinite (121.4–infinite) | Infinite (121.4–infinite) |
| C Wallonia | 20 | 82.8 (35.9–infinite) | 114.3 (45.6–infinite) | 114.3 (45.6–infinite) |
| W Wallonia | 20 | 126.2 (44.2–infinite) | 167.5 (57.4–infinite) | 167.5 (57.4-infinite) |
Discussion
Our analyses of more than 600 individual multi-locus genotypes of European red deer have uncovered substantial structuring across the continent and, as expected given the higher mutation rates in microsatellites, the overall nuclear genetic structure was more complex than that found in phylogeographic studies based on mtDNA. Microsatellites are generally more appropriate for the detection of small-scale and/or more recent structuring but the European data set once more confirmed the genetic uniqueness of both the Sardinian and the Mesola red deer and also yielded similar patterns to those uncovered by mtDNA phylogeography.
European Genetic Structure and Phylogeography
Due to convergence problems, the STRUCTURE results were inconclusive. Re-running the analysis using the substantially longer burn-in of 106 did not solve the issue (results not shown). To the best of our knowledge, STRUCURE does not allow a formal assessment of the convergence of the MCMC chains, via the statistic by Gelman and Rubin (1992), for example. While similar problems have been reported (e.g. Frantz et al. 2014), the issue is particularly striking here. We therefore limited our inferences to the results obtained by the BAPS algorithm. Contrary to STRUCTURE, the overall picture provided by BAPS was consistent in that Sardinian, Mesola, Norwegian and, to a lesser extent, Swedish red deer were the most divergent of the European populations. Also, the BAPS and the DAPC analyses retrieved a clear signal of genetic divergence between western and eastern Europe.
The BAPS analysis yielded 26 distinct genetic clusters across Europe. Even if this is an overestimate (BAPS can overestimate the number of genetic clusters because it has a tendency to identify populations consisting of only a few individuals, as observed here as well; Latch et al. 2006), it clearly shows the differentiation at a comparatively small geographical scale in European red deer. In line with this, both F ST and Jost’s D est showed significant differentiation at a rather high level. Our F ST of 0.166 is almost exactly the same as that found for another cervid species with a similar distribution range in Europe, the roe deer (Capreolus capreolus; F ST across Europe based on 704 specimens and 11 microsatellite loci was found to be 0.16 by Randi et al. 2004). As expected, overall F ST was much higher for mtDNA control region sequences (because within-population diversity is lower; F ST = 0.84, Skog et al. 2009).
Most BAPS clusters comprise local populations and/or mostly geographically adjacent sampling sites. Given the mutation rates of microsatellites, this is an expected outcome and in line with many microsatellite studies on red deer at local or regional scales (e.g. Feulner et al. 2004; Nielsen et al. 2008; Haanes et al. 2011; Niedziałkowska et al. 2012; Höglund et al. 2013; Karaiskou et al. 2014; Krojerová-Prokesová et al. 2015). However, if low K values of the BAPS analysis are considered, the geographical distribution of the clusters are very interesting. Indeed, if all European red deer are clustered into only 3 groups, the geographical pattern bears a striking resemblance to the phylogeographic pattern derived from mtDNA: 2 main groups (west and east, respectively) that show an overlap in Central Europe and Poland, and a minor group containing the red deer from Sardinia. These groups correspond to the mtDNA lineages A (west), C (east), and B (Sardinia and North Africa). Instead of the North-African Barbary deer (of which unfortunately no samples were available for the present study), the microsatellite analysis groups the Sardinian red deer with Mesola whose mtDNA affinities are somewhat intermediate between the western and eastern clade (Skog et al. 2009; Niedziałkowska et al. 2011), with the most recent study favoring closer relationships to the eastern group (Lorenzini and Garofalo 2015). Norway becomes separated from this group at the next higher level of K = 4. The DAPC confirmed these results in that after the removal of the outlier populations there was a clear differentiation between Western, Central and Central-Eastern Europe on the one hand and South-Eastern and southern Central Europe on the other. In red deer, concordance between mtDNA phylogroup distribution and microsatellite structuring has been found before, albeit at a smaller geographical scale, in the Czech Republic (Krojerová-Prokesová et al. 2015) and in Greece (Karaiskou et al. 2014).
The most convincing explanation for the biogeographic pattern observed in the present study is that the microsatellite structure of red deer across Europe still carries a signature of the postglacial recolonization process from 2 main glacial refugia (Iberia/southern France in the west, the Balkans and possibly the Carpathian region in the east, Sommer et al. 2008). In line with this, values of allelic richness are highest where the 2 main BAPS clusters and DAPC groups (west and east) meet—in Central Europe and Poland, which is also where the western and eastern mtDNA lineages co-occur (Niedziałkowska et al. 2011; Fickel et al. 2012; Krojerová-Prokesová et al. 2015). To what extent this zone of overlap is natural, however, cannot be definitively answered due to the high number of translocations that are known to have occurred. It would be interesting to see whether phylogeographic data from one or more nuclear markers with lower mutation rates than microsatellites also confirm the large-scale pattern of 3 groups found in mtDNA and microsatellites.
Within the West-Palaearctic red deer, the Tyrrhenian red deer from Corsica and Sardinia (C. e. corsicanus) and the North-African Barbary red deer (C. e. barbarus) comprise a distinct mtDNA lineage (B), and their phylogeographic history is not entirely clear, e. g. it is still being debated whether the Tyrrhenian deer are derived from introduced Barbary deer or vice versa (see Zachos and Hartl 2011 and references therein). If the Tyrrhenian deer are descendants of Italian mainland deer, then they should be closely related to the red deer from Mesola (recently described as C. e. italicus, Zachos et al. 2014) which are the last surviving native Italian red deer. While this is not supported by mtDNA studies, close affinities between the 2 have been found based on microsatellites (Hajji et al. 2008). It is interesting to note that the present data set of 13 microsatellites—none of which is identical to the 8 loci used by Hajji et al. (2008)—yielded the same result of a close relationship between C. e. corsicanus and C. e. italicus. It seems therefore unlikely that these results are simply an artefact due to drift effects in 2 recently severely bottlenecked populations—for this to be true the bottlenecks would have to have resulted in similar and unique allele frequencies in 2 completely non-overlapping sets of altogether 21 loci. Rather, it seems more likely that the result is indicative of a true signal of phylogeographic relationships between mainland Italian and Sardinian/Corsican red deer, a question that ultimately only ancient DNA studies will be able to answer.
Genetic Diversity and Effective Population Sizes
Genetic diversity values were in the range known for microsatellite loci in red deer (see Table 1 in Zachos and Hartl 2011 for a compilation of values from all over Europe). The lowest values when considering both allelic diversity/richness and heterozygosities were expectedly found in the severely bottlenecked populations from Sardinia and Mesola in Italy, and known bottlenecks can also account for the low diversity (at least in terms of allelic richness) found in Scandinavian red deer from Sweden and Norway which is in accordance with findings from other studies (Haanes et al. 2010a, Haanes et al. 2010b, Haanes et al. 2011).
We present here also the first estimation of effective population sizes in red deer across a large area of their distribution. Overall, N e values were in the range of, although with lower maximum values than, those previously calculated for German and Spanish red deer based on genetic and demographic data (Martinez et al. 2002; Kuehn et al. 2003). The calculations have also confirmed the genetic depletion of the red deer from Sardinia and Mesola as a consequence of past bottlenecks and near-extinction. Values between 2 and 8 are the lowest in Europe and even considerably lower than the N e = 20 calculated with the same approach (LDNe) for the endangered Kashmir red deer or hangul (C. e. hanglu) whose total census population size is estimated at just above 200 (Mukesh et al. 2015). While both the Sardinian and Mesola red deer have increased in numbers recently and are not threatened with immediate extinction anymore, the long-term consequences of low genetic diversity and inbreeding remain unclear. While overall the LD approach is viewed as a reliable method, there are many unknowns in any calculation of effective population size (Luikart et al. 2010). The values therefore might best be viewed in a comparative context rather than as absolute values for each of the populations separately.
Conclusion
Although a common and locally abundant game animal today, the red deer faced extirpation in many parts of its range during past centuries. Documentation on recolonization—whether natural or human-mediated—is usually scarce, and what is known on translocations from the literature (e.g. Niethammer 1963, Apollonio et al. 2014) is almost certainly only the tip of the iceberg. In fact, it is believed that the present gene pool of many if not most free-living populations of red deer in Europe contains at least some genetic material that goes back to introductions (Hartl et al. 2003). Evidence for purely autochthonous populations is rare (and usually not conclusive), with some possible examples being red deer in Mesola (Zachos et al. 2014), Skåne in southern Sweden (Höglund et al. 2013) , some areas in Spain (Carranza et al. 2016) and the Scottish Highlands (Pérez-Espona et al. 2009). Although genetic analyses are a powerful means to elucidate the status of populations with respect to their natural or anthropogenic origin (see Kuehn et al. 2004; Frantz et al. 2006), such analyses have not been carried out for most of the distribution range. Many of the populations analyzed in the present study will therefore not be completely natural units (it is known, e.g. that the Châteauroux red deer have partly been introduced from the Domaine National de Chambord). However, a “purist” approach allowing only completely native populations in a species as deeply impacted anthropogenically as the red deer in Europe is neither feasible nor would it, in our view, be desirable, because human impacts are one of the most important factors in shaping genetic structure in red deer and as such are also a relevant aspect of their present-day biology. Our analysis has presented evidence that not only with respect to mtDNA but also for microsatellite DNA presumably natural patterns are still visible in red deer across Europe, and our data set, being the most comprehensive of its kind so far, may serve as a continent-wide comparison for geographically more restricted studies in the future.
Supplementary Material
Supplementary material can be found at http://www.jhered.oxfordjournals.org/.
Data Availability
Data (microsatellite genotypes) deposited at Dryad: http://dx.doi.org/doi:10.5061/dryad.1v6p1
Supplementary Material
References
- Apollonio M, Scandura M, Sprem N. 2014. Reintroductions as a management tool for European Ungulates. In: Putman R, Apollonio M, editors. Behaviour and management of European Ungulates. Dunbeath: Whittles Publishing; p. 46–77. [Google Scholar]
- Belkhir K, Borsa P, Chikhi L, Raufaste N, Bonhomme F. 2004. Genetix 4.05.2, logiciel sous Windows™ pour la genetique des populations. Laboratoire Génome, Populations, Interactions, CNRS UMR 5000, Université de Montpellier II. [Google Scholar]
- Carden RF, McDevitt AD, Zachos FE, Woodman PC, O’Toole P, Rose H, Monaghan NT, Campana MG, Bradley DG, Edwards CJ. 2012. Phylogeographic, ancient DNA, fossil and morphometric analyses reveal ancient and modern introductions of a large mammal: the complex case of red deer (Cervus elaphus) in Ireland. Quat Sci Rev. 42:74–84. [Google Scholar]
- Carranza J, Salinas M, de Andrés D, Pérez-González J. 2016. Iberian red deer: paraphyletic nature at mtDNA but nuclear markers support its genetic identity. Ecol Evol 6 (4):905–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corander J, Waldmann P, Marttinen P, Sillanpää MJ. 2004. BAPS 2: enhanced possibilities for the analysis of genetic population structure. Bioinformatics. 20:2363–2369. [DOI] [PubMed] [Google Scholar]
- Dellicour S, Frantz AC, Colyn M, Bertouille S, Chaumont F, Flamand MC. 2011. Population structure and genetic diversity of red deer (Cervus elaphus) in forest fragments in north-western France. Conserv Genet. 12:1287–1297. [Google Scholar]
- Do C, Waples RS, Peel D, Macbeth GM, Tillett BJ, Ovenden JR. 2014. NeEstimator v2: re-implementation of software for the estimation of contemporary effective population size (Ne) from genetic data. Mol Ecol Resour. 14:209–214. [DOI] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL. (1990) Isolation of plant DNA from fresh tissue. Focus. 12:13–15. [Google Scholar]
- Excoffier L, Lischer HEL. 2010. Arlequin suite version 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Res. 10:564–567. [DOI] [PubMed] [Google Scholar]
- Fernández-García JL, Carranza J, Martínez JG, Randi E. 2014. Mitochondrial D-loop phylogeny signals two native Iberian red deer (Cervus elaphus) lineages genetically different to Western and Eastern European red deer and infers human-mediated translocations. Biodivers Conserv. 23:537–554. [Google Scholar]
- Feulner PG, Bielfeldt W, Zachos FE, Bradvarovic J, Eckert I, Hartl GB. 2004. Mitochondrial DNA and microsatellite analyses of the genetic status of the presumed subspecies Cervus elaphus montanus (Carpathian red deer). Heredity (Edinb). 93:299–306. [DOI] [PubMed] [Google Scholar]
- Fickel J, Bubliy OA, Stache A, Noventa T, Jirsa A, Heurich M. 2012. Crossing the border? Structure of the red deer (Cervus elaphus) population from the Bavarian-Bohemian forest ecosystem. Mamm Biol. 77: 211–220. [Google Scholar]
- Frantz AC, Hamman J-L, Klein F. 2008. Fine-scale genetic structure of red deer (Cervus elaphus) in a French temperate forest. Eur J Wildl Res. 54:44–52. [Google Scholar]
- Frantz AC, McDevitt AD, Pope LC, Kochan J, Davison J, Clements CF, Elmeros M, Molina-Vacas G, Ruiz-Gonzalez A, Balestrieri A, et al. (2014) Revisiting the phylogeography and demography of European badgers (Meles meles) based on broad sampling, multiple markers and simulations. Heredity 113:443–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frantz AC, Pourtois JT, Heuertz M, Schley L, Flamand MC, Krier A, Bertouille S, Chaumont F, Burke T. 2006. Genetic structure and assignment tests demonstrate illegal translocation of red deer (Cervus elaphus) into a continuous population. Mol Ecol. 15:3191–3203. [DOI] [PubMed] [Google Scholar]
- Gelman A, Rubin DB. 1992. Inference from iterative simulation using multiple sequences. Stat Sci. 7:457–511. [Google Scholar]
- Goudet J. 1995. FSTAT: a computer program to calculate F-statistics. J Hered. 86:485–486. [Google Scholar]
- Haanes H, Røed KH, Flagstad Ø, Rosef O. 2010. a. Genetic structure in an expanding cervid population after population reduction. Conserv Genet. 11:11–20. [Google Scholar]
- Haanes H, Røed KH, Mysterud A, Langvatn R, Rosef O. 2010. b. Consequences for genetic diversity and population performance of introducing continental red deer into the northern distribution range. Conserv Genet. 11:1653–1665. [Google Scholar]
- Haanes H, Røed KH, Pérez-Espona S, Rosef O. 2011. Low genetic variation support bottlenecks in Scandinavian red deer. Eur J Wildl Res. 57:1137–1150. [Google Scholar]
- Hajji GM, Charfi-Cheikrouha F, Lorenzini R, Vigne J-D, Hartl GB, Zachos FE. 2008. Phylogeography and founder effect of the endangered Corsican red deer (Cervus elaphus corsicanus). Biodivers Conserv. 17:659–673. [Google Scholar]
- Hartl GB, Zachos F, Nadlinger K. 2003. Genetic diversity in European red deer (Cervus elaphus L.): anthropogenic influences on natural populations. CR Biologies. 326:S37–S42. [DOI] [PubMed] [Google Scholar]
- Hewitt G. 2000. The genetic legacy of the Quaternary ice ages. Nature. 405:907–913. [DOI] [PubMed] [Google Scholar]
- Hmwe SS, Zachos FE, Eckert I, Lorenzini R, Fico R, Hartl GB. (2006. a). Conservation genetics of the endangered red deer from Sardinia and Mesola with further remarks on the phylogeography of Cervus elaphus corsicanus . Biol J Linn Soc. 88:691–701. [Google Scholar]
- Hmwe SS, Zachos FE, Sale JB, Rose HR, Hartl GB. (2006. b). Genetic variability and differentiation in red deer (Cervus elaphus) from Scotland and England. J Zool. 270:479–487. [Google Scholar]
- Höglund J, Cortazar-Chinarro A, Jarnemo A, Thulin C-G. 2013. Genetic variation and structure in Scandinavian red deer (Cervus elaphus): influence of ancestry, past hunting, and restoration management. Biol J Linn Soc. 109:43–53. [Google Scholar]
- Jombart T. 2008. adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics. 24:1403–1405. [DOI] [PubMed] [Google Scholar]
- Jombart T, Devillard S, Balloux F. 2010. Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genet. 11:94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaiskou N, Tsakogiannis A, Gkagkavouzis K, Papika S, Latsoudis P, Kavakiotis I, Pantis J, Abatzopoulos TJ, Triantaphyllidis C, Triantafyllidis A; Operator of Parnitha National Park 2014. Greece: a Balkan subrefuge for a remnant red deer (Cervus elaphus) population. J Hered. 105:334–344. [DOI] [PubMed] [Google Scholar]
- Krojerová-Prokešová J, Barančeková M, Koubek P. 2015. Admixture of Eastern and Western European Red Deer Lineages as a Result of Postglacial Recolonization of the Czech Republic (Central Europe). J Hered. 106:375–385. [DOI] [PubMed] [Google Scholar]
- Kuehn R, Schroeder W, Pirchner F, Rottmann O. 2003. Genetic diversity, gene flow and drift in Bavarian red deer populations (Cervus elaphus). Conserv Genet. 4:157–166. [Google Scholar]
- Latch EK, Dharmarajan G, Glaubitz JC, Rhodes OE Jr. 2006. Relative performance of Bayesian clustering software for inferring population substructure and individual assignment at low levels of population differentiation. Conserv Genet. 7:295–302. [Google Scholar]
- Kuehn R, Haller H, Schroeder W, Rottmann O. 2004. Genetic roots of the red deer (Cervus elaphus) population in Eastern Switzerland. J Hered. 95:136–143. [DOI] [PubMed] [Google Scholar]
- Linnell JDC, Zachos FE. 2011. Status and distribution patterns of European ungulates: genetics, population history and conservation. In: Putman R, Apollonio M, Andersen R, editors. Ungulate Management in Europe: Problems and Practices. Cambridge: Cambridge University Press, p. 12–53. [Google Scholar]
- Lorenzini R, Garofalo L. 2015. Insights into the evolutionary history of Cervus (Cervidae, tribe Cervini) based on Bayesian analysis of mitochondrial marker sequences, with first indications for a new species. J Zool Syst Evol Res 53:340–349 [Google Scholar]
- Luikart G, Ryman N, Tallmon DA, Schwartz MK, Allendorf FW. 2010. Estimation of census and effective population sizes: the increasing usefulness of DNA-based approaches. Conserv Genet. 11:355–373. [Google Scholar]
- Martínez JG, Carranza J, Fernández-García JL, Sánchez-Prieto CB. 2002. Genetic variation of red deer populations under hunter exploitation in southwestern Spain. J Wildlife Manage. 66:1273–1282. [Google Scholar]
- Mukesh Kumar VP, Sharma LK, Shukla M, Sathyakumar S. 2015. Pragmatic perspective on conservation genetics and demographic history of the last surviving population of Kashmir red deer (Cervus elaphus hanglu) in India. PLoS One. 10:e0117069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M. 1978. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics. 89:583–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedziałkowska M, Jędrzejewska B, Honnen A-C, Otto T, Sidorovich VE, Perzanowski K, Skog A, Hartl GB, Borowik T, Bunevich AN, et al. 2011. Molecular biogeography of red deer Cervus elaphus from eastern Europe: insights from mitochondrial DNA sequences. Acta Theriol. 56: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedziałkowska M, Jędrzejewska B, Wójcik JM, Goodman SJ. 2012. Genetic structure of red deer population in Northeastern Poland in relation to the history of human interventions. J Wildlife Manage. 76:1264–1276. [Google Scholar]
- Nielsen EK, Olesen CR, Pertoldi C, Gravlund P, Barker JSF, Mucci N, Randi E, Loeschcke V. 2008. Genetic structure of the Danish red deer (Cervus elaphus). Biol J Linn Soc. 95:688–701. [Google Scholar]
- Niethammer G. 1963. Die Einbürgerung von Säugetieren und Vögeln in Europa. Hamburg, Berlin: Paul Parey. [Google Scholar]
- Nussey DH, Pemberton J, Donald A, Kruuk LE. 2006. Genetic consequences of human management in an introduced island population of red deer (Cervus elaphus). Heredity (Edinb). 97:56–65. [DOI] [PubMed] [Google Scholar]
- Peakall R, Smouse PE. 2012. GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research–an update. Bioinformatics. 28:2537–2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Espona S, Pérez-Barbería FJ, Goodall-Copestake WP, Jiggins CD, Gordon IJ, Pemberton JM. 2009. Genetic diversity and population structure of Scottish Highland red deer (Cervus elaphus) populations: a mitochondrial survey. Heredity (Edinb). 102:199–210. [DOI] [PubMed] [Google Scholar]
- Pérez-González J, Frantz AC, Torres-Porras J, Castillo L, Carranza J. (2012) Population structure, habitat features and genetic structure of managed red deer populations. Eur J Wildl Res. 58:933–943. [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. 2000. Inference of population structure using multilocus genotype data. Genetics. 155:945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randi E, Alves PC, Carranza J, Milosevic-Zlatanovic S, Sfougaris A, Mucci N. 2004. Phylogeography of roe deer (Capreolus capreolus) populations: the effects of historical genetic subdivisions and recent nonequilibrium dynamics. Mol Ecol. 13:3071–3083. [DOI] [PubMed] [Google Scholar]
- Raymond M, Rousset F. 1995. GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. J Hered. 86:248–249. [Google Scholar]
- Rosenberg NA. (2004) DISTRUCT: a program for the graphical display of population structure. Mol Ecol Notes. 4:137–138. [Google Scholar]
- Scandura M, Iacolina L, Crestanello B, Pecchioli E, Di B, enedetto MF, Russo V, Davoli R, Apollonio M, Bertorelle G. 2008. Ancient vs. recent processes as factors shaping the genetic variation of the European wild boar: are the effects of the last glaciation still detectable? Mol Ecol. 17:1745–1762. [DOI] [PubMed] [Google Scholar]
- Skog A Zachos FE Rueness EK Feulner PGD Mysterud A Langvatn R Lorenzini R Hmwe SS Lehoczky I Hartl GB Stenseth NC and Jakobsen KS. 2009. Phylogeography of red deer (Cervus elaphus) in Europe. J Biogeogr. 36:66–77. [Google Scholar]
- Sommer RS, Zachos FE, Street M, Jöris O, Skog A, Benecke N. (2008) Late Quaternary distribution dynamics and phylogeography of the red deer (Cervus elaphus) in Europe. Quat Sci Rev. 27:714–733. [Google Scholar]
- Verhoeven KJF, Simonsen KL, McIntyre LM. (2005) Implementing false discovery rate control: increasing your power. Oikos. 108:643–647. [Google Scholar]
- Waples RS, DO C. 2008. ldne: a program for estimating effective population size from data on linkage disequilibrium. Mol Ecol Resour. 8:753–756. [DOI] [PubMed] [Google Scholar]
- Zachos FE, Althoff C, Steynitz Y, Eckert I, Hartl GB. 2007. Genetic analysis of an isolated red deer (Cervus elaphus) population showing signs of inbreeding depression. Eur J Wildl Res. 53:61–67. [Google Scholar]
- Zachos FE, Hartl GB. 2011. Phylogeography, population genetics and conservation of the European red deer Cervus elaphus . Mammal Rev. 41:138–150. [Google Scholar]
- Zachos FE, Mattioli S, Ferretti F, Lorenzini R. 2014. The unique Mesola red deer of Italy: taxonomic recognition (Cervus elaphus italicus nova ssp., Cervidae) would endorse conservation. Ital J Zool. 81:136–143. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data (microsatellite genotypes) deposited at Dryad: http://dx.doi.org/doi:10.5061/dryad.1v6p1