Abstract
MicroRNAs (miRNAs) are a class of small noncoding RNAs of ~22 nt in length which are involved in the regulation of gene expression at the posttranscriptional level by degrading their target mRNAs and/or inhibiting their translation. Expressed ubiquitously or in a tissue-specific manner, miRNAs are involved in the regulation of many biological processes such as cell proliferation, differentiation, apoptosis, and the maintenance of normal cellular physiology. Many miRNAs are expressed in embryonic, postnatal, and adult hearts. Aberrant expression or genetic deletion of miRNAs is associated with abnormal cardiac cell differentiation, disruption of heart development, and cardiac dysfunction. This chapter will summarize the history, biogenesis, and processing of miRNAs as well as their function in heart development, remodeling, and disease.
1. Introduction
Gene expression, as a basic biological process, can be characterized as a response to stimuli. However, the level of change in gene expression needs to be carefully controlled to ensure the proper cellular response; similarly, a lack of adjustment may lead to abnormal cellular function. Thus, the regulation of gene expression and its mechanisms of action are fundamental to all organisms and have been thoroughly studied in multiple species and organ systems. Therefore, the recent determination of a new class of small RNAs (microRNAs, miRNAs), as key elements in this regulatory repertoire, has generated enormous interest in this field. It is now known that miRNAs function within the genetic regulatory network at the posttran-scriptional level. By imperfect base pairing with mRNAs in a sequence-dependent manner, these miRNAs repress gene expression by degrading target mRNAs and/or inhibiting their translation. Some miRNAs are expressed broadly, while others are restricted to specific tissues or cell types. Roles for miRNAs have been demonstrated in the regulation of a broad range of biological activities and their aberrant expression has been correlated with embryonic malformations and organ dysfunction. Heart function, in particular, is essential for proper vertebrate embryogenesis, and the early morphogenesis and development of this organ are dependent on tightly regulated genetic networks. This chapter is intended to offer a summarized review of the history, biogenesis, and processing of miRNAs, and their function during heart development.
2. An Abridged Overview of Heart Development
Eukaryotic cell functions, including growth, proliferation, differentiation, and survival, rely on several mechanisms including respiration and energy production. Respiration is the diffusion of oxygen into cells, where it will be used as a final acceptor of protons, in the respiratory chain, for the production of energy in the form of ATP (Alberts et al., 1994). In “simple” multicellular organisms, such as the worm (Caenorhabditis elegans) oxygen can freely diffuse into the cells; however, in higher invertebrates and vertebrates, oxygen has to be brought directly into contact with the internal tissues and cells by the lymph or blood through a circulatory system. Directional flow within a circulatory system allows the exchange of oxygen and nutrients for waste produced by cells and tissues (Harvey, 1999, 2002). The directionality of the lymph or blood flow is produced by the pumping action of the heart, which beats rhythmically in an organized and regular manner to force fluid movement from the caudal to the cranial end of an organism (Challice and Virágh, 1974; Nishii and Shibata, 2006). This cardiac pump takes multiple forms, from a simple single cardiac tube in invertebrates (i.e., the “fruit fly” Drosophila melanogaster) to a two-chambered structure in fish (i.e., the “zebrafish” Danio rerio), a three-chambered heart in amphibians (i.e., the “frog” Xenopus laevis) and reptiles, and finally to a four-chambered heart in birds (i.e., the “chicken” Gallus gallus) and mammals (i.e., the “mouse” Mus musculus) (Harvey, 1999, 2002; Kirby, 2007).
Heart function is required early during embryogenesis for survival and the subsequent growth of other tissues and organs. The cells that form the heart originate from cardiac progenitors specified during embryonic gastrulation, a dynamic cellular process which results in the formation of the three germ tissue layers: ectoderm, mesoderm, and endoderm. The concerted and simultaneous activity of endodermal and ectodermal signaling permits the formation of the mesoderm and restricts the location of the cardiogenic cells, as well as their specification. Several molecules with inducing or inhibiting properties are involved in each of the different stages of development (Harvey, 1999; Kirby, 2007; Olson, 2006). For cardiac development, these different stages include (1) specification of cardiac progenitors, (2) initial migration of these cells during gastrulation, (3) further cellular migration to form the cardiac fields, (4) formation of the cardiac crescent, (5) formation of the tubular heart, (6) cellular recruitment at the poles of the heart, and (7) cardiac chamber differentiation (Kirby, 2007).
Tracing analyses utilizing vital dyes and cell/tissue explant experiments have shown that early cardiac progenitor cells are located in the anterior region of the primitive streak in the chick and mouse embryo (Buckingham et al., 2005; Harvey, 2002; Srivastava, 2006). At the onset of gastrulation, initial posterior to anterior migration of these cells is regulated by the activity of the FGF family of signaling factors including FGF2, FGF4, and FGF8 which have been proposed to signal through FGFR1 (Ciruna and Rossant, 2001; Harvey, 2002). Similarly, bone morphogenic protein 2 (BMP2), a member of the TGF-β superfamily of growth factors, is initially expressed in the posterior end of the primitive streak by the onset of gastrulation and implicated in early cell migrations (Andrée et al., 1998).
At embryonic day 6.5 (E6.5), as a result of FGF-8-dependent expression of Mesp-1 and Mesp-2, cardiac progenitor cells migrate bilaterally to the anterior and lateral region of the embryo colonizing the splanchnic layer of the lateral plate mesoderm and form two early lateral cardiac fields (Buckingham et al., 2005; Harvey, 1999; Kitajima et al., 2000; Saga et al., 1999). Canonical Wnt/β-catenin signaling within the neural ectoderm induces the Mesp-1 positive mesodermal lineage during early gastrulation (Bondue et al., 2008; Saga et al., 1999). These data confirm crucial roles for the early action of Bmp2 and FGF signaling in conferring cardiogenic potential (Lough et al., 1996), guiding the migration of progenitor cells, and restricting the location of the cardiac fields. It is also aided by the activity of a concentration gradient of the Wnt/β-catenin, which will induce the underlying anterior mesoderm into head tissue (Brade et al., 2006; van de Schans et al., 2008).
Approximately a day later (E7.5), these early cardiogenic progenitor cells migrate cranially and converge across the midline region of the embryo to form a horseshoe-shaped epithelial fold, the cardiac crescent. The lateral cardiac fields in mouse and chicken embryos were initially identified by the expression of Nkx2-5, a vertebrate homeodomain transcription factor that is the homologue of the D. melanogaster tinman gene (Bodmer et al., 1990; Kasahara et al., 1998; Lyons et al., 1995; Tanaka et al., 1999). In mouse and chick embryos, Nkx2-5 is the Nkx factor most widely expressed in the early lateral cardiac fields and cardiac crescent during cardiogenesis (Lyons et al., 1995; Tanaka et al., 1999). However, in contrast to the function of tinman in flies, no Nkx gene acts a master regulator of cardiogenesis (Newman and Krieg, 1998). In a dynamic process, the medial region of the cardiac crescent subsequently bulges and moves cranially bringing the lateral regions toward the midline; these two lateral cell populations eventually fuse to form the heart tube (E8.0). It is during the heart tube forming stage that the primitive heart regions can be anatomically and molecularly identified for the first time. From caudal to cranial, these structures are: (1) The inflow tract, where blood is collected to be directed into the heart and which is composed of the right and left horns of the sinus venosus which are the continuation of the common cardinal veins; (2) the primitive atrial chamber; (3) the primitive ventricular chamber; and (4) the outflow tract, which at this stage is known as the conotruncus or bulbus cordis (Buckingham et al., 2005; Kirby, 2007; Srivastava, 2006). During the formation of the heart tube, Wnt/β-catenin signaling, especially Wnt-3a and Wnt-8, is necessary for the induction and maintenance of the expression of Isl-1 (Cohen et al., 2007) and Fgf10 (Lizhu et al., 2007). These factors are crucial to the correct formation of the second heart field and the expression of Shh, Bmp4, and Bmp7 (Tzahor, 2007).
At E9.5, the heart begins a substantial remodeling process with the rightward bending of the heart tube to realign the cardiac chambers (looping); the caudal region of the cardiac tube, including the two sinus venosus horns, is brought cranially to position them dorsally to the outflow tract. By E10.5, the process of cardiac looping is nearly complete as the sinus venosus is shifted to the right side of the heart. The appearance of two outgrowing structures, called the sinus valves, demarcates the junction of the right common cardinal vein and the right atrium. Also, the first signs of septation of the cardiac chambers are evident at E10.5 (Challice and Virágh, 1974; Van Mierop and Gessner, 1970). By E11.5, the caudal region of the heart is finally positioned dorsal to the arterial pole and the subsequent septation within the cardiac chambers, in oxygen breathing vertebrate species, are the final steps in their specification. Initial septation of the outflow tract, by formation of internal cushions, divides it into a pulmonary and arterial arch. The right sinus venosus horn is brought in juxtaposition to the right atrium, and the left horn reduces in size and becomes the coronary artery. The atria and ventricles are separated by the interatrial and interventricular septa, respectively (Buckingham et al., 2005; Harvey, 2002; Kirby, 2007; Van Mierop and Gessner, 1970).
Differentiation of cells within the heart is regulated by a variety of transcription factors which are again dependent on the expression of first or second heart field factors. The bHLH transcription factors, Hand1 and Hand2, whose expression marks the left and right ventricles, respectively, are key examples. Hand2 is dependent on the expression of Isl-1, while Hand1 is dependent on the expression of Nkx2-5 (Buckingham et al., 2005; Cai et al., 2003). Nkx2-5 interacts with the T-box family of transcription factors and is involved in the activation or repression of gene expression required for the differentiation of numerous cardiac structures. Several members of the T-box family have been identified to be expressed in cardiac tissue including Tbx-1, -2, -3, -5, -18, and Tbx-20 (Plageman and Yutzey, 2005). Interaction of Nkx2-5 and Tbx5 results in activation of the Atrial Natriuretic Factor (ANF) gene and the differentiation of the cardiac chambers and cardiac trabeculation in the ventricles (Hiroi et al., 2001). Nkx2-5 expression is dispensable for the initial formation of the sinoatrial node (SAN) (Blaschke et al., 2007; Espinoza-Lewis et al., 2009, 2011; Kasahara et al., 1998). However, the interaction of Nkx2-5 with Tbx2 or Tbx3 results in the repression of the ANF gene and Connexin-40 (Cx40) during differentiation of the cardiac conduction system, including the SAN (Habets et al., 2002; Hoogaars et al., 2004). Later in development, cardiac outgrowth takes place and by E14.5 the final shape of the heart is achieved; the sinus venosus has regressed and slowly integrated into the dorsal wall of the right atrium, the outflow tract has divided into the pulmonary and arterial arches, septation of the cardiac chambers is complete, and the ventricular chambers have become the major volumetric components of the heart (Buckingham et al., 2005; Harvey, 1999, 2002; Kirby, 2007; Srivastava, 2006).
3. The World of the miRNAs
We have briefly summarized some aspects involved in embryonic cardiac morphogenesis and the molecules involved in its regulation. As noted above, cardiac progenitor cells arise from the specification and determination of a special region of the mesoderm. Early determination is driven by the activity of the transcription factors Nkx2-5 and Isl-1 (Cai et al., 2003; Kasahara et al., 1998; Lyons et al., 1995; Tanaka et al., 1999). Also, it is implied that the final cellular response in the form of expression of a genetic profile is a result of not only the activity of transcription factors, but also the concerted activity of extracellular molecules, their signal transduction, and fine-tuning regulatory mechanisms. Gene regulation is achieved by the direct binding of transcription factors (activators or repressors) to cis-regulatory elements in order to up- or downregulate gene expression. This process can be enhanced or inhibited by the activity of extracellular signals, the activity of intracellular mediators, the status of chromosomal structure, the activity of transcription cofactors, and posttranslational modification of the protein product. In short, gene expression is regulated at the transcriptional, translational, and/or posttranslational level (Alberts et al., 1994; Srivastava, 2006). In recent years, a novel mechanism has been described which functions at the posttranscriptional level to modulate and fine-tune gene expression by targeting the mRNA. Such mRNA posttran-scriptional regulation results in either mRNA degradation or mRNA translation inhibition. This elegant mechanism is carried out by a newly described class of small RNA molecules, the miRNAs.
3.1. miRNAs: A brief history
Embryonic development is a well-organized, highly regulated event with strict spatiotemporal requirements. Stages are categorized by the observation of characteristic anatomical and molecular changes. In the “worm” C. elegans, several stages of development can be easily observed by simple visual inspection. From fertilized egg, to embryo, to larva, to adult, the worm’s life cycle is completed in about 2 days. The larva stage is the lengthiest and is divided into four stages named L1–L4 (Anderson, 1995). Defects in any of these stages through the disruption of the temporal patterns of cell division and differentiation conducted by regulatory “heterochronic genes” result in larval or adult abnormalities (Chalfie et al., 1981). Mutations in these heterochronic genes result in the induction of cell fate transformations, such as recapitulation of an earlier phenotype at late stages or by adopting late stage phenotypes prematurely (Ambros and Horvitz, 1987; Chalfie et al., 1981).
Initially, heterochronic mutations were related to and identified in four C. elegans genes (lin-4, lin-14, lin-28, and lin-29) (Ambros and Horvitz, 1984). The lin-4 mutation resulted in cell lineage reiterations which main-tained earlier larval phenotypes. This resulted in supernumerary moults and the continuous production of larval-specific cuticle with the extension of late larval stages (Ambros and Horvitz, 1984; Chalfie et al., 1981). The lin-14 gain-of-function mutation, which produces a semidominant allele, results in cell lineage retarded development; certain late stage cells adopt fates expressed in cells at earlier stages. In contrast, lin-14 loss-of-function mutations, which result in null alleles, generate precocious cell lineage development, with certain early stage cells adopting late stage fates prematurely (Ambros and Horvitz, 1987). Interestingly, the lin-4 and lin-14 mutant phenotypes were found to occur in the same cell lineages (Ambros and Horvitz, 1987). Additionally, it was found that lin-14 mutant phenotypes were lin-4 dependent (Ambros and Horvitz, 1987; Arasu et al., 1991; Ruvkun and Giusto, 1989; Wightman et al., 1993). Those observations suggested a direct regulatory mechanism between these two genes (Ambros and Horvitz, 1987; Arasu et al., 1991; Chalfie et al., 1981). The lin-14 gene product is a protein that is detected at high levels only in the early larval L1 stage. Surprisingly, however, RNA protection assays revealed that lin-14 mRNA was expressed and stable in all larval stages (Ruvkun and Giusto, 1989). Extensive screening for lin-14 mutants facilitated a large analysis of the lin-14 gene structure and determined a regulatory sequence in the lin-14 3′UTR as responsible for the negative regulation of gene expression, specifically in inhibiting mRNA translation (Lee et al., 1993; Wightman et al., 1993). Similarly, gene structure analysis, in addition to a highly laborious gene cloning strategy, suggested that the lin-4 gene product is not a protein. Indeed, RNA protection analysis demonstrated that this locus produced two small RNA products, a 69-nt-long product (lin-4L) and a 21-nt-long product (lin-4S) (Lee et al., 1993). Computational analysis indicated that the lin-4L was a precursor-like molecule which includes the lin-4S product in a hairpin or stem-loop secondary structure and that both were complementary to conserved repeated sequences in the lin-14 negative regulatory 3′UTR in a nonperfect manner. However, the 5′ 6–8nt of lin-4S displayed an exact match with that of the lin-14 3′UTR (Lee et al., 1993; Wightman et al., 1993). Simultaneously, it was reported that the lin-14 3′UTR could mediate the downregulation of the expression of an unrelated protein (Wightman et al., 1993). A construct comprising the LacZ-lin-14_3′UTR driven by the Collagen-10 promoter (Col10-LacZ-lin-14_3′UTR) was engineered and injected into wild-type C. elegans embryos. X-gal staining showed the expected fading coloration of cells after the L2 larval stage. In contrast, injection of the construct into a lin-4 null mutant line (e912) resulted in the maintenance of the X-gal staining in all four larval stages, thus demonstrating the mechanism of the novel predicted regulatory mechanism (Wightman et al., 1993).
Similarly, genetic screening for mutations to suppress the synthetic sterile phenotype in C. elegans was performed. Several candidate genes were identified, including let-7 (Reinhart et al., 2000). let-7 mutations result in supernumerary moults in the L4-adult stage reiterating earlier larval patterns of cell division. This defect was partially suppressed by mutations in the lin-41, lin-14, lin-28, and lin-42 genes. Gene structure, Northern blot, and molecular analyses showed that the let-7 product was not a protein but a 21-nt-long small RNA molecule. Additionally, computational analysis predicted and determined that the lin-14, lin-28, lin-41, lin-42, and daf-12 heterochronic genes contain nonperfect complementary sequences in their 3′UTR. Transgenic C. elegans containing a Col10-LacZ-lin-41_3′’UTR construct demonstrated that let-7 acts in similar fashion to lin-4 (Reinhart et al., 2000). Due to their nature and their specific function in regulating the temporal patterns of cell division in the C. elegans larvae, both lin-4 and let-7 gene products were termed small temporal RNAs (stRNAs) (Lee et al., 1993; Pasquinelli et al., 2000).
In order to provide a complete overview of this new regulatory mechanism, we believe it is necessary to briefly describe a few experimental landmarks. These discoveries were reported in between the determination of the lin-4 and the let-7 mechanism of action and helped shape the idea that small RNAs were indeed important players in the regulation of gene expression.
It was observed that overexpression of a sense or antisense RNA in plants (Hammond et al., 2001b; Jorgensen et al., 1996; Que and Jorgensen, 1998) or the injection of antisense RNA in C. elegans resulted in down-regulation of gene expression (Fire et al., 1991; Guo and Kemphues, 1995). Based on those empirical results, it was hypothesized that the regulation of gene expression by overexpression of a sense RNA was effective due to the quenching of the necessary factors needed for translation of the targeted mRNA molecule (cosuppression). However, a different mechanism was believed to be responsible for the downregulation of expression by over-expression of an antisense RNA; it was thought to induce the formation of a long double-stranded RNA (dsRNA) molecule, thus inhibiting translation (Fire et al., 1991; Hammond et al., 2001b; Jorgensen et al., 1996; Que and Jorgensen, 1998). It was known that dsRNA could affect gene expression by triggering interferon inducible pathways that inhibit translation, known as a “panic response,” through the activation of protein kinases. However, the effects observed when using antisense RNA were gene specific differing from the broad translation inhibition due to kinase activation (Hammond et al., 2001b; Williams, 1999). Strikingly, more dramatic effects in down-regulation of gene expression and higher gene specificity were found in C. elegans when sense and antisense RNA molecules were introduced at the same time (Fire et al., 1998) leading to the thought that a specific mechanism for RNA silencing must exist. This mechanism was termed RNA interference or RNAi (Fire et al., 1998).
Independently, a similar regulatory system, drawing parallelism to the RNAi mechanism, was previously described in plants, termed posttran-scriptional gene silencing (PTGS). This process, which is guided by small ~22–25nt RNA molecules, was identified after the introduction of a long antisense RNA (Hammond et al., 2001b; Jorgensen et al., 1996; Que and Jorgensen, 1998). Following the methodology described in PTGS studies, similar results were later obtained in D. melanogaster S2-cultured cells (Fire et al., 1991) and in the “worm” C. elegans (Guo and Kemphues, 1995).
Thus, the description of ~22–25nt small RNA molecules which guide the PTGS/RNAi mechanism resembled the earlier description of the small temporal genes lin-4 and let-7. stRNAs and RNAi molecules share bio-chemical and mechanistic characteristics; they are small in size, double stranded, and possess a 5′-phosphate group and a 3′-hydroxyl group in a 2-nt tail (Bartel, 2004; Bartel and Bartel, 2003). It was inferred, from the latter characteristic, that RNase III must be responsible for such end-product. Indeed, Dicer, previously identified to be a component of the PTGS complex, was also found to be involved in the maturation of the stRNA (Grishok et al., 2001; Hammond et al., 2000). Ironically, it was by using RNAi against the human Dicer gene in mammalian cell cultures that it was demonstrated that let-7 maturation was DICER dependent (Hutvágner et al., 2001). The analysis of DICER-produced biochemical products, along with an elegant cloning strategy and bioinformatic predictions, led almost simultaneously and fortuitously to the discovery of small RNAs resembling stRNAs in several model systems including D. melanogaster, C. elegans, and in HeLa-cultured cells. Although these new small RNAs were not expressed in a timely fashion as the stRNAs, biochemical similarities proved sufficient to classify them into a new and large class of small RNAs which include lin-4 and let-7 as founding members. Due to their short length, they were termed miRNAs (Lagos-Quintana et al., 2001; Lau et al., 2001; Lee and Ambros, 2001).
3.2. miRNA: Biogenesis
The small ~22-nt-long single-strand noncoding miRNA molecules are first expressed as a long transcript known as primary miRNA (pri-RNA) either as a single independent amplicon, as part of a polycistronic RNA molecule, or from intronic sequences within a protein-coding gene host (Fig. 10.1) (Bartel, 2004; Sayed and Abdellatif, 2011). The single or polycistronic molecule is characterized by the adoption of a secondary structure known as the stem-loop or hairpin (Bartel, 2004; Sayed and Abdellatif, 2011). Initial nuclear processing of the long pri-RNA molecule is performed by the microprocessor complex composed of the Class I RNAse type III Drosha and the stabilizing protein Pasha (Partner of Drosha, in C. elegans), or Dgcr8 (DiGeorge critical region gene 8, in mice and humans). This process generates a ~70-nt-long RNA molecule in a two-step highly consistent, characteristic, and sequence-independent manner (Denli et al., 2004; Han et al., 2006; Lee et al., 2003; Wu et al., 2000). dsRNA is recognized by Dgcr8 and cleaved by Drosha in a staggered fashion, ~11nt from the base (Gregory et al., 2004; Han et al., 2006). The result of the staggered cleavage is a hairpin-like molecule, also known as precursor-miRNA (pre-miRNA), with a 5′-phosphate in the forward strand and a 3′-hydroxyl group in the 2 nt-overhang tail on the reverse strand (Han et al., 2006). Intronic miRNAs, known as miRtrons, are expressed along with their host gene and the pre-miRNA is a product of splicing. These miRtrons have been shown to bypass Drosha/Dgcr8 initial processing and have been reported in D. melanogaster, C. elegans, and mammals (Ruby et al., 2007).
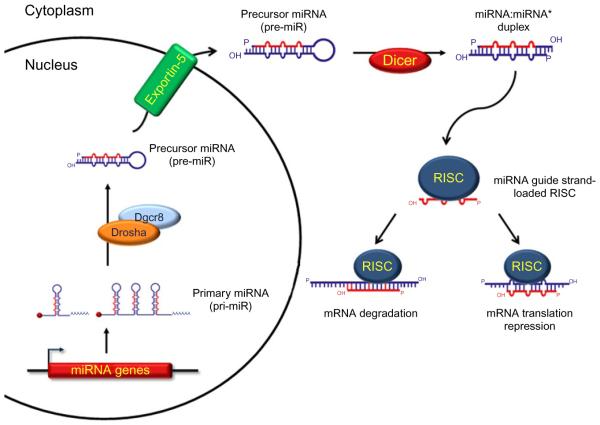
Figure 10.1.
MicroRNA Biogenesis
In the nucleus, a miRNA gene is transcribed into a long RNA molecule containing a single miRNA sequence or several miRNA sequences (polycistron or cluster), known as primary miRNA (pri-miRNA). Pri-miR-NAs are protected by the addition of a 5′-m7G cap (red ball) and a 3′ poly-A tail. Pre-miRNAs are processed by the microprocessor machinery composed of Drosha and the stabilizing protein Dgcr8, thus producing a precursor-miRNA molecule (pre-miRNA). Pre-miRNAs are transported out of the nucleus to the cytoplasm by the active trans-porter Exportin-5 and further processed by Dicer into a miRNA:miRNA* duplex. The RNA-induced silencing complex (RISC) is assembled and loaded with the miRNA guide strand due to the unwinding of the miRNA:miRNA* duplex. A loaded RISC will facilitate the recognition of mRNA targets leading to the mRNA degradation or the inhibition of translation.
Importins and Exportins are involved in the active nuclear transport of proteins and small RNAs such as pre-rRNA and tRNA (Bohnsack et al., 2004). Exportin-5 is a Ras–GTP-dependent active transporter involved in the nuclear export of adenovirus VA1 (a small 160-nt small noncoding RNA) and in tRNA transport (Bohnsack et al., 2004; Yi et al., 2003). Exportin-5 binds to the 3′ 2nt-overhang in tRNAs (Bohnsack et al., 2004; Calado et al., 2002), a characteristic also found in Drosha/Dgcr8-processed miRNAs. Indeed, Exportin-5 was identified as the nuclear receptor which mediates miRNA export to the cytoplasm by recognizing and binding to the 3′ 2-nt overhang and dsRNA region of the pre-miRNA in a sequence-independent manner (Bohnsack et al., 2004; Lee et al., 2011; Lund et al., 2004; Yi et al., 2003).
In the cytoplasm, the pre-miRNA is further processed by excision of the loop structure in order to convert the hairpin or stem-loop small RNA into a short dsRNA molecule. Dicer was initially identified as an RNase type III involved in the processing of large dsRNA molecules for the production of siRNA in PTGS in plants (Bartel and Bartel, 2003; Hammond et al., 2001b; Jorgensen et al., 1996; Que and Jorgensen, 1998) and RNAi in C. elegans and D. melanogaster (Bernstein et al., 2001; Fire et al., 1998; Grishok et al., 2001; Hutvágner et al., 2001). DICER, as an RNase type III, recognizes the double-stranded nature of the pre-miRNA and cleaves in a staggered manner at about two helical turns from the base of the hairpin excising the loop from the hairpin and producing a mature imperfectly matched dsRNA molecule (Grishok et al., 2001; Hutvágner et al., 2001; Ketting and Plasterk, 2000). Dicer homologues have been found in several organisms and are known as Dicer-like (Dcl-1-4) in Arabidopsis thaliana; Dcr (Dcr-1 and Dcr-2) in D. melanogaster; and Dicer-1 in mice, rats, and humans (Bartel, 2004).
The mature dsRNA or RNA duplex is composed of the mature miRNA as one of the strands (guide strand), while the other strand is known as miRNA* (passenger strand). Small RNA high-throughput cloning has identified that the majority of the strands screened correspond to miRNAs, while a low number of clones represent miRNA*s, indicating that miRNA:miRNA* duplexes are short lived and that miRNA*s are quickly degraded. However, even though miRNA*s are found in low numbers, a few have been reported to be functional (Bartel, 2004; Sayed and Abdellatif, 2011).
3.3. miRNAs, RISC, and gene silencing mechanism
The two-step RNA processing driven by Drosha and Dicer produces a dsRNA molecule or miRNA:miRNA* duplex which is recognized and bound by a large protein complex known as the RNA-induced silencing complex (RISC). “Loaded” into the RISC complex, the miRNA: miRNA* duplex is unwound and the miRNA guide strand is separated from the miRNA* passenger strand. The passenger strand is readily degraded while the guide strand directs mRNA targeting.
Several genetic screens in A. thaliana, C. elegans, and D. melanogaster suggested the existence of a conserved protein complex implicated in silencing activity induced by small dsRNA. In A. thaliana, mutations in the AGO-1 gene result in aberrations during leaf development and defects in the floral organs. The leaves are smaller and their outgrowth is retarded and their shape resembles a small squid; thus the mutants were known as Argonautes. Additionally, AGO-1 mutants are ineffective in PTGS (Bohmert et al., 1998). A similar phenotype was previously observed in the suppressor of gene silencing (SGS-2/SDE-1) mutant plants (Bartel, 2004; Bartel and Bartel, 2003; Hammond et al., 2001b). Additionally, RNAi deficiencies were found in Neurospora crassa QDE-2 mutants (Catalanotto et al., 2000), and in C. elegans rde-1, rde-4, and mut-7 mutants (Ketting et al., 1999; Tabara et al., 1999) among others (Fagard et al., 2000; Hammond et al., 2001b). Interestingly, these latter genes show shared homology to the A. thaliana AGO-1 product (Fagard et al., 2000), indicating the presence of a conserved molecular mechanism involved in gene silencing. In D. melanogaster, S2-cultured cells transfected with a mixture of a Luciferase expressing construct and dsRNA targeting the Luciferase ORF were subjected to lysate fractionation (Hammond et al., 2000, 2001a). Certain fractions possessed RNAi processing ability, determined by the presence of small RNAs and absence of the Luciferase mRNA, and also coprecipitated a low-abundance protein. Protein sequence homology searches again determined that this protein showed a high homology to AGO-1; thus it was termed AGO-2 and classified as a member of the Argonaute superfamily of proteins (Hammond et al., 2000, 2001a). Several other Ago proteins have been identified; C. elegans contains 27 members, while only four Ago proteins (AGO-1, AGO-2, AGO-3, AGO-4) are found in mice and humans (Pratt and MacRae, 2009).
The Argonaute superfamily of proteins can be subclassified into three groups: (1) the Ago subfamily, to which siRNA and miRNA bind; (2) the PIWI subfamily, to which piRNAs bind (piRNAs are a subclass of PIWI interacting small RNAs (rv27nt in length) expressed in the germ line regulating gene silencing of retrotransposons particularly during spermatogenesis); and (3) a class of C. elegans Ago-like proteins (worm Ago’s or WAGO’s) (Kapoor et al., 2008). Argonaute proteins are ~100kDa in average and are at the core of every RISC (Kawamata and Tomari, 2010). Argonaute proteins are also known as PPD proteins due to the conservation of the PAZ and PIWI domains. The PAZ domain is an independently stable domain with a β-barrel core which weakly binds single-stranded RNA as well as dsRNA (Cerutti et al., 2000; Yan et al., 2003). The PIWI domain, also involved in single-stranded RNA binding, recognizes the 5′-phosphate of the guide strand (Cerutti et al., 2000). Human AGO-2 was found to be sufficient and necessary for processing of small RNAs (Hammond et al., 2001a; Meister et al., 2004). Interestingly, although all Ago proteins are capable of binding small dsRNA, it is only AGO-2, in humans and mice, which possesses the catalytic activity known as “slicing” (Kawamata and Tomari, 2010).
In D. melanogaster, the Ago-2-containing lysate fraction was determined to be ~500KDa, much larger than the predicted molecular weight for Ago-2 (Hammond et al., 2001a). This discrepancy indicates that Ago-2, albeit being highly important, is only one component of a larger protein complex. Indeed, several other components of this large protein complex have been discovered and described. TRBP (TAR RNA-binding proteins) has been found and reported to be a Dicer protein partner involved in the “loading” of the RISC (Chendrimada et al., 2005). TRBP functions in a similar manner to D. melanogaster R2D2 and Loquacious; however, TRBP, R2D2, and Loquacious are not homologous proteins (Czech et al., 2008; Liu et al., 2003). GW182/Tnrc6 proteins as well as the helicases Rck and MOV10 initially identified to be a component of the RNA processing bodies (P-bodies) recruit “loaded” Ago-2 to P-bodies where final unwinding and RNA processing occurs (Kulkarni et al., 2010). Dcp-1 and Dcp-2 (decapping enzymes) are also present in P-bodies and are involved in RNA processing by excising the 5′-GpC-RNA cap to initiate RNA degradation (Kawamata and Tomari, 2010; Kulkarni et al., 2010).
As noted above, the miRNA:miRNA* duplex is produced in a two-step process directed by the activity of Drosha/Dgcr8 in the nucleus and Dicer in the cytoplasm, respectively. This miRNA:miRNA* duplex is bound by Ago-2 (this complex is also known as a loaded RISC) and further processed. This involves unwinding of the duplex and selection of the functional miRNA guide strand, followed by degradation of the nonfunctional passenger strand and final assembly of the complete RISC (also known as holoenzymatic-RISC or holo-RISC). This process prepares the miRNA for the recognition of and annealing to its target mRNA (Bartel, 2004; Kawamata and Tomari, 2010; Sayed and Abdellatif, 2011).
Loading of the miRNA:miRNA* duplex into Ago-2 is not a random process. First, it is hypothesized that the transfer of the duplex from Dicer to Ago-2 is guided by direct contact. Both Dicer and Ago-2 contain a conserved PAZ domain, involved in weakly binding RNA molecules, and believed to be involved in Dicer:Ago-2 protein–protein interaction. Indeed, Dicer coimmunoprecipitates with Ago-2 in D. melanogaster S2-cultured cells (Hammond et al., 2001a). However, Ago proteins are unable to load miRNAs without the involvement of stabilizing factors. In D. melanogaster, this Dicer-interacting protein is known as R2D2, while in humans, a factor with a similar function is called the TAR-binding protein 2 (TRBP2); however, these two proteins are not homologous and a true R2D2 homologue has not yet been found in humans (Czech et al., 2008; Kawamata and Tomari, 2010; Liu et al., 2003).
Additionally, loading of the miRNA:miRNA* duplex is directed by the thermodynamic stability of the duplex itself and is sequence dependent, including center mismatches near or in guide positions. Guide positions vary and are specific for the type of Dicer or Ago proteins involved in the loading process; generally, guide positions involve nucleotides at residues 7–11. Each Ago protein has a preference for the 5′end nucleotide present in the miRNA:miRNA* duplex. This 5′nucleotide, along with sequence mismatches, confers the duplex with asymmetry in the thermodynamic stability of the strands directing the loading of the guide strand. The orientation of the duplex is also sensed by the three-dimensional conformation of the recognizing protein. Interestingly, for the A. thaliana AGO-1 and AGO-2 proteins, the positional swapping of the MID and PIWI domains switches their nucleotide preference resulting in a preferential loading of the passenger strand. However, in humans, AGO proteins do not have a preference for the 5′nucleotide and it seems that loading is a simpler process. Nevertheless, human AGO proteins appear to have a preference for duplexes with center mismatches and disfavor duplexes with mismatches at the ends (Kawamata and Tomari, 2010; Kawamata et al., 2009).
Two processes have been described for the unwinding and separation of the miRNA:miRNA* duplex. First, Ago-2 (in flies as well as humans) has been shown to cleave and nick the passenger strand (Leuschner et al., 2006; Matranga et al., 2005; Miyoshi et al., 2005; Rand et al., 2005). In an Ago-2 slicing activity-dependent manner, separation by degradation of the passenger strand has been reported as a result of the activity of the Mg2+-dependent endonuclease known as C3PO (component 3 promoter of RISC), in D. melanogaster, as well as the activity of the exonuclease QIP (QDE-1 interacting protein), in N. crassa (Liu et al., 2009; Maiti et al., 2007).
Second, a slower slicer-independent unwinding and separation of the passenger strand has been reported. This process is accelerated by and is the effect of the lower thermodynamic stability of the duplex compared to that of the miRNA:target duplex in a sequence-dependent manner. Additionally, in an ATP-independent manner, sequence pairing and center sequence mismatches have been shown to be essential for the unwinding of the miRNA:miRNA* duplex. Thus, in this case, it could be noted that the passenger strand is simply replaced by the target mRNA due to a “mirror image” effect in which the passenger strand acts as a first “target” sequence replaced by a more stable mRNA target sequence (Kawamata and Tomari, 2010; Kawamata et al., 2009). This hypothesis is reinforced by the enhanced stability of the miRNA–RISC provided by the target mRNA; in other words, abundance of target mRNA increases accumulation of the cognate miRNA within the RISC. Moreover, enhanced accumulation of passenger strands is observed when a synthetic target is introduced in the system (Chatterjee et al., 2011).
miRNA complementary sequences are most commonly located in the 3′- or 5′UTR of the target mRNA molecules. The miRNA pairing to the mRNA target sequence occurs in a nonperfect manner, commonly presenting central sequence mismatches. However, miRNAs are perfectly complementary to their target sequence at nucleotide ~2–8 in the 5′ of the miRNA, known as the “seed” sequence (Lee et al., 1993; Wightman et al., 1993). The presence of the miRNA in the cellular cytoplasm triggers the aggregation of the RISC. The RISC associates with several proteins involved in mRNA decapping to induce recircularization of the mRNA molecule; these include exonucleases to induce deadenylation and/or polyribosomes in order to block the translation of the mRNA target (Bartel, 2009).
3.4. miRNA expression and regulation
Initially, mutations in the lin-4 gene were rescued by the introduction of a 683bp genomic DNA fragment, indicating that all or mostly all the regulatory elements for transcription and regulation of the lin-4 gene were located within this fragment. Also, following gene structure analysis and cloning strategies, it was discovered that the lin-4 gene was transcribed and expressed as a single amplicon (Ambros and Horvitz, 1987). RNA polymerase III drives the transcription of small RNAs including tRNAs, snRNAs, and U6 RNAs, and it was believed to be the miRNA transcription initiator. However, evidence accumulated to suggest that RNA polymerase II was also involved in the transcription of miRNA genes. Currently, it is believed that RNA polymerase II is the major player responsible for the transcription of miRNAs with few being transcribed by RNA polymerase III (Bartel, 2004; Sayed and Abdellatif, 2011).
Computational prediction and cloning analyses have shown that miR-NAs are highly conserved and can be located in the genome embedded within an intron of a host gene (miRtron) or as a single- or polycistronic unit. The location of the miRNA gene is an important determinant of its expression and regulation. As a miRtron, miRNA expression is dictated by regulatory elements present in the promoter region of the host gene. miR-208a and miR-208b are located within intron-31 of the α-MHC and intron-29 of the β-MHC gene, respectively (Callis et al., 2009; van Rooij et al., 2009). As a single or a polycistronic unit, miRNA genes have their own cis-regulatory sequences and transcription is modified in a tissue- and cell-specific manner drawing similarities to transcription regulation of protein-coding genes. miR-1 and miR-133 contain conserved CArG boxes in their promoter regions and are directly regulated by SRF (Liu et al., 2008; Zhao et al., 2005). Indeed, in an elegant model of interregulation, it has been shown that in skeletal muscle, miR-133-modulated SRF expression represses cell proliferation, while miR-1-modulated HDAC4 expression represses MEF2-activated genes and induces cell differentiation. In a feedback regulatory loop, MyoD (a skeletal muscle transcription master regulator) and SRF regulate the expression of the miR-1/miR-133 polycistron (Fig. 10.2) (Chen et al., 2006). Similarly, SRF, Myocardin, and Nkx2-5 directly regulate the expression of miR145/miR143 in cardiac cells as well as in smooth muscle cells (Cordes and Srivastava, 2009; Liu et al., 2007).
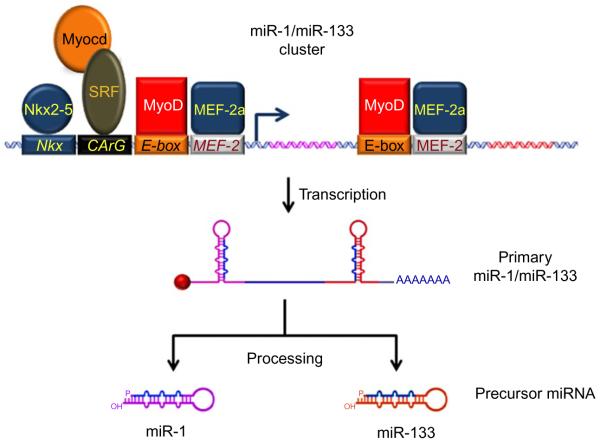
Figure 10.2. Regulation and processing of the miR-1/miR-133 cluster.
The miR-1/miR-133 clusters are present in different chromosomes (see text). Several transcription factor response elements are shown in the 5′upstream promoter region as well as in the “intergeneic” region. As shown, Nkx, CArG, E-box, and MEF-2 boxes represent recognition sites for Nkx2-5, SRF, Myogenin/MyoD, and MEF-2a binding, respectively. Further processing of the cluster results in the formation of two hairpin precursor miRNAs, miR-1 (in purple) and miR-133 (in red). Mature miRNA is represented in blue in both hairpins. Primary miRNAs are protected from early degradation by the addition of a 5′-m7G cap (red ball) and a 3′ poly-A tail.
Posttranscriptional regulation of miRNA expression has recently been reported in C. elegans and involves the activity of the Terminal Urydil Transferase 4 protein (TUT4). TUT4 is a noncanonical poly (A) polymerase which is recruited to the pre-miRNA by Lin28 protein due to the recognition of a GGAG motif in the terminal loop; it adds a poly (U) tail at the 3′end of the stem-loop, thus inhibiting the processing by the Dicer protein (Heo et al., 2009). Additionally, it has been shown that terminal loop sequences direct the binding of the RNA-binding protein hnRNP A1. Binding of hnRNP A1 to the terminal loop of pri-miRNAs remodels the three-dimensional conformation of the hairpin, thereby modulating miRNA processing. Thus, hnRNP A1 acts as a regulator of Drosha-mediated miRNA processing (Guil and Caceres, 2007; Michlewski and Caceres, 2010; Michlewski et al., 2008, 2010). Similarly, posttranscriptional processing of miRNAs is affected by extracellular signaling. Indeed, pri-miR-21 to pre-miR-21 processing is enhanced by the recruitment of SMAD proteins (the TGF-β and BMP signal transducers) to the Drosha miRNA microprocessor complex by the RNA helicase p68 and the consequent accumulation of pre-miRNA molecules in the complex (Davis et al., 2008).
Additionally, chromatin structure and three-dimensional configuration have also been shown to contribute to the regulation of miRNA expression. Gene compartmentalization in various genomic domains is achieved by the periodic looping of chromatin also known as scaffold/matrix-attachment regions (S/MARs). S/MARs expose or hide specific cis-regulatory sequences in a cell- and tissue-specific manner where exposed elements are bound by the transcriptional machinery to up- or downregulate gene expression. Disruption in the expression of S/MAR-binding proteins, such as SATB1 and SMAR1, among others, included within the transcriptional machinery, leads to chromatin modification and gene regulation alteration. Accumulation of S/MAR-binding proteins has been shown in upstream regions of the miR-17-92 cluster, as well as in the individual miRNAs let-7b, miR-17, miR-93, and miR-221 (Chavali et al., 2011).
4. miRNAs in Cardiac Development, Function, and Disease
Retrospectively, the first indication that miRNAs play an important role in embryonic development came to light with the genetic evidence that the small RNAs lin-4 and let-7 played a role in the posttranscriptional regulation of the expression of other heterochronic genes (Lee et al., 1993; Reinhart et al., 2000; Wightman et al., 1993). Subsequently, the discovery of miRNAs in other organisms and the description of the molecular machinery responsible for miRNA processing and function consolidated the notion that miRNAs indeed play an important role in several cellular processes. To date, cloning of small RNA molecules, quantitative RT-PCR screening, as well as microarray expression profiling have identified a large number of miRNAs in many tissues and organisms (Glazov et al., 2008; Lagos-Quintana et al., 2001, 2002, 2003; Lau et al., 2001; Lee and Ambros, 2001). Detailed information about miRNAs is accumulated in the miRNA database known as “miRBase.” According to its most recent release (Release 18, November 2011), there are 18, 226 entries expressing 21, 643 mature miRNA products in 168 species. In the mouse, there are 741 entries expressing 1283 mature miRNA sequences; in the human database, there are 1527 entries expressing 2108 mature miRNA sequences (miRBase: http://www.mirbase.org).
Expression and functional dissection of the mechanism of miRNA fine-tuning of gene expression has revealed considerable intersection and cross talk with well-described signaling and transcriptional networks for the regulation of development, morphogenesis, cell fate determination, and other cellular processes. In this section, we will discuss the function of miRNAs in heart development and summarize their role during cardiac remodeling and in disease.
4.1. miRNA function during embryonic stem cell differentiation into cardiomyocytes
Processing of mature miRNAs is a multistep biological process driven by the activity of the small RNA microprocessor (Drosha/Dgcr8) in the nucleus and Dicer in the cytoplasm. In flies, depletion of the Dicer homologue Dcr-1 results in the derepression of target genes with noticeable effects during development and patterning (Lee et al., 2004). Similarly, targeted mutation of the mouse Dicer gene resulted in early embryonic lethality as a result of the depletion of the embryonic stem cell (ESC) pool (Bernstein et al., 2003). In tissue culture, mouse ES cells carrying a conditional mutation of the Dicer gene display severe defects in differentiation, aberrant expression of stem cell markers, reduction in epigenetic silencing, and absence of all small dsRNA molecules (Kanellopoulou et al., 2005). Although these cells maintain a slow rate of proliferation, they do not contribute to the formation of embryonic chimeras when injected into blastocysts (Kanellopoulou et al., 2005). Similarly, a Dgcr8 null mutation in mouse ES cells results in the accumulation of pri-miRNAs and the absence of precursors and mature miRNA. However, Dgcr8 null ES cells differ from Dicer null ES cells in that the former express stem cell molecular markers normally (Wang et al., 2007).
miRNA and mRNA expression profiles have documented that mouse and human ES-derived cardiomyocytes, compared to fetal and adult heart tissue, exhibit a correlation between miRNA expression and the up- or downregulation of mRNA expression. Although it has been demonstrated that miRNAs target mostly 3′UTR mRNA sequences, such an inverse correlation in miRNA and mRNA expression indicates that miRNAs are surely involved in the cardiac differentiation regulatory network (Synnergren et al., 2011). Indeed, cardiac differentiation of human fetal cardiomyocyte progenitor cells, or mouse ES cells, is accompanied by an upregulation of several miRNAs including miR-1, miR-133, and miR-499. Correspondently, adenoviral overexpression of miR-1 and miR-499 reduced progenitor cell proliferation by 25% and 15%, respectively, while enhancing differentiation (Sluijter et al., 2011). Simultaneously, lower protein levels of several progenitor cell or stem cell molecular markers were observed (Ivey et al., 2008; Sluijter et al., 2011). Downregulation of the Notch signaling pathway has been demonstrated to be necessary for normal muscle differentiation, while its overexpression inhibits such lineage fate. During cardiac differentiation, Delta, a Notch ligand, is targeted by miR-1. miR-1 lowers the protein levels of Delta as hESC cardiac differentiation progresses (Ivey et al., 2008). Similarly, miR-1 targets and induces the translational repression of HDAC4, a transcriptional repressor of Mef2-dependent activation of muscle-specific genes. Meanwhile, miR-499 has been shown to repress the translation of Sox6 (Sluijter et al., 2011), the sex-determining region-Y box 6 transcription factor which is involved in the maintenance of cardiac development (Cohen-Barak et al., 2003). Moreover, knockdown experiments in ESCs demonstrate that miR-1 is required for normal smooth muscle and cardiac muscle differentiation, at least in part, by modulating the expression of the Kruppel-like factor 4 (Klf4) (Xie et al., 2011). In addition, miR-1 overexpression in ESC attenuates apoptosis by indirectly elevating the levels of p-Akt (which, in turn, diminishes PTEN and Caspase-3 levels) and also by modulating the expression of Hsp60 (Glass and Singla, 2011; Shan et al., 2010). It is predicted that still more miRNAs remain to be identified with significant roles in the regulation of cardiomyocyte cell fate.
4.2. miRNAs during cardiac development
As noted above, miRNAs have been revealed to have an enormous impact during embryonic development, as observed in Dicer null mice (Bernstein et al., 2003), and Dgcr8 null mice (Wang et al., 2007). In the heart, conditional mutation of Dicer by employing cardiac tissue-specific Cre lines has revealed that miRNAs are not only necessary for cardiac embryonic development but also essential for postnatal cardiac maintenance and function (Chen et al., 2008; da Costa Martins et al., 2008; Huang et al., 2010b; Saxena and Tabin, 2011). Surprisingly, it has been shown that the developmental program is unaffected when individual miRNAs are mutated. Nevertheless, such miRNAs appear to be powerful mediators in the modulation of gene expression under stress conditions (Cordes and Srivastava, 2009; Huang et al., 2010a; Tatsuguchi et al., 2007; van Rooij et al., 2007).
Myo-miRs or muscle miRNAs is a term used to describe those miR-NAs specifically expressed or enriched in smooth, skeletal, and/or cardiac muscle (van Rooij et al., 2009). In particular, miR-1, miR-133, miR-208, and miR-499 have been reported to be highly expressed in cardiac tissue from the early stages of heart development to the adult. These specific myo-miRs have been shown to be involved in cardiac development, regulating cell proliferation, differentiation, and apoptosis, and also playing an important role in several cardiac diseases, such as cardiac hypertrophy, myocardial infarction (MI), cardiac arrhythmia, and heart failure (Sayed and Abdellatif, 2011; van Rooij et al., 2009). In addition, many non-myo-miRs, those expressed in multiple tissues as well as heart muscle, are also involved in the development and differentiation of this organ (Sayed and Abdellatif, 2011).
miR-1 and the closely related miR-206 possess an identical seed sequence and are related in function and expression; however, they are present in different genomic loci and their expression and transcriptional regulation differs. miR-1 is predominantly enriched in cardiac and skeletal muscle tissues, while miR-206 is a skeletal muscle-specific miRNA (Chen et al., 2010). Both miR-1 and miR-206 are expressed along with miR-133 as a bicistronic unit from three different genomic loci: in mice, miR-1-1/ miR-133a-2 (chromosome 2), miR-1-2/miR-133a1 (chromosome 18), and miR-206/miR-133b (chromosome 1) (Liu et al., 2007). Cloning screening determined that miR-1 expression accounts for ~45% of the miRNAs found in the adult mouse heart and ~24% in the adult human heart, suggesting its participation in cardiac formation and function (Lagos-Quintana et al., 2002). In chick embryos, in situ hybridization shows that miR-1 is expressed in early stages of cardiac development, at the onset of cardiomyocyte differentiation and during the formation of the heart tube, as well as in the developing somites where skeletal muscle progenitors are located (Darnell et al., 2006). Similar results were obtained by X-gal staining in transgenic mice in which the LacZ cassette is driven by miR-1 upstream fragments (Zhao et al., 2005). Sequence analysis of this upstream fragment revealed conserved consensus response elements corresponding to the early cardiac determination transcription factor Nkx2-5. Accordingly, the fly transcription factor Tinman and its homologue Nkx2-5 in mice have been shown to regulate the expression of miR-1 by directly binding upstream-specific cis-regulatory response elements (Qian et al., 2011). The Rho-GTPase CDC42 interacts with Tinman/Nkx2-5 and cooperates to regulate miR-1 expression; more interestingly, CDC42 itself is a miR-1 target. This feedback regulatory network is involved in cardiac output and the formation of a normal myofibrillar architecture (Qian et al., 2011). Similarly, SRF, Mef2c, MyoD, and Myogenin have been shown to directly bind consensus-conserved response elements in order to regulate miR-1 expression in cardiac and skeletal muscle tissues (Liu et al., 2007; Rao et al., 2006). Interestingly, myocardin, a well-known transcription cofactor of SRF, has been recently reported to regulate the expression of miR-1 in vascular smooth muscle cells (Wang et al., 2001). Inducible overexpression of myocardin results in an upregulation of the expression of miR-1 which, in turn, inhibits cell proliferation due to the downregulation of the serine/ threonine kinase Pim-1 (Chen et al., 2011; Jiang et al., 2010).
The early embryonic expression, together with its regulation by early cardiac differentiation determinants, suggests that miR-1 may play an important role in cardiac development and function. Indeed, miR-1 loss-of-function in D. melanogaster is embryonic lethal due to a disruption in the patterning of the dorsal vessel as a result of a defect in muscle differentiation. This results in a failure in cardiac progenitor cell determination, leading to an increase in undifferentiated progenitor cells (Kwon et al., 2005; Sokol and Ambros, 2005). This phenotype is strongly related to the miR-1-mediated downregulation of Delta, a Notch ligand involved in cell–cell signaling required for the differentiation and maintenance of skeletal and cardiac muscle (Kwon et al., 2005). Similarly, in mice, null mutation of miR-1-2 results in noticeable embryonic lethality from E15.5 up to 50% lethality at weaning time. miR-1-2 null mutant embryos show pericardial edema and cardiac dysfunction. Juvenile null mice present cardiac hyper-plasia due to cell proliferation up to three times higher than normal, ventricular hypertrophy, or dilated cardiomyopathy, in addition to defects in heart function related to defects in the cardiac conduction system. In the surviving mice, higher levels of Irx5 (a miR-1 target which is involved in the regulation of the cardiac repolarization gradient through Kcnd2/Kv4.2) and higher protein levels of Hand2 are observed (Costantini et al., 2005; Zhao et al., 2007). In contrast, miR-1 overexpression driven by the β-MHC promoter in transgenic mice during early cardiac formation results in stunted growth and cardiac development arrest at embryonic day E13.5 (E13.5). The hearts show thin-walled ventricles and heart failure due to lower levels of proliferation compared to wild-type controls (Zhao et al., 2005). miR-1 was shown to target the 3′UTR of Hand2, a bHLH tran-scription factor expressed early in development beginning at E7.75–E8.0 in the linear heart tube and further regulating differentiation of the right ventricle (Srivastava et al., 1997).
miR-133 is a component of the bicistronic unit along with miR-1, yet miR-133 expression is not as abundant (Lagos-Quintana et al., 2002). However, miR-133 has been shown as an important player during cardiac development. In an effort to uncover miR-133 functions in the heart, miR-133a-1 and miR-133a-2 were targeted for null mutation. Analyses of single miR-133 gene mutation resulted in no differences when compared to wild-type controls. When analyzing double null mutation of miR-133a-1 and miR-133a-2, it was observed that the double mutation resulted in partial embryonic or postnatal lethality. This lethality is likely related to cardiovascular defects including ventricular septal defects (VSDs) and dilation in the atria and ventricles as a result of increased proliferation and, intriguingly, increased apoptosis near the interventricular septum (Liu et al., 2008; Sayed and Abdellatif, 2011). Mutant animals that escape the early lethality develop dilated cardiomyopathy and present thinner ventricular walls and cardiac failure. However, cardiac hypertrophy and VSD are not observed in mutant mice (Liu et al., 2008). These latter results are in contradiction to previous observations where downregulation of miR-133 by specific antagomirs induces cardiac hypertrophy (Carè et al., 2007). These phenotype discrepancies might be attributed to the imperfect downregulation of miR-133 by the antagomir and/or residual expression of miR-133b since it is not targeted in the miR-133a-1/miR-133a-2 null mice. On the other hand, overexpression of miR-133 during mouse embryonic stages driven by the β-MHC promoter results in a phenotype similar to that found in β-MHC-miR-1 overexpressing mice (including embryonic lethality at mid-gestational stages (rvE15.5), reduced proliferation inducing thinning of the ventricular walls, VSD, and heart failure) (Liu et al., 2008).
As a member of the bicistronic unit with miR-1, miR-133 expression is also regulated by several transcription factors including SRF. Interestingly, the miR-133a-1/miR-133a-2 double knockout mice show a twofold increase in SRF expression and the activation of a smooth muscle genetic network in cardiac tissue (Liu et al., 2008). SRF overexpression in postnatal hearts driven by the α-MHC promoter induces cardiac dilation, in addition to increased heart weight due to hypertrophy and the reactivation of a fetal genetic program. Moreover, the 3′UTRs of SRF, as well as CyclinD2 and CDC42, are targeted by miR-133 indicating that miR-133 is involved in the control of cell proliferation and cell cycle (Carè; et al., 2007; Liu et al., 2008).
Although miR-1 and miR-133 are transcribed as a single molecule, their maturation and function differ. As noted, miR-1 is highly expressed in cardiac and skeletal muscle tissues, while miR-133 appears less abundant. Both miRNAs play important roles in the development of the cardiac tissue, albeit with opposite functions; miR-1 promotes and regulates cell differentiation, while miR-133 induces cell proliferation and the maintenance of the progenitor cell pool (Chen et al., 2006). A similar result is observed when miR-1 is overexpressed in Xenopus embryos where myoblast and cardiac precursor proliferation is reduced while differentiation is enhanced. However, injection of miR-133 results in augmented cell proliferation with a reduction in cell differentiation, more so in myoblast than in cardiac progenitor cells (Chen et al., 2006). This apparent contradiction in the effects of miR-1 and miR-133 in cell proliferation and cell differentiation might be explained by the differences in the nature of their targets and their distinct and possible independent transcription regulation, even though both belong to a bicistronic unit. As noted, miR-1 targets HDAC4, which represses myoblast differentiation, while miR-133 has been shown to target SRF, which is involved in transcriptional regulation of proliferation and differentiation of muscle cells (Chen et al., 2006). Interestingly, SRF directly binds to upstream regions of the miR-1/miR-133 bicistronic unit, which contain consensus CArG boxes, to activate its transcription (Zhao et al., 2005). Moreover, MEF2 and MyoD also directly regulate the transcription of miR-1/miR-133 by binding to an intergenic enhancer containing MEF2 response elements and E-box binding sequences. These intergenic enhancers, present in the miR-1-1/miR-133a-2 and miR-1-2/miR-133a-1 bicistrons, are hypothesized to drive miR-1/miR-133 differential expression (Fig. 10.2). Taken together, these results indicate that regulatory feedback networks are in place in order to regulate cell proliferation versus cell differentiation, in part mediated by miR-1 and miR-133 (Liu et al., 2007).
During cardiac development, not only miR-1/miR-133 but also other miRNAs have been shown to exert an effect. Recently, miR-130a was reported to be involved in cardiac development through targeting the transcription factor Friend-of-GATA 2 (Fog-2). miR-130a is broadly expressed in kidney, liver, testis, and brain but highly enriched in lung and heart. Embryonic overexpression of miR-130a, driven by the β-MHC promoter, results in ventricular hypoplasia and embryonic lethality at mid-gestation stages (E13.5–E14.5), a phenotype resembling that described for Fog-2 null mutant mice (Kim et al., 2009). Similarly in mice, miR-195, a member of the miR-15 family, was shown to induce ventricular hypoplasia and VSD when overexpressed in embryonic stages under the β-MHC promoter, in part by affecting the expression of the Check point 1 protein (Check1). Knockdown experiments using locked nucleic acid oligomers against miR-15 derepresses Check1 and induces postnatal myocytes to reenter the cell cycle (Porrello et al., 2011).
In zebrafish, cardiac expression of miR-138 is found to be restricted to the ventricle. Knockdown experiments targeting miR-138 result in pericardial edema at late cardiac stages (48hpf ) reflecting cardiac dysfunction. However, initial cardiac development and formation appears undisturbed. Further, miR-138 knockdown resulted in the expansion of the expression fields of aldh1a2 (retinoic acid dehydrogenase 2-Raldh2), a putative miR-138 target that is involved in retinoic acid synthesis. Derepression of Raldh2 and versican (another putative miR-138 target that encodes a versatile extracellular matrix proteoglycan) in the heart induces a defect in cardiac patterning characterized by the aberrant formation of the atrio-ventricular canal (AVC) (Morton et al., 2008). Knockdown of miR-143, a miRNA highly expressed in the cardiovascular system, derepresses the RA signaling pathway which profoundly influences cardiac patterning (Miyasaka et al., 2011). Similarly, miR-143 regulates ventricular cardiomyocyte F-actin remodeling. Downregulation of miR-143 results in ventricular collapse by targeting adducin3 (add3), an F-capping protein, affecting F-actin dynamics, cell morphology and contractility (Deacon et al., 2010). Further, in an interesting twist, the mechanical input of the heartbeat itself induces the expression of miR-143 although the molecular pathway is not fully known (Miyasaka et al., 2011). Additionally, in a screen performed in Dicer mutant zebrafish hearts, Has2, a molecular marker of AVC formation, was found to be upregulated. In turn, overexpression of miR-23, a putative targeting miRNA, was found to repress Has2 expression and to reduce extracellular matrix remodeling and endocardial jelly formation, resulting in impaired cardiac valve formation. Also, miR-23 blocks the EMT (endothelial-to-mesenchymal transformation) induced by the TGF-β signaling pathway possibly due to targeting Has2, icat, and tmem2 3′UTR sequences (Lagendijk et al., 2011). These studies indicate that miRNAs are novel regulators of EMT, cellular migration driven by signaling cues, and are essential for the normal formation of the cardiac tube. The Slit/Robo signaling pathway has been shown to be one of the most important migration/repression signals during development (Kidd et al., 1999). miR-218 is expressed from intronic sequences in SLIT2 and SLIT3 and targets the ROBO1 and ROBO2 mRNA to repress their translation. Overexpression of miR-218 and the consequent repression of the ROBO proteins reduced the activation of the MAPK pathway and disrupted the VEGF-induced migration of endocardial and myocardial cells, leading to an aberrant heart (Fish et al., 2011)
The polycistronic miR-17-92 cluster, also called Oncomir-1, comprises seven miRNAs and related miRNA clusters are ubiquitously expressed. The miR-17-92 cluster has been shown to be related to the enhancement of cell proliferation in several cancers, reduction of apoptosis, and increasing tumor angiogenesis (He et al., 2005). During cardiac development, null mutation of the related miR-106arv363 and miR-106brv25 clusters resulted in no observable phenotype and mice are viable and seemingly normal; however, null mutation of the miR-17-92 cluster has been reported to result in lung hypoplasia and induce the appearance of VSD at late developmental stages leading to immediate postnatal lethality (Ventura et al., 2008).
4.3. miRNAs in cardiac remodeling and pathological cardiac processes
Pathological processes are the cause or the effect of a disruption of the normal physiological status of a cell, tissue, organ, and ultimately an organism. These changes are also seen at the molecular level as a dynamic change in gene expression and manifested by altered genetic interactions. In the heart, cardiac pathologies often refer to stress-related disorders as well as congenital diseases which ultimately lead to cardiac tissue remodeling. These pathological defects include cardiac hypertrophy, cardiac dilation, and MI. Genome-wide analyses have uncovered significant changes in genetic expression signatures when the cardiac tissue is under stress conditions, showing a change in mRNA expression along with up- or down-regulation of miRNAs. In particular, under stress conditions, the 3′UTRs of mRNAs were shortened and the repressive function of miRNAs was reduced. Also, there is an activation of a fetal genetic program, which is indicative of a return to a developmental program in order to mitigate the effects of such a stress (Fig. 10.3) (Park et al., 2011; Thum et al., 2007).
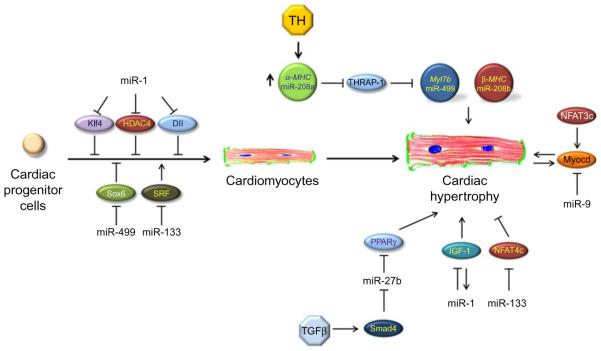
Figure 10.3. miRNA function and integration in the regulatory networks to regulate cardiomyocyte differentiation and disease.
Graphic representation of several regulatory networks targeted by miRNAs in cardiomyocyte differentiation from cardiac progenitor cells, cardiomyocytes, and their function in the regulation of cardiac hypertrophy. Also, the integration of miRNAs in such regulatory networks by targeting transcription factors, signaling molecules, and/or structural proteins is shown.
miR-208 is expressed specifically in cardiac tissue and is detected as two isoforms, miR-208a and miR-208b. They are transcribed from intronic sequences in the myosin heavy chain (MHC) genes Myh6 (α-MHC) and Myh7 (β-MHC), respectively. These two miRNAs differ only in 3nt but they share an identical “seed” sequence, indicating that both miRNAs might target the same genes. During cardiac development, the MHC genes are expressed at different developmental stages; the slow contractile β-MHC is predominantly expressed in adult skeletal muscle and in the heart during embryonic stages, whereas the fast contractile α-MHC is expressed at late embryonic stages and in the adult heart, becoming the predominant cardiac MHC shortly after birth. The switch from β-MHC to α-MHC is dependent on the activity of thyroid hormone (TH) signaling at birth which directly regulates α-MHC and β-MHC cis-response elements, repressing β-MHC and activating α-MHC expression (Weiss and Leinwand, 1996). Due to their location within introns, miR-208b and miR-208a are coexpressed with their host genes. Therefore, the TH signaling also regulates a miR-208b to miR-208a switch, which was previously unrecognized when investigating the MHC switch, at birth (Callis et al., 2009; van Rooij et al., 2007). In stress models which induce cardiac hypertrophy, such as thoracic aortic banding or transgenic overexpression of calcineurin in the heart (Molkentin et al., 1998), the β-MHC to α-MHC switch at birth is not observed and the β-MHC embryonic levels persist postnatally and in the adult heart (Callis et al., 2009).
In mice, miR-208 null mutations do not show any severe developmental phenotypes. The mice are viable although they exhibit some abnormalities in the cardiac conduction system as adults, including atrial conduction deficiencies and atrial fibrillation. This is consistent with the observation that miR-208a targets GATA4, an early cardiac differentiation marker and a direct regulator of Cx40 expression in the conduction system (Callis et al., 2009). Although miR-208a null mice show normal α-MHC and β-MHC expression levels, under stress conditions, the signature persistence of β-MHC in postnatal stages is not observed (van Rooij et al., 2007). Nevertheless, other cardiac hypertrophy markers such as ANF are upregulated, indicating that the effect of miR-208a mutation is specific in the modulation of β-MHC levels. Indeed, overexpression of miR-208a driven by the α-MHC promoter induces cardiac hypertrophy and the induction of β-MHC but not ANF (Callis et al., 2009). Through bioinformatic screens, it was predicted and later experimentally demonstrated that the thyroid receptor associated protein 1 (THRAP1) was a miR-208a target. THRAP-1 is necessary for TH signaling by recruiting RNA pol II to cis-regulatory elements of target genes, in this case, to downregulate β-MHC and upregulate α-MHC at birth when TH levels increase (Canepari et al., 1998). Thus, THRAP-1 expression modulation is determined by miR-208a, providing a direct mechanism for regulation of β-MHC expression. Consequently, the expression of miR-208b is also disturbed in miR-208a null mutant mice under stress conditions. Indeed, miR-208a overexpression represses the expression of miR-208b, and in hypertrophied miR-208a null mice hearts, miR-208b upregulation is stunted (Callis et al., 2009). A third member of the miR-208 family, miR-499, is encoded by intron 19 of the mouse Myh7b gene. Similar to β-MHC, Myh7b and miR-499 are highly enriched in skeletal muscle as well as in the embryonic heart and their expression is absent in miR-208a overexpressing mice. Interestingly, over-expression of miR-499 is sufficient to counteract the effects of loss of miR-208a in mice, including the restoration of β-MHC and miR-208b expression to normal levels, and the normal response to TH inhibition (Fig. 10.3) (van Rooij et al., 2009).
The expression of miR-1 was reduced in hypertrophic hearts induced by transgenic overexpression of calcineurin as well as in the hearts of human patients presenting with cardiac hypertrophy or aortic stenosis (Sayed and Abdellatif, 2011). On the other hand, knockdown of miR-1 in mice proves efficient in induction of cardiac hypertrophy. Intriguingly, therapeutic delivery of miR-1 has been shown to reverse hypertrophy and preserve cardiac function and result in the upregulation of SERCA2a, ANF, and β-MHC (Karakikes et al., 2010). However, results obtained from human patients complicated this issue. It was shown that the expression of miR-1 is up- or downregulated depending on the type of cardiac stress (i.e., cardiac hypertrophy vs. ischemia) indicating that a variety of factors are in play during such events. Indeed, several genes that play an important role in these processes have been shown to be targeted by miR-1, indicating that miR-1 regulates several genetic networks independently in the heart. These genes include Cdk9, involved in the cell cycle; Mef2a, involved in myoblast differentiation; calmodulin, a Ca2+-binding protein involved in several cellular processes; insulin-like growth factor-1, involved in cell growth; and the heat shock proteins Hsp60 and Hsp70, which are involved in apoptosis (Fig. 10.4) (Elia et al., 2009; Sayed et al., 2007; Shan et al., 2010.)
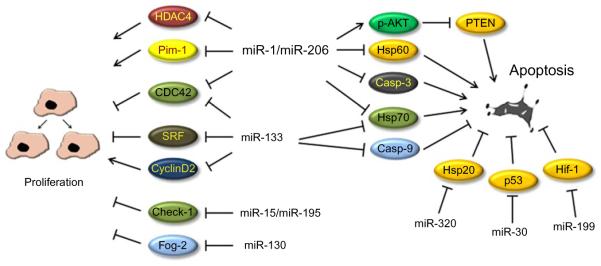
Figure 10.4. miRNA regulatory effects on cellular processes.
Graphic representation of several miRNAs’ regulatory effects on two opposing cellular processes such as proliferation and apoptosis. It is worthy to note that one miRNA might be involved in both processes by targeting different molecules which, in turn, induce or inhibit such process.
In addition to miR-1, miR-133 has also been shown to be downregulated during the onset of cardiac hypertrophy. Indeed, knockdown of miR-133 was sufficient to induce cardiac hypertrophy; however, as described previously, miR-133a-1/miR-133a-2 double ablation results in aberrant myocyte proliferation and apoptosis, VSDs, and partial embryonic lethality without induction of hypertrophy (Liu et al., 2008). The difference in the results might be explained by the residual miR-133b expression in the double knockout model and the difference in the timing of onset of hypertrophy (Abdellatif, 2010; Carè et al., 2007). Alternatively, it is formally possible that the oligonucleotide-based miR-133 knockdown could result in “off-target” effects. Additionally, overexpression of miR-133 under the α-MHC promoter shows that miR-133 transgenic mice are protected from stress-induced cardiac fibrosis, although the transgenic mice also present with some cardiac conduction deficiencies (Matkovich et al., 2010). However, transgenic overexpression of miR-133 under the β-MHC promoter results in early embryonic lethality. Similar to miR-1, the effects of miR-133 during cardiac development and cardiac hypertrophy are results of the interplay between a wide variety of mRNA targets, including Caspase-9 and Hsp70 in apoptosis, SRF in myocyte differentiation, Cdc42 in the regulation of the cell cycle, among others (Abdellatif, 2010; Carè et al., 2007).
miR-21, another miRNA which is dramatically upregulated during cardiac hypertrophy, has been shown to repress the expression of the sprouty gene 1 (Spry-1), a known inhibitor of the ERK–MAP kinase pathway, thus enhancing cell survival (Thum et al., 2008). Knockdown of endogenous miR- 21 in mice under pressure overload conditions results in attenuation of the hypertrophic response measured in terms of collagen fiber deposition and cardiac function (Thum et al., 2008). Conversely, overexpression of miR-21 in mice shows that it exerts a protective effect as determined by the reduction of the myocardial infarct size and reduced apoptosis in the infarct border zone under ischemia conditions. miR-21 was proposed to repress the expression of PDCD24 in this setting (Dong et al., 2009). However, these observations do not correlate with results reported for miR-21 null mutant mice. Deletion of miR-21 does not result in any phenotype and mice do not show any differences compared to wild-type controls, when under stress conditions (Patrick et al., 2010). Certain differences in experimental details might account for the observed contradictory results, highlighting the critical impact of genetic approaches in deciphering the function of individual miRNAs.
A molecular signature of cardiac hypertrophy is the upregulation of myocardin expression. Indeed, myocardin overexpression in cultured cardiomyocytes is sufficient to induce cardiomyocyte hypertrophy (Xing et al., 2006). During hypertrophy, the nuclear factor activator of T cells (NFAT3c and NFATc4) directly regulates myocardin expression by binding to the upstream regions of the myocardin promoter. In turn, miR-9 suppresses myocardin expression by targeting the myocardin 3′UTR, attenuating the effects of hypertrophy. Similar effects are observed upon miR-133a repression of NFAT4c and lowering of its protein levels (Li et al., 2010; Wang et al., 2010).
During cardiac remodeling, fibrosis or fibrotic deposition is enhanced, resulting in an increase of muscular thickening and the loss of flexibility. Fibrotic deposition involves the enhanced proliferation of cardiac fibroblasts and the enhanced production of extracellular matrix components. Several molecular pathways, including the TGF-β signaling pathway, are involved in these processes. Generation of mice with a null mutation in Smad-4 results in animals with hypertrophic hearts and is correlated with an upregulation of miR-27b. Silencing of miR-27b is sufficient to mitigate the effects of Smad-4 null mutation, or the hypertrophy induced in a pressure overload model (Wang et al., 2011). Conversely, TGF-β1, an inhibitor of myoblast differentiation, downregulates miR-24 expression by direct binding of Smad-3 and Smad-4 to upstream promoter sequences. In Smad-4 null hearts, miR-24 is upregulated which then induces endothelial cell apoptosis under cardiac ischemic conditions by targeting the endothelium-rich transcription factor GATA2 and the p21-activated kinase PAK4 (Sun et al., 2008). Overexpression of miR-24 in zebrafish impairs normal cardiac angiogenesis, and knockdown experiments result in a reduced infarct size by vascularization enhancement (Fiedler et al., 2011).
Higher levels of cellular apoptosis have been observed following MI and ischemia/reperfusion (I/R) injury and several miRNAs have been shown to regulate such cellular process. In mice, miR-320 expression is decreased following I/R injury. miR-320 overexpression increases apoptosis and increases the extent of the infarct size after I/R, while miR-320 knockdown shows cell-protective effects through the derepression of Hsp20 (Dong et al., 2009; Ren et al., 2009). Similarly, miR-1/206 has been shown to function as a cardioprotective factor through the repression of Hsp60 and Hsp70 (Fig. 10.4). Additionally, miR-30, miR-133, and miR-199a were shown to function as apoptotic factors through targeting p53, Caspase-9, and Hif-1a and Sirt-1, respectively (Li, 2010).
5. Final Conclusions
In the past decade, miRNAs have clearly established themselves as important manipulators of gene expression and key regulators of cell fate determination, proliferation, differentiation, and cell death during embryogenesis as well as in postnatal life. Yet, only a fraction of the total number of miRNAs have been studied in detail and the true magnitude of their regulatory ability remains to be determined. Indeed, the miRNA’s characteristic “seed” sequence along with its imperfect base pairing allows for one miRNA to target multiple mRNAs; as a corollary, a single mRNA could be targeted by many miRNAs, increasing the level of complexity in the regulation of these intertwined genetic networks. We are confident that we will see more and more reports on the roles of miRNAs in the regulation of a variety of essential biological processes.
Human disease, in general, and cardiac diseases, specifically, are commonly associated with genetic haploinsufficiencies and/or point mutations. Such mutations often lead to the reduction of gene expression and/or function. Therefore, miRNAs, which function as reagents for fine-tuning levels of mRNAs and their protein products, are potential pharmacological targets that can be exploited for therapeutic purposes. With further characterization and the elucidation of context-dependent functions, miRNAs will provide new avenues for the diagnosis, prognosis, and treatment of cardiovascular diseases.
ACKNOWLEDGMENTS
We thank members of the Wang laboratory for discussion and support. We thank Dr. John Mably for careful reading of the chapter and stimulating discussion. Research in the Wang lab was supported by the March of Dimes Birth Defect Foundation and the National Institutes of Health. R. A. E. L. is a postdoctoral fellow grantee of the American Heart Association. D. Z. W. is an established investigator of the American Heart Association.
REFERENCES
- Abdellatif M. The role of microRNA-133 in cardiac hypertrophy uncovered. Circ. Res. 2010;106:16–18. doi: 10.1161/CIRCRESAHA.109.212183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberts B, Bray D, Lewis J, Raff M, Roberts K, Watson J. Molecular Biology of the Cell. 3rd. Garland publishing; New York & London: 1994. pp. 72–73. 292–334. [Google Scholar]
- Ambros V, Horvitz HR. Heterochronic mutants of the Nematode Caenorhabditis elegans. Science. 1984;226:409–416. doi: 10.1126/science.6494891. [DOI] [PubMed] [Google Scholar]
- Ambros V, Horvitz HR. The lin-14 locus of Caenorhabditis elegans controls the time of expression of specific postembryonic developmental events. Genes Dev. 1987;1:398–414. doi: 10.1101/gad.1.4.398. [DOI] [PubMed] [Google Scholar]
- Anderson P. Mutagenesis. In: Epstein F, Shakes C, editors. Caenorhabditis elegans: Modern Biological Analysis of an Organism. Academic press; California: 1995. pp. 31–54. [Google Scholar]
- Andrée B, Duprez D, Vorbusch B, Arnold HH, Brand T. BMP-2 induces ectopic expression of cardiac lineage markers and interferes with somite formation in chicken embryos. Mech. Dev. 1998;70:119–131. doi: 10.1016/s0925-4773(97)00186-x. [DOI] [PubMed] [Google Scholar]
- Arasu P, Wightman B, Ruvkun G. Temporal regulation of lin-14 by the antagonistic action of two other heterochronic genes, lin-4 and lin-28. Genes Dev. 1991;5:1825–1833. doi: 10.1101/gad.5.10.1825. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel B, Bartel DP. MicroRNAs: At the root of plant development? Plant Physiol. 2003;132:709–717. doi: 10.1104/pp.103.023630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, Li MZ, Mills AA, Elledge SJ, Anderson KV, Hannon GJ. Dicer is essential for mouse development. Nat. Genet. 2003;35:215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- Blaschke RJ, Hahurij ND, Kuijper S, Just S, Wisse LJ, Deissler K, Maxelon T, Anastassiadis K, Spitzer J, Hardt SE, Scholer H, Feitsma H, et al. Targeted mutation reveals essential functions of the homeodomain transcription factor Shox2 in sinoatrial and pacemaking development. Circulation. 2007;115:1830–1838. doi: 10.1161/CIRCULATIONAHA.106.637819. [DOI] [PubMed] [Google Scholar]
- Bodmer R, Jan LY, Jan YN. A new homeobox-containing gene, msh-2, is transiently expressed early during mesoderm formation of Drosophila. Development. 1990;110:661–669. doi: 10.1242/dev.110.3.661. [DOI] [PubMed] [Google Scholar]
- Bohmert K, Camus I, Bellini C, Bouchez D, Caboche M, Benning C. AGO1 defines a novel locus of Arabidopsis controlling leaf development. EMBO J. 1998;17:170–180. doi: 10.1093/emboj/17.1.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohnsack MT, Czaplinski K, Görlich D. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA. 2004;10:185–191. doi: 10.1261/rna.5167604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondue A, Lapouge G, Paulissen C, Semeraro C, Iacovino M, Kyba M, Blanpain C. Mesp1 acts as a master regulator of multipotent cardiovascular progenitor specification. Cell Stem Cell. 2008;3:69–84. doi: 10.1016/j.stem.2008.06.009. [DOI] [PubMed] [Google Scholar]
- Brade T, Manner J, Kuhl M. The role of Wnt signalling in cardiac development and tissue remodelling in the mature heart. Cardiovasc. Res. 2006;72:198–209. doi: 10.1016/j.cardiores.2006.06.025. [DOI] [PubMed] [Google Scholar]
- Buckingham M, Meilhac S, Zaffran S. Building the mammalian heart from two sources of myocardial cells. Nat. Rev. Genet. 2005;6:826–835. doi: 10.1038/nrg1710. [DOI] [PubMed] [Google Scholar]
- Cai C-L, Liang X, Shi Y, Chu P-H, Pfaff SL, Chen J, Evans S. Isl1 identifies a cardiac progenitor population that proliferates prior to differentiation and contributes a majority of cells to the heart. Dev. Cell. 2003;5:877–889. doi: 10.1016/s1534-5807(03)00363-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calado A, Treichel N, Muller E-C, Otto A, Kutay U. Exportin-5-mediated nuclear export of eukaryotic elongation factor 1A and tRNA. EMBO J. 2002;21:6216–6224. doi: 10.1093/emboj/cdf620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callis TE, Pandya K, Seok HY, Tang RH, Tatsuguchi M, Huang ZP, Chen JF, Deng Z, Gunn B, Shumate J. MicroRNA-208a is a regulator of cardiac hypertrophy and conduction in mice. J. Clin. Invest. 2009;119:2772–2786. doi: 10.1172/JCI36154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canepari M, Cappelli V, Pellegrino MA, Zanardi MC, Reggiani C. Thyroid hormone regulation of MHC isoform composition and myofibrillar ATPase activity in rat skeletal muscles. Arch. Physiol. Biochem. 1998;106:308–315. doi: 10.1076/apab.106.4.308.4373. [DOI] [PubMed] [Google Scholar]
- Carè A, Catalucci D, Felicetti F, Bonci D, Addario A, Gallo P, Bang M-L, Segnalini P, Gu Y, Dalton ND, Elia L, Latronico MVG, et al. MicroRNA-133 controls cardiac hypertrophy. Nat. Med. 2007;13:613–618. doi: 10.1038/nm1582. [DOI] [PubMed] [Google Scholar]
- Catalanotto C, Azzalin G, Macino G, Cogoni C. Transcription: Gene silencing in worms and fungi. Nature. 2000;404:245. doi: 10.1038/35005169. [DOI] [PubMed] [Google Scholar]
- Cerutti L, Mian N, Bateman A. Domains in gene silencing and cell differentiation proteins: The novel PAZ domain and redefinition of the Piwi domain. Trends Biochem. Sci. 2000;25:481–482. doi: 10.1016/s0968-0004(00)01641-8. [DOI] [PubMed] [Google Scholar]
- Chalfie M, Horvitz HR, Sulston JE. Mutations that lead to reiterations in the cell lineages of C. elegans. Cell. 1981;24:59–69. doi: 10.1016/0092-8674(81)90501-8. [DOI] [PubMed] [Google Scholar]
- Challice CE, Virágh S. The architectural development of the early mammalian heart. Tissue Cell. 1974;6:447–462. doi: 10.1016/0040-8166(74)90037-8. [DOI] [PubMed] [Google Scholar]
- Chatterjee S, Fasler M, Büssing I, Großhans H. Target-mediated protection of endogenous microRNAs in C. elegans. Dev. Cell. 2011;20:388–396. doi: 10.1016/j.devcel.2011.02.008. [DOI] [PubMed] [Google Scholar]
- Chavali PL, Funa K, Chavali S. Cis-regulation of microRNA expression by scaffold/matrix-attachment regions. Nucleic Acids Res. 2011;39:6908–6918. doi: 10.1093/nar/gkr303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JF, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM, Conlon FL, Wang DZ. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat. Genet. 2006;38:228–233. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J-F, Murchison EP, Tang R, Callis TE, Tatsuguchi M, Deng Z, Rojas M, Hammond SM, Schneider MD, Selzman CH, Meissner G, Patterson C, et al. Targeted deletion of Dicer in the heart leads to dilated cardiomyopathy and heart failure. Proc. Natl. Acad. Sci. USA. 2008;105:2111–2116. doi: 10.1073/pnas.0710228105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JF, Tao Y, Li J, Deng Z, Yan Z, Xiao X, Wang DZ. MicroRNA-1 and microRNA-206 regulate skeletal muscle satellite cell proliferation and differentiation by repressing Pax7. J. Cell Biol. 2010;190:867–879. doi: 10.1083/jcb.200911036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Yin H, Jiang Y, Radhakrishnan SK, Huang Z-P, Li J, Shi Z, Kilsdonk EPC, Gui Y, Wang D-Z, Zheng X-L. Induction of microRNA-1 by myocardin in smooth muscle cells inhibits cell proliferation. Arterioscler. Thromb. Vasc. Biol. 2011;31:368–375. doi: 10.1161/ATVBAHA.110.218149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, Shiekhattar R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciruna B, Rossant J. FGF signaling regulates mesoderm cell fate specification and morphogenetic movement at the primitive streak. Dev. Cell. 2001;1:37–49. doi: 10.1016/s1534-5807(01)00017-x. [DOI] [PubMed] [Google Scholar]
- Cohen ED, Wang Z, Lepore JJ, Lu MM, Taketo MM, Epstein DJ, Morrisey EE. Wnt/beta-catenin signaling promotes expansion of Isl-1-positive cardiac progenitor cells through regulation of FGF signaling. J. Clin. Invest. 2007;117:1794–1804. doi: 10.1172/JCI31731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen-Barak O, Yi Z, Hagiwara N, Monzen K, Komuro I, Brilliant MH. Sox6 regulation of cardiac myocyte development. Nucleic Acids Res. 2003;31:5941–5948. doi: 10.1093/nar/gkg807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordes KR, Srivastava D. MicroRNA regulation of cardiovascular development. Circ. Res. 2009;104:724–732. doi: 10.1161/CIRCRESAHA.108.192872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costantini DL, Arruda EP, Agarwal P, Kim K-H, Zhu Y, Zhu W, Lebel M, Cheng CW, Park CY, Pierce SA, Guerchicoff A, Pollevick GD, et al. The homeodomain transcription factor Irx5 establishes the mouse cardiac ventricular repolarization gradient. Cell. 2005;123:347–358. doi: 10.1016/j.cell.2005.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czech B, Malone CD, Zhou R, Stark A, Schlingeheyde C, Dus M, Perrimon N, Kellis M, Wohlschlegel JA, Sachidanandam R, Hannon GJ, Brennecke J. An endogenous small interfering RNA pathway in Drosophila. Nature. 2008;453:798–802. doi: 10.1038/nature07007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Costa Martins PA, Bourajjaj M, Gladka M, Kortland M, van Oort RJ, Pinto YM, Molkentin JD, De Windt LJ. Conditional Dicer gene deletion in the postnatal myocardium provokes spontaneous cardiac remodeling. Circulation. 2008;118:1567–1576. doi: 10.1161/CIRCULATIONAHA.108.769984. [DOI] [PubMed] [Google Scholar]
- Darnell DK, Kaur S, Stanislaw S, Konieczka JK, Yatskievych TA, Antin PB. MicroRNA expression during chick embryo development. Dev. Dyn. 2006;235:3156–3165. doi: 10.1002/dvdy.20956. [DOI] [PubMed] [Google Scholar]
- Davis BN, Hilyard AC, Lagna G, Hata A. SMAD proteins control DROSHA-mediated microRNA maturation. Nature. 2008;454:56–61. doi: 10.1038/nature07086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deacon DC, Nevis KR, Cashman TJ, Zhou Y, Zhao L, Washko D, Guner-Ataman B, Burns CG, Burns CE. The miR-143-adducin3 pathway is essential for cardiac chamber morphogenesis. Development. 2010;137:1887–1896. doi: 10.1242/dev.050526. [DOI] [PubMed] [Google Scholar]
- Denli AM, Tops BBJ, Plasterk RHA, Ketting RF, Hannon GJ. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- Dong S, Cheng Y, Yang J, Li J, Liu X, Wang X, Wang D, Krall TJ, Delphin ES, Zhang C. MicroRNA expression signature and the role of microRNA-21 in the early phase of acute myocardial infarction. J. Biol. Chem. 2009;284:29514–29525. doi: 10.1074/jbc.M109.027896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elia L, Contu R, Quintavalle M, Varrone F, Chimenti C, Russo MA, Cimino V, De Marinis L, Frustaci A, Catalucci D, Condorelli G. Reciprocal regulation of microRNA-1 and insulin-like growth factor-1 signal transduction cascade in cardiac and skeletal muscle in physiological and pathological conditions. Circulation. 2009;120:2377–2385. doi: 10.1161/CIRCULATIONAHA.109.879429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinoza-Lewis RA, Yu L, He F, Liu H, Tang R, Shi J, Sun X, Martin JF, Wang D, Yang J, Chen Y. Shox2 is essential for the differentiation of cardiac pacemaker cells by repressing Nkx2-5. Dev. Biol. 2009;327:376–385. doi: 10.1016/j.ydbio.2008.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinoza-Lewis RNA, Liu H, Sun C, Chen C, Jiao K, Chen Y. Ectopic expression of Nkx2.5 suppresses the formation of the sinoatrial node in mice. Dev. Biol. 2011;356:359–369. doi: 10.1016/j.ydbio.2011.05.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagard M, Boutet SP, Morel J-B, Bellini C, Vaucheret H. AGO1, QDE-2, and RDE-1 are related proteins required for post-transcriptional gene silencing in plants, quelling in fungi, and RNA interference in animals. Proc. Natl. Acad. Sci. USA. 2000;97:11650–11654. doi: 10.1073/pnas.200217597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiedler J, Jazbutyte V, Kirchmaier BC, Gupta SK, Lorenzen J, Hartmann D, Galuppo P, Kneitz S, Pena JTG, Sohn-Lee C, Loyer X, Soutschek J, et al. MicroRNA-24 regulates vascularity after myocardial infarction/clinical perspective. Circulation. 2011;124:720–730. doi: 10.1161/CIRCULATIONAHA.111.039008. [DOI] [PubMed] [Google Scholar]
- Fire A, Albertson D, Harrison SW, Moerman DG. Production of antisense RNA leads to effective and specific inhibition of gene expression in C. elegans muscle. Development. 1991;113:503–514. doi: 10.1242/dev.113.2.503. [DOI] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Fish JE, Wythe JD, Xiao T, Bruneau BG, Stainier DYR, Srivastava D, Woo S. A Slit/miR-218/Robo regulatory loop is required during heart tube formation in zebrafish. Development. 2011;138:1409–1419. doi: 10.1242/dev.060046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass C, Singla DK. MicroRNA-1 transfected embryonic stem cells enhance cardiac myocyte differentiation and inhibit apoptosis by modulating PTEN/Akt pathway in the infarcted heart. Am. J. Physiol. Heart Circ. Physiol. 2011;301:H2038–H2049. doi: 10.1152/ajpheart.00271.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazov EA, Cottee PA, Barris WC, Moore RJ, Dalrymple BP, Tizard ML. A microRNA catalog of the developing chicken embryo identified by a deep sequencing approach. Genome Res. 2008;18:957–964. doi: 10.1101/gr.074740.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory RI, Yan K-P, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R. The Microprocessor complex mediates the genesis of micro-RNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- Grishok A, Pasquinelli AE, Conte D, Li N, Parrish S, Ha I, Baillie DL, Fire A, Ruvkun G, Mello CC. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001;106:23–34. doi: 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- Guil S, Caceres JF. The multifunctional RNA-binding protein hnRNP A1 is required for processing of miR-18a. Nat. Struct. Mol. Biol. 2007;14:591–596. doi: 10.1038/nsmb1250. [DOI] [PubMed] [Google Scholar]
- Guo S, Kemphues KJ. par-1, a gene required for establishing polarity in C. elegans embryos, encodes a putative Ser/Thr kinase that is asymmetrically distributed. Cell. 1995;81:611–620. doi: 10.1016/0092-8674(95)90082-9. [DOI] [PubMed] [Google Scholar]
- Habets PEMH, Moorman AFM, Clout DEW, van Roon MA, Lingbeek M, van Lohuizen M, Campione M, Christoffels VM. Cooperative action of Tbx2 and Nkx2.5 inhibits ANF expression in the atrioventricular canal: Implications for cardiac chamber formation. Genes Dev. 2002;16:1234–1246. doi: 10.1101/gad.222902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond SM, Bernstein E, Beach D, Hannon GJ. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000;404:293–296. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- Hammond SM, Boettcher S, Caudy AA, Kobayashi R, Hannon GJ. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science. 2001a;293:1146–1150. doi: 10.1126/science.1064023. [DOI] [PubMed] [Google Scholar]
- Hammond SM, Caudy AA, Hannon GJ. Post-transcriptional gene silencing by double-stranded RNA. Nat. Rev. Genet. 2001b;2:110–119. doi: 10.1038/35052556. [DOI] [PubMed] [Google Scholar]
- Han J, Lee Y, Yeom K-H, Nam J-W, Heo I, Rhee J-K, Sohn SY, Cho Y, Zhang B-T, Kim VN. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- Harvey RP. Heart Development. Academic Press; London: 1999. [Google Scholar]
- Harvey RP. Patterning the vertebrate heart. Nat. Rev. Genet. 2002;3:544–556. doi: 10.1038/nrg843. [DOI] [PubMed] [Google Scholar]
- He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, Hammond SM. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo I, Joo C, Kim Y-K, Ha M, Yoon M-J, Cho J, Yeom K-H, Han J, Kim VN. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell. 2009;138:696–708. doi: 10.1016/j.cell.2009.08.002. [DOI] [PubMed] [Google Scholar]
- Hiroi Y, Kudoh S, Monzen K, Ikeda Y, Yazaki Y, Nagai R, Komuro I. Tbx5 associates with Nkx2-5 and synergistically promotes cardiomyocyte differentiation. Nat. Genet. 2001;28:276–280. doi: 10.1038/90123. [DOI] [PubMed] [Google Scholar]
- Hoogaars WMH, Tessari A, Moorman AFM, de Boer PAJ, Hagoort J, Soufan AT, Campione M, Christoffels VM. The transcriptional repressor Tbx3 delineates the developing central conduction system of the heart. Cardiovasc. Res. 2004;62:489–499. doi: 10.1016/j.cardiores.2004.01.030. [DOI] [PubMed] [Google Scholar]
- Huang Z-P, Neppl R, Wang D-Z. MicroRNAs in cardiac remodeling and disease. J. Cardiovasc. Transl. Res. 2010a;3:212–218. doi: 10.1007/s12265-010-9165-y. [DOI] [PubMed] [Google Scholar]
- Huang ZP, Chen JF, Regan JN, Maguire CT, Tang RH, Dong XR, Majesky MW, Wang DZ. Loss of microRNAs in neural crest leads to cardiovascular syndromes resembling human congenital heart defects. Arterioscler. Thromb. Vasc. Biol. 2010b;30:2575–2586. doi: 10.1161/ATVBAHA.110.213306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvágner G, McLachlan J, Pasquinelli AE, Bálint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- Ivey KN, Muth A, Arnold J, King FW, Yeh R-F, Fish JE, Hsiao EC, Schwartz RJ, Conklin BR, Bernstein HS, Srivastava D. MicroRNA regulation of cell lineages in mouse and human embryonic stem cells. Cell Stem Cell. 2008;2:219–229. doi: 10.1016/j.stem.2008.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Yin H, Zheng X-L. MicroRNA-1 inhibits myocardin-induced contractility of human vascular smooth muscle cells. J. Cell. Physiol. 2010;225:506–511. doi: 10.1002/jcp.22230. [DOI] [PubMed] [Google Scholar]
- Jorgensen RA, Cluster PD, English J, Que Q, Napoli CA. Chalcone synthase cosuppression phenotypes in petunia flowers: Comparison of sense vs. antisense constructs and single-copy vs. complex T-DNA sequences. Plant Mol. Biol. 1996;31:957–973. doi: 10.1007/BF00040715. [DOI] [PubMed] [Google Scholar]
- Kanellopoulou C, Muljo SA, Kung AL, Ganesan S, Drapkin R, Jenuwein T, Livingston DM, Rajewsky K. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 2005;19:489–501. doi: 10.1101/gad.1248505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor M, Arora R, Lama T, Nijhawan A, Khurana J, Tyagi A, Kapoor S. Genome-wide identification, organization and phylogenetic analysis of Dicer-like, Argonaute and RNA-dependent RNA polymerase gene families and their expression analysis during reproductive development and stress in rice. BMC Genomics. 2008;9:451. doi: 10.1186/1471-2164-9-451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karakikes I, Chaanine A, Kim J, Lebeche D, Hajjar R. Therapeutic cardiac-targeted delivery of mir-1 reverses hypertrophy and preserves cardiac function in a pressure overload animal model. Circulation. 2010:A20916. [Google Scholar]
- Kasahara H, Bartunkova S, Schinke M, Tanaka M, Izumo S. Cardiac and extracardiac expression of Csx/Nkx2.5 homeodomain protein. Circ. Res. 1998;82:936–946. doi: 10.1161/01.res.82.9.936. [DOI] [PubMed] [Google Scholar]
- Kawamata T, Tomari Y. Making RISC. Trends Biochem. Sci. 2010;35:368–376. doi: 10.1016/j.tibs.2010.03.009. [DOI] [PubMed] [Google Scholar]
- Kawamata T, Seitz H, Tomari Y. Structural determinants of miRNAs for RISC loading and slicer-independent unwinding. Nat. Struct. Mol. Biol. 2009;16:953–960. doi: 10.1038/nsmb.1630. [DOI] [PubMed] [Google Scholar]
- Ketting RF, Plasterk RHA. A genetic link between co-suppression and RNA interference in C. elegans. Nature. 2000;404:296–298. doi: 10.1038/35005113. [DOI] [PubMed] [Google Scholar]
- Ketting RF, Haverkamp THA, van Luenen HGAM, Plasterk RHA. Mut-7 of C. elegans, required for transposon silencing and RNA interference, is a homolog of Werner syndrome helicase and RNaseD. Cell. 1999;99:133–141. doi: 10.1016/s0092-8674(00)81645-1. [DOI] [PubMed] [Google Scholar]
- Kidd T, Bland KS, Goodman CS. Slit is the midline repellent for the robo receptor in Drosophila. Cell. 1999;96:785–794. doi: 10.1016/s0092-8674(00)80589-9. [DOI] [PubMed] [Google Scholar]
- Kim GH, Samant SA, Earley JU, Svensson EC. Translational control of FOG-2 expression in cardiomyocytes by microRNA-130a. PLoS One. 2009;4:e6161. doi: 10.1371/journal.pone.0006161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby ML. Cardiac Development. 1st Oxford University Press; New York: 2007. [Google Scholar]
- Kitajima S, Takagi A, Inoue T, Saga Y. MesP1 and MesP2 are essential for the development of cardiac mesoderm. Development. 2000;127:3215–3226. doi: 10.1242/dev.127.15.3215. [DOI] [PubMed] [Google Scholar]
- Kulkarni M, Ozgur S, Stoecklin G. On track with P-bodies. Biochem. Soc. Trans. 2010;38:242–251. doi: 10.1042/BST0380242. [DOI] [PubMed] [Google Scholar]
- Kwon C, Han Z, Olson EN, Srivastava D. MicroRNA1 influences cardiac differentiation in Drosophila and regulates Notch signaling. Proc. Natl. Acad. Sci. USA. 2005;102:18986–18991. doi: 10.1073/pnas.0509535102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagendijk AK, Goumans MJ, Burkhard SB, Bakkers J. MicroRNA-23 restricts cardiac valve formation by inhibiting Has2 and extracellular hyaluronic acid production/novelty and significance. Circ. Res. 2011;109:649–657. doi: 10.1161/CIRCRESAHA.111.247635. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr. Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Meyer J, Borkhardt A, Tuschl T. New microRNAs from mouse and human. RNA. 2003;9:175–179. doi: 10.1261/rna.2146903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, Kim VN. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- Lee YS, Nakahara K, Pham JW, Kim K, He Z, Sontheimer EJ, Carthew RW. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell. 2004;117:69–81. doi: 10.1016/s0092-8674(04)00261-2. [DOI] [PubMed] [Google Scholar]
- Lee SJ, Jiko C, Yamashita E, Tsukihara T. Selective nuclear export mechanism of small RNAs. Curr. Opin. Struct. Biol. 2011;21:101–108. doi: 10.1016/j.sbi.2010.11.004. [DOI] [PubMed] [Google Scholar]
- Leuschner PJF, Ameres SL, Kueng S, Martinez J. Cleavage of the siRNA passenger strand during RISC assembly in human cells. EMBO Rep. 2006;7:314–320. doi: 10.1038/sj.embor.7400637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P. MicroRNAs in cardiac apoptosis. J. Cardiovasc. Transl. Res. 2010;3:219–224. doi: 10.1007/s12265-010-9175-9. [DOI] [PubMed] [Google Scholar]
- Li Q, Lin X, Yang X, Chang J. NFATc4 is negatively regulated in miR-133a-mediated cardiomyocyte hypertrophic repression. Am. J. Physiol. Heart Circ. Physiol. 2010;298:H1340–H1347. doi: 10.1152/ajpheart.00592.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Rand TA, Kalidas S, Du F, Kim H-E, Smith DP, Wang X. R2D2, a bridge between the initiation and effector steps of the Drosophila RNAi pathway. Science. 2003;301:1921–1925. doi: 10.1126/science.1088710. [DOI] [PubMed] [Google Scholar]
- Liu N, Williams AH, Kim Y, McAnally J, Bezprozvannaya S, Sutherland LB, Richardson JA, Bassel-Duby R, Olson EN. An intragenic MEF2-dependent enhancer directs muscle-specific expression of microRNAs 1 and 133. Proc. Natl. Acad. Sci. USA. 2007;104:20844–20849. doi: 10.1073/pnas.0710558105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Bezprozvannaya S, Williams AH, Qi X, Richardson JA, Bassel-Duby R, Olson EN. MicroRNA-133a regulates cardiomyocyte proliferation and suppresses smooth muscle gene expression in the heart. Genes Dev. 2008;22:3242–3254. doi: 10.1101/gad.1738708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Ye X, Jiang F, Liang C, Chen D, Peng J, Kinch LN, Grishin NV, Liu Q. C3PO, an endoribonuclease that promotes RNAi by facilitating RISC activation. Science. 2009;325:750–753. doi: 10.1126/science.1176325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lizhu L, Li C, Wenlai Z, Daniel D, Xiaoxue Z, Chen-Leng C, Lei B, Lei Y, Jody M, Rolf K, Michael GR, Ju C, et al. β-Catenin directly regulates Islet1 expression in cardiovascular progenitors and is required for multiple aspects of cardiogenesis. Proc. Natl. Acad. Sci. USA. 2007;104:9313–9318. doi: 10.1073/pnas.0700923104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lough J, Barron M, Brogley M, Sugi Y, Bolender DL, Zhu X. Combined BMP-2 and FGF-4, but neither factor alone, induces cardiogenesis in non-precardiac embryonic mesoderm. Dev. Biol. 1996;178:198–202. doi: 10.1006/dbio.1996.0211. [DOI] [PubMed] [Google Scholar]
- Lund E, Güttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- Lyons I, Parsons LM, Hartley L, Li R, Andrews JE, Robb L, Harvey RP. Myogenic and morphogenetic defects in the heart tubes of murine embryos lacking the homeo box gene Nkx2-5. Genes Dev. 1995;9:1654–1666. doi: 10.1101/gad.9.13.1654. [DOI] [PubMed] [Google Scholar]
- Maiti M, Lee H-C, Liu Y. QIP, a putative exonuclease, interacts with the Neurospora Argonaute protein and facilitates conversion of duplex siRNA into single strands. Genes Dev. 2007;21:590–600. doi: 10.1101/gad.1497607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matkovich SJ, Wang W, Tu Y, Eschenbacher WH, Dorn LE, Condorelli G, Diwan A, Nerbonne JM, Dorn GW. MicroRNA-133a protects against myocardial fibrosis and modulates electrical repolarization without affecting hypertrophy in pressure-overloaded adult hearts. Circ. Res. 2010;106:166–175. doi: 10.1161/CIRCRESAHA.109.202176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matranga C, Tomari Y, Shin C, Bartel DP, Zamore PD. Passengerstrand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell. 2005;123:607–620. doi: 10.1016/j.cell.2005.08.044. [DOI] [PubMed] [Google Scholar]
- Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell. 2004;15:185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Michlewski G, Caceres JF. Antagonistic role of hnRNP A1 and KSRP in the regulation of let-7a biogenesis. Nat. Struct. Mol. Biol. 2010;17:1011–1018. doi: 10.1038/nsmb.1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michlewski G, Guil S, Semple CA, Cãceres JF. Posttranscriptional regulation of miRNAs harboring conserved terminal loops. Mol. Cell. 2008;32:383–393. doi: 10.1016/j.molcel.2008.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michlewski G, Guil S, Cáceres JF, Großhans H. Regulation of MicroRNAs, Regulation of MicroRNAs. Springer; New York: 2010. Stimulation of pri-miR-18a processing by hnRNP A1; pp. 28–35. [Google Scholar]
- Miyasaka KY, Kida YS, Banjo T, Ueki Y, Nagayama K, Matsumoto T, Sato M, Ogura T. Heartbeat regulates cardiogenesis by suppressing retinoic acid signaling via expression of miR-143. Mech. Dev. 2011;128:18–28. doi: 10.1016/j.mod.2010.09.002. [DOI] [PubMed] [Google Scholar]
- Miyoshi K, Tsukumo H, Nagami T, Siomi H, Siomi MC. Slicer function of Drosophila Argonautes and its involvement in RISC formation. Genes Dev. 2005;19:2837–2848. doi: 10.1101/gad.1370605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molkentin JD, Lu JR, Antos CL, Markham B, Richardson J, Robbins J, Grant SR, Olson EN. A calcineurin-dependent transcriptional pathway for cardiac hypertrophy. Cell. 1998;93:215–228. doi: 10.1016/s0092-8674(00)81573-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton SU, Scherz PJ, Cordes KR, Ivey KN, Stainier DYR, Srivastava D. MicroRNA-138 modulates cardiac patterning during embryonic development. Proc. Natl. Acad. Sci. USA. 2008;105:17830–17835. doi: 10.1073/pnas.0804673105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman CS, Krieg PA. Tinman-related genes expressed during heart development in Xenopus. Dev. Genet. 1998;22:230–238. doi: 10.1002/(SICI)1520-6408(1998)22:3<230::AID-DVG5>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Nishii K, Shibata Y. Mode and determination of the initial contraction stage in the mouse embryo heart. Anat. Embryol. 2006;211:95–100. doi: 10.1007/s00429-005-0065-x. [DOI] [PubMed] [Google Scholar]
- Olson EN. Gene regulatory networks in the evolution and development of the heart. Science. 2006;313:1922–1927. doi: 10.1126/science.1132292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JY, Li W, Zheng D, Zhai P, Zhao Y, Matsuda T, Vatner SF, Sadoshima J, Tian B. Comparative analysis of mRNA isoform expression in cardiac hypertrophy and development reveals multiple post-transcriptional regulatory modules. PLoS One. 2011;6:e22391. doi: 10.1371/journal.pone.0022391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B, Hayward DC, Ball EE, Degnan B, Muller P, Spring J, Srinivasan A, et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:86–89. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- Patrick DM, Montgomery RL, Qi X, Obad S, Kauppinen S, Hill JA, van Rooij E, Olson EN. Stress-dependent cardiac remodeling occurs in the absence of microRNA-21 in mice. J. Clin. Invest. 2010;120:3912–3916. doi: 10.1172/JCI43604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plageman TF, Jr., Yutzey KE. T-box genes and heart development: Putting the “T” in heart. Dev. Dyn. 2005;232:11–20. doi: 10.1002/dvdy.20201. [DOI] [PubMed] [Google Scholar]
- Porrello ER, Johnson BA, Aurora AB, Simpson E, Nam Y-J, Matkovich SJ, Dorn GW, Van Rooij E, Olson EN. MiR-15 family regulates postnatal mitotic arrest of cardiomyocytes. Circ. Res. 2011;109:670–679. doi: 10.1161/CIRCRESAHA.111.248880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt AJ, MacRae IJ. The RNA-induced silencing complex: A versatile gene-silencing machine. J. Biol. Chem. 2009;284:17897–17901. doi: 10.1074/jbc.R900012200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian L, Wythe JD, Liu J, Cartry J, Vogler G, Mohapatra B, Otway RT, Huang Y, King IN, Maillet M, Zheng Y, Crawley T, et al. Tinman/Nkx2-5 acts via miR-1 and upstream of Cdc42 to regulate heart function across species. J. Cell Biol. 2011;193:1181–1196. doi: 10.1083/jcb.201006114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Que Q, Jorgensen RA. Homology-based control of gene expression patterns in transgenic petunia flowers. Dev. Genet. 1998;22:100–109. doi: 10.1002/(SICI)1520-6408(1998)22:1<100::AID-DVG10>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- Rand TA, Petersen S, Du F, Wang X. Argonaute2 cleaves the anti-guide strand of siRNA during RISC activation. Cell. 2005;123:621–629. doi: 10.1016/j.cell.2005.10.020. [DOI] [PubMed] [Google Scholar]
- Rao PK, Kumar RM, Farkhondeh M, Baskerville S, Lodish HF. Myogenic factors that regulate expression of muscle-specific microRNAs. Proc. Natl. Acad. Sci. USA. 2006;103:8721–8726. doi: 10.1073/pnas.0602831103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- Ren X-P, Wu J, Wang X, Sartor MA, Qian J, Jones K, Nicolaou P, Pritchard TJ, Fan G-C. MicroRNA-320 is involved in the regulation of cardiac ischemia/reperfusion injury by targeting heat-shock protein 20. Circulation. 2009;119:2357–2366. doi: 10.1161/CIRCULATIONAHA.108.814145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby JG, Jan CH, Bartel DP. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007;448:83–86. doi: 10.1038/nature05983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruvkun G, Giusto J. The Caenorhabditis elegans heterochronic gene lin-14 encodes a nuclear protein that forms a temporal developmental switch. Nature. 1989;338:313–319. doi: 10.1038/338313a0. [DOI] [PubMed] [Google Scholar]
- Saga Y, Miyagawa-Tomita S, Takagi A, Kitajima S, Miyazaki J, Inoue T. MesP1 is expressed in the heart precursor cells and required for the formation of a single heart tube. Development. 1999;126:3437–3447. doi: 10.1242/dev.126.15.3437. [DOI] [PubMed] [Google Scholar]
- Saxena A, Tabin CJ. miRNA-processing enzyme Dicer is necessary for cardiac outflow tract alignment and chamber septation. Proc. Natl. Acad. Sci. USA. 2011;107:87–91. doi: 10.1073/pnas.0912870107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayed D, Abdellatif M. MicroRNAs in development and disease. Physiol. Rev. 2011;91:827–887. doi: 10.1152/physrev.00006.2010. [DOI] [PubMed] [Google Scholar]
- Sayed D, Hong C, Chen I-Y, Lypowy J, Abdellatif M. MicroRNAs play an essential role in the development of cardiac hypertrophy. Circ. Res. 2007;100:416–424. doi: 10.1161/01.RES.0000257913.42552.23. [DOI] [PubMed] [Google Scholar]
- Shan Z-X, Lin Q-X, Deng C-Y, Zhu J-N, Mai L-P, Liu J-L, Fu Y-H, Liu X-Y, Li Y-X, Zhang Y-Y, Lin S-G, Yu X-Y. miR-1/miR-206 regulate Hsp60 expression contributing to glucose-mediated apoptosis in cardiomyocytes. FEBS Lett. 2010;584:3592–3600. doi: 10.1016/j.febslet.2010.07.027. [DOI] [PubMed] [Google Scholar]
- Sluijter JPG, van Mil A, van Vliet P, Metz CHG, Liu J, Doevendans PA, Goumans M-J. MicroRNA-1 and -499 regulate differentiation and proliferation in human-derived cardiomyocyte progenitor cells. Arterioscler. Thromb. Vasc. Biol. 2011;30:859–868. doi: 10.1161/ATVBAHA.109.197434. [DOI] [PubMed] [Google Scholar]
- Sokol NS, Ambros V. Mesodermally expressed Drosophila microRNA-1 is regulated by Twist and is required in muscles during larval growth. Genes Dev. 2005;19:2343–2354. doi: 10.1101/gad.1356105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava D. Making or breaking the heart: From lineage determination to morphogenesis. Cell. 2006;126:1037–1048. doi: 10.1016/j.cell.2006.09.003. [DOI] [PubMed] [Google Scholar]
- Srivastava D, Thomas T, Lin Q, Kirby ML, Brown D, Olson EN. Regulation of cardiac mesodermal and neural crest development by the bHLH transcription factor, dHAND. Nat. Genet. 1997;16:154–160. doi: 10.1038/ng0697-154. [DOI] [PubMed] [Google Scholar]
- Sun Q, Zhang Y, Yang G, Chen X, Zhang Y, Cao G, Wang J, Sun Y, Zhang P, Fan M, Shao N, Yang X. Transforming growth factor-b-regulated miR-24 promotes skeletal muscle differentiation. Nucleic Acids Res. 2008;36:2690–2699. doi: 10.1093/nar/gkn032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Synnergren J, Améen C, Lindahl A, Olsson B, Sartipy P. Expression of microRNAs and their target mRNAs in human stem cell-derived cardiomyocyte clusters and in heart tissue. Physiol. Genomics. 2011;43:581–594. doi: 10.1152/physiolgenomics.00074.2010. [DOI] [PubMed] [Google Scholar]
- Tabara H, Sarkissian M, Kelly WG, Fleenor J, Grishok A, Timmons L, Fire A, Mello CC. The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell. 1999;99:123–132. doi: 10.1016/s0092-8674(00)81644-x. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Chen Z, Bartunkova S, Yamasaki N, Izumo S. The cardiac homeobox gene Csx/Nkx2.5 lies genetically upstream of multiple genes essential for heart development. Development. 1999;126:1269–1280. doi: 10.1242/dev.126.6.1269. [DOI] [PubMed] [Google Scholar]
- Tatsuguchi M, Seok HY, Callis TE, Thomson JM, Chen JF, Newman M, Rojas M, Hammond SM, Wang DZ. Expression of microRNAs is dynamically regulated during cardiomyocyte hypertrophy. J. Mol. Cell. Cardiol. 2007;42:1137–1141. doi: 10.1016/j.yjmcc.2007.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thum T, Galuppo P, Wolf C, Fiedler J, Kneitz S, van Laake LW, Doevendans PA, Mummery CL, Borlak J, Haverich A. MicroRNAs in the human heart: A clue to fetal gene reprogramming in heart failure. Circulation. 2007;116:258–267. doi: 10.1161/CIRCULATIONAHA.107.687947. [DOI] [PubMed] [Google Scholar]
- Thum T, Gross C, Fiedler J, Fischer T, Kissler S, Bussen M, Galuppo P, Just S, Rottbauer W, Frantz S, Castoldi M, Soutschek J, et al. MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature. 2008;456:980–984. doi: 10.1038/nature07511. [DOI] [PubMed] [Google Scholar]
- Tzahor E. Wnt/beta-catenin signaling and cardiogenesis: Timing does matter. Dev. Cell. 2007;13:10–13. doi: 10.1016/j.devcel.2007.06.006. [DOI] [PubMed] [Google Scholar]
- van de Schans VAM, Smits JFM, Blankesteijn WM. The Wnt/frizzled pathway in cardiovascular development and disease: Friend or foe? Eur. J. Pharmacol. 2008;585:338–345. doi: 10.1016/j.ejphar.2008.02.093. [DOI] [PubMed] [Google Scholar]
- Van Mierop LH, Gessner IH. The morphologic development of the sinoatrial node in the mouse. Am. J. Cardiol. 1970;25:204–212. doi: 10.1016/0002-9149(70)90580-1. [DOI] [PubMed] [Google Scholar]
- van Rooij E, Sutherland LB, Qi X, Richardson JA, Hill J, Olson EN. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science. 2007;316:575–579. doi: 10.1126/science.1139089. [DOI] [PubMed] [Google Scholar]
- van Rooij E, Quiat D, Johnson BA, Sutherland LB, Qi X, Richardson JA, Kelm RJ, Olson EN. A family of microRNAs encoded by myosin genes governs myosin expression and muscle performance. Dev. Cell. 2009;17:662–673. doi: 10.1016/j.devcel.2009.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, Erkeland SJ, Newman J, Bronson RT, Crowley D, Stone JR, Jaenisch R, Sharp PA, et al. Targeted deletion reveals essential and overlapping functions of the miR-17rv92 family of miRNA clusters. Cell. 2008;132:875–886. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Chang PS, Wang Z, Sutherland L, Richardson JA, Small E, Krieg PA, Olson EN. Activation of cardiac gene expression by myocardin, a transcriptional cofactor for serum response factor. Cell. 2001;105:851–862. doi: 10.1016/s0092-8674(01)00404-4. [DOI] [PubMed] [Google Scholar]
- Wang Y, Medvid R, Melton C, Jaenisch R, Blelloch R. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat. Genet. 2007;39:380–385. doi: 10.1038/ng1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Long B, Zhou J, Li P-F. miR-9 and NFATc3 regulate myocardin in cardiac hypertrophy. J. Biol. Chem. 2010;285:11903–11912. doi: 10.1074/jbc.M109.098004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Song Y, Zhang Y, Xiao H, Sun Q, Hou N, Guo S, Wang Y, Fan K, Zhan D, Zha L, Cao Y, et al. Cardiomyocyte overexpression of miR-27b induces cardiac hypertrophy and dysfunction in mice. Cell Res. 2011 Aug 16; doi: 10.1038/cr.2011.132. 2011. doi:10.1038/cr.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss A, Leinwand LA. The mammalian myosin heavy chain gene family. Annu. Rev. Cell Dev. Biol. 1996;12:417–439. doi: 10.1146/annurev.cellbio.12.1.417. [DOI] [PubMed] [Google Scholar]
- Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- Williams BR. PKR: A sentinel kinase for cellular stress. Oncogene. 1999;18:6112–6120. doi: 10.1038/sj.onc.1203127. [DOI] [PubMed] [Google Scholar]
- Wu H, Xu H, Miraglia LJ, Crooke ST. Human RNase III Is a 160-kDa protein involved in preribosomal RNA processing. J. Biol. Chem. 2000;275:36957–36965. doi: 10.1074/jbc.M005494200. [DOI] [PubMed] [Google Scholar]
- Xie C, Huang H, Sun X, Guo Y, Hamblin M, Ritchie RP, Garcia-Barrio MT, Zhang J, Chen YE. MicroRNA-1 regulates smooth muscle cell differentiation by repressing Kruppel-like factor 4. Stem Cells Dev. 2011;20:205–210. doi: 10.1089/scd.2010.0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing W, Zhang TC, Cao D, Wang Z, Antos CL, Li S, Wang Y, Olson EN, Wang DZ. Myocardin induces cardiomyocyte hypertrophy. Circ. Res. 2006;98:1089–1097. doi: 10.1161/01.RES.0000218781.23144.3e. [DOI] [PubMed] [Google Scholar]
- Yan KS, Yan S, Farooq A, Han A, Zeng L, Zhou M-M. Structure and conserved RNA binding of the PAZ domain. Nature. 2003;426:469–474. doi: 10.1038/nature02129. [DOI] [PubMed] [Google Scholar]
- Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Samal E, Srivastava D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature. 2005;436:214–220. doi: 10.1038/nature03817. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Ransom JF, Li A, Vedantham V, Von M, Muth AN, Tsuchihashi T, McManus MT, Schwartz RJ, Srivastava D. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2. Cell. 2007;129:303–317. doi: 10.1016/j.cell.2007.03.030. [DOI] [PubMed] [Google Scholar]