Abstract
MicroRNAs (miRNAs) control gene expression by binding to their target mRNAs for degradation and/or translation repression and are implicated in many aspects of cellular physiology. Our previous study shows that miR-29b acts as a biological repressor of intestinal mucosal growth, but its exact downstream targets remain largely unknown. In the present study, we found that mRNAs, encoding Wnt co-receptor LRP6 (low-density lipoprotein-receptor-related protein 6) and RNA-binding protein (RBP) HuR, are novel targets of miR-29b in intestinal epithelial cells (IECs) and that expression of LRP6 and HuR is tightly regulated by miR-29b at the post-transcriptional level. miR-29b interacted with both Lrp6 and HuR mRNAs via their 3′-UTRs and inhibited LRP6 and HuR expression by destabilizing Lrp6 and HuR mRNAs and repressing their translation. Studies using heterologous reporter constructs revealed a greater repressive effect of miR-29b through a single binding site in the Lrp6 or HuR 3′-UTR, whereas deletion mutation of this site prevented miR-29b-induced repression of LRP6 and HuR expression. Repression of HuR by miR-29b in turn also contributed to miR-29b-induced LRP6 inhibition, since ectopic overexpression of HuR in cells overexpressing miR-29b restored LRP6 expression to near normal levels. Taken together, our results suggest that miR-29b inhibits expression of LRP6 and HuR post-transcriptionally, thus playing a role in the regulation of IEC proliferation and intestinal epithelial homoeostasis.
Keywords: intestinal epithelial homoeostasis, mRNA stability, non-coding RNAs, translation, Wnt signalling
INTRODUCTION
In response to stressful environmental conditions, post-transcriptional regulation, particularly altered mRNA stability and translation, is a major mechanism by which mammalian cells control gene expression [1,2]. mRNAs are targeted for rapid degradation and/or translational repression through a process involving the interaction of specific mRNA sequences (cis-elements) with specific trans-acting factors such as microRNAs (miRNAs) and RNA-binding proteins (RBPs) [3–5]. miRNAs are a class of short non-coding RNAs spanning ∼22 nt, which interact with the 3′-UTRs of target mRNAs in the miRNA-induced silencing complex (miRISC), leading to translational repression or mRNA decay or both [2,6,7]. High-throughput and functional studies show that miRNAs play important roles in many aspects of cellular physiology and pathological processes such as inflammation and tumorigenesis [6,8,9]. miRNAs have also emerged as master regulators of the homoeostasis of normal gastrointestinal (GI) mucosa [3,10–14], the tissue with the most rapid turnover rate in the body [15]. Our previous study demonstrates that inhibition of mucosal growth in the small intestine by fasting or polyamine depletion is associated with a significant increase in the level of miR-29b, whereas locked nucleic acid (LNA)-modified mediated miR-29b silencing stimulates intestinal mucosal renewal [10,12]. To identify miR-29b target mRNAs implicated in these processes, two mRNAs encoding cyclin-dependent kinase 2 (CDK2) and menin proteins are directly associated with and tightly regulated by miR-29b in intestinal epithelial cells (IECs) [12,13], but the complete set of mRNAs controlled by miR-29b in the regulation of IEC proliferation and apoptosis remains to be fully elucidated.
Wnt signalling plays a pivotal role in gut development and mucosal regeneration through its regulation of diverse biological functions, including cell proliferation, apoptosis and migration [16–18]. The canonical Wnt signalling pathway consists of a cascade of events that initiate after binding of a Wnt protein ligand to a Frizzled (Fz) family receptor and its co-receptors LRP5/6 (low-density lipoprotein-receptor-related protein 5/6). This Fz–LRP complex is necessary for effective Wnt signalling, which leads to an accumulation of dephosphorylated β-catenin and its stabilization [19,20]. Subsequently, the stabilized β-catenin undergoes nuclear translocation and association with T-cell factor (TCF) transcription factors, enabling transactivation of Wnt target genes. Target deletion of the Wnt gene or inactivation of Wnt signalling by overexpressing the Wnt natural inhibitor Dikkopf1 disrupts gut development, represses mucosal growth and delays healing after mucosal injury [21,22]. In studies in vitro, co-culture of IECs with Wnt-overexpressing fibroblasts increases Wnt-dependent transcriptional activity, stimulates IEC proliferation and enhances epithelial repair after wounding [17,23]. Several recent studies have demonstrated that maintenance of normal cellular abundance of LRP proteins is critical for biological functions of Wnt signals [24–26] and that LRP6 expression is tightly regulated at the post-transcriptional level by tissue-specific miRNAs such as miR-513c in human glioblastoma cells [27], miR-126 in papillary thyroid carcinoma cells [28] and miR-19 in endothelial cells [29]. To our best knowledge, there are no studies available showing the exact role of miR-29b in the regulation of LRP6 expression in IECs.
HuR is among the most prominent translation and turnover regulatory RBPs, and it associates with U-rich elements located in the 3′-UTRs of labile mRNAs [30]. Upon binding to a target mRNA, HuR stabilizes it, alters its translation or performs both functions [31]. Our previous studies show that HuR stabilizes mRNAs encoding nucleophosmin and p53 [32], activating transcription factor 2 (ATF2) [33], X-linked inhibitor of apoptosis (XIAP) [34] and JunD [35] and enhances translation of mRNAs encoding Myc [22], stromal interaction molecule 1 (Stim1) [11] and occludin [36], thus modulating IEC proliferation, apoptosis and cell-to-cell interaction. We have recently reported that tissue-specific HuR deletion in IECs decreases regenerative potential of crypt progenitors in the small intestine and causes significant mucosal atrophy by decreasing LRP6 expression [37]. At the molecular level, HuR was found to bind the Lrp6 mRNA via its 3′-UTR and enhanced LRP6 expression by stabilizing Lrp6 mRNA and stimulating its translation. In the present study, we found further that both HuR and Lrp6 mRNAs are targets of miR-29b in IECs and these associations destabilize HuR and Lrp6 mRNAs and repress their translation. Moreover, forced expression of HuR rescues LRP6 expression in cells overexpressing miR-29b, suggesting that the down-regulation of HuR by miR-29b also contributes to miR-29b-induced LRP6 repression.
MATERIALS AND METHODS
Chemicals and cell culture
Tissue culture medium and dialysed FBS were from Invitrogen and biochemicals were from Sigma. The antibodies recognizing LRP6, HuR and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were from Cell Signaling Technology and Santa Cruz Biotechnology. The secondary antibody conjugated to horseradish peroxidase was from Sigma. Pre-miR™ miRNA precursors of miR-29b (pre-miR-29b) were from Ambion and biotin-labelled miR-29b was custom made by Dharmacon. The IEC-6 cell line, derived from normal rat intestinal crypt cells, was purchased from the A.T.C.C. at passage 13 and was maintained in T-150 flasks in Dulbecco's modified Eagle's medium (DMEM) supplemented with 5% heat-inactivated FBS. IEC-6 cells were used at passages 15–20 [38,39]. Caco-2 cells (a human colon carcinoma cell line) were also purchased from the A.T.C.C. and cultured similarly to the IEC-6 cells with slight modification [40].
Plasmid construction
The chimaeric firefly luciferase reporter construct of the Lrp6 coding region (CR), 5′-UTR or 3′-UTR was generated as described in [37]. The partial transcripts of Lrp6 CR (spanning positions 1502–2064), full-length 5′-UTR or partial 3′-UTR (spanning positions 8661–9870) were amplified and subcloned into the pmirGLO Dual-Luciferase miRNA Target Expression Vector (Promega) to generate the pmirGLO-Luc-Lrp6-CR-F1, pmirGLO-Luc-Lrp6-5′-UTR and pmirGLO-Luc-Lrp6-3′-UTR-F1; all cloning was confirmed by DNA sequencing and enzyme digestion. The chimaeric firefly luciferase reporter constructs of the full-length HuR CR (spanning positions 211–910), and partial transcripts of HuR 3′-UTR, F1 (spanning positions 4061–4270 and F2 (spanning positions 4200–4550), were generated similarly to the Lrp6. Transient transfections were performed using the Lipofectamine reagent as recommended by the manufacturer (Invitrogen). The luciferase reporter constructs were transfected into cells along with phRL-null, a Renilla luciferase control reporter vector from Promega, to monitor transfection efficiencies as described previously [22]. Luciferase activity was measured using the Dual Luciferase Assay System, and the level of pmirGLO-Luc-Lrp6 or Luc-HuR luciferase activity were normalized to Renilla luciferase activity and were further compared with the levels of luciferase mRNA in every experiment. All of the primer sequences for generating these constructs are provided in Supplementary Table S1.
Recombinant adenoviral plasmids containing human HuR were constructed by using the Adeno-X Expression System according to the protocol provided by the manufacturer (Clontech). Briefly, the full-length cDNA of human wild-type HuR was cloned into pShuttle; pAdeno-HuR (AdHuR) was constructed by digesting the pShuttle construct with PI-SceI/I-CeuI and ligating the resultant fragment into the PI-SceI/I-CeuI sites of the pAdeno-X adenoviral vector as described previously [11,39]. Recombinant adenoviral plasmids were packaged into infectious adenoviral particles by transfecting human embryonic kidney (HEK)-293 cells using LipofectAMINE Plus reagent (Gibco). Titres of the adenoviral stock were determined by a standard plaque-forming assay. Recombinant adenoviruses were screened for the expression of the introduced gene by Western blot analysis using anti-HuR antibody. pAdeno-X, which was the recombinant replication-incompetent adenovirus carrying no HuR cDNA insert (Adnull), was grown and purified as described above and served as a control adenovirus. Cells were infected with AdHuR or Adnull, and expression of HuR was assayed 48 h after the infection.
Reverse transcription (RT) and quantitative real-time PCR analyses
Total RNA was isolated by using RNeasy mini kit (Qiagen) and used in RT and PCR amplification as described in [41]. The levels of Gapdh PCR product were assessed to monitor the evenness in RNA input in RT–PCR samples. Quantitative real-time PCR (Q-PCR) analysis was performed using 7500-Fast Real-Time PCR Systems with specific primers, probes and software (Applied Biosystems). For miRNA studies, the levels of miR-29b were also quantified by Q-PCR by using Taqman MicroRNA assay; levels of small nuclear RNA (snRNA) U6 were measured as an endogenous control.
Western blotting analysis
Whole-cell lysates were prepared using 2% SDS, sonicated and centrifuged (15000 g) at 4°C for 15 min. The supernatants were boiled for 5 min and size-fractionated by SDS/PAGE (10% acrylamide). After transferring proteins on to nitrocellulose filters, the blots were incubated with primary antibodies recognizing LRP6, HuR or GAPDH; following incubation with secondary antibodies, immunocomplexes were developed by using chemiluminescence.
Analysis of newly translated protein
Nascent LRP6 and HuR were detected by Click-iT protein analysis detection kit (Life Technologies) and performed following the company's instructions with minor modification [12]. Briefly, cells were incubated in methionine-free medium and then exposed to L-azidohomoalanine (AHA). Cell lysates were mixed with reaction buffer containing biotin/alkyne reagent and CuSO4 for 20 min, and the biotin/alkyne/azide-modified protein complex was pulled down using paramagnetic Streptavidin-conjugated Dynabeads. The pull-down material was resolved by SDS/PAGE (10% gel) and analysed by Western immunoblotting analysis using antibodies against LRP6, HuR or GAPDH.
Polysome analysis was performed as described in [41]. Briefly, cells at ∼70% confluence were incubated for 15 min in 0.1 mg/ml cycloheximide, lifted by scraping in 1 ml of polysome extraction buffer and lysed on ice for 10 min. Nuclei were pelleted, and the resulting supernatant was fractionated through a 10–50% linear sucrose gradient to fractionate cytoplasmic components according to their molecular mass. The eluted fractions were prepared with a fraction collector (Brandel), and their quality was monitored at 254 nm using a UV-6 detector (ISCO). After RNA in each fraction was extracted, the levels of Lrp6, HuR and Gapdh mRNAs were quantified by Q-PCR in each of the fractions.
Biotin-labelled miR-29b pull-down assays
Binding of miR-29b to target mRNAs was examined by biotin-labelled miR-29b as described in [12,13]. Briefly, biotin-labelled miR-29b was transfected into cells and 24 h later whole-cell lysates were collected, mixed with Streptavidin–Dynabeads (Invitrogen) and incubated at 4°C with rotation overnight. After the beads were washed thoroughly, the bead-bound RNAs were isolated and subjected to RT followed by Q-PCR analysis. Input RNA was extracted and served as control.
Statistics
Values are means ± S.E.M. from three to six samples. Immunoblotting results were repeated three times. The significance of the difference between means was determined by ANOVA. The level of significance was determined by using Duncan's multiple-range test [42].
RESULTS
Lrp6 and HuR mRNAs are novel targets of miR-29b
Using the standard online software (TargetScan and RNA22), we found that there are predicted binding sites of miR-29b within the 3′-UTRs of the Lrp6 and HuR mRNAs (Figure 1A), suggesting that miR-29b directly interacts with the Lrp6 and HuR mRNAs and is involved in the regulation of their expression. To test this possibility, the following two sets of experiments were performed. First, we examined the association of miR-29b with the Lrp6 or HuR mRNA by RNA pull-down assays using biotin-labelled miR-29b (custom-made by Dharmacon, shown in Figure 1B, panel a). To assess the transfection efficiency, the levels of miR-29b and small nuclear RNA U6 (which served as control) were examined by Q-PCR analysis 24 h after transfection. As shown in Figure 1B, panel b, cells transfected with the biotin-labelled miR-29b exhibited elevated miR-29b levels but displayed no changes in RNA U6 levels (Figure 1B, panel c). When the presence of Lrp6 and HuR mRNAs in the materials pulled down by biotin-miR-29b was examined, the levels of Lrp6 and HuR mRNAs were highly enriched in the materials from cells transfected with the biotin-labelled miR-29b but not from cells transfected with scrambled control miRNA (Figure 1C, panel a). The interaction of miR-29b with Lrp6 or HuR mRNA is specific, because increasing the levels of biotin-miR-29b failed to increase its binding to the mRNAs that encode Fz-7 and DNMT1 (DNA methyltransferase 1). The levels of Fz-7 and Dnmt1 mRNAs in the pulled-down materials were indistinguishable in cells transfected with biotin-labelled miR-29b and cells transfected with scrambled miRNA. In addition, transfection with biotin-labelled miR-29b did not alter the steady-state levels of total Lrp6, HuR, Fz-7 and Dnmt1 mRNAs (Figure 1C, panel b). These results strongly suggest that Lrp6 and HuR mRNAs are novel targets of miR-29b and that miR-29b is able to form the miR-29b–Lrp6 mRNA or miR-29b–HuR mRNA complex.
Figure 1. miR-29b directly interacts with the Lrp6 and HuR mRNAs.
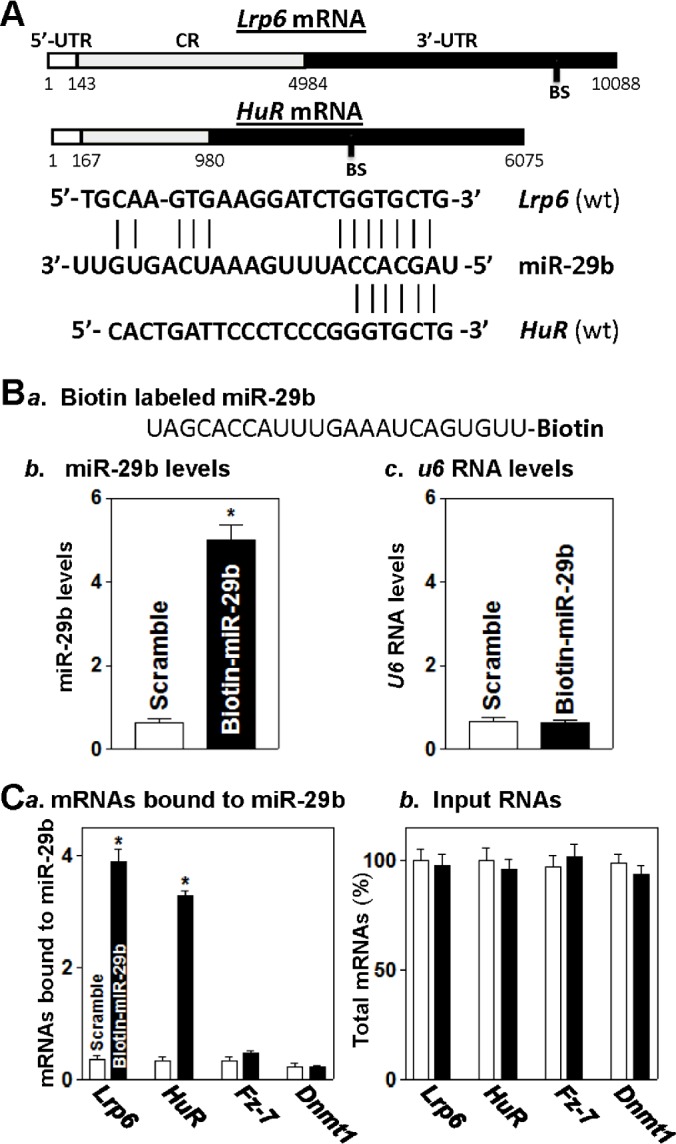
(A) Schematic representations of the Lrp6 and HuR mRNAs depicting predicted binding sites (BS) for miR-29b in their 3′-UTRs. (B) Levels of biotinylated miR-29b after transfection: (panel a) schematic representation of biotinylated miR-29b; and (panels b and c) levels of biotinylated miR-29b and small RNA U6 as measured by Q-PCR analysis 24 h after transfection. Values are means ± S.E.M. from three separate experiments. *P<0.05 compared with cells transfected with control scrambled oligomer. (C) Association of biotinylated miR-29b with mRNAs encoding LRP6, HuR, Fz-7 and DNMT1: (panel a) Levels of Lrp6, HuR, Fz-7 and Dnmt1 mRNAs in the materials pulled down by biotinylated miR-29b and (panel b) levels of total input mRNAs. *P<0.05 compared with cells transfected with control scrambled oligomer.
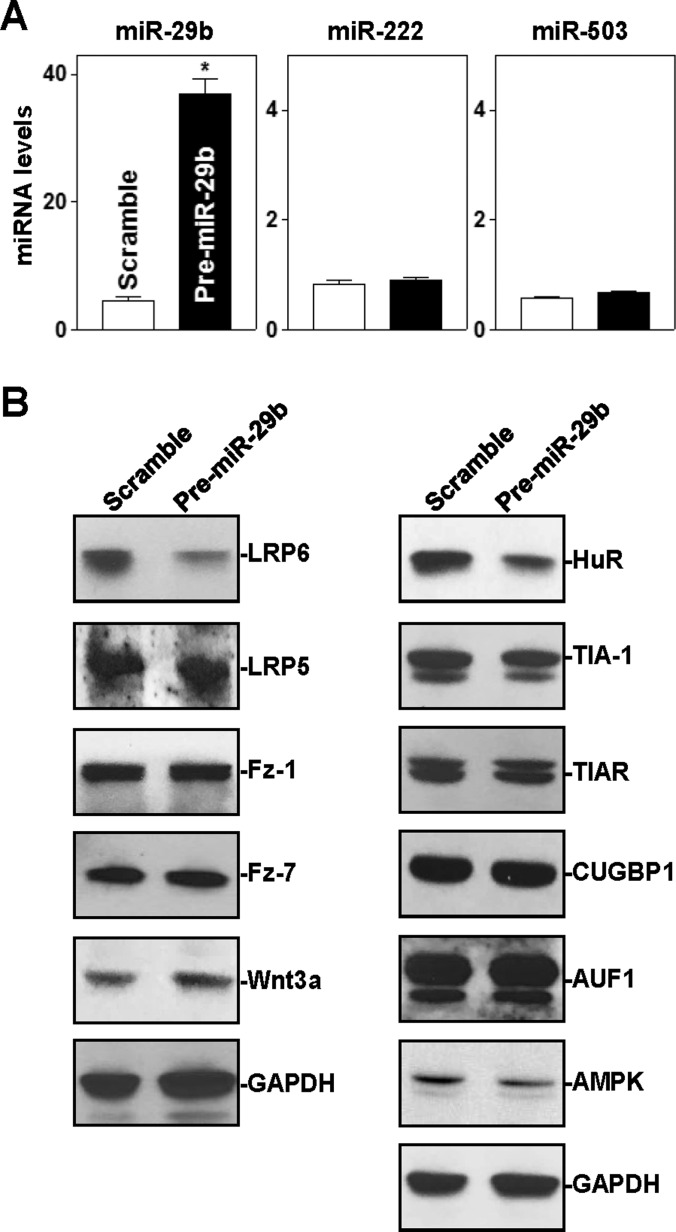
Secondly, we defined the functional consequences of miR-29b association with Lrp6 or HuR mRNA. In this study, miR-29b levels were increased by transfection with the miR-29b precursor (pre-miR-29b). As shown in Figure 2(A), transfection with pre-miR-29b increased miR-29b levels remarkably without effect on the levels of miR-222 and miR-503. Interestingly, increased levels of miR-29b by pre-miR-29b transfection specifically decreased LRP6 protein levels (Figure 2B, left), but it did not reduce the levels of other Wnt signalling-associated proteins including LRP5, Fz-1, Fz-7 and Wnt3a. Moreover, increased miR-29b also inhibited expression of the RBP HuR, although it failed to alter the cellular abundance of other RBPs such as TIA-1 (T-cell intracellular antigen-1), TIAR (TIA-1-related protein), CUGBP1 (CUG-binding protein 1), AUF1 (AU-binding factor 1) and HuR-regulatory protein AMPK (AMP-activated protein kinase) (Figure 2B, right). The levels of LRP6 and HuR proteins in pre-miR-29b-transfected cells decreased by ∼71% and ∼63% (n=3; P<0.05), respectively, compared with those in cells transfected with scrambled control miRNA. On the other hand, transfection with pre-miR-29b did not affect cell viability as measured by Trypan Blue staining (results not shown). These results indicate that increasing the levels of miR-29b specifically represses expression of LRP6 and HuR.
Figure 2. Ectopic overexpression of miR-29b inhibits expression levels of LRP6 and HuR.
(A) Levels of miR-29b (left), miR-222 (middle) and miR-503 (right) 48 h after transfection with pre-miR-29b or control scramble. Values are means ± S.E.M. from three separate experiments. *P<0.05 compared with cells transfected with control scramble. (B) Representative immunoblots of Wnt signal-associated factors (left) and RBPs (right) in cells described in (A). Whole-cell lysates were prepared for Western blotting; equal loading was monitored by assessing GAPDH levels.
miR-29b decreases the stability and translation of Lrp6 and HuR mRNAs via their 3′-UTRs
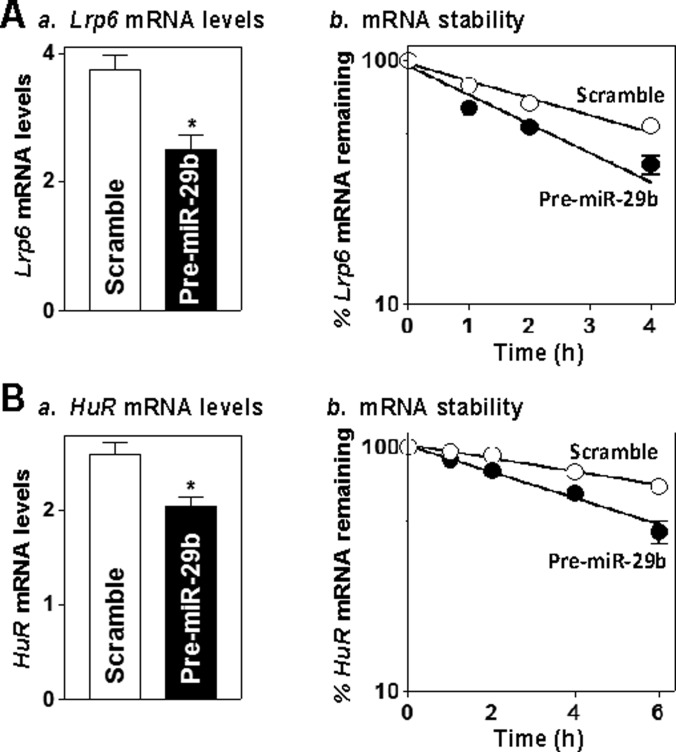
To investigate the mechanism underlying miR-29b in regulating LRP6 and HuR expression, we examined changes in the stability and translation of Lrp6 and HuR mRNAs after miR-29b overexpression. Increased levels of miR-29b by transfection with pre-miR-29b decreased the levels of Lrp6 (Figure 3A, left) and HuR (Figure 3B, left) mRNAs as a result of increases in their degradation rates (Figures 3A and 3B, right). The half-lives of Lrp6 and HuR mRNAs in cells overexpressing miR-29b were decreased compared with those measured in cells transfected with scrambled control miRNA. In contrast, miR-29b overexpression did not alter the half-life of Gapdh mRNA and its cellular content (results not shown). However, quantitative analysis of these results indicated that the levels of LRP6 and HuR protein expression were decreased by ∼71 and ∼63% in pre-miR-29b-transfected cells (Figure 1B), whereas the levels of their respective mRNAs were only reduced by ∼40 and ∼25% respectively.
Figure 3. miR-29b destabilizes Lrp6 and HuR mRNAs.
(A) Half-life of Lrp6 mRNA 48 h after transfection with pre-miR-29b. Total cellular RNA was isolated at the indicated times after administration of actinomycin D (5 mg/ml), and remaining levels of Lrp6 and Gapdh mRNAs were measured by Q-PCR analysis. Values are means ± S.E.M. from three separate experiments. (B) Half-life of the HuR mRNA in cells described in (A).
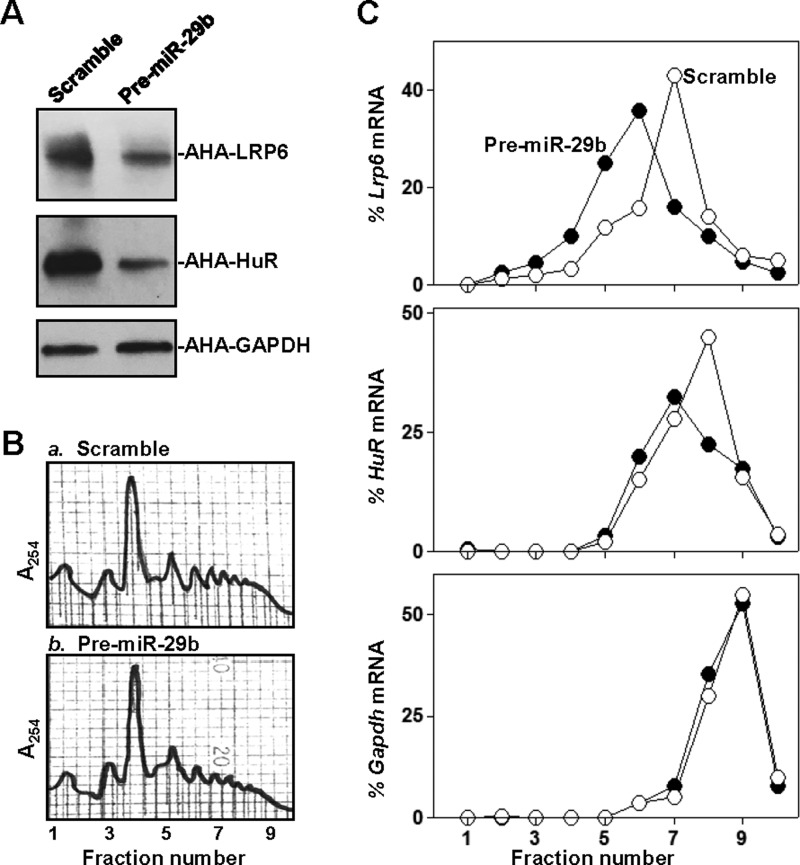
Therefore, we examined the involvement of miR-29b in the regulation of LRP6 and HuR translation. De novo synthesis of LRP6 and HuR proteins was analysed by measuring AHA incorporation after miR-29b overexpression. As shown in Figure 4(A), the levels of newly synthesized LRP6 and HuR proteins decreased significantly in pre-miR-29b-transfected cells compared with cells transfected with scrambled control miRNA. The levels of newly synthesized LRP6 and HuR proteins in pre-miR-29b-transfected cells decreased by ∼67 and ∼78% (n=3; P<0.05), respectively, compared with those of cells transfected with scrambled control miRNA. Inhibition of LRP6 and HuR protein synthesis by miR-29b was specific, since there was no change in nascent GAPDH synthesis after miR-29b overexpression. Changes in translation of these proteins after miR-29b overexpression were further studied by examining the sizes of polysomes. We fractionated cytoplasmic components through sucrose gradients and analysed polysome distribution profiles. Although increasing the levels of miR-29b did not affect global polysomal profiles (Figure 4B), the abundance of each of Lrp6 and HuR mRNAs associated with actively translating fractions (peaking at fractions 7–9) decreased in pre-miR-29b-transfected cells, with a moderate leftward shift of the mRNAs towards lower-translating fractions (peaking at fractions 5–6; Figure 4C). On the other hand, Gapdh mRNA, encoding a housekeeping protein, was distributed similarly in both groups. These results indicate that miR-29b inhibits expression of LRP6 and HuR by both destabilizing Lrp6 and HuR mRNAs and repressing their translation.
Figure 4. miR-29b inhibits translation of LRP6 and HuR.
(A) Newly synthesized proteins of LRP6 and HuR as measured by AHA incorporation assays. At 48 h after transfection with pre-miR-29b, cells were exposed to AHA, and then cell lysates were incubated with the reaction buffer containing biotin/alkyne reagent. The biotin/alkyne/azide-modified protein complex was pulled down by paramagnetic Streptavidin-conjugated Dynabeads. (B) Polysomal profiles in cells described in (A). Nuclei were pelleted, and the resulting supernatants were fractionated through a 10–50% linear sucrose gradient. (C) Distributions of Lrp6 (top), HuR (middle) and Gapdh (bottom) mRNAs in each gradient fraction prepared from cells described in (B). The levels of Lrp6, HuR and Gapdh mRNAs were plotted as a percentage of the total Lrp6, HuR or Gapdh mRNA levels in the samples. Three separate experiments were performed and showed similar results.
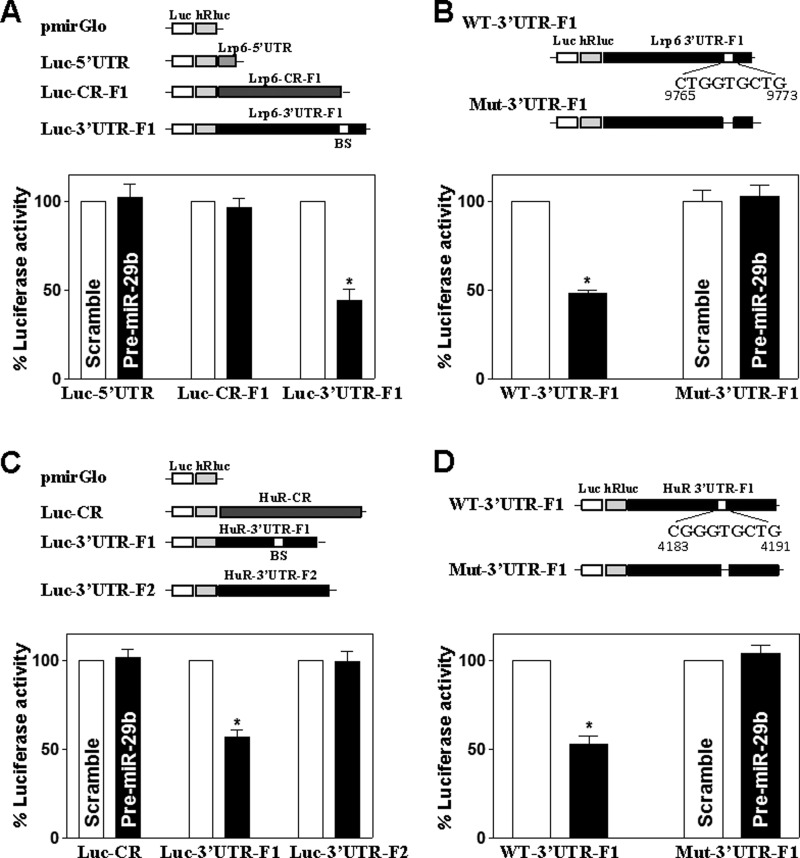
To determine whether the inhibitory effect of miR-29b on LRP6 expression was mediated through the 5′-UTR, CR or 3′-UTR, fractions of the Lrp6 5′-UTR, CR and 3′-UTR were subcloned into the pmirGLO dual-luciferase miRNA target expression vector to generate pmirGlo-Lrp6-5′-UTR, pmirGLO-Lrp6-CR-F1 and pmirGLO-Lrp6-3′-UTR-F1 reporter constructs (Figure 5A, top). miR-29b overexpression by transfection with pre-miR-29b selectively decreased the levels of pmirGLO-Lrp6-3′-UTR-F1 (with predicted miR-29b-binding site) luciferase reporter activity (Figure 5A, bottom), but it failed to inhibit the activities of pmirGLO-Lrp6-5′-UTR and -CR-F1 (without the potential miR-29b-binding site) reporters. To further characterize the specific binding site of miR-29b in the Lrp6 3′-UTR, the reporter construct that expressed chimaeric RNA containing the luciferase and partial transcripts spanning the Lrp6 3′-UTR without the potential binding site was prepared by eliminating nucleotides spanning positions 9765–9773 of the Lrp6 3′-UTR-F1 (Figure 5B, top). Ectopic miR-29b overexpression decreased the levels of luciferase reporter gene activity when cells were transfected with wild-type 3′-UTR-F1, but repression by miR-29b was completely prevented when this specific site from the Lrp6 3′-UTR-F1 was deleted (Figure 5B, bottom). Similarly, the luciferase reporter constructs containing full-length HuR CR or partial transcripts of HuR 3′-UTR, the fractions (F) 1 and 2, were also generated. miR-29b overexpression decreased the levels of luciferase reporter activity of pmirGLO-HuR-3′-UTR-F1 that contains the potential miR-29b-binding site (Figure 5C), but it failed to inhibit the activity of pmirGLO-HuR-3′-UTR-F2 and pmirGLO-HuR-CR reporters that have no predicted binding site for miR-29b (Figure 5C). Deletion mutation of the binding site for miR-29b from the HuR-3′-UTR by eliminating nucleotides spanning positions 4183–4191 from the HuR 3′-UTR-F1 (Figure 5D, top) prevented the repression by miR-29b (Figure 5D, bottom). Taken together, these results strongly suggest that increasing the levels of miR-29b destabilizes Lrp6 and HuR mRNAs and inhibits their translation through interaction with the specific cis-elements located at Lrp6 and HuR 3′-UTRs.
Figure 5. miR-29b represses LRP6 and HuR expression via interaction with their 3′-UTRs.
(A) Changes in activities of Lrp6 CR, 5′-UTR or 3′-UTR luciferase (Luc) reporters after ectopic overexpression of miR-29b. Top, schematic illustrations of plasmids of different chimaeric firefly Lrp6 luciferase reporters. Bottom, levels of activities of luciferase reporters containing the Lrp6 5′-UTR, or fraction (F) of its CR or 3′-UTR. At 24 h after transfection with pre-miR-29b, cells were transfected with different Lrp6 luciferase reporter plasmids. Levels of firefly and Renilla luciferase activities were assayed 24 h later. Results were normalized to the Renilla luciferase activities and expressed as the means ± S.E.M. data from three separate experiments. *P<0.05 compared with cells transfected with scrambled RNA. (B) Effect of deletion mutation of specific miR-29b-binding site (schematic diagram) in the Lrp6 3′-UTR on luciferase reporter activities after ectopic overexpression of miR-29b. *P<0.05 compared with cells transfected with control scrambled oligomer. (C and D) Activities of HuR CR or 3′-UTR-F1 or -F2 luciferase reporters in the presence or absence of the miR-29b-binding site in cells described in (A).
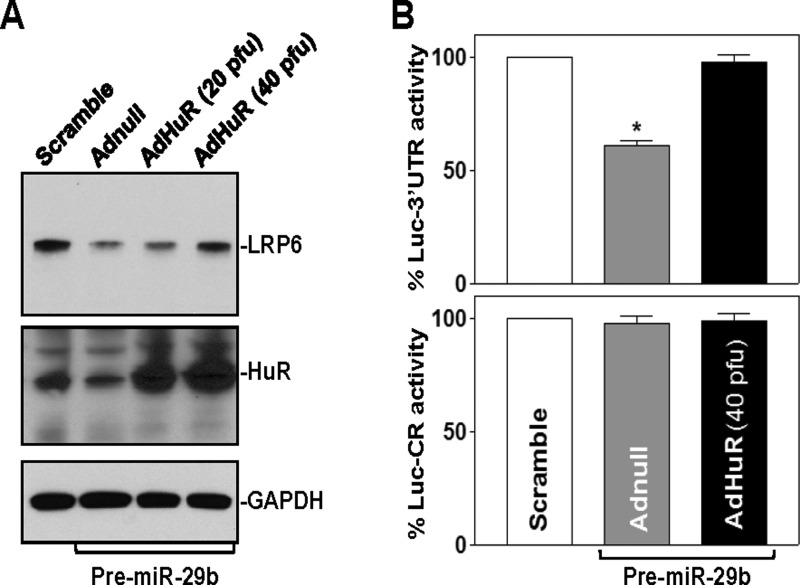
Increased HuR rescues LRP6 expression in cells overexpressing miR-29b
To determine the role of decreased levels of HuR in the miR-29b-induced repression of LRP6, we examined the effect of overexpressing wild-type HuR on LRP6 expression in cells transfected with pre-miR-29b. The adenoviral vector containing the corresponding human HuR cDNA under the control of the human cytomegalovirus immediate early gene promoter (AdHuR) was constructed as described previously [39]. The adenoviral vectors used in this study infect IECs with near 100% efficiency [43]; >95% of Caco-2 cells were positive when they were infected for 24 h with the adenoviral vector encoding GFP (results not shown). As noted in Figure 6(A), the levels of HuR protein increased with viral load and reached ∼6- and ∼10-fold higher levels (n=3; P<0.05) than pre-miR-29b-transfected cells when AdHuR was used at 20 and 40 plaque-forming units (pfu)/cell respectively. Transient infection of pre-miR-29b-transfected cells with AdHuR (40 pfu/cell) significantly increased LRP6 expression; the level of LRP6 protein increased from 0.16±0.03 in cells transfected with pre-miR-29b alone to 0.96±0.11 in cells co-transfected with pre-miR-29b and HuR (n=3; P<0.05). A control adenovirus that lacked exogenous HuR cDNA (Adnull) failed to alter HuR and LRP6 levels. Consistently, HuR overexpression also induced the levels of Lrp6-3′-UTR-F1 luciferase reporter gene activity (Figure 6B, top), but it did not affect the activity of Lrp6-CR-F1 luciferase reporter (Figure 6A, bottom). These results indicate that the reduction in HuR levels contributes to miR-29b-induced LRP6 repression, suggesting that miR-29b regulates LRP6 post-transcriptionally by both directly interacting with the Lrp6 mRNA and down-regulating HuR abundance.
Figure 6. Increased HuR rescuers LRP6 expression in cells overexpressing miR-29b.
(A) Representative immunoblots of LRP6 and HuR in miR-29b-overexpressing cells infected with the recombinant adenoviral encoding HuR cDNA (AdHuR) or adenoviral vector lacking HuR cDNA (Adnull) at a multiplicity of infection of 20 or 40 pfu/cells; the expression of HuR and LRP6 was examined 48 h after co-transfection with pre-miR-29b and AdHuR or Adnull. GAPDH immunoblotting was performed as an internal control for equal loading. (B) Changes in activities of Lrp6 3′-UTR (top) or its CR (bottom) luciferase reporters in cells described in (A). Values are means ± S.E.M. from three experiments. *P<0.05 compared with scramble control miRNA or cells co-transfected with pre-miR-29b and AdHuR.
DISCUSSION
Although miR-29b functions as a biological repressor of intestinal mucosal growth [12,13,44], little is known about its exact target mRNAs implicated in the regulation of IEC proliferation, differentiation, apoptosis and migration. In the present study, we identify the mRNAs encoding LRP6 and HuR proteins as novel targets of miR-29b in IECs and demonstrate that formation of the miR-29b–Lrp6 mRNA or miR-29b–HuR mRNA complex destabilizes Lrp6 and HuR mRNAs and represses their translation. Since HuR is a ubiquitous RBP and regulates gene expression at the post-transcriptional level, our findings provide insight into the regulation of one type of post-transcriptional regulator (an RBP) by another type of post-transcriptional regulator (a miRNA). Inhibition of HuR by miR-29b is specific, since ectopic overexpression of miR-29b fails to alter expression levels of other RBPs including TIA-1, TIAR, CUGBP1 and AUF1. Interestingly, increasing the level of HuR rescues LRP6 expression in cells overexpressing miR-29b, indicating that LRP6 repression by miR-29b results not only from the direct interaction of miR-29b with the Lrp6 mRNA but also from the down-regulation of HuR. These findings advance our understanding of the molecular mechanisms underlying the regulation of Lrp6 and HuR gene expression and strongly suggest that miR-29b inhibits intestinal mucosal growth at least in part by inactivating the Wnt signalling pathway through repression of LRP6 expression.
Our results indicate that miR-29b interacted with the Lrp6 and HuR mRNAs via their 3′-UTRs, but not with the CRs in IECs. Studies using various ectopic reporters bearing partial transcripts spanning the Lrp6 or HuR 3′-UTR with or without the miR-29b-binding site further demonstrated that the sequence spanning positions 9765–9773 in the Lrp6 3′-UTR or the sequence spanning positions 4183–4191 in the HuR 3′-UTR was the predominant and functional site through which miR-29b interacted and repressed their expression post-transcriptionally. The inhibition of LRP6 or HuR expression by miR-29b was completely prevented when the specific binding site was deleted from the Lrp6- or HuR-3′-UTR. Consistent with these findings, several studies have shown that miR-29b directly interacts with the 3′-UTRs of Cdk2 [12], Dnmt3a and Dnmt3b [45], and Bcr/Abl1 (encoding BCR/ABL1 protein) mRNAs [46] and thus destabilizes them and/or represses their translation. In some instances, miR-29b was also found to associate with the CR of target mRNA for its regulatory actions; for example, our previous study revealed that miR-29b represses menin translation by directly interacting with the Men1 CR rather than its 3′-UTR [13].
The control of HuR expression by miR-29b raises an intriguing possibility that the miR-29b regulon co-ordinates gene expression at multiple post-transcriptional levels. HuR stabilizes and modulates the translation of numerous target mRNAs involved in cellular responses to stress agents, proliferative signals, immune triggers and developmental cues [31,47,48]. If miR-29b decreases the stability or translation of a putative target mRNA, it is possible that the effect is indirect, should HuR be a stabilizing and/or translation stimulatory RBP for that particular mRNA. In support of the notion, Abdelmohsen et al. [49] reported that miR-519 represses HuR translation and reduces HuR-regulated gene expression, thus inhibiting cell proliferation. In addition, our previous study [50] shows that miR-503 represses translation of the RBP CUGBP1 that destabilizes and represses translation of target mRNAs, whereas increasing the levels of miR-503 enhances expression of inhibitor of apoptosis (IAP) family of proteins c-IAP1 and c-IAP2 by decreasing CUGBP1 levels, because this stimulatory effect is prevented by CUGBP1 overexpression. In broad terms, our findings advise caution regarding the inhibitory functions of some miRNAs upon some target mRNAs, since they could result from an indirect suppressive effect of the miRNA on the stabilizing and translational regulatory RBPs such as HuR and CUGBP1 which may control the same target mRNAs.
Although the specific molecular mechanism by which miR-29b destabilizes and represses translation of the Lrp6 and HuR mRNAs remains unknown, it has been reported that several miRNAs repress gene expression by altering the subcellular location of target mRNAs [10,11,50]. For example, miR-195 destabilizes Stim1 mRNA by enhancing the recruitment of Stim1 mRNA to processing bodies (P-bodies) [11], where mRNAs are sorted for translation repression and/or degradation [51]. Silencing P-body-resident proteins such as Argonaute 2 (Ago2) or RCK prevents miR-195-induced destabilzation of the Stim1 mRNA, whereas Ago2 overexpression enhances the inhibitory effect of miR-195 [11]. Similarly, the miR-503-induced Cugbp1 mRNA destabilization and the miR-222-induced repression of CDK4 translation were also directly linked to increased translocation of Cugbp1 and Cdk4 mRNAs to P-bodies [10,50]. However, it is unclear at present whether miR-29b alters the subcellular localization of the Lrp6 and/or HuR mRNAs to P-bodies in IECs and whether an increase in the recruitment of the Lrp6 and/or HuR mRNAs to P-bodies affects their stability and translation. These possibilities are actively investigated in our ongoing studies.
Because maintenance of normal cellular abundances of LRP6 and HuR are essential for dynamic intestinal mucosal regeneration [22,37], the repression of LRP6 and HuR by miR-29b is of biological significance. The epithelium of the intestinal mucosa is a rapidly self-renewing tissue and its homoeostasis is tightly regulated at multiple levels by numerous intracellular and extracellular factors [15,19]. IECs continuously replicate within the intestinal crypts, and this process is counterbalanced by apoptosis [3]. Inhibition of intestinal mucosal growth occurs commonly in various pathological conditions, which are associated with an increase in miR-29b levels [10,13,44]. Our previous study [12] shows that ectopic miR-29b overexpression causes IEC growth arrest in G1-phase, whereas LNA-mediated miR-29b silencing results in mucosal hyperplasia in mice. The present study provides new evidence suggesting that miR-29b inhibits IEC proliferation and subsequently mucosal growth by decreasing LRP6 and HuR levels. In summary, our results indicate that miR-29b destabilizes and suppresses translation of the Lrp6 and HuR mRNAs through interaction with their 3′-UTRs rather than CRs. Our findings also suggest that control of cellular LRP6 and HuR expression by miR-29b in IECs plays an important role in maintaining intestinal epithelial homoeostasis under biological conditions.
Acknowledgments
J.-Y. Wang is a Senior Research Career Scientist, Biomedical Laboratory Research and Development Service, U.S. Department of Veterans Affairs.
Abbreviations
- Ago2
Argonaute 2
- AHA
L-azidohomoalanine
- AUF1
AU-binding factor 1
- CDK2
cyclin-dependent kinase 2
- CR
coding region
- CUGBP1
CUG-binding protein 1
- DNMT1
DNA methyltransferase 1
- Fz
Frizzled
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- IAP
inhibitor of apoptosis
- IEC
intestinal epithelial cell
- LNA
locked nucleic acid
- LRP
low-density lipoprotein-receptor-related protein
- P-body
processing body
- pfu
plaque-forming units
- Q-PCR
quantitative real-time PCR
- RBP
RNA-binding protein
- RT
reverse transcription
- Stim1
stromal interaction molecule 1
- TIA-1
T-cell intracellular antigen-1
- TIAR
TIA-1-related protein
AUTHOR CONTRIBUTION
Yanwu Li and Gang Chen performed most experiments and summarized data. Jun-Yao Wang performed some experiments regarding luciferase reporters. Tongtong Zou and Lan Liu contributed to studies involving translation assays. Lan Xiao and Hee Kyoung Chung performed studies using HuR overexpression. Jaladanki Rao contributed to data analysis. Jian-Ying Wang designed experiments, analysed data, and prepared manuscript.
FUNDING
This work was supported by the U.S. Department of Veterans Affairs [grant numbers BX-332 and BX-713 (to J.-Y.W. and J.N.R.)]; and the National Institutes of Health [grant numbers DK57819, DK61972 and DK68491 (to J.-Y.W.)].
References
- 1.Garneau N.L., Wilusz J., Wilusz C.J. The highways and byways of mRNA decay. Nat. Rev. Mol. Cell Biol. 2007;8:113–126. doi: 10.1038/nrm2104. [DOI] [PubMed] [Google Scholar]
- 2.Omer A.D., Janas M.M., Novina C.D. The chicken or the egg: microRNA-mediated regulation of mRNA translation or mRNA stability. Mol. Cell. 2009;35:739–740. doi: 10.1016/j.molcel.2009.09.003. [DOI] [PubMed] [Google Scholar]
- 3.Chandrasekharappa S.C., Guru S.C., Manickam P., Olufemi S.E., Collins F.S., Emmert-Buck M.R., Debelenko L.V., Zhuang Z., Lubensky I.A., Liotta L.A., et al. Positional cloning of the gene for multiple endocrine neoplasia-type 1. Science. 1997;276:404–407. doi: 10.1126/science.276.5311.404. [DOI] [PubMed] [Google Scholar]
- 4.Leung A.K., Sharp P.A. MicroRNA functions in stress responses. Mol. Cell. 2010;40:205–215. doi: 10.1016/j.molcel.2010.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Kouwenhove M., Kedde M., Agami R. MicroRNA regulation by RNA-binding proteins and its implications for cancer. Nat. Rev. Cancer. 2011;11:644–656. doi: 10.1038/nrc3107. [DOI] [PubMed] [Google Scholar]
- 6.Mendell J.T., Olson E.N. MicroRNAs in stress signaling and human disease. Cell. 2012;148:1172–1187. doi: 10.1016/j.cell.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ebert M.S., Sharp P.A. Roles for microRNAs in conferring robustness to biological processes. Cell. 2012;149:515–524. doi: 10.1016/j.cell.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eulalio A., Huntzinger E., Izaurralde E. Getting to the root of miRNA-mediated gene silencing. Cell. 2008;132:9–14. doi: 10.1016/j.cell.2007.12.024. [DOI] [PubMed] [Google Scholar]
- 9.Leung A.K., Sharp P.A. microRNAs: a safeguard against turmoil? Cell. 2007;130:581–585. doi: 10.1016/j.cell.2007.08.010. [DOI] [PubMed] [Google Scholar]
- 10.Xiao L., Cui Y. H., Rao J.N., Zou T., Liu L., Smith A., Turner D.J., Gorospe M., Wang J.Y. Regulation of cyclin-dependent kinase 4 translation through CUG-binding protein 1 and microRNA-222 by polyamines. Mol. Biol. Cell. 2011;22:3055–3069. doi: 10.1091/mbc.E11-01-0069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhuang R., Rao J.N., Zou T., Liu L., Xiao L., Cao S., Hansraj N. Z., Gorospe M., Wang J.Y. miR-195 competes with HuR to modulate stim1 mRNA stability and regulate cell migration. Nucleic Acids Res. 2013;41:7905–7919. doi: 10.1093/nar/gkt565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xiao L., Rao J.N., Zou T., Liu L., Cao S., Martindale J.L., Su W., Chung H.K., Gorospe M., Wang J.Y. miR-29b represses intestinal mucosal growth by inhibiting translation of cyclin-dependent kinase 2. Mol. Biol. Cell. 2013;24:3038–3046. doi: 10.1091/mbc.E13-05-0287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ouyang M., Su W., Xiao L., Rao J.N., Jiang L., Li Y., Turner D.J., Gorospe M., Wang J.Y. Modulation by miR-29b of intestinal epithelium homoeostasis through the repression of menin translation. Biochem. J. 2015;465:315–323. doi: 10.1042/BJ20141028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cao S., Xiao L., Rao J.N., Zou T., Liu L., Zhang D., Turner D.J., Gorospe M., Wang J.Y. Inhibition of Smurf2 translation by miR-322/503 modulates TGF-β/Smad2 signaling and intestinal epithelial homeostasis. Mol. Biol. Cell. 2014;25:1234–1243. doi: 10.1091/mbc.E13-09-0560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krausova M., Korinek V. Wnt signaling in adult intestinal stem cells and cancer. Cell. Signal. 2014;26:570–579. doi: 10.1016/j.cellsig.2013.11.032. [DOI] [PubMed] [Google Scholar]
- 16.van Es J.H., Jay P., Gregorieff A., van Gijn M.E., Jonkheer S., Hatzis P., Thiele A., van den Born M., Begthel H., Brabletz T., et al. Wnt signalling induces maturation of Paneth cells in intestinal crypts. Nat. Cell Biol. 2005;7:381–386. doi: 10.1038/ncb1240. [DOI] [PubMed] [Google Scholar]
- 17.Liu L., Rao J.N., Zou T., Xiao L., Smith A., Zhuang R., Turner D.J., Wang J.Y. Activation of Wnt3a signaling stimulates intestinal epithelial repair by promoting c-Myc-regulated gene expression. Am. J. Physiol. Cell Physiol. 2012;302:C277–C285. doi: 10.1152/ajpcell.00341.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu L., Ouyang M., Rao J.N., Zou T., Xiao L., Chung H.K., Wu J., Donahue J.M., Gorospe M., Wang J.Y. Competition between RNA-binding proteins CELF1 and HuR modulates MYC translation and intestinal epithelium renewal. Mol. Biol. Cell. 2015;26:1797–1810. doi: 10.1091/mbc.E14-11-1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clevers H. Wnt/β-catenin signaling in development and disease. Cell. 2006;127:469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 20.He X., Semenov M., Tamai K., Zeng X. LDL receptor-related proteins 5 and 6 in Wnt/β-catenin signaling: arrows point the way. Development. 2004;131:1663–1677. doi: 10.1242/dev.01117. [DOI] [PubMed] [Google Scholar]
- 21.Koch S., Capaldo C.T., Samarin S., Nava P., Neumaier I., Skerra A., Sacks D.B., Parkos C.A., Nusrat A. Dkk-1 inhibits intestinal epithelial cell migration by attenuating directional polarization of leading edge cells. Mol. Biol. Cell. 2009;20:4816–4825. doi: 10.1091/mbc.E09-05-0415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu L., Rao J.N., Zou T., Xiao L., Wang P.Y., Turner D.J., Gorospe M., Wang J.Y. Polyamines regulate c-Myc translation through Chk2-dependent HuR phosphorylation. Mol. Biol. Cell. 2009;20:4885–4898. doi: 10.1091/mbc.E09-07-0550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chung H.K., Chen Y., Rao J.N., Liu L., Xiao L., Turner D.J., Yang P., Gorospe M., Wang J.Y. Transgenic expression of miR-222 disrupts intestinal epithelial regeneration by targeting multiple genes including Frizzled-7. Mol. Med. 2015;21:676–687. doi: 10.2119/molmed.2015.00147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lara-Castillo N., Johnson M.L. LRP receptor family member associated bone disease. Rev. Endocr. Metab. Disord. 2015;16:141–148. doi: 10.1007/s11154-015-9315-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arensman M.D., Nguyen P., Kershaw K.M., Lay A.R., Ostertag-Hill C.A., Sherman M.H., Downes M., Liddle C, Evans R.M., Dawson D.W. Calcipotriol targets LRP6 to inhibit Wnt signaling in pancreatic cancer. Mol. Cancer Res. 2015;13:1509–1519. doi: 10.1158/1541-7786.MCR-15-0204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Srivastava R., Zhang J., Go G.W., Narayanan A., Nottoli T.P., Mani A. Impaired LRP6-TCF7L2 activity enhances smooth muscle cell plasticity and causes coronary artery disease. Cell Rep. 2015;13:746–759. doi: 10.1016/j.celrep.2015.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu J., Sun T., Hu X. microRNA-513c suppresses the proliferation of human glioblastoma cells by repressing low-density lipoprotein receptor-related protein 6. Mol. Med. Rep. 2015;12:4403–4409. doi: 10.3892/mmr.2015.3913. [DOI] [PubMed] [Google Scholar]
- 28.Wen Q., Zhao J., Bai L., Wang T., Zhang H., Ma Q. miR-126 inhibits papillary thyroid carcinoma growth by targeting LRP6. Oncol. Rep. 2015;34:2202–2210. doi: 10.3892/or.2015.4165. [DOI] [PubMed] [Google Scholar]
- 29.Landskroner-Eiger S., Qiu C., Perrotta P., Siragusa M., Lee M.Y., Ulrich V., Luciano A.K., Zhuang Z.W., Corti F., Simons M., et al. Endothelial miR-17∼92 cluster negatively regulates arteriogenesis via miRNA-19 repression of Wnt signaling. Proc. Natl. Acad. Sci. U.S.A. 2015;112:12812–12817. doi: 10.1073/pnas.1507094112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abdelmohsen K., Pullmann R., Jr, Lal A., Kim H.H., Galban S., Yang X., Blethrow J.D., Walker M., Shubert J., Gillespie D.A., et al. Phosphorylation of HuR by Chk2 regulates SIRT1 expression. Mol. Cell. 2007;25:543–557. doi: 10.1016/j.molcel.2007.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Papadaki O., Milatos S., Grammenoudi S., Mukherjee N., Keene J.D., Kontoyiannis D.L. Control of thymic T cell maturation, deletion and egress by the RNA-binding protein HuR. J. Immunol. 2009;182:6779–6788. doi: 10.4049/jimmunol.0900377. [DOI] [PubMed] [Google Scholar]
- 32.Zou T., Mazan-Mamczarz K., Rao J.N., Liu L., Marasa B.S., Zhang A.H., Xiao L., Pullmann R., Gorospe M., Wang J.Y. Polyamine depletion increases cytoplasmic levels of RNA-binding protein HuR leading to stabilization of nucleophosmin and p53 mRNAs. J. Biol. Chem. 2006;281:19387–19394. doi: 10.1074/jbc.M602344200. [DOI] [PubMed] [Google Scholar]
- 33.Xiao L., Rao J.N., Zou T., Liu L., Marasa B.S., Chen J., Turner D.J., Zhou H., Gorospe M., Wang J.Y. Polyamines regulate the stability of activating transcription factor-2 mRNA through RNA-binding protein HuR in intestinal epithelial cells. Mol. Biol. Cell. 2007;18:4579–4590. doi: 10.1091/mbc.E07-07-0675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang X., Zou T., Rao J.N., Liu L., Xiao L., Wang P.Y., Cui Y.H., Gorospe M., Wang J.Y. Stabilization of XIAP mRNA through the RNA binding protein HuR regulated by cellular polyamines. Nucleic Acids Res. 2009;37:7623–7637. doi: 10.1093/nar/gkp755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zou T., Rao J.N., Liu L., Xiao L., Yu T.X., Jiang P., Gorospe M., Wang J.Y. Polyamines regulate the stability of JunD mRNA by modulating the competitive binding of its 3′ untranslated region to HuR and AUF1. Mol. Cell. Biol. 2010;30:5021–5032. doi: 10.1128/MCB.00807-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu T.X., Wang P.Y., Rao J.N., Zou T., Liu L., Xiao L., Gorospe M., Wang J.Y. Chk2-dependent HuR phosphorylation regulates occludin mRNA translation and epithelial barrier function. Nucleic Acids Res. 2011;39:8472–8487. doi: 10.1093/nar/gkr567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu L., Christodoulou-Vafeiadou E., Rao J.N., Zou T., Xiao L., Chung H.K., Yang H., Gorospe M., Kontoyiannis D., Wang J.Y. RNA-binding protein HuR promotes growth of small intestinal mucosa by activating the Wnt signaling pathway. Mol. Biol. Cell. 2014;25:3308–3318. doi: 10.1091/mbc.E14-03-0853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang J.Y., McCormack S.A., Viar M.J., Wang H., Tzen C.Y., Scott R.E., Johnson L.R. Decreased expression of protooncogenes c-fos, c-myc, and c-jun following polyamine depletion in IEC-6 cells. Am. J. Physiol. 1993;265:G331–G338. doi: 10.1152/ajpgi.1993.265.2.G331. [DOI] [PubMed] [Google Scholar]
- 39.Liu L., Guo X., Rao J.N., Zou T., Marasa B.S., Chen J., Greenspon J., Casero R.A., Jr, Wang J.Y. Polyamine-modulated c-Myc expression in normal intestinal epithelial cells regulates p21Cip1 transcription through a proximal promoter region. Biochem. J. 2006;398:257–267. doi: 10.1042/BJ20060217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xiao L., Rao J.N., Cao S., Liu L., Chung H.K., Zhang Y., Zhang J., Liu Y., Gorospe M., Wang J.Y. Long noncoding RNA SPRY4-IT1 regulates intestinal epithelial barrier function by modulating the expression levels of tight junction proteins. Mol. Biol. Cell. 2016;27:617–626. doi: 10.1091/mbc.E15-10-0703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu T. X., Rao J.N., Zou T., Liu L., Xiao L., Ouyang M., Cao S., Gorospe M., Wang J.Y. Competitive binding of CUGBP1 and HuR to occludin mRNA controls its translation and modulates epithelial barrier function. Mol. Biol. Cell. 2013;24:85–99. doi: 10.1091/mbc.E12-07-0531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Harter J.L. Critical values for Duncan's new multiple range tests. Biometric. 1960;16:671–685. doi: 10.2307/2527770. [DOI] [Google Scholar]
- 43.Liu L., Li L., Rao J.N., Zou T., Zhang H.M., Boneva D., Bernard M.S., Wang J.Y. Polyamine-modulated expression of c-myc plays a critical role in stimulation of normal intestinal epithelial cell proliferation. Am. J. Physiol. Cell Physiol. 2005;288:C89–C99. doi: 10.1152/ajpcell.00326.2004. [DOI] [PubMed] [Google Scholar]
- 44.Zou T, Rao J.N., Liu L., Xiao L., Chung H.K., Li Y., Chen G., Gorospe M., Wang J.Y. JunD enhances miR-29b levels transcriptionally and posttranscriptionally to inhibit proliferation of intestinal epithelial cells. Am. J. Physiol. Cell Physiol. 2015;308:C813–C824. doi: 10.1152/ajpcell.00027.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guo X., Liu Q., Wang G., Zhu S., Gao L., Hong W., Chen Y., Wu M., Liu H., Jiang C., et al. microRNA-29b is a novel mediator of Sox2 function in the regulation of somatic cell reprogramming. Cell Res. 2013;23:142–156. doi: 10.1038/cr.2012.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li Y., Wang H., Tao K., Xiao Q., Huang Z., Zhong L., Cao W., Wen J., Feng W. miR-29b suppresses CML cell proliferation and induces apoptosis via regulation of BCR/ABL1 protein. Exp. Cell Res. 2013;319:1094–1101. doi: 10.1016/j.yexcr.2013.02.002. [DOI] [PubMed] [Google Scholar]
- 47.Ghosh M., Aguila H.L., Michaud J., Ai Y., Wu M.T., Hemmes A., Ristimaki A., Guo C., Furneaux H., Hla T. Essential role of the RNA-binding protein HuR in progenitor cell survival in mice. J. Clin. Invest. 2009;119:3530–3543. doi: 10.1172/JCI38263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Katsanou V., Milatos S., Yiakouvaki A., Sgantzis N., Kotsoni A., Alexiou M, Harokopos V., Aidinis V., Hemberger M., Kontoyiannis D.L. The RNA-binding protein Elav1/HuR is essential for placental branching morphogenesis and embryonic development. Mol. Cell. Biol. 2009;29:2762–2776. doi: 10.1128/MCB.01393-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Abdelmohsen K., Srikantan S., Kuwano Y., Gorospe M. miR-519 reduces cell proliferation by lowering RNA-binding protein HuR levels. Proc. Natl. Acad. Sci. U.S.A. 2008;105:20297–20302. doi: 10.1073/pnas.0809376106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cui Y.H., Xiao L., Rao J.N., Zou T., Liu L., Chen Y., Turner D.J., Gorospe M., Wang J.Y. miR-503 represses CUG-binding protein 1 translation by recruiting CUGBP1 mRNA to processing bodies. Mol. Biol. Cell. 2012;23:151–162. doi: 10.1091/mbc.E11-05-0456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kulkarni M., Ozgur S., Stoecklin G. On track with P-bodies. Biochem. Soc. Trans. 2010;38:242–251. doi: 10.1042/BST0380242. [DOI] [PubMed] [Google Scholar]