We describe the first human RNase 6 crystal structure in complex with sulfate anions. Kinetic analysis, site-directed mutagenesis and molecular dynamics simulations identified novel substrate recognition and cleavage sites.
Keywords: kinetic characterization, molecular dynamics, protein crystallography, RNase A superfamily, RNase k6, sulfate anion
Abstract
Human RNase 6 is a cationic secreted protein that belongs to the RNase A superfamily. Its expression is induced in neutrophils and monocytes upon bacterial infection, suggesting a role in host defence. We present here the crystal structure of RNase 6 obtained at 1.72 Å (1 Å=0.1 nm) resolution, which is the first report for the protein 3D structure and thereby setting the basis for functional studies. The structure shows an overall kidney-shaped globular fold shared with the other known family members. Three sulfate anions bound to RNase 6 were found, interacting with residues at the main active site (His15, His122 and Gln14) and cationic surface-exposed residues (His36, His39, Arg66 and His67). Kinetic characterization, together with prediction of protein–nucleotide complexes by molecular dynamics, was applied to analyse the RNase 6 substrate nitrogenous base and phosphate selectivity. Our results reveal that, although RNase 6 is a moderate catalyst in comparison with the pancreatic RNase type, its structure includes lineage-specific features that facilitate its activity towards polymeric nucleotide substrates. In particular, enzyme interactions at the substrate 5′ end can provide an endonuclease-type cleavage pattern. Interestingly, the RNase 6 crystal structure revealed a novel secondary active site conformed by the His36–His39 dyad that facilitates the polynucleotide substrate catalysis.
INTRODUCTION
Human RNase 6 is a protein belonging to the bovine pancreatic ribonuclease A (RNase A) superfamily, a vertebrate-specific family comprising small secretory proteins, sharing a common overall 3D structure and displaying a variety of properties. Together with the first ascribed function of pancreatic RNases to digest RNA, several family members were reported to be involved in innate immunity, showing toxicity towards a wide spectrum of pathogens, from viruses, bacteria, fungi and protozoa to helminth parasites [1–3]. An unusually high evolution rate within the family and the antimicrobial properties of distantly related members suggested a common ancestral innate immunity role [4,5]. In humans, the family includes eight known members, also called the ‘canonical RNases’ (Figure 1A). Despite their low sequence identity, ranging from 30% to 70%, we observe the conservation of the disulfide bonding pattern and the catalytic triad. All members are highly cationic and are localized at the long arm (q) of human chromosome 14 [6,7].
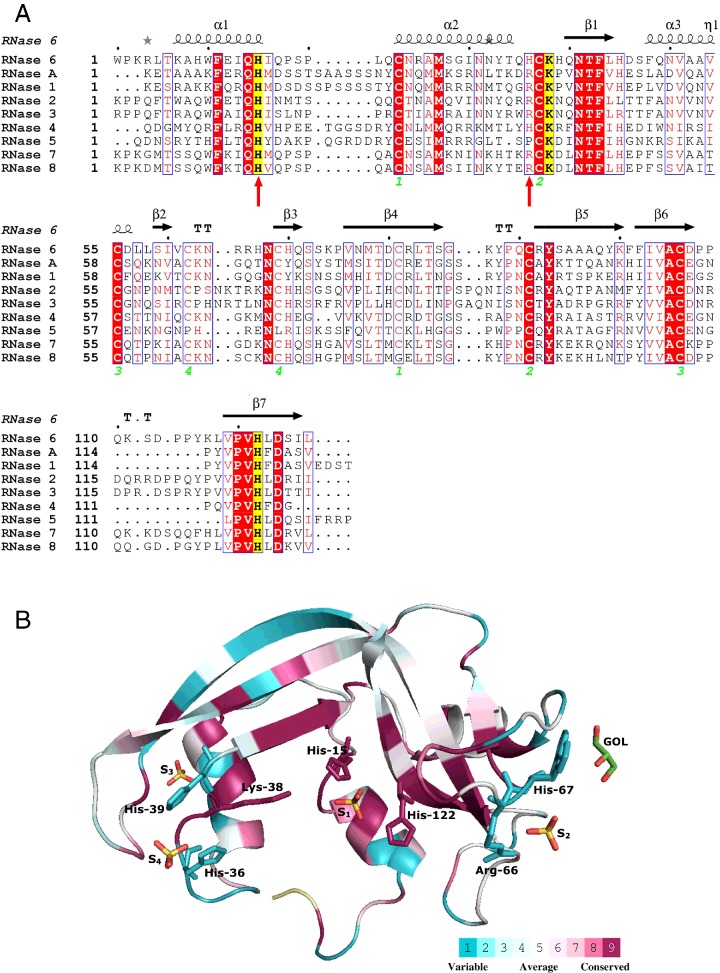
Figure 1. Primary structure of human RNases and 3D structure of RNase 6 coloured by residue conservation score.
(A) Structure-based sequence of the eight canonical human RNases together with RNase A. The active sites are highlighted in yellow. The four disulfide bonds are labelled with green numbers. Tested mutations on RNase 6 are indicated with red arrows. The alignment was performed using ClustalW, and drawn using ESPript (http://espript.ibcp.fr/ESPript/). Labels are as follows: red box, white character for strict identity; red character for similarity in a group and character with a blue frame for similarity across groups. (B) RNase 6 3D structure surface representation using the CONSURF web server (http://consurf.tau.ac.il/) featuring the relationships among the evolutionary conservation of amino acid positions within the RNase A family. The 3D structure shows residues coloured by their conservation score using the colour-coding bar at the bottom. Sulfate anions (S1–S4) and the glycerol (GOL) molecule found in the crystal structure are depicted. Conserved residues belonging to the RNase catalytic site and interacting with bound sulfate anions are labelled.
RNase 6, also named RNase k6, was first to be identified during a genomic search for a homologous protein of bovine kidney RNase (RNase k2) and localized on q11 region of chromosome 14 [8,9]. The newly identified human mature protein sequence was found to share 72% identity with its bovine RNase k2 counterpart. Divergence of the kidney RNases in comparison with the prototype reference family pancreatic type RNases supported their involvement in a differentiated biological role. Due to the presence of human RNase 6 in a large variety of tissues and its expression in monocytes and neutrophils, it was proposed that it could play a role in host defence. Indeed, recent studies by Becknell et al. [10] showed the protein expression in macrophages and epithelial cells at the urinary tract in response to exposure of uropathogenic bacteria. Spencer and co-workers also reported a potent antimicrobial activity in vitro against Gram-negative and Gram-positive bacteria for RNase 6, together with its closest homologue, RNase 7, and proposed both proteins as being responsible for the mammalian urinary tract sterility maintenance [11,10]. Experimental evidence was also provided by the reported down-regulation of RNase 6 together with other host innate immunity proteins induced by the human immunodeficiency virus (HIV) [12]. RNase 6 displays 55% amino acid identity with RNase 7, and belongs to the RNase 6, 7 and 8 cluster, sharing with them common structural features (Figure 1A). Interestingly, even though it has been found that eosinophil RNase 2 and RNase 3 gene lineages have undergone one of the highest rates of divergent evolution to produce paralogous genes [13,14], RNase 6 primate gene lineages appear to have evolved in a more conservative mode [9]. On the other hand, a contradictory scenario has been reported in rodents, in which the evolution of RNase 6 presents a substantially higher rate [15]. All in all, a similar tendency towards an isoelectric point increase is shared within the eosinophil lineage.
The RNase A superfamily members share a conserved catalytic mechanism that was thoroughly characterized thanks to the pioneering enzymology studies during the first half of the XX's century [16–18]. RNase A catalyses the cleavage of the 3′5′ phosphodiester bond of single polynucleotide substrates, showing selectivity for pyrimidines at the main base subsite (B1) and a preference for purines at the secondary base site (B2). Degradation of polynucleotide substrate is also assisted by additional binding sites at both sides of the catalytic centre, referred to as Bn, Rn and pn for bases, ribose and phosphate binding respectively [19].
Preliminary kinetic characterization of RNase 6 upon its discovery indicated a moderate catalytic efficiency with respect to the family reference member RNase A. Estimation of kinetic parameters using yeast tRNA as a substrate reported approximately a 40-fold reduced catalytic rate in comparison with RNase 2 [8]. Further side-by-side comparison of RNase 6 catalytic efficiency confirmed an overall moderate relative catalytic efficiency, higher than that of RNase 3 but significantly lower than that of RNase 7 [20–22].
In the present paper, we describe the first crystal structure of RNase 6. The protein structural analysis is complemented by its enzymatic characterization to highlight RNase 6's singularity within the RNase A family context.
MATERIALS AND METHODS
Expression and purification of the recombinant proteins
A plasmid containing the gene of recombinant human RNase 6 was transformed in a prokaryote expression system. The cDNA encoding RNase 6 sequence was a gift from Dr Helene Rosenberg (National Institutes of Health, Bethesda, MD, U.S.A.). Mutant variants were constructed using the Quik Change Site-Directed Mutagenesis kit (Stratagene). All constructs were confirmed by DNA sequencing and the purified protein was analysed by MALDI–TOF-MS and N-terminal sequencing.
The genes were subcloned in plasmid pET11c for prokaryote high yield expression. Escherichia coli BL21(DE3) competent cells were transformed with the pET11c/RNase 6 plasmid. The expression protocol was optimized from the previously described procedure [20] to optimize the RNase 6 final recovery yield. For high yield expression, bacteria were grown in Terrific broth (TB), containing 400 μg/ml ampicillin. Recombinant protein was expressed after cell induction with 1 mM IPTG added when the culture showed a D600 of 0.6. The cell pellet was collected after 4 h of culture at 37°C. Cells were resuspended in 10 mM Tris/HCl and 2 mM EDTA, pH 8, and sonicated at 50 W for 10 min with 30-s cycles. After centrifugation at 15000 g for 30 min, the pellet fraction containing inclusion bodies was processed as follows: the pellet fraction was washed with 50 mM Tris/HCl, 2 mM EDTA and 0.3 M NaCl, pH 8, and after centrifugation at 20000 g for 30 min, the pellet was dissolved in 12 ml of 6 M guanidinium chloride, 0.1 M Tris/acetate and 2 mM EDTA, pH 8.5, containing 80 mM GSH, and incubated under nitrogen for 2 h at room temperature. The protein was then refolded by a rapid 100-fold dilution into 0.1 M Tris/HCl, pH 7.5, containing 0.5 M L-arginine, and GSSG was added to obtain a GSH/GSSG ratio of 4. Dilution in the refolding buffer was adjusted to obtain a final protein concentration of 30–150 μg/ml. The protein was incubated in refolding buffer for 48–72 h at 4°C. The folded protein was then concentrated, buffer-exchanged against 0.015 M Tris/HCl, pH 7, and purified by cation-exchange chromatography on a Resource S column equilibrated with the same buffer. The protein was eluted with a linear NaCl gradient from 0 to 2 M in 0.015 M Tris/HCl, pH 7, buffer. Further purification was achieved by reverse-phase chromatography on a Vydac C4 column, using an acetonitrile gradient. The homogeneity of the purified proteins was checked by SDS/15% PAGE and Coomassie Blue staining and by N-terminal sequencing. RNase 3 and RNase 7 were expressed as previously described [20,24].
Spectrophotometric kinetic analysis
Polycytidylic acid [poly(C)], polyuridylic acid [poly(U)], polyadenylic acid [poly(A)], poly(A): poly(U), CpA, UpA, UpG and cytidine 2′,3′-cyclic phosphate (C>p) (Sigma–Aldrich) were used as substrates, and the kinetic parameters were determined by a spectrophotometric method as described in [20]. RNase A, used as a control protein, was purchased from Sigma. Assays were carried out in 50 mM sodium acetate and 1 mM EDTA, pH 5.5, at 25°C, using 1-cm pathlength cells. Substrate concentration was determined spectrophotometrically using the following molar absorption coefficients: ε268=8400 M−1·cm−1 for C>p; ε265=21000 M−1·cm−1 for CpA, ε261=23500 M−1·cm−1 for UpA, ε261=20600 M−1·cm−1 for UpG, ε268=6200 M−1·cm−1 for poly(C), ε260=9430 M−1·cm−1 for poly(U) and ε260=4430 M−1·cm−1 for poly(A):poly(U) for each nucleotide unit. The activity was measured by following the initial reaction velocities using the difference molar absorption coefficients, in relation to cleaved phosphodiester bonds: Δε286=1450 M−1·cm−1 for CpA, Δε286=570 M−1·cm−1 for UpA, Δε280=480 M−1·cm−1 for UpG, Δε250=2380 M−1·cm−1 for poly(C), Δε282=829 M−1·cm−1 for poly(U) and Δε260=3400 M−1·cm−1 for poly(A):poly(U) for transphosphorylation reaction, and Δε286=1450 M−1 cm−1 for C>p hydrolysis reaction [20,25]. Duplicates of seven substrate concentrations (ranging from 0.1 to 2 mM) were tested for each condition. Final enzyme concentrations were selected from 0.1 to 10 μM depending on the RNase activity for each assayed substrate. Kinetic parameters were obtained by the non-linear regression GraFit data analysis program (Erithacus Software). Relative activity of RNase mutants was calculated by comparison of initial velocities (V0), using a substrate concentration of 0.1 mM for dinucleotides and 0.5 mg/ml for polynucleotides.
Activity staining gel
Zymograms were performed following the method previously described [26]. SDS/15% polyacrylamide gels were cast with 0.3 mg/ml poly(C) (Sigma–Aldrich) and run at a constant current of 40 mA for 1.5 h. Then, the SDS was extracted from the gel with 10 mM Tris/HCl, pH 8, and 10% (v/v) propan-2-ol. The gel was then incubated in the activity buffer (0.1 M Tris/HCl, pH 8) to allow enzymatic digestion of the embedded substrate and then stained with 0.2% (w/v) Toluidine Blue (Merck) in 10 mM Tris/HCl, pH 8, for 10 min. Positive bands appeared white against the blue background. The loading buffer had no 2-mercaptoethanol to facilitate recovery of active enzymes. RNase A (Sigma–Aldrich) was used as a control.
Analysis of polynucleotide cleavage pattern
The characterization of the RNases, substrate cleavage patterns was carried out by studying the digestion product profiles, as previously described [27]. The poly(C) substrate (Sigma–Aldrich) was dissolved at a concentration of 0.5 mg/ml in 10 mM HEPES/KOH at pH 7.5. Then, 50 μl of the poly(C) solution was digested with 10 μl of enzyme solution at 25°C for 1 h. Enzyme final concentrations were adjusted for each RNase: 50 nM for RNase 6 and RNase 6-H36R and 1.4 μM for RNase 6-H15A and RNase 7-H15A. At different digestion times the products of the reaction were separated by reverse-phase HPLC (Nova Pak C18, Waters) according to the previously described procedure [27,28]. Briefly, the RNase/poly(C) reaction mixtures (50 μl and 15 μl for wild-type and mutant RNase 6 respectively) were injected on to the column equilibrated with solvent A (10% (w/v) ammonium acetate and 1% (v/v) acetonitrile) and the elution was carried out by an initial 10-min wash and 50-min gradient from 100% solvent A to 10% solvent A plus 90% solvent B (10% (w/v) ammonium acetate and 11% (v/v) acetonitrile). Product elution was detected from the absorbance at 260 nm, and peak identification was performed according to previous characterization of oligocytidylic acids [29].
Protein crystallization
RNase 6 crystals were obtained after high-throughput screening of available commercial kits by the hanging-drop vapour-diffusion methodology at 20°C. In one of these kits, JCSG-plus™ HT-96 (Molecular Dimensions), RNase 6 at 10 mg/ml was able to crystallize under one condition (0.2 M NaCl, 0.1 M sodium cacodylate, and 2 M (NH4)2SO4, pH 6.5). This condition was optimized to improve the crystal size by the hanging-drop methodology by mixing 1 μl of the protein sample with 1 μl of the crystallization buffer. The best condition resulting from this optimization was 0.05 M NaCl, 0.1 M sodium cacodylate (pH 6.5) and 2 M (NH4)2SO4. Cubic crystals appeared after 10 days of incubation at 20°C and were soaked in the cryoprotectant solution by adding 15% glycerol to the crystallization buffer prior to X-ray exposure.
Data collection, processing and protein structure solving
Data were collected at the XALOC BL13 beamline station of ALBA synchrotron (Spain) using a wavelength of 0.9795 Å. Data collection was performed at 100 K using a Pilatus 6M detector (Dectris®), 800 images were taken at texp=0.2 s, Δφ=0.2°. The data obtained were processed with XDS (MPI for Medical Research) [30]. The structure was solved by molecular replacement with Phenix Phaser-MR program using an RNase 6 model constructed upon NMR structure of RNase 7 (PDB ID: 2HKY) [31]. Iterative cycles of refinement and manual building were applied using PHENIX [32] and Coot [33] respectively until no further improvement of Rfree could be achieved. Finally, the stereochemistry of the structure was validated with SFCHECK [34] and WHAT_CHECK [35]. Table 1 shows the data collection and structure refinement statistics.
Table 1. Data collection, processing and structure refinement parameters for the RNase 6 crystal structure (PDB ID: 4X09).
*Rmerge = Σhkl Σj-1 to N|Ihkl-Ihkl(j)|/ Σhkl Σj-1 to N Ihkl(j), where N is the redundancy of the data. †Outermost shell is 1.78–1.72 Å. ‡Rcrystal=Σh|Fo-Fc|/ΣhFo, where Fo and Fc are the observed and calculated structure factor amplitudes of reflection h respectively. §Rfree is equal to Rcryst for a randomly selected 5% subset of reflections not used in the refinement.
| Parameter | Value |
|---|---|
| Data collection | |
| Space group | P212121 |
| Unit cell | |
| a, b, c (Å) | 27.73, 38.86, 97.97 |
| α, β, γ (°) | 90.0, 90.0, 90.0 |
| Number of molecules in asymmetric unit | 1 |
| Resolution (Å) | 1.72 |
| Number of total reflections | 22981 |
| Number of unique reflections | 11717 |
| Rmerge*,† (%) | 2.8 (23.4) |
| I/σI† | 13.0 (2.4) |
| Completeness for range† (%) | 99.2 (99.0) |
| Wilson B factor (Å2) | 24.7 |
| Matthews coefficient (Å3/Da) | 1.80 |
| Solvent content (%) | 31.71 |
| Refinement | |
| Resolution range (Å) | 48.98–1.72 |
| Rcryst‡/Rfree§ (%) | 19.28/22.67 |
| Number of protein atoms | 1068 |
| Number of water molecules | 124 |
| Number of bound anions | 4 |
| RMSD from ideality | |
| In bond lengths (Å) | 0.004 |
| In bond angles (deg) | 0.908 |
| Average B factors (Å2) | |
| All protein atoms | 30.34 |
| Main-chain atoms | 27.30 |
| Side-chain atoms | 33.23 |
| Sulfate anion atoms | 57.60 |
| Glycerol atoms | 49.55 |
| Water molecules | 40.67 |
Structure modelling
Molecular modelling predictions were carried out using protein–nucleotide docking and molecular dynamics (MD) simulations. Docking simulations were conducted with AutoDock 4.2.6 (Scripps Research Institute) and MD simulations were performed with GROMACS 4.5.5 [36]. RNase A and RNase 6 complexes with dinucleotides (CpA, UpA and UpG) were predicted. The initial RNase A–dinucleotides’ positions were determined on the basis of crystallographic data of RNase A bound to d(CpA) [37]. For RNase 6–dinucleotide complexes, the position of the S1 sulfate was taken as reference. Due to the inactive position of His122 in the RNase 6 crystal, the position of the histidine was adjusted to the ‘active’ conformation taking RNase A as a reference (PDB ID: 1RPG).
For MD simulations the force field AMBER99SB-ILDN [38] was used both for protein and RNA components. All of the complexes were centred in a cubic cell with a minimum distance of box to solute of 1.0 nm. The unit cell was filled with transferable intermolecular potential 3P (TIP3P) water [39] in neutral conditions with 150 mM NaCl. Neighbour search was performed using a group cut-off scheme with a cut-off of 1.4 nm for van der Waals interactions and 0.9 nm for the other short-range Lennard–Jones interactions. For long range interactions, smooth particle mesh of Ewald (PME) [40,41] was used with a fourth-order interpolation scheme and 0.16-nm grid spacing for FFT. The bonds were constrained with the P-LINCS algorithm [42], with an integration time step of 2 fs. The energy of the system was minimized using the steepest descendant algorithm and equilibrated in two steps. First, an initial constant volume equilibration (NVT) of 100 ps was performed with a temperature of 300 K using a modified Berendsen thermostat. Then, 100 ps of constant pressure equilibration (NPT) was run at 1 bar (100 kPa) with a Parrinello–Rahman barostat [43,44] at 300 K and the same thermostat. Finally, 20 ns production runs were performed under an NPT ensemble without applying restraints. Three independent simulations in periodic boundary conditions were conducted for each complex. The evolution of the average RMSD for all non-hydrogen ligand atoms after least-squares fitting to the original position was calculated.
For prediction of the RNase 6–heptanucleotide complex, the RNase A–d(ATAA) crystal structure was taken as a reference (PDB ID: 1RCN [45]). First, the d(ATAA) co-ordinates were used to build an AUAA ribonucleotide. His122 of RNase 6 was fixed in the corresponding active conformation. Local search docking with 2000 cycles and 2000 iterations was performed with AutoDock 4.2.6 [46] to adjust the AUAA position to RNase 6 active site. Then, three cytidines were added to the 5′ end of the tetranucleotide. The sulfate positions of the RNase 6 structures were taken as a reference to place the phosphates corresponding to the extended nucleotide. Then, a steepest descent energy minimization of the complex was performed with GROMACS 4.5.5. MD simulations were also applied using the same protocol described for dinucleotides.
Prediction of pKa values
Prediction of pKa values of selected protein residues was performed using the Rosetta online server ROSIE [47]. The estimated pKa values of selected histidine residues were calculated by using a neighbour sphere of 15 Å and considering the protonation state of ionizable residues. A starting pKa reference value of 6.3 for each histidine residue was ascribed. The program evaluates all potential conformational rotamers together with the influence of side chain and backbone mobility. RNase A (PDB ID: 7RSA) co-ordinates were used as a reference control. Predicted pKa values for His12, His105 and His119 in RNase A were found in accordance with the previously reported experimental values [48]. His105 in RNase A and His67 in RNase 6 were selected as control solvent-exposed residues, not involved in Coulombic interactions with nearby residues. The RNase A double mutant (RNase A-H7H10) [49] crystal structure (PD ID: 5ET4; Blanco, Salazar, Moussaoui and Boix unpublished results) was also analysed to evaluate the predicted pKa values for an engineered secondary site located at RNase A secondary phosphate-binding site.
RESULTS
RNase 6 3D structure
RNase 6 crystals diffracted to 1.72 Å. The structure was solved using RNase 7 structure as a model (PDB ID: 2HKY [31]). The crystallographic statistics for the data collection, processing and structure solving are provided in Table 1. Structural data for RNase 6 structure are available in the PDB under the accession number 4X09. The RNase 6 3D structure (Figure 1B) complies with the RNase A superfamily overall conformation, with a kidney-shaped structure formed by seven β-strands and three α-helices cross-linked by four conserved disulfide bonds, as listed in Supplementary Table S1. Loop residues Trp1–Lys3, Gln17–Leu21, Lys63–Arg66, Gly86–Gln90 and Pro108–Ser112 are partially disordered. In particular, practically no electron density was visualized for residues Pro2, Lys3, Gln17, Leu21, Gly86 and Lys87 that could not be properly modelled. Alternative side-chain conformations were modelled for some residues (Gln14, Asn32, Ser59, His67, Met78 and Thr79). Specifically, high motion values were observed in loops L1, L2 and L8, where 4% of the residues are disordered, as reported for the RNase 7 structure [31]. Residues involved in crystal packing were analysed by the PISA web server [50]. The intermolecular contacts are listed in Supplementary Table S2. Interactions are found mostly between β3, β4, β5 and β7 strand residues (Gln71, Arg82, Ala97–Tyr99, Ser125 and Ile126) and loop residues. No packing contacts are seen in the environment of the active site, therefore enabling further substrate-binding studies.
An overall comparison of RNase 6 crystal structure with previously solved structures of RNase A superfamily members highlighted some interesting particularities. First, we observed a distinct conformation of the RNase 6 N-terminus, where Trp1, unique in the RNase A superfamily, folds back towards the protein core. Next, we found several non-conserved histidine residues, unique to RNase 6, that contribute to the protein structure peculiarities. His9 was observed to interact with Glu12 by a salt bridge, which would participate in the stabilization of the first α-helix, as reported in RNase A for the Glu2–Arg10 salt bridge [51,52]. Unusual rotamer conformations, showing an unambiguous electron density, were observed for two non-conserved histidines: His36 and His67.
RNase 6 sulfate-binding sites
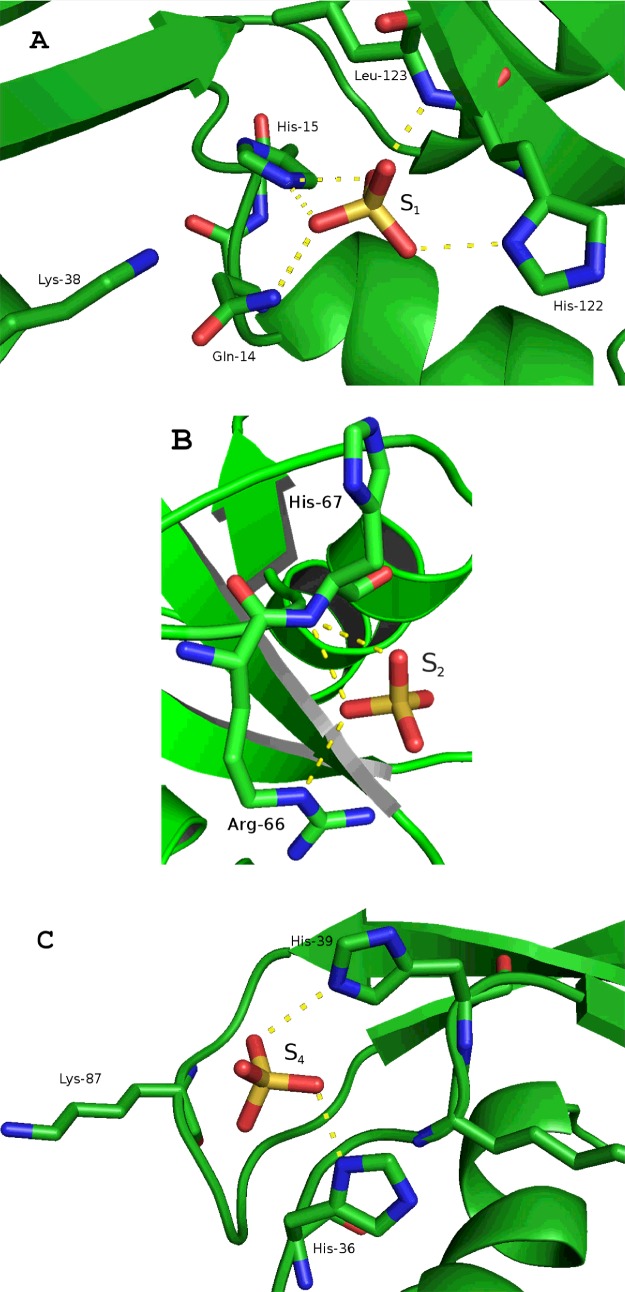
The protein was crystallized in the presence of ammonium sulfate and four sulfate anions were identified in the crystal asymmetric unit (Figure 1B and Supplementary Figure S1). Three of these sulfates corresponded to defined cationic regions exposed at the protein surface and correlate to putative RNA phosphate-binding sites, whereas the fourth sulfate is involved in the crystal packing. Sulfate anion interactions with nearby residues are listed in Supplementary Table S3 and illustrated in Figure 2. Sulfate labelled S1 corresponds to the active site of the enzyme, conserved in all canonical RNases. The presence of either a sulfate or phosphate anion at the enzyme active site was already reported for RNase A [53,54] and other family members: RNase 2–EDN (eosinophil-derived neurotoxin) [55], RNase 3–ECP (eosinophil cationic protein) [56] and RNase 5–angiogenin [57]. On the other hand, the second sulfate (S2) found in the RNase 6 structure binds to a distinct region not reported in any other family members (Arg66/His67). Comparative studies of our structure with the RNase A–tetranucleotide analogue [45] suggested that this region may represent a distinct phosphate-interaction subsite located at the 5′ end of the RNA substrate. In addition, a third sulfate anion was visualized in the RNase 6 structure bound to two histidine residues (His36 and His39), His39 being unique to RNase 6 among all RNase A members. Interestingly, both histidine residues together with a close by lysine residue (Lys87) mimic the disposition of a putative RNase active site (Figure 3), as also identified using the PDBeMotif analysis tool available at the PDBe server (http://www.ebi.ac.uk/pdbe-site/pdbemotif/).
Figure 2. Detail of sulfate binding interactions in the RNase 6 crystal structure.
Atoms involved in the protein–anion interactions are listed in Supplementary Table S3. The sulfate involved in the crystal packing (S3) is not shown. The structure was drawn with PyMol 1.7.2 (DeLano Scientific).
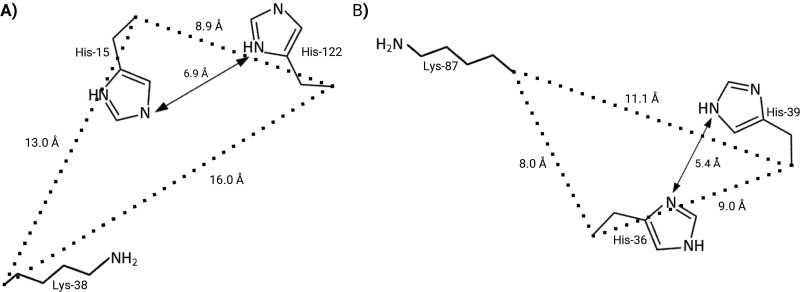
Figure 3. Illustrative scheme of RNase 6 main and putative secondary active sites.
Illustrative scheme of the RNase 6 main active site (A) and the putative secondary site (B). CpA atom distances are labelled together with the hisitidine ND1 to NE2 respective distances. The figure was created with PyMol (DeLano Scientific).
RNase 6 main active site
The RNase 6 crystal structure illustrates the conservation of the active-site architecture within the RNase A family. Residues His15, Lys38 and His122 (His12, Lys41 and His119 RNase A counterparts) build the active-site groove, with His122 adopting the so-called ‘inactive’ orientation [58], a conformation reported to be favoured in acidic and high ionic salt solutions [59]. Comparison of atomic crystal structures for RNase A indicated that His119 can adopt two conformations: A (active) and B (inactive), where the ‘active’ ring orientation is required for adenine binding at the secondary base B2 site [37,60]. On the other hand, the active orientation of the RNase A His119 ring was reported to be favoured by the hydrogen bond interaction with the vicinal Asp121 residue, whose interaction would account for the correct His119 tautomer in catalysis [61]. RNase 6 counterpart (Asp124), located in an equivalent conformation, could also perform an equivalent role. However, the inactive His122 conformation is fixed in our RNase 6 structure by a hydrogen bond interaction with Lys7. Therefore, the close proximity of Lys7 might alter significantly the properties of the His122 catalytic residue. Likewise, we considered the potential influence of the nearby His36/His39 RNase 6 residues on the other active-site histidine residue (His15). The presence of cationic residues near to the RNase 6 active site could shape the enzyme's performance, and may account for a reduction in its catalytic efficiency in comparison with RNase A (Table 2 and Supplementary Table S4). Although the RNase 6 structure conserves equivalent positions for some key residues at the active-site environment, such as Gln14 and Asn41 (Gln11 and Asn44 respectively in RNase A), the close proximity of residues such as Lys7, His36 or His39 should not be disregarded.
Table 2. Kinetic parameters of RNases for dinucleotide and mononucleotide cyclic phosphate substrates.
ND: not detected at the assayed conditions. *All assays were carried out in duplicate by a spectrophotometric assay as described in the Materials and methods section. Kinetic parameters were estimated from non-linear regression data using the GraFit program. †Values shown for RNase A activity are taken from [73–75].
| UpA | UpG | CpA | C>p | ||
|---|---|---|---|---|---|
| RNase 6 | Km (mM)* | 2.63±0.3 | ND | 1.22±0.2 | 2.06±0.3 |
| kcat (s−1) | 12.9±1.1 | ND | 1.08±0.1 | 3.25×10−3 ± 0.06 | |
| kcat /Km (s−1·M−1) | 4.90×103 | ND | 8.85×102 | 1.60 | |
| RNase 3 | Km (mM) | 2.7±0.66 | ND | 1.7±0.3 | 3±0.53 |
| kcat (s−1) | 1.22±0.12 | ND | 0.55±0.06 | 3.2×10−3 ± 5.1×10−4 | |
| kcat /Km (s−1·M−1) | 4.47×102 | ND | 3.23×102 | 1.07 | |
| RNase A† | Km (mM) | 0.7 | 2.0 | 0.5 | 1.06±0.1 |
| kcat (s−1) | 2.69×103 | 1.38×102 | 2.3×103 | 2.28±0.18 | |
| kcat /Km (s−1·M−1) | 3.84×106 | 6.9×104 | 4.59×106 | 2.15×103 |
RNase 6 nucleotide-binding sites
The RNase 6 crystal structure was further analysed to elucidate the structural basis for the protein substrate specificity and kinetic properties. Comparison of the RNase 6 structure with RNase A indicated conservation of most residues at the B1 site. In contrast, non-conserved substitutions were found for Phe120 and Ser123 RNase A counterpart residues. Phe120 is observed in RNase A to contribute stacking interactions to fix the pyrimidine ring [37]. Nevertheless, a leucine residue in RNase 6, also present in RNase 3, might also contribute to shape the protein base binding hydrophobic cavity. In any case, the presence of Phe120 in both RNase A and RNase 4, two very efficient RNase A family members, might also explain their relative higher catalytic activity. On the other hand, B1 selectivity is considered to be dependent on a Thr–Asp dyad relative position. Thr45–Asp83 interactions were attributed in RNase A to shift from cytidine to uridine preference [17]. However, in RNase 6, the presence of Gln40, hydrogen-bonded to Asp80 (Asp83 counterpart), fixes the latter in a distinct orientation that might interfere in its interaction with Thr42 (Thr45 counterpart), and modifies the pyrimidine-binding mode. Additionally, RNase A and RNase 4 (an RNase with an unusually strong uridine preference [62]), have an hydrophobic residue at the Gln40 position, which was considered to help locating the Thr45 residue at the most favoured conformation for uridine base binding [63]. To note, Gln40–Asp80 pair is unique to RNase 6 among the eight human canonical RNases (Figure 1). This scenario might explain in RNase 6 the observed increase Km value for both UpA and CpA with respect to RNase A (Table 2). On the other hand, the orientation of Asp80, closer to RNase A counterpart but clearly distinct from RNase 4, may explain RNase 6's moderate preference for uridine at the B1 site (Table 3). Interestingly, the uridine predilection of RNase 4 would be mostly determined by Arg101 that can directly interact with the uridine carbonyl group and bringes Asp80 closer to the vicinity of Thr45 [64]. Noteworthily, the presence of Arg101 is characteristic of the RNase 4 lineage, whereas RNase A and RNase 6 have a lysine at this position which is pointing in the opposite direction.
Table 3. Catalytic activity ratio of RNases for the assayed nucleotides.
ND: not detected at the assayed conditions. *Data for Poly(U):Poly(A) were taken from [76].
| CpA/UpA | CpA/C>p | Poly(C)/poly(U) | Poly(U)/poly(U):poly(A) | |
|---|---|---|---|---|
| RNase 6 | 0.18 | 5.5×102 | 1.02 | 0.43 |
| RNase 3 | 0.72 | 3.0×102 | 1.74 | ND |
| RNase A* | 1.20 | 2.1×103 | 8.14 | 307 |
Next, we analysed the RNase 6 structure at the secondary base site (B2). Conservation of Asn68 and Asn64 (Asn71 and Asn67 in RNase A respectively) is observed. Notwithstanding, the presence of a non-conserved cationic residue at position 66 might alter significantly the region. Additionally, although there is a conservative substitution for Glu111 in RNase A (Asp107 in RNase 6), the distinct loop orientation, due to the presence of a proline residue in RNase 6, modifies considerably the region's putative interactions. Altogether, the B2 site architecture would provide reduced binding interactions to the purine base, retaining a preference for adenine compared with guanine, as confirmed by our kinetic results (Tables 2 and 3, and Supplementary Table S4) and reported for most of the other tested mammalian RNase A family members [65–67]. Finally, we searched the RNase 6 structure for phosphate-binding sites that could contribute to the recognition of an RNA polymeric substrate, as described for RNase A [19]. Towards this aim, the protein nucleotide-binding mode was further analysed by molecular modelling.
Structure analysis by molecular modelling simulations
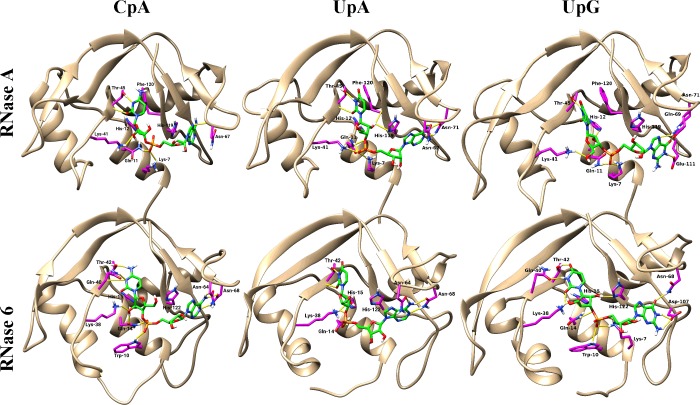
MD simulations were performed to predict the overall RNase 6 substrate-binding mode. Three independent runs were carried out for a total of 20 ns. All ligand positions were fully stabilized after a time lapse of 5 ns, showing a final average RMSD ranging from 0.15 to 0.3 nm. First of all, MD simulations were applied to RNase dinucleotide complexes taking as a model reference the RNase A–d(CpA) crystal structure [37]. Results indicated that the CpA and UpA dinucleotides could accommodate in a similar orientation into the RNase 6 catalytic cleft, showing no significant displacement from the original location (Figure 4). A close inspection of predicted RNase 6–dinucleotide interactions corroborated some of the structural features inferred from the crystal structure analysis. Equivalent interactions at the phosphate position suggested a conserved binding mode at the main phosphate-binding site (p1). Noteworthily, although the starting position of His122 in the RNase 6 crystal was found in the inactive conformation, the residue was adjusted to its active orientation before modelling, remaining in the favoured orientation for catalysis after all MD simulations. Interestingly, together with equivalent relative positioning of Gln14, His15 and His122, the presence in the RNase 6 active-site neighbourhood of Lys7 and Trp10 (Ala4 and Lys7 in RNase A) would account for significant differences at the p1 environment.
Figure 4. Predicted RNase 6 and RNase A structures in complex with dinucleotides.
Predicted structure of RNase 6 and RNase A in complex with CpA, UpA and UpG dinucleotides after MD simulations, as detailed in the Materials and methods section. Nucleotides are coloured green. RNases interacting residues are coloured magenta. Hydrogen bonds are coloured yellow. Structures were drawn with UCSF Chimera 1.10 [77].
The binding pattern described for RNase A was also mostly conserved for RNase 6 at the main pyrimidine base site (B1). Leu123 in RNase 6 was observed to partially supply the Phe120-stabilizing role in RNase A, but might induce a minor tilt of the ring plane. Interestingly, together with the conserved bidentate hydrogen bond between Thr42 and the pyrimidine base, the close-by Gln40 could directly interact with the Asp80 and the N3 atom of the pyrimidine base.
On the other hand, the comparison between the secondary base-binding site (B2) of both RNases showed minor differences in the relative position of the adenine base, but a more pronounced base displacement for the guanine-containing dinucleotides. Equivalent hydrogen bond interactions to adenine were found in RNase 6 for Asn64 and Asn68 (Asn67 and Asn71 in RNase A), together with base-stacking interactions with His122 (His119 in RNase A). However, although the main determinants for adenine binding were conserved, we observed how the presence of Arg66, unique to RNase 6, is significantly altering the environment. Indeed, in all of our predicted protein–dinucleotide complexes, we observed a salt bridge between Arg66 and Asp107, which shifted the aspartate position from the corresponding RNase A anionic residue (Glu111).
In particular, a major displacement from the original position was observed for the UpG dinucleotide in complex with both RNases. MD simulations indicated that the guanine base could not properly fit into the B2 site, promoting non-canonical orientations, where the guanine base was partially displaced. Moreover, in both RNases, the bidentate hydrogen bond to Asn68 (Asn71 in RNase A) was partially lost. Variability among replicates in the dinucleotide positioning reminded the reported productive and non-productive binding mode for the crystallographic complexes of RNase A with 2′5′-UpG [68]. Additionally, interactions in both RNases between the guanine C2 amino group and the Asp107 (Glu111 in RNase A) residue would fix the base in a less favoured orientation, pushing away the phosphate from the main active site. Nonetheless, the relative UpG displacement from the original position was less pronounced in the predicted RNase A complex, probably due to the contribution of Phe120-stacking interactions at B1, that provide a better fixation of the pyrimidine base. On its side, the presence in RNase 6 of Lys7 close to the active-site environment would favour the displacement of the phosphate towards the p2 region.
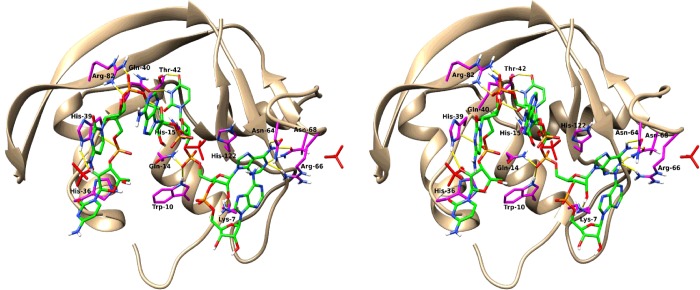
Complementarily, to gain further insight into the protein nucleotide-binding mode we modelled an heptanucleotide complex taking the RNase A–d(ApTpApA) crystal structure [45] as a starting reference model (Figure 5). The oligonucleotide fitted nicely into the enzyme active cleft showing only significant differences from the RNase A-binding mode at the secondary substrate-binding sites. Figure 5 highlights the protein residues at hydrogen bond distance observed in the protein–heptanucleotide complex. Putative RNase 6 substrate-binding sites were ascribed for base and phosphate recognition (Supplementary Table S5). Together with conserved equivalent sites to RNase A, the predicted complex illustrated the presence in RNase 6 structure of unique interacting residues at the RNA 5′ end. A novel specific region at p−2 and p−3 sites would be conformed by His36, His39 and Lys87. In addition, stacking interactions between His36 and the base located at the B−2 position were also identified in the model.
Figure 5. Stereo view of predicted RNase 6 heptanucleotide complex.
RNase 6 in complex with CCCAUAA heptanucleotide after a MD simulation, as described in the Materials and methods section. The heptanucleotide is coloured green. Interacting residues of RNase 6 are coloured turquoise. Protein-interacting residues and ligand atoms are coloured according to their element. Hydrogen bonds are coloured yellow. Overlapped sulfate ions of the original coordinates of the crystal are coloured magenta. The structures were drawn with UCSF Chimera 1.10 [77].
Kinetic characterization of RNase 6
Next, we determined the RNase 6 enzymatic activity against a variety of RNA substrates to correlate the protein structure with its enzymatic properties. RNase 6 catalytic efficiency ratio was compared with RNase 3 and RNase A (Supplementary Table S4), as representative family members for low and high catalytic efficiency respectively [18,23,66].
First, kinetic parameters for RNase 6 were calculated for dinucleotide and cyclic mononucleotide substrates (Table 2). Comparison of Km and kcat values for cyclic mono- and di-nucleotides indicated that most of the decrease in the catalytic efficiency of RNase 6 in comparison with RNase A (from 100- to 1000-fold) is due to a reduction in the catalytic constant value. A side-by-side comparison of substrate relative ratio (Table 3) highlighted for RNase 6 a preference for uridine at the B1 site and selectivity for adenine at B2. Additionally, the enzyme activity was assayed against polymeric substrates (Table 3 and Supplementary Table S4). RNase 6 displayed a low catalytic efficiency for polynucleotides in comparison with RNase A (Supplementary Figure S2), showing a similar reduced relative catalytic activity as previously reported for RNase 3 [20]. Interestingly, we observed that RNase 6 had the ability to degrade the double stranded poly(U):poly(A) substrate, a property not shared by RNase 3.
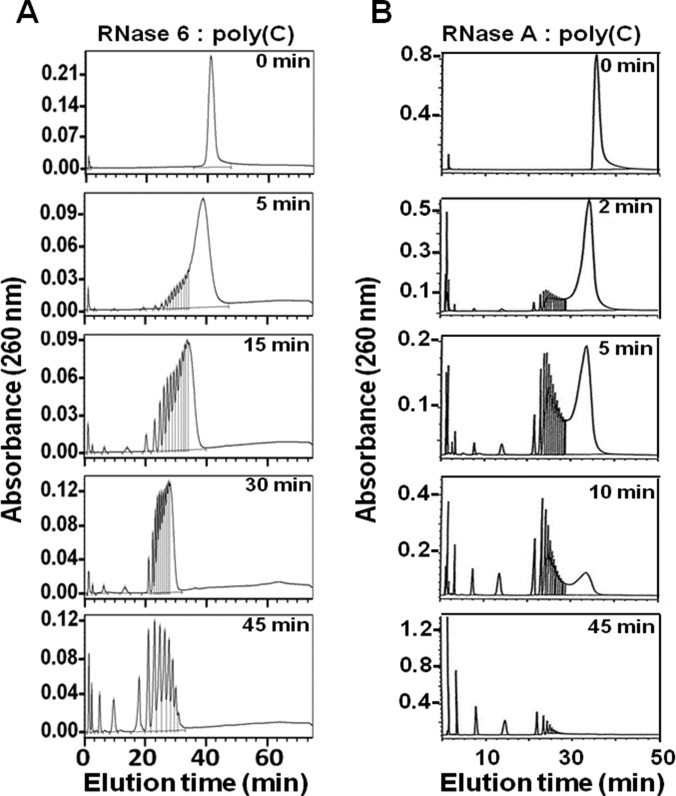
Complementarily, to evaluate the contribution of the RNase 6-specific subsite arrangement, we analysed the enzyme poly(C) cleavage pattern, as a model of polymeric substrate (Figure 6). The oligonucleotides obtained from the substrate digestion were eluted by reverse-phase chromatography, where the mononucleotide fraction is eluted at the initial conditions and oligonucleotides of increasing size are eluted at increasing retention times. The previously optimized methodology distinguishes between consecutive sizes up to eight or nine nucleotides, whereas the peaks corresponding to higher-molecular-mass components contain more than one oligonucleotide size due to the column discrimination power limitations [29]. Our previous work on RNase A cleavage pattern showed a decrease in the initial poly(C) peak followed by the formation of intermediate oligonucleotides with an average size of around six or seven residues, a pattern that supported the enzyme multisubsite structure providing a characteristic endonuclease-type activity [29]. Noteworthily, RNase 6 displayed a characteristic digestion profile differentiated from that previously obtained for RNase A [27], where there is first the formation of considerably large intermediates which are subsequently digested, giving rise to relatively shorter intermediates. Interestingly, the decrease in the original polynucleotide substrate was enhanced for RNase 6 at short incubation times. On the other hand, the predominance of smaller oligonucleotides did not take place until most of the high-molecular-mass polymeric substrate has been degraded, revealing a singularized cleavage pattern, that could be ascribed to a pronounced endonuclease mechanism.
Figure 6. Analysis of polynucleotide cleavage pattern by RNase 6 and RNase A.
Poly(C) cleavage pattern obtained by RNase 6 (A) compared with RNase A [27] (B). Chromatography profiles of poly(C) digestion products are shown at selected incubation times corresponding to representative steps of the catalysis process. See the Materials and methods section for substrate digestion conditions.
Kinetic characterization of RNase 6 mutants
Analysis of the RNase 6 three-dimensional structure and molecular modelling predictions suggested that the enzyme facility to cleave polymeric substrates could be related to the presence of surface-exposed cationic residues that might facilitate the RNA anchorage and degradation. Noteworthily, the presence in RNase 6 of a histidine pair (His36/His39) (Figure 1 and Supplementary Figure S3) that adopts a configuration equivalent to the His15/His122 dyad at the RNase main active site, suggested the existence of a secondary active centre (Figure 3). To evaluate this hypothesis, we designed mutant variants at His15 and His36. First, the enzyme main active site was removed by His15 mutation to alanine. His15 in RNase 6 was selected as the RNase A His12 counterpart, where the corresponding H12A mutant was reported by Raines and co-workers to totally abolish the RNase A catalytic activity [69]. Additionally, His36 was mutated to arginine to remove the putative secondary catalytic histidine residue while retaining an exposed cationic charge. In addition, most RNase A family members, including RNase 7, the closest RNase 6 homologue, display an arginine residue at an equivalent position (Figure 1 and Supplementary Figure S3). Eventually, a double mutant (His15A/His36A) was engineered to evaluate simultaneously the removal of the main and secondary active sites.
Kinetic characterization of point mutants confirmed the key role of both His15 and His36 (Table 4). The contribution of His36 was found to be mostly critical for polymeric substrates. On the other hand, we observed how the RNase 6-H15A mutant was retaining a significantly high activity for polymeric substrates (approximately 35–40% relative activity respect to wild-type RNase). Interestingly, for the assayed dinucleotides, a residual relative activity of approximately 15% was observed, which was abolished by the double mutation. Moreover, the RNase 6-H15A activity was further compared with the corresponding RNase 7-H15A mutant (Table 4). The complete abolishment of activity for the RNase 7 active site mutant for dinucleotides, in contrast with RNase 6-H15A, corroborated the hypothesis. On the other hand, a remnant activity for polynucleotides is observed for both the RNase 6 double mutant and the RNase 7-H15A mutant. Likewise, residual catalytic activity for the RNase A-H12K/H119Q mutant was attributed to be solely promoted by the RNA structure distortion induced by the enzyme interaction [49].
Table 4. Relative catalytic activities for wild-type RNases 6 and 7 and mutant variants.
ND: not detected at the assayed conditions. *Data expressed in percentage of activity in relation to the wild-type protein (%). Mean values were calculated from triplicate assays, showing in all cases a standard error below 10%.
| UpA | CpA | Poly(U) | Poly(U):poly(A) | |
|---|---|---|---|---|
| RNase 6 | 100 | 100 | 100 | 100 |
| RNase 6-H15A* | 13 | 14 | 77 | 63 |
| RNase 6-H36R | 94 | 86 | 36 | 40 |
| RNase 6-H15A/H36A | ND | ND | 15 | 5 |
| RNase 7 | 100 | 100 | 100 | 100 |
| RNase 7-H15A | ND | ND | 3 | 7 |
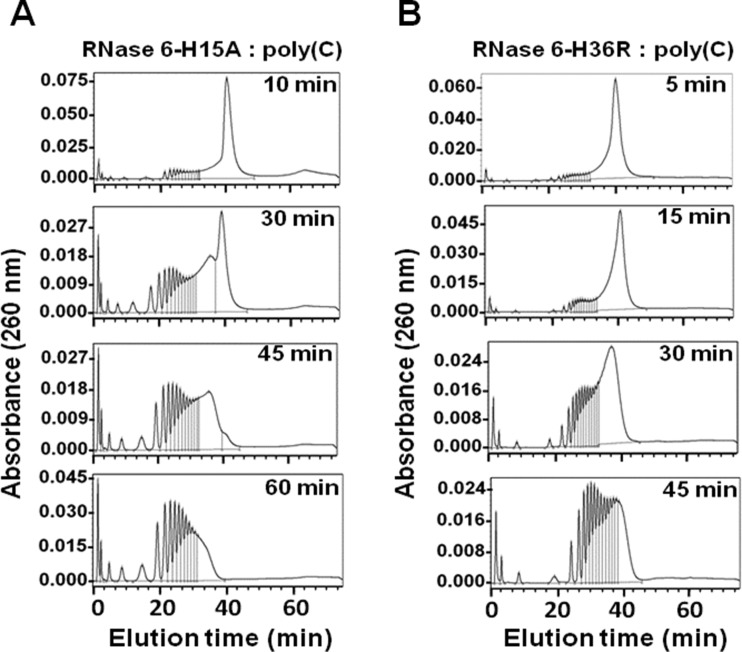
Complementarily, the analysis of RNase 6 activity on polymeric substrates was also assayed on an activity staining gel and by the analysis of the polynucleotide digestion products. The side-by-side comparison of the polynucleotide cleavage product profiles obtained by incubation with both RNase 6-H15A and RNase 7-H15A mutants, devoid of the main active-site histidine residue, confirmed the presence in RNase 6 of another cleavage site. We observed that, using the same assay conditions, a considerable amount of activity is achieved with the RNase 6-H15A mutant, whereas non-significant activity is observed for the corresponding RNase 7-H15A mutant (Supplementary Figure S4). Comparison of both RNases activity on poly(C) together with RNase 6 mutants was also analysed by the zymogram technique (Supplementary Figure S2), where the RNase 6 double mutant displayed no detectable activity at even a 5-fold protein concentration. Interestingly, when comparing the poly(C) digestion pattern along with the His15 and 36 respective mutants’ profiles we observe how both mutants shifted their product elution profile towards a more exonuclease-type pattern, accumulating shorter intermediates than the native enzyme at an earlier stage of the reaction. A similar profile progression was previously reported for RNase 3, described to present a subsites arrangement that would favour an exonuclease cleavage pattern [20].
DISCUSSION
We report in the present paper the first crystal structure RNase 6 in complex with sulfate anions (Table 1). The location of putative phosphate-binding sites was deduced from the position of the sulfate anions in the crystal structure (Figure 2 and Supplementary Table S3). Structural analysis and kinetic characterization were carried out to outline the protein nucleotide-binding sites arrangement. The first sulfate was ascribed to the main RNase phosphate-binding site, shared with the other RNase A family members [54,55,67]. Two additional sulfate anions were located at the protein-exposed cationic residues (His36/His39 and Arg66/His67) and would correspond to secondary phosphate-binding sites. In particular, the His36/His39 site is proposed as a novel and unique site within the RNase A family (Figures 1 and 3). Kinetic studies corroborated the involvement of His36 in enzyme catalysis (Table 4).
The contribution of RNase 6 novel secondary catalytic site in the catalytic mechanism was further analysed by the characterization of the enzyme polynucleotide cleavage pattern (Figure 6). The distribution of substrate digestion products indicated that the breakdown of the polymeric substrate was not a random process. The chromatography profiles revealed the preference of RNase 6 for the binding and cleavage of long RNA strands, where the broken phosphodiester bonds would be located considerably spaced apart from the end of the chain. By comparison with the previously characterized RNase A cleavage pattern [29], we concluded that the RNase 6 activity towards the polymeric substrate was even more endonucleolytic than the previously described for RNase A [19]. On the other hand, we observed how the poly(C) digestion profile underwent a pronounced shift in its cleavage pattern towards an exonuclease-type propensity when the main active-site histidine was mutated (Figure 7).
Figure 7. Analysis of polynucleotide cleavage pattern by RNase 6 mutants.
Poly(C) cleavage pattern by RNase 6-H15A (A) and RNase 6-H36R (B) mutants. Chromatography profiles of poly(C) digestion products are shown at selected incubation times corresponding to representative steps of the catalysis process.
Complementarily, structural analysis also illustrated a well-defined substrate multisubsite arrangement for RNase 6. Interestingly, the predicted RNase 6 complex with a heptanucleotide by MD (Figure 5) revealed the presence of novel sites at the RNA 5′ end that would be ascribed to B−2 and p−2/p−3 sites (Supplementary Table S5).
Interestingly, the RNase 6 structure revealed equivalent histidine ND1 to NE2 atomic distances for His15/His122 and His36/His39, suggesting the presence of a secondary catalytic site. In addition, we observed stacking interactions between the B−2 base of the oligonucleotide and His39 in the predicted complex. However, structural analysis identified no additional residues for binding of adjacent bases at both phosphate sides. Notwithstanding, kinetic data on dinucleotides indicated a reduced efficiency performance for this novel active site. Indeed, the percentage of remnant catalytic activity displayed by the RNase 6-H15A mutant is similar to the activity displayed by a previously engineered RNase A construct with an additional active site created at the p2 phosphate-binding site position (RNase A-H7H10 variant) [49]. Likewise, a similar scenario might be envisaged for RNase 6 (see Supplementary Table S6 for comparison of estimated pKa values for both RNase A His7/His10 and RNase 6 His36/His39 pairs). The calculated pKa values for the corresponding histidine residues exposed at the protein surface indicated that neither of them underwent a significant decrease from the reference value that could provide an efficient base catalyst. Therefore, the predicted pKa for His36 and His39 could not reproduce properly the efficient RNase A catalytic site [18,48]. And last, but not least, the presence of a third residue that could stabilize the transition state intermediate is not obvious. Interestingly, we find a neighbouring lysine residue (Lys87) at the novel site environment that could meet the required RNase active site geometry (see Figure 3 for a schematic illustration). Unfortunately, the Lys87 side chain is disordered in the present RNase 6 crystal structure, providing no information on its proper orientation. However, the Lys87 side-chain conformation predicted by MD in the RNase 6–heptanucleotide complex did provide interactions with a phosphate located at the p−3 site (Supplementary Table S5). Noteworthily, Lys87 is found in all primate RNase 6 counterparts, but is absent from the murine sequence [9,15]. Interestingly, even if the RNase A mutant at Lys41 was shown to drastically reduce the enzyme catalytic efficiency [70], the insertion of a secondary catalytic site for RNase A at the p2 location conformed by only a histidine dyad was also able to provide approximately 10–15% of remnant activity in the absence of the main active site [49]. Likewise, the proposed novel RNase 6 secondary site would behave as a poor catalyst. In any case, the present kinetic data indicate that the presence of an anchoring site at the 36–39 region enhances significantly the enzyme catalysis of polymeric substrates. Interestingly, Sorrentino and co-workers’ analysis of the enzymatic properties of RNase A family members suggested that the presence of a cationic cluster at that region, combined with an anionic residue at RNase A residue at the 83 position (Asp80 in RNase 6), correlated to the facility to destabilize dsRNA, exposing single-stranded stretches to the enzyme for cleavage [65,71,72]. Interestingly, RNase 6 shows a particular overabundance of histidine residues at its polypeptide sequence, in comparison with the other family homologues. As previously mentioned, most of these histidine residues are unique to the RNase 6 lineage (Figure 1B) [9,15]. In particular, we observed two cationic clusters (His36/His39) and (Arg66/His67) fully conserved among the more evolved primate members. To note, whereas His36 and His67 are found in all known primate primary structures, His39 and Arg66 are only present in the more evolved primates. Overall, evolutionary pressure would have drifted RNase 6 towards a slightly more cationic primary structure with an overabundance of histidine residues, providing a novel secondary active site. Future research should explore the ultimate implications in the protein's physiological function.
Conclusions
The RNase 6 first crystal structure has provided us the opportunity to explore its structure–function relationship. By combining structural analysis together with molecular modelling and kinetic characterization, we were able to spot key regions contributing to the enzyme's substrate specificity and catalytic properties. Results highlighted the RNase 6 multisubsite arrangement for substrate binding and the contribution of a secondary catalytic site that facilitates the cleavage of polynucleotide substrates. Further work is needed to fully characterize RNase 6's enzymatic properties towards the understanding of its specific mechanism of action.
Acknowledgments
We thank all of the staff at the beamline BL13 (XALOC) at the ALBA Synchrotron Light Facility (Cerdanyola del Vallès, Spain) for their support during data collection. Heartfelt thanks go to Jordi Joanhuix and Fernando Gil for all of the help provided.
Abbreviations
- C>p
cytidine 2′,3′-cyclic phosphate
- poly(A)
polyadenylic acid
- poly(C)
polycytidylic acid
- poly(U)
polyuridylic acid
AUTHOR CONTRIBUTION
Ester Boix and Mohammed Moussaoui conceived and designed the experimental work. Guillem Prats-Ejarque, Javier Arranz-Trullen, Jose Blanco, Mohammed Moussaoui and David Pulido performed the experiments. Guillem Prats-Ejarque, Mohammed Moussaoui, David Pulido, Victòria Nogués and Ester Boix analysed the data. Guillem Prats-Ejarque, Javier Arranz-Trullén and Ester Boix drafted the paper. Victòria Nogués, Mohammed Moussaoui, Guillem Prats-Ejarque and Ester Boix revised the final paper.
FUNDING
This work was supported by the Ministerio de Economía y Competitividad cofinanced by FEDER funds [grant numbers BFU2012-38695 and BES-2010-036238 (predoctoral fellowship to J.A.B.)]; the Generalitat de Catalunya [grant number 2014-SGR-728]; and the Universitat Autònoma de Barcelona [grant number 406-02-02/2013 (predoctoral fellowship to J.A.)].
References
- 1.Boix E., Nogués M.V. Mammalian antimicrobial proteins and peptides: overview on the RNase A superfamily members involved in innate host defence. Mol. Biosyst. 2007;3:317–335. doi: 10.1039/b617527a. [DOI] [PubMed] [Google Scholar]
- 2.Rosenberg H.F. RNase A ribonucleases and host defense: an evolving story. J. Leukoc. Biol. 2008;83:1079–1087. doi: 10.1189/jlb.1107725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gupta S.K., Haigh B.J., Griffin F.J., Wheeler T.T. The mammalian secreted RNases: mechanisms of action in host defence. Innate Immun. 2012;19:86–97. doi: 10.1177/1753425912446955. [DOI] [PubMed] [Google Scholar]
- 4.Zhang J., Rosenberg H.F. Complementary advantageous substitutions in the evolution of an antiviral RNase of higher primates. Proc. Natl. Acad. Sci. U.S.A. 2002;99:5486–5491. doi: 10.1073/pnas.072626199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pizzo E., D'Alessio G. The success of the RNase scaffold in the advance of biosciences and in evolution. Gene. 2007;406:8–12. doi: 10.1016/j.gene.2007.05.006. [DOI] [PubMed] [Google Scholar]
- 6.Hamann K.J., Ten R.M., Loegering D.A., Jenkins R.B., Heise M.T., Schad C.R., Pease L.R., Gleich G.J., Barker R.L. Structure and chromosome localization of the human eosinophil-derived neurotoxin and eosinophil cationic protein genes: evidence for intronless coding sequences in the ribonuclease gene superfamily. Genomics. 1990;7:535–546. doi: 10.1016/0888-7543(90)90197-3. [DOI] [PubMed] [Google Scholar]
- 7.Zhang J., Dyer K.D., Rosenberg H.F. RNase 8, a novel RNase A superfamily ribonuclease expressed uniquely in placenta. Nucleic Acids Res. 2002;30:1169–1175. doi: 10.1093/nar/30.5.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rosenberg H.F., Dyer K.D. Molecular cloning and characterization of a novel human ribonuclease (RNase k6): increasing diversity in the enlarging ribonuclease gene family. Nucleic Acids Res. 1996;24:3507–3513. doi: 10.1093/nar/24.18.3507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deming M.S., Dyer K.D., Bankier A.T., Piper M.B., Dear P.H., Rosenberg H.F. Ribonuclease k6: chromosomal mapping and divergent rates of evolution within the RNase A gene superfamily. Genome Res. 1998;8:599–607. doi: 10.1101/gr.8.6.599. [DOI] [PubMed] [Google Scholar]
- 10.Becknell B., Eichler T.E., Beceiro S., Li B., Easterling R.S., Carpenter A.R., James C.L., McHugh K.M., Hains D.S., Partida-Sanchez S., Spencer J.D. Ribonucleases 6 and 7 have antimicrobial function in the human and murine urinary tract. Kidney Int. 2015;87:151–161. doi: 10.1038/ki.2014.268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Spencer J.D., Schwaderer A.L., Wang H., Bartz J., Kline J., Eichler T., DeSouza K.R., Sims-Lucas S., Baker P., Hains D.S. Ribonuclease 7, an antimicrobial peptide upregulated during infection, contributes to microbial defense of the human urinary tract. Kidney Int. 2013;83:615–625. doi: 10.1038/ki.2012.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jelicic K., Cimbro R., Nawaz F., Huang D.W., Zheng X., Yang J., Lempicki R.A., Pascuccio M., Van Ryk D., Schwing C., et al. The HIV-1 envelope protein gp120 impairs B cell proliferation by inducing TGF-beta1 production and FcRL4 expression. Nat. Immunol. 2013;14:1256–1265. doi: 10.1038/ni.2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang J., Dyer K.D., Rosenberg H.F. Evolution of the rodent eosinophil-associated RNase gene family by rapid gene sorting and positive selection. Proc. Natl. Acad. Sci. U.S.A. 2000;97:4701–4706. doi: 10.1073/pnas.080071397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McDevitt A.L., Deming M.S., Rosenberg H.F., Dyer K.D. Gene structure and enzymatic activity of mouse eosinophil-associated ribonuclease 2. Gene. 2001;267:23–30. doi: 10.1016/S0378-1119(01)00392-4. [DOI] [PubMed] [Google Scholar]
- 15.Dyer K.D., Rosenberg H.F., Zhang J. Isolation, characterization, and evolutionary divergence of mouse RNase 6: evidence for unusual evolution in rodents. J. Mol. Evol. 2004;59:657–665. doi: 10.1007/s00239-004-2657-0. [DOI] [PubMed] [Google Scholar]
- 16.Richards F.M., Wyckoff H.W. Bovine pancreatic ribonuclease. Enzymes IV. 1971:647–806. doi: 10.1016/S1874-6047(08)60384-4. [DOI] [Google Scholar]
- 17.Raines R.T. Ribonuclease A. Chem. Rev. 1998;98:1045–1065. doi: 10.1021/cr960427h. [DOI] [PubMed] [Google Scholar]
- 18.Cuchillo C.M., Nogués M.V., Raines R.T. Bovine pancreatic ribonuclease: fifty years of the first enzymatic reaction mechanism. Biochemistry. 2011;50:7835–7841. doi: 10.1021/bi201075b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nogués M.V., Moussaoui M., Boix E., Vilanova M., Ribó M., Cuchillo C.M. The contribution of noncatalytic phosphate-binding subsites to the mechanism of bovine pancreatic ribonuclease A. Cell. Mol. Life Sci. 1998;54:766–774. doi: 10.1007/s000180050205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boix E., Nikolovski Z., Moiseyev G., Rosenberg H.F., Cuchillo C.M., Nogues M.V. Kinetic and product distribution analysis of human eosinophil cationic protein indicates a subsite arrangement that favors exonuclease-type activity. J. Biol. Chem. 1999;274:15605–15614. doi: 10.1074/jbc.274.22.15605. [DOI] [PubMed] [Google Scholar]
- 21.Rosenberg H.F. Recombinant human eosinophil cationic protein. Ribonuclease activity is not essential for cytotoxicity. J. Biol. Chem. 1995;270:7876–7881. doi: 10.1074/jbc.270.14.7876. [DOI] [PubMed] [Google Scholar]
- 22.Harder J., Schroder J.-M. RNase 7, a novel innate immune defense antimicrobial protein of healthy human skin. J. Biol. Chem. 2002;277:46779–46784. doi: 10.1074/jbc.M207587200. [DOI] [PubMed] [Google Scholar]
- 23.Boix E., Salazar V.A., Torrent M., Pulido D., Nogués M.V., Moussaoui M. Structural determinants of the eosinophil cationic protein antimicrobial activity. Biol. Chem. 2012;393:801–815. doi: 10.1515/hsz-2012-0160. [DOI] [PubMed] [Google Scholar]
- 24.Torrent M., Sanchez D., Buzon V., Nogues M.V, Cladera J., Boix E. Comparison of the membrane interaction mechanism of two antimicrobial RNases: RNase 3/ECP and RNase 7. Biochim. Biophys. Acta. 2009;1788:1116–1125. doi: 10.1016/j.bbamem.2009.01.013. [DOI] [PubMed] [Google Scholar]
- 25.Libonati M., Sorrentino S. Degradation of double-stranded RNA by mammalian pancreatic-type ribonucleases. Methods Enzymol. 2001;341:234–248. doi: 10.1016/S0076-6879(01)41155-4. [DOI] [PubMed] [Google Scholar]
- 26.Bravo J., Fernández E., Ribó M., Dellorens R., Cuchillo C.M. A versatile negative-staining ribonuclease zymogram. Anal. Biochem. 1994;219:82–86. doi: 10.1006/abio.1994.1234. [DOI] [PubMed] [Google Scholar]
- 27.Moussaoui M., Guasch A., Boix E., Cuchillo C.M., Nogués M.V. The role of non-catalytic binding subsites in the endonuclease activity of bovine pancreatic ribonuclease A. J. Biol. Chem. 1996;271:4687–4692. doi: 10.1074/jbc.271.9.4687. [DOI] [PubMed] [Google Scholar]
- 28.Nogues M.V., Cuchillo C.M. Analysis by HPLC of distributive activities and the synthetic (back) reaction of pancreatic-type ribonucleases. Methods Mol. Biol. 2001;160:15–24. doi: 10.1385/1-59259-233-3:015. [DOI] [PubMed] [Google Scholar]
- 29.Cuchillo C.M., Moussaoui M., Barman T., Travers F., Nogues M.V. The exo- or endonucleolytic preference of bovine pancreatic ribonuclease A depends on its subsites structure and on the substrate size. Protein Sci. 2002;11:117–128. doi: 10.1110/ps.ps.13702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kabsch W. XDS. Acta Crystallogr. D Biol. Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang Y.C., Lin Y.M., Chang T.W., Wu S.H., Lee Y.S., Chang M.D., Chen C., Wu S.J., Liao Y.D. The flexible and clustered lysine residues of human ribonuclease 7 are critical for membrane permeability and antimicrobial activity. J. Biol. Chem. 2007;282:4626–4633. doi: 10.1074/jbc.M607321200. [DOI] [PubMed] [Google Scholar]
- 32.Adams P.D., Afonine P.V, Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.-W., Kapral G.J., Grosse-Kunstleve R.W., et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 34.Vaguine A.A., Richelle J., Wodak S.J. SFCHECK: a unified set of procedures for evaluating the quality of macromolecular structure-factor data and their agreement with the atomic model. Acta Crystallogr. D Biol. Crystallogr. 1999;55:191–205. doi: 10.1107/S0907444998006684. [DOI] [PubMed] [Google Scholar]
- 35.Hooft R.W., Vriend G., Sander C., Abola E.E. Errors in protein structures. Nature. 1996;23:381. doi: 10.1038/381272a0. [DOI] [PubMed] [Google Scholar]
- 36.Pronk S., Páll S., Schulz R., Larsson P., Bjelkmar P., Apostolov R., Shirts M.R., Smith J.C., Kasson P.M., Van Der Spoel D., et al. GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics. 2013;29:845–854. doi: 10.1093/bioinformatics/btt055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zegers I., Maes D., Poortmans F., Palmer R., Wyns L. The structures of RNase A complexed with 3′ -CMP and d(CpA): active site conformation and conserved water molecules. Protein Sci. 1994;3:2322–2339. doi: 10.1002/pro.5560031217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lindorff-Larsen K., Piana S., Palmo K., Maragakis P., Klepeis J.L., Dror R.O., Shaw D.E. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins. 2010;78:1950–1958. doi: 10.1002/prot.22711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jorgensen W.L., Chandrasekhar J., Madura J.D., Impey R.W., Klein M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926. doi: 10.1063/1.445869. [DOI] [Google Scholar]
- 40.Darden T., York D., Pedersen L. Particle mesh Ewald: an N·log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089. doi: 10.1063/1.464397. [DOI] [Google Scholar]
- 41.Essmann U., Perera L., Berkowitz M.L., Darden T., Lee H., Pedersen L.G. A smooth particle mesh Ewald method. J. Chem. Phys. 1995;103:8577. doi: 10.1063/1.470117. [DOI] [Google Scholar]
- 42.Hess B. P-LINCS: a parallel linear constraint solver for molecular simulation. J. Chem. Theory Comput. 2008;4:116–122. doi: 10.1021/ct700200b. [DOI] [PubMed] [Google Scholar]
- 43.Parrinello M., Rahman A. Polymorphic transitions in single crystals: a new molecular dynamics method. J. Appl. Phys. 1981;52:7182–7190. doi: 10.1063/1.328693. [DOI] [Google Scholar]
- 44.Nosé S., Klein M.L. Constant pressure molecular dynamics for molecular systems. Mol. Phys. 1983;50:1055–1076. doi: 10.1080/00268978300102851. [DOI] [Google Scholar]
- 45.Fontecilla-Camps J.C., de Llorens R., le Du M.H., Cuchillo C.M. Crystal structure of ribonuclease A.d(ApTpApApG) complex. Direct evidence for extended substrate recognition. J. Biol. Chem. 1994;269:21526–21531. doi: 10.2210/pdb1rcn/pdb. [DOI] [PubMed] [Google Scholar]
- 46.Morris G.M., Huey R., Lindstrom W., Sanner M.F., Belew R.K., Goodsell D.S., Olson A.J. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comput. Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kilambi K.P., Gray J.J. Rapid calculation of protein pKa values using Rosetta. Biophys. J. 2012;103:587–595. doi: 10.1016/j.bpj.2012.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fisher B.M., Schultz L.W., Raines R.T. Coulombic effects of remote subsites on the active site of ribonuclease A. Biochemistry. 1998;37:17386–17401. doi: 10.1021/bi981369s. [DOI] [PubMed] [Google Scholar]
- 49.Moussaoui M., Cuchillo C.M., Nogués M.V. A phosphate-binding subsite in bovine pancreatic ribonuclease A can be converted into a very efficient catalytic site. Protein Sci. 2007;16:99–109. doi: 10.1110/ps.062251707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Krissinel E., Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 51.Rico M., Gallego E., Santoro J., Bermejo F.J., Nieto J.L., Herranz J. On the fundamental role of the Glu 2- … Arg 10+ salt bridge in the folding of isolated ribonuclease A S-peptide. Biochem. Biophys. Res. Commun. 1984;123:757–763. doi: 10.1016/0006-291X(84)90294-8. [DOI] [PubMed] [Google Scholar]
- 52.Chatani E., Hayashi R. Functional and structural roles of constituent amino acid residues of bovine pancreatic ribonuclease A. J. Biosci. Bioeng. 2001;92:98–107. doi: 10.1016/S1389-1723(01)80208-5. [DOI] [PubMed] [Google Scholar]
- 53.Berisio R., Sica F., Lamzin V.S., Wilson K.S., Zagari A., Mazzarella L. Atomic resolution structures of ribonuclease A at six pH values. Acta Crystallogr. D Biol. Crystallogr. 2002;58:441–450. doi: 10.1107/S0907444901021758. [DOI] [PubMed] [Google Scholar]
- 54.Fedorov A.A., Joseph-McCarthy D., Fedorov E., Sirakova D., Graf I., Almo S.C. Ionic interactions in crystalline bovine pancreatic ribonuclease A. Biochemistry. 1996;35:15962–15979. doi: 10.1021/bi961533g. [DOI] [PubMed] [Google Scholar]
- 55.Leonidas D.D., Boix E., Prill R., Suzuki M., Turton R., Minson K., Swaminathan G.J., Youle R.J., Acharya K.R. Mapping the ribonucleolytic active site of eosinophil-derived neurotoxin (EDN): high resolution crystal structures of EDN complexes with adenylic nucleotide inhibitors. J. Biol. Chem. 2001;276:15009–15017. doi: 10.1074/jbc.M010585200. [DOI] [PubMed] [Google Scholar]
- 56.Boix E., Pulido D., Moussaoui M., Nogues M.V., Russi S. The sulfate-binding site structure of the human eosinophil cationic protein as revealed by a new crystal form. J. Struct. Biol. 2012;179:1–9. doi: 10.1016/j.jsb.2012.04.023. [DOI] [PubMed] [Google Scholar]
- 57.Holloway D.E., Chavali G.B., Hares M.C., Subramanian V., Acharya K.R. Structure of murine angiogenin: features of the substrate- and cell-binding regions and prospects for inhibitor-binding studies. Acta Crystallogr. D Biol. Crystallogr. 2005;61:1568–1578. doi: 10.1107/S0907444905029616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Borkakoti N. The active site of ribonuclease A from the crystallographic studies of ribonuclease-A-inhibitor complexes. Eur. J. Biochem. 1983;132:89–94. doi: 10.1111/j.1432-1033.1983.tb07329.x. [DOI] [PubMed] [Google Scholar]
- 59.Berisio R., Lamzin V.S., Sica F., Wilson K.S., Zagari A., Mazzarella L. Protein titration in the crystal state. J. Mol. Biol. 1999;292:845–854. doi: 10.1006/jmbi.1999.3093. [DOI] [PubMed] [Google Scholar]
- 60.deMel V.S., Martin P.D., Doscher M.S., Edwards B.F. Structural changes that accompany the reduced catalytic efficiency of two semisynthetic ribonuclease analogs. J. Biol. Chem. 1992;267:247–256. [PubMed] [Google Scholar]
- 61.Schultz L.W., Quirk D.J., Raines R.T. His…Asp catalytic dyad of ribonuclease A: structure and function of the wild-type, D121N, and D121A enzymes. Biochemistry. 1998;37:8886–8898. doi: 10.1021/bi972766q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hofsteenge J., Vicentini A., Zelenko O. Ribonuclease 4, an evolutionarily highly conserved member of the superfamily. Cell. Mol. Life Sci. 1998;54:804–810. doi: 10.1007/s000180050209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vicentini A.M., Kote-Jarai Z., Hofsteenge J. Structural determinants of the uridine-preferring specificity of RNase PL3. Biochemistry. 1996;35:9128–9132. doi: 10.1021/bi960457e. [DOI] [PubMed] [Google Scholar]
- 64.Terzyan S.S., Peracaula R., de Llorens R., Tsushima Y., Yamada H., Seno M., Gomis-Ruth F.X., Coll M. The three-dimensional structure of human RNase 4, unliganded and complexed with d(Up), reveals the basis for its uridine selectivity. J. Mol. Biol. 1999;285:205–214. doi: 10.1006/jmbi.1998.2288. [DOI] [PubMed] [Google Scholar]
- 65.Sorrentino S. Human extracellular ribonucleases: multiplicity, molecular diversity and catalytic properties of the major RNase types. Cell. Mol. Life Sci. 1998;54:785–794. doi: 10.1007/s000180050207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sorrentino S. The eight human “canonical” ribonucleases: molecular diversity, catalytic properties, and special biological actions of the enzyme proteins. FEBS Lett. 2010;584:2194–2200. doi: 10.1016/j.febslet.2010.04.018. [DOI] [PubMed] [Google Scholar]
- 67.Boix E., Blanco J.A., Nogués M.V., Moussaoui M. Nucleotide binding architecture for secreted cytotoxic endoribonucleases. Biochimie. 2013;95:1087–1097. doi: 10.1016/j.biochi.2012.12.015. [DOI] [PubMed] [Google Scholar]
- 68.Vitagliano L., Merlino A., Zagari A., Mazzarella L. Productive and nonproductive binding to ribonuclease A: X-ray structure of two complexes with uridylyl(2’,5')guanosine. Protein Sci. 2000;9:1217–1225. doi: 10.1110/ps.9.6.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Park C., Schultz L.W., Raines R.T. Contribution of the active site histidine residues of ribonuclease A to nucleic acid binding. Biochemistry. 2001;40:4949–4956. doi: 10.1021/bi0100182. [DOI] [PubMed] [Google Scholar]
- 70.Trautwein K., Holliger P., Stackhouse J., Benner S.A. Site-directed mutagenesis of bovine pancreatic ribonuclease: lysine-41 and aspartate-121. FEBS Lett. 1991;281:275–277. doi: 10.1016/0014-5793(91)80410-5. [DOI] [PubMed] [Google Scholar]
- 71.Libonati M., Sorrentino S. Revisiting the action of bovine ribonuclease A and pancreatic-type ribonucleases on double-stranded RNA. Mol. Cell. Biochem. 1992;117:139–151. doi: 10.1007/BF00230753. [DOI] [PubMed] [Google Scholar]
- 72.Yakovlev G., Moiseyev G.P., Sorrentino S., De Prisco R., Libonati M. Single-strand-preferring RNases degrade double-stranded RNAs by destabilizing its secondary structure. J. Biomol. Struct. Dyn. 1997;15:243–250. doi: 10.1080/07391102.1997.10508189. [DOI] [PubMed] [Google Scholar]
- 73.Shapiro R., Fett J.W., Strydom D.J., Vallee B.L. Isolation and characterization of a human colon carcinoma-secreted enzyme with pancreatic ribonuclease-like activity. Biochemistry. 1986;25:7255–7264. doi: 10.1021/bi00371a002. [DOI] [PubMed] [Google Scholar]
- 74.Follmann H., Wieker H.J., Witzel H. On the mechanism of the ribonuclease reaction. 2. The pre-ordering in the substrate as the accelerating factor in cinucleoside phosphates and analagous compounds. Eur. J. Biochem. 1967;1:243–250. doi: 10.1111/j.1432-1033.1967.tb00068.x. [DOI] [PubMed] [Google Scholar]
- 75.Boix E., Nogues M.V., Schein C.H., Benner S.A., Cuchillo C.M. Reverse transphosphorylation by ribonuclease A needs an intact p2-binding site. J. Biol. Chem. 1994;269:2529–2534. [PubMed] [Google Scholar]
- 76.Sorrentino S., Glitz D.G. Ribonuclease activity and substrate preference of human eosinophil cationic protein (ECP) FEBS Lett. 1991;288:23–26. doi: 10.1016/0014-5793(91)80994-E. [DOI] [PubMed] [Google Scholar]
- 77.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]