Abstract
The Saccharomyces Genome Database (SGD; http://www.yeastgenome.org/) is the authoritative community resource for the Saccharomyces cerevisiae reference genome sequence and its annotation. To provide a wider scope of genetic and phenotypic variation in yeast, the genome sequences and their corresponding annotations from 11 alternative S. cerevisiae reference strains have been integrated into SGD. Genomic and protein sequence information for genes from these strains are now available on the Sequence and Protein tab of the corresponding Locus Summary pages. We illustrate how these genome sequences can be utilized to aid our understanding of strain-specific functional and phenotypic differences.
Database URL: www.yeastgenome.org
Introduction
The genome of the budding yeast Saccharomyces cerevisiae was the first available complete eukaryotic genome sequence. The reference genome for S. cerevisiae was determined from the strain S288C. This sequence has been fully annotated and maintained at the Saccharomyces Genome Database (SGD) for close to two decades. SGD has served as the repository of Saccharomyces cerevisiae genomic and biological data since that time (1, 2). Although dozens of S. cerevisiae strain sequences have been made accessible through SGD for the utility of sequence analysis tools such as the BLAST search (3), these sequence data were not available from SGD Sequence pages and not integrated into the database due to the uncertainty associated with the data (e.g. poor sequence coverage). With the advent of high-throughput Next Generation Sequencing technology, the genomic sequence of 337 S. cerevisiae strains at deep sequence fold coverage has been released by several laboratories (see references 4–15). In addition, one group will soon release over a 1000 additional yeast genome sequences (e.g. The 1002 Yeast Genome Project, http://1002genomes.u-strasbg.fr/). To move forward in the era of big genomic data, SGD has integrated genome sequences and annotations from 11 strains of S. cerevisiae with a substantial history of use over past decades and large body of published experimental data. Users can easily access these genomes via Sequence and Strain pages, as well as through the sequence analysis tools provided by SGD.
Integration of Alternative Reference Genome Sequences
At SGD, we have incorporated these non-S288C alternative strain sequences into the database. The alternative reference strains, chosen based on the availability of substantial amounts of published experimental data, include: CEN.PK (16), D273-10B (17), FL100 (18), JK9-3d (19), RM11-1a (20), SEY6210 (21), SK1 (22), Sigma1278b (23), W303 (24), X2180-1A (25) and Y55 (26) (see Table 1). These are the genomes for which we have the most curated phenotype data, and for which we aim to curate specific functional information. For each of the 11 non-S288C sequences, there is an annotation available for each open reading frame (ORF) based on the annotation of the reference S288C strain. The genomic, coding and protein sequences for the ORFs in these other strains are available to view and download, on the Sequence page of the corresponding ORF in the reference strain S288C (see Figure 1). In addition to these 11 alternative strain sequences, 14 other strain genomes were processed using Automated Genome Analysis PipelinE, the automatic annotation pipeline (8), and are available for download from the Sequence page. All are accessible for sequence analysis tools provided by SGD.
Table 1.
Summary information on the 11 alternative reference strains
| Strain | Description | Source (ATCC ID) | NCBI BioSample Accession | Number of ORFs | Phenotype count per strain (%) | References |
|---|---|---|---|---|---|---|
| RM11-1a | A natural isolate collected from a California vineyard | UCD 2788 (UC Davis culture collection ID) | SAMN03020228 | 5323 | 2 (0.002) | (20) |
| Y55 | Laboratory strain originally isolated from wine grapes | ATCC: 52530 | SAMN03020218 | 5359 | 18 (0.015) | (26) |
| FL100 | Laboratory strain | ATCC: 28383 | SAMN03020232 | 5366 | 57 (0.046) | (18) |
| JK9-3d | Laboratory strain | ATCC: MYA-555 | SAMN03020238 | 5385 | 111 (0.09) | (19) |
| CEN.PK | Laboratory strain popular for use in systems biology studies | ATCC: MYA-1108 | SAMN03020234 | 5379 | 213 (0.174) | (16) |
| X2180-1A | S288C derivative laboratory strain | ATCC: 204504 | SAMN03020236 | 5387 | 276 (0.225) | (25) |
| D273-10B | Lab strain used for mitochondrial studies | ATCC: 24657 | SAMN03020237 | 5383 | 278 (0.227) | (17) |
| SEY6210 | Lab strain used in studies of autophagy and protein sorting | ATCC: 96099 | SAMN03020235 | 5400 | 414 (0.337) | (21) |
| SK1 | Lab strain used for studying sporulation and meiosis | ATCC: 204720 | SAMN03020220 | 5350 | 859 (0.7) | (22) |
| Sigma1278b | Used in pseudohyphal growth studies | ATCC: 42800 | SAMN03020229 | 5358 | 2170 (1.768) | (23) |
| W303 | Laboratory strain used for research into aging | ATCC: 20060 | SAMN03020233 | 5397 | 3158 (2.573) | (24) |
We chose 11 non-S288C strains as alternative reference strains based on the number of phenotype annotations curated in SGD. Phenotype studies are most frequently reported using the S288C reference strain, i.e. 84.78% of phenotypic counts in SGD are based on work in S288C. Other than S288C, the 11 alternative reference strains have been most frequently used for yeast phenotype studies. The alternative strains are used for specific areas of biology (e.g. CEN.PK for systems biology, D273-10B for mitochondrial studies, SEY6210 for autophagy and protein sorting, SK1 for sporulation and meiosis, Sigma1278b for pseudohyphal growth, and W303 for aging). The source of the sequenced strain genome is summarized with ATCC ID. The assembly and raw sequence data of each strain has been deposited in NCBI and can be found with NCBI BioSample accession numbers.
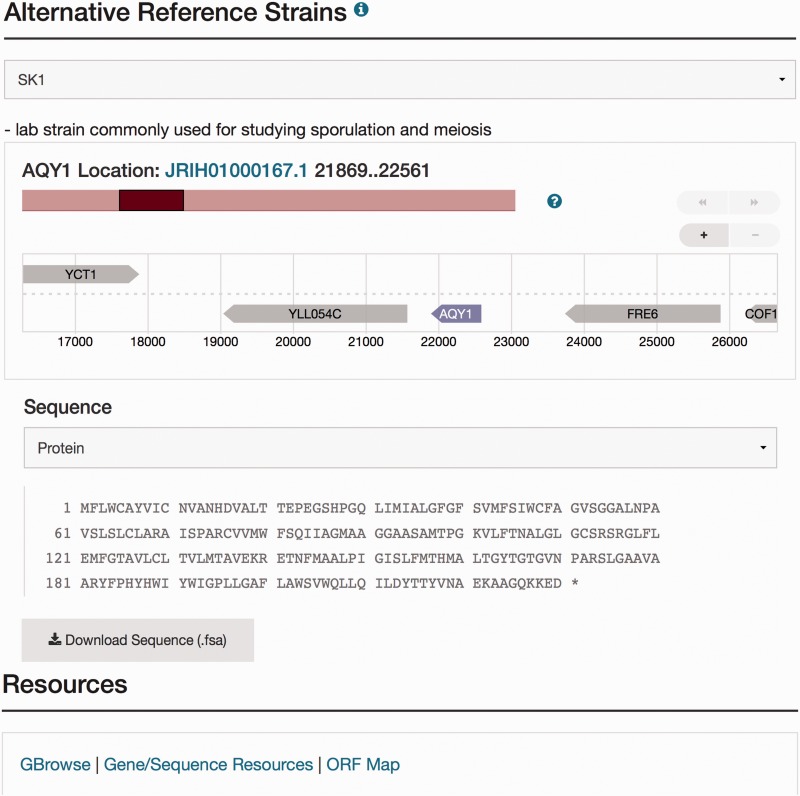
Figure 1.
Visualization of the new sequence data for AQY1 in the database. The Sequence page can be accessed by selecting the tab at the top of the AQY1 LSP, labeled ‘Sequence.’ There are six sections within the Sequence page including the ‘Sequence Overview’ for descriptive information, ‘Alternative Reference Strains’ for viewing the DNA or protein sequence in a selected alternative reference strain, and ‘Resources’ for access to processed results using sequence analysis tools (e.g. a new tool called SGD Variant Viewer (28), updated BLAST search, and ClustalW multiple sequence alignments). Selection of SK1 as an alternative strain is illustrated in this figure. Neighbor genes of AQY1 in SK1 are also shown in the visualization. When the user clicks a sequence analysis tool (URL in blue) in the resources tab, they will view the gene-specific results using that tool.
Utility of the New Sequence Data Demonstrated by a Use-Case Example
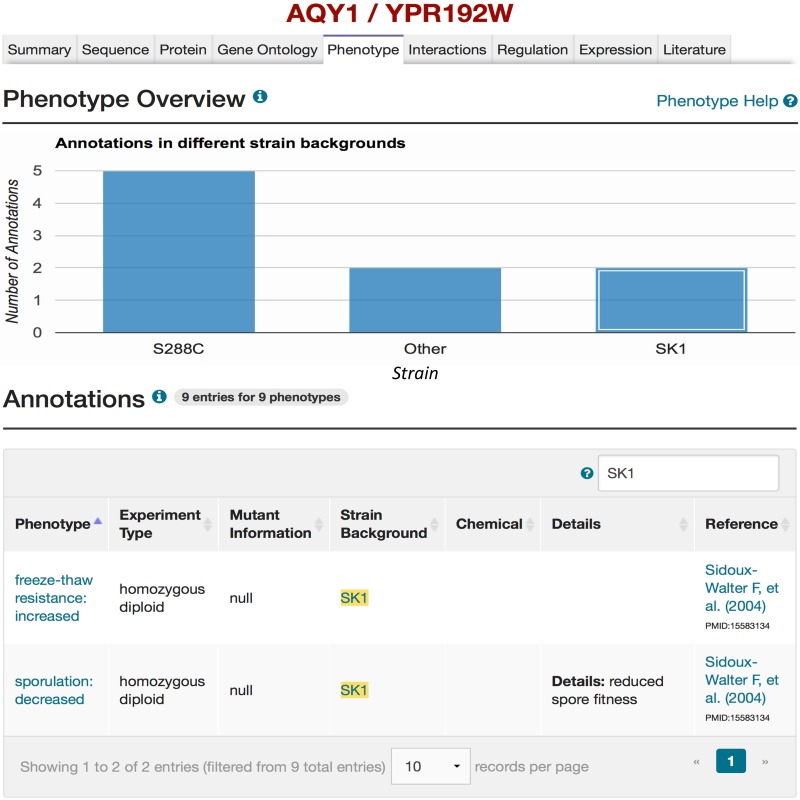
In the following section, we illustrate how a researcher can use these new genome sequences in conjunction with other features in SGD. For example, if a user wishes to investigate the contribution of genetic variation in the aquaporin encoding gene AQY1 (YPR192W) and its relevance to alterations in sporulation efficiency, represented by phenotype changes, they can start by visiting the corresponding Locus Summary Page (LSP) for AQY1. This page can be reached by searching for ‘AQY1’ via the quick search located at the upper right side on most pages of the SGD website (http://www.yeastgenome.org). A page with all phenotype data available for AQY1, accessible from the AQY1 LSP, lists the phenotypes displayed by various mutants of AQY1 along with relevant details such as the strain background, the type of mutant, and references (Figure 2). The bar charts available at the top of the Phenotype page summarize the breakdown of phenotypes by type of mutation and strain background. Clicking on the SK1 bar from this chart anchors to the annotation table below, providing access to the relevant annotations and associated detail (Figure 2). More information about the strain itself (i.e. genotype and assembly of the genome) can be found by selecting the strain name in the table of the Annotations section (Figure 3).
Figure 2.
Mutant phenotypes for AQY1 gene in SGD. By querying for ‘AQY1’ using the SGD search box, and selecting the Phenotype tab on the AQY1 LSP, phenotype information for AQY1 can be viewed. A bar chart summarizes how many phenotypic annotations have been curated in different strain backgrounds (e.g. two mutant phenotypes in the SK1 strain background). If the box for SK1 in the bar chart is selected, the details of the two mutant phenotypes for AQY1 in the SK1 strain background will be listed in the Annotations section. Users also can refer to the relevant literature (27) that describes studies on the mutant phenotypes resulted from polymorphisms in the AQY1 gene in strain SK1 as shown in the “Reference” column of the table. Users can also access more information of the alternative strain (SK1) by selecting the strain name (highlighted in yellow) in the Annotations table (see Figure 3).
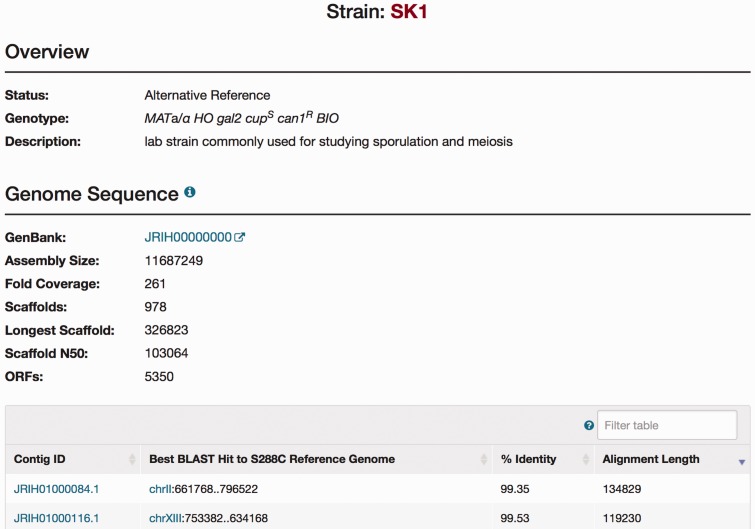
Figure 3.
The SGD SK1 Strain page. Strain pages are available for each of the alternative strains. The Strain page includes an Overview section for descriptive information of each strain, and a Genome Sequence section for brief statistics of the new sequence integrated in the database (NCBI accession number, the number of scaffolds, and the number of ORFs) and a table for information on each contig.
A study of sporulation-specific phenotypes within the AQY1 gene of SK1 (27) described strain-background specific variation in two residues (V121 and P255) that contribute to the activation of AQY1 in both SK1 and Sigma1278b. In other strain backgrounds, including S288C, AQY1 is inactive due to mutations at these positions (M121 and T225) within the coding sequence. To explore the polymorphisms in these critical residues relevant to the phenotypic variations in all alternative strains, users can move from the Phenotype page to the Sequence page of AQY1 by selecting the Sequence tab available at the top of the page (Figure 1). The Sequence page offers options to display the genomic and coding sequence of the gene in the reference strain S288C or in one of the alternative reference strains or other strains. If users wish to view and obtain the protein sequence of AQY1 in SK1 strain, they can make SK1 the alternative reference strain, and simply select the protein sequence (Figure 1) from the pull-down menu. Sequences in 14 strains other than the alternative references are listed in the Other Strains section of the Sequence page where sequence is available for download. Sequence tools containing the new reference genome sequence such as BLAST and sequence alignment options with other S. cerevisiae and fungal sequences are accessible in the Resources section of the Sequence page. A link to Variant Viewer, a new visualization tool within SGD is also in the Resource section of the Sequence page (28).
Variant Viewer can be use to visualize variation within AQY1 in the alternative strain genomes (Figure 4). For example, the two critical mutations located at position V121 (guided in a yellow line in Figure 4) and P255 in strains Sigma1278b, RM11-1a, SK1 and Y55 that are associated with the activation of Aqy1p can be visualized. The other eight laboratory strains show M121 and T255 in these positions, which cause the inactivation of Aqy1p in these laboratory strains (29). The Aqy1p C-terminus in eight laboratory strains is conserved while Sigma1278b, RM11-1a and Y55 show a longer C-terminus and SK1, a shorter C-terminus. The extended C-terminus of Sigma1278b is known to reduce the expression of the Aqy1p (30). We can predict that the expression of Aqy1p may also be reduced in Y55 and RM11-1a and the short C-terminus in SK1 may enhance Aqy1p expression. Other than these two residues (V121 and P255) and the extended C-terminus, Sigma1278b, RM11-1a and Y55 show strong conservation at the amino acid level with the eight laboratory strains. Unlike Sigma1278b, RM11-1a and Y55, SK1 shows more variation in AQY1 compared with the other strains. These additional mutations in SK1 may be also relevant to AQY1 protein function. By studying the variation within the sequence of AQY1 in the alternative reference strains, researchers can raise several important scientific questions concerning the phenotypic variation and the relationship with sequence variation in the alternative strains. This is an example of a study on a single gene. We expect that researchers studying other yeast genes can make use of these new sequence data to study variation in a similar manner.
Figure 4.
Visualization of genetic variation within AQY1 gene across alternative reference strains in SGD. Variations within AQY1 among 11 alternative strains and the S288C reference strain are depicted in the SGD Variant Viewer. Non-synonymous mutations and deletions/insertions with their location information in AQY1 are shown in the viewer. We can see that AQY1 is conserved in most laboratory strains. SK1 shows the most non-synonymous mutations relative to the S288C reference unlike the other strains. There are two common mutations that appear in four strains Sigma1278b, RM11-1a, SK1 and Y55: V121 (guided in a yellow line in the viewer) and P255. SK1 has a shorter C-terminus than other strains whereas Sigma1278b, RM11-1a and Y55 have longer C-termini than the eight laboratory strains. Other than the C-terminus and two mutations (V121 and P255), Sigma1278b, RM11-1a and Y55 show conserved amino acids with the eight laboratory strains unlike SK1.
Future directions
The integration of the alternative strain genomes in SGD will accelerate yeast genetics and population genomics studies by providing a user-friendly environment for the use of sequence data. To increase the accuracy of users’ studies, we plan to improve the quality of genome assemblies and annotations associated with these sequence data. As updated information becomes available and errors are corrected, they will be incorporated into future genome releases. We also anticipate expanding the reference genome panel in the future to include additional strains in order to accommodate emerging or underserved areas of study. The goal of these challenging, on-going efforts is to empower yeast research as the big genomic data era of yeasts continues to emerge.
Funding
This work is supported by a grant from the National Human Genome Research Institute at the United States National Institutes of Health (U41 HG001315). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Human Genome Research Institute or the National Institutes of Health. The funders had no role in design, data processing, implementation, decision to publish or preparation of the article. Funding for open access fee is provided by U41 HG001315.
Conflict of interest. None declared.
References
- 1.Cherry J.M., Ball C., Weng S. et al. (1997) Genetic and physical maps of Saccharomyces cerevisiae. Nature, 387, 67–73. [PMC free article] [PubMed] [Google Scholar]
- 2.Cherry J.M., Adler C., Ball C. et al. (1998) SGD: saccharomyces genome database. Nucleic Acids Res., 26, 73–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Engel S.R., Cherry J.M. (2013) The new modern era of yeast genomics: community sequencing and the resulting annotation of multiple Saccharomyces cerevisiae strains at the Saccharomyces Genome Database. Database (Oxford), 2013, bat012.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Strope P.K., Skelly D.A., Kozmin S.G. et al. (2015) The 100-genomes strains, an S. cerevisiae resource that illuminates its natural phenotypic and genotypic variation and emergence as an opportunistic pathogen. Genome Res., 25, 762–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bergström A., Simpson J.T., Salinas F. et al. (2014) A high-definition view of functional genetic variation from natural yeast genomes. Mol. Biol. Evol., 31, 872–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wohlbach D.J., Rovinskiy N., Lewis J.A. et al. (2014) Comparative genomics of Saccharomyces cerevisiae natural isolates for bioenergy production. Genome Biol. Evol., 6, 2557–2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Y., Zhang W., Zheng D. et al. (2014) Genomic evolution of Saccharomyces cerevisiae under Chinese rice wine fermentation. Genome Biol. Evol., 6, 2516–2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Song G., Dickins B., Demeter J. et al. (2015) AGAPE (Automated Genome Analysis PipelinE) for pan-genome analysis of Saccharomyces cerevisiae. PLoS One, 10, e0120671.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Engel S., Dietrich F., Fisk D. et al. (2013) The reference genome sequence of Saccharomyces cerevisiae: then and now. G3, 4, 389–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wei W., McCusker J., Hyman R. et al. (2007) Genome sequencing and comparative analysis of Saccharomyces cerevisiae strain YJM789. Proc. Natl. Acad. Sci. U S A, 104, 12825–12830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Argueso J., Carazzolle M., Mieczkowski P. et al. (2009) Genome structure of a Saccharomyces cerevisiae strain widely used in bioethanol production. Genome Res., 19, 2258–2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Akao T., Yashiro I., Hosoyama A. et al. (2011) Whole-genome sequencing of sake yeast Saccharomyces cerevisiae Kyokai no. 7. DNA Res., 18, 423–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Novo M., Bigey F., Beyne E. et al. (2009) Eukaryote-to-eukaryote gene transfer events revealed by the genome sequence of the wine yeast Saccharomyces cerevisiae EC1118. Proc. Natl. Acad. Sci. U S A, 106, 16333–16338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Borneman A., Desany B., Riches D. et al. (2011) Whole-genome comparison reveals novel genetic elements that characterize the genome of industrial strains of Saccharomyces cerevisiae. PLoS Genet., 7, e1001287.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Borneman A., Forgan A., Kolouchova R. et al. (2016) Whole genome comparison reveals High levels of inbreeding and strain redundancy across the spectrum of commercial wine strains of Saccharomyces cerevisiae. G3 (Bethesda), doi:10.1534/g3.115.025692 vol. 6 no. 4957-971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nijkamp J., van den Broek M., Datema E. et al. (2012) De novo sequencing, assembly and analysis of the genome of the laboratory strain Saccharomyces cerevisiae CEN.PK113-7D, a model for modern industrial biotechnology. Microb. Cell. Fact, 11, 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sherman F. (1963) Respiration-deficient mutants of yeast. Genetics, 48, 375–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Casaregola S., Nguyen H., Lepingle A. et al. (1998) A family of laboratory strains of Saccharomyces cerevisiae carry rearrangements involving chromosomes I and III. Yeast, 14, 551–564. [DOI] [PubMed] [Google Scholar]
- 19.Heitman J., Movva N.R., Hall M.N. (1991) Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science, 253, 905–909. [DOI] [PubMed] [Google Scholar]
- 20.Brem R., Yvert G., Clinton R., Kruglyak L. (2002) Genetic dissection of transcriptional regulation in budding yeast. Science, 296, 752–755. [DOI] [PubMed] [Google Scholar]
- 21.Robinson J.S., Klionsky D.J., Banta L.M. et al. (1988) Protein sorting in Saccharomyces cerevisiae: isolation of mutants defective in the delivery and processing of multiple vacuolar hydrolases. Mol. Cell. Biol., 8, 4936–4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kane S.M., Roth R. (1974) Carbohydrate metabolism during ascospore development in yeast. J. Bacteriol., 118, 8–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dowell D., Ryan O., Jansen A. et al. (2010) Genotype to phenotype: a complex problem. Science, 328, 469.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ralser M., Kuhl H., Ralser M. et al. (2012) The Saccharomyces cerevisiae W303-K6001 cross-platform genome sequence: insights into ancestry and physiology of a laboratory mutt. Open Biol., 2, 120093.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mortimer R., Johnston J. (1986) Genealogy of principal strains of the yeast genetic stock center. Genetics, 113, 35–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Borts R.H., Lichten M., Hearn M. et al. (1984) Physical monitoring of meiotic recombination in Saccharomyces cerevisiae. Cold Spring Harb. Symp. Quant. Biol., 49, 67–76. [DOI] [PubMed] [Google Scholar]
- 27.Sidoux-Walter F., Pettersson N., Hohmann S. (2004) The Saccharomyces cerevisiae aquaporin AQY1 is involved in sporulation. Proc. Natl. Acad. Sci. USA, 101, 17422–17427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sheppard T., Hitz B., Engel S. et al. (2016) The Saccharomyces genome database variant viewer. Nucleic Acids Res., 44, D698–D702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bonhivers M., Carbrey J.M., Gould S.J., Agre P. (1998) Aquaporins in Saccharomyces genetic and functional distinctions between laboratory and wild-type strains. J. Biol. Chem., 273, 27565–27572. [DOI] [PubMed] [Google Scholar]
- 30.Laizé V., Tacnet F., Ripoche P., Hohmann S. (2000) Polymorphism of Saccharomyces cerevisiae aquaporins. Yeast, 16, 897–903. [DOI] [PubMed] [Google Scholar]