Summary
During the past decade, advanced techniques in structural biology have provided atomic level information on the platelet integrin αIIbβ3 activation mechanism that results in it adopting a high-affinity ligand-binding conformation(s). This review focuses on advances in imaging intact αIIbβ3 in a lipid bilayer in the absence of detergent and new structural insights into the changes in the ligand-binding pocket with receptor activation and ligand binding. It concludes with descriptions of novel therapeutic αIIbβ3 antagonists being developed based on an advanced knowledge of the receptor’s structure.
Keywords: electron microscopy, integrin alphaIIbbeta3, platelet, thrombosis, x-ray crystallography
αIIbβ3 structure in a lipid bilayer
αIIbβ3 is the paradigmatic integrin receptor, with studies of its structure and function, including inside-out signaling, ligand binding, and outside-in signaling, leading the way in understanding the biology of the entire family of receptors [1]. It is required for normal hemostasis as demonstrated by the lifelong bleeding diathesis suffered by patients with Glanzmann thrombasthenia, who lack the receptor or receptor function on a genetic basis [1]. It is also a validated drug target because it plays a central, non-redundant role in uncontrolled platelet aggregation leading to thrombosis and ischemic cardiovascular disease [2]. Some genetic variants of αIIbβ3 are immunogenic and lead to the alloimmune disorders neonatal alloimmune thrombocytopenia and post-transfusion purpura [3]. This review will focus on recent structural findings related to the conformation of αIIbβ3 in a lipid bilayer and the changes in the ligand-binding pocket with ligand binding, along with implications of the latter for the development of novel αIIbβ3 antagonists for clinical use.
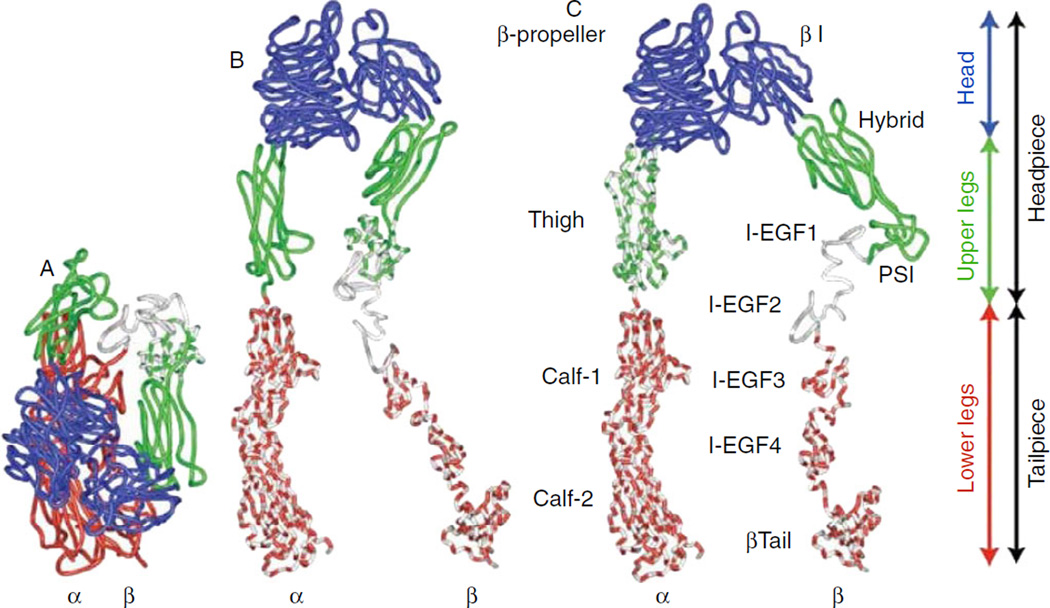
A number of αIIbβ3 structures have been reported, and the results have dramatically improved our understanding of the relationship between structure and function. These include (i) crystal structures of the unliganded αIIbβ3 ectodomain and the liganded αIIbβ3 headpiece [4–8], (ii) NMR structures of the transmembrane and cytoplasmic domains of αIIb and β3 [9–12], (iii) rotary shadowing and/or negative stain 2-dimensional (2D) images of αIIbβ3 in detergent, with or without ligands or mAbs [13–15], (iv) transmission electron cryomicroscopy (cryo-EM) of αIIbβ3 in detergent [16], (v) small-angle neutron scattering (SANS) of αIIbβ3 in detergent [17], (vi) small-angle x-ray scattering (SAXS) of αIIbβ3 in detergent [18], (vii) cryoelectron tomography of αIIbβ3 in liposomes [19], (viii) electron tomography of activated αIIbβ3 in detergent [20], and (ix) negative stain EM of αIIbβ3 in lipid bilayer nanodiscs, with and without the talin head domain [19,21,22]. While each structure has provided very valuable information, the data are not concordant. In particular: (i) the crystal structure of the unliganded receptor does not fit snuggly in the cryo-EM, SAXS, or SANS maps (Fig. 1); (ii) the 3D reconstruction of the cryo-EM maps shows the head domain pointing away from the putative membrane, whereas the reconstructions of the SANS and SAXS images were interpreted as being most consistent with the crystal structure oriented so that the head points toward the membrane; and (iii) the cryo-EM data suggest that the lower legs are bent, whereas the crystal structure of the ectodomain and the SANS and SAXS studies were interpreted as consistent with the straight, parallel, and adjacent lower leg conformations seen in the crystal structure [4].
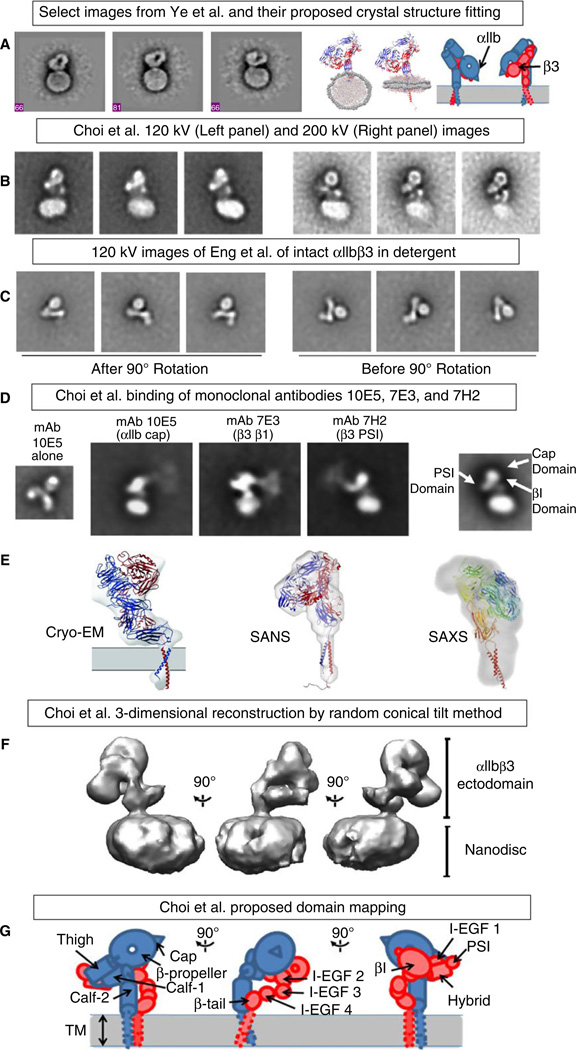
Fig. 1.
(A) Ye et al.’s [19] negative stain EM images of αIIbβ3 nanodiscs and model. (B) Choi et al.’s [22] negative stain EM images at 120 (left) and 200 kV (right). (C) Negative stain EM images of Eng et al. [18] after (left) and before (right) rotating to facilitate comparison with Choi et al.’s images. (D) Choi et al. negative stain EM images of αIIbβ3 nanodiscs reacted with mAbs 10E5 (αIIb cap), 7E3 (β3 βI), and 7H2 (β3 PSI). (E) Cryo-EM [16], small-angle neutron scattering (SANS) [17] and small-angle x-ray scattering (SAXS) [18] maps with fitted crystal structures. (F, G) Choi et al. 3D reconstruction map and model (Reproduced from Refs [16–19] and [22] with permission).
Nanodisc technology enabled studies of intact, purified αIIbβ3 in a lipid bilayer in the absence of detergent, an important intermediate step in understanding the conformation of the receptor in a platelet membrane. Ye et al. [19] performed 2D EM imaging of αIIbβ3 in nanodiscs and found a compact structure for the receptor in the absence of activation (Fig. 1); the addition of the talin head domain resulted in an increase in the percentage of receptors adopting either a fully extended conformation or a slightly less compact shape. They proposed a model in which the head domain points down toward the membrane. Our laboratory built on these pioneering studies and analyzed more than 12 000 pairs of tilted and untilted negative stain images of unactivated full length αIIbβ3 in nanodiscs to develop a 3D reconstruction of the electron density map at 20.5 A resolution (Fig. 1) [22]. We then obtained a first approximation of a 3D molecular model of αIIbβ3 in the nanodisc by molecular docking of the αIIbβ3 ectodomain crystal structure domains into the EM electron density map, using the binding of monoclonal antibodies with known epitopes to guide the process (Fig. 1). The most striking feature of our 2D images and the resulting 3D reconstruction was the orientation of the head domain away from the membrane. In addition, we did not find the leg domains to be straight and parallel; instead, there was a bend between the calf-1 and calf-2 domains of αIIb and a coiled path for the β3 IEGF-2–4 domains, with the latter entering into relatively few contacts with neighboring structures rather than lying in a cleft between the β3 headpiece and the αIIb lower leg. These data suggest an important role for the region that links the distal calf-2 and β-tail domains to their respective TM domains in transmitting the conformational changes in the TM domains associated with inside-out activation. This interpretation is supported by studies demonstrating that alanine mutations of key residues in the β3 I-EGF-4 and β-tail domains produce constitutively active receptors [23]. The 3D reconstruction also suggests that receptor extension requires changes in multiple interdomain articulations rather than a change in a single fulcrum in each subunit. Advances in cryo-EM technology, especially improvements in detectors, should allow for higher resolution images of unliganded and liganded αIIbβ3, providing better domain fitting and insights into how inside-out signaling initiates the conformational changes responsible for ligand binding. The resulting predictions will, however, require independent confirmation through mutagenesis and functional studies.
Ligand binding to the RGD binding pocket
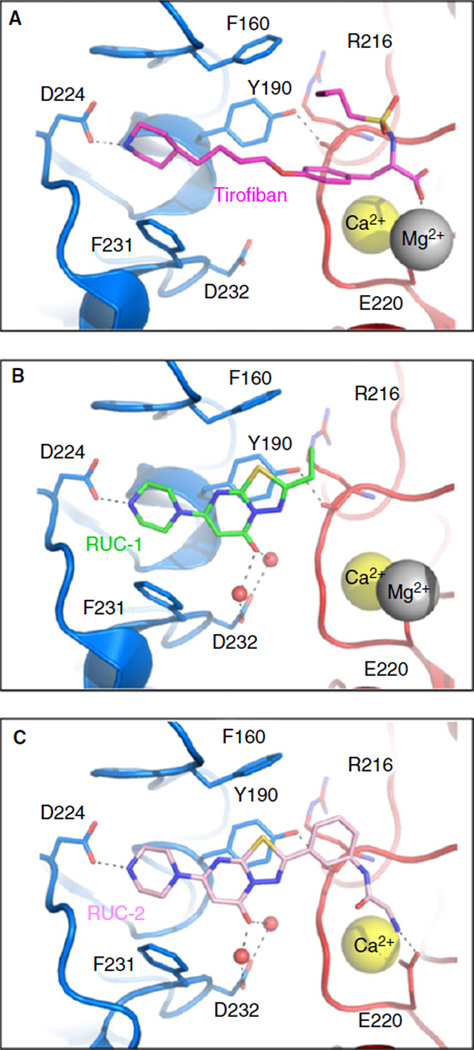
The pioneering crystallographic studies of Xiong et al. [24] on RGD peptide binding to αVβ3 provided the first atomic resolution description of ligand binding to a β3 integrin receptor, but the study was limited because the ligand was soaked into the crystal. Xiao et al. [5] provided the first description of ligand binding to the αIIbβ3 headpiece at the atomic level based on crystal structures, including the mechanisms of binding of the two small molecule αIIbβ3 antagonists in clinical use, eptifibatide and tirofiban (Fig. 2A). The crystal structures of the unliganded αIIbβ3 ectodomain [4], the complex of αIIbβ3 with the fibrinogen γ chain dodecapeptide [7], and intermediate states of αIIbβ3 between the closed, inactive conformation and the open, active conformation [8] provided a wealth of information on the changes that accompany ligand binding. Three metal ions in the β3 I domain play important roles in ligand binding, the MIDAS, SyMBS, and ADMIDAS, and each is discussed below. When crystalized in the presence of both Ca 2+ and Mg 2+, Ca2+ occupies both the SyMBS and ADMIDAS and Mg2+ occupies the MIDAS [6]. The central features of ligand binding are the reorganization of the amino acid residues that coordinate the MIDAS and ADMIDAS metal ions (Fig. 3) and a swing-out motion of the β3 hybrid and PSI domains away from the β3 I domain and the αIIb subunit as a result of a pistonlike displacement of the β3 I domain α7-helix that causes a 62° reorientation between the β3 I and hybrid domains and a 70Å separation between the knees of the αIIb and β3 subunit legs (Fig. 4) [5].
Fig. 2.
Crystal structures of (A) tirofiban, (B) RUC-1, and (C) RUC-2 in complex with αIIbβ3 (PDBs 3NIF, 2VDM, and 3T3M, respectively). RUC-2’s interaction with E220 leads to loss of the MIDAS Mg2+ (silver ball).
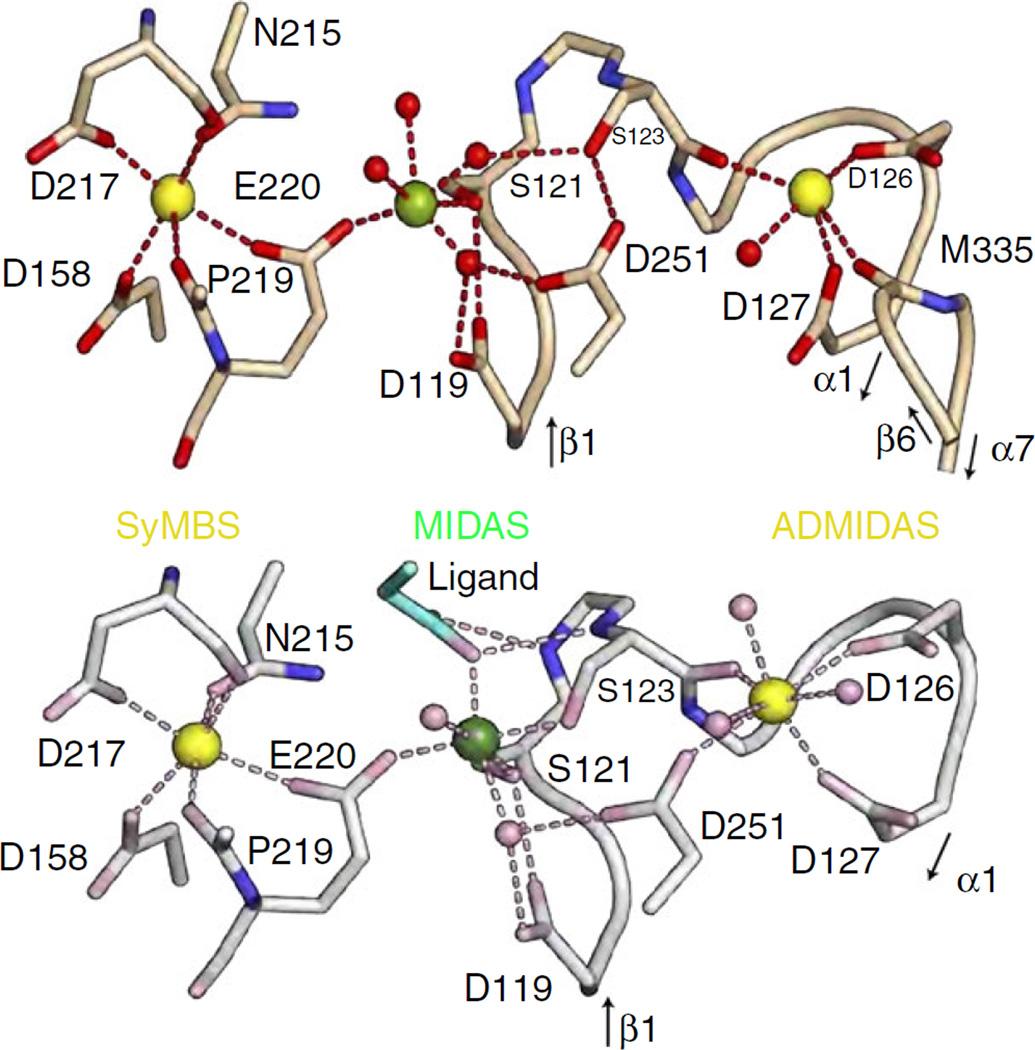
Fig. 3.
Metal ion coordination sites in the β3 βI domain in unliganded-closed αIIbβ3 (upper panel) and liganded-open αIIbβ3 (lower panel) [4]. Ca2+ ions are gold and Mg2+ ions are green. Note that only the ligand Asp carboxyl is depicted and that it interacts with the two backbone nitrogens in the β1-α1 loop and stabilizes a conformation in which the ADMIDAS is closer to the MIDAS, setting in motion a series of events that lead to both swing-out of the β3 hybrid domain and extension of the αIIbβ3 headpiece away from the leg regions (Reproduced from Ref. [4] with permission).
Fig. 4.
αIIbβ3 in its compact, bent conformation (A) and models of its structure after extension (B) and after both extension and swing-out of the β3 hybrid domain (C). (Reproduced from Ref. [5] with permission).
The MIDAS
The events at the MIDAS that accompany ligand binding can be briefly summarized as follows [4,5,7]: (i) The ligand’s positively charged arginine or lysine binds to the negatively charged Asp residue 224 in αIIb. (ii) Although both Mg2+ and Ca2+ can support ligand binding to αIIbβ3, the only crystal structures available have Mg2+ in the MIDAS. The Mg2+ is held relatively weakly in the MIDAS by four water molecules, the S121 hydroxyl oxygen, and one of the two E220 carboxyl oxygens (Fig. 3, top panel), as evidenced by the need for millimolar concentrations of Mg2+ to support ligand binding and platelet aggregation [25]. The weak binding of the Mg2+ to the β3 MIDAS residues allows it to retain more of its electropositivity to capture the ligand through one of the latter’s Asp carboxyl oxygens [26]. (iii) The ligand carboxyl oxygens not only help coordinate the Mg2+, but also interact with the backbone nitrogens of Y122 and S123 in the β1-α1 loop in the β3 I domain and the side-chain of N215, which also participates in coordinating the SyMBS (Fig. 2, bottom panel). These interactions stabilize conformations in which the β1-α1 loop moves closer to the MIDAS and thus trigger the swing-out motion described above. (iv) The αIIbβ3 headpiece undergoes extension away from the legs, either before or after the swing-out motion (Fig. 4).
Factors other than the presence of a ligand carboxyl affect the swing-out process as not all RGD-based antagonists induce major conformational changes [27]. One possible explanation for the differences comes from studies of the binding of fibronectin fragments to αVβ3, in which a high-affinity fragment with a Trp substitution for Ser after the RGD sequence did not induce a major conformational change with binding [28]. The crystal structures of the complexes showed that the Trp interacted with β3 Tyr122 in the β1-α1 loop and thus prevented the motion of the loop toward the MIDAS. In addition, Tyr1446 in the fibronectin fragment now interacted with the ADMIDAS metal ion via a water molecule [28].
The SyMBS
The MIDAS is connected to the synergistic metal ion-binding site (SyMBS [4]; termed ligand-associated metal ion-binding site [LIMBS] in αVβ3) [24], which contains a Ca2+ ion in crystal structures when the receptor is crystalized in the presence of both Ca2+ and Mg2+. Unlike the MIDAS Mg2+, the SyMBS Ca2+ is relatively inaccessible to solvent [29]. It is required for ligand binding to αIIbβ3 [29], but its precise role is still under investigation. Variations in SyMBS coordinating residues among integrin β subunits can alter both ligand binding and the impact of pH on metal ion binding to the SyMBS [30]. In fact, differences in the reported presence or absence of divalent cations in the SyMBS (LIMBS) in crystal structures of αIIbβ3 and αVβ3 most likely reflect technical differences in the pH and divalent cation concentrations used during crystal formation [31]. β3 E220 plays an important role in both the SyMBS and MIDAS as one of its carboxyl oxygens coordinates the Ca2+ in the SyMBS and the other coordinates the Mg2+ in the MIDAS (Fig. 3). The coordination of the SyMBS metal ion by one of the E220 oxygens probably reduces the strength of binding of the other E220 oxygen to the MIDAS metal ion, thus enhancing the MIDAS metal ion’s electropositivity and its ability to bind a ligand’s carboxyl oxygen [26]. The SyMBS and LIMBS residues make contact with the integrin αIIb and αV subunits, respectively, and so differences between the αIIb and αV contacts may contribute to differences in ligand binding between the two receptors despite their sharing the same β3 subunit [32].
The ADMIDAS
In the unliganded crystal structures of αVβ3 and αIIbβ3, the ADMIDAS metal ion is coordinated by the residues from the βb1-α1 loop, the α1 helix, and the β6-α7 loop, including the carbonyl oxygen of S123, carboxyl oxygens of D126 and D127, and the carbonyl oxygen of M335 (Fig. 3) [4,33]. In contrast to the RGD-based ligands that bind to the β3 subunit of αVβ3 and αIIbβ3 by one of the ligand’s carboxyl group oxygens completing the coordination of the metal ion of the MIDAS, the fibrinogen c chain C-terminal dodecapeptide (HHLGGAKQAGDV) binds to αIIbβ3 by a combination of the coordination of the MIDAS metal ion by the Asp carboxyl and the coordination of the ADMIDAS metal ion by the C-terminal Val carboxyl through an intermediate water molecule [7]. The importance of the latter interaction is supported by the observation that amidation of the C-terminal Val carboxyl reduces the affinity of the peptide for the receptor by more than 80% [34].
Ligand binding leads to reorganization of the ADMI-DAS, with the ADMIDAS metal ion, the β1-α1 loop, and the α1 helix moving closer to the MIDAS; the coordination of the ADMIDAS ion by the M335 carbonyl being replaced by a carboxyl oxygen of D251; and the movement of the β6-α7 loop further from the ADMI-DAS. The reorientation of D251 away from the MIDAS and toward the ADMIDAS has been proposed to reduce the binding of the MIDAS metal ion, thereby increasing its electropositivity and thus its ability to bind the ligand Asp [35,36]. The movement of the β6-α7 loop is associated with a downward motion of the α7 helix and the dramatic swing-out motion of the hybrid domain [5]. The movement of the β1-α1 loop toward the MIDAS is also thought to contribute to enhanced ligand binding by orienting the two backbone nitrogens in residues β3 Y122 and S123 so that they interact with the ligand’s carboxyl oxygen that does not coordinate the MIDAS metal ion and the ligand’s carboxyl oxygen that does coordinate the metal ion, respectively (Fig. 3).
Based on studies of ligand binding to αVβ3 and other integrin receptors, the ADMIDAS has been considered a negative regulatory site responsible for integrin inhibition by high concentration of Ca2+ [37–39]. Unlike αVβ3, however, αIIbβ3 -mediated binding of fibrinogen is not inhibited by Ca2+ [25]. Studies conducted with other integrin receptors or the isolated β3 βA (I-like) domain identified variable effects of altering the coordination of the ADMIDAS metal ion, with alterations in surface expression and receptor conformation [40], enhanced or reduced adhesion to immobilized ligands [35,36,41,42], decreased lymphocyte migration in vitro and abnormal lymphocyte homing in vivo [43], diminished adhesion to immobilized ligand [44], and diminished ability to bind soluble ligand [45]. In contrast, a β3 D251A mutation did not affect either αIIbβ3 surface expression or binding of the ligand-mimetic antibodies PAC-1 and OPG2 [46]. β3 D126A and D127A mutations in αIIbβ3 did not decrease surface expression of the receptor or cell adhesion to immobilized fibrinogen in the presence of Ca2+ and Mg2+ [47]. An isolated recombinant β3 βI domain containing a D126A mutation in the ADMIDAS was, however, reported to have decreased binding of soluble fibrinogen [48]. It has been proposed that the β3 residue Ala252, and the corresponding Ala in β1, distinguish these β integrin families from the β2 and β7 families. The latter have instead an Asp residue that enhances the electronegativity near the MIDAS, reduces ligand binding affinity, and defines the response to the loss of the ADMIDAS coordinating residues [35,36].
While reconciling these data is challenging, especially the dramatic difference in cation preference for ligand binding to αVβ3 and αIIbβ3 despite their sharing the same β3 subunit that contains the cations directly engaged in ligand binding, it is likely that the ability of the fibrinogen γ chain dodecapeptide (which interacts with αIIbβ3, but not αVβ3) to bind to both the MIDAS and ADMIDAS metal ions [7] is important. Crystal structures of α5β1 in the presence and absence of Ca2+ demonstrate that loss of the ADMIDAS Ca2+ facilitates the movement of the β1-α1 loop toward the MIDAS in response to the binding of an RGD peptide, leading to higher ligand binding affinity as the ligand Asp carboxyl gains additional interactions with the backbone nitrogens in the loop [39]. As αVβ3 interacts with the 572RGD574 sequence in the fibrinogen a chain rather than the γ chain dodecapeptide [49], loss of the αVβ3 ADMIDAS Ca2+ would be expected to enhance its affinity for fibrinogen; in contrast, the effect of loss of the ADMIDAS Ca2+ in αIIbβ3 would reflect the balance from gaining higher affinity for the Asp carboxyl that binds to the MIDAS, but losing the interaction of the terminal Val carboxyl with the ADMIDAS Ca2+. Support for this interpretation comes from the studies of the binding of the high-affinity fibronectin fragment to αVβ3 because the binding of this fragment was not inhibited by Ca2+ in association with it developing a water-mediated interaction with the ADMI-DAS metal ion [28]. Further support comes from studies of the monoclonal antibody AP7, which contains an RGDGGN sequence in its heavy chain CDR3 region [38]. This antibody binds to both αIIbβ3 and αVβ3; Ca2+ inhibits its binding to αVβ3, but not αIIbβ3. Changing the sequence to RGDGGA resulted in no effect on the its binding to αVβ3 or its inhibition by Ca2+, but led to complete loss of binding to αIIbβ3, presumably due to loss of Asn-mediated binding to the ADMIDAS.
The structural basis of new αIIbβ3 antagonists
Three αIIbβ3 antagonists have been approved for human use in the USA, starting with abciximab, the chimeric Fab fragment of the murine monoclonal antibody 7E3, in 1994, followed in 1998 by eptifibatide, modeled on the KGD sequence, and tirofiban, modeled on the RGD sequence. These drugs have demonstrated efficacy in reducing death and ischemic complications of percutaneous coronary artery interventions in a large number of randomized studies [2], but they are associated with an increased risk of major bleeding and thrombocytopenia. As a result, their use is restricted to situations in which there is a high risk of thrombosis. Attempts to develop orally active αIIbβ3 antagonists based on the RGD sequence failed because the agents were not efficacious and several were associated with increased mortality [50,51]; they also caused thrombocytopenia [52–54]. The R(K)GD-based drugs all bind by the same fundamental mechanism in which there are two major points of attachment, one via a positively charged residue interacting with the αIIb D224 and the other via a ligand aspartic acid carboxyl oxygen coordinating the MIDAS Mg2+ (Fig. 2A). As a result, all of these agents induce conformational changes in the receptor and induce the receptor to adopt a high-affinity ligand-binding state, that is, they are partial agonists. Thus, it has been hypothesized that the increased mortality with the oral agents was due to their ‘priming’ the αIIbβ3 receptor to adopt a high-affinity ligand-binding state, resulting in platelet aggregation [50,51]. In fact, eptifibatide and tirofiban also prime the receptor, which may limit their efficacy [21,55–57]. Thrombocytopenia produced by both the oral and intravenous agents may also result from their inducing conformational changes that expose regions of the receptor to which some patients have preformed antibodies [52–54]. Thus, there are theoretical reasons to try to develop αIIbβ3 antagonists that do not induce receptor extension and swing-out.
Ur-3216/2922
This small molecule antagonist does not induce the conformational changes in the receptor produced by RGD-based compounds [58]. Molecular docking suggests that while it binds to αIIb D224, its carboxyl does not engage the MIDAS metal ion; rather it appears to form a salt bridge with β3 Arg 165 and a hydrogen bond with Tyr 166 [59].
Activation-specific single-chain antibodies
Schwarz et al. [60,61] produced human single-chain antibodies specific for activated αIIbβ3 and showed that they do not induce LIBS epitope expression or platelet adhesion to fibrinogen. These antibodies inhibit platelet thrombus formation in vitro and in animal models with less prolongation of the bleeding time than other αIIbβ3 antagonists. They almost all contain an RXD sequence, which would be expected to induce the active conformation of the receptor, so it is likely that the surrounding amino acids modify the binding so as to prevent the conformational change.
The RUC compounds
We identified RUC-1 (Fig. 2B) by screening 33 264 small molecules for their ability to inhibit platelet adhesion to fibrinogen [57,62]. It inhibits ADP-induced platelet aggregation with an IC50 of ~13 µm and soluble fibrinogen binding to platelets and purified αIIbβ3, but does not inhibit ligand binding to αVβ3, GPIb, or α2β1. Unlike tirofiban and/or eptifibatide, RUC-1 binding does not induce conformational changes in the β3 subunit detectable by LIBS mAbs, Stokes radius changes, or electron microscopy, and pretreatment of purified αIIbβ3 with RUC-1 does not enhance fibrinogen binding (‘priming’) [6,57]. Molecular docking and x-ray crystallography indicated that RUC-1 is unique among αIIbβ3 antagonists in binding exclusively to αIIb; moreover, it does not induce the rearrangement of the β3 βI subunit or the extensive hybrid domain swing-out we previously found associated with the binding of RGD-based agents [5,6]. With Dr. Craig Thomas’ medicinal chemistry group at NIH, we synthesized RUC-2 (Fig. 2C), which is ~100-fold higher in affinity than RUC-1 but equally selective for αIIbβ3 compared to αVβ3 [21]. It has antithrombotic properties in the carotid artery FeCl3 injury model using mice developed by Poncz’s group expressing human αIIβ and mouse β3 [62]. It also does not induce major conformational changes in β3 as judged by mAb binding, light scattering, or gel chromatography, nor does it prime the receptor to bind ligand [21]. Unlike eptifibatide and tirofiban, neither RUC-1 nor RUC-2 induces extension of intact αIIbβ3 molecules inserted into lipid bilayer nanodiscs [21]. X-ray crystallographic analysis of the RUC-2-αIIbβ3 headpiece complex in 1 mm Ca2+/5 mm Mg2+ at 2.6 Å revealed that RUC-2 binds to αIIb the way RUC-1 does, but in addition, it binds to the β3 MIDAS residue E220, thus displacing Mg2+ from the MIDAS [21]. Thus, RUC-2 locks the receptor in an inactive conformation by displacing the Mg + required for ligand binding, eliminating the ability of a ligand Asp carboxyl to bind and initiate the movement of the β1-α1 loop. More recently, we synthesized a much more water-soluble derivative of RUC-2, RUC-4, and it shows very similar binding and antithrombotic properties. It currently is under development for the prehospital therapy of ST segment elevated myocardial infarction [63].
Altering intracellular signaling of a αIIbβ3
Although space limitations do not permit a detailed description of this topic, an alternative approach to inhibiting αIIbβ3 is to alter the intracellular signaling that leads to activation of αIIbβ3. There have been dramatic advances in understanding the structural details of this process, providing a wide range of potential targets [59,64–66].
Acknowledgments
Supported in part by grant HL19278 from the Heart, Lung and Blood Institute, grant TR000043 from the National Center for Advancing Translational Science (NCATS), and funds from Stony Brook University. I want to acknowledge my outstanding collaborators who contributed to the studies of the RUC compounds, including the research groups led by Dr. Marta Filizola of the Icahn School of Medicine at Mount Sinai, Dr. Craig Thomas of the National Center for Advancing Translational Science (NCATS), Dr. Timothy Springer of Harvard University, Dr. Mort Poncz of the University of Pennsylvania, and Dr. Thomas Diacovo of Columbia University. I want to thank Suzanne Rivera for outstanding administrative assistance and Dr. Lorena Buitrago for assistance with the figures.
Footnotes
Disclosure of Conflict of Interests
In accord with federal law and institutional policies, B. Coller has royalty interests in abciximab (Centocor) and the VerifyNow assays (Accumetrics). Rockefeller University has applied for patents on the RUC compounds.
References
- 1.Coller BS, Shattil SJ. The GPIIb/IIIa (integrin alphaIIbbeta3) odyssey: a technology-driven saga of a receptor with twists, turns, and even a bend. Blood. 2008;112:3011–3025. doi: 10.1182/blood-2008-06-077891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bosch X, Marrugat J, Sanchis J. Platelet glycoprotein IIb/IIIa blockers during percutaneous coronary intervention and as the initial medical treatment of non-ST segment elevation acute coronary syndromes. Cochrane Database Syst Rev. 2013;11:CD002130. doi: 10.1002/14651858.CD002130.pub4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peterson JA, McFarland JG, Curtis BR, Aster RH. Neonatal alloimmune thrombocytopenia: pathogenesis, diagnosis and management. Br J Haematol. 2013;161:3–14. doi: 10.1111/bjh.12235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhu J, Luo BH, Xiao T, Zhang C, Nishida N, Springer TA. Structure of a complete integrin ectodomain in a physiologic resting state and activation and deactivation by applied forces. Mol Cell. 2008;32:849–861. doi: 10.1016/j.molcel.2008.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xiao T, Takagi J, Coller BS, Wang J, Springer TA. Structural basis for allostery in integrins and binding to fibrinogen-mimetic therapeutics. Nature. 2004;432:59–67. doi: 10.1038/nature02976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu J, Zhu J, Negri A, Provasi D, Filizola M, Coller BS, Springer TA. Closed headpiece of integrin αIIbβ3 and its complex with an αIIbβ3-specific antagonist that does not induce opening. Blood. 2010;116:5050–5059. doi: 10.1182/blood-2010-04-281154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Springer TA, Zhu J, Xiao T. Structural basis for distinctive recognition of fibrinogen gammaC peptide by the platelet integrin alphaIIbbeta3. J Cell Biol. 2008;182:791–800. doi: 10.1083/jcb.200801146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu J, Zhu J, Springer TA. Complete integrin headpiece opening in eight steps. J Cell Biol. 2013;201:1053–1068. doi: 10.1083/jcb.201212037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim C, Schmidt T, Cho EG, Ye F, Ulmer TS, Ginsberg MH. Basic amino-acid side chains regulate transmembrane integrin signalling. Nature. 2012;481:209–213. doi: 10.1038/nature10697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang J, Ma YQ, Page RC, Misra S, Plow EF, Qin J. Structure of an integrin alphaIIb beta3 transmembrane-cytoplasmic hetero-complex provides insight into integrin activation. Proc Natl Acad Sci U S A. 2009;106:17729–17734. doi: 10.1073/pnas.0909589106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma YQ, Yang J, Pesho MM, Vinogradova O, Qin J, Plow EF. Regulation of integrin alphaIIbbeta3 activation by distinct regions of its cytoplasmic tails. Biochemistry. 2006;45:6656–6662. doi: 10.1021/bi060279h. [DOI] [PubMed] [Google Scholar]
- 12.Huang H, Vogel HJ. Structural basis for the activation of platelet integrin alphaIIbbeta3 by calcium- and integrin-binding protein 1. J Am Chem Soc. 2012;134:3864–3872. doi: 10.1021/ja2111306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carrell NA, Fitzgerald LA, Steiner B, Erickson HP, Phillips DR. Structure of human platelet membrane glycoproteins IIb and IIIa as determined by electron microscopy. J Biol Chem. 1985;260:1743–1749. [PubMed] [Google Scholar]
- 14.Weisel JW, Nagaswami C, Vilaire G, Bennett JS. Examination of the platelet membrane glycoprotein IIb-IIIa complex and its interaction with fibrinogen and other ligands by electron microscopy. J Biol Chem. 1992;267:16637–16643. [PubMed] [Google Scholar]
- 15.Du X, Gu M, Weisel JW, Nagaswami C, Bennett JS, Bowditch R, Ginsberg MH. Long range propagation of conformational changes in integrin alpha IIb beta 3. J Biol Chem. 1993;268:23087–23092. [PubMed] [Google Scholar]
- 16.Adair BD, Yeager M. Three-dimensional model of the human platelet integrin alpha IIbbeta 3 based on electron cryomicroscopy and x-ray crystallography. Proc Natl Acad Sci USA. 2002;99:14059–14064. doi: 10.1073/pnas.212498199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nogales A, Garcia C, Perez J, Callow P, Ezquerra TA, Gonzalez-Rodriguez J. Three-dimensional model of human platelet integrin alphaIIb beta3 in solution obtained by small angle neutron scattering. J Biol Chem. 2010;285:1023–1031. doi: 10.1074/jbc.M109.050039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eng ET, Smagghe BJ, Walz T, Springer TA. Intact alphaIIbbe-ta3 integrin is extended after activation as measured by solution X-ray scattering and electron microscopy. J Biol Chem. 2011;286:35218–35226. doi: 10.1074/jbc.M111.275107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ye F, Hu G, Taylor D, Ratnikov B, Bobkov AA, McLean MA, Sligar SG, Taylor KA, Ginsberg MH. Recreation of the terminal events in physiological integrin activation. J Cell Biol. 2010;188:157–173. doi: 10.1083/jcb.200908045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iwasaki K, Mitsuoka K, Fujiyoshi Y, Fujisawa Y, Kikuchi M, Sekiguchi K, Yamada T. Electron tomography reveals diverse conformations of integrin alphaIIbbeta3 in the active state. J Struct Biol. 2005;150:259–267. doi: 10.1016/j.jsb.2005.03.005. [DOI] [PubMed] [Google Scholar]
- 21.Zhu J, Choi WS, McCoy J, Negri A, Zhu J, Naini S, Li J, Shen M, Huang W, Bougie D, Rasmussen M, Aster R, Thomas CJ, Filizola M, Springer TA, Coller BS. Structure-guided design of a high affinity platelet integrin αIIbβ3 receptor antagonist that disrupts Mg2+ binding to the MIDAS. Sci Transl Med. 2012;4:1–12. doi: 10.1126/scitranslmed.3003576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Choi WS, Rice WJ, Stokes DL, Coller BS. Three-dimensional reconstruction of intact human integrin alphaIIbbeta3: new implications for activation-dependent ligand binding. Blood. 2013;122:4165–4171. doi: 10.1182/blood-2013-04-499194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Donald JE, Zhu H, Litvinov RI, DeGrado WF, Bennett JS. Identification of interacting hot spots in the beta3 integrin stalk using comprehensive interface design. J Biol Chem. 2010;285:38658–38665. doi: 10.1074/jbc.M110.170670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xiong JP, Stehle T, Zhang R, Joachimiak A, Frech M, Goodman SL, Arnaout MA. Crystal structure of the extracellular segment of integrin alpha Vbeta3 in complex with an Arg-Gly-Asp ligand. Science. 2002;296:151–155. doi: 10.1126/science.1069040. [DOI] [PubMed] [Google Scholar]
- 25.Bennett JS, Vilaire G. Exposure of platelet fibrinogen receptors by ADP and epinephrine. J Clin Invest. 1979;64:1393–1401. doi: 10.1172/JCI109597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vorup-Jensen T, Waldron TT, Astrof N, Shimaoka M, Springer TA. The connection between metal ion affinity and ligand affinity in integrin I domains. Biochim Biophys Acta. 2007;1774:1148–1155. doi: 10.1016/j.bbapap.2007.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Honda S, Tomiyama Y, Aoki T, Shiraga M, Kurata Y, Seki J, Matsuzawa Y. Association between ligand-induced conformational changes of integrin IIbbeta3 and IIbbeta3-mediated intra-cellular Ca2+ signaling. Blood. 1998;92:3675–3683. [PubMed] [Google Scholar]
- 28.van Agthoven JF, Xiong JP, Alonso JL, Rui X, Adair BD, Goodman SL, Arnaout MA. Structural basis for pure antagonism of integrin alphaVbeta3 by a high-affinity form of fibronectin. Nat Struct Mol Biol. 2014;21:383–388. doi: 10.1038/nsmb.2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murcia M, Jirouskova M, Li J, Coller BS, Filizola M. Functional and computational studies of the ligand-associated metal binding site of beta3 integrins. Proteins. 2008;71:1779–1791. doi: 10.1002/prot.21859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dong X, Hudson NE, Lu C, Springer TA. Structural determinants of integrin beta-subunit specificity for latent TGF-beta. Nat Struct Mol Biol. 2014;21:1091–1096. doi: 10.1038/nsmb.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dong X, Mi LZ, Zhu J, Wang W, Hu P, Luo BH, Springer TA. alpha(V)beta(3) integrin crystal structures and their functional implications. Biochemistry. 2012;51:8814–8828. doi: 10.1021/bi300734n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rui X, Mehrbod M, van Agthoven JF, Anand S, Xiong JP, Mofrad MR, Arnaout MA. The alpha-subunit regulates stability of the metal ion at the ligand-associated metal ion-binding site in beta3 integrins. J Biol Chem. 2014;289:23256–23263. doi: 10.1074/jbc.M114.581470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xiong JP, Stehle T, Diefenbach B, Zhang R, Dunker R, Scott DL, Joachimiak A, Goodman SL, Arnaout MA. Crystal structure of the extracellular segment of integrin alphaVbeta3. Science. 2001;294:339–345. doi: 10.1126/science.1064535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kloczewiak M, Timmons S, Bednarek MA, Sakon M, Hawiger J. Platelet receptor recognition domain on the gamma chain of human fibrinogen and its synthetic peptide analogues. Biochemistry. 1989;28:2915–2919. doi: 10.1021/bi00433a025. [DOI] [PubMed] [Google Scholar]
- 35.Raborn J, Luo BH. Mutagenesis studies of the beta I domain metal ion binding sites on integrin alphaVbeta3 ligand binding affinity. J Cell Biochem. 2012;113:1190–1197. doi: 10.1002/jcb.23448. [DOI] [PubMed] [Google Scholar]
- 36.Raborn J, Fu T, Wu X, Xiu Z, Li G, Luo BH. Variation in one residue associated with the metal ion-dependent adhesion site regulates alphaIIbbeta3 integrin ligand binding affinity. PLoS One. 2013;8:e76793. doi: 10.1371/journal.pone.0076793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hu DD, Barbas CF, Smith JW. An allosteric Ca2+ binding site on the beta3-integrins that regulates the dissociation rate for RGD ligands. J Biol Chem. 1996;271:21745–21751. doi: 10.1074/jbc.271.36.21745. [DOI] [PubMed] [Google Scholar]
- 38.Kunicki TJ, Annis DS, Felding-Habermann B. Molecular determinants of arg-gly-asp ligand specificity for beta3 integrins. J Biol Chem. 1997;272:4103–4107. doi: 10.1074/jbc.272.7.4103. [DOI] [PubMed] [Google Scholar]
- 39.Xia W, Springer TA. Metal ion and ligand binding of integrin alpha5beta1. Proc Natl Acad Sci U S A. 2014;111:17863–1788. doi: 10.1073/pnas.1420645111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goodman TG, Bajt ML. Identifying the putative metal ion-dependent adhesion site in the beta2 (CD18) subunit required for alphaLbeta2 and alphaMbeta2 ligand interactions. J Biol Chem. 1996;271:23729–23736. doi: 10.1074/jbc.271.39.23729. [DOI] [PubMed] [Google Scholar]
- 41.Chen J, Salas A, Springer TA. Bistable regulation of integrin adhesiveness by a bipolar metal ion cluster. Nat Struct Biol. 2003;10:995–1001. doi: 10.1038/nsb1011. [DOI] [PubMed] [Google Scholar]
- 42.Chen J, Takagi J, Xie C, Xiao T, Luo BH, Springer TA. The relative influence of metal ion binding sites in the I-like domain and the interface with the hybrid domain on rolling and firm adhesion by integrin alpha4beta7. J Biol Chem. 2004;279:55556–55561. doi: 10.1074/jbc.M407773200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Park EJ, Mora JR, Carman CV, Chen J, Sasaki Y, Cheng G, von Andrian UH, Shimaoka M. Aberrant activation of integrin alpha4beta7 suppresses lymphocyte migration to the gut. J Clin Invest. 2007;117:2526–2538. doi: 10.1172/JCI31570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kamata T, Tieu KK, Tarui T, Puzon-McLaughlin W, Hogg N, Takada Y. The role of the CPNKEKEC sequence in the beta(2) subunit I domain in regulation of integrin alpha(L)beta(2) (LFA-1) J Immunol. 2002;168:2296–2301. doi: 10.4049/jimmunol.168.5.2296. [DOI] [PubMed] [Google Scholar]
- 45.Mould AP, Barton SJ, Askari JA, Craig SE, Humphries MJ. Role of ADMIDAS cation-binding site in ligand recognition by integrin alpha 5 beta 1. J Biol Chem. 2003;278:51622–51629. doi: 10.1074/jbc.M306655200. [DOI] [PubMed] [Google Scholar]
- 46.Tozer EC, Liddington RC, Sutcliffe MJ, Smeeton AH, Loftus JC. Ligand binding to integrin alphaIIbbeta3 is dependent on a MIDAS-like domain in the beta3 subunit. J Biol Chem. 1996;271:21978–21984. doi: 10.1074/jbc.271.36.21978. [DOI] [PubMed] [Google Scholar]
- 47.Bajt ML, Loftus JC. Mutation of a ligand binding domain of beta 3 integrin Integral role of oxygenated residues in alpha IIb beta 3 (GPIIb-IIIa) receptor function. J Biol Chem. 1994;269:20913–20919. [PubMed] [Google Scholar]
- 48.Pesho MM, Bledzka K, Michalec L, Cierniewski CS, Plow EF. The specificity and function of the metal-binding sites in the integrin beta3 A-domain. J Biol Chem. 2006;281:23034–23041. doi: 10.1074/jbc.M602856200. [DOI] [PubMed] [Google Scholar]
- 49.Cheresh DA, Berliner SA, Vicente V, Ruggeri ZM. Recognition of distinct adhesive sites on fibrinogen by related integrins on platelets and endothelial cells. Cell. 1989;58:945–953. doi: 10.1016/0092-8674(89)90946-x. [DOI] [PubMed] [Google Scholar]
- 50.Cox D. Oral GPIIb/IIIa antagonists: what went wrong? Curr Pharm Des. 2004;10:1587–1596. doi: 10.2174/1381612043384673. [DOI] [PubMed] [Google Scholar]
- 51.Chew DP, Bhatt DL, Topol EJ. Oral glycoprotein IIb/IIIa inhibitors: why don’t they work? Am J Cardiovasc Drugs. 2001;1:421–428. doi: 10.2165/00129784-200101060-00002. [DOI] [PubMed] [Google Scholar]
- 52.Brassard JA, Curtis BR, Cooper RA, Ferguson J, Komocsar W, Ehardt M, Kupfer S, Maurath C, Swabb E, Cannon CP, Aster RH. Acute thrombocytopenia in patients treated with the oral glycoprotein IIb/IIIa inhibitors xemilofiban and orbofiban: evidence for an immune etiology. Thromb Haemost. 2002;88:892–897. [PubMed] [Google Scholar]
- 53.Aster RH, Curtis BR, Bougie DW, Dunkley S, Greinacher A, Warkentin TE, Chong BH. Thrombocytopenia associated with the use of GPIIb/IIIa inhibitors: position paper of the ISTH working group on thrombocytopenia and GPIIb/IIIa inhibitors. J Thromb Haemost. 2006;4:678–679. doi: 10.1111/j.1538-7836.2006.01829.x. [DOI] [PubMed] [Google Scholar]
- 54.Scirica BM, Cannon CP, Cooper R, Aster RH, Brassard J, McCabe CH, Charlesworth A, Skene AM, Braunwald E. Drug-induced thrombocytopenia and thrombosis: evidence from patients receiving an oral glycoprotein IIb/IIIa inhibitor in the Orbofiban in Patients with Unstable coronary Syndromes-(OPUS-TIMI 16) trial. J Thromb Thrombolysis. 2006;22:95–102. doi: 10.1007/s11239-006-8669-4. [DOI] [PubMed] [Google Scholar]
- 55.Bassler N, Loeffler C, Mangin P, Yuan Y, Schwarz M, Hagemeyer CE, Eisenhardt SU, Ahrens I, Bode C, Jackson SP, Peter K. A mechanistic model for paradoxical platelet activation by ligand-mimetic alphaIIb beta3 (GPIIb/IIIa) antagonists. Arterioscler Thromb Vasc Biol. 2007;27:e9–e15. doi: 10.1161/01.ATV.0000255307.65939.59. [DOI] [PubMed] [Google Scholar]
- 56.Hantgan RR, Stahle MC. Integrin priming dynamics: mechanisms of integrin antagonist-promoted alphaIIbbeta3:PAC-1 molecular recognition. Biochemistry. 2009;48:8355–8365. doi: 10.1021/bi900475k. [DOI] [PubMed] [Google Scholar]
- 57.Blue R, Murcia M, Karan C, Jirouskova M, Coller BS. Application of high throughput screening to identify a novel αIIβ-specific small molecule inhibitor of αIIbβ3-mediated platelet interaction with fibrinogen. Blood. 2008;111:1248–1256. doi: 10.1182/blood-2007-08-105544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Aga Y, Baba K, Tam S, Nakanishi T, Yoneda K, Kita J, Ueno H. UR-3216: a new generation oral platelet GPIIb/IIIa antagonist. Curr Pharm Des. 2004;10:1597–1601. doi: 10.2174/1381612043384592. [DOI] [PubMed] [Google Scholar]
- 59.Cox D, Brennan M, Moran N. Integrins as therapeutic targets: lessons and opportunities. Nat Rev Drug Discov. 2010;9:804–820. doi: 10.1038/nrd3266. [DOI] [PubMed] [Google Scholar]
- 60.Schwarz M, Rottgen P, Takada Y, Le Gall F, Knackmuss S, Bassler N, Buttner C, Little M, Bode C, Peter K. Single-chain antibodies for the conformation-specific blockade of activated platelet integrin alphaIIbbeta3 designed by subtractive selection from naive human phage libraries. FASEB J. 2004;18:1704–1706. doi: 10.1096/fj.04-1513fje. [DOI] [PubMed] [Google Scholar]
- 61.Schwarz M, Meade G, Stoll P, Ylanne J, Bassler N, Chen YC, Hagemeyer CE, Ahrens I, Moran N, Kenny D, Fitzgerald D, Bode C, Peter K. Conformation-specific blockade of the integrin GPIIb/IIIa: a novel antiplatelet strategy that selectively targets activated platelets. Circ Res. 2006;99:25–33. doi: 10.1161/01.RES.0000232317.84122.0c. [DOI] [PubMed] [Google Scholar]
- 62.Blue R, Kowalska MA, Hirsch J, Murcia M, Janczak CA, Harrington A, Jirouskova M, Li J, Fuentes R, Thornton MA, Filizola M, Poncz M, Coller BS. Structural and therapeutic insights from the species specificity and in vivo antithrombotic activity of a novel αIIb-specific αIIbβ3 antagonist. Blood. 2009;114:195–201. doi: 10.1182/blood-2008-08-169243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li J, Vootukuri S, Shang Y, Negri A, Jiang JK, Nedelman MA, Diacovo TG, Filizola M, Thomas CJ, Coller BS. RUC-4: a novel αIIbβ3 antagonist for pre-hospital therapy of myocardial infarction. Arterioscler Thromb Vasc Biol. 2014;34:2321–2329. doi: 10.1161/ATVBAHA.114.303724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ye F, Snider AK, Ginsberg MH. Talin and kindlin: the one-two punch in integrin activation. Front Med. 2014;8:6–16. doi: 10.1007/s11684-014-0317-3. [DOI] [PubMed] [Google Scholar]
- 65.Das M, Subbayya IS, Qin J, Plow EF. Mechanisms of talin-dependent integrin signaling and crosstalk. Biochim Biophys Acta. 2014;1838:579–588. doi: 10.1016/j.bbamem.2013.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Provasi D, Negri A, Coller BS, Filizola M. Talin-driven inside-out activation mechanism of platelet αIIbβ3 integrin probed by multimicrosecond, all-atom molecular dynamics simulations. Proteins. 2014;82:3231–3240. doi: 10.1002/prot.24540. [DOI] [PMC free article] [PubMed] [Google Scholar]