Summary
A fundamental challenge in calcium imaging has been to infer spike rates of neurons from the measured noisy fluorescence traces. We systematically evaluate different spike inference algorithms on a large benchmark dataset (>100.000 spikes) recorded from varying neural tissue (V1 and retina) using different calcium indicators (OGB-1 and GCaMP6). In addition, we introduce a new algorithm based on supervised learning in flexible probabilistic models and find that it performs better than other published techniques. Importantly, it outperforms other algorithms even when applied to entirely new datasets for which no simultaneously recorded data is available. Future data acquired in new experimental conditions can be used to further improve the spike prediction accuracy and generalization performance of the model. Finally, we show that comparing algorithms on artificial data is not informative about performance on real data, suggesting that benchmarking different methods with real-world datasets may greatly facilitate future algorithmic developments in neuroscience.
Introduction
Over the past two decades, two-photon imaging has become one of the most widely used techniques for studying information processing in neural populations in vivo (Denk et al., 1990; Kerr and Denk, 2008). Typically, a calcium indicator such as the synthetic dye Oregon green BAPTA-1 (OGB-1) (Stosiek et al., 2003) or the genetically encoded GCaMP6 (Chen et al., 2013) is used to image a large fraction of cells in a neural tissue. Individual action potentials lead to a fast rise in fluorescence, followed by a slow decay with a time constant of hundreds of milliseconds (Chen et al., 2013; Kerr et al., 2005). Commonly, neural population activity from dozens or hundreds of cells is imaged using relatively slow scanning speeds (<15 Hz), but novel fast scanning methods (Cotton et al., 2013; Grewe et al., 2010; Valmianski et al., 2010) (up to several 100 Hz) have opened additional opportunities for studying neural population activity at increased temporal resolution.
A fundamental challenge has been to infer the time-varying spike rate of neurons from the measured noisy calcium fluorescence traces. To solve this problem of spike inference, several different approaches have been proposed, including template-matching (Greenberg et al., 2008; Grewe et al., 2010; Oñativia et al., 2013), deconvolution (Park et al., 2013; Yaksi and Friedrich, 2006) and approximate Bayesian inference (Pnevmatikakis et al., 2016, 2013; Vogelstein et al., 2010, 2009). These methods have in common that they assume a forward generative model of calcium signal generation which is then inverted to infer spike times. Forward models incorporate strong a-priori assumptions about the shape of the calcium fluorescence signal induced by a single spike and the statistics of the noise. Alternatively, simple supervised learning techniques have been used to learn the relationship between calcium signals and spikes from data (Sasaki et al., 2008).
However, it is currently not known which approach is most successful at inferring spikes under typical experimental conditions used for population imaging, as a detailed quantitative comparison of different algorithms on large datasets has been lacking. Rather, most published algorithms have only been evaluated on relatively small experimental datasets often collected zooming in on individual cells. Also, performance measures differ between studies. In addition, the question of how well we can reconstruct the spikes of neurons given calcium measurements has been studied theoretically or using simulated datasets (Lütcke et al., 2013; Wilt et al., 2013). While such studies offer the advantage that many model parameters are under the control of the investigator, they still rely on model assumptions and thus do not answer the question of how well we can reconstruct spikes from actual measurements.
Here, we pursue two goals: (1) we systematically evaluate a range of spike inference algorithms on a large dataset including simultaneous measurements of spikes and calcium signals in primary visual cortex and the retina of mice using OGB-1 and GCaMP6 as calcium indicators collected ex-vivo and in anesthetized and awake animals and (2) introduce a new data-driven approach based on supervised learning in flexible probabilistic models to infer spikes from calcium fluorescence traces. We show that our new method outperforms all previously published techniques even when tested on data collected under new experimental conditions not used for training.
Results
A flexible probabilistic model for spike inference
Here we introduce a new algorithm for spike inference from calcium data. We propose to model the probabilistic relationship between a segment of the fluorescence trace xt and the number of spikes kt in a small time bin, assuming they are Poisson distributed with rate λ(xt):
Instead of relying on a specific forward model, we parameterize the firing rate λ(xt) using a recently introduced extension of generalized linear models, the factored spike-triggered mixture (STM) model (Theis et al., 2013) (Fig. 1a; see Methods):
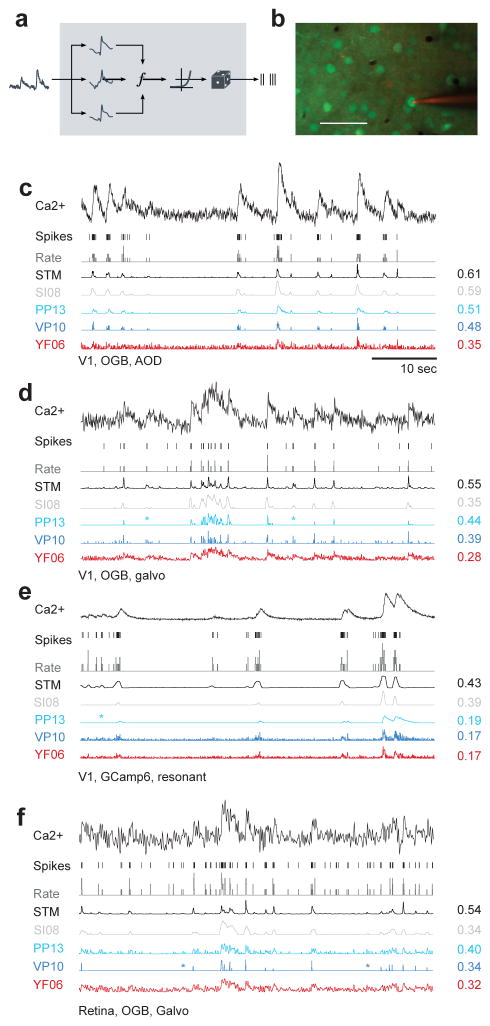
Figure 1. Spike inference from calcium measurements.
a) Schematic of the probabilistic STM model.
b) Simultaneous recording of spikes and calcium fluorescence traces in primary visual cortex of anesthetized mice. Green: Cells labeled with OGB-1 indicator. Red: Patch pipette filled with Alexa Fluor 594. Scale bar: 50 μm.
c) Example cell recorded from mouse V1 under anesthesia using AOD scanner and OGB-1 as indicator. From top to bottom: Calcium fluorescence trace, spikes, spike rate in bins of 40 ms (grey), inferred spike rate using the STM model (black), SI08, PP13, VP14 and YF06. All traces were scaled independently for clarity. On the right, correlation between the inferred and the original spike rate.
d) Example cell recorded from mouse V1 under anesthesia using galvanometric scanners and OGB-1 as indicator. For legend, see c).
e) Example cell recorded from mouse V1 under anesthesia using resonance scanner and GCaMP6s as indicator. Note the different indicator dynamics. For legend, see c).
f) Example cell recorded from the ex-vivo mouse retina using galvanometric scanners and OGB-1 as indicator. For legend, see c).
We train this model on simultaneous recordings of spikes and calcium traces to learn a set of K linear features wk and M quadratic features um (‘supervised learning’), which are predictive of the occurrence of spikes in the fluorescence trace. Importantly, this model is sufficiently flexible to capture non-linear relationships between fluorescence traces and spikes, but at the same time is sufficiently restricted to avoid overfitting when little data is available. Below we will evaluate whether this model is too simple or already more complex than necessary by comparing its performance to that of multi-layer neural networks and simple LNP-type models.
In contrast to many methods that result in a single most likely spike train (a ‘point estimate’) using a probabilistic model provides us with an estimate of the expected firing rate, λ(xt), and a distribution over spike counts, as fully Bayesian methods do (Pnevmatikakis et al., 2013; Vogelstein et al., 2009). An advantage of access to a distribution over spike trains is that it allows us, for example, to estimate the uncertainty in the predictions. Example spikes trains consistent with the calcium measurements can be easily generated from our model without spending considerable computational resources. While generating a single ‘most likely spike train’ is also possible, its interpretation is less clear, as the result depends on the parametrization.
Benchmarking spike inference algorithms on experimental data
To quantitatively evaluate different spike inference approaches including our model, we acquired a large benchmark dataset with a total of 90 traces from 73 neurons, in which we simultaneously recorded calcium signals and spikes (Fig. 1b; in total >100,000 spikes). These cells were recorded with different scanning methods, different calcium indicators, in different brain states and at different sampling rates (see Table 1 and Methods). We used four datasets for our main analysis: Dataset 1 consisted of 16 neurons recorded in-vivo in V1 of anesthetized mice using fast 3D AOD-based imaging (Cotton et al., 2013) at ~320 Hz with OGB-1 as indicator. Dataset 2 consisted of 31 neurons recorded in-vivo in anesthetized mouse V1 using raster scanning at ~12 Hz with OGB-1 as indicator. Dataset 3 consisted of 19 segments recorded from 11 neurons in-vivo in anesthetized mouse V1 using the genetic calcium indicator GCaMP6s with a resonance scanner at ~59 Hz. Finally, dataset 4 consisted of 9 retinal ganglion cells recorded ex-vivo at ~8 Hz using raster scanning with OGB-1 as indicator (Briggman and Euler, 2011). In addition, we collected a small dataset of 6 cells from V1 of awake mice using again the genetic calcium indicator GCaMP6s (Reimer et al., 2014) to demonstrate the performance during awake imaging (see below). We resampled the calcium traces from all datasets to a common resolution of 100 Hz. Importantly, all of our datasets were acquired at a zoom factor commonly used in population imaging such that the signal quality should match well that commonly encountered in these preparations (see Table 1).
Table 1.
Datasets
| Set | Area | Brain state | n | Indicator | Scan frequency | Scanning method | #spikes | sp/s | Field of view |
|---|---|---|---|---|---|---|---|---|---|
| 1 | V1 | AN | 16 | OGB-1 | 322.5 ± 53.2 | 3D AOD | 19,876 | 1.86 | 200×200×100 μm2 |
| 2 | V1 | AN | 31 | OGB-1 | 11.8 ± 0.9 | 2D galvo scan | 32,385 | 2.47 | 250×250 μm2 |
| 3 | V1 | AN | 19* (11) | GCamp6s | 59.1 | 2D resonant | 23,974 | 2.58 | 265×265 μm2135×135 μm2 |
| 4 | Retina | Ex vivo | 9 | OGB-1 | 7.8 | 2D galvo scan | 12,488 | 4.36 | 100×100 μm2 |
| 5 | V1 | AWK | 15 (6)** | GCamp6s | 59.1 | 2D resonant | 12,413 | 4.87 | 265×265 μm2 |
For this dataset, 19 recordings were performed on 11 neurons
For this dataset, 15 recordings were performed on 6 neurons
AN: in-vivo anesthetized, AWK: in-vivo awake
We compared the performance of our algorithm (STM) and that of algorithms representative of the different approaches (see Table 2 and Methods), including simple deconvolution (YF06, Yaksi and Friedrich, 2006), MAP (VP10, known as ‘fast-oopsi’, Vogelstein et al., 2010) and Bayesian inference (PP13, (Pnevmatikakis et al., 2013); VP09, Vogelstein et al., 2009) in generative models, template-matching by finite rate of innovation (OD13, Oñativia et al., 2013) and supervised learning using a support vector machine (SI08, Sasaki et al., 2008). To provide a baseline level of performance, we evaluated how closely the calcium trace followed the spike train without any further processing (raw).
Table 2.
Algorithms
| Algorithm | Approach | Technique | Reference |
|---|---|---|---|
| STM | Supervised | STM | This paper |
| SI08 | Supervised | PCA+SVM | (Sasaki et al., 2008) |
| PP13 | Generative | MCMC sampling | (Pnevmatikakis et al., 2013) |
| OD13 | Template matching | Finite rate innovation | (Oñativia et al., 2013) |
| VP10 | Generative | MAP estimation | (Vogelstein et al., 2010) |
| VP09 | Generative | SMC sampling | (Vogelstein et al., 2009) |
| YF06 | Generative | Deconvolution | (Yaksi and Friedrich, 2006) |
We focus on two measures of spike reconstruction performance to provide a quantitative evaluation of the different techniques: (i) the correlation between the original and the reconstructed spike train and (ii) the information gained about the spike train based on the calcium signal (see Methods). For completeness, we computed (iii) the area under the ROC curve (AUC), which has also been used in the literature. The AUC score is a less sensitive measure of spike reconstruction performance, as e.g. an algorithm could consistently overestimate high rates compared to low rates and yet yield the same AUC (for a more technical discussion, see Methods).
To provide a fair comparison between the different algorithms, we evaluated their performance using leave-one-out cross-validation: we estimated the parameters of the algorithms on all but one cell from a dataset and tested them on the one remaining cell, repeating this procedure for each cell in the dataset (see Methods). For the algorithms based on generative models, we selected the hyperparameters during cross-validation (VP10, VP09) or using a sampling based approach (PP13; see Methods).
Supervised learning sets benchmark
We found that the spike rates predicted by our algorithm matched the true spike train closely, for cells from each dataset including both indicators OGB-1 and GCaMP6 (Fig. 1c–f). The other tested algorithms generally showed worse prediction performance: For example, YF06 typically resulted in very noisy estimates of the spike density function (Fig. 1c–f) and both VP10 and PP13 missed single spikes (Fig. 1d–f, marked by asterisk) and had difficulties modeling the dynamics of the GCaMP6 indicator (Fig. 1e).
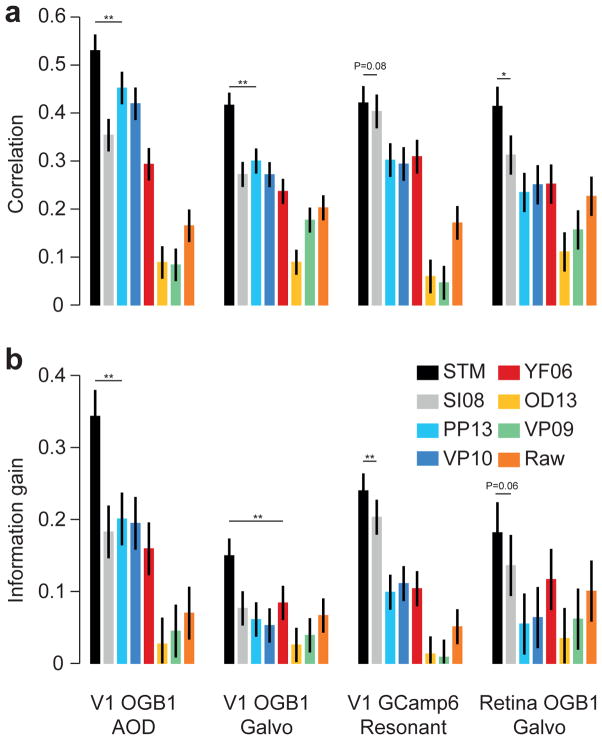
A quantitative comparison revealed that our STM method reconstructed the true spike trains better than its competitors, yielding a consistently higher correlation and information gain for all four datasets (Fig. 2a, b; evaluated at 25 Hz; for statistics, see figure). The median improvement in correlation across all recordings achieved by the STM over its two closest competitors was 0.12 (0.07–0.14; median and bootstrapped 95%-confidence interval, N=75) for SI08 – the other supervised learning approach based on SVMs – and 0.1 (0.08–0.13) for PP13 – Bayesian inference in a generative model – yielding a median improvement of 33% and 32%, respectively. Similarly, the STM explained 6.8 (5.0–7.7; SI08) and 9.6 (8.1–12.1; PP13) percent points more marginal entropy (measured by the relative information gain).
Figure 2. Quantitative evaluation of spike inference performance.
a) Correlation (mean± 2 SEM for repeated measure designs) between the true spike rate and the inferred spike rate for different algorithms (see legend for color code) evaluated on the four different datasets with anesthetized/ex-vivo data (with n=16, 31, 19 and 9, respectively). Markers above bars show the result of a Wilcoxon sign rank test between the STM model and its closest competitor (see Methods, * denotes P<0.05, ** denotes P<0.01). The evaluation was performed in bins of 40 ms.
b) As in a) but for information gained about the true spike train by observing the calcium trace.
When evaluated with respect to AUC, the performance of the STM model and these two algorithms was about the same (Suppl. Fig. 1), yielding a median difference in AUC of −0.01 (−0.02–0.01) and 0.01 (−0.01–0.02). This is likely because the AUC is the least sensitive of the three measures, as discussed above. As a side remark, note that AUC is closely related to the cost function optimized by SI08, which is based on a support vector machine. To show that the features extracted by our STM algorithm are more informative about the spike rate than those used by SI08, one can use a SVM on top of these features and obtain on 3 out of 4 datasets higher performance than SI08 (Suppl. Fig. 1).
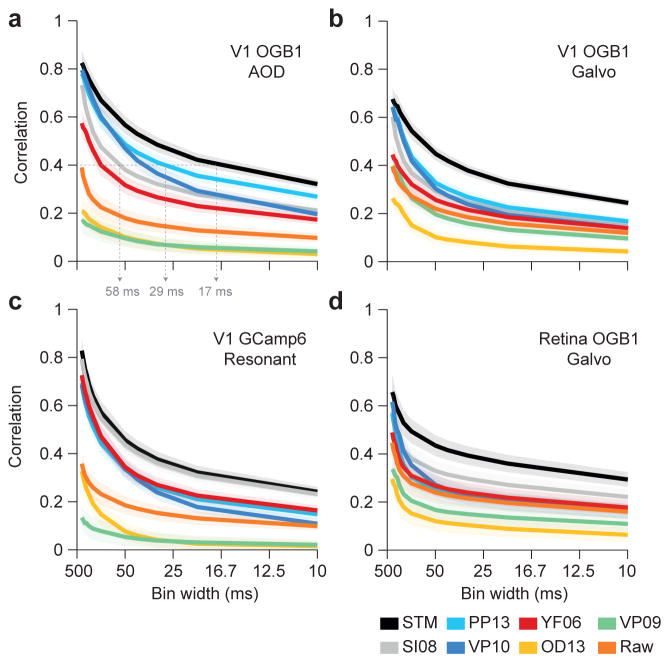
To evaluate timing accuracy, we asked what correlation between the inferred and true rate was achieved when ignoring timing details finer than a certain bin width (between 10 and several hundreds of milliseconds; Fig. 3): the correlation value reported for a bin width of 50 ms reflects only firing rate changes at a time scale larger than 50 ms as it compares observed spike counts with average predicted firing rates in 50 ms bins, while finer variations are ignored. In contrast, achieving a similar correlation value for 10 ms bins requires much higher timing accuracy, as the relative rate fluctuations in the finer time bins matter. This method is similar to the one used in (Greenberg et al., 2008), but in addition takes false positives/false negatives into account. Note that the binning affects the evaluation of the algorithm, not the spike inference. For all bin widths, the inference step was performed at the common sampling rate of 100 Hz (independent of scanning rate).
Figure 3. Timing accuracy of spike rate inference.
Correlation (mean ± 2 SEM for repeated measure designs) between the true and inferred spike rate as a function of temporal resolution for all four datasets with anesthetized/ex-vivo data (a–d) with n=16, 31,19 and 9, respectively. Grey dashed arrows in a) highlight the temporal resolution needed to achieve a correlation of 0.4 with different algorithms (see text).
Not surprisingly, correlation decayed as a function of bin width for all algorithms, as the resolution of increasingly fine detail becomes an increasingly challenging problem. However, the STM model performed better than the other algorithms in particular for small bin widths, providing higher temporal resolution (Fig. 3; also Suppl. Fig. 2). Consequently, if the desired average correlation between inferred and true spike rates deemed acceptable was 0.4, our method was able to achieve that using time bins of ~17 ms, whereas competing methods required ~29 and ~58 ms (PP13 and SI08, respectively; evaluated on dataset 1, Fig. 3a). Interestingly, VP10 (‘fast-oopsi’) performed similar to our method for low sampling rates, but its performance deteriorated consistently on all datasets to the performance level of VF06 with increasing sampling rates (Fig. 3).
The performance of the STM model could not be further improved using a more flexible multilayer neural network for modeling the non-linear rate function λt (Fig. 4 and Suppl. Fig. 3). To test this, we replaced the STM model by a neural network with two hidden layers, but found that this change resulted in only marginal performance improvement (Fig. 4). In addition, we tested whether a much simpler linear-nonlinear model would suffice to model λt. We found that the STM model performed significantly better than the simple LNP model (Fig. 4 and Suppl. Fig. 3). Therefore, the choice of the STM seems to provide a good compromise between flexibility of the model structure and generalization performance. In comparison to the neural network, the STM is derived from a fully interpretable probabilistic generative model (Theis et al., 2013).
Figure 4. Evaluating model complexity.
a) Correlation (mean ± 2 SEM for repeated measure designs) between the true and inferred spike rate comparing the STM model (black) with a flexible multilayer neural network (dark grey) and a simple LNP model (light grey) evaluated on the four different datasets collected under anesthesia/ex-vivo (with n=16, 31, 19 and 9, respectively). Markers above bars show the result of a Wilcoxon signed rank test between the STM model and the LNP model (see Methods, * denotes P<0.05, ** denotes P<0.01). The evaluation was performed in bins of 40 ms.
b) Information gained about the true spike train by observing the calcium trace performing the same model comparison described in a).
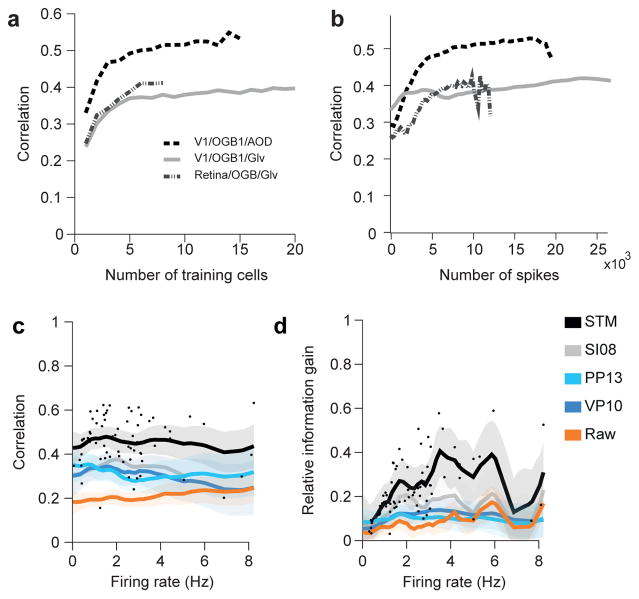
Importantly, already a small training set about 5–10 cells or 10.000 spikes was sufficient to achieve good performance with the STM model trained de novo (Fig. 5a,b and Suppl. Fig. 4a–d). We tested the prediction performance of the STM model with training sets of various sizes and found that it saturated between 5 and 10 cells for all datasets, arguing that a few simultaneously recorded cells may suffice to directly adapt the algorithms to new datasets acquired in other laboratories or with new imaging methods. In addition, we analyzed the training performance as a function of the number of spikes used for training and found that beyond ~10.000 spikes in the training set predictions do not improve much (Fig. 5b and Suppl. Fig 4c,d). Of course, these two factors are not independent: Recording 10.000 spikes from a single neuron will likely not yield the same quality predictions as recording 1.000 spikes from 10 neurons each. Finally, the superior performance of the STM was largely independent of the firing rate of the neuron within the limited range of firing rate in our sample of cells (Fig. 5c,d and Suppl. Fig. 4e).
Figure 5. Dependence on training set size and firing rate.
a) Mean correlation for STM model on the four different datasets collected under anesthesia/ex-vivo as a function of the number of neurons/segments in the training set.
b) Mean correlation for STM model as a function of the number of neurons/segments in the training set as a function of the number of spikes in the training set. Large training sets (on the right) lead to less spikes in the test set, making the evaluation noisier.
c) Correlation as a function of average firing rate of a cell. Dots mark correlation of STM model for individual traces. Solid lines indicate mean of a Gaussian process fit to correlation values for each of the indicated algorithms. Shaded areas are 95%-CI.
d) As in c. for relative information gain.
Generalization of performance to new datasets
In addition, we tested how well our algorithm performs if no simultaneous spike-calcium recordings are available for a new preparation or if a researcher wants to apply our algorithm without collecting simultaneous spike-calcium recordings, such that de-novo training of the model is impossible.
Remarkably, the STM model was able to generalize to new data sets that were recorded under different conditions than the data used for training. To test this, we trained the algorithms on three of the datasets and evaluated it on the remaining one (Fig. 6a) – that is, we applied the algorithm to an entirely new set of cells not seen during training. The STM algorithm still showed better performance than the other algorithms (Fig. 6b,c and Suppl. Fig. 5a), including superior performance on the GCamp6 dataset when trained solely on the three OGB datasets (Fig. 6b,c).
Figure 6. Spike inference without training data.
a) Schematic illustrating the setup: The algorithms are trained on all cells from three datasets (here: all but the GCaMP dataset) and evaluated on the remaining dataset (here: the GCaMP dataset), testing how well it generalizes to settings it has not seen during training.
b) Correlation (mean± 2 SEM for repeated measure designs) between the true spike rate and the inferred spike density function for a subset of the algorithms (see legend for color code) evaluated on each of the four different datasets collected under anesthesia/ex-vivo (with n=16, 31, 19 and 9, respectively), trained on the remaining three. Markers above bars show the result of a Wilcoxon sign rank test between the STM model and its closest competitor (see Methods, * denotes P<0.05, ** denotes P<0.01). The evaluation was performed in bins of 40 ms.
c) Information gained about the true spike train by observing the calcium trace performing the generalization analysis described in a).
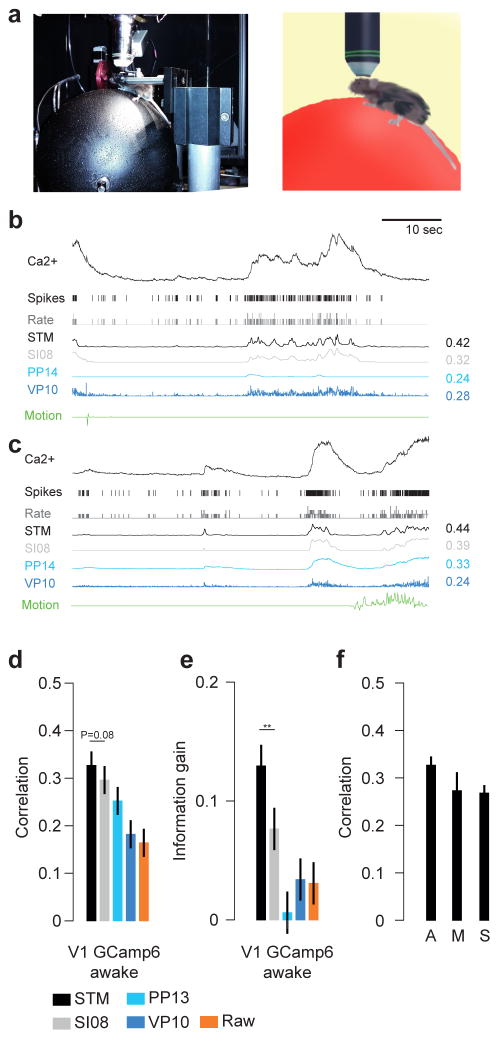
Next, we tested whether the algorithm’s performance would also transfer to recordings in head-fixed awake animals running on a Styrofoam ball (Fig. 7a) (Reimer et al., 2014). Brain movements and brain state fluctuations caused by the animal running on the ball may induce additional variability in the recordings, which renders spike inference under these conditions more difficult. Example neurons showed good spike inference performance for the STM model in periods without (Fig. 7b) and with movement (Fig. 7c). Overall, the STM trained on all neurons recorded in anesthetized animals or ex-vivo retina (n=75 traces from 70 cells) performed better than or comparable to the other algorithms on the awake data recorded using GCamp6s (n=15 traces from 6 cells; Fig. 7d,e and Suppl. Fig. 5b), further underscoring its generalization abilities. In addition, when we split the data into parts with and without motion (410.1 s vs. 2056.9 s), we found that the STM model’s performance was not impaired during periods where the mouse moved (Fig. 7f, correlation 0.27±0.03 vs. 0.27± 0.02, mean ± SEM).
Figure 7. Spike inference on awake data.
a) Photograph and illustration of a mouse sitting on a Styrofoam ball during a combined imaging/electrophysiology experiment.
b) Example recording as in Fig. 1 but for data recorded in awake animals using GCaMP6s as indicator. During this recording, the mouse moved very little (green trace). Algorithms were trained on anesthetized data and tested on awake data.
c) As in b) but for a period with substantial movement of the mouse (right).
d) Correlation (mean ± 2 SEM for repeated measure designs) between the true spike rate and the inferred spike density function for a subset of the algorithms (see legend for color code) evaluated on awake data (n=15 segements), trained on all anesthetized data. Markers above bars show the result of a Wilcoxon sign rank test between the STM model and its closest competitor (see Methods, * denotes P<0.05, ** denotes P<0.01). The evaluation was performed in bins of 40 ms.
e) As in d) but for information gain.
f) Evaluation of the effect of movement for the STM model. Recordings were separated into periods with and without motion (A: all, M: Moving, S: stationary). Mouse movement left the performance unchanged.
We finally tested the different algorithms on three data sets using different GCamp-indicators acquired focusing on individual cells (in contrast to our population imaging dataset; n=29 cells; data publicly available from Svoboda lab, see Methods). Similarly to above, our algorithm was trained on two of these datasets and tested on the third. In addition, we included all cells from datasets 1–4 in to the training set, as there are only comparably few spikes in the Svoboda lab datasets. Focusing on individual cells makes the data less noisy, resulting on overall much higher correlation and AUC values (Suppl. Fig. 6). The STM algorithm performed well and on a par with VP10 regarding all three measures used for evaluation (Suppl. Fig. 6).
Taken together, our analysis indicates that good performance can be expected for our algorithm when it is directly applied on novel datasets without further training (see Discussion). A pre-trained version of our algorithm is available for download (see Methods).
Comparisons on artificial data
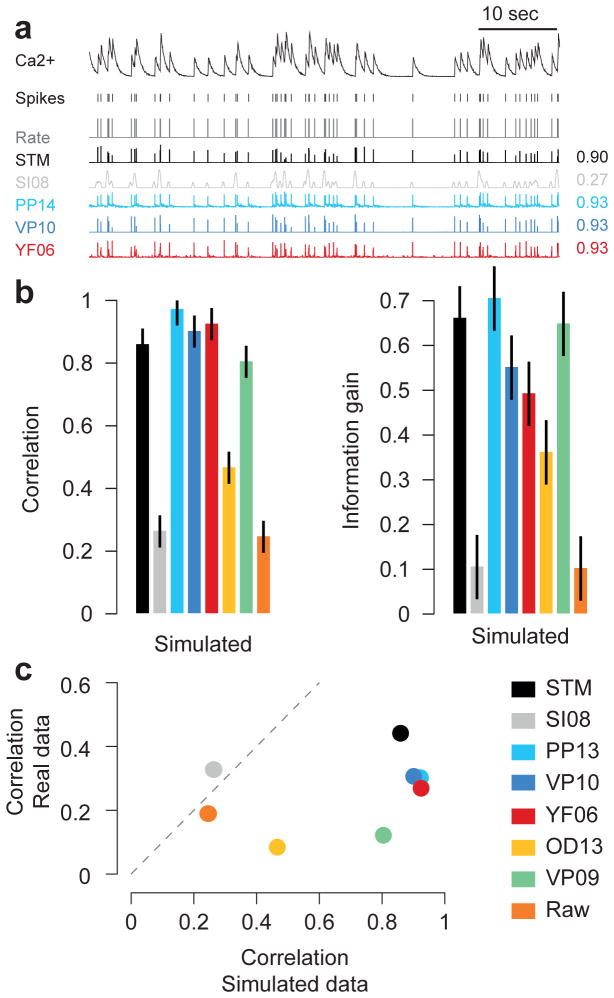
Finally, we evaluated the performance of the algorithms on simulated data and show that this was not predictive of the performance of the algorithms on the real datasets (Fig. 7). To test this, we simulated data from a simple biophysical model of calcium fluorescence generation (Fig. 7a, see Methods, Vogelstein et al., 2009). We then applied the same cross-validation procedure as before to evaluate the performance of the algorithms (Fig. 7b). Not surprisingly, we found that all algorithms based on this or a similar generative model (PP13, VP10, YF06) performed well. Interestingly, even the algorithms that performed least well for the real data (OD13, VP09) showed good performance on the artificial data. The STM model was among the top-performing algorithms, in contrast to the other supervised learning algorithm (SI08). A direct comparison of the performance on the simulated dataset and the experimental data clearly illustrates that the former is not a good predictor of the latter (Fig. 7c).
Discussion
Here we provide a benchmark comparison of different algorithms for spike rate inference from calcium imaging recordings on ground truth data. We evaluate the algorithms for a wide range of recording conditions including OGB-1 and GCamp6 as calcium indicators, anesthetized and awake imaging, different scanning techniques, neural tissues, and with respect to different metrics. In addition, we introduced a new algorithm for inferring spikes from calcium traces based on supervised training of a flexible probabilistic model and showed that this model performs currently better than all previously published algorithms for this problem under most conditions. Importantly, once trained, inferring spike rates using our algorithm is very fast, so even very large datasets can be processed rapidly. Interestingly, two of the three best algorithms rely on supervised learning to infer the relationship between calcium signal and spikes, suggesting that a data-driven approach offers distinct advantages over approaches based on forward models of the relationship between the two signals.
The superior performance of our algorithm carried over to new datasets not seen during training, promising good spike inference performance even when applied to a new dataset where no simultaneous recordings are available. To use the algorithm ‘out of the box’, we provide it for download pre-trained with all experimental data used in this paper (see Methods). In particular, its performance carried over to data recorded in awake animals, where brain movements or brain state fluctuations may render spike inference more difficult. In our recordings, motion in the Z-axis was small, on the order of 1–2 μm (Reimer et al., 2014 their Supplementary Information); if there was more brain movement in a given preparation and thus more neuropil contamination, generalization may be impaired. In addition, changing brain states during movement of the mouse compared to quiet restfulness (Niell and Stryker, 2010) may change the relationship between spikes and calcium signals. While we did not observe such effects in our data (Fig. 7), it is certainly possible that they will become apparent with more data from awake animals with more frequent periods of running (here only ~20% of the data).
The fact that our algorithm can be used without extra training data is crucial, as this is often considered an important advantage of algorithms based on generative models. Note that for entirely new experimental conditions (e.g. a new calcium indicator), the performance of neither class of algorithms is guaranteed, however, and both need to be evaluated on a dataset with simultaneous recordings. For unsupervised methods, if such an evaluation reveals poor performance, e.g. because the assumed generative model does not match the structure of the dataset at hand (as seen e.g. with the GCamp6 data; Fig. 1e and 2), the only way to improve the algorithm would be to adapt the generative model and modify the inference procedures accordingly. In contrast, any simultaneous data collected in the future can be readily used to retrain our supervised algorithm and further improve its spike prediction and generalization performance. In fact, our choice of the spike triggered mixture model for estimating spike rates from calcium traces is motivated by its ability to automatically switch between different sub-models whenever the statistics of the data change (Theis et al., 2013). This property of the model might also allow the algorithm to accommodate different spike-calcium relationships in different brain states in awake animals, if they were to be found with more data from awake animals.
Interestingly, our evaluation shows that the correlation between inferred and real spike rates obtained at a temporal resolution of 40 ms is at best 0.4–0.6, depending on the dataset with substantial variability between cells (Fig. 5c–d). This means that so far even the best spike inference algorithms make a substantial amount of errors, and one should be aware that for population imaging the inferred rates correspond to fairly coarse estimates of the true spike trains. It will be an interesting question whether new algorithmic ideas, new indicators (Chen et al., 2013; Inoue et al., 2014; St-Pierre et al., 2014; Thestrup et al., 2014) or scanning techniques will bring these values closer to 1, or whether these low correlations reflect a general limitation of population imaging approaches. Factors contributing to this limitation may include technical aspects of the imaging procedure such as neuropil contamination or activity-induced changes in blood vessel diameter and biophysical issues connected to the intracellular calcium dynamics. Our evaluation further shows that good spike inference performance on model data by no means guarantees good performance on real population imaging data (Fig. 8). We believe theoretical model based studies (Lütcke et al., 2013; Wilt et al., 2013) will remain useful to systematically explore how performance depends on model parameters, such as noise level or violations of the generative model, but will need to be followed up by systematic quantitative benchmark comparisons on datasets such as provided here.
Figure 8. Evaluating algorithms on artificial data.
a) Example trace sampled from a generative model, true spikes and binned rate as well as reconstructed spike rate from four different algorithms (conventions as in Fig. 1). Numbers on the right denote correlations between true and inferred spike trains.
b) Correlation (mean ± 2 SEM for repeated measure designs) and information gain computed on a simulated dataset with 20 traces. For algorithms see legend.
c) Scatter plot comparing performance on simulated data with that on real data (averaged over cells from all datasets collected under anesthesia/ex-vivo), suggesting little predictive value of performance on simulated data.
Our proposed method is solely concerned with the problem of spike inference, and does not infer the regions of interests (ROIs) from observed data or infers tuning properties of neurons simultaneously. Recently, several methods have been proposed to jointly infer ROIs and spikes (Diego and Hamprecht, 2014; Maruyama et al., 2014; Pnevmatikakis et al., 2016). These methods have the benefit that they exploit the full spatio-temporal structure of the problem of spike inference in calcium imaging and offer an unbiased approach for ROI placement. Since ROIs can also be placed using supervised learning (Valmianski et al., 2010), it should be feasible to develop supervised paradigms for simultaneous ROI placement and spike inference or combinations of unsupervised and supervised methods. Likewise, a recent study has combined spike rate inference with the estimation of response properties of neurons, such as tuning functions (Ganmor et al., 2016) and it would be interesting to evaluate the use of supervised techniques for this problem as well.
We presented the first quantitative benchmarking approach to evaluating spike inference algorithms on a large dataset of population imaging data. We believe that such a benchmarking approach can also be an important catalyst for improvements on various computational problems in neuroscience, from systems identification to neuron reconstruction, as it is already used successfully in machine learning and related fields to drive new algorithmic developments. To catalyze the development of better spike inference algorithms for calcium imaging data, we will organize a competition, which will be announced separately.
Methods
Experimental procedures
Datasets 1 and 2: Primary visual cortex (V1) – OGB-1
We recorded calcium traces from neural populations loaded with Oregon green BAPTA-1 (OGB-1, Invitrogen) as calcium indicator in layer 2/3 of anesthetized wild type mice (male C57CL/6J, age: p40–p60) with a custom-built two-photon microscope using previously described methods (Cotton et al., 2013; Froudarakis et al., 2014). We used glass pipettes for targeted two-photon-guided loose cell patching of single cells. More details are provided in the Supplementary Material. All procedures performed on mice were conducted in accordance with the ethical guidelines of the National Institutes of Health and were approved by the Baylor College of Medicine IACUC.
Datasets 3 and 5: Primary visual cortex (V1) – GCaMP6
We recorded calcium traces from neural populations in layer 2/3 of (1) isoflurane-anesthetized and (2) awake wild type mice (male C57CL/6J, age: 2–8 months; N=2 and N=1 mice for anesthetized and awake, respectively) using a resonant scanning microscope (ThorLabs) using methods described previously (Reimer et al., 2014). During awake experiments, the mouse was placed on a treadmill with its head restrained beneath the microscope objective (Reimer et al., 2014). Simultaneous loose-patch and two-photon calcium imaging recordings were conducted as described above. Data was split into segments involving movement or no movement by thresholding velocity traces. More details are provided in the Supplementary Material.
Dataset 4: Retina
Imaging experiments in whole-mount retina of dark-adapted wild-type mice (both genders, C57BL/6J, p21–42) electroporated with OGB-1 were performed as described previously (Briggman and Euler, 2011) using a MOM-type two-photon microscope (Euler et al., 2009). For juxtacellular spike recordings, OGB-1 labeled somata were targeted with a glass-pipette under dim IR illumination to establish a loose (<1GΩ) seal. All procedures were performed in accordance with the law on animal protection (Tierschutzgesetz) issued by the German Federal Government and were approved by the institutional animal welfare committee of the University of Tübingen. More details are provided in the Supplementary Material.
Dataset from Svoboda lab
We used a publicly available dataset provided by the GENIE project, Svoboda lab, at Janelia farm on crcns.org (Akerboom et al., 2012; Chen et al., 2013; Svoboda, 2014). This dataset contains 9 cells recorded with GCaMP5, 11 cells recorded with GCaMP6f and 9 cells recorded with GCaMP6s. The total number of spikes was 2735, 4536 and 2123, respectively, and therefore much lower than for our datasets. Typically, these cells were recorded focusing on a single cell rather than recording from an entire population with lower zoom as in our dataset. For a detailed description of the data, see (Akerboom et al., 2012; Chen et al., 2013).
Preprocessing
We resampled all fluorescence traces and spike trains to 100 Hz (using scipy.signal.resample from the SciPy Python package). This allowed us to apply models across datasets independent of which dataset was used for training. We removed linear trends from the fluorescence traces by fitting a robust linear regression with Gaussian scale mixture residuals. That is, for each fluorescence trace Ft, we found parameters a, b, πk, and σk with maximal likelihood under the model
and computed F̃t = Ft − at − b. We used three different noise components (K = 3). Afterwards, we normalized the traces such that the 5th percentile of each trace’s fluorescence distribution is at zero, and the 80th percentile is at 1. Normalizing by percentiles instead of the minimum and maximum is more robust to outliers and less dependent on the firing rate of the neuron producing the fluorescence.
Supervised learning in flexible probabilistic models for spike inference
We predict the number of spikes kt falling in the t-th time bin of a neuron’s spike train based on 1000 ms windows of the fluorescence trace centered around t (preprocessed fluorescence snippets xt). We reduced the dimensionality of the fluorescence windows via PCA, keeping at least 95% of the variance (resulting in 8 to 20 dimensions). Keeping 99% of the variance and slightly regularizing the model’s parameters gave similar results. Only for the Svoboda dataset we found it was necessary to keep 99% of the variance to achieve optimal results.
We assume that the spike counts kt given the preprocessed fluorescence snippets xt can be modeled using a Poisson distribution,
We tested three models for the firing rate λ(xt) function:
-
A spike-triggered mixture (STM) model (Theis et al., 2013) with exponential nonlinearity,
where wk are linear filters, um are quadratic filters weighted by βkm for each of K components, and bk is a offset for each component. We used three components and two quadratic features (K = 3, M = 2). The performance of the algorithm was not particularly sensitive to the choice of these parameters (we evaluated K = 1, … 4 and M = 1, …, 4 in a grid search using one dataset).
-
As a simpler alternative, we use the linear-nonlinear-Poisson (LNP) neuron with exponential nonlinearity,
where w is a linear filter and b is an offset.
-
As a more flexible alternative, we used a multi-layer neural network (ML-NN) with two hidden layers,
where g(y) = max(0, y) is a point-wise rectifying nonlinearity and W1 and W2 are matrices. We tested MLPs with 10 and 5 hidden units, and 5 and 3 hidden units for the first and second hidden layer, respectively. Again, the performance of the algorithm was not particularly sensitive to these parameters.
Parameters of all models were optimized by maximizing the average log-likelihood for a given training set,
using limited-memory BFGS (Byrd et al., 1995), a standard quasi-Newton method. To increase robustness against potential local optima in the likelihood of the STM and the ML-NN, we trained four models with randomly initialized parameters and geometrically averaged their predictions. The geometric average of several Poisson distributions again yields a Poisson distribution whose rate parameter is the geometric average of the rate parameters of the individual Poisson distributions.
Other algorithms
SI08
This approach is based on applying a support-vector machine (SVM) on two PCA features of preprocessed segments of calcium traces. We re-implemented the features following closely the procedures described in (Sasaki et al., 2008). As the prediction signal, we used the distance of the input features to the SVM’s separating hyperplane, setting negative predictions to zero. We cross-validated the regularization parameter of the SVM but found that it had little impact on performance.
PP13
The algorithm performs Bayesian inference in a generative model, using maximum a posteriori (MAP) estimates for spike inference and MCMC on a portion of the calcium trace for estimating hyperparameters. We used a Matlab implementation provided by the authors of (Pnevmatikakis et al., 2013), which has contributed to the later published (Pnevmatikakis et al., 2016). We also tried selecting the hyperparameters through cross-validation, which did not substantially change the overall results.
VP10
The fast-oopsi or non-negative deconvolution technique constrains the inferred spike rates to be positive (Vogelstein et al., 2010), performing approximate inference in a generative model. We used the implementation provided by the author 1. We adjusted the hyperparameters using cross-validation by performing a search over a grid of 54 parameter sets controlling the degree of assumed observation noise and the expected number of spikes (Fig. 2a–b). In Fig. 5b–c the hyperparameters were instead directly inferred from the calcium traces by the algorithm.
YF06
The deconvolution algorithm (Yaksi and Friedrich, 2006) removes noise by local smoothing and the inverse filter resulting from the calcium transient. We used a Matlab implementation provided by the authors. Using the cross-validation procedure outlined above, we automatically tuned the algorithm by testing 66 different parameter sets. The parameters controlled the cutoff frequency of a low-pass filter, a time constant of the filter used for deconvolution, and whether or not an iterative smoothing procedure was applied to the fluorescence traces.
OD13
This algorithm performs a template-matching based approach by using the finite rate of innovation-theory as described in (Oñativia et al., 2013). We used the implementation provided on the author’s homepage2. We adjusted the exponential time constant parameter using cross-validation.
VP09
This algorithm performs Bayesian inference in a generative model as described in (Vogelstein et al., 2009). We used the implementation provided by the author3. Since this algorithm is based on the same generative model as fast-oopsi but is much slower, we used the hyperparameters inferred by cross-validating fast-oopsi in Fig. 2a–b and the hyperparameters automatically inferred by the algorithm in Fig. 5b–c.
Performance evaluation
Unless otherwise noted, we evaluated the performance of the algorithms on spike trains binned at 40 ms resolution. For Fig. 3 and Suppl. Fig. 2, we changed the bin width between 10 ms and 500 ms. We used cross-validation to evaluate the performance of our framework, i.e. we estimated the parameters of our model on a training set, typically consisting of all but one cell for each dataset, and evaluated its performance on the remaining cell. This procedure was iterated such that each cell was held out as a test cell once. Results obtained using the different training and test sets were subsequently averaged.
Correlation
We computed the linear correlation coefficient between the true binned spike train and the inferred one. This is a widely used measure with a simple and intuitive interpretation, taking the overall shape of the spike density function into account. However, the correlation coefficient is invariant under affine transformations, which means that predictions optimized for this measure cannot be directly interpreted as spike counts or firing rates. In further contrast to information gain, it also does not take the uncertainty of the predictions into account. That is, a method which predicts the spike count to be 5 with absolute certainty will be treated the same as a method which experts the spike count to be somewhere between 0 and 10 assigning equal probability to each possible outcome.
Information gain
The information gain provides a model based estimate of the amount of information about the spike train extracted from the calcium trace. Unlike AUC and correlation, it takes into account the uncertainty of the prediction.
Assuming an average firing rate of λ and a predicted firing rate of λt at time t, the expected information gain (in bits per bin) can be estimated as
assuming Poisson statistics and independence of spike counts in different bins. The estimated information gain is bounded from above by the (unknown) amount of information about the spike train contained in the calcium trace, as well as by the marginal entropy of the spike train, which can be estimated using
We computed a relative information gain by dividing the information gain averaged over all cells by the average estimated entropy,
where is the information gain measured for the n-th cell in the dataset.
This can be interpreted as the fraction of entropy in the data explained away by the model (measured in percent points). Since only our method was optimized to yield Poisson firing rates, we allowed all methods a single monotonically increasing nonlinear function, which we optimized to maximize the average information gain over all cells. That is, we evaluated
where f is a piecewise linear monotonically increasing function optimized to maximize the information gain averaged over all cells (using an SLSQP implementation in SciPy).
AUC
The AUC score can be computed as the probability that a randomly picked prediction for a bin containing a spike is larger than a randomly picked prediction for a bin containing no spike (Fawcett, 2006). While this is a commonly used score for evaluating spike inference procedures (Vogelstein et al., 2010), it is not sensitive to changes in the relative height of different parts of the spike density function, as it is invariant under arbitrary strictly monotonically increasing transformations. For example, if predicted rates were squared, high rates would be over proportionally boosted compared to low rates, while yielding equivalent AUC scores.
Statistical analysis
We used generalized Loftus & Masson standard errors of the means for repeated measure designs (Franz and Loftus, 2012) and report the mean ± 2 SEM. To assess statistical significance, we compare the performance of the STM model to the performance of its next best competitor, performing a one-sided Wilcoxon signed rank test and report significance or the respective p-value above a line spanning the respective columns. If the STM is not the best model, we perform the comparison between the best model and the STM. We fitted a Gaussian Process model with a Gaussian kernel in Fig. 5c and d using the implementation provided by scikit-learn. The kernel width is chosen automatically via maximum-likelihood estimate (Pedregosa et al., 2011).
Generation of artificial data
We simulated data by sampling from the generative model used by Vogelstein et al. (2010). That is, we first generated spike counts by independently sampling each bin of a spike train from a Poisson distribution, then convolving the spike train with an exponential kernel to arrive at an artificial calcium concentration, and finally adding Poisson noise to generate a Fluorescence signal xt.
The firing rate λ for each cell was randomly chosen to be between 0 and 400 spikes per second. The parameters γ, a, and b were fixed to 0.98, 100 and 1, respectively, and data was generated at a sampling rate of 100 Hz.
Code availability
We provide a Python implementation of our algorithm online (https://github.com/lucastheis/c2s). The package includes a pre-trained version of our algorithm, which is readily usable even without simultaneous recordings and has been trained on our entire dataset. The pre-trained algorithm has been trained on all five datasets presented in this paper as well as the publicly available data from the Svoboda lab. To accommodate the wider range of data, we made the model slightly more flexible allowing 6 linear and 4 quadratic components as well as accounting for 99% of the variance in the dimensionality reduction step.
Supplementary Material
Highlights.
We evaluate algorithms for spike inference from two-photon calcium recordings.
A new supervised algorithm performs best across neural tissues and indicators.
Its performance transfers to new datasets without a need for retraining.
Simulated data is not informative about performance on real data.
Acknowledgments
We thank J. Vogelstein, R. Friedrich, E. Pnevtmatikakis and P. Dragotti for making the code for their algorithms available.
This work was supported by the German Federal Ministry of Education and Research (BMBF) through the Bernstein Center for Computational Neuroscience (FKZ 01GQ1002 to T.E., M.B. and A.S.T.); the Deutsche Forschungsgemeinschaft (DFG) through grant BE3848-1 to M.B., BE 5601/1-1 to PB and BA 5283/1-1 to T.B.; the Werner Reichardt Centre for Integrative Neuroscience Tübingen (EXC307); grants DP1EY023176, DP1OD008301, P30EY002520, T32EY07001, the McKnight Scholar Award and the Arnold and Mabel Beckman Foundation Young Investigator Award to A.S.T.
Footnotes
Author contributions
PB, MB and LT designed the project. LT analyzed the data with input from PB. MF, JR and AST acquired V1 data. MR, TB and TE acquired retinal data. PB wrote the paper with input from all authors. PB and MB supervised the project.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akerboom J, Chen TW, Wardill TJ, Tian L, Marvin JS, Mutlu S, Calderon NC, Esposti F, Borghuis BG, Sun XR, Gordus A, Orger MB, Portugues R, Engert F, Macklin JJ, Filosa A, Aggarwal A, Kerr RA, Takagi R, Kracun S, Shigetomi E, Khakh BS, Baier H, Lagnado L, Wang SSH, Bargmann CI, Kimmel BE, Jayaraman V, Svoboda K, Kim DS, Schreiter ER, Looger LL. Optimization of a GCaMP Calcium Indicator for Neural Activity Imaging. J Neurosci. 2012;32:13819–13840. doi: 10.1523/JNEUROSCI.2601-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggman KL, Euler T. Bulk electroporation and population calcium imaging in the adult mammalian retina. J Neurophysiol. 2011;105:2601–9. doi: 10.1152/jn.00722.2010. [DOI] [PubMed] [Google Scholar]
- Byrd RH, Lu P, Nocedal J, Zhu C. A Limited Memory Algorithm for Bound Constrained Optimization. SIAM J Sci Comput. 1995 doi: 10.1137/0916069. [DOI] [Google Scholar]
- Chen TW, Wardill TJ, Sun Y, Pulver SR, Renninger SL, Baohan A, Schreiter ER, Kerr Ra, Orger MB, Jayaraman V, Looger LL, Svoboda K, Kim DS. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature. 2013;499:295–300. doi: 10.1038/nature12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotton RJ, Froudarakis E, Storer P, Saggau P, Tolias AS. Three-dimensional mapping of microcircuit correlation structure. Front Neural Circuits. 2013;7:151. doi: 10.3389/fncir.2013.00151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denk W, Strickler J, Webb W. Two-photon laser scanning fluorescence microscopy. Science (80- ) 1990;248:73–76. doi: 10.1126/science.2321027. [DOI] [PubMed] [Google Scholar]
- Diego F, Hamprecht FA. Sparse space-time deconvolution for calcium image analysis. Neural Information Processing Systems. 2014:1–9. [Google Scholar]
- Euler T, Hausselt SE, Margolis DJ, Breuninger T, Castell X, Detwiler PB, Denk W. Eyecup scope--optical recordings of light stimulus-evoked fluorescence signals in the retina. Pflugers Arch. 2009;457:1393–414. doi: 10.1007/s00424-008-0603-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawcett T. An introduction to ROC analysis. Pattern Recognit Lett. 2006;27:861–874. doi: 10.1016/j.patrec.2005.10.010. [DOI] [Google Scholar]
- Franz VH, Loftus GR. Standard errors and confidence intervals in within-subjects designs: generalizing Loftus and Masson (1994) and avoiding the biases of alternative accounts. Psychon Bull Rev. 2012;19:395–404. doi: 10.3758/s13423-012-0230-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froudarakis E, Berens P, Ecker AS, Cotton RJ, Sinz FH, Yatsenko D, Saggau P, Bethge M, Tolias AS. Population code in mouse V1 facilitates readout of natural scenes through increased sparseness. Nat Neurosci. 2014;17:851–857. doi: 10.1038/nn.3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganmor E, Krumin M, Rossi LF, Carandini M, Simoncelli EP. Direct Estimation of Firing Rates from Calcium Imaging Data. 2016:1–34. arXiv/q-bio.NC. [Google Scholar]
- Greenberg DS, Houweling AR, Kerr JND. Population imaging of ongoing neuronal activity in the visual cortex of awake rats. Nat Neurosci. 2008;11:749–51. doi: 10.1038/nn.2140. [DOI] [PubMed] [Google Scholar]
- Grewe BF, Langer D, Kasper H, Kampa BM, Helmchen F. High-speed in vivo calcium imaging reveals neuronal network activity with near-millisecond precision. Nat Methods. 2010;7:399–405. doi: 10.1038/nmeth.1453. [DOI] [PubMed] [Google Scholar]
- Inoue M, Takeuchi A, Horigane S, Ohkura M, Gengyo-Ando K, Fujii H, Kamijo S, Takemoto-Kimura S, Kano M, Nakai J, Kitamura K, Bito H. Rational design of a high-affinity, fast, red calcium indicator R-CaMP2. Nat Methods. 2014;12 doi: 10.1038/nmeth.3185. [DOI] [PubMed] [Google Scholar]
- Kerr JND, Denk W. Imaging in vivo: watching the brain in action. Nat Rev Neurosci. 2008;9:195–205. doi: 10.1038/nrn2338. [DOI] [PubMed] [Google Scholar]
- Kerr JND, Greenberg D, Helmchen F. Imaging input and output of neocortical networks in vivo. Proc Natl Acad Sci U S A. 2005;102:14063–8. doi: 10.1073/pnas.0506029102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lütcke H, Gerhard F, Zenke F, Gerstner W, Helmchen F. Inference of neuronal network spike dynamics and topology from calcium imaging data. Front Neural Circuits. 2013;7:201. doi: 10.3389/fncir.2013.00201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama R, Maeda K, Moroda H, Kato I, Inoue M, Miyakawa H, Aonishi T. Detecting cells using non-negative matrix factorization on calcium imaging data. Neural Netw. 2014;55:11–9. doi: 10.1016/j.neunet.2014.03.007. [DOI] [PubMed] [Google Scholar]
- Niell CM, Stryker MP. Modulation of visual responses by behavioral state in mouse visual cortex. Neuron. 2010;65:472–9. doi: 10.1016/j.neuron.2010.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oñativia J, Schultz SR, Dragotti PL. A finite rate of innovation algorithm for fast and accurate spike detection from two-photon calcium imaging. J Neural Eng. 2013;10:046017. doi: 10.1088/1741-2560/10/4/046017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park IJ, Bobkov YV, Ache BW, Principe JC. Quantifying bursting neuron activity from calcium signals using blind deconvolution. J Neurosci Methods. 2013;218:196–205. doi: 10.1016/j.jneumeth.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V, Vanderplas J, Passos A, Cournapeau D, Brucher M, Perrot M, Duchesnay É. Scikit-learn: Machine Learning in Python. J Mach Learn Res. 2011;12:2825–2830. [Google Scholar]
- Pnevmatikakis EA, Merel J, Pakman A, Paninski L. Bayesian spike inference from calcium imaging data, in: 2013 Asilomar Conference on Signals, Systems and Computers. IEEE. 2013:349–353. doi: 10.1109/ACSSC.2013.6810293. [DOI] [Google Scholar]
- Pnevmatikakis EA, Soudry D, Gao Y, Machado TA, Merel J, Pfau D, Reardon T, Mu Y, Lacefield C, Yang W, Ahrens M, Bruno R, Jessell TM, Peterka DS, Yuste R, Paninski L. Simultaneous Denoising, Deconvolution, and Demixing of Calcium Imaging Data. Neuron. 2016:1–15. doi: 10.1016/j.neuron.2015.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimer J, Froudarakis E, Cadwell CR, Yatsenko D, Denfield GH, Tolias AS. Pupil Fluctuations Track Fast Switching of Cortical States during Quiet Wakefulness. Neuron. 2014;84:355–362. doi: 10.1016/j.neuron.2014.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki T, Takahashi N, Matsuki N, Ikegaya Y. Fast and accurate detection of action potentials from somatic calcium fluctuations. J Neurophysiol. 2008;100:1668–76. doi: 10.1152/jn.00084.2008. [DOI] [PubMed] [Google Scholar]
- Stosiek C, Garaschuk O, Holthoff K, Konnerth A. In vivo two-photon calcium imaging of neuronal networks. Proc Natl Acad Sci U S A. 2003;100:7319–24. doi: 10.1073/pnas.1232232100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Pierre F, Marshall JD, Yang Y, Gong Y, Schnitzer MJ, Lin MZ. High-fidelity optical reporting of neuronal electrical activity with an ultrafast fluorescent voltage sensor. Nat Neurosci. 2014;17:884–9. doi: 10.1038/nn.3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svoboda K GENIE P. at J.F. Simultaneous imaging and loose-seal cell-attached electrical recordings from neurons expressing a variety of genetically encoded calcium indicators. 2014 http://dx.doi.org/10.6080/K02R3PMN.
- Theis L, Chagas AM, Arnstein D, Schwarz C, Bethge M. Beyond GLMs: a generative mixture modeling approach to neural system identification. PLoS Comput Biol. 2013;9:e1003356. doi: 10.1371/journal.pcbi.1003356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thestrup T, Litzlbauer J, Bartholomäus I, Mues M, Russo L, Dana H, Kovalchuk Y, Liang Y, Kalamakis G, Laukat Y, Becker S, Witte G, Geiger A, Allen T, Rome LC, Chen TW, Kim DS, Garaschuk O, Griesinger C, Griesbeck O. Optimized ratiometric calcium sensors for functional in vivo imaging of neurons and T lymphocytes. Nat Methods. 2014;11:175–82. doi: 10.1038/nmeth.2773. [DOI] [PubMed] [Google Scholar]
- Valmianski I, Shih AY, Driscoll JD, Matthews DW, Freund Y, Kleinfeld D. Automatic identification of fluorescently labeled brain cells for rapid functional imaging. J Neurophysiol. 2010;104:1803–1811. doi: 10.1152/jn.00484.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein JT, Packer AM, Machado Ta, Sippy T, Babadi B, Yuste R, Paninski L. Fast nonnegative deconvolution for spike train inference from population calcium imaging. J Neurophysiol. 2010;104:3691–704. doi: 10.1152/jn.01073.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein JT, Watson BO, Packer AM, Yuste R, Jedynak B, Paninski L. Spike inference from calcium imaging using sequential Monte Carlo methods. Biophys J. 2009;97:636–55. doi: 10.1016/j.bpj.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilt Ba, Fitzgerald JE, Schnitzer MJ. Photon shot noise limits on optical detection of neuronal spikes and estimation of spike timing. Biophys J. 2013;104:51–62. doi: 10.1016/j.bpj.2012.07.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaksi E, Friedrich RW. Reconstruction of firing rate changes across neuronal populations by temporally deconvolved Ca2+ imaging. Nat Methods. 2006;3:377–83. doi: 10.1038/nmeth874. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.