SUMMARY
Dietary fructose is primarily metabolized in the liver. Here we demonstrated that hepatocellular carcinoma (HCC) cells compared to normal hepatocytes largely reduce fructose metabolism rate and reactive oxygen species level, resulting from c-Myc–dependent and heterogeneous nuclear ribonucleoprotein (hnRNP) H1 and H2-mediadited switch from high-activity fructokinase (KHK)-C to low-activity KHK-A isoform expression. Importantly, KHK-A acts as a protein kinase, phosphorylating and activating phosphoribosyl pyrophosphate synthetase 1 (PRPS1) to promote the pentose phosphate pathway-dependent de novo nucleic acid synthesis and HCC formation. Furthermore, c-Myc, hnRNPH1/2, and KHK-A expression levels and PRPS1 T225 phosphorylation levels correlate with each other in HCC specimens and are associated with poor prognosis for HCC. These findings reveal a pivotal mechanism underlying the distinct fructose metabolism between HCC cells and normal hepatocytes and highlight the instrumental role of KHK-A protein kinase activity in promoting de novo nucleic acid synthesis and HCC development.
Keywords: KHK-A, KHK-C, PRPS1, hnRNPH1/2, c-Myc, phosphorylation, glycolysis, fructose, nucleotide acid synthesis, hepatocellular carcinomagenesis
Dietary fructose, derived from fruits and vegetables and present in added sugars such as sucrose and high-fructose corn syrup, is metabolized primarily in liver and epidemiologically linked with obesity and metabolic syndrome1–3. Fructose is metabolized by fructokinase (also known as ketohexokinase; KHK). KHK uses adenosine triphosphate (ATP) as a phosphate donor and phosphorylates fructose to produce fructose 1-phosphate (F1P). F1P is subsequently metabolized by aldolase B and triokinase to dihydroxyacetone phosphate and glyceraldehyde-3-phosphate, which enter the latter stages of glycolysis. Fructose catabolism initiated by KHK therefore bypasses important glycolytic regulatory steps in glycolysis that generate fructose 1,6-bisphosphate via the action of the energy-sensitive enzyme phosphofructokinase (PFK), resulting in greater lipogenesis than that resulting from glycolysis-regulated lipid production. In addition, fructose catabolism bypasses the glucose-6-phosphate and fructose-6-phosphate-derived pentose phosphate pathway (PPP), which produces ribose-5-phosphate (R5P) for de novo synthesis of nucleotides and nucleic acids (Supplementary Fig. 1)3, 4. Although glucose and fructose catabolism in a normal liver have been well studied, it is unknown whether these sugars are distinctly metabolized to promote development of hepatocellular carcinoma (HCC), one of the most aggressive cancers with a median survival time shorter than 1 year5–7.
Regulation of mutually exclusive splicing of the adjacent exons 3C and 3A of the KHK gene by an unknown mechanism leads to alternative expression of KHK-C and KHK-A isoform, respectively8, 9. KHK-C, which is expressed primarily in the liver, has much greater affinity and a lower Michaelis constant (Km) for fructose than does KHK-A10, 11. KHK-C rapidly metabolizes fructose to F1P, resulting in transient depletion of intracellular phosphate and ATP10. Thus, KHK-C is considered the primary enzyme involved in fructose metabolism. In contrast, KHK-A is expressed at low levels in a wide range of tissues, and its physiological substrate remains unknown10. The physiological significance of these two isoforms is demonstrated by the distinct effects of KHK-C and KHK-A on fructose-induced metabolic syndrome and heart disease observed in mice4, 12–14.
Tumor cells predominantly produce energy via a high rate of glycolysis followed by lactic acid fermentation in the cytosol, even in the presence of ample oxygen. This tumor-specific Warburg effect promotes tumor progression15, 16. The enhanced production of glycolytic intermediates thereby can be used for synthesis of cellular building blocks (nucleotides, amino acids, and lipids) to meet the demands of cell proliferation. Phosphoribosyl pyrophosphate (PRPP) synthetases (PRPSs) are enzymes that catalyze the first and rate-limiting reaction for nucleotide synthesis, producing PRPP from R5P by transferring the β,γ-diphosphoryl moiety of ATP to the C1-hydroxy group of R5P (Supplementary Fig. 1)17, 18. PRPP is then used for synthesis of purine and pyrimidine nucleotides, the pyridine nucleotide cofactors NAD and NADP, and the amino acids histidine (His) and tryptophan19. Human PRPSs have three isoforms that share very high sequence identity: PRPS1 and PRPS2 genes are expressed in a wide range of tissues, whereas PRPS3 is expressed specifically in the testis. PRPSs require Mg2+, sulfate (SO4−), and phosphate for enzymatic activity. Despite having a 94% amino acid sequence identity, PRPS1 and PRPS2 differ significantly in their regulation. PRPS1 is inhibited by the nucleotide biosynthesis products ADP, AMP, and GDP, while PRPS2 is largely resistant to this feedback inhibition20, 21. Mutations of PRPS1, which reduced feedback inhibition of purine biosynthesis, were identified in relapsed childhood B cell acute lymphoblastic leukemia (ALL)22, whereas PRPS2 and enhanced PRPS2 expression were shown to be crucial for lymphoma and melanoma cell survival23–25. However, the mechanism through which the aberrant posttranslational regulation of PRPS contributes to tumor development is unknown.
In this report, we demonstrated that HCC cells reduce the fructose metabolism rate, which is mediated by heterogeneous nuclear ribonucleoprotein (hnRNP) H1 and H2-dependent alternative splicing of the KHK gene by exon 3C exclusion and exon 3A inclusion, resulting in a switch from KHK-C to KHK-A expression. KHK-A acts as a protein kinase to phosphorylate and activate PRPS1 by prevention of inhibitory nucleotide binding and facilitation of ATP binding, leading to enhanced nucleic acid synthesis for tumorigenesis.
hnRNPH1/2 expression switches KHK-C expression to KHK-A expression in HCC cells
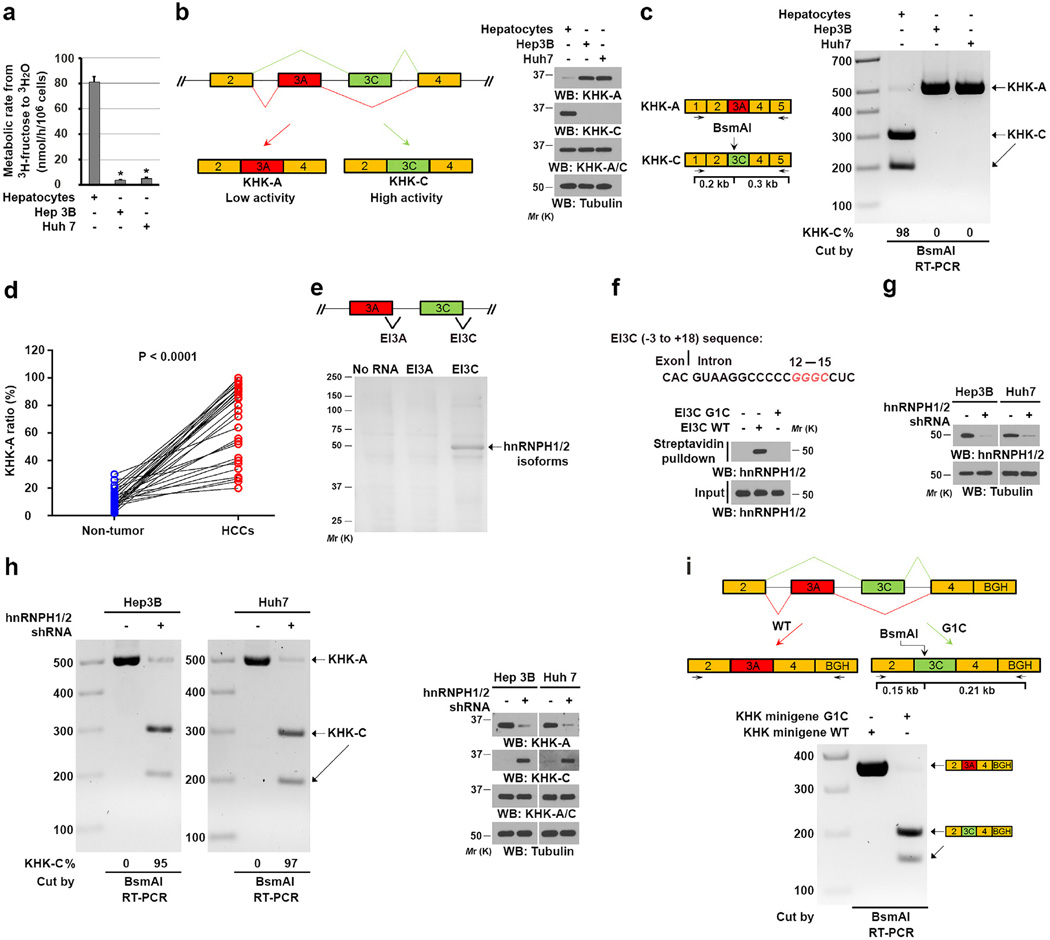
To determine whether normal hepatocytes and HCC cells differ in their fructose consumption, we cultured Hep3B and Huh7 HCC cells and normal hepatocytes in the presence of D-[5-3H]-fructose tracer. Tumor cells had much dramatically reduced rates of fructose metabolism compared to the normal hepatocytes (Fig. 1a). Immunoblot analysis with KHK isoform-specific antibodies revealed that Hep3B and Huh7 cells exclusively expressed the KHK-A isoform, while KHK-C was predominantly expressed in normal hepatocytes (Fig. 1b). These results suggested an altered splicing of KHK pre-mRNA in HCC cells, which promotes expression of low-activity KHK-A rather than active KHK-C contributing to reduced fructose metabolism in HCC cells. Consistent with a mechanism of aberrant splicing, a quantitative RT-PCR analysis followed by exon 3C-specific digestion by the restriction enzyme BsmAI demonstrated that the KHK-A mRNA isoform was predominantly expressed in HCC cells, whereas normal hepatocytes primarily expressed KHK-C (Fig. 1c). Consistent results were obtained by analyses of 30 paired human HCC and matched adjacent non-tumor tissue samples (Fig. 1d and Supplementary Fig. 2a). These results implicated an aberrant splicing change associated with hepatocellular transformation.
Figure 1. hnRNPH1/2 expression switches KHK-C expression to KHK-A expression in HCC cells.
(a) The indicated cells were incubated with 10 µCi of 5-3H-fructose (1 µM). The fructose metabolic rate was measured by monitoring the conversion of D-[5-3H]-fructose to 3H2O. The data represent the mean ± s.d. from n = 3 independent experiments. A two-tailed Student’s t test was used. * represents P < 0.01 between the hepatocellular carcinoma cells and the hepatocytes.
(b) Schematic diagram of splicing of KHK (left panel). Immunoblot analyses were performed using the indicated antibodies (right panel).
(c) Schematic diagram of the restriction site of BsmAI in exon 3C of KHK (left panel). Total RNA was extracted from the indicated cells. The reverse transcription-polymerase chain reaction (RT-PCR) products were digested by BsmAI and resolved on an agarose gel (right panel).
(d) Total RNA was extracted from human HCC samples containing tumor and non-tumor tissue. The RT-PCR products were digested by BsmAI and resolved on agarose gels (Supplementary Fig. 2a). The KHK-A:KHK-C ratios in HCC and matched non-tumor liver tissue samples obtained from n = 30 patients were calculated. A paired two-tailed Student’s t test was used. P < 0.0001.
(e) The indicated streptavidin bead-immobilized biotin-labeled RNA probes (60 nucleotides) spanning the EI3A and EI3C splice sites (top panel) were incubated with Hep3B cell lysates. The associated proteins were separated using SDS-PAGE and stained with Coomassie Brilliant Blue (bottom panel). The indicated protein band was excised for mass spectrometric analysis and identified as hnRNPH1/2 (peptide hits, 66; P < 0.0001). P values were calculated using a paired Student t-test.
(f) The putative hnRNPH1/2 binding residues (red) in the EI3C sequence (exon 3C and its 3' adjacent intron) are shown (top panel). Streptavidin bead-immobilized WT or mutated EI3C RNA was incubated with Hep3B cell lysates, which was followed by immunoblot analyses with the indicated antibodies (bottom panel).
(g and h) The indicated cells did or did not express hnRNPH1/2 shRNA (g). The RT-PCR products were digested by BsmAI (h left panel). Immunoblot analyses were performed using indicated antibodies (g, h right panel).
(i) Schematic diagram of the KHK minigene constructs with or without G1C mutation (top panel), which were expressed in Hep3B cells. The RT-PCR products were digested by BsmAI and resolved on an agarose gel (bottom panel). BGH, bovine growth hormone polyadenylation signal.
In b, c, and e–i, data represent 1 out of 3 experiments. Unprocessed original scans of blots are shown in Supplementary Fig. 7.
To identify the mechanism underlying KHK splicing, we synthesized biotin-labeled EI3A and EI3C RNA nucleotides spanning boundary regions of exons 3A-3' and 3C-3' with their adjacent introns. A pull-down assay of Hep3B cell lysates with these immobilized RNA nucleotides demonstrated that the EI3C but not the EI3A nucleotide was associated with major proteins with molecular masses of approximately 50,000 (Mr 50K) identified as hnRNPH1 and hnRNPH2 through mass spectrometry (Fig. 1e). HnRNPH1 and hnRNPH2 are RNA-binding proteins with well-established roles as sequence-specific repressors of splicing, and they recognize the GGGC/GGGA consensus sequence26. Analysis of the EI3C sequence revealed a GGGC motif adjacent to exon 3C (Fig. 1f top panel). Mutation of G1 in this motif into C abolished the binding of hnRNPH1/2 to EI3C (Fig. 1f bottom panel). In addition, immunoprecipitation of hnRNPH1/2 and RT-PCR analysis of hnRNPH1/2-associated RNA demonstrated association of hnRNPH1/2 with an RNA fragment transcribed from exon 3C and the GGGC motif (Supplementary Fig. 2b). These results strongly suggested that hnRNPH1 and hnRNPH2 bind to the GGGC motif in the intron immediately adjacent to exon 3C-3'.
HnRNPH1 and hnRNPH2 are more than 98% homologous (Supplementary Fig. 2c), suggesting that they have functional redundancy. We depleted both hnRNPH1 and hnRNPH2 by expressing a short hairpin RNA (shRNA) against hnRNPH1/2 in Hep3B and Huh7 cells (Fig. 1g). RT-PCR analyses demonstrated that hnRNPH1/2 depletion resulted in a switch of expression of KHK from BsmAI-resistant KHK-A to BsmAI-sensitive KHK-C (Fig. 1h left panel), and immunoblot analyses detected altered corresponding protein expression (Fig. 1h right panel). These results indicated that binding of hnRNPH1/2 to the exon 3C-3'/intron region leads to exclusion of exon 3C and inclusion of exon 3A for KHK-A expression.
The 5' and 3' splicing sites of genes are recognized by U1-type small nuclear ribonucleic protein (U1 snRNP) and U2AF, respectively. HnRNPH1 and hnRNPH2 repress a splicing site and promote alternative 5' splicing selection26, 27. We next synthesized pre-mRNAs by transcribing a plasmid DNA carrying the T7 promoter and a truncated adenovirus 2 major late (AdML) transcription unit (exons 1 and 2), in which we replaced exon 1 and the intron with KHK exon 3C and its downstream intron with or without the G1C mutation (Supplementary Fig. 2d left panel). Incubation of these pre-mRNAs with nuclear extracts of Hep3B cells with or without hnRNPH1/2 depletion and PCR analysis demonstrated that G1C mutation and hnRNPH1/2 depletion promoted the splicing and removal of the intron (Supplementary Fig. 2d right panel). These results further supported that hnRNPH1/2 binding blocks the recognition of the exon 3C 5' splice site (immediately downstream from exon 3C), which prevents recognition of exon 3C and favors the inclusion of exon 3A.
To further validate this finding in vivo, we transfected Hep3B cells with a plasmid containing a minigene including the gene fragment of KHK from exon 2 to exon 4 with or without G1C mutation (Fig. 1i top panel). RT-PCR analyses demonstrated that wild-type (WT) KHK minigene expression resulted in inclusion of exon 3A rather than exon 3C, whereas KHK minigene expression with G1C mutation led to inclusion of exon 3C but not exon 3A in mRNA (Fig. 1i bottom panel). These results provided additional evidence supporting the finding that binding of hnRNPH1/2 to the GGGC motif in the intron adjacent to KHK exon 3C leads to exclusion of exon 3C and inclusion of exon 3A.
c-Myc–enhanced hnRNPH1/2 expression switches KHK-C expression to KHK-A expression in HCC cells
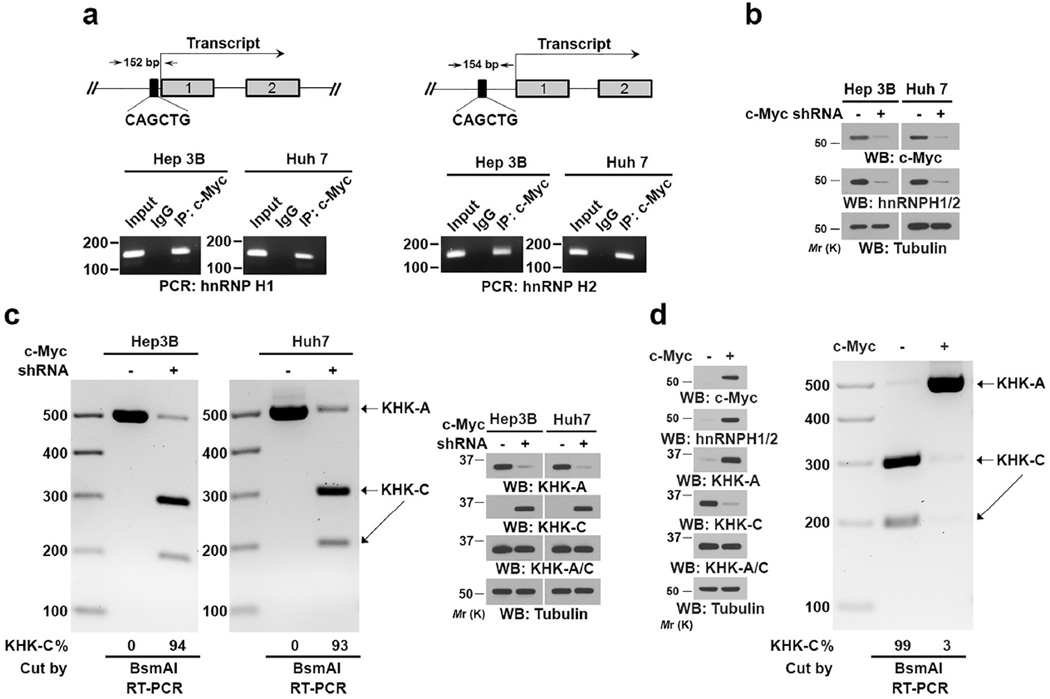
It was reported that c-Myc, which is commonly overexpressed in primary and advanced HCC, regulates the expression of hnRNPH1 by binding to a noncanonical E-box region in the promoters of the hnRNPH1 gene28, 29. Consistent with this observation, chromatin immunoprecipitation (ChIP) showed that c-Myc bound to the promoter regions of both hnRNPH1 and hnRNPH2 in Hep3B and Huh7 cells (Fig. 2a) and shRNA-mediated depletion of c-Myc reduced expression of hnRNPH1/2 (Fig. 2b). Importantly, c-Myc depletion resulted in a concomitant decrease in KHK-A mRNA and increase in KHK-C mRNA expression with corresponding protein expression (Fig. 2c). In addition, overexpression of c-Myc in normal hepatocytes resulted in increased expression of hnRNPH1/2 and KHK-A and decreased KHK-C expression (Fig. 2d). These results indicated that c-Myc upregulates hnRNPH1/2 expression, which in turn promotes a switch from KHK-C to KHK-A expression.
Figure 2. C-Myc–enhanced hnRNPH1/2 expression switches from KHK-C expression to KHK-A expression in HCC cells.
(a) ChIP analyses of the indicated cells were performed with an anti-c-Myc antibody and indicated primers for a promoter region of hnRNPH1 (left panel) or hnRNPH2 (right panel).
(b) The indicated HCC cells with or without c-Myc depletion were analyzed using an immunoblot assay with the indicated antibodies.
(c) The indicated cells did or did not have c-Myc shRNA expression. The RT-PCR products were digested by BsmAI and resolved on an agarose gel (left panel). Immunoblot analysis was performed using the indicated antibodies (right panel). The band intensity was quantified using the Image Lab software program.
(d) Normal hepatocytes were infected with or without lentiviruses expressing c-Myc. Immunoblot assay was performed with the indicated antibodies (left panel). In addition, total RNA was extracted, and the RT-PCR products were digested by BsmAI and resolved on an agarose gel (right panel).
Data represent 1 out of 3 experiments. Unprocessed original scans of blots are shown in Supplementary Fig. 7.
KHK-A phosphorylates PRPS1 at Thr225
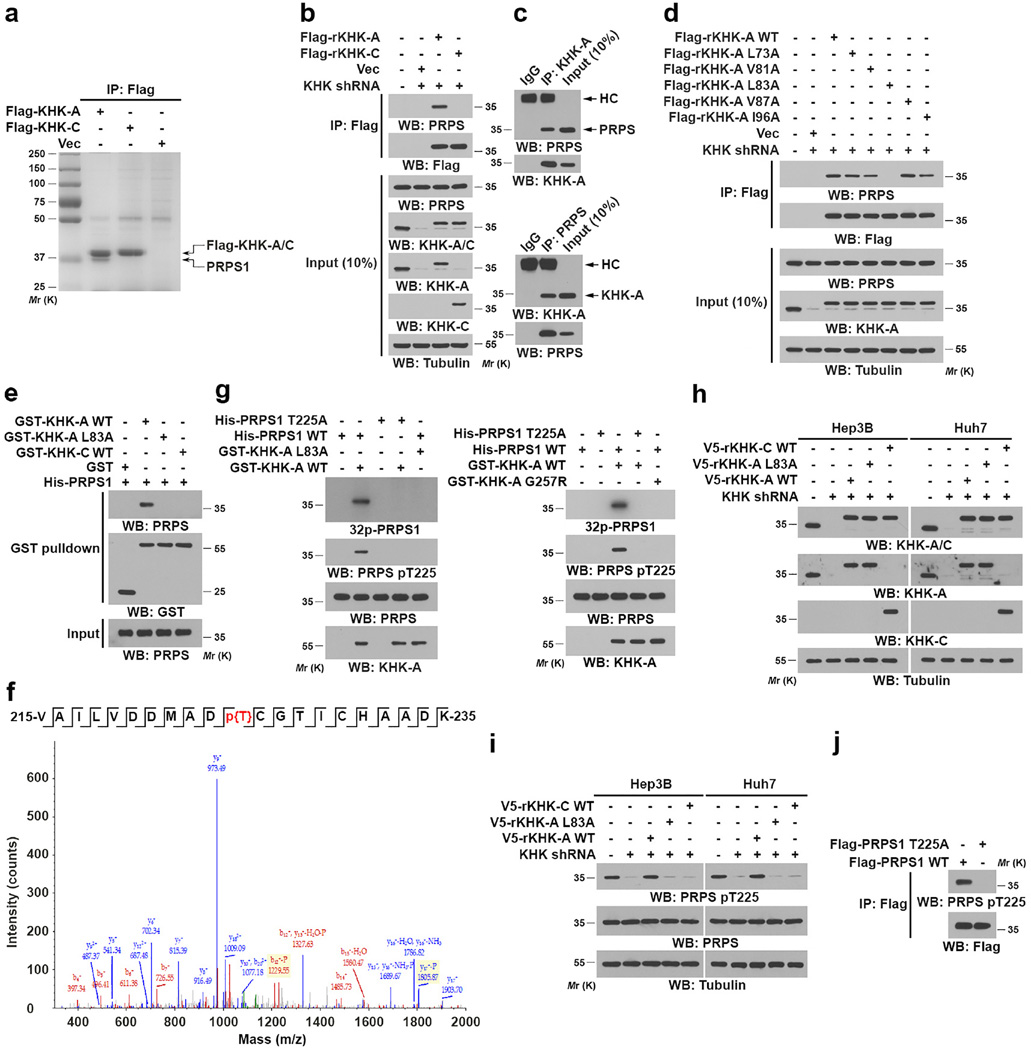
KHK-A poorly phosphorylates fructose, and its physiological substrate remains unidentified10. To this end, we immunoprecipitated KHK-C and KHK-A from Hep3B cells followed by Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Fig. 3a shows that a protein (about 35-kDa) was specifically associated with KHK-A but not KHK-C. Mass spectrometric analysis identified this protein as PRPS1. Immunoprecipitation analyses revealed that Flag-tagged KHK-A but not KHK-C binds to endogenous PRPS1 (Fig. 3b). We also detected binding of endogenous KHK-A to endogenous PRPS1 (Fig. 3c). In addition, purified GST-KHK-A directly bound to purified His-PRPS1, but not PRPS2 (Supplementary Fig. 3a), and the expression of PRPS2 was barely detectable in normal hepatocytes and HCC cells (Supplementary Fig. 3b). To uncover the interaction site of KHK-A, we mutated hydrophobic surface residues among 45 amino acids coded by exon 3A. Only mutation of L83A was found to abolish the interaction between KHK-A and PRPS1 detected via co-immunoprecipitation assay in vivo (Fig. 3d), which was further validated using a glutathione S-transferase (GST) pull-down assay in vitro (Fig. 3e). In contrast, KHK-C, which shares 37.8% protein sequence identity comparing the exon 3A- and 3C-coding regions, failed to directly bind to PRPS1 (Fig. 3e). These results indicated that L83 of KHK-A interacts directly with PRPS1.
Figure 3. KHK-A phosphorylates PRPS1 at Thr225.
(a) Immunoprecipitates of Flag-KHK-A and Flag-KHK-C from Hep3B cell lysate with corresponding antibodies were separated via SDS-PAGE and stained with Coomassie Brilliant Blue. The indicated protein band was excised for mass spectrometric analysis and identified as PRPS1 (peptide hits, 121).
(b) Lysates of Hep3B cells expressing Flag-KHK-C or Flag-KHK-A protein were immunoprecipitated with an anti-Flag antibody.
(c) Hep3B cell lysates were immunoprecipitated with an anti-KHK-A antibody (top panel) or an anti-PRPS1 antibody (bottom panel). HC, heavy chain of IgG.
(d) Lysates of Hep3B cells expressing the indicated Flag-KHK-A proteins were immunoprecipitated with an anti-Flag antibody.
(e) GST pull-down analysis was performed by mixing purified immobilized WT GST-KHK-A, GST-KHK-A L83A, or GST-KHK-C on glutathione agarose beads with purified His-PRPS1.
(f) In vitro phosphorylation analysis was performed by mixing bacterially purified GST-KHK-A and His-PRPS1 in the presence of ATP. Mass spectrometric analysis of a tryptic fragment of PRPS1 at mass-to-charge ratio (m/z) 1150.479 (mass error, −6 ppm) matched the doubly charged peptide 215-VAILVDDMADTCGTICHAADK-235 suggesting that T225 was phosphorylated. The Mascot score was 43, and the expectation value was 0.0062.
(g) In vitro phosphorylation analysis, SDS-PAGE and autoradiography were performed by mixing purified WT KHK-A, KHK-A L83A (left panel), or KHK-A G257R (right panel) with purified WT PRPS1 or PRPS1 T225A in the presence of [γ32P]-ATP.
(h and i) Hep3B or Huh7 cells with or without expression of KHK shRNA were reconstituted with or without expression of the indicated KHK proteins.
(j) Lysates of Hep3B cells expressing the indicated Flag-PRPS1 proteins were immunoprecipitated with an anti-Flag antibody.
In a–e and g–j, data represent 1 out of 3 experiments. Unprocessed original scans of blots are shown in Supplementary Fig. 7.
KHK-A, which has greatly reduced affinity for fructose (Km: 9.41 mM) and does not efficiently use ATP to phosphorylate fructose10, interacts with PRPS1 with much high affinity (Km: 0.228 µM) (Supplementary Fig. 3c). Of note, the intracellular concentration of PRPS-1 is about 1.8 µM (Supplementary Fig. 3d), which is much higher than the Km of KHK-A for PRPS1. This finding led us to determine whether KHK-A can function as a protein kinase to phosphorylate PRPS1 rather than fructose in HCC cells. We performed an in vitro phosphorylation assay by mixing purified recombinant KHK-A and PRPS1 (Supplementary Fig. 3e) in the presence of ATP. Liquid chromatography-coupled Orbitrap tandem mass spectrometric analysis demonstrated that KHK-A phosphorylates PRPS1 at T225 (Fig. 3f). The phosphorylation of PRPS1, but not PRPS2 (Supplementary Figs. 3f, 3g), by WT KHK-A, but not KHK-A L83A (Fig. 3g, left panel), was also detected using autoradiography with [γ32P]-ATP and a specific anti-phospho-PRPS1 T225 antibody. In addition, we created a KHK-A ATP-binding site mutation by converting the conserved Gly257 in the ATP-γ-phosphate binding motif (GAGD) to Arg30. KHK-A G257R, which was still able to bind to PRPS1 (Supplementary Fig. 3h), lost its binding to ATP (Supplementary Fig. 3i) and failed to phosphorylate PRPS1 at T225 (Fig. 3g, right panel). As expected, KHK-C, did not phosphorylate PRPS1 (Supplementary Fig. 3j). These results indicated that the interaction between KHK-A and PRPS1 and the ATP-binding ability of KHK-A are required for PRPS1 phosphorylation.
We next expressed KHK shRNA and reconstituted the expression of RNA interference-resistant (r) WT KHK-A, rKHK-A L83A, or WT rKHK-C in both Hep3B and Huh7 cells (Fig. 3h). Immunoblot analysis demonstrated that KHK depletion abrogated PRPS1 T225 phosphorylation, which was rescued by expression of WT rKHK-A but not WT rKHK-C or rKHK-A L83A (Fig. 3i). In line with this finding, PRPS1 T225A expressed in Hep3B cells was resistant to phosphorylation (Fig. 3j). Notably, PRPS1 T225 phosphorylation was not affected by increased concentrations of fructose (up to 20 mM) in cultured medium, further demonstrating the high affinity of KHK-A for PRPS1 (Supplementary Fig. 3k). These results indicated that KHK-A phosphorylates PRPS1 at T225.
KHK-A-mediated PRPS1 phosphorylation activates PRPS1
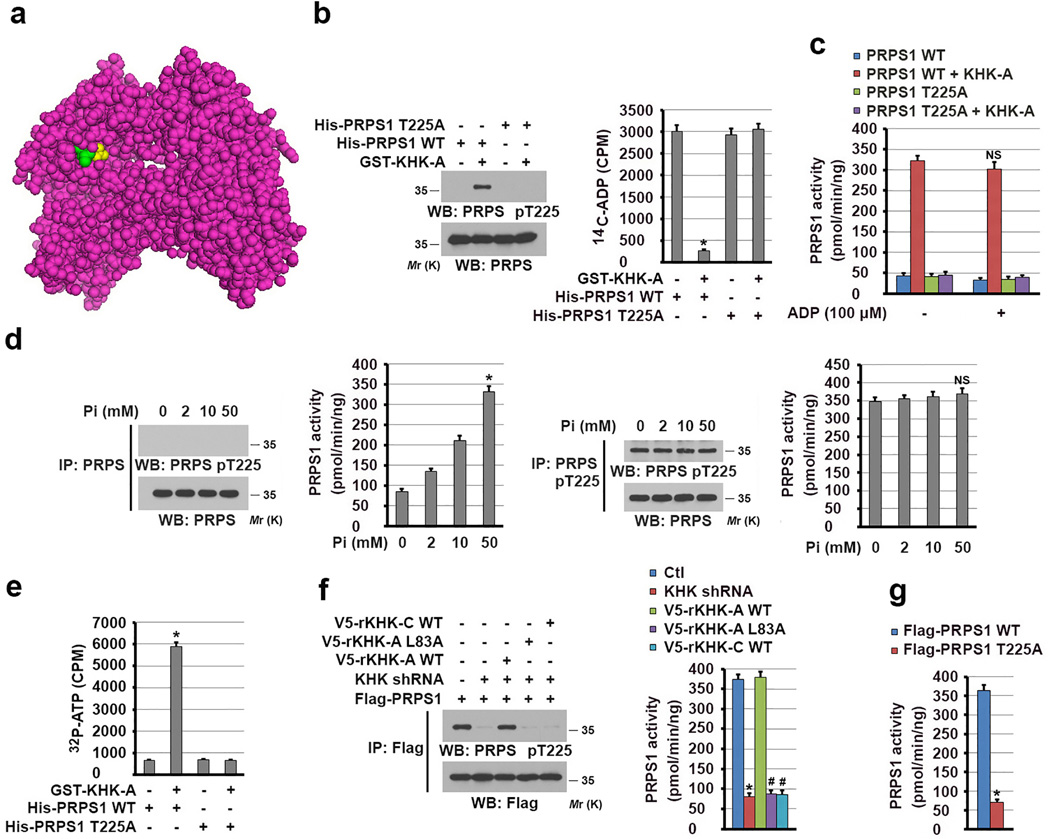
The activators (sulfate and phosphate) and inhibitors (ADP, AMP, IMP, and GMP) of PRPS1 compete for the same allosteric regulatory site for regulation of PRPS1 activity in vitro21, 31, 32. Our structure analysis of PRPS1 revealed that T225 of PRPS1 is in the allosteric site (Fig. 4a) and that T225 phosphorylation may physically block the binding of ADP to PRPS1. To examine this possibility, we mixed purified His-PRPS1 proteins with GST-KHK-A to phosphorylate PRPS1 in vitro (Fig. 4b left panel) followed by incubation of 14C-ADP. Phosphorylation of WT PRPS1, but not PRPS1 T225A mutant, largely blocked the binding of ADP to PRPS1 (Fig. 4b right panel). These results strongly suggested that PRPS1 T225 phosphorylation sterically blocked the access of the allosteric site of PRPS1 to ADP. In line with this finding, we revealed that PRPS1 T225 phosphorylation significantly increased PRPS1 activity (Fig. 4c). In addition, inclusion of ADP at a high dose, which reduced phosphate-dependent PRPS1 activity (Supplementary Fig. 4a), had a limited effect on the activity of phosphorylated PRPS1 (Fig. 4c). Similar results were also observed using AMP, IMP, and GMP, which did not affect the phosphorylated PRPS activity, but inhibited phosphate-dependent PRPS1 activity to a variable extent (Supplementary Figs. 4b, 4c).
Figure 4. KHK-A-mediated PRPS1 phosphorylation activates PRPS1.
(a) The structure of PRPS1 (PDB code 2HCR), exhibiting binding of sulfate (yellow) to the allosteric site of PRPS1. T225 is shown in green color.
(b) In vitro phosphorylation analysis was performed by mixing bacterially purified GST-KHK-A and the indicated immobilized His-PRPS1 proteins in the presence of ATP. 14C-ADP was then added to the mixture, and His-PRPS1–bound 14C-ADP was measured (right panel). The data represent the mean ± s.d. from n = 3 independent experiments. Immunoblot analysis with the indicated antibodies demonstrated the status of PRPS1 T225 phosphorylation (left panel). A two-tailed Student’s t test was used. * represents P < 0.01 between the phosphorylated and non-phosphorylated PRPS1.
(c) The activity of bacterially purified WT His-PRPS1 or the PRPS1 T225A mutant with or without phosphorylation by GST-KHK-A was measured in the presence or absence of ADP. The data represent the mean ± SD from n = 3 independent experiments. A two-tailed Student’s t test was used. NS represents not significant difference between the indicated sample and the counterpart in the absence of ADP.
(d) The activity of immunoprecipitated PRPS1 from normal hepatocytes (two left-hand panels) and Hep3B cells (two right-hand panels) was measured in the presence of indicated concentrations of phosphate. The data represent the mean ± s.d. from n = 3 independent experiments. A two-tailed Student’s t test was used. * represents P < 0.01; NS represents not significant difference between the indicated sample and the counterpart in the absence of Pi.
(e) [γ32P]-ATP was mixed with WT PRPS1 or the PRPS1 T225A mutant with or without phosphorylation by GST-KHK-A. His-PRPS1–bound [γ32P]-ATP was measured. The data represent the mean ± s.d. from n = 3 independent experiments. A two-tailed Student’s t test was used. * represents P < 0.01 between the phosphorylated and non-phosphorylated PRPS1.
(f) The activity of Flag-PRPS1 immunoprecipitated from Hep3B cells expressing KHK shRNA with or without reconstituted expression of the indicated rKHK proteins was measured (right panel). The data represent the mean ± s.d. from n = 3 independent experiments. Immunoblot analysis with the indicated antibodies demonstrated the status of PRPS1 T225 phosphorylation (left panel). A two-tailed Student’s t test was used. * represents P < 0.01 between the cells with or without KHK depletion. # represents P < 0.01 between the KHK-depleted cells with reconstituted expression of WT rKHK-A and the KHK-depleted cells with reconstituted expression of rKHK-A L83A and WT rKHK-C.
(g) The activity of the indicated Flag-PRPS1 proteins immunoprecipitated from Hep3B cells was measured. The data represent the mean ± s.d. from n = 3 independent experiments. A two-tailed Student’s t test was used. * represents P < 0.01 between the cells expressing WT PRPS1 and PRPS1 T225A.
In b, d, and f, data represent 1 out of 3 experiments. Unprocessed original scans of blots are shown in Supplementary Fig. 7.
To further understand how PRPS1 is distinctly regulated in normal hepatocytes and HCC cells, we immunoprecipitated PRPS1 and found that PRPS1 from normal hepatocyte was activated by phosphate in a dosage-dependent manner (Fig. 4d left panel). In contrast, phosphorylated PRPS1, which had high activity and was more than 90% of total PRPS1 in Hep3B cells (Supplementary Fig. 4d), was resistant to be regulated by phosphate (Fig. 3d right panel). These results strongly suggested that PRPS1 activity in normal cells is dynamically regulated and can be activated by phosphate. Activated PRPS1 increases production of the nucleotides, such as ADP AMP, IMP and GMP, which will inhibit PRPS1 in a feedback manner. In contrast, phosphorylation of PRPS1 by KHK-A in HCC cells abrogates the feedback inhibition by these nucleotides and results in a constantly high level of PRPS1 activity.
Given that regulation of the allosteric site of PRPS1 affects the binding of ATP to PRPS1 and subsequent its activation21, 31, 32, we performed an in vitro ATP-binding assay and demonstrated that phosphorylation of PRPS1 T225 greatly enhanced the binding of [γ32P]-ATP to WT PRPS1 compared to non-phosphorylated PRPS1 and the PRPS1 T225A mutant (Fig. 4e). Taken together, these results strongly suggested that PRPS1 T225 phosphorylation results in formation of an active, open conformation for binding of ATP and activation of PRPS1.
To determine the role of KHK-A in regulation of PRPS1 in HCC cells, we immunoprecipitated Flag-PRPS1 from Hep3B cells and found that KHK depletion reduced the T225 phosphorylation (Fig. 4f left panel) and activity (Fig. 4f right panel) of PRPS1, and this inhibitory effect was rescued by reconstituted expression of WT rKHK-A but not rKHK-A L83A or WT rKHK-C (Fig. 4f). In addition, immunoprecipitated Flag-PRPS1 T225A, which was resistant to phosphorylation (Fig. 4e), had much lower activity than WT PRPS1 (Fig. 4g). These results indicated that KHK-A phosphorylates and activates PRPS1.
KHK-A promotes de novo nucleic acid synthesis and reduces ROS level
To determine the role of KHK-A in PRPS1-dependent nucleic acid synthesis, we incubated Hep3B and Huh7 cells with D-[6-14C]-glucose and revealed that hnRNPH1/2 depletion largely reduced glucose-derived production of both 14C-RNA (Fig. 5a) and the RNA precursor inosine monophosphate (IMP) (Supplementary Fig. 5a). In addition, reconstituted expression of rKHK-A L83A and WT rKHK-C failed to rescue this effect (Fig. 5b and Supplementary Fig. 5b).
Figure 5. KHK-A promotes de novo nucleic acid synthesis and reduces ROS production.
(a) Hep3B and Huh7 cells with or without expression of hnRNPH1/2 shRNA were labeled with D-[6-14C]-glucose for 6 h. Influx of glucose into RNA extracted from the cells was measured. The data represent the mean ± s.d. from n = 3 independent experiments. A two-tailed Student’s t test was used. * represents P < 0.01 between the cells with or without hnRNPH1/2 depletion.
(b) Hep3B and Huh7 cells expressing KHK shRNA with or without reconstituted expression of the indicated rKHK proteins were labeled with D-[6-14C]-glucose for 6 h. Influx of glucose into RNA extracted from the cells was measured. The data represent the mean ± SD from n = 3 independent experiments. A two-tailed Student’s t test was used. * represents P < 0.01 between the cells with or without KHK depletion. # represents P < 0.01 between the KHK-depleted cells with reconstituted expression of WT rKHK-A and the KHK-depleted cells with reconstituted expression of rKHK-A L83A and WT rKHK-C.
(c) Hep3B and Huh7 cells expressing PRPS1 shRNA were reconstituted the expression of the indicated rPRPS1 proteins. Immunoblot analysis was performed with the indicated antibodies (left panel). These cells were labeled with D-[6-14C]-glucose for 6 h. Influx of glucose into RNA extracted from the cells was measured. The data represent the mean ± s.d. from n = 3 independent experiments (right panel). A two-tailed Student’s t test was used. * represents P < 0.01 between the cells with or without PRPS1 depletion. # represents P < 0.01 between the PRPS1-depleted cells with reconstituted expression of WT rPRPS1 and rPRPS1 T225A.
(d) Normal hepatocytes (left panel) and Hep3B cells (right panel) were labeled with D-[6-14C]-glucose for 1 h in the presence or absence of Pi (50 mM) for the indicated periods of time. Influx of glucose into RNA extracted from the cells was measured. The data represent the mean ± s.d. from n = 3 independent experiments.
(e) Hep3B cells with reconstituted expression of rKHK-C or rKHK-A were cultured in the presence or absence of the indicated dosage of fructose for 2 h. Levels of cellular ROS were measured. The data represent the mean ± s.d. from n = 3 independent experiments. A two-tailed Student’s t test was used. * represents P < 0.01 between the indicated cells and the counterparts without fructose treatment. In c, data represent 1 out of 3 experiments. Unprocessed original scans of blots are shown in Supplementary Fig. 7.
PRPS1 and PRPS2 have more than 94% homology and share conserved T225. In line with the finding that the HCC cell lines primarily express PRPS1 with barely detectable PRPS2 mRNA (Supplementary Fig. 3b), PRPS1 depletion in Hep3B and Huh7 cells by expressing a PRPS1 shRNA greatly reduced the total expression of PRPS proteins, as detected by immunoblot analyses with an antibody recognizing both PRPS1 and PRPS2 (Fig. 5c left panel). Notably, PRPS1 depletion inhibited production of RNA and IMP, which was rescued by reconstituted expression of WT rPRPS1 but not rPRPS1 T225A (Fig. 5c right panel and Supplementary Fig. 5c). These results indicated that KHK-A–dependent PRPS1 phosphorylation promote glucose-derived de novo nucleotide and nucleic acid synthesis.
PRPS1 T225 phosphorylation abrogates the allosteric feedback inhibition by ADP and other nucleotides. As expected, reconstituted expression of active PRPS1 A190T mutant, which lost the binding of the nucleotide inhibitors at the allosteric sites and was identified in relapsed childhood B cell ALL22, had a comparable IMP production compared to expression of WT PRPS1 (Supplementary Fig. 5c), which is predominately phosphorylated in Hep3B cells. In line with the finding that PRPS1 activity in normal cells but not HCC cells is regulated by phosphate (Fig. 4d), increased phosphate concentration in the culture medium enhanced de novo RNA production in normal hepatocytes, but not in Hep3B cells (Fig. 5d). However, this increased RNA production in normal hepatocytes was transient and downregulated after prolonged incubation with phosphate, suggesting a negative feedback inhibition of PRPS1 by newly synthetized nucleotides. These results indicate that PRPS1-dependent nucleic acid synthesis in normal hepatocytes is dynamically regulated; in contrast, cancer cells acquire constantly high level of PRPS1 activity for nucleic acid production.
Fructose metabolism reduces intracellular phosphate and ATP levels and generates many folds more reactive oxygen species (ROS) than glucose metabolism, which are disadvantages to cell growth and survival10, 33–35. The switch from high-activity KHK-C expression to low-activity KHK-A expression may prevent fructose metabolism-induced these disadvantages. To test this hypothesis, we cultured Hep3B cells with reconstituted expression of KHK-C or KHK-A in the presence or absence of fructose. We found that fructose significantly reduced the levels of ATP (Supplementary Fig. 5d) and phosphate (Supplementary Fig. 5e) and increased ROS levels in Hep3B cells expressing KHK-C but not in Hep3B cells expressing KHK-A (Fig. 5e). Taken together, these results indicated that aberrant splicing of KHK transcript in HCC cells results in enhanced nucleic acid synthesis and inhibited the effect of fructose metabolism on the regulation of phosphate, ATP, and ROS.
KHK-A–mediated PRPS1 phosphorylation promotes tumorigenesis and is associated with HCC pathogenesis
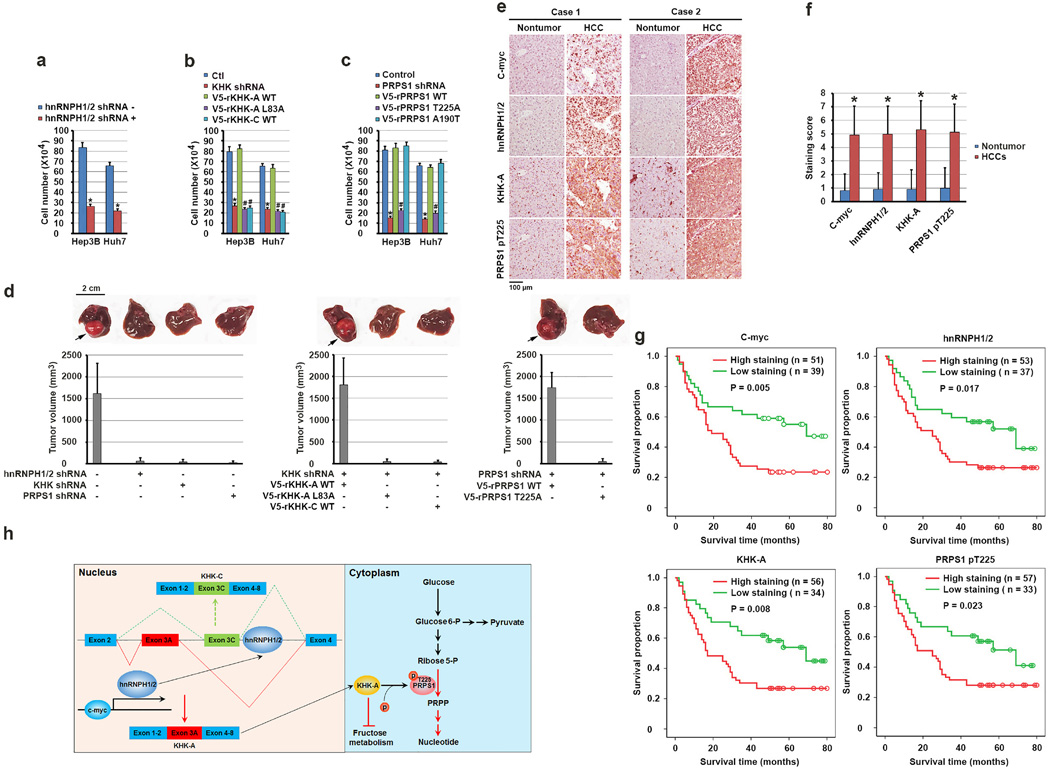
Enhanced de novo nucleic acid synthesis and ATP production and reduced ROS level promote tumor cell proliferation. As expected, hnRNPH1/2 depletion greatly inhibited the proliferation of Hep3B and Huh7 cells (Fig. 6a). In addition, compared to the HCC cells with reconstituted expression of rKHK-A, rPRPS1, and rPRPS1 A190T, the cells with KHK depletion or reconstituted rKHK-A L83A or WT rKHK-C expression (Fig. 6b) and the cells with PRPS1 depletion or reconstituted expression of rPRPS1 T225A expression (Fig. 6c) exhibited much reduced proliferation without obviously altered apoptosis (Supplementary Fig. 6a). In addition, increased fructose concentration in medium inhibited the proliferation of Hep3B with reconstituted expression of rKHK-C, but not rKHK-A, and this inhibition was partially alleviated by treatment with reducing reagent n-acetylcysteine (NAC) (Supplementary Fig. 6b).
Figure 6. KHK-A–dependent phosphorylation of PRPS1 promotes hepatocellular tumorigenesis and is associated with the pathogenesis of HCC.
(a, b, c) A total of 1 × 105 Hep3B and Huh7 cells with or without expression of hnRNPH1/2 shRNA (a), KHK shRNA (b), PRPS1 shRNA (c) and with or without reconstituted expression of the indicated WT or mutated proteins were plated for 3 days. The cells were then collected and counted. The data represent the mean ± s.d. from n = 3 independent experiments. A two-tailed Student’s t test was used. * represents P < 0.01 between the cells with or without hnRNPH1/2 depletion (a), the cells with or without KHK depletion (b), the cells with or without PRPS1 depletion (c). # represents P < 0.01 between the KHK-depleted cells with reconstituted expression of WT rKHK-A and the KHK-depleted cells with reconstituted expression of rKHK-A L83A and WT rKHK-C (b), and between PRPS1-depleted cells with reconstituted expression of WT rPRPS1 and rPRPS1 T225A (c).
(d) A total of 1 × 106 Huh7 cells with or without expression of hnRNPH1/2 shRNA, KHK shRNA, or PRPS1 shRNA and with or without reconstituted expression of their WT counterparts and the indicated mutants were intrahepatically injected into athymic nude mice (n = 5 mice/group). The mice were sacrificed and examined for tumor growth 28 days after injection. The arrows point to the tumors.
(e) Immunohistochemical staining of 90 human HCC and matched non-tumor tissue samples for the indicated antibodies was performed. Representative photos of stains in two cases are shown.
(f) c-Myc, hnRNPH1/2, and KHK-A expression levels and PRPS1 T225 phosphorylation levels in HCC and matched non-tumor liver samples were obtained from n = 90 patients. The indicated staining scores in HCC and matched non-tumor liver samples were compared using a paired Student t-test. *p < 0.001.
(g) Kaplan-Meier plots of the overall survival rates in the patients (n = 90) with HCC in the groups with high (staining score, 5–8.0) and low (staining score, 0–4.0) expression of c-Myc, hnRNPH1/2, and KHK-A and phosphorylation of PRPS1 T225. The P values were calculated using the log-rank test.
(h) A mechanism of KHK-A–promoted de novo nucleotide synthesis. c-Myc enhances the expression of hnRNPH1 and hnRNPH2, which bind to the exon 3C-3'/intron region of KHK, leading to alternative splicing of KHK pre-mRNA for KHK-A expression and reduced fructose metabolism. KHK-A phosphorylates and activates PRPS1, resulting in enhanced production of nucleotide and nucleotide acid derived from glycolysis and promotion of hepatocellular tumorigenesis.
We next intra-hepatically injected Huh7 cells depleted of hnRNPH1/2, KHK, or PRPS1 into nude mice. Depletion of these proteins markedly inhibited hepatocellular tumor growth (Fig. 6d). Notably, reconstituted expression of WT rKHK-A or WT rPRPS1, but not rKHK-A L83A, WT rKHK-C, or rPRPS1 T225A, rescued tumor growth (Fig. 6d). Immunoblot analysis demonstrated strong phosphorylation of PRPS1 T225 and high expression of proliferating cell nuclear antigen (PCNA) in the tissue of tumors derived from Hep3B cells with reconstituted expression of WT rKHK-A or rPRPS1 compared to tumor tissues with reconstituted expression of rKHK-A L83A, WT rKHK-C, or rPRPS1 T225A (Supplementary Fig. 6c). These results indicated that hnRNPH1/2-regulated KHK-A expression and KHK-A–mediated PRPS1 phosphorylation promote HCC development.
To determine the clinical relevance of our findings, we performed immunohistochemical (IHC) analysis of 90 primary HCC samples and matched non-tumor liver tissue samples with validated antibodies recognizing c-Myc, hnRNPH1/2, KHK-A, and phosphorylated PRPS1 T225 proteins (Supplementary Fig. 6d). We revealed that the levels of c-Myc, hnRNPH1/2, and KHK-A expression and PRPS1 T225 phosphorylation were markedly higher in HCC samples than in matched non-tumor tissue samples (Fig.4e and 4f) and correlated with each other in HCC samples (Supplementary Fig. 6e).
We accessed the survival durations in the 90 HCC patients, all of whom underwent standard surgical resection of HCC. Fig. 6g shows that high levels of c-Myc, hnRNPH1/2, KHK-A, and PRPS1 T225 phosphorylation in HCC samples correlated with poor overall survival durations in the patients and vice versa. In addition, expression levels of these proteins were independent prognostic factors for overall survival (Supplementary Table 1). These results supported a critical role for KHK-A–dependent PRPS1 phosphorylation in the clinical aggressiveness of human HCC.
DISCUSSION
Fructose is a major form of dietary sugar that is catalyzed in liver for lipogenesis1–3. However, compared with normal liver cells, HCC cells have much low levels of fructose metabolism, which may prevent enhanced ROS production and uncontrolled lipid production3, 4, 33. The low fructose metabolism is mediated by hnRNPH1/2 expression induced by c-Myc, whose overexpression is prevalent in HCC cells36.
We demonstrated that hnRNPH1/2 bind to a consensus sequence located immediately adjacent to the exon 3C 5' splice site. Mutation of the consensus site or hnRNPH1/2 depletion results in switching from KHK-A to KHK-C isoform expression. These results indicate that hnRNPH1/2 control the outcome of a mutually exclusive splicing event by binding and repressing the recognition of exon 3C to allow exon 3A inclusion by potentially inducing intronic structures conducive to inclusion of exon 3A and exclusion of exon 3C26, 27.
Tumor cells have enhanced glycolysis for anabolic synthesis. In the present study, we identified a critical mechanism underlying glucose-derived de novo nucleic acid synthesis via KHK-A–mediated phosphorylation and activation of PRPS1 (Fig. 6h). The understanding that an hnRNPH1/2-dependent switch from KHK-C to KHK-A expression and KHK-A–mediated phosphorylation of PRPS1 are required for HCC development and that PRPS1 T225 phosphorylation and hnRNPH1/2 expression correlate with prognosis for HCC may provide a molecular basis for improved diagnosis and treatment of HCC.
In summary, our findings demonstrated that HCC cells have a distinct regulatory mechanism of fructose catabolism that differs from the mechanisms in their normal counterparts. We showed that coordinated regulation of glucose and fructose metabolism in supporting HCC cell proliferation by KHK-A is essential for HCC development. The demonstration that KHK-A functions as a protein kinase highlights the dual roles of KHK-A as both a metabolic enzyme and a protein kinase in cell metabolism and proliferation greatly impacts the understanding of protein enzymes with multiple roles in controlling cellular functions.
Supplementary Material
Acknowledgments
We thank Bih-Fang Pan for technical support and Donald Norwood for critical reading of this manuscript. This work was supported by National Cancer Institute grants 2R01 CA109035 (Z.L.) and 1R0 CA169603 (Z.L.), National Institute of Neurological Disorders and Stroke grant 1R01 NS089754 (Z.L.), MD Anderson Support Grant CA016672, the James S. McDonnell Foundation 21st Century Science Initiative in Brain Cancer Research Award 220020318 (Z.L.), 2P50 CA127001 (Brain Cancer SPORE), a Sister Institution Network Fund from MD Anderson (Z.L; CN.Q.), NIH High-End Instrumentation program grant 1S10OD012304-01 (D.H.); CPRIT Core Facility Grant RP130397 (D.H.); the Odyssey Fellowship from MD Anderson (X.L.); and research grants 81272340 (CN.Q.) and 81472386 (CN.Q.) from the National Natural Science Foundation of China. Z.L. is a Ruby E. Rutherford Distinguished Professor.
Footnotes
The authors declare no competing financial interests.
CONTRIBUTIONS
This study was conceived by Z.L. and X.L.; Z.L. and X.L. designed the study; X.L., X.Q., L.-X.P., Y.J., D.H., Y.Z., Y.X., J.L., and G.C. performed experiments; L.W., X.W., and C.-N.Q. provided reagents and technical assistance; Z.L. and X.L. wrote the paper with comments from all the authors.
REFERENCES
- 1.Havel PJ. Dietary fructose: implications for dysregulation of energy homeostasis and lipid/carbohydrate metabolism. Nutrition reviews. 2005;63:133–157. doi: 10.1301/nr.2005.may.133-157. [DOI] [PubMed] [Google Scholar]
- 2.Tappy L, Le KA. Metabolic effects of fructose and the worldwide increase in obesity. Physiological reviews. 2010;90:23–46. doi: 10.1152/physrev.00019.2009. [DOI] [PubMed] [Google Scholar]
- 3.Lyssiotis CA, Cantley LC. Metabolic syndrome: F stands for fructose and fat. Nature. 2013;502:181–182. doi: 10.1038/502181a. [DOI] [PubMed] [Google Scholar]
- 4.Ishimoto T, et al. Opposing effects of fructokinase C and A isoforms on fructose-induced metabolic syndrome in mice. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:4320–4325. doi: 10.1073/pnas.1119908109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fong ZV, Tanabe KK. The clinical management of hepatocellular carcinoma in the United States, Europe, and Asia: a comprehensive and evidence-based comparison and review. Cancer. 2014;120:2824–2838. doi: 10.1002/cncr.28730. [DOI] [PubMed] [Google Scholar]
- 6.Chan SL, et al. International validation of the Chinese university prognostic index for staging of hepatocellular carcinoma: a joint United Kingdom and Hong Kong study. Chinese journal of cancer. 2014;33:481–491. doi: 10.5732/cjc.014.10133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zuo TT, Zheng RS, Zhang SW, Zeng HM, Chen WQ. Incidence and mortality of liver cancer in China in 2011. Chinese journal of cancer. 2015;34:56. doi: 10.1186/s40880-015-0056-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bonthron DT, Brady N, Donaldson IA, Steinmann B. Molecular basis of essential fructosuria: molecular cloning and mutational analysis of human ketohexokinase (fructokinase) Human molecular genetics. 1994;3:1627–1631. doi: 10.1093/hmg/3.9.1627. [DOI] [PubMed] [Google Scholar]
- 9.Hayward BE, Bonthron DT. Structure and alternative splicing of the ketohexokinase gene. European journal of biochemistry / FEBS. 1998;257:85–91. doi: 10.1046/j.1432-1327.1998.2570085.x. [DOI] [PubMed] [Google Scholar]
- 10.Diggle CP, et al. Ketohexokinase: expression and localization of the principal fructose-metabolizing enzyme. The journal of histochemistry and cytochemistry : official journal of the Histochemistry Society. 2009;57:763–774. doi: 10.1369/jhc.2009.953190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Asipu A, Hayward BE, O'Reilly J, Bonthron DT. Properties of normal and mutant recombinant human ketohexokinases and implications for the pathogenesis of essential fructosuria. Diabetes. 2003;52:2426–2432. doi: 10.2337/diabetes.52.9.2426. [DOI] [PubMed] [Google Scholar]
- 12.Lanaspa MA, et al. Endogenous fructose production and metabolism in the liver contributes to the development of metabolic syndrome. Nature communications. 2013;4:2434. doi: 10.1038/ncomms3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ishimoto T, et al. High-fat and high-sucrose (western) diet induces steatohepatitis that is dependent on fructokinase. Hepatology. 2013;58:1632–1643. doi: 10.1002/hep.26594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mirtschink P, et al. HIF-driven SF3B1 induces KHK-C to enforce fructolysis and heart disease. Nature. 2015;522:444–449. doi: 10.1038/nature14508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang W, et al. ERK1/2-dependent phosphorylation and nuclear translocation of PKM2 promotes the Warburg effect. Nat Cell Biol. 2012;14:1295–1304. doi: 10.1038/ncb2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang W, Lu Z. Nuclear PKM2 regulates the Warburg effect. Cell Cycle. 2013;12:3154–3158. doi: 10.4161/cc.26182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kornberg A, Lieberman I, Simms ES. Enzymatic synthesis and properties of 5-phosphoribosylpyrophosphate. The Journal of biological chemistry. 1955;215:389–402. [PubMed] [Google Scholar]
- 18.Khorana HG, Fernandes JF, Kornberg A. Pyrophosphorylation of ribose 5-phosphate in the enzymatic synthesis of 5-phosphorylribose 1-pyrophosphate. The Journal of biological chemistry. 1958;230:941–948. [PubMed] [Google Scholar]
- 19.Hove-Jensen B. Mutation in the phosphoribosylpyrophosphate synthetase gene (prs) that results in simultaneous requirements for purine and pyrimidine nucleosides, nicotinamide nucleotide, histidine, and tryptophan in Escherichia coli. Journal of bacteriology. 1988;170:1148–1152. doi: 10.1128/jb.170.3.1148-1152.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nosal JM, Switzer RL, Becker MA. Overexpression, purification, and characterization of recombinant human 5-phosphoribosyl-1-pyrophosphate synthetase isozymes I and II. The Journal of biological chemistry. 1993;268:10168–10175. [PubMed] [Google Scholar]
- 21.Li S, Lu Y, Peng B, Ding J. Crystal structure of human phosphoribosylpyrophosphate synthetase 1 reveals a novel allosteric site. The Biochemical journal. 2007;401:39–47. doi: 10.1042/BJ20061066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li B, et al. Negative feedback-defective PRPS1 mutants drive thiopurine resistance in relapsed childhood ALL. Nat Med. 2015;21:563–571. doi: 10.1038/nm.3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cunningham JT, Moreno MV, Lodi A, Ronen SM, Ruggero D. Protein and nucleotide biosynthesis are coupled by a single rate-limiting enzyme, PRPS2, to drive cancer. Cell. 2014;157:1088–1103. doi: 10.1016/j.cell.2014.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mannava S, et al. Direct role of nucleotide metabolism in C-MYC-dependent proliferation of melanoma cells. Cell Cycle. 2008;7:2392–2400. doi: 10.4161/cc.6390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McMahon SB. Control of nucleotide biosynthesis by the MYC oncoprotein. Cell Cycle. 2008;7:2275–2276. [PMC free article] [PubMed] [Google Scholar]
- 26.Martinez-Contreras R, et al. hnRNP proteins and splicing control. Advances in experimental medicine and biology. 2007;623:123–147. doi: 10.1007/978-0-387-77374-2_8. [DOI] [PubMed] [Google Scholar]
- 27.Keller W. The RNA lariat: a new ring to the splicing of mRNA precursors. Cell. 1984;39:423–425. doi: 10.1016/0092-8674(84)90449-5. [DOI] [PubMed] [Google Scholar]
- 28.Rauch J, et al. c-Myc regulates RNA splicing of the A-Raf kinase and its activation of the ERK pathway. Cancer research. 2011;71:4664–4674. doi: 10.1158/0008-5472.CAN-10-4447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zimonjic DB, Popescu NC. Role of DLC1 tumor suppressor gene and MYC oncogene in pathogenesis of human hepatocellular carcinoma: potential prospects for combined targeted therapeutics (review) International journal of oncology. 2012;41:393–406. doi: 10.3892/ijo.2012.1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trinh CH, Asipu A, Bonthron DT, Phillips SE. Structures of alternatively spliced isoforms of human ketohexokinase. Acta Crystallogr D Biol Crystallogr. 2009;65:201–211. doi: 10.1107/S0907444908041115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eriksen TA, Kadziola A, Bentsen AK, Harlow KW, Larsen S. Structural basis for the function of Bacillus subtilis phosphoribosyl-pyrophosphate synthetase. Nature structural biology. 2000;7:303–308. doi: 10.1038/74069. [DOI] [PubMed] [Google Scholar]
- 32.Willemoes M, Hove-Jensen B, Larsen S. Steady state kinetic model for the binding of substrates and allosteric effectors to Escherichia coli phosphoribosyl-diphosphate synthase. The Journal of biological chemistry. 2000;275:35408–35412. doi: 10.1074/jbc.M006346200. [DOI] [PubMed] [Google Scholar]
- 33.Lim JS, Mietus-Snyder M, Valente A, Schwarz JM, Lustig RH. The role of fructose in the pathogenesis of NAFLD and the metabolic syndrome. Nature reviews. Gastroenterology & hepatology. 2010;7:251–264. doi: 10.1038/nrgastro.2010.41. [DOI] [PubMed] [Google Scholar]
- 34.Bose T, Chakraborti AS. Fructose-induced structural and functional modifications of hemoglobin: implication for oxidative stress in diabetes mellitus. Biochimica et biophysica acta. 2008;1780:800–808. doi: 10.1016/j.bbagen.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 35.Bunn HF, Higgins PJ. Reaction of monosaccharides with proteins: possible evolutionary significance. Science. 1981;213:222–224. doi: 10.1126/science.12192669. [DOI] [PubMed] [Google Scholar]
- 36.Wang Y, et al. Prognostic significance of c-myc and AIB1 amplification in hepatocellular carcinoma. A broad survey using high-throughput tissue microarray. Cancer. 2002;95:2346–2352. doi: 10.1002/cncr.10963. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.