Abstract
Characterizing the abundance and genomic distribution of repetitive DNAs provides information on genome evolution, especially regarding the origin and differentiation of sex chromosomes. Triportheus fishes offer a useful model to explore the evolution of sex chromosomes, since they represent a monophyletic group in which all species share a ZZ/ZW sex chromosome system. In this study, we analyzed the distribution of 13 classes of repetitive DNA sequences by FISH, including microsatellites, rDNAs, and transposable elements in 6 Triportheus species, in order to investigate the fate of the sex-specific chromosome among them. These findings show the dynamic differentiation process of the W chromosome concerning changes in the repetitive DNA fraction of the heterochromatin. The differential accumulation of the same class of repeats on this chromosome, in both nearby and distant species, reflects the inherent dynamism of the microsatellites, as well as the plasticity that shapes the evolutionary history of the sex chromosomes, even among closely related species sharing a same sex chromosome system.
Key words: FISH, microsatellites, retrotransposons, sex chromosomes differentiation.
Sex chromosomes and their differentiation are valuable tools for evolutionary genetics. Well-differentiated sex chromosomes, comprising male and female heterogamety, are found in mammals and birds, respectively, contrasting with the wide range of sex chromosome systems found in fishes, lizards, plants, and insects (Graves 2008). Indeed, fishes display a broad scenario, which includes undifferentiated, early or highly differentiated sex chromosomes, simple and multiple sex chromosome systems and male or female heterogamety (Thorgaard 1983; Bertollo et al. 2000; Artoni et al. 2001; Phillips and Ráb 2001; Silva et al. 2009; Cioffi et al. 2011a, 2011b).
Sex chromosomes are thought to evolve from an ancestral autosomal pair, in which structural and/or DNA changes occur in the sex specific chromosome. As a general rule, partial or complete suppression of recombination in the sex pair is required as an essential step in this process (Kobayashi et al. 2013). With the absence of recombination, heterochromatin and repetitive sequences can be amplified, playing an important role in the differentiation of the sex-specific chromosome (Charlesworth et al. 2005).
Relevant evolutionary information has been provided by repetitive DNA analyzes when focusing the origin and differentiation of sex chromosomes (Itoh et al. 2008; Cioffi et al. 2011b; Kejnovský et al. 2013). This DNA fraction is composed of satellite, minisatellite, and microsatellite-DNA organized in tandem and dispersed repeats, which include transposable elements (TEs) (Jurka et al. 2007; López-Flores and Garrido-Ramos 2012). Microsatellites are dynamic components of prokaryotic and eukaryotic genomes, mainly composed of mono-, di-, tri-, and tetranucleotide repeats (Ellegren 2004). On the other hand, TEs have approximately 20–30kb in size and are inserted randomly into the DNA (Jurka et al. 2007).
Fishes from the Triportheus genus (Characiformes, Triportheidae) offer a useful model to analyze sex chromosomes evolution, since they represent a particular group in which all analyzed species share a ZZ/ZW sex chromosome system (Artoni et al. 2001). The basal origin of this system is reinforced by the interspecific homology of the Z chromosome within the genus, as evidenced by whole chromosome painting in 5 Triportheus species using the Z chromosome of T. nematurus as a probe (Diniz et al. 2008). In all Triportheus species, the W chromosome is always smaller than the Z one, almost entirely heterochromatic (Artoni et al. 2001) and carrying 18S rDNA sites on their long arms (Marquioni et al. 2013), although differing in sizes and amounts of heterochromatin (Artoni et al. 2001; de Bello Cioffi et al. 2012; Yano et al. 2014a).
In this study, we analyzed the distribution of 13 classes of repetitive DNA sequences, including microsatellites, rDNAs, and TEs in the sex chromosomes of 6 Triportheus species, some of them now analyzed for the first time. The differential distribution of repetitive elements on the W chromosomes indicated that it has differentiated from the Z in a different fashion in each species, highlighting the ongoing dynamic differentiation process of this sex-specific chromosome.
Materials and Methods
Material Collection and Conventional Chromosome Analysis
Females and males of Triportheus albus, T. guentheri, T. nematurus, T. pantanensis, T. aff. rotundatus, and T. signatus from different Brazilian river basins were analyzed. The samples were collected with the authorization of the Brazilian environmental agency ICMBIO/SISBIO (License number 48628-2). All species were properly identified by morphological criteria and deposited in the fish museum of the Laboratory of Biology and Genetic of Fishes of the Universidade Estadual Paulista (UNESP—Botucatu—SP), with the respective deposit numbers (Table 1). The experiments followed ethical conducts, in accordance with the Ethics Committee on Animal Experimentation of the Universidade Federal de São Carlos (Process number CEUA 1853260315). Mitotic chromosomes were obtained from cells of the anterior portion of the kidney, according to Bertollo et al. (2015). In addition to standard Giemsa staining, C-banding method was also applied to detect C-positive heterochromatin (Sumner 1972).
Table 1.
Brazilian collection sites of the Triportheus species analyzed, with the sample sizes
| Species | Site | Basin | N | Deposit number |
|---|---|---|---|---|
| Triportheus albus | Araguaia river | Araguaia-Tocantins | (04 ♀; 04 ♂) | LBP18620 |
| Triportheus guentheri | Inhuma lake | São Francisco | (12 ♀; 06 ♂) | LBP18628 |
| Triportheus nematurus | Paraguai river | Paraguai | (09 ♀; 07 ♂) | LBP18624 |
| Triportheus pantanensis | Paraguai river | Paraguai | (01 ♀; 01 ♂) | LBP18623 |
| Triportheus aff. rotundatus | Paraguai river | Paraguai | (19 ♀; 21 ♂) | LBP18625 |
| Triportheus signatus | Piracicaba river | Tietê | (13 ♀; 24 ♂) | LBP18619 |
Probe Preparation
Oligonucleotide probes containing microsatellite sequences (CA)15, (GA)15, (GC)15, (TA)15, (CAA)10, (CAC)10, (CAG)10, (CAT)10, (CGG)10, and (GAA)10 were directly labeled with Cy5 during synthesis (Sigma, St. Louis, MO, USA), as described by Kubat et al. (2008). The retrotransposable elements Rex1 and Rex6 were obtained by PCR according to Volff et al. (1999). The 18S rDNA probe, corresponding to a 1400-bp segment of the 18S rRNA gene, was obtained via PCR from the nuclear DNA of T. nematurus, according to Cioffi et al. (2009). All these probes were labeled with Digoxigenin, DIG-11-dUTP (2′-Deoxyuridine-5′-Triphosphate) using DIG-Nick-translation Mix (Roche) and used for the fluorescence in situ hybridization (FISH) experiments.
Fluorescence In Situ Hybridization and Signal Detection
For FISH method, the slides with the fixed chromosomes were first incubated at 37 ºC for 1h. Subsequently, they were treated with RNAse (10mg/ml) for 1h at 37 °C in a moist chamber. Next a 5-min wash using 1× PBS was performed followed by adding of 0.005% pepsin solution to the slides for 10min at room temperature (RT). The slides were then washed again with 1× PBS. As final step of pretreatment, the material was fixed with 1% formaldehyde at RT for 10min. After further washing, the slides were dehydrated with 70, 85, and 100% ethanol, 2min each. Afterwards, chromosomal DNA was denatured in 70% formamide/2× SSC for 3min at 72 °C. The slides were dehydrated again in a cold ethanol series (70, 85, and 100%), 5min each. The hybridization mixture, containing 100ng of denatured probe, 10mg/ml dextran sulfate, 2× SSC and 50% formamide (final volume of 30 μl), were heated to 95 °C for 10min and then applied on the slides. Hybridization was performed for a period of 16–18h at 37 °C in a moist chamber. After hybridization, the slides were washed for 5min with 2× SSC and then rinsed quickly in 1× PBS. The detection was performed using anti-digoxigenin-FITC (Roche) for the Rex1 probe and anti-digoxigenin-rhodamine (Roche) for the Rex6 probe. The signal for the 18S rDNA probe was detected using anti-digoxigenin-FITC (Roche) for simultaneous use with Rex6 probe or anti-digoxigenin-rhodamine (Roche) for simultaneous use with Rex1 probe. After the detection, the slides were washed for 5min in 4× SSCT using a shaker at RT, for 3 times. Subsequently, the slides were dehydrated again in an ethanol series (70, 85, and 100%), 2min each. After the complete drying of the slides, the chromosomes were counterstained with DAPI (4′, 6-diamidino-2-phenylindole)/antifade (1.2mg/ml, Vector Laboratories).
Microscopy Analyses
At least 30 metaphase spreads were analyzed to confirm the diploid chromosome numbers, karyotype structure and FISH results. Images were captured by the CoolSNAP system software, Image Pro Plus, 4.1 (Media Cybernetics, Silver Spring, MD, USA), coupled to an Olympus BX50 microscope (Olympus Corporation, Ishikawa, Japan). The chromosomes were classified as metacentric (m), submetacentric (sm) or subtelocentric (st) according to their arm ratios (Levan et al. 1964).
Data Archiving
In fulfilment of data archiving guidelines (Baker 2013), the data in this work is presented in the micrographs in Figures 1, 2 and 3.
Figure 1.
Female karyotypes of Triportheus aff. rotundatus and T. pantanensis arranged from Giemsa-stained (above) and C-banded chromosomes (below). The ZW sex chromosomes are boxed. Note the conspicuous C positive heterochromatin accumulated on the W chromosomes. Bar = 5 µm.
Figure 2.
Distribution of microsatellites on the Z and W chromosomes of 6 Triportheus species. Note the preferential accumulation of some repeats on the W chromosomes.
Figure 3.
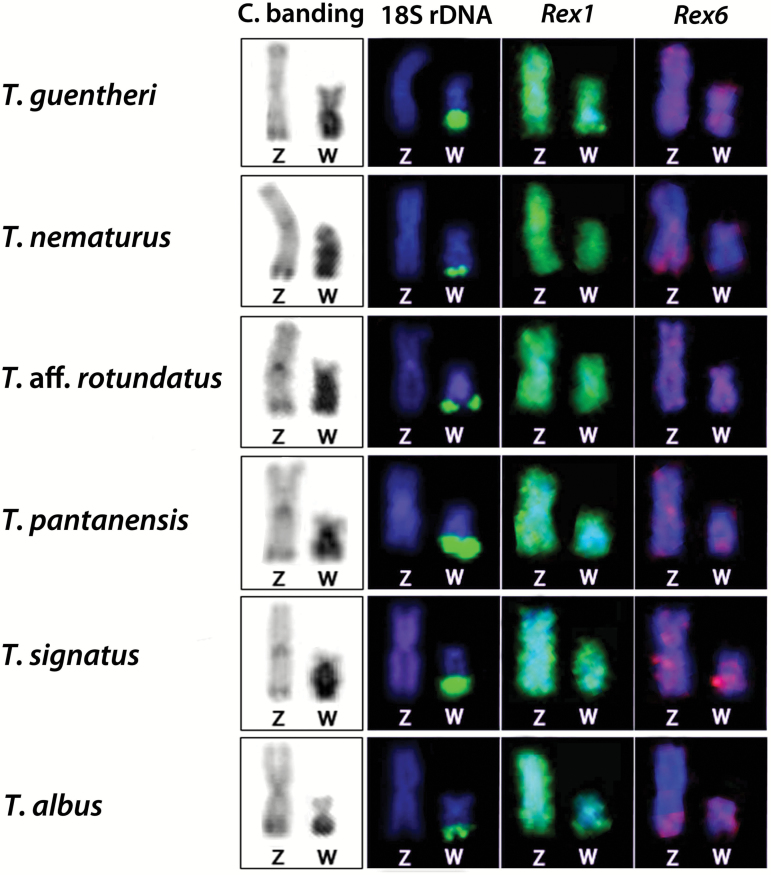
C-banding and distribution of repetitive DNA sequences on the Z and W chromosomes of 6 Triportheus species. Note the occurrence of 18S rDNA gene and the preferential accumulation of heterochromatin on the W chromosomes.
Results
Karyotyping and C-Banding of T. pantanensis and T. aff. rotundatus
T. pantanensis and T. aff. rotundatus were cytogenetically analyzed for the first time in this study. They have identical karyotype features to the other Triportheus species, i.e. 2n = 52 in males and females, composed mainly by m/sm and some st chromosomes, with a clear heteromorphic ZZ/ZW sex chromosome system. The Z chromosome is m and the largest one of the karyotype, while the W is sm and smaller than Z (Figure 1). The C-positive heterochromatin has a predominant localization on the centromeric region of majority of chromosome pairs. The Z chromosome also has additional heterochromatin in both telomeric regions, while the W chromosome is almost entirely heterochromatic, except for its short arms (Figures 1 and 3). These general patterns of Z and W chromosomes are also shared by the other Triportheus species until now analyzed.
Chromosomal Mapping of Repetitive Elements
Several microsatellites showed preferential accumulation on the W chromosome. Indeed, this chromosome presents a remarkable accumulation of the (CA)n and (GA)n microsatellites in all species (Figure 2). In a particular way, differential signals on the W chromosome was showed by (CAC)n and (CG)n microsatellites in T. albus and (CAT)n in T. nematurus, while (CAG)n and (GAA)n showed large accumulation in T. signatus (Figure 2). In relation to the Z chromosome, only (GAA)n repeats had a differential accumulation in T. albus, T. aff. rotundatus, and T. signatus.
Concerning autosomal pairs, the (CA)n and (GA)n microsatellites showed a general accumulation in the terminal position of all chromosomes of the 6 species. In addition, a compartmentalized accumulation of (CAT)n was found on 4 chromosomal pairs of T. guentheri. The remaining microsatellites had a well-dispersed pattern or very small signals in the terminal region of some specific chromosomes (Supplementary Figures T1–T6).
The retrotransposons Rex1 and Rex6 did not show preferential accumulation on the W chromosome (Figure 3). In all species, scattered signals of both TEs were observed throughout the length of all chromosomes, in addition to small Rex6 terminal clusters in some other ones (Supplementary Figure T7).
18S rDNA cistrons also occur on the long arms of the W chromosome in all Triportheus species (Figure 3). The mapping of this rDNA was simultaneously applied in all FISH experiments in order to properly identify this chromosome.
Discussion
Distribution of Repetitive DNAs in the Sex Chromosomes
The importance of using microsatellites investigations to analyze sex chromosome differentiation has been emphasized in several groups of plants and animals (Schlötterer 2000; Pokorná et al. 2011; Yano et al. 2014b; MacDonald et al. 2014). The intrinsic ability of microsatellites for expansion in the non-recombining chromosome regions has been supported in some species, such as the plant belonging to the order Caryophyllales, Rumex acetosa, where the accumulation of (CA)n and (CAA)n repeats in the young Y chromosome was associated with its differentiation process (Kejnovský et al. 2013). Although not yet completely enlightened, the DNA slippage, or the misalignment of repetitive DNAs, has been usually linked with the initial microsatellite expansion (Kejnovský et al. 2013). Such slippage errors may be corrected during recombination, but if recombination is suppressed, this provides the opportunity for further expansion of larger arrays of microsatellites (Charlesworth et al. 1994; Kejnovský et al. 2013). Thus, it is possible that microsatellite accumulation represents one of the factors acting on the differentiation process of the sex-specific chromosome.
In Triportheus species, a pronounced expansion of (CA)n and (GA)n repeats generally occurs in both sex chromosomes. In addition, a large accumulation of some specific microsatellites is also found in the W chromosome, as (CAC)n and (CG)n in T. albus, (CAG)n and (GAA)n in T. signatus and (CAT)n in T. nematurus, indicating that the differentiation of this chromosome is clearly associated with microsatellites accumulation (Figure 2). Similarly, the Z chromosome of T. albus, T. aff. rotundatus, and T. signatus, shares a differential accumulation of (GAA)n repeats, as also previously found in T. trifurcatus (Yano et al. 2014a).
Dinucleotides are the most frequent short repeats and, among them, (CA)n microsatellite prevails in all vertebrates and arthropods (Tóth et al. 2000), followed by (GA)n and (TA)n (Ellegren 2004). The larger distribution of dinucleotides than other repeats is probably due to their higher instability, allowing more slippage mechanisms (Katti et al. 2001). This particular condition is probably associated with the strong (CA)n and (GA)n signals on the W chromosome of Triportheus species. However, although microsatellite density is influenced by base composition, species-specific molecular features, such as enzymes related to replication or DNA repair, or even the proper euchromatin/heterochromatin chromosomal organization can, altogether, influence the density of microsatellites in the genome (Bachtrog et al. 1999; Toth et al. 2000; Katti et al. 2001).
Transposable elements also play an important role in the evolutionary processes of the sex chromosomes (Ferreira et al. 2011). Rex are among the most investigated TEs in fishes due to their wide distribution in the genome, allowing a comparative analysis among different species (Splendore de Borba et al. 2013). Although in some cases Rex retrotransposons have been mapped in the heterochromatic regions, they show a dispersed distribution throughout the whole genome in approximately 60% of the fish families (reviewed in Ferreira et al. 2011), as also observed for Rex1 and Rex6 sequences in all Triportheus species yet analyzed. However, no differential accumulation of Rex transposons was observed on the W chromosomes of Triportheus, contrasting with Leporinus genus, where Rex1 was accumulated on the W chromosome in L. obtusidens, and L. macrocephalus and on the W1 chromosome of L. elongatus (Splendore de Borba et al. 2013). Although microsatellites are usually associated with TEs (Kejnovský et al. 2013), this is not the case for Triportheus. In fact, the accumulation of microsatellites contrasts with the uniform distribution of Rex elements on the W chromosome, reflecting the independent path of these repetitive DNA classes in the sex-specific chromosome, probably due to the faster dynamism of microsatellites.
Species-Specific Dynamics of Triportheus Sex Chromosomes
The Triportheidae family was established by Oliveira et al. (2011), encompassing the genera Agoniates, Clupeacharax, Engraulisoma, Lignobrycon, and Triportheus. All previously analyzed Triportheus species share a ZZ/ZW sex chromosome system, in which the W chromosome is smaller than the Z one and almost entirely heterochromatic (Artoni et al. 2001). Accordingly, the same cytogenetic traits are present in T. pantanensis and T. aff. rotundatus, reinforcing the hypothesis that this sex chromosome system represent a genus-specific ancestral trait (Artoni et al. 2001).
According to the recent phylogenetic study, Triportheidae and Triportheus represents monophyletic groups, in which Triportheus originated at 26.2±6.5 Myr, having T. auritus as the most ancestral species (20.7±6.5) (Mariguela et al. 2016). In this way, the ZZ/ZW sex chromosome system is a basal trait for Triportheus and, perhaps, also present in other Triportheidae species. In fact, despite the lack of additional studies, a similar ZZ/ZW sex chromosome system is also found in Lignobrycon, a sister group of Triportheus (Rodrigues et al. 2013).
This study, with Triportheus as a model, highlights how the sex chromosomes evolution is a labile process, even among related species. Indeed, the W chromosome is subjected to particular evolutionary processes among species, as evidenced by its unequal accumulation of microsatellites. These findings are also supported by some previous studies in other Triportheus species, such as T. auritus (de Bello Cioffi et al. 2012), and T. trifurcatus (Yano et al. 2014a), in which the accumulation of the (CA)n and (GA)n microsatellites on the W chromosome is similar to the pattern found in this study. In 2 of the most recent originated species, T. trifurcatus and T. signatus (Mariguela et al. 2016), more classes of microsatellites had substantial accumulation on the W chromosome. However, it is not possible to have a correlation between age and accumulation of repeats sequences on the sex chromosomes, since in other more recently originated species, such as T. pantanensis, T. nematurus, and T. aff. rotundatus no preferential microsatellites accumulation was verified. Thus, despite the role of these repetitive sequences in the differentiation process of the sex-specific chromosome, it is put in evidence that this chromosome is subjected to distinctive evolutionary mechanisms, maybe in view of a free selective pressure. Such plasticity of the differentiation process of the sex-specific chromosome was also found in Leporinus (Anostomidae, Characiformes), where the heterochromatic W chromosome also has a differential distribution of some repeats among species (Poltronieri et al. 2004).
Among other lower vertebrates, morphological variations in the sex chromosomes of closely related lizard species or populations (Ezaz et al. 2009), with large or no accumulation of microsatellites (Pokorná et al. 2011), were also reported. Similar scenarios can also be found in several other animal and plant species, indicating that the sex chromosomes differentiation is an ongoing and dynamic process. Indeed, a high degree of differentiation is frequently observed in these chromosomes, appearing to have evolved independently many times among animal and plant species (Schlötterer 2000; Graves 2008). Notably, undifferentiated sex chromosomes, together with different sex chromosomes systems, can coexist in a same fish family or genus, or even in different populations of a same species, providing effective models to explore evolutionary events linked to sex chromosomes (reviewed in Cioffi et al. 2011a). For instance, in the Neotropical Parodontidae family, several species have no differentiated heteromorphic sex chromosomes while some Apareiodon and Parodon species share a same ZZ/ZW sex system, but with variations in size and morphology of the W chromosome. In addition, A. affinis shows a distinct and particular ZZ/ZW1W2 multiple sex chromosome system (reviewed in Bellafronte et al. 2011). Accordingly, in salmonid fishes simple and multiple sex chromosome systems can also be found in the same genus, as in Oncorhynchus (Phillips and Ráb 2001). In addition, some populations of rainbow trout display a heteromorphic Y chromosome, which is not observed in other ones (Thorgaard 1983).
Our current findings show the dynamic differentiation process of the sex-specific chromosome concerning the repetitive DNA fraction of the heterochromatin. Besides standing out the involvement of heterochromatin on the differentiation of the sex pair, it is highlighted that repetitive DNAs play a differential role in this heterochromatinization process. The differential accumulation of the same class of repeats on the W chromosome of both close and distant species reflects the inherent dynamism of microsatellites, as well as the plasticity that shapes the evolutionary history of the sex chromosomes.
Supplementary Material
Supplementary material can be found at http://www.jhered.oxfordjournals.org/.
Funding
This study was supported by the Conselho Nacional de Desenvolvimento Científico e Tecnológico - CNPq (Proc. nº 306896/2014-1); Fundação de Amparo à Pesquisa do Estado de São Paulo – FAPESP (Proc. nº 2014/22532–7); and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior – CAPES (Bolsista da CAPES (PDSE): Proc. nº BEX 11744/13–8).
Supplementary Material
References
- Artoni RF, Falcão JN, Moreira-Filho O, Bertollo LA. 2001. An uncommon condition for a sex chromosome system in Characidae fish. Distribution and differentiation of the ZZ/ZW system in Triportheus. Chromosome Res. 9:449–456. [DOI] [PubMed] [Google Scholar]
- Bachtrog D, Weiss S, Zangerl B, Brem G, Schlötterer C. 1999. Distribution of dinucleotide microsatellites in the Drosophila melanogaster genome. Mol Biol Evol. 16:602–610. [DOI] [PubMed] [Google Scholar]
- Baker CS. 2013. Journal of heredity adopts joint data archiving policy. J Hered. 104:1. [DOI] [PubMed] [Google Scholar]
- Bellafronte E, Schemberger MO, Moreira-Filho O, Almeida MC, Artoni RF, Margarido VP, Vicari MR. 2011. Chromosomal markers in Parodontidae: An analysis of new and reviewed data with phylogenetic inferences. Rev Fish Biol Fisher. 21:559–570. [Google Scholar]
- Bertollo LA, Born GG, Dergam JA, Fenocchio AS, Moreira-Filho O. 2000. A biodiversity approach in the neotropical erythrinidae fish, Hoplias malabaricus. Karyotypic survey, geographic distribution of cytotypes and cytotaxonomic considerations. Chromosome Res. 8:603–613. [DOI] [PubMed] [Google Scholar]
- Bertollo LAC, Cioffi MB, Moreira-Filho O. 2015. Direct chromosome preparation from Freshwater Teleost Fishes. In: Ozouf-Costaz C Pisano E, Foresti F Almeida Toledo LF, editors. Fish cytogenetic techniques (Chondrichthyans and Teleosts), 1st ed. Enfield: CRC Press; p. 21–26. [Google Scholar]
- Charlesworth B, Sniegowski P, Stephan W. 1994. The evolutionary dynamics of repetitive DNA in eukaryotes. Nature. 371:215–220. [DOI] [PubMed] [Google Scholar]
- Charlesworth D, Charlesworth B, Marais G. 2005. Steps in the evolution of heteromorphic sex chromosomes. Heredity. 95:118–128. [DOI] [PubMed] [Google Scholar]
- Cioffi MB, Martins C, Centofante L, Jacobina U, Bertollo LA. 2009. Chromosomal variability among allopatric populations of Erythrinidae fish Hoplias malabaricus: mapping of three classes of repetitive DNAs. Cytogenet Genome Res. 125:132–141. [DOI] [PubMed] [Google Scholar]
- Cioffi MB, Camacho JP, Bertollo LA. 2011. a. Repetitive DNAs and differentiation of sex chromosomes in neotropical fishes. Cytogenet Genome Res. 132:188–194. [DOI] [PubMed] [Google Scholar]
- Cioffi MB, Kejnovsky E, Bertollo LA. 2011. b. The chromosomal distribution of microsatellite repeats in the genome of the wolf fish Hoplias malabaricus, focusing on the sex chromosomes. Cytogenet Genome Res. 132:289–296. [DOI] [PubMed] [Google Scholar]
- de Bello Cioffi M, Kejnovský E, Marquioni V, Poltronieri J, Molina WF, Diniz D, Bertollo LA. 2012. The key role of repeated DNAs in sex chromosome evolution in two fish species with ZW sex chromosome system. Mol Cytogenet. 5:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diniz D, Laudicina A, Cioffi MB, Bertollo LA. 2008. Microdissection and whole chromosome painting. Improving sex chromosome analysis in Triportheus (Teleostei, Characiformes). Cytogenet Genome Res. 122:163–168. [DOI] [PubMed] [Google Scholar]
- Ellegren H. 2004. Microsatellites: simple sequences with complex evolution. Nat Rev Genet. 5:435–445. [DOI] [PubMed] [Google Scholar]
- Ezaz T, Sarre SD, O’Meally D, Marshall Graves JA, Georges A. 2009. Sex chromosome evolution in lizards: independent origins and rapid transitions. Cytogenet Genome Res. 127:249–260. [DOI] [PubMed] [Google Scholar]
- Ferreira DC, Porto-Foresti F, Oliveira C, Foresti F. 2011. Transposable elements as a potential source for understanding the fish genome. Mob Genet Elements. 1:112–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves JAM. 2008. Weird animal genomes and the evolution of vertebrate sex and sex chromosomes. Ann Rev Genet. 42:565–586. [DOI] [PubMed] [Google Scholar]
- Itoh Y, Kampf K, Arnold AP. 2008. Molecular cloning of zebra finch W chromosome repetitive sequences: evolution of the avian W chromosome. Chromosoma. 117:111–121. [DOI] [PubMed] [Google Scholar]
- Jurka J, Kapitonov VV, Kohany O, Jurka MV. 2007. Repetitive sequences in complex genomes: structure and evolution. Annu Rev Genomics Hum Genet. 8:241–259. [DOI] [PubMed] [Google Scholar]
- Katti MV, Ranjekar PK, Gupta VS. 2001. Differential distribution of simple sequence repeats in eukaryotic genome sequences. Mol Biol Evol. 18:1161–1167. [DOI] [PubMed] [Google Scholar]
- Kejnovský E, Michalovova M, Steflova P, Kejnovska I, Manzano S, Hobza R, Kubat Z, Kovarik J, Jamilena M, Vyskot B. 2013. Expansion of microsatellites on evolutionary young Y chromosome. PLoS One. 8:e45519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Y, Nagahama Y, Nakamura M. 2013. Diversity and plasticity of sex determination and differentiation in fishes. Sex Dev. 7:115–125. [DOI] [PubMed] [Google Scholar]
- Kubat Z, Hobza R, Vyskot B, Kejnovsky E. 2008. Microsatellite accumulation on the Y chromosome in Silene latifolia. Genome. 51:350–356. [DOI] [PubMed] [Google Scholar]
- Levan A, Fredga K, Sandberg AA. 1964. Nomenclature for centromeric position on chromosomes. Hereditas. 52:201–220. [Google Scholar]
- López-Flores I, Garrido-Ramos MA. 2012. The repetitive DNA content of eukaryotic genomes. In: Garrido Ramos MA, editor. Repetitive DNA, Genome Dynamics, Basel, SU: Karger; p. 1–28. [DOI] [PubMed] [Google Scholar]
- MacDonald AJ, Fitzsimmons NN, Chambers B, Renfree MB, Sarre SD. 2014. Sex-linked and autosomal microsatellites provide new insights into island populations of the tammar wallaby. Heredity (Edinb). 112:333–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariguela TC, Roxo FF, Foresti F, Oliveira C. 2016. Phylogeny and biogeography of Triportheidae (Teleostei: Characiformes) based on molecular data. Mol Phylogenet Evol. 96:130–139. [DOI] [PubMed] [Google Scholar]
- Marquioni V, Bertollo LA, Diniz D, Cioffi Mde B. 2013. Comparative chromosomal mapping in Triportheus fish species. Analysis of synteny between ribosomal genes. Micron. 45:129–135. [DOI] [PubMed] [Google Scholar]
- Oliveira C, Avelino GS, Abe KT, Mariguela TC, Benine RC, Ortí G, Vari RP, Corrêa e Castro RM. 2011. Phylogenetic relationships within the speciose family Characidae (Teleostei: Ostariophysi: Characiformes) based on multilocus analysis and extensive ingroup sampling. BMC Evol Biol. 11:275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips RB, Ráb P. 2001. Chromosome evolution in the Salmonidae (Pisces): an update. Biol Rev. 76:1–25. [DOI] [PubMed] [Google Scholar]
- Pokorná M, Kratochvíl L, Kejnovský E. 2011. Microsatellite distribution on sex chromosomes at different stages of heteromorphism and heterochromatinization in two lizard species (Squamata: Eublepharidae: Coleonyx elegans and lacertidae: Eremias velox). BMC Genet. 12:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poltronieri J, Marquioni V, Bertollo LA, Kejnovsky E, Molina WF, Liehr T, Cioffi MB. 2014. Comparative chromosomal mapping of microsatellites in Leporinus species (Characiformes, Anostomidae): unequal accumulation on the W chromosomes. Cytogenet Genome Res. 142:40–45. [DOI] [PubMed] [Google Scholar]
- Rodrigues AS, Diniz D, Affonso PRAM. 2013. Análise citogenética preliminar da piaba-facão Lignobrycon myersi (Osteichthyes, Characiformes), espécie ameaçada do estado da Bahia. XV Simpósio de Citogenética e Genética de Peixes Jequié, BA. [Google Scholar]
- Schlötterer C. 2000. Microsatellite analysis indicates genetic differentiation of the neo-sex chromosomes in Drosophila americana americana . Heredity. 85:610–616. [DOI] [PubMed] [Google Scholar]
- Silva DS, Milhomem SS, Pieczarka JC, Nagamachi CY. 2009. Cytogenetic studies in Eigenmannia virescens (Sternopygidae, Gymnotiformes) and new inferences on the origin of sex chromosomes in the Eigenmannia genus. BMC Genet. 10:74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Splendore de Borba R, Lourenço da Silva E, Parise-Maltempi PP. 2013. Chromosome mapping of retrotransposable elements Rex1 and Rex3 in Leporinus Spix, 1829 species (Characiformes: Anostomidae) and its relationships among heterochromatic segments and W sex chromosome. Mob Genet Elements. 3:e27460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumner AT. 1972. A simple technique for demonstrating centromeric heterochromatin. Exp Cell Res. 75:304–306. [DOI] [PubMed] [Google Scholar]
- Thorgaard GH. 1983. Chromosomal differences among rainbow trout populations. Copeia. 3:650–662. [Google Scholar]
- Tóth G, Gáspári Z, Jurka J. 2000. Microsatellites in different eukaryotic genomes: survey and analysis. Genome Res. 10:967–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volff JN, Körting C, Sweeney K, Schartl M. 1999. The non-LTR retrotransposon Rex3 from the fish Xiphophorus is widespread among teleosts. Mol Biol Evol. 16:1427–1438. [DOI] [PubMed] [Google Scholar]
- Yano CF, Poltronieri J, Bertollo LA, Artoni RF, Liehr T, de Bello Cioffi M. 2014. a. Chromosomal mapping of repetitive DNAs in Triportheus trifurcatus (Characidae, Characiformes): insights into the differentiation of the Z and W chromosomes. PLoS One. 9:e90946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano CF, Bertollo LA, Molina WF, Liehr T, Cioffi Mde B. 2014. b. Genomic organization of repetitive DNAs and its implications for male karyotype and the neo-Y chromosome differentiation in Erythrinus erythrinus (Characiformes, Erythrinidae). Comp Cytogenet. 8:139–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.