Abstract
Background and Purpose:
Definitive diagnosis of Creutzfeldt–Jakob disease (CJD) requires demonstration of infective prion protein (PrPSc) in brain tissues by immunohistochemistry or immunoblot, making antemortem diagnosis of CJD difficult. The World Health Organization (WHO) recommends detection of 14-3-3 protein in cerebrospinal fluid (CSF) in cases of dementia, with clinical correlation, as a useful diagnostic marker for CJD, obviating the need for brain biopsy. This facility is currently available in only a few specialized centers in the West and no commercial kit is available for clinical diagnostic use in India. Hence the objective of this study was to develop an in-house sensitive assay for quantitation of 14-3-3 protein and to evaluate its diagnostic potential to detect 14-3-3 proteins in CSF as a biomarker in suspected cases of CJD.
Materials and Methods:
A minigene expressing the “core” 14-3-3 protein was synthesized by overlapping polymerase chain reaction (PCR) and the recombinant protein was produced by employing a bacterial expression system. Polyclonal antibodies raised in rabbit against the purified recombinant protein were used for developing a dot blot assay with avidin-biotin technology for signal amplification and quantitation of 14-3-3 protein in CSF.
Results:
The results in the present study suggest the diagnostic potential of the dot blot method with about 10-fold difference (P< 0.001) in the CSF levels of 14-3-3 protein between the CJD cases (N= 50) and disease controls (N= 70). The receiver operating characteristic (ROC) analysis of the results suggested an optimal cutoff value of 2 ng/mL.
Conclusions:
We have developed an indigenous, economical, and sensitive dot blot method for the quantitation of 14-3-3 protein in CSF.
Keywords: Core 14-3-3 protein, Creutzfeldt–Jakob disease (CJD), dot blot, overlapping polymerase chain reaction (PCR), synthetic gene
Introduction
The 14-3-3 protein belongs to a family of conserved, dimeric proteins with a monomeric molecular mass of about 30 kDa, and it is ubiquitously expressed in various mammalian tissues.[1] There are seven known mammalian 14-3-3 isotypes (β, γ, ε, ζ, η, σ, and θ/τ).[2] The highest tissue concentration of 14-3-3 proteins is found in the brain, comprising about 1% of its total soluble protein.[1] Although the function of this family of highly conserved proteins is not completely known, recent evidence indicates their involvement in multiple cellular processes[3,4,5] such as activators of neurotransmitter synthesis, signalling molecules, tumor suppressors, and also interacting with various protein kinases, receptor proteins, enzymes, structural and cytoskeletal proteins, proteins involved in cell cycle and transcriptional control, and proteins modulating apoptosis. The multitude of binding partners and their key roles in different physiological processes make 14-3-3 proteins an interesting target to investigate their role in pathological processes.[6] The 14-3-3 proteins have been detected in the cerebrospinal fluid (CSF) in various neurological disorders.[7] but they are more elevated in Creutzfeldt–Jakob disease (CJD). The methods of detection included qualitative study by immunohistochemistry,[8] semiquantitative evaluation by immunoblotting,[9,10] or quantitation by immunoassays.[11,12]
CJD is a rare form of rapidly progressive neurological disorder with dementia, myoclonus, and characteristic electroencephalogram (EEG) findings.[13] Due to the few relatively specific antemortem diagnostic signs, it is difficult to distinguish the sporadic form of CJD (sCJD) from rapidly evolving Alzheimer's disease (AD) and other dementing illnesses.[14] Definitive diagnosis of CJD is possible by immunostaining for the infective prion protein (PrPSc) on brain tissue collected at biopsy or autopsy or immunoblot using fresh brain tissue.[15,16] This is practiced at national CJD registries in the West and diagnosis is offered. The potential transmissibility of the disease while handling the neural tissue poses risk to the scientist/technician and the attending staff. Though the prevalence of CJD in India is found to be low in comparison to the West,[17] the cases are diagnosed with serious public health concern. The World Health Organization (WHO) has recommended detection of 14-3-3 protein in CSF in cases of rapidly progressive dementia, with clinical correlation, as a useful diagnostic marker for CJD, minimizing the need for brain biopsy.[18] Though 14-3-3 protein as a generic protein is detected in a few other neurological disorders, it plays a role as biomarker for the diagnosis of CJD with high probability. Over the past 2-3 years, with increased awareness, more and more possible and probable cases of CJD have been referred to the CJD Registry at the National Institute of Mental Health and Neurosciences (NIMHANS), Bengaluru, Karnataka, South India, seeking diagnosis, with a need to tailor the clinical and nursing management strategies. Such diagnostic testing is available currently in only a few specialized centers in the West at a high cost (100-150 USD per test) and no kit for diagnostic purpose is available for testing the patient samples in India.
The objective of the proposed study was to develop an in-house, sensitive assay for quantitation of 14-3-3 protein and evaluate its specificity and sensitivity in diagnosis of CJD (prion disease). Toward this, a minigene expressing the “core” 14-3-3 protein was synthesized by overlapping polymerase chain reaction (PCR) and the recombinant protein was produced by employing a bacterial expression system. Polyclonal antibodies were raised in the rabbit against the purified recombinant protein and used for cost-effective dot blot assay following avidin-biotin technology, using diaminobenzidine as the chromogen. This method was used for quantitation of 14-3-3 protein in CSF samples from cases of CJD and disease controls from cases of other neurodegenerative diseases such AD and Parkinson disease (PD).
Materials and Methods
Animals
New Zealand White rabbits from the Central Animal Research Facility, NIMHANS, Bengaluru, India were used for the production of polyclonal antibodies. They received pellet diet and water ad libitum. They were maintained at 12 h light and 12 h dark cycles under controlled conditions of humidity and ambient temperature. This study was approved by the Institutional Animal Ethics Committee.
Materials
Oligonucleotides were custom-synthesized by Sigma-Aldrich (Bengaluru, India). Deep-vent DNA polymerase, plasmid pMAL-c4X, Bam H1 and Eco R1 restriction enzymes, T4 DNA ligase, amylose resin, factor Xa, and EK12 cells were purchased from New England Biolabs (Hitchin, Hertfordshire, UK). Isopropyl thiogalactoside (IPTG), avidin-peroxidase, 3,3'-diaminobenzidine, gelatin, tricine, triton X-100, biotin N-hydroxysuccinimide ester, protein A-Sepharose, polyvinylidine difluoride (PVDF) membrane, Freund's Complete and Incomplete Adjuvants were procured from Sigma Aldrich (Bengaluru). Centrifugal concentrator units were purchased from Sartorius (Gottingen, Germany). All other reagents used were of analytical grade and obtained locally. CSF samples were collected into sterile tubes and placed on ice, and within 1 h of collection, frozen and preserved at −80°C until analyses were obtained from the archives of the Human Brain Tissue Repository (HBTR), NIMHANS. This study was approved by the Institutional Human Ethics Committee.
Synthesis and cloning of minigene encoding “core” 14-3-3 protein and expression of recombinant plasmid
The approach adopted in the synthesis of “core” 14-3-3 gene and the methodology followed for its expression in Escherichia coli (E. coli) was similar to that described earlier.[19,20] For this purpose, E. coli optimized 335 nucleotide sequences encompassing the open reading frame (ORF) of core 14-3-3 was designed. The sequence was then divided into 8 primers with overlaps of 16-18 nt. The minigene was synthesized over 5 rounds of overlapping PCR. The products of each round were analyzed by 2% agarose gel electrophoresis. Bacterial cultures, plasmid purification, and transformations were performed following standard protocols.[21] After ascertaining the correct nucleotide sequence, the final PCR product was double digested with Eco R1 and Bam H1 and cloned into pMAL-c4X vector. Expression of core 14-3-3 protein was achieved by transformation and induction of EK12 cells with 1 mM IPTG. Using amylose affinity chromatography, the recombinant protein was purified to homogeneity from 8M urea insoluble fraction. Vector-encoded fusion tag of the recombinant protein was cleaved by factor Xa digestion.[20] Homogeneity of the purified peptide was ascertained by tricine–sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis.[22]
Production of polyclonal antibodies
Core 14-3-3 protein was emulsified in Freund's Complete Adjuvant and injected subcutaneously into rabbits at a dose of 1 mg per animal for primary injection. For subsequent boosters at 4-week intervals, 500 µg of the antigen emulsified in Freund's Incomplete Adjuvant was administered subcutaneously. The immune recognition patterns of the antisera were determined by direct enzyme-linked immunosorbent assay (ELISA).[23]
Development of dot blot assay and its validation
Toward this, immunoglobulins (IgG) were purified from antiserum by protein A-Sepharose chromatography.[24] The purified IgG were biotinylated by the N-hydroxysuccinimide coupling method.[25] In the dot blot, for the standards, “core” 14-3-3 protein [10-1.25 ng in 10 μL of phosphate-buffered saline (PBS)] was spotted on PVDF membrane, allowed to air-dry, and the unoccupied area was blocked for 3 h in 3% bovine serum albumin (BSA) in PBS. Next, the membranes were incubated with biotinylated anti-14-3-3 antibody (200 ng/mL) at 4°C overnight. The blot was washed 6 times (10 min for each change of wash) with the buffer [50 mM phosphate, pH 7.0 containing 0.25% gelatin, 1M NaCl, 0.05% triton X-100, and 1 mM thylenediaminetetraacetic acid (EDTA)]. The blots were placed in 1:1000 diluted avidin-peroxidase for 1 h at room temperature (RT) and washed again, as described earlier. The antigen-antibody complex was visualized by using 3,3'-diaminobenzidine (1 mg/mL, in 50 mM citrate buffer, pH 5.5) as the chromogen.
For the CSF samples, 1 mL of CSF was concentrated to 10 µL (100-fold) using the centrifugal concentrators, by spinning at 5000 rpm for 90 min at 4° C in a refrigerated centrifuge. The CSF sample was then loaded on PVDF membrane and processed as described above. The intensity of the color developed was quantitated by densitometric scanning and calculated from the graph constructed with the concentration of the antigen standards.
For validation of the test, CSF samples from CJD cases were retrieved from the archives of the HBTR, NIMHANS. This included 50 cases. Four were definite CJD (confirmed by immunohistochemical demonstration of PrPsc in brain tissues examined postmortem using KG9 monoclonal antibody procured from the Roslin Institute, University of Edinburgh, UK) and 46 were probable CJD, classified based on Master's criteria for diagnosis of CJD.[13] CSF samples collected from patients who were suffering from the following disorders: AD, PD, leukodystrophy, chronic alcoholism, Down syndrome, multiple sclerosis, head injury, or Wilson disease served as disease controls.
Statistical analysis
Results were analyzed with the aid of GraphPad Version 3 (Prizm; GraphPad Software Inc., San Diego, California, USA). The results were expressed as the mean ± standard error of the mean (S.E.M). The differences were analyzed using Mann–Whitney test, assuming Gaussian distribution. Differences were considered significant if P < 0.05. Standard measures of statistical significance were used to identify true-positive, true-negative, false-positive, and false-negative results. The levels of 14-3-3 in 50 CJD cases and 70 disease controls were compared using receiver operating characteristic (ROC) analysis.
Results
Choice and preparation of the antigen
For the development of dot blot method for quantitation of 14-3-3 protein, the amino acid sequence of all the seven isoforms of 14-3-3 protein were scrutinized using Clustal W program.[26] The analysis revealed the presence of 5 clusters exhibiting 100% homology among them. Next, these sequences were subjected to computer algorithm AMPHI for indicating the propensity to harbor relevant T-helper cell recognition motifs.[27] Based on these predictions, the sequences were selected for their immunogenic potential, while eliminating the irrelevant sequences from the full length 14-3-3 protein. These 5 sequences were aligned with gly-gly linker sequence [Figure 1] and considered as the core protein (103 amino acids long). The synthetic gene encoding the “core” sequence of 14-3-3 protein was constructed using the overlapping PCR and the products were analyzed by 2% agarose gel electrophoresis [Figure 2]. A stepwise elongation of the core template (83 nt) occurred after the second (153 nt), third (219 nt), and fourth (276 nt) rounds of PCR, finally yielding a 335-nt long product. The purified PCR product was cloned into pMAL-c4X vector. The DNA sequence of the recombinant plasmid revealed that the inserted sequence was appropriate and corresponded to the gene as designed.
Figure 1.
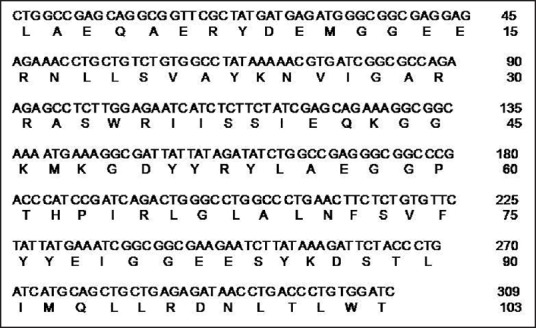
Complete DNA sequence of the synthetic gene encoding the core sequence of 14-3-3 protein and its primary amino acid sequence represented in single letter code
Figure 2.
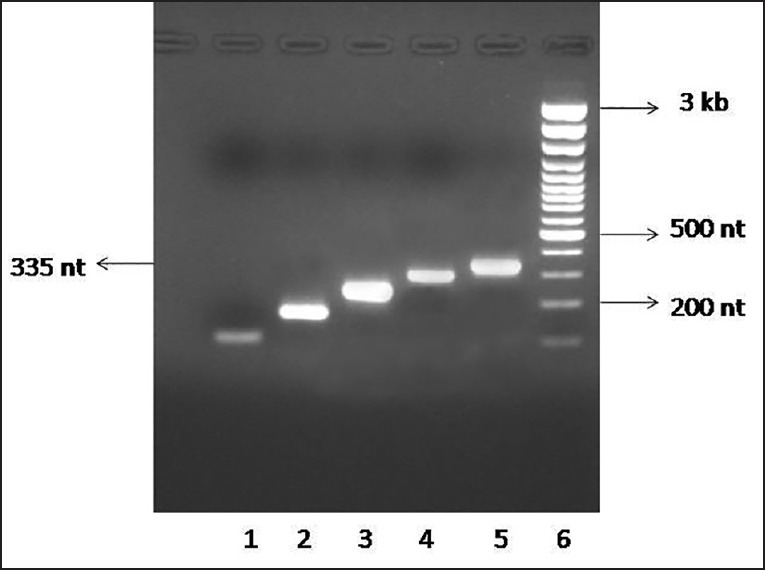
Analysis of PCR products by 2% agarose gel electrophoresis. The following samples were loaded in the gel. Lane 1: 5 μ L of 1st PCR product; Lane 2: 5 μ L of 2nd PCR product; Lane 3: 5 μ L of 3rd PCR product; Lane 4: 5 μ L of 4th PCR product; Lane 5: 5 μ L of 5th PCR product; Lane 6: 4 μ L of 100-bp DNA ladder
The expression of the recombinant “core”14-3-3 protein (11 kDa) was achieved by induction with IPTG as an extension of maltose binding protein (MBP) fusion protein (43 kDa) corresponding to the 54KDa band on SDS-PAGE. As the fusion protein was accumulated as insoluble aggregates, it was first solubilized in 8 M urea and further purified by passing through amylose affinity chromatography. After cleaving the fusion tag, the recombinant “core” 14-3-3 protein was analyzed by tricine–SDS-PAGE [Figure 3]. The yield was about 1 mg of recombinant protein per liter of bacterial culture.
Figure 3.
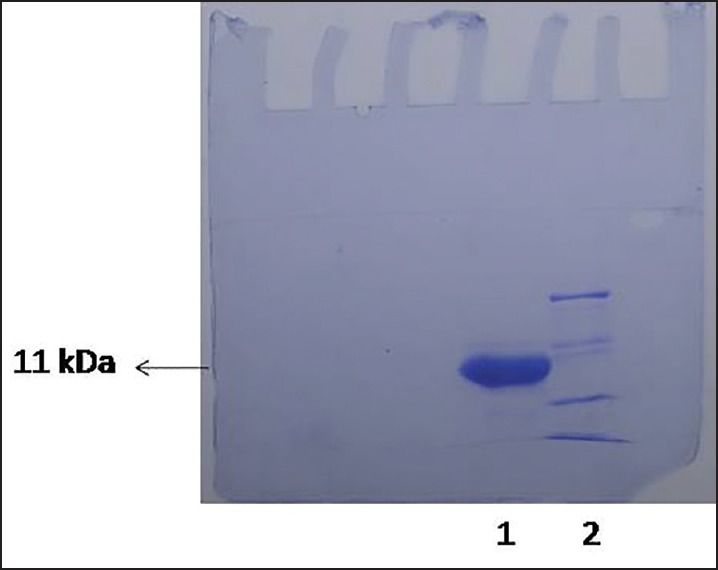
Tricine–SDS-PAGE analysis of purified recombinant core 14-3-3 protein. Shown are the 50 μg of purified protein in lane 1 and molecular weight markers in lane 2
Production of polyclonal antibodies to recombinant “core” 14-3-3 protein
The immune recognition pattern of the antiserum toward “core” 14-3-3 protein and the antibody titer was determined by direct ELISA [Figure 4]. The dilution of peptide-specific sera at 50% Bmax was found to be approx. 1:10,000.
Figure 4.
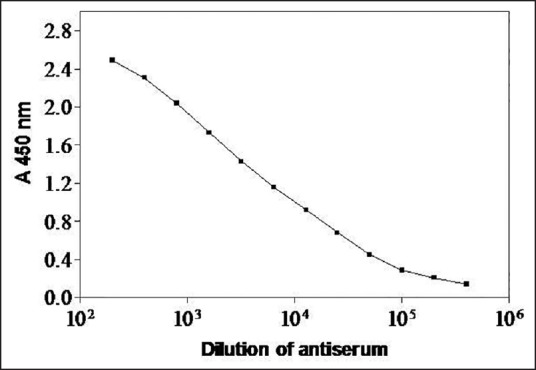
Immunoreactivity of rabbit anti-core protein serum. Binding curve obtained with serially diluted rabbit anti-core 14-3-3 protein against 1 μ g per well of the antigen is shown
Quantitation of 14-3-3 protein in human CSF samples by dot blot method
For the standards, “core” 14-3-3 protein was spotted (10-1.25 ng in 10 μL of PBS) on PVDF membrane for immunoblotting [Figure 5a]. A total of 120 samples were analyzed by this method and the data are presented in Table 1. A representative immunostained section of the brain tissue from a definite case of CJD, with PrPSc-specific monoclonal KG9 antibody. served as the positive reference [Figure 5b]. The dot blot analyses of the CSF samples from the 4 definite CJD cases are shown in Figure 5c and some of the probable CJD samples in Figure 5d, and the pattern obtained with disease controls in Figure 5e. The results obtained in the present study strongly affirm high diagnostic utility of the dot blot method with almost 10-fold difference in the levels of 14-3-3 protein between the CJD cases and controls [Table 2].
Figure 5.
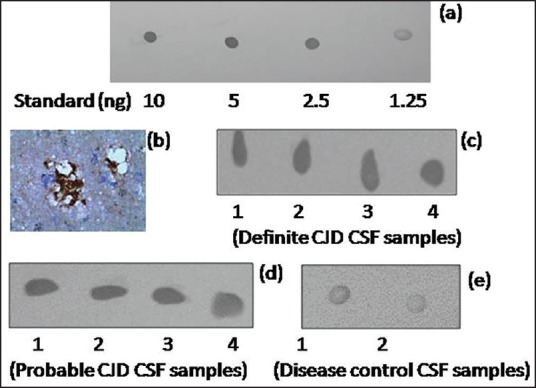
Dot blot detection sensitivity of biotinylated anti-core 14-3-3 antibody. The immunoreactivity obtained with varying concentrations of 14-3-3 protein (10-1.25 ng) is shown in panel (a). Photographs of the representative PrPSc-immunostained brain section (magnification: 400×) and the dot blots corresponding to definite CJD samples shown in panels (b) and (c) respectively. Representative dot blots of probable CJD and disease control samples shown in panels (d) and (e) respectively
Table 1.
Details of the samples analyzed by dot blot
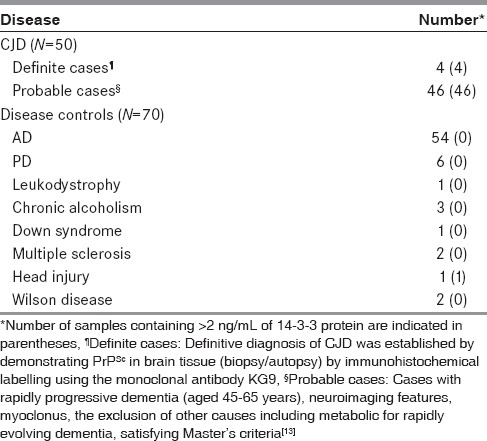
Table 2.
The results of the 14-3-3 assay

For evaluating test utility, ROC analysis was carried out. The results suggested an optimal cutoff value of 2 ng/mL. Using this cutoff, we obtained a sensitivity of 100% for definite and probable cases and a specificity of 86% when other neurological disorders were considered, with a positive predictive score of 88% and a negative predictive score of 100%.
Discussion
Presence of 14-3-3 protein, as individual isoforms or as a combination of the isoforms have been reported to be a biomarker in the CSF of CJD cases, by ELISA and western blotting methods, albeit with conflicting reports.[9,10,11,12,28,29] However, the analytic mean of the cutoff data between CJD and case controls is not very clear, often with low specificity. With a view to enhance the specificity of the 14-3-3 antiserum for selective recognition in CSF samples, we developed a solid phase immunoassay using antibodies generated against immunologically relevant “core” 14-3-3 protein.
The analysis of CSF samples using this indigenously developed method indicated a significantly higher level of 14-3-3 in definite as well as probable CJD samples, compared to the CSF from patients with other neurological disorders. These findings highlight the utility of the assay, in clinical practice for the diagnosis of CJD. The use of a core protein encompassing the epitopes shared by all the isoforms as the antigen contributed to the high specificity and sensitivity of the assay. The antibodies to this protein used in this assay detect total 14-3-3 content, recognizing the isoforms present in CSF. Although further evaluation of the 14-3-3 isoform pattern by using isoform-specific antibodies may still be required, this technique may be useful in differentiating CJD from other diseases including rapidly progressive central nervous system disorders. Because of the ubiquitous nature of 14-3-3 protein in the brain, it is expected to find traceable amounts of this protein in CSF from cases of head injury. However, this presence did not alter the specificity and sensitivity of the assay, which retained its diagnostic usefulness. Further validation of this test requires large-scale studies of larger numbers of samples.
This study has great financial implications for the patient as, at present, the analysis of 14-3-3 protein is being offered by only a few centers such as the National CJD Research and Surveillance Unit at the University of Edinburgh (http://www.cjd.ed.ac.uk/diagnosis), UK, the National Prion Disease Pathology Surveillance Center at Case Western Reserve University, Ohio (http://case.edu/med/pathology/centers/npdpsc/) USA, the Mayo Clinic, USA (www.mayomedicallaboratories.com), and the Australian National CJD Registry, Australia (http://ancjdr.path.unimelb.edu.au/diagnostic-testing/14-3-3-csf-protein-test), often at an approximate cost of 100-150 USD equivalent' per sample. In contrast, our indigenously developed 14-3-3 assay incurred the reagent cost of <20 USD equivalent. In view of this economic advantage, we are working toward offering a cost-effective, sensitive diagnostic test from the Department of Neurochemistry at NIMHANS at an affordable price, to the clinicians managing cases of CJD-prion disease.
Financial support and sponsorship
Financial Support from ICMR, New Delhi.
Conflicts of interest
There are no conflicts of interest.
Acknowledgment
The authors acknowledge the financial assistance received from Indian Council of Medical Research, New Delhi, India [Grant no. 5/4-5/128/Neuro/CAR/2013-NCD-I] to establish this facility.
References
- 1.Moore BW, Perez VJ. Specific acidic proteins in the nervous system. In: Carlson FD, editor. Physiological and Biochemical Aspects of Nervous Integration. Englewood Cliffs, NJ: Prentice-Hall Publishers; 1967. pp. 343–59. [Google Scholar]
- 2.Berg D, Holzmann C, Reiss O. 14-3-3 proteins in the nervous system. Nat Rev Neurosci. 2003;4:752–62. doi: 10.1038/nrn1197. [DOI] [PubMed] [Google Scholar]
- 3.Yaffe MB. How do 14-3-3 proteins work.— Gatekeeper phosphorylation and the molecular anvil hypothesis? FEBS Lett. 2002;513:53–7. doi: 10.1016/s0014-5793(01)03288-4. [DOI] [PubMed] [Google Scholar]
- 4.van Hemert MJ, Steensma HY, van Heusden GP. 14-3-3 proteins: Key regulators of cell division, signaling and apoptosis. Bioessays. 2001;23:936–46. doi: 10.1002/bies.1134. [DOI] [PubMed] [Google Scholar]
- 5.Fu H, Subramanian RR, Masters SC. 14-3-3 proteins: Structure, function, andregulation. Ann Rev Pharmacol Toxicol. 2000;40:617–47. doi: 10.1146/annurev.pharmtox.40.1.617. [DOI] [PubMed] [Google Scholar]
- 6.Foote M, Zhou Y. 14-3-3 proteins in neurological disorders. Int J Biochem Mol Biol. 2012;3:152–64. [PMC free article] [PubMed] [Google Scholar]
- 7.Layfield R, Fergusson J, Aitken A, Lowe J, Landon M, Mayer RJ. Neurofibrillary tangles of Alzheimer's disease brains contain 14-3-3 proteins. Neurosci Lett. 1996;209:57–60. doi: 10.1016/0304-3940(96)12598-2. [DOI] [PubMed] [Google Scholar]
- 8.Cao L, Cao W, Zhang W, Lin H, Yang X, Zhen H, et al. Identification of 14-3-3 protein isoforms in human astrocytoma by immunohistochemistry. Neurosci Lett. 2008;432:94–9. doi: 10.1016/j.neulet.2007.11.071. [DOI] [PubMed] [Google Scholar]
- 9.Satoh K, Tobiume M, Matsui Y, Mutsukura K, Nishida N, Shiga Y, et al. Establishment of a standard 14-3-3 protein assay of cerebrospinal fluid as a diagnostic tool for Creutzfeldt-Jakob disease. Lab Invest. 2010;90:1637–44. doi: 10.1038/labinvest.2009.68. [DOI] [PubMed] [Google Scholar]
- 10.Shiga Y, Wakabayashi H, Miyazawa K, Kido H, Itoyama Y. 14-3-3 protein levels and isoform patterns in the cerebrospinal fluid of Creutzfeldt-Jakob disease patients in theprogressive and terminal stages. J Clin Neurosci. 2006;13:661–5. doi: 10.1016/j.jocn.2005.09.004. [DOI] [PubMed] [Google Scholar]
- 11.Van Everbroeck BRJ, Boons J, Cras P. 14-3-3 {gamma}-isoform detection distinguishes sporadic Creutzfeldt-Jakob disease from other dementias. J NeurolNeurosurg Psychiatry. 2005;76:100–2. doi: 10.1136/jnnp.2003.032037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matsui Y, Satoh K, Miyazaki T, Shirabe S, Atarashi R, Mutsukura K, et al. High sensitivity of an ELISA kit for detection of the gamma-isoform of 14-3-3 proteins: Usefulness in laboratory diagnosis of human prion disease. BMC Neurol. 2011;11:120. doi: 10.1186/1471-2377-11-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Masters CL, Harris JO, Gajdusek DC, Gibbs CJ, Jr, Bernoulli C, Asher DM. Creutzfeldt-Jakob disease: Patterns of worldwide occurrence and the significance of familial and sporadic clustering. Ann Neurol. 1979;5:177–88. doi: 10.1002/ana.410050212. [DOI] [PubMed] [Google Scholar]
- 14.Satishchandra P, Sharma P, Chatterji P, Shankar SK. Psychiatric manifestations of Creutzfeldt Jakob disease: Probable neuropathological correlates. Neurol India. 1996;44:43–6. [PubMed] [Google Scholar]
- 15.Parchi P, Giese A, Capellari S, Brown P, Schulz-Schaeffer W, Windl O, et al. Classification of sporadic Creutzfeldt-Jakob disease based on molecular and phenotypic analysis of 300 subjects. Ann Neurol. 1999;46:224–33. [PubMed] [Google Scholar]
- 16.Mahadevan A, Shankar SK, Yasha TC, Santosh V, Sarkar C, Desai AP, et al. Brain biopsy in Creutzfeldt-Jakob disease: Evolution of pathological changes by prion protein immunohistochemistry. Neuropathol Appl Neurobiol. 2002;28:314–24. doi: 10.1046/j.1365-2990.2002.00399.x. [DOI] [PubMed] [Google Scholar]
- 17.Satishchandra P, Shankar SK. Creutzfeldt-Jakob disease in India (1971-1990) Neuroepidemiology. 1991;10:27–32. doi: 10.1159/000110244. [DOI] [PubMed] [Google Scholar]
- 18.Diagnosis and Therapy of Human Transmissible Spongiform Encephalopathies: Report of a WHO Consultation WHO/EMC/ZDI/98.9. Geneva: World Health Organization; 1998. Global Surveillance; pp. 1–29. [Google Scholar]
- 19.DiDonato A, deNigris M, Russo N, DiBiase S, D'Alessio G. A method for synthesizing genes and cDNAs by the polymerase chain reaction. Anal Biochem. 1993;212:291–3. doi: 10.1006/abio.1993.1328. [DOI] [PubMed] [Google Scholar]
- 20.Subramanian S, Madhavadas S, Balasubramanian P. Influence of conformational antibodies on dissociation of fibrillar amyloid beta (A beta 1-42) in vitro. Indian J Exp Biol. 2009;47:309–13. [PubMed] [Google Scholar]
- 21.Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. NY: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 22.Schägger H, von Jagow G. Tricine-sodium dodecyl sulphate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100kDa. Anal Biochem. 1987;166:368–79. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 23.Subramanian S, Divya Shree AN. Enhanced Th2 immunity after DNA prime-protein boost immunization with amyloid beta (1-42) plus CpG Oligodeoxynucleotides in aged rats. Neurosci Lett. 2008;436:219–22. doi: 10.1016/j.neulet.2008.03.024. [DOI] [PubMed] [Google Scholar]
- 24.Subramanian S, Bandopadhyay D, Mishra PK, Mathew M, John M. Design anddevelopment of non-fibrillar amyloid beta as a potential Alzheimer vaccine. BiochemBiophys Res Commun. 2010;394:393–7. doi: 10.1016/j.bbrc.2010.03.031. [DOI] [PubMed] [Google Scholar]
- 25.Sélo I, Négroni L, Créminon C, Grassi J, Wal JM. Preferential labeling of alpha-amino N-terminal groups in peptides by biotin: Application to the detection of specific anti-peptide antibodies by enzyme immunoassays. J Immunol Methods. 1996;99:127–38. doi: 10.1016/s0022-1759(96)00173-1. [DOI] [PubMed] [Google Scholar]
- 26.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acid Res. 1994;22:4673–80. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Margalit H, Spouge JL, Cornett JL, Cease KB, Delisi C, Berzofsky JA. Prediction of immunodominant helper T-cell antigenic sites from the primary sequence. J Immunol. 1987;138:2213–29. [PubMed] [Google Scholar]
- 28.Muayqil T, Gronseth G, Camicioli R. Evidence-based guideline: Diagnostic accuracy of CSF 14-3-3 protein in sporadic Creutzfeldt-Jakob disease: Report of the guideline development subcommittee of the American Academy of Neurology. Neurology. 2012;79:1499–506. doi: 10.1212/WNL.0b013e31826d5fc3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paterson RW, Torres-Chae CC, Kuo AL, Ando T, Nguyen EA, Wong K, et al. Differential diagnosis of Jakob-Creutzfeldt disease. Arch Neurol. 2012;69:1578–82. doi: 10.1001/2013.jamaneurol.79. [DOI] [PMC free article] [PubMed] [Google Scholar]


