Abstract
In recent years, long noncoding RNAs (lncRNAs) have been demonstrated to play important roles in the development of human cancer. We assessed the role of lncRNA ANRIL in non-small-cell lung cancer (NSCLC). Quantitative real-time polymerase chain reaction was employed to detect the expression of ANRIL in NSCLC tissues and paired nontumor tissues. The high expression level of ANRIL was positively correlated with advanced tumor–node–metastasis stage and greater tumor diameter. Furthermore, chromatin immunoprecipitation assays confirmed the physical interaction between c-Myc and ANRIL. ANRIL silencing significantly inhibited NSCLC cell proliferation. Together, we showed that ANRIL is involved in the oncogenesis of NSCLC, and ANRIL may be a potential therapeutic target for patients with NSCLC.
Keywords: c-Myc, ANRIL, cell proliferation, NSCLC
Introduction
Lung cancer remains the leading cause of cancer-related deaths around the world.1 Non-small-cell lung cancer (NSCLC), including adenocarcinoma, squamous cell carcinoma, and large cell carcinoma account for >85% of all lung cancer cases.2 Despite the promising advances made in therapeutic technologies, the prognosis of this disease remains dismal, with the 5-year survival rate of NSCLC (all stages combined) being only ~13%.3 Tumor metastasis and recurrence are common and represent major obstacles to the improvement of patient survival. The long-term survival rate is <50% even after curative resections in NSCLC.4 Therefore, illumination of the mechanisms underlying metastasis and recurrence is the prerequisite for the development of novel therapeutics in NSCLC.
Long noncoding RNAs (lncRNAs) are a class of noncoding RNAs longer than 200 nucleotides and with limited protein-coding potential. Studies revealed that lncRNAs could interact with DNA, RNA, or protein and regulate a variety of genes with diversified mechanisms, thereby having an effect on a large number of cellular pathways, including carcinogenesis.5–10 Aberrantly expressed lncRNAs have been demonstrated to play key roles in the progression of NSCLC.11–13
lncRNA ANRIL (CDKN2B antisense RNA 1) is a 3.8 kb lncRNA transcribed from the INK4B-ARF-INK4A gene cluster in the opposite direction.14 ANRIL has been demonstrated to exert oncogenic activity in a variety of carcinomas, including NSCLC13 and gastric cancer.15 However, the factors that lead to its upregulation and the biological roles of ANRIL in NSCLC remain to be explored.
c-Myc is a well-noted transcriptional factor and has been shown to be abnormally overexpressed in a variety of carcinomas, including NSCLC.16 As a vital transcription regulator, c-Myc plays an essential role in the regulation of many biological processes, including cell cycle, proliferation, apoptosis, protein synthesis, and cell metabolism.17 It can bind to the cis-regulatory element E-box (CACGTG) to transactivate targeted genes.18 In this study, we have shown that c-Myc can directly transactivate ANRIL via interacting with putative c-Myc target response element in the promoter region of ANRIL. Moreover, we examined the effect of ANRIL on the biological behaviors of NSCLC cells. Our results demonstrated that ANRIL could promote the proliferation of NSCLC cells.
Materials and methods
Patient samples
Fifty-six NSCLC tissues and their paired adjacent normal tissues in this study were obtained from patients who underwent radical resections at the Sixth Hospital (Shanghai Jiaotong University, Shanghai, People’s Republic of China). The study was approved by Research Ethics Committee of Shanghai Jiaotong University (Shanghai, People’s Republic of China). Informed consent was obtained from all patients.
Cell culture
An NSCLC squamous carcinoma cell line (SK-MES-1), three NSCLC adenocarcinoma cell lines (A549, SPC-A1, and NCI-H1975), and a normal human bronchial epithelial cell line (16HBE) were purchased from the Institute of Biochemistry and Cell Biology of the Chinese Academy of Sciences (Shanghai, People’s Republic of China). The cells were cultured in Roswell Park Memorial Institute 1640 or Dulbecco’s Modified Eagle’s Medium (DMEM) (Gibco-BRL; Grand Island, NY, USA) medium supplemented with 10% fetal bovine serum (Thermo Fisher Scientific, Waltham, MA, USA), 100 U/mL penicillin, and 100 mg/mL streptomycin (Invitrogen) in incubator at 37°C with 5% CO2.
RNA extraction and quantitative real-time polymerase chain reaction
Total RNA was extracted from cancer cells utilizing the Trizol reagent (Invitrogen) following the manufacturer’s instructions. First strand complementary DNA (cDNA) was generated with the Reverse Transcription System Kit (Invitrogen). The cDNA template was then amplified using the standard SYBR Green polymerase chain reaction (PCR) kit protocol with real-time (RT)-PCR. The quantitative real-time polymerase chain reaction (qRT-PCR) was performed on ABI 7500 system (Applied Biosystems, Foster City, CA, USA). β-Actin was used as the internal control, and messenger RNA (mRNA) values were normalized to that of β-actin. The relative expression of mRNA was calculated with the 2−ΔΔCt method. Each sample was repeated in triplicate. qRT-PCR data were analyzed and converted to relative fold changes. The primer sequences used in the present study were as follows: ANRIL, 5′-TTGTGAAGCCCAAGTACTGC-3′ (forward), 5′-TTCACTGTGGAGACGTTGGT-3′ (reverse); c-Myc, 5′-CACCGAGTCGTAGTCGAGGT-3′ (forward), 5′-GCTGCTTAGACGCTGGATTT-3′ (reverse); GAPDH, (glyceraldehyde-3-phosphate dehydrogenase) 5′-AGAAGGCTGGGGCTCATTTG-3′ (forward), 5′-AGGGGCCATCCACAGTCTTC-3′ (reverse); and β-actin, 5′-CTGGGACGACATGGAGAAAA-3′ (forward), 5′-AAGGAAGGCTGGAAGAGTGC-3′ (reverse).
Plasmid construction
The ANRIL-specific small interfering RNAs, c-Myc-specific siRNA, and control siRNA were purchased from GenePharma (Shanghai, People’s Republic of China). The c-Myc-specific siRNA sequences were 5′-GGUGAUCCAGACUCUGACCUU-3′; for ANRIL, 5′-GGUCAUCUCAUUGCUCUAU-3′; and for siRNA control, 5′-UUCUCCGAACGUGUCACGUTT-3′. The cDNA of c-Myc was PCR-amplified and subcloned into the pcDNA3.1 vector (Invitrogen).
The primers were c-Myc: 5′-CCGGGAATTCCTGGATTTTTTTCGGGTAGTG-3′ and 5′-CCGGCTCGAGTTACGCACAAGAGTTCCGTAG-3′. Transfections were carried out utilizing the Lipofectamine 2000 kit (Invitrogen). The transfected cells were harvested at 48 hours after transfection.
The promoter region of ANRIL (−862 to −325, 537 bp) was PCR-amplified with the primers 5′-GGGGTACCAT GTCCGCTGCACTTT-3′ (forward) and 5′-GCAAAGCTTACCCAACCTGGTAAGTGG-3′ (reverse) and then subcloned into basic firefly luciferase reporter (pGL3). pGL3 construct containing the ANRIL promoter region with point mutations in the E-box element was synthesized with the QuikChange Site-Directed Mutagenesis kit (St Agilent Technologies, Santa Clara, CA, USA) using the primer 5′-ACTGGTACCTAAGCGGAGAGAGTCCCACACAGG-3′ (forward) and 5′-ACTAAGCTTGAGTCAGAGTTCCCCAC-3′ (reverse).
Chromatin immunoprecipitation assay
We performed chromatin immunoprecipitation (ChIP) assay using EZ-CHIP KIT (Millipore, Billerica, MA, USA). Quantification of immunoprecipitated DNA was performed using qPCR with SYBR Green Mix (Takara, Dalian, People’s Republic of China) with ChIP primers: 5′-AAGATCTCGGAACGGCTCT-3′ (forward), 5′-TCAGGTGACGGATGTAGCTA-3′ (reverse). ChIP data were calculated as a percentage relative to the input DNA.
Luciferase reporter assay
pcDNA3.1-c-Myc or c-Myc siRNA and the luciferase reporter construct were cotransfected into NSCLC cells. The construct was cotransfected with the Renilla luciferase plasmid to monitor transfection efficiency (Promega, Madison, WI, USA). The relative luciferase activity was normalized with Renilla luciferase activity.
Cell viability assay and ethynyl deoxyuridine incorporation analysis
Cell viability was assessed utilizing the Cell Counting Kit 8 (CCK-8, Dojindo, Tokyo, Japan). Control and NSCLC cells transfected with desired vector were seeded into the 96-well plates at an initial density of 105 cells/mL. After indicated periods of cultivation, CCK-8 solution was added into each well (10 μL per 100 μL of medium in each well) and incubated for 2 hours. The absorbance was determined using a microplate reader (MRX; Dynex Technologies, West Sussex, UK) at 450 nm. Data were expressed as the percentage of viable cells over control. Each experiment was repeated in triplicate. Ethynyl deoxyuridine (EdU) assay kit (Ribobio, Guangzhou, People’s Republic of China) was used to analyze S-phase cells proportion. Briefly, control and treated cells were incubated with DMEM (Gibco-BRL) media containing 5 μM EdU, 10% fetal bovine serum (Invitrogen), and 1% penicillin/streptomycin (Gibco) for 2 hours. The control and treated cells were then fixed with 4% formaldehyde at room temperature for 30 minutes and incubated with Triton X-100 (0.5%) for 20 minutes. After that, the cells were washed with phosphate-buffered saline (3×5 minutes) and treated with 1× Apollo® reaction cocktail (Ribobio) for 30 minutes. Finally, the DNA were stained with Hoechst 33342 (5 μg/mL) for 30 minutes and visualized under a fluorescent microscope.
Western blot analysis
Total cell lysates were lysed in radioimmunoprecipitation assay buffer (Cell Signaling Technology, Beverly, MA, USA) and prepared in a 1× sodium dodecyl sulfate buffer. Equal quantities of proteins were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis and transferred onto a polyvinylidene fluoride Immobilon-P membrane (Millipore). The membrane was blocked with 5% nonfat milk and then incubated with indicated primary antibodies overnight at 4°C. After incubation with primary antibodies specific for proliferating cell nuclear antigen (PCNA), c-Myc, or β-actin (Abcam, Cambridge, UK), the blots were incubated with goat antirabbit secondary antibody (Abcam) and visualized with enhanced chemiluminescence.
Statistical analysis
All statistical analyses were performed utilizing SPSS 17.0 (SPSS, Chicago, IL, USA). All data were expressed as mean ± standard deviation from three independent experiments. Differences between the expression levels of ANRIL in NSCLC patients were compared with the Wilcoxon signed-rank test. The associations between the expression level of ANRIL and clinical parameters were calculated with the Mann–Whitney U-test. The correlation between ANRIL and c-Myc mRNA expression level was analyzed by Pearson’s correlation. Statistical evaluations were analyzed with independent Student’s t-test unless otherwise indicated. A P-value (two-sided) <0.05 was considered to be statistically significant.
Results
ANRIL is upregulated in NSCLC tissues
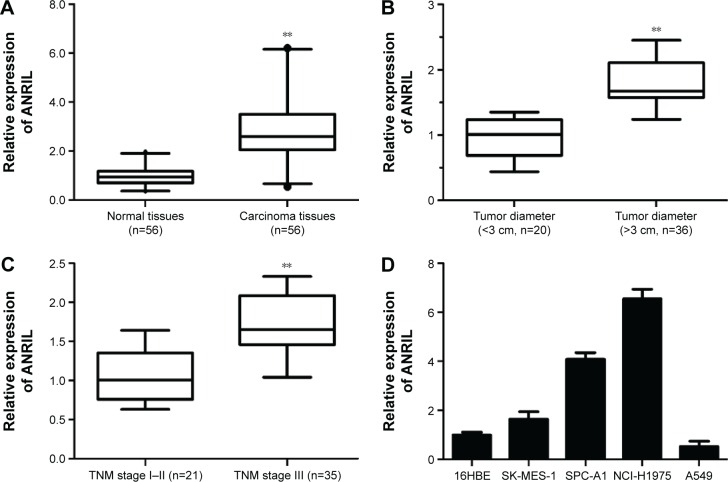
To determine whether ANRIL is differentially expressed in NSCLC tissues, qRT-PCR analysis was performed to determine the expression level of ANRIL. Among the 56 pairs of NSCLC patients, the expression level of ANRIL was significantly increased in NSCLC tissues compared with the adjacent normal tissues on the whole (P<0.001, Wilcoxon signed-rank test). Then, we examined the correlation between the expression level of ANRIL and the clinicopathological parameters in NSCLC (Figure 1A). As illustrated in Figure 1B and C and Table 1, ANRIL upregulation was positively correlated with greater tumor diameter (P=0.024, P<0.05) and advanced tumor–node–metastasis stage (P=0.035, P<0.05). However, no correlation between ANRIL expression and other clinical factors, such as sex, tumor histological grade, and lymph node metastasis was found in our study. We utilized qRT-PCR to determine the expression level of ANRIL in a series of lung cancer cell lines, including adenocarcinoma and squamous carcinoma subtypes. When normalized to 16HBE, the expression level of ANRIL was upregulated in NSCLC cell lines (SK-MES-1, SPC-A1, and NCI-H1975) but downregulated in A549 (Figure 1D).
Figure 1.
ANRIL expression in NSCLC and its clinical significance.
Notes: (A) Difference in expression levels of ANRIL expression levels between NSCLC tissues and matched normal tissues. The expression of ANRIL was normalized to β-actin. The statistical differences between samples were analyzed with Wilcoxon signed-rank test (n=56, P<0.01). (B) Relationship between ANRIL expression and primary tumor growth (P<0.01). (C) Relationship between ANRIL expression and TNM stage (P<0.01). (D) qRT-PCR analysis of ANRIL expression levels in NSCLC cell lines (SK-MES-1, SPC-A1, NCI-H1975, and A549) compared with the normal bronchial epithelial cell line (16HBE). **P<0.01.
Abbreviations: NSCLC, non-small-cell lung cancer; qRT-PCR, quantitative real-time polymerase chain reaction; TNM, tumor–node–metastasis.
Table 1.
Correlation between ANRIL expression and clinicopathologic characteristics
| Clinicopathologic parameters | ANRIL expression | Number of cases | P-value | |
|---|---|---|---|---|
| High | Low | |||
| Age (years) | ||||
| 60 | 10 | 14 | 24 | 0.230 |
| >60 | 14 | 18 | 32 | |
| Sex | ||||
| Male | 13 | 11 | 24 | 0.404 |
| Female | 13 | 19 | 32 | |
| Tumor diameter (cm) | ||||
| ≤3 | 11 | 9 | 20 | 0.024a |
| >3 | 24 | 12 | 36 | |
| Lymph node metastasis | ||||
| Yes | 7 | 11 | 18 | 0.405 |
| No | 18 | 21 | 39 | |
| TNM stage | ||||
| I–II | 5 | 16 | 21 | 0.035a |
| III | 25 | 10 | 35 | |
| Degree of differentiation | ||||
| Well and moderate | 14 | 16 | 30 | 0.378 |
| Poor | 15 | 11 | 26 | |
Note:
Statistically significant (P<0.05).
Abbreviation: TNM, tumor–node–metastasis.
c-Myc physically binds to the promoter region of ANRIL and induces its upregulation
In order to explore how the transcription of ANRIL was regulated, we performed a search for possible transcription factor-binding sites in the promoter region of ANRIL with the JASPAR database (http://jaspar.genereg.net) and found one E-box element at ~589 upstream of ANRIL, which may be recognized by c-Myc.
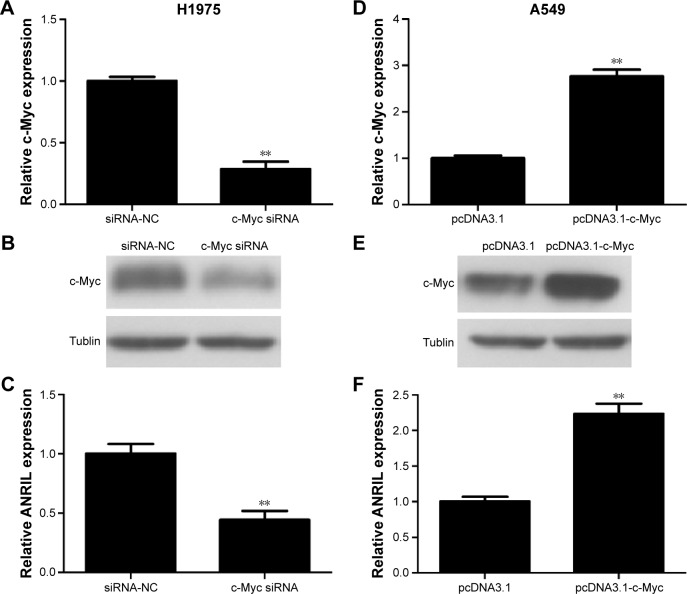
In the first place, we would like to investigate whether c-Myc indeed regulates the expression of ANRIL. We transfected c-Myc plasmid or siRNA into NSCLC cell lines (A549 and H1975). We used qPCR and Western blot assay to evaluate the efficiency of transfection (Figure 2A, B, D, and E). Forty-eight hours after treatment, we examined the expression level of ANRIL. Our results demonstrated that inhibition of c-Myc decreased the expression of ANRIL in H1975 cells (Figure 2C) while exogenous c-Myc increased the expression of ANRIL in A549 cells (Figure 2F).
Figure 2.
The expression changes of c-Myc or ANRIL after transfection of c-Myc siRNA, siRNA-NC or pcDNA3.1-c-Myc, pcDNA3.1 in NSCLC cells.
Notes: The relative mRNA expression levels were evaluated with real-time qPCR. c-Myc protein levels were determined using Western blot assay. Each experiment was performed in triplicate. (A) c-Myc siRNA significantly downregulated the expression of c-Myc at mRNA levels in H1975 cells. (B) Representative images of Western blot results indicated c-Myc siRNA significantly downregulated the expression of c-Myc at protein level in H1975 cells. (C) c-Myc siRNA significantly downregulated the expression of ANRIL in H1975 cells. (D) pcDNA3.1-c-Myc markedly upregulated the expression of c-Myc at mRNA levels in A549 cells. (E) Representative images of Western blot results indicated pcDNA3.1-c-Myc significantly upregulated the expression of c-Myc at protein levels in A549 cells. (F) pcDNA3.1-c-Myc significantly upregulated the expression of ANRIL in A549 cells. **P<0.01.
Abbreviations: NC, negative control; NSCLC, non-small-cell lung cancer; qPCR, quantitative polymerase chain reaction; siRNA, small interfering RNA; mRNA, messenger RNA.
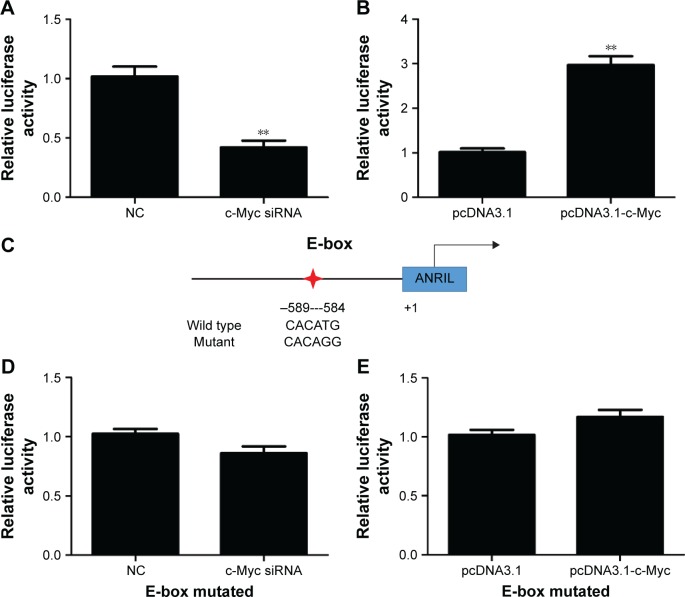
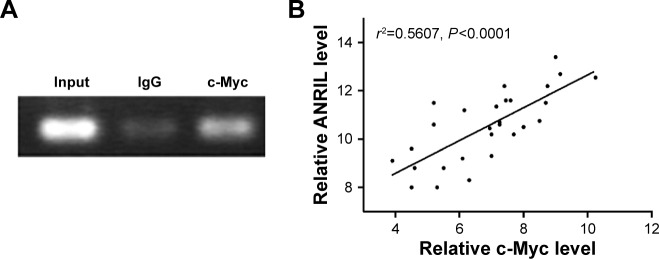
To explore the molecular mechanisms of the effects that c-Myc had on ANRIL, we cloned the promoter region of ANRIL into the basic firefly luciferase reporter (pGL3) and cotransfected the construct with pcDNA3.1-c-Myc into the A549 or c-Myc siRNA into H1975 cells. Ectopic expression of c-Myc dramatically enhanced the luciferase activity of the construct, while inhibition of c-Myc suppressed the luciferase activity of the construct (Figure 3A and B). To further confirm the specificity of the interaction, we synthesized the E-box element with point mutations (Figure 3C) and cotransfected cells with c-Myc siRNA or pcDNA3.1-c-Myc. Our data showed that mutations in the E-box element attenuated the effects of c-Myc on the promoter activity of ANRIL (Figure 3D and E). To further confirm the physical interaction between c-Myc and the E-box element in the promoter region of ANRIL, we performed ChIP assay. c-Myc immunoprecipitates were obviously enriched in the DNA fragments compared with the negative control immunoglobulin G immunoprecipitates (Figure 4A). Thus, our results reveal that c-Myc activates the transcription of ANRIL via physically interacting with the c-Myc-responsive element (E-box) in the ANRIL promoter region. Furthermore, a positive correlation was found between the expression levels of ANRIL and c-Myc (r2=0.5607, P<0.001, Figure 4B).
Figure 3.
c-Myc regulates the ANRIL promoter activity, depending on the E-box element.
Notes: (A) Dual luciferase assay on H1975 cells cotransfected with firefly luciferase constructs containing the ANRIL promoter and c-Myc siRNA or the control siRNA. (B) Dual luciferase assay on A549 cells cotransfected with firefly luciferase constructs containing the ANRIL promoter and pcDNA3.1-c-Myc or pcDNA3.1. (C) Schematic of the ANRIL-promoter-luciferase construct is depicted with the locations of the E-box element and the sequences of point mutation. The red star indicates the location of the putative E-box in the promoter region of ANRIL. (D) Dual luciferase assay of H1975 cells cotransfected with the ANRIL promoter reporter constructs (mutant at the E-box element) and c-Myc siRNA or siRNA NC. (E) Dual luciferase assay of A549 cells cotransfected with the ANRIL promoter reporter constructs (mutant at the E-box element) and pcDNA3.1-c-Myc or pcDNA3.1. All of the transfections were performed in triplicate. The values are presented as the mean ± standard deviation of the ratio of firefly luciferase activity to Renilla luciferase activity and are representative of at least three independent experiments. **P<0.01.
Abbreviations: NC, negative control; siRNA, small interfering RNA.
Figure 4.
Identification of the binding activity of c-Myc and the ANRIL promoter and the correlation between ANRIL and c-Myc.
Notes: (A) ChIP was used to confirm the c-Myc-binding activity. The c-Myc antibody effectively enriched the DNA sequence covering the putative binding element. Normal rabbit IgG was used as a negative control and an anti-c-Myc polymerase antibody was used as a positive control. (B) The correlation between ANRIL and c-Myc expression levels in NSCLC tissues (n=28). qRT-PCR was performed in triplicate for each sample and assays were repeated once. The relative levels were normalized to β-actin. Each point in the scatter graph represents an individual sample, in which relative c-Myc levels indicate on x-axis and ANRIL levels on y-axis. The x-axis shows normalized Ct values for c-Myc determined by qRT-PCR. The y-axis shows normalized Ct values for ANRIL determined by qRT-PCR.
Abbreviations: ChIP, chromatin immunoprecipitation; Ct, cycle threshold; IgG, immunoglobulin G; NSCLC, non-small-cell lung cancer; qRT-PCR, quantitative real-time polymerase chain reaction.
ANRIL promotes the proliferation of NSCLC cells
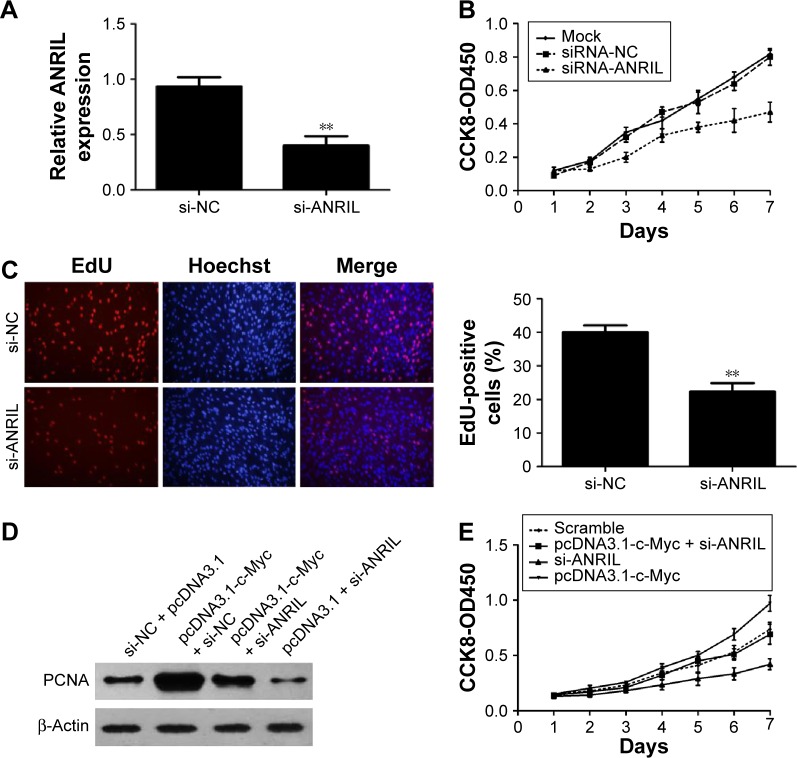
To explore the biological role of ANRIL in NSCLC development, we knocked down ANRIL by transfecting ANRIL siRNA into H1975 cells (Figure 5A). CCK-8 assays demonstrated that suppression of ANRIL inhibited the cell proliferation in H1975 cells (Figure 5B). Furthermore, the percentage of EdU-positive cells was reduced with ANRIL knockdown (Figure 5C). PCNA is deemed as an important index to estimate cell proliferation. Consistent with the attenuated proliferation, H1975 cells had markedly decreased expression of PCNA with ANRIL knockdown (Figure 5D). Furthermore, ANRIL silencing attenuated c-Myc-induced cell growth promotion (Figure 5E). Western blot analysis of PCNA confirmed the results from cell viability assays (Figure 5D).
Figure 5.
Effect of ANRIL on cell proliferation.
Notes: (A) qRT-PCR analysis of ANRIL expression following treatment of H1975 cells with specific siRNA targeting ANRIL. (B) H1975 cells were transfected with si-ANRIL or si-NC. CCK8 assays were performed to determine the proliferation of H1975 cells. Data were expressed as the percentage of viable cells as follows: relative viability (%) = (A450treated − A450blank)/(A450control − A450blank) ×100%. (C) The ANRIL silencing inhibited DNA replication in H1975 cells compared to controls as determined by the EdU incorporation assay, Original magnifications: ×200. EdU-positive cell counts were analyzed by using software of the light microscope. (D) Changes in the proliferation marker, PCNA, were shown by Western blotting analysis and normalized to β-actin after si-ANRIL transfection. Relative protein expression was identified (n=3). (E) Knockdown of ANRIL could partly reverse c-Myc-induced growth promotion as determined with CCK-8 assays. Data represent the mean ± SD from three independent experiments. **P<0.01.
Abbreviations: CCK-8, Cell Counting Kit 8; EdU, ethynyl deoxyuridine; NC, negative control; OD, optical density; PCNA, proliferating cell nuclear antigen; qRT-PCR, quantitative real-time polymerase chain reaction; SD, standard deviation; si, small interfering.
Discussion
It is known to all that human genomes encode thousands of lncRNAs.5 More and more studies are demonstrating that lncRNAs could be used as biomarkers and therapeutic targets in cancer.9–12 In the present study, we first explored the expression pattern of ANRIL in NSCLC. Our study identified that ANRIL is upregulated in NSCLC tissues than in the paired normal tissues. Also, we demonstrated that increased ANRIL expression level was positively correlated with greater tumor diameter and advanced tumor–node–metastasis stage.
Similar to the protein-coding counterparts, lncRNAs are also subject to typical transcription-factor-mediated and epigenetics-mediated regulation. For example, EZH2 could epigenetically suppress lncRNA SPRY4-IT1 expression in NSCLC,19 ANRIL is transactivated by the transcription factor E2F1 following DNA damage,20 HOTAIR could be transactivated by hypoxia inducible factor-1α.21 c-Myc has been shown to be overexpressed in a wide range of human tumors.16 c-Myc, a key transcription factor, plays a vital role in regulating many cellular biological processes, including cell proliferation, cell cycle, apoptosis, and cell metabolism.17 It can transactivate targeted genes via binding to the E-box element (CACGTG).18 We revealed that c-Myc directly binds to the E-box in the ANRIL promoter region and induced ANRIL expression.
In addition to ANRIL, which could be induced by c-Myc, previous studies have characterized a number of c-Myc-responsive lncRNAs, including HOTAIR,22 H19,23 and MINCR.24 These studies, in addition to ours, suggest that lncRNAs may also play a role in c-Myc-induced signaling.
In the present study, the function of ANRIL was investigated by loss-of-functions. As a result, RNA interference-mediated downregulation of ANRIL obviously inhibited NSCLC cell proliferation. c-Myc-induced cell growth promotion could be attenuated by ANRIL silencing. These results revealed that ANRIL may function as a downstream effector of c-Myc.
In conclusion, our study results suggest that ANRIL is a malignant driver in NSCLC that can be activated by transcription factor c-Myc. These data implicate that ANRIL may be a potential target of NSCLC therapy.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63(1):11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Taylor MD, Nagji AS, Bhamidipati CM, et al. Tumor recurrence after complete resection for non-small cell lung cancer. Ann Thorac Surg. 2012;93(6):1813–1820. doi: 10.1016/j.athoracsur.2012.03.031. discussion 1820–1821. [DOI] [PubMed] [Google Scholar]
- 3.Boolell V, Alamgeer M, Watkins DN, Ganju V. The evolution of therapies in non-small cell lung cancer. Cancers (Basel) 2015;7(3):1815–1846. doi: 10.3390/cancers7030864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vijayvergia N, Shah PC, Denlinger CS. Survivorship in non-small cell lung cancer: challenges faced and steps forward. J Natl Compr Canc Netw. 2015;13(9):1151–1161. doi: 10.6004/jnccn.2015.0140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang P, Xue Y, Han Y, et al. The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation. Science. 2014;344:310–313. doi: 10.1126/science.1251456. [DOI] [PubMed] [Google Scholar]
- 6.Cao C, Sun J, Zhang D, et al. The long intergenic noncoding RNA UFC1, a target of MicroRNA 34a, interacts with the mRNA stabilizing protein HuR to increase levels of β-catenin in HCC cells. Gastroenterology. 2015;148:415–426. doi: 10.1053/j.gastro.2014.10.012. [DOI] [PubMed] [Google Scholar]
- 7.Hu Y, Wang J, Qian J, et al. Long noncoding RNA GAPLINC regulates CD44-dependent cell invasiveness and associates with poor prognosis of gastric cancer. Cancer Res. 2014;74:6890–6902. doi: 10.1158/0008-5472.CAN-14-0686. [DOI] [PubMed] [Google Scholar]
- 8.Li Z, Chao TC, Chang KY, et al. The long noncoding RNA THRIL regulates TNFα expression through its interaction with hnRNPL. Proc Natl Acad Sci U S A. 2014;111:1002–1007. doi: 10.1073/pnas.1313768111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang F, Zhang L, Huo XS, et al. Long noncoding RNA high expression in hepatocellular carcinoma facilitates tumor growth through enhancer of zeste homolog 2 in humans. Hepatology. 2011;54:1679–1689. doi: 10.1002/hep.24563. [DOI] [PubMed] [Google Scholar]
- 10.Pandey GK, Mitra S, Subhash S, et al. The risk-associated long noncoding RNA NBAT-1 controls neuroblastoma progression by regulating cell proliferation and neuronal differentiation. Cancer Cell. 2014;26:722–737. doi: 10.1016/j.ccell.2014.09.014. [DOI] [PubMed] [Google Scholar]
- 11.Zhang E, He X, Yin D, et al. Increased expression of long noncoding RNA TUG1 predicts a poor prognosis of gastric cancer and regulates cell proliferation by epigenetically silencing of p57. Cell Death Dis. 2016;7:e2109. doi: 10.1038/cddis.2015.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Han L, Zhang EB, Yin DD, et al. Low expression of long noncoding RNA PANDAR predicts a poor prognosis of non-small cell lung cancer and affects cell apoptosis by regulating Bcl-2. Cell Death Dis. 2015;6:e1665. doi: 10.1038/cddis.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nie FQ, Sun M, Yang JS, et al. Long noncoding RNA ANRIL promotes non-small cell lung cancer cell proliferation and inhibits apoptosis by silencing KLF2 and P21 expression. Mol Cancer Ther. 2015;14(1):268–277. doi: 10.1158/1535-7163.MCT-14-0492. [DOI] [PubMed] [Google Scholar]
- 14.Yap KL, Li S, Muñoz-Cabello AM, et al. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol Cell. 2010;38(5):662–674. doi: 10.1016/j.molcel.2010.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang EB, Kong R, Yin DD, et al. Long noncoding RNA ANRIL indicates a poor prognosis of gastric cancer and promotes tumor growth by epigenetically silencing of miR-99a/miR-449a. Oncotarget. 2014;5(8):2276–2292. doi: 10.18632/oncotarget.1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen BJ, Wu YL, Tanaka Y, Zhang W. Small molecules targeting c-Myc oncogene: promising anti-cancer therapeutics. Int J Biol Sci. 2014;10(10):1084–1096. doi: 10.7150/ijbs.10190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dang CV. MYC on the path to cancer. Cell. 2012;149(1):22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Solomon DL, Amati B, Land H. Distinct DNA binding preferences for the c-Myc/Max and Max/Max dimers. Nucleic Acids Res. 1993;21(23):5372–5376. doi: 10.1093/nar/21.23.5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun M, Liu XH, Lu KH, et al. EZH2-mediated epigenetic suppression of long noncoding RNA SPRY4-IT1 promotes NSCLC cell proliferation and metastasis by affecting the epithelial-mesenchymal transition. Cell Death Dis. 2014;5:e1298. doi: 10.1038/cddis.2014.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wan G, Mathur R, Hu X, et al. Long non-coding RNA ANRIL (CDKN2B-AS) is induced by the ATM-E2F1 signaling pathway. Cell Signal. 2013;25(5):1086–1095. doi: 10.1016/j.cellsig.2013.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou C, Ye L, Jiang C, Bai J, Chi Y, Zhang H. Long noncoding RNA HOTAIR, a hypoxia-inducible factor-1α activated driver of malignancy, enhances hypoxic cancer cell proliferation, migration, and invasion in non-small cell lung cancer. Tumour Biol. 2015;36(12):9179–9188. doi: 10.1007/s13277-015-3453-8. [DOI] [PubMed] [Google Scholar]
- 22.Ma MZ, Li CX, Zhang Y, et al. Long non-coding RNA HOTAIR, a c-Myc activated driver of malignancy, negatively regulates miRNA-130a in gallbladder cancer. Mol Cancer. 2014;13:156. doi: 10.1186/1476-4598-13-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barsyte-Lovejoy DL, Lau SK, Boutros PC, et al. The c-Myc oncogene directly induces the H19 noncoding RNA by allele-specific binding to potentiate tumorigenesis. Cancer Res. 2006;66(10):5330–5337. doi: 10.1158/0008-5472.CAN-06-0037. [DOI] [PubMed] [Google Scholar]
- 24.Doose G, Haake A, Bernhart SH, et al. ICGC MMML-Seq Consortium MINCR is a MYC-induced lncRNA able to modulate MYC’s transcriptional network in Burkitt lymphoma cells. Proc Natl Acad Sci U S A. 2015;112(38):E5261–E5270. doi: 10.1073/pnas.1505753112. [DOI] [PMC free article] [PubMed] [Google Scholar]