Abstract
Background
Literature-based discovery (LBD) is characterized by uncovering hidden associations in non-interacting scientific literature. Prior approaches to LBD include use of: 1) domain expertise and structured background knowledge to manually filter and explore the literature, 2) distributional statistics and graph-theoretic measures to rank interesting connections, and 3) heuristics to help eliminate spurious connections. However, manual approaches to LBD are not scalable and purely distributional approaches may not be sufficient to obtain insights into the meaning of poorly understood associations. While several graph-based approaches have the potential to elucidate associations, their effectiveness has not been fully demonstrated. A considerable degree of a priori knowledge, heuristics, and manual filtering is still required.
Objectives
In this paper we implement and evaluate a context-driven, automatic subgraph creation method that captures multifaceted complex associations between biomedical concepts to facilitate LBD. Given a pair of concepts, our method automatically generates a ranked list of subgraphs, which provide informative and potentially unknown associations between such concepts.
Methods
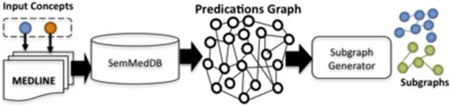
To generate subgraphs, the set of all MEDLINE articles that contain either of the two specified concepts (A, C) are first collected. Then binary relationships or assertions, which are automatically extracted from the MEDLINE articles, called semantic predications, are used to create a labeled directed predications graph. In this predications graph, a path is represented as a sequence of semantic predications. The hierarchical agglomerative clustering (HAC) algorithm is then applied to cluster paths that are bounded by the two concepts (A, C). HAC relies on implicit semantics captured through Medical Subject Heading (MeSH) descriptors, and explicit semantics from the MeSH hierarchy, for clustering. Paths that exceed a threshold of semantic relatedness are clustered into subgraphs based on their shared context. Finally, the automatically generated clusters are provided as a ranked list of subgraphs.
Results
The subgraphs generated using this approach facilitated the rediscovery of 8 out of 9 existing scientific discoveries. In particular, they directly (or indirectly) led to the recovery of several intermediates (or B-concepts) between A- and C-terms, while also providing insights into the meaning of the associations. Such meaning is derived from predicates between the concepts, as well as the provenance of the semantic predications in MEDLINE. Additionally, by generating subgraphs on different thematic dimensions (such as Cellular Activity, Pharmaceutical Treatment and Tissue Function), the approach may enable a broader understanding of the nature of complex associations between concepts. Finally, in a statistical evaluation to determine the interestingness of the subgraphs, it was observed that an arbitrary association is mentioned in only approximately 4 articles in MEDLINE on average.
Conclusion
These results suggest that leveraging the implicit and explicit semantics provided by manually assigned MeSH descriptors is an effective representation for capturing the underlying context of complex associations, along multiple thematic dimensions in LBD situations.
Keywords: Literature-based discovery (LBD), Graph mining, Path clustering, Hierarchical agglomerative clustering, Semantic Similarity, Semantic relatedness, Medical Subject Headings (MeSH)
Graphical abstract
1. Introduction
Literature-based discovery (LBD) refers to the process of uncovering hidden connections that are implicit in scientific literature. Numerous hypotheses have been generated from scientific literature, using the LBD paradigm, which influenced innovations in diagnosis, treatment, preventions, and overall public health. The notion of LBD was proposed by Don R. Swanson (1924–2012) in 1986, through the well-known Raynaud Syndrome–Dietary Fish Oils Hypothesis (RS-DFO) [1]. By reading the titles of more than 4000 MEDLINE articles, Swanson serendipitously discovered that Dietary Fish Oils (DFO) lower Blood Viscosity, reduce Platelet Aggregation and inhibit Vascular Reactivity (specifically Vasoconstriction). Concomitantly, he observed that a reduction in both Blood Viscosity and Platelet Aggregation, as well as the inhibition of Vasoconstriction, appeared to prevent Raynaud Disease; a circulatory disorder that causes periods of severely restricted blood flow to the fingers and toes [2]. Swanson therefore postulated that “dietary fish oil might ameliorate or prevent Raynaud’s syndrome.” This hypothesis was clinically confirmed by DiGiacomo et al. [3] in 1989.
Swanson’s discovery is interesting because explicit associations between DFO and these intermediate concepts (i.e., Blood Viscosity, Platelet Aggregation and Vasoconstriction) had long existed in the literature [4, 5, 6, 7, 8]. Likewise, explicit associations between the intermediates and RS had been well documented [9, 2]. The serendipity in Swanson’s Hypothesis lies in the fact that no explicit associations linking DFO and RS directly had been previously articulated in a single document.
To develop this hypothesis, Swanson performed a Dialog® Scisearch using Raynaud and Fish Oil terms, on titles and abstracts of MEDLINE and Em-base (Excepta Medica) citations, in November 1985. There were approximately 1000 articles in the Raynaud set and 3000 in the Fish Oil set. He found that only four articles among a reduced set of 489 articles (after filtering), contained cross-references spanning both sets. Among these four articles, only two articles [10, 11] discussed relevant aspects of RS with DFO; although not in the context of Swanson’s discovery. Swanson speculated that this phenomenon of logically related but noninteracting literatures alludes to the existence of undiscovered public knowledge [1]. Logically related information fragments may exist in the literature, but may have never been connected, or fully elucidated. He subsequently exploited his awareness of the existence of such undiscovered associations and investigated several other scenarios (three with Smalheiser [12, 13, 14]) that later led to new scientific discoveries [15, 16]. Swanson grounded his observations in a paradigm now commonly known as the ABC model [1] for LBD, which is an integral part of LBD research, facilitating the generation of several hypotheses [1, 15, 16, 12, 13, 14, 17, 18, 19, 20, 21, 22, 23, 24, 25].
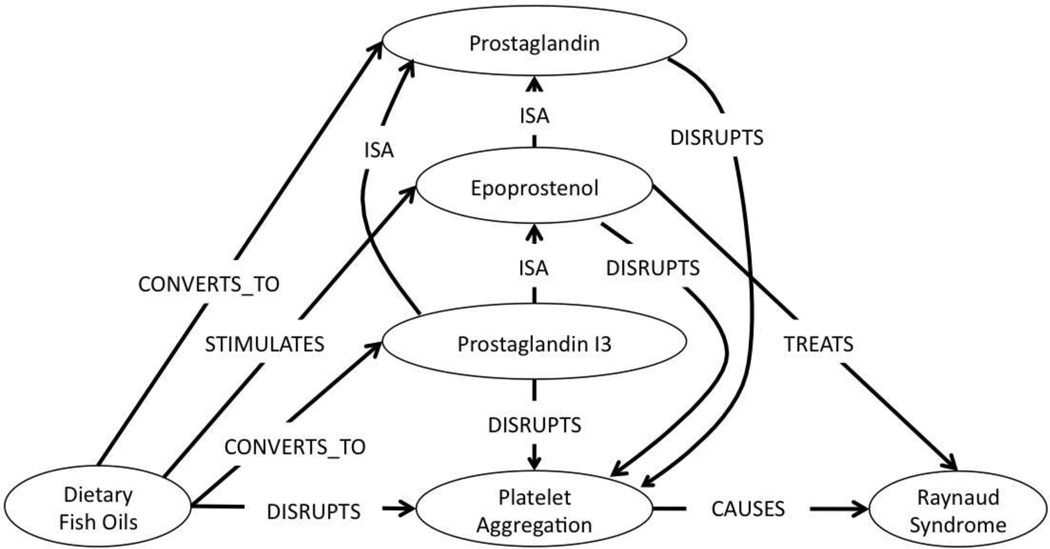
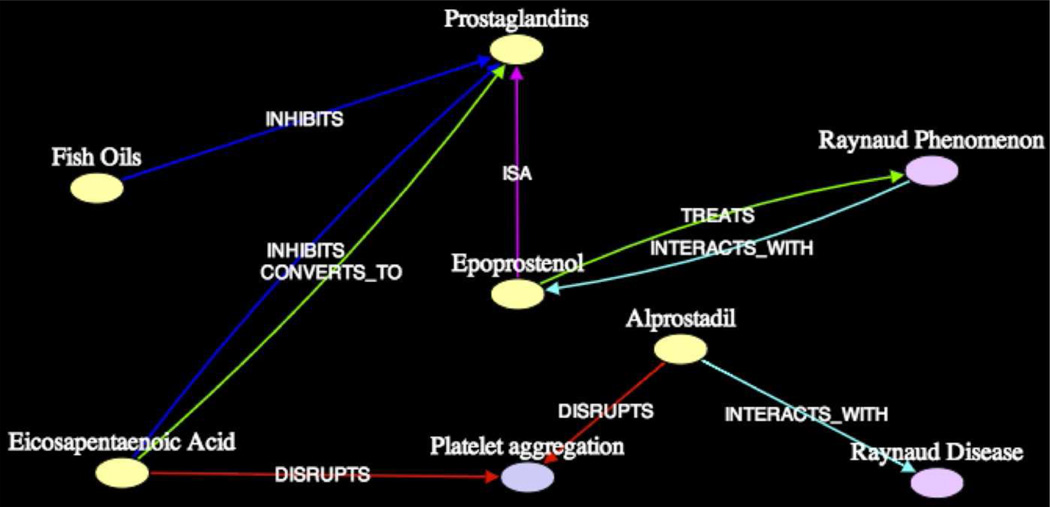
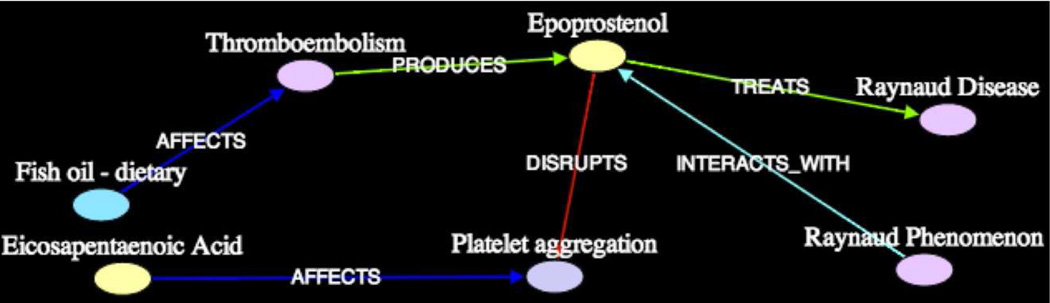
In current biomedical research, while finding unknown intermediates is an important task, domain scientists are often interested in developing a deeper understanding of causal relationships and mechanisms of interaction among concepts. For example, consider the complex scenario depicted in Figure 1, in which Dietary Fish Oils produce several Prostaglandins, including Prostaglandin I3 (PGI3) and Epoprostenol (PGI2, the synthetic form of Prostacyclin). The latter of these Prostaglandins (Epoprostenol) was known to treat Raynaud Syndrome. It was also known to disrupt Platelet Aggregation. Since Platelet Aggregation is deemed a cause of Raynaud Syndrome, one can reasonably conclude that a plausible mechanism by which Dietary Fish Oils treat Raynaud Syndrome is through the production of Prostaglandins, which actively disrupt Platelet Aggregation.
Figure 1.
Complex association between Dietary Fish Oils and Raynaud Syndrome
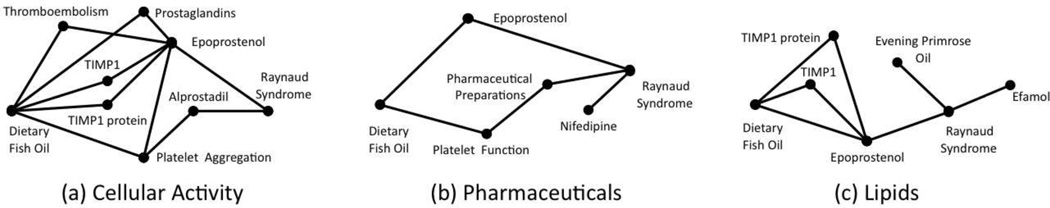
Aside from detecting such causal associations, it is known that complex associations may exist between concepts, in many different ways. For example, Figure 2 shows that Dietary Fish Oils and Raynaud Syndrome are associated in at least the following three ways: 1) in terms of Cellular Activity involving Blood platelets/Prostaglandins, as shown in Figure 2a, 2) through Pharmaceuticals that contain calcium channel blockers, such as Nifedipine and Verapamil, as shown in Figure 2b, and 3) through Lipids/Fatty Acids from Efamol and Evening primrose oil, as shown in Figure 2c.
Figure 2.
Thematic dimensions of association for Raynaud Syndrome and Dietary Fish Oil
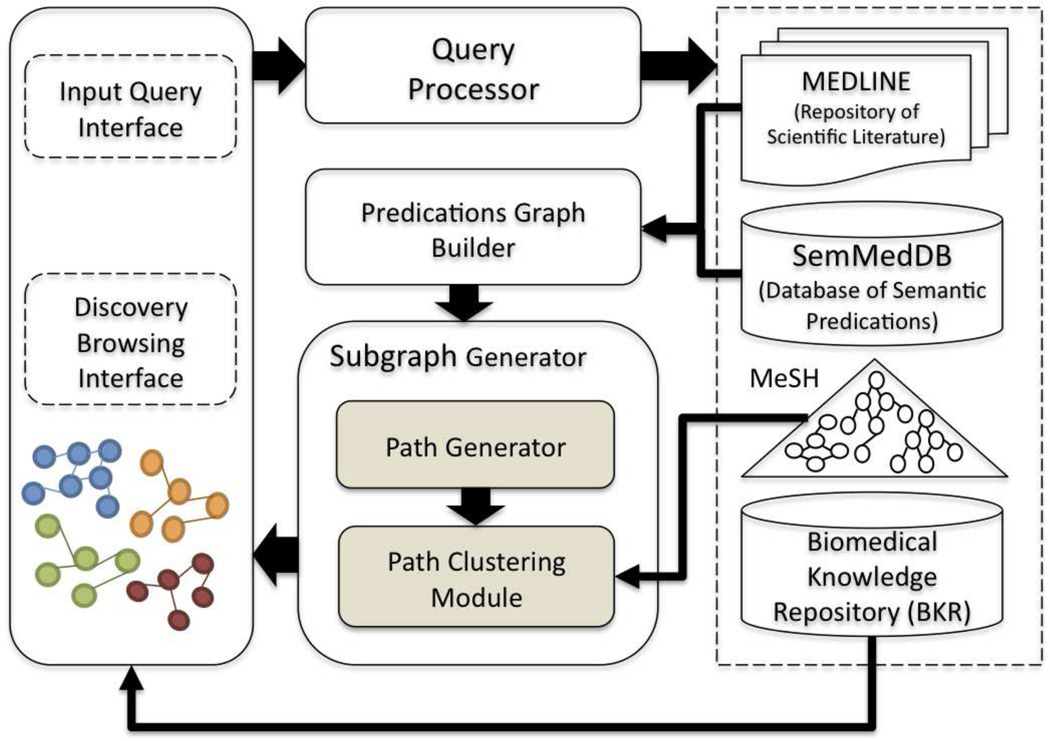
Figure 3.
System Architecture
In this paper, we build on our previous approach [26], in which we rediscovered and decomposed the Raynaud Syndrome – Dietary Fish Oils discovery. In our previous work, we manually created the multi-faceted subgraphs, by grouping together paths of semantic predications. Recall that a semantic predication is a binary relation between two concepts, expressed in the form (subject, predicate, object). Here, we present a method that uses rich representations to automatically create such subgraphs, by leveraging implicit and explicit semantics provided by MeSH descriptors1. To create the subgraphs, we first specify the context of a semantic predication and then use it to infer the context of a path. Paths are then clustered into coherent subgraphs on multiple thematic dimensions, based on their shared context.
The approach requires only three items from the user as input: 1) a list of concept labels for source (A) and target (C), 2) the maximum path length k of paths to be generated (default k = 2, for ABC associations), and 3) a cut-off date dt for articles to be included from the scientific literature. If no cut-off date is provided then all MEDLINE articles are used. The output of the approach is a ranked list of subgraphs S – i.e., create a function ℱ : q→ S, where q = {A, C, dt, k}.
To facilitate understanding the meaning of associations present in the subgraphs, the predicates of the semantic predications and their provenance in MEDLINE are provided (see Section 4). Relationships that are not explicit in the subgraphs, but are inferred, can be explored by composing MEDLINE queries (as we will show). The collective use of predicates, provenance and MEDLINE queries for knowledge exploration constitute the notion of discovery browsing, introduced by Wilkowski et al. [27] and extended by Cairelli et al. [28]. Discovery browsing is enabled when a system guides the user through their exploration of the literature in a process of cooperative reciprocity. The “user iteratively focuses system output, thus controlling the large number of relationships often generated in literature-based discovery systems.”
To assess the efficacy of our approach, two forms of evaluation were conducted: 1) an evidence-based evaluation and 2) a statistical evaluation. The evidence-based evaluation showed that the generated subgraphs could facilitate the re-discovery of 8 out of 9 existing discoveries [1, 15, 16, 12, 13, 14, 29, 30] (not recovered [28]). The statistical evaluation showed that an arbitrary association occurs only in approximately 4 articles in MEDLINE on average. This evaluation determines the interestingness of the subgraphs in general, as a way to assess whether a domain scientist might be interested in an arbitrary subgraph in the first place (see in Section 4.2). These results suggest that the subgraphs created using our approach provide an effective way of finding and elucidating poorly understood associations and may be of interest to domain scientists. In this paper we make the following specific contributions:
We develop a novel context-driven subgraph creation method for closed LBD (both A and C are known), capable of finding complex associations. Our approach is distinct from previous approaches, which are mainly based on statistical frequency, graph metrics, and specificity.
We implement an unsupervised clustering algorithm to automatically create complex subgraphs using implicit and explicit semantics, without the need for complex heuristics for filtering.
We illustrate the role of discovery browsing, through the use of predicates and provenance to supplement the subgraphs with insights from the scientific literature.
We show the effectiveness of this approach in facilitating the rediscovery of 8 out of 9 existing scientific discoveries.
The rest of this paper is organized as follows: Related Work is covered in Section 2. The approach to automatic subgraph creation is discussed in Section 3. Experimental Results are presented in Section 4 and a thorough discussion on limitations and future work are presented in Section 5. Conclusions are presented in Section 6.
2. Related Work
Leveraging rich representations of textual content from scientific literature could be effective for finding and elucidating complex associations. Rich representations exploit implicit, formal (or explicit) and powerful semantics [31] to capture context, which may be important in providing deeper insights into the nature of associations. Gordon and Dumais made this crucial observation in [32] after successfully applying the popular technique of Latent Semantic Indexing (LSI) for LBD. The authors reported that LSI was only slightly more effective than traditional frequency-based metrics, such as token frequency, record frequency, and term frequency-inverse global frequency (tf-igf) [33] for finding intermediates. While LSI was successful for knowledge rediscovery, the authors speculated that richer representations of textual content are needed to capture “evidence suggestive of ‘causal’ relationships in the literature (which may be revealed independently of their statistical prominence).” Moreover, they stressed the need for “semantic and category knowledge to improve the step of identifying [intermediate and] terminal concepts.”
Many techniques for finding hidden connections (or associations) between biomedical concepts from scientific literature however, utilize frequency-based and graph-theoretic metrics. Few methods have been developed to ‘seamlessly’ find and elucidate complex associations, by going beyond reliance on implicit semantics. Instead, the conventional wisdom has been that discoveries are likely to arise from logical connections between source (A) concepts, intermediates (B) and targets (C) that frequently or rarely (co)occur in the literature, or are highly or rarely connected in a knowledge base.
The earliest frequency-based approaches utilized ‘frequency of occurrence’ mainly through measures of term (and concept) frequency [34, 17]. Other measures such as relative frequency, token frequency, term frequency-inverse global frequency (tf-igf) [33], and term frequency-inverse document frequency (tf-idf) [35, 36] were also used to rank intermediates. Subsequent approaches utilized ‘frequency of co-occurrence’ using techniques such as LSI [32], association rules [37, 38, 39, 20], and probability distributions [20, 40, 41, 29]. Torvik et al. [40, 42] even used an ensemble approach to find intermediates that combined statistical and temporal features.
While distributional approaches have been successful for some LBD situations, the underlying frequencies only provide an indirect way of capturing the meaning of associations among concepts. For instance, consider the association in which Dietary Fish Oils (A) inhibit Platelet Aggregation (B) and the aggregation of blood platelets causes Raynaud Disease (C). While Dietary Fish Oils, Platelet Aggregation, and Raynaud Disease may frequently co-occur in the literature, their precise association is not explicitly captured by their co-occurrence. A second issue is that the underlying frequency distribution may not be adequate for capturing related concepts, which may be important in elucidating causal relationships and mechanisms of interaction.
To address these problems, several relations-based techniques [19, 22, 43] have been developed, which use the explicit relationships (or predicates) between concepts. Such predicates are typically obtained from structured background knowledge or known a priori by domain experts. For example, Hristovski et al. in [22], developed a relations-based approach that used ordered alternating sequences of predicates and classes (or semantic types) called discovery patterns. These patterns are specified a priori using insights from background knowledge. Using discovery patterns, Hristovski argues that if a Disease causes a change in a Substance/Body Function and a Drug inhibits this change, then the Drug MAYBE TREATS the Disease. The CAUSES-INHIBITS sequence is used to uncover potentially new Drug treatments for the Disease.
While intuitive, the relations-based approach is mainly applicable in scenarios where both predicates and semantic types are known, or can be easily obtained. This is not always trivial, as illustrated in the scenario from Figure 1. Additionally, it can be argued that hierarchical relations from the schema of a domain specific knowledge base, such as the Unified Medical Language System (UMLS) can also be used to create such complex subgraphs, using measures like specificity. However, the semantic types for Prostaglandins and Platelet Aggregation are Eicosanoids and Cell Function, respectively. These semantic types share no common ancestors in their lineage in the UMLS Semantic Network (https://uts.nlm.nih.gov/semanticnetwork.html). And while associative relations can be used instead, a proven and repeatable schema-driven approach that captures this level of complexity has not been forthcoming.
Contemporary approaches to LBD focus on creating subgraphs, which comprise of binary relations among concepts, called semantic predications. These predications are extracted directly from assertions in scientific literature, using SemRep [44]. Wilkowski et al. [27] developed a graph-theoretic approach based on semantic predications that iteratively (and manually) uses a greedy strategy to create the ‘best’ subgraph, by weighting edges using degree centrality. This approach was used to elucidate the association among Norepinephrine, Depression, and Sleep.
Wilkowski’s approach is similar to the approach by Ramakrishnan et al. [45], in which a greedy strategy is applied, using an ensemble of features, to generate complex associations. Ramakrishnan’s approach is fully automatic and uses class and property specificity, instance-level rarity, and refraction to find hidden connections. However, this approach was used on a synthetically generated dataset, instead of a real dataset consisting of semantic predications. Ramakrishnan notes that this approach was used, in exploratory research, to recover the connections from the Raynaud Syndrome – Dietary Fish Oils discovery. However its broader applicability for LBD in general has not been fully demonstrated. Reliance on hierarchical relationships in the UMLS Semantic Network is subject to inconsistencies since the UMLS is a terminology and not a formal ontology. Also, by design, the trees in the UMLS Semantic Network are fairly disjoint, as for Prostaglandins and Platelet Aggregation.
Goodwin et al. [46] developed a hybrid approach that uses spreading activation for LBD, deriving weights from relative frequencies (of concepts and semantic predications) and degree centrality. This approach was used to successfully recover the intermediate Cortisol in the Testosterone – Sleep discovery [30], and also to elucidate the Norepinephrine, Depression, and Sleep scenario from [27]. However, Goodwin generates a list of intermediates instead of a graph. It is therefore unclear how the spreading activation algorithm might be adapted to capture the context of complex associations. In [47] van der Eijk et al. clustered only MeSH descriptors (not semantic predications) into subgraphs, based on frequency of co-occurrence and Hebbian Learning. This approach provided new insights into the association between Deafness and Macular Dystrophy, and between Insulin and Ferritin. In recent work, Spangler et al. [48] also used distributional statistics (tf-idf) to weight edges in a kinase network, using graph diffusion applied to a Laplacian Matrix. The approach creates an n-ary similarity tree in which 7 new p53 kinases were discovered, which could revolutionize Cancer treatments. The approach for clustering of cliques developed by Zhang et al. [49, 50] may be used to capture subgraphs on multiple thematic dimensions. However, the approach is based on degree centrality and is therefore more likely to create subgraphs that only consist of highly connected concepts from the literature.
In spite of the successes of and frequency-, relations-, and graph- based approaches to LBD, more effective methods for capturing the context of associations are desired. Gordon and Dumais suggested a possible independence between frequency and causality for LBD in [32]. We believe that complex associations that elucidate the relationships among concepts depend both on implicit and explicit context. Further, we believe that capturing such context may be the important in segregating complex associations along multiple thematic dimensions. In this paper, we explore the idea that hidden connections, and their related concepts, which help elucidate underlying complex associations, are more dependent on context than frequency, connectivity or specificity. In the next section, the approach for automatic subgraph creation based on this premise is presented.
3. Approach
To automatically create complex subgraphs our approach relies on three datasets. The first dataset is MEDLINE, which is a repository of more than 23 million bibliographic citations maintained by the National Library of Medicine (NLM). The second is SemMedDB [51], a database of more than 65 million semantic predications extracted from MEDLINE. Semantic predications are extracted using a tool called SemRep2, developed at NLM. The third is the Biomedical Knowledge Repository (BKR), a knowledge base consisting of statements from the UMLS Metathesaurus together with semantic predications extracted using SemRep. These datasets are used for automatic subgraph creation in five steps: 1) Query Specification, 2) Candidate Graph Generation, 3) Path Context Representation, 4) Path Clustering, and 5) Subgraph Ranking. Each step is discussed in the following subsections, and also outlined in Algorithm 1:
Algorithm 1. autoSubGen (Set A, Set C, Integer k, Date dt).
|
3.1. Query Specification
The system (called Obvio3, see Appendix A) first requires a query, denoted q, which can be specified initially by providing the labels of two concepts of interest (A, C). These terms are manually mapped to concept unique identifiers (or CUIs), using the UMLS Semantic Navigator4. For example, the A-term Dietary Fish Oil, maps to the UMLS concept C0016157, whose label is also Fish Oils. Initial A- and C-terms are also manually augmented with other closely related concepts. For example, the concepts Fish oil – dietary (C0016157) and Eicosapentaenoic Acid (C0000545) are closely related to Fish Oils (C0016157) and are therefore added to the query. Next, the cut-off date dt for the literature to be included may be optionally provided. If no cut-off date is given the system uses the entire MEDLINE database. The maximum path length k, of paths to be generated between A and C may then also be optionally provided. If none is given, the system defaults to a maximum path length of k = 2. An example query for Raynaud Syndrome – Dietary Fish Oils is as follows: q = ({Fish Oils, Fish oil – dietary, Eicosapentaenoic Acid}, {Raynaud Phenomenon, Raynaud Disease}, 11/01/1985, 3). In plain English, ‘get me all subgraphs between dietary fish oils and raynaud syndrome, using scientific literature published before November 1985, and consisting of paths up to length 3.’
3.2. Candidate Graph Generation
Given a query q = (A, C, dt, k), the Query Processor (Figure 3, top center) then retrieves the set of MEDLINE documents D that contain any of the terms (i.e., labels) in the A- and C- sets (Algorithm 1, line 1). These documents form the corpus from which semantic predications will subsequently be obtained. To obtain the predications, the set of PubMed identifiers (or PMIDs) for each article in D is processed by the Predications Graph Builder (Figure 3, middle center), which creates a labeled directed predications graph, denoted G. To achieve this, the graph builder collects the semantic predications for each document in D that are also present in SemMedDB5 (Algorithm 1, line 4). The graph builder then creates a predications graph (Algorithm 1, line 5) in which nodes are UMLS concepts and edges are UMLS predicates. This graph is delivered as input to the Subgraph Generator (Figure 3, bottom center), which first uses the Path Generator to extract all paths between (A, C) up to length k, using the Depth First Search (DFS) algorithm (Algorithm 1, line 8). DFS is selected because both A and C are known. However, the choice of Breadth First Search (BFS) may be equally effective for graph traversal, but has not been explored, since performance is not the primary focus at this point. Using DFS, the path generator effectively uses the predications graph to produce paths (or ρ-path associations from[52]), except that edges are oriented in either direction, as we previously noted in [26]. This restricted set of paths is called the reachability relation R [53] (or candidate graph) between A and C at length k, and date range dt. This candidate graph represents a more likely set from which discoveries will arise.
3.3. Path Context Representation
The candidate graph is provided as input to the Path Clustering Module (Figure 3, bottom center), which requires a definition for the context of a path p to cluster related paths into subgraphs. To specify path context, denoted C(p), we first specify the context of a semantic predication t, denoted c(t). The context of each predication in the path is then aggregated to obtain overall path context.
To define the context of a semantic predication, we make two assumptions, based on observations about MEDLINE articles. The first observation is that MeSH descriptors are manually assigned to MEDLINE articles (titles and abstract only) by MeSH indexers, based on human interpretation of the meaning of the entire article. These descriptors provide a concept-level semantic summary of the full text. Similarly, semantic predications also provide a semantic summary of the meaning of the content. However, semantic predications provide a relational semantic summary, by linking concepts using explicit predicates.
We therefore assume that the MeSH descriptors and the semantic predications of an article capture its implicit context. This context is shared across the two abstractions of the meaning of the content. A semantic predication may therefore be represented in terms of the MeSH descriptors assigned to the article in which the predication occurs. This is the basis for our interchange-ability assumption for subgraph creation, which states that the concept-level semantic summary and relational semantic summary of a MEDLINE article, are interchangeable. More specifically, given a semantic predication t and a MEDLINE article d such that t is extracted from d, the context of the semantic predication c(t) = M(d), where M(d) is the set of MeSH descriptors assigned to d. Likewise, the context of a MeSH descriptor m, denoted c(m), is the set of semantic predications T(d), assigned to the article d in which m occurs (i.e., c(m) = T(d)
If this assumption holds, then we can make a second assumption, which is that the implicit context of a semantic predication t across the entire corpus can be represented as a vector of MeSH descriptors aggregated from each document containing t (based on distributional semantics). This is the basis for our context distribution assumption for subgraph creation, which states that the implicit context of a semantic predication can be expressed as the distribution of all MeSH descriptors associated with all articles in which the predication occurs.
Since our fundamental premise for subgraph creation is that relatedness among concepts is independent of statistical frequency (as noted by Gordon and Dumais [32]), graph metrics or specificity, our vector representation is downgraded to the Boolean-valued set representation (i.e., the equivalent of a binary vector), in which a MeSH descriptor is either present or absent in the distribution. The context of a path
| (1) |
is therefore the aggregation of its predication context sets.
3.4. Path Clustering
The Path Clustering Module uses the context set C(p) for each path p in the candidate graph R to cluster related paths pi and pj, based on their shared context. To compute this shared context between paths, the system initially computes the intersection s″(pi,pj) = C(pi) ∩ C(pj) of their shared MeSH descriptors. However, to account for inexact matches between MeSH descriptors across the two sets, this intersection is enhanced using the MeSH hierarchy, which provides explicit (or formal) semantics. Specifically, we use the Cartesian product of the two context sets C(pi) × C(pj) to determine which pairs of MeSH descriptors adequately indicate relatedness between the paths. Pairs of descriptors (mi, mj), whose similarity is above some threshold of MeSH semantic similarity are retained, while those below are discarded. The key idea is to maximize the weights of the in-context descriptors and minimize the weights of the out-of-context descriptors.
To compute semantic similarity between MeSH descriptors the measure of dice similarity is used. Dice similarity computes the proportion of common ancestors between descriptors in the MeSH hierarchy (MH). For two MeSH terms mi and mj the dice similarity is computed as
| (2) |
where ancestor s (mi)M H is the set of all ancestors of mi in MeSH. The maximum similarity between two descriptors computed using dice similarity is 1. This maximum value occurs when the descriptors are equal. (i.e., mi = mj). The range of similarity values is [0,1].
In this computation, pairs of descriptors, whose dice similarity exceed the threshold of semantic similarity (manually assigned as τsim = 0.75) are normalized to a value of 1. This normalized dice similarity
| (3) |
is therefore computed conditionally. The initial overall semantic relatedness
| (4) |
between pi and pj is the sum of the normalized pairwise dice similarity scores that exceed the threshold of semantic similarity, across the Cartesian Product of the context sets C(pi) × C(pj).
A consequence of this semantics-enhanced shared context metric, is that a broad range of relatedness scores may exist. Paths that are very similar, which have many exact (and inexact) MeSH descriptors in common will have very scores, while others may have low scores. To dampen the major differences in similarity scores of different path pairs, we apply a log reduction on the normalized dice similarity scores. This is achieved by first computing the relatedness score between a given MeSH descriptor a in context set C(pi) against the entire set of descriptors in the context set C(pj). This computation yields the similarity score
| (5) |
The log reduction
| (6) |
is then applied to sim′(a,C(pj)), and the overall semantic relatedness between the two paths is the aggregate of the log-reduced scores for each descriptor in C(pi) and the entire set in C(pj). This metric , is our basis for finding and elucidating complex associations among concepts, along multiple thematic dimensions, based on implicit and explicit semantics, alluded to by Gordon and Dumaisin[32].
In the next step the hierarchical agglomerative clustering (HAC) algorithm is used to create subgraphs by clustering related paths (Algorithm 1, line 11, getClusters(R)). In the bucket population step, the algorithm initializes |R| buckets, one for each path in the candidate graph. For a given path, the relatedness score is computed for each of the remaining |R| −1 paths. If two paths are sufficiently related, they must be placed in the same cluster. To achieve this, a method to automatically determine the threshold for path relatedness denoted τrel , is required.
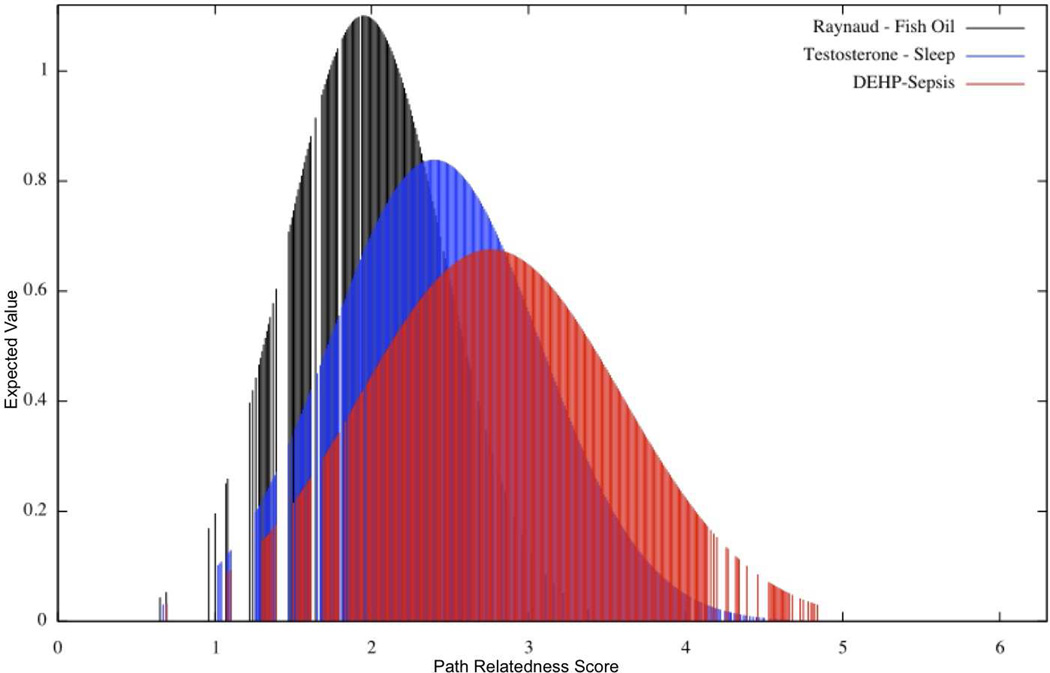
To obtain the threshold for path relatedness the distribution of path relatedness scores between all pairs of paths in the candidate graph was pre-computed (i.e., (|R| × |R – 1|)/2 scores). Figure 4 shows the distribution of relatedness scores for three experiments in the initial stages of our research. Each distribution approximates to a Gaussian (or normal) distribution.
Figure 4.
Gaussian Distribution of Path Relatedness scores for three rediscovery scenarios
In statistics, the first standard deviation (−σ, +σ) from the mean of a Gaussian distribution corresponds to the point of inflection. This point likely indicates a shift in a trend or phenomenon. When the manually determined thresholds for path relatedness for the same three experiments were compared to the σ, 2σ, and 3σ of the Gaussian distribution, it was observed that the manual thresholds were consistently between the 2σ and 3σ, as shown in Table 1. The second deviation from the mean of the Gaussian distribution (τrel = 2σ) was therefore selected as the path relatedness threshold for clustering. During clustering, all pairs of paths with relatedness scores above this automatically determined threshold were added to the same cluster.
Table 1.
Threshold Comparisons
| Scenario | Path Relatedness Scores | Max | ||
|---|---|---|---|---|
| 2 Std. Dev. | Manual | 3 Std. Dev. | ||
| Raynaud- Fish Oil | 2.68 | 3.0 | 3.04 | 3.38 |
| Testosterone-Sleep | 3.35 | 3.5 | 3.83 | 6.22 |
| DEHP-Sepsis | 3.94 | 4.0 | 4.53 | 4.84 |
In the next phase of HAC (bucket merging), buckets that contain multiple paths were merged if their inter-cluster similarity exceeded the threshold for path relatedness. That is, for each pair of paths (pi,pj) across a pair of buckets Ba and Bb, the inter-cluster similarity
| (7) |
was computed as the sum of the semantic relatedness scores, normalized by the sizes of the two buckets. The clustering algorithm terminated when the number of clusters between successive iterations remained unchanged.
3.5. Subgraph Ranking
The generated subgraphs were then ranked (Algorithm 1, line 11, rankClusters(S′)) where S′ is the unranked list of subgraphs from getClusters(R). Subgraphs containing more than one path are ranked in descending order, based on their intra-cluster similarity, which measures the compactness of the cluster. To compute this measure
| (8) |
the aggregate of the relatedness score for each pair of paths (pi ≠ pj) in a given cluster B is obtained and then normalized.
Singleton clusters consisting of only one path are ranked in ascending order using the measure of association rarity. Given a path pi, we define an association A(pi) as the set of unique concepts in the path. Association rarity is therefore the number of MEDLINE articles f(A(pi)) that contain only the concepts in the path. For singleton buckets, bucket rarity
| (9) |
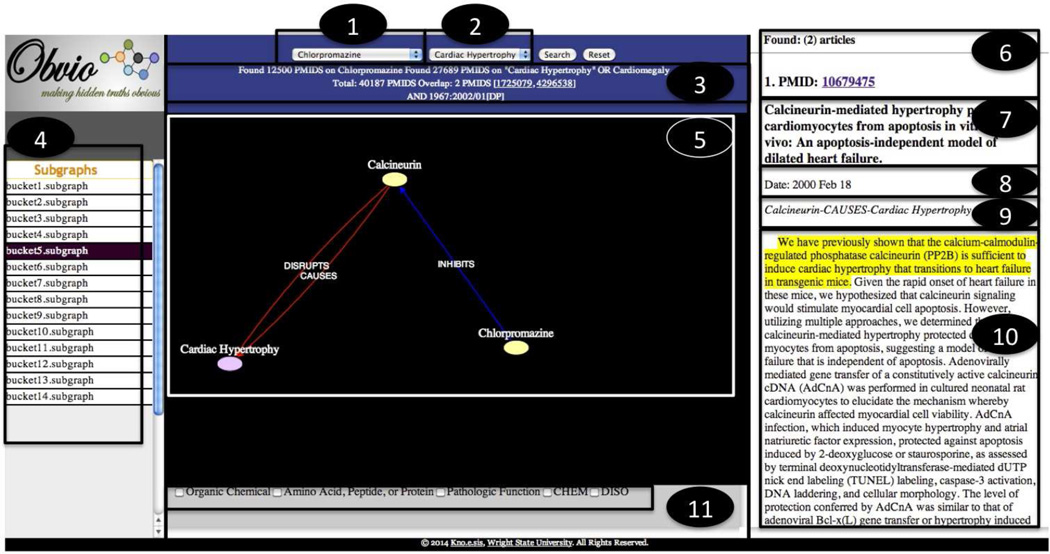
is the same as association rarity, since B = {pi} and |B| = 1. The ranked list of clusters is rendered to the user for inspection in the Discovery Browsing Interface (Figure 3, middle left). This interface is shown in Appendix A and also available online (live tool - http://knoesis-hpco.cs.wright.edu/obvio/, video demo - http://bit.ly/obviodemo). Concepts are color-coded based on semantic groups obtained from the BKR, while predicates are color-coded based on a locally developed coding scheme, since none exists for predicates in the BKR.
Using this approach, 8 out of 9 existing scientific discoveries were recovered. These well-known discoveries are: 1) Raynaud - Fish Oil (1986) [1], 2) Magnesium - Migraine (1988) [15], 3) Somatomedin C - Arginine (1990) [16], 4) Indomethacin - Alzheimer’s Disease (1996) [12], 5) Estrogen - Alzheimer’s Disease (1996) [13] 6) Calcium-Independent Phospholipase A2 - Schizophrenia (1998) [14], 7) Chlorpromazine - Cardiac Hypertrophy (2004) [29], 8) Testosterone - Sleep (2012) [30] and 9) Diethylhexyl (DEHP) - Sepsis (2013) [28]. In the next section the application of this approach for the rediscoveries is discussed.
4. Experimental Results
Given the absence of a gold standard dataset in LBD research, knowledge rediscovery is considered a de facto standard for evaluating LBD systems. To assess the effectiveness of our context-driven subgraph method, both an evidence-based evaluation and a statistical evaluation were conducted. The evidence-based evaluation qualitatively determines the extent to which our approach is capable of rediscovering the known knowledge, while the statistical evaluation is intended to measure the likelihood that a domain expert might be motivated to explore an arbitrary subgraph generated by the system. The latter achieves this by measuring the ‘interestingness’ of a subgraph, by quantifying the rarity of its associations in MEDLINE. Associations that have never been mentioned in any MEDLINE article are considered rare and most interesting. These are called zero-rarity associations (ZR). The obvious caveat is that rare associations are not necessarily all interesting. The next section discusses the evidence-based evaluation.
4.1. Evidence-Based Evaluation
The first aspect of the evidence-based evaluation reports on the number of intermediates from a discovery that could be retrieved by our system. The second aspect substantiates the meaning of each association using evidence from the literature. Such evidence can be derived first using the predicates of the semantic predications in the subgraph. When this is insufficient or contradictory, evidence can be obtained using the provenance of the predications in MED-LINE. Additionally, queries can be composed and executed in PubMed6 to explore inferred associations, not explicitly stated in the subgraphs, as commonly practiced.
For each rediscovery scenario, no concept filters were specified to exclude concepts based on semantic types or groups. A generic predicate filter, called the STRICT filter was applied uniformly by the system (not the user), across some experiments, to exclude less informative UMLS predicates, such as ASSOCIATED_WITH, INTERACTS_WITH, and AFFECTS. This limited degree of manual filtering is the extent of a priori knowledge required for subgraph generation in the system.
Due to space limitations, only three experiments are discussed in detail: 1) Raynaud - Fish Oil, 2) Magnesium - Migraine and 3) Somatomedin C - Arginine. The six remaining experiments are discussed briefly in Section 4.1.4. Further details on each experiment are available in [54] and in the following online supplementary materials: 1) the Obvio wiki page - (http://wiki.knoesis.org/index.php/Obvio, section on Automatic Subgraph Creation), 2) a video demo - http://bit.ly/obviodemo and 3) a beta-version of the Obvio web application - http://knoesis-hpco.cs.wright.edu/obvio/. Also note that in the following tables, the letter Y (for yes) is used to indicate that the status S of an intermediate as ‘found directly in a subgraph’ at position P in the list of subgraphs. The symbol Y* indicates that an intermediate was found through discovery browsing. The next section discusses the application of our approach to the Raynaud - Fish Oil discovery.
4.1.1. Raynaud Syndrome – Dietary Fish Oils
In November 1985, American Information Scientist Don R. Swanson (1924 – 2012) explored the research question of the role of Dietary Fish Oils (from salmon, mackerel, albacore, etc.) in Raynaud Syndrome. Through the methods described in [1], Swanson discovered that “dietary fish oil might ameliorate or prevent Raynaud’s syndrome.” This is because Dietary Fish Oils: 1) inhibit Platelet Aggregation, 2) increase the flow of blood (by reducing Blood Viscosity), and 3) also have a regulatory effect on the smooth muscle (thereby preventing Vasoconstriction and stimulating Vasodilation). Each of these concepts is causally implicated in Raynaud.
We seeded our algorithm with three concepts as sources: 1) Fish Oils (C0016157), 2) Fish oil - dietary (C0556145), and 3) Eicosapentaenoic Acid (C0000545), and two concepts as targets: 1) Raynaud Disease (C0034734) and 2) Raynaud Phenomenon (C0034735). The corpus consisted of the relevant 61 full text articles discussed by Swanson [1] in the pre-November 1985 period. There were only 4 articles from the Dietary Fish Oil set, which were in the Raynaud set. The path length was set to 3 and no predicate filter was specified. These choices are consistent with the choices in our earlier experiments in [26], in which we rediscovered and decomposed this hypothesis by manually constructing the subgraphs, using domain expertise as context.
The algorithm terminated in less than 5 minutes (on a 64-bit Linux Virtual Machine, 8 Intel 2.4GHz processors, 32GB RAM, and 1.5TB hard drive), producing 4 subgraphs (and 134 singletons) at 2σ and one subgraph (and 164 singletons) at 3σ. There were 1035 unique concepts and 4143 unique predications in the predications graph and the candidate graph contained 171 paths of length 3. Figure 5 shows that at 3σ, subgraph1 (the only subgraph produced) directly contains the intermediate Platelet Aggregation, which many rediscovery approaches consider sufficient to constitute a rediscovery. However, to better substantiate the association, we utilize the predicates in the subgraph, together with the provenance of the predications in MEDLINE, along with traditional PubMed search, to provide evidence.
Figure 5.
Subgraph1 (k = 3, 3σ) on Eicosapentaenoic Acid, Platelet Aggregation and Raynaud Syndrome
The predication which states that [Eicosapentaenoic Acid CONVERTS.TO Prostaglandins] was extracted from the following corroborating sentence, in the full text of the following article [PMID6827988] by Harris et al. The authors state that the “recent discovery that the prostaglandins derived from eicosapentaenoic acid have biological effects different than those derived from arachidonic acid (C20:4w6) has generated further interest in fish oils.” Two of the other 61 articles [PMID6321621, PMID6314583] contained this predication. Harris also refers to the 1979 article [PMID218223] by Needleman et al., which suggests further that [Eicosapentaenoic Acid CONVERTS TO Prostaglandin (PGI3)] in its metabolic pathway. And the full text of 1985 article [PMID2997286] by von Schaky et al. confirms that Eicosapentaenoic Acid produces Prostaglandin (PGI3) and Epoprostenol (Prostacyclin (PGI2)). von Schaky notes that “dietary EPA is transformed in vivo in humans into prostaglandins I3, which is as active … as the vasodilatory and antiaggregatory prostaglandin I2.”
The subgraph also contains the predication which states that [Eicosapentaenoic Acid DISRUPTS Platelet Aggregation]. This predication was extracted from the full text of the article [PMID6320840] by Saynor et al., who refers to the “Mechanisms underlying the inhibition of platelet aggregation by eicosapentaenoic acid and its metabolites.” The predication [Alprostadil DISRUPTS Platelet Aggregation] was extracted from the full text of the article [PMID6302714] by Dyerberg et al., who pointed out that another author7 “was the first to show that [Prostaglandin E1] PGE1 inhibited platelet aggregation.” The previously mentioned article by von Schaky also alludes to this point.
Conversely, the predication [Epoprostenol TREATS Raynaud’s Phenomenon] was correctly extracted from two articles; by Dowd et al. [PMID7037038], who discusses “Treatment of Raynaud’s phenomenon by intravenous infusion of prostacyclin (PGI2)” and by Belch et al. [PMID3883365], who discusses “Increased prostacyclin metabolites and decreased red cell deformability in patients with systemic sclerosis and Raynauds syndrome.” Since both Alprostadil (P GE1) and Epoprostenol (P GI2) are synthetic forms of Prostaglandins, it is plausible that both Alprostadil and Epoprostenol actually treat Raynaud’s Syndrome by disrupting Platelet Aggregation. Indeed, the 1982 article [PMID6890719] by Pardy et al., obtained through a date-restricted MEDLINE query8, confirms that Alprostadil (PGE1) treats Raynaud Phenomenon, instead of the weaker INTERACTS WITH relationship, present in the subgraph. The role of Platelet Aggregation in causing Raynaud, which is inferred and not explicit in the subgraph, is easily confirmed using another MEDLINE query (Platelet Aggregation AND Raynaud AND 1865:1985/11[DP]), which yields the 1985 article [PMID3985417] by Soro et al.
This subgraph together with discovery browsing suggest a richer relationship among Eicosapentaenoic Acid, Platelet Aggregation, and Raynaud Syndrome than would be provided by their co-occurrence. Rather, it appears that one mechanism by which [Eicosapentaenoic Acid TREATS Raynaud Syndrome] is by stimulating a series of Prostaglandins (namely, Prostaglandin I3 (P GI3), Prostaglandin E1(PGE1), and Prostacyclin (PGI2)), which actually disrupt Platelet Aggregation. This observation was first articulated by Swanson in [1].
An important observation is that the subgraph contains contradicting semantic predications. For example, the two predications [Eicosapentaenoic Acid CONVERTS TO Prostaglandins] and [Eicosapentaenoic Acid INHIBITS Prostaglandins] are opposing. The full text of the article [PMID6827988] by Harris et al., from which the predication [Eicosapentaenoic Acid CONVERTS TO Prostaglandins] was extracted supports its claim. However, the full text of the lone article [PMID6301111] by Moncada from which the predication [Eicosapentaenoic Acid INHIBITS Prostaglandins] was extracted states that “It is clear, therefore, that both prostaglandin dependent and independent pathways of platelet aggregation are inhibited by EPA in vitro.” This is an incorrect extraction from SemRep. The author is noting that [Eicosapentaenoic Acid INHIBITS Platelet Aggregation], not Prostaglandins as the predication suggests. It is important to note that resolution of such discrepancies is part of the discovery browsing process, which requires adjudication by domain experts. We provide the infrastructure for achieving this through provenance.
The second intermediate Blood Viscosity, was found in the list of zero-rarity singletons (result #15 in Table 2). The actual singleton, which states that [Eicosapentaenoic Acid DISRUPTS Blood Viscosity], [Ketanserin DISRUPTS Blood Viscosity], [Ketanserin TREATS Raynaud Disease], suggests a causal relationship between Blood Viscosity and Raynaud Syndrome. This inferred relation that [Blood Viscosity CAUSES Raynaud Syndrome] is confirmed in the 1984 article [PMID6707529] by Larcan et. al through a MEDLINE search. The statement [Ketanserin DISRUPTS Blood Viscosity] is verified in the following articles [PMID401574], [PMID6303363] and [PMID2412054]. Likewise, the predication [Ketanserin TREATS Raynaud Disease] can be verified in the article [PMID6432198] by Roald et al. and also [PMID6209510] by Bounameaux et al.
Table 2.
Comparison of rediscoveries with other approaches for Raynaud Syndrome - Dietary Fish Oils
Table 2 shows the number of intermediates rediscovered for this experiment compared with 4 other approaches. The intermediate Vascular Reactivity (in reference to Vasoconstriction) was not found explicitly by our approach (although can be inferred from the article [PMID2997286] by von Schacky et al.). This result is not completely unexpected, since it is known from our reports in [26] that SemRep interprets “Vascular” and “Reactivity” as separate concepts. Hristovski in [22] was also subject to the same limitation.
Srinivasan [36] found all three intermediates in the top 2 of the top 30. However, note that Srinivasan’s approach relies on a priori knowledge of the semantic types of the intermediates for filtering and is manually intensive. Additionally, that approach does not create complex subgraphs, nor does it provide evidence for the meaning of associations using predicates. Hristovski et al. [22] and Weeber et al. [34] also require considerable domain expertise, particularly for specification of a priori relations (i.e., semantic types and discovery patterns). Gordon and Lindsay [35] find intermediates but make no attempt to elucidate the meaning of the associations.
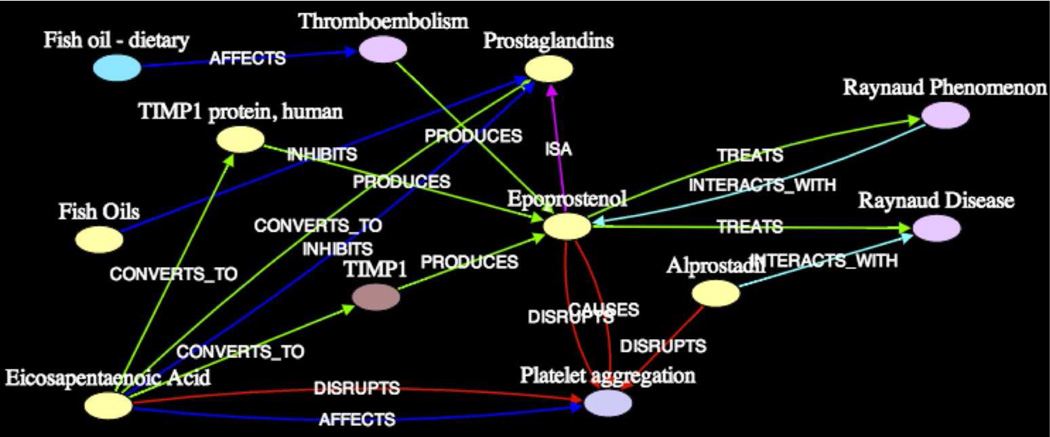
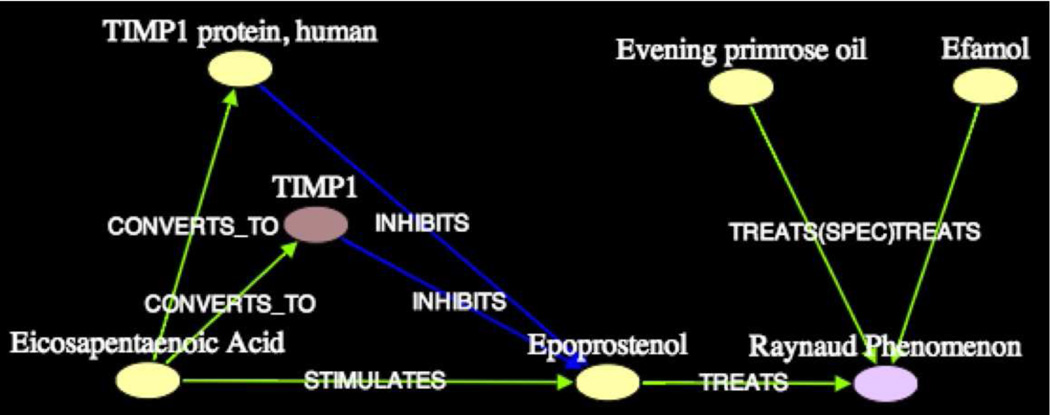
To illustrate that our subgraphs capture different thematic dimensions of association between two concepts, consider the four subgraphs using τrel = 2σ as the threshold for clustering. Subgraph1 in Figure 6a is similar to subgraph1 (at 3σ) except that it includes the three additional intermediates, TIMP1, TIMP1 protein, human, and Thromboembolism. This is naturally due to a lower threshold for path relatedness. By inspection, this subgraph elucidates the association between Dietary Fish Oils and Raynaud Syndrome through Blood Platelet-s/Prostaglandins, similar to the previous subgraph.
Figure 6.
Subgraph1 (k = 3, 2σ) on Dietary Fish Oils - Raynaud Syndrome (Blood Platelet-s/Prostaglandins)
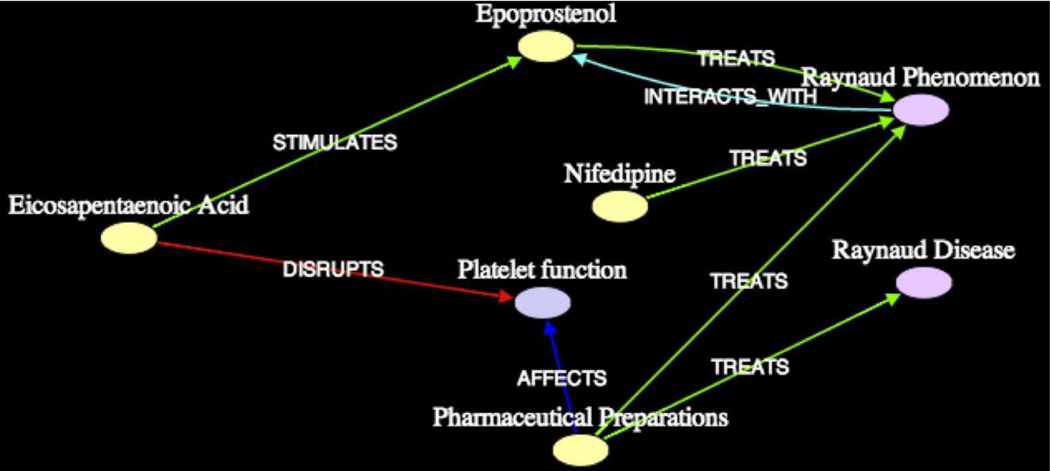
Subgraph2 (shown in Figure 7) associates Dietary Fish Oils and Raynaud Syndrome from the perspective of Pharmaceuticals, including Nifedipine, Pentifylline, Thyrocalcitonin, and Trinitrin detailed especially in the article [PMID6352267] by Kahan et al., from which the predication [Nifedipine TREATS Raynaud Phenomenon] was extracted.
Figure 7.
Subgraph2 (k = 3, 2σ) on Dietary Fish Oils - Raynaud Syndrome (Pharmaceuticals)
Subgraph3 in Figure 8 discusses the role of various Fatty Acids, which associate TIMP1, Epoprostenol, Efamol and Evening Primrose (see [PMID4082084, PMID6318123, PMID6321621]).
Figure 8.
Subgraph3 (k = 3, 2σ) on Dietary Fish Oils - Raynaud Syndrome (Lipids/Fatty Acids)
Subgraph4 in Figure 9, which focuses more on Cellular Activity at the level of Blood Platelets involving Thromboembolism, is subsumed by subgraph1. Currently, subgraph subsumption has not been addressed in this work and remains a system limitation, discussed in Section 5. In the next section, the Migraine -Migraine experiment is discussed.
Figure 9.
Subgraph4 (k = 3, 2σ) on Eicosapentaenoic Acid, Platelet Aggregation and Raynaud Syndrome (Blood Platelets)
4.1.2. Magnesium - Migraine
In August 1987, Swanson explored the research question of the role of Magnesium in Migraine Disorder. Through the methods described in [15] he discovered 11 neglected connections between Magnesium and Migraine. He found that Magnesium deficiency might exacerbate Migraine due to complications involving Stress (Type A personality), Spreading Cortical Depression, Epilepsy, Platelet Aggregation, Serotonin, Substance P, Inflammation, Vasoconstriction, Prostaglandin formation, and Hypoxia. Also, as a natural calcium channel blocker, Magnesium may prevent Migraine attacks.
We seeded our algorithm with Magnesium (C0024467) as the source and Migraine Disorders (C0149931) as the target. The path length was set to 2 and no predicate filter was used, to be more consistent with the discovery. The corpus consisted of more than 47,000 articles from the pre-August 1987 period (i.e., 41,507 abstracts on Magnesium and 6,171 on Migraine, 7 overlapping). There were 14697 unique concepts, 73,960 predications in the predications graph and 256 distinct paths of length 2 in the candidate graph. The algorithm terminated in less than one hour, producing 25 subgraphs (and 151 singletons) at 2σ and 6 subgraphs (and 231 singletons) at 3σ.
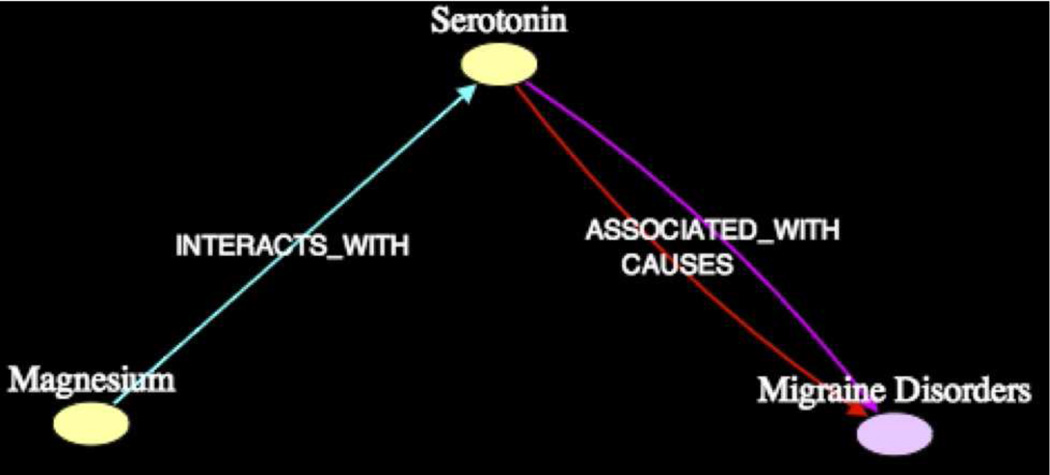
With regards to Serotonin, it was known from the 1973 article [PMID4725298] by Vosgeru (one of the 7 overlapping) that Magnesium Glutamate was used to treat Migraine. Figure 10 shows that the intermediate Serotonin was found in subgraph1 at 2σ. The lone article [PMID3629724] by Pertseva et al. from which the predication [Magnesium INTERACTS.WITH Serotonin] was extracted, is inconclusive. According to Swanson, [Magnesium INHIBITS Serotonin]. The article [PMID3512233] by Houston et al. from which the predication [Serotonin CAUSES Migraine] was extracted (among three others), suggested that elevated levels of Serotonin can induce Vasoconstriction, which causes Migraine. Houston explicitly states that “much evidence has implicated serotonin (5-hydroxytryptamine) in the pathogenesis of migraine.” The article further notes that Serotonin is released from Platelet Aggregation and might reach sufficient levels to exacerbate Migraine, as noted by Swanson. The 1987 article [PMID2440758] by Briel et al. (through a MEDLINE search) confirms that Magnesium inhibits Platelet Aggregation. It follows that elevated Magnesium levels may inhibit both Serotonin and Platelet Aggregation, and so treat Migraine.
Figure 10.
Subgraph1 (k = 2, 2σ) Magnesium - Migraine
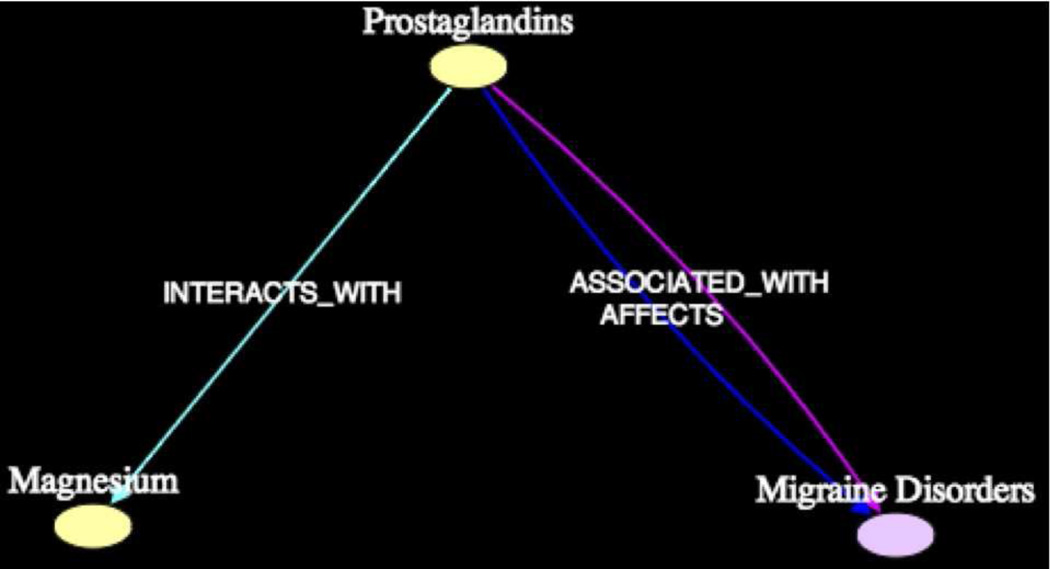
Figure 11 shows subgraph4, which contains the intermediate Prostaglandins between Magnesium and Migraine. The lone article [PMID3871957] by Fried-lander et al. from which the predication [Prostaglandins INTERACTS.WITH Magnesium] was extracted, suggested that a decrease in prostaglandin synthesis is accompanied by lower levels of magnesium (and calcium). This conclusion is based on the title: “Decreased calcium and magnesium urinary excretion during prostaglandin synthesis inhibition in the rat” as noted by Swanson. The 1986 article [PMID3016750] by Nigam et al. confirms that [Magnesium STIMULATES Prostaglandins] as suggested by Swanson. The article [PMID89390] by Hakkarainen et al. from which the predication [Prostaglandins ASSOCI-ATED.WITH Migraine Disorders] was extracted (among only three others) states that “Tolfenamic acid (a potent inhibitor of prostaglandin biosynthesis) was effective in treating acute migraine attacks.” The specific role of Prostaglandins in Migraine was unclear however, even after discovery browsing. Swanson suggested that [Prostaglandins INHIBITS Migraine].
Figure 11.
Subgraph4 (k = 2, 2σ) Magnesium - Migraine
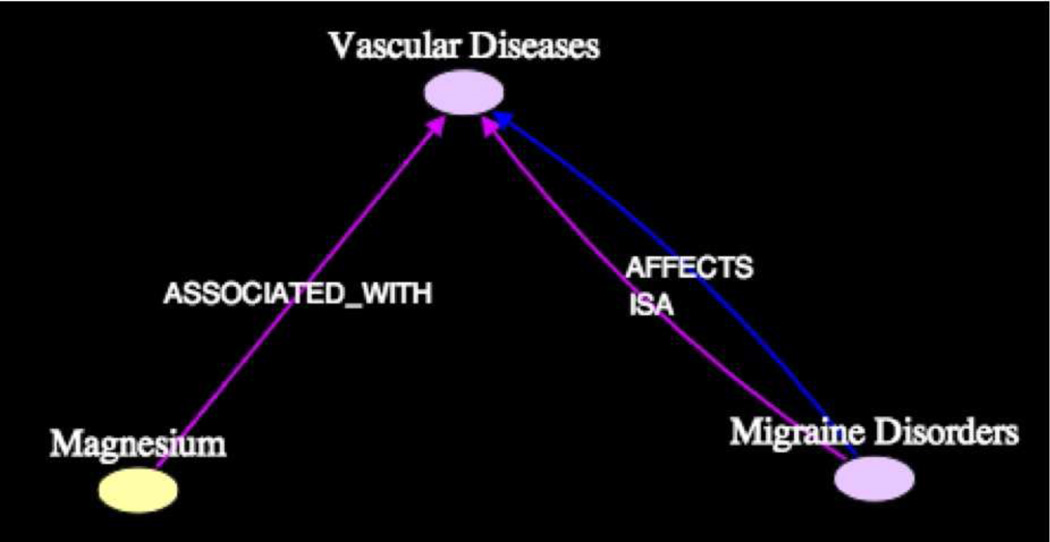
Figure 12 shows that the intermediate Vascular Disease was found explicitly in subgraph9. The title of the article [PMID4260015] by Wustenberg et al. from which the predication [Magnesium ASSOCIATED.WITH Vascular Disease] was extracted, suggests a role for magnesium in vascular reactivity. The title of the article reads in part, “… Findings in magnesium metabolism in vascular diseases.” Similar to the predication with Serotonin, it is unclear from this title that [Magnesium INHIBITS Vasoconstriction] as noted by Swanson. On the other hand, the article [PMID1153064] by Domzal, from which the predication [Migraine Disorders ISA Vascular Diseases] was extracted (among three others), suggests that migraine is also a vascular disorder, although primarily a cerebral disorder. The lone article [PMID3945397] by Coppeto et al. from which the predication [Migraine Disorders AFFECTS Vascular Diseases] was extracted provides more compelling evidence by linking migraine and vascular retinopathy as suggested by Swanson. Coppeto reported that “two migraineurs suffered sudden, persisting loss of vision from retinal vascular occlusion.” This effect is consistent with the observation by Houston et al. from the article [PMID3512233] on Serotonin from subgraph1. Salati et al. in [PMID6225285], from which the predication [Migraine Disorders ISA Vascular Diseases] was extracted, noted a dependency among Migraine, Vascular diseases, Epilepsy, and Autoscopy (outer-body hallucination).
Figure 12.
Subgraph9 (k = 2, 2σ) Magnesium - Migraine Table 3: Comparison of rediscoveries with other approaches for Magnesium - Migraine
The two calcium channel blockers, Nifedipine and Verapamil were the only intermediates in subgraph22 (not shown). All three articles [PMID2425960, PMID3673084, PMID6539877] confirmed that these calcium channel blockers treat Migraine as suggested by Swanson. The article [PMID537283] by Khoda et al. from which the predication [Verapamil INTERACTS.WITH Magnesium] was extracted suggested that Magnesium inhibits Verapamil as noted by Swan-son.
The intermediate Hydrocephalus (accumulation of fluid in the brain), which leads to Brain Edema (referred to as Inflammation by Swanson), was found among the zero-rarity associations (see Table 3). The remaining intermediates Hypoxia, Spreading Cortical Depression, Stress (Type A Personality), and Substance P were not found among the subgraphs.
Table 3.
Comparison of rediscoveries with other approaches for Magnesium – Migraine
| Scenario | Intermediate(s) | Cameron | Srinivasan [7] |
Weeber [4] |
Blake [37] |
Gordon [3] |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| S | P | S | P | S | P | S | P | S | P | ||
| Magnesium - Migraine |
Calcium Channel Blockers | Y | 22 | Y | 3 | Y | - | Y | 10 | Y | 1 |
| Epilepsy | Y* | 9 | - | - | Y | - | Y | 8 | Y | 3 | |
| Hypoxia | - | - | Y | 5 | - | - | Y | 6 | Y | 77 | |
| Inflammation | Y* | ZR-3 | Y | 2 | Y | - | Y | 170 | Y | 82 | |
| Platelet Activity | Y* | 1 | Y | 2 | Y | - | Y | 2 | Y | 8 | |
| Prostaglandins | Y | 4 | Y | 1 | Y | - | Y | 42 | Y | 27 | |
| Stress/Type A Personality | - | - | Y | 1 | Y | - | Y | 23 | - | - | |
| Serotonin | Y | 1 | Y | 1 | Y | - | Y | 5 | Y | 1 | |
| Spreading Cortical Depression | - | - | Y | 6 | - | - | Y | 45 | - | - | |
| Substance P | - | - | Y | 18 | Y | - | Y | 38 | Y | 23 | |
| Vascular Mechanisms | Y | 9 | Y | 1 | Y | - | Y | 46 | Y | 16 | |
Interestingly, only subgraph22 on the calcium channel blockers was a complex subgraph in which existing knowledge was recovered. While several intermediates related to Vascular Reactivity, such as Vasospasm, Vascular Function, Vasoconstriction, and Vascular Disease exists, their shared context did not meet our threshold for path relatedness and hence they were not grouped into the same cluster. The shortcomings of SemRep in extracting Vascular Reactivity may also have been a limiting factor. Still, altogether 10 out of the 25 subgraphs contained complex associations.
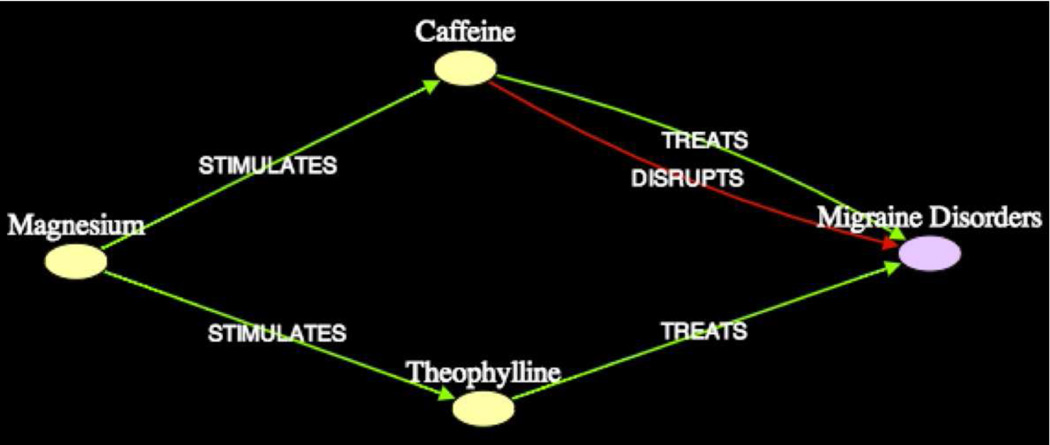
Subgraph7 (shown in Figure 13) for example, links Theophylline and Caffeine, with Magnesium and Migraine, which have different semantic types, but belong to the general group of Stimulants. Subgraph6 (not shown) associates Epinephrine and Glucose from the perspective of Metabolism. Table 3 shows that ultimately, 7 out of the 11 associations found by Swanson could be found using our approach.
Figure 13.
Subgraph7 (k = 2, 2σ) Magnesium - Migraine
4.1.3. Somatomedin C - Arginine
In April 1989, Swanson explored the research question of the role of the dietary amino acid Arginine and the protein Somatomedin C (also called Insulin-Like Growth Factor 1 (IGF1)) in Growth. Through the methods discussed in [16], Swanson discovered 4 implicit connections between Somatomedin C and Arginine. He found that Arginine intake could: 1) stimulate Growth and protein synthesis, 2) promote Wound Healing and cell regeneration, 3) facilitate nutritional repletion and overcome Malnutrition, and 4) improve Body Mass (and Weight), especially in the elderly and debilitated.
We seeded our algorithm with Somatomedins (C0037657) and Insulin-Like Growth Factor I (C0021665) as the sources, and Arginine (C0003765) as the target. The corpus consisted of more than 11,000 articles (819 on Somatomedins and 10,698 on Arginine (with 53 overlapping), in the pre-April 1989 period. The path length was set to 2, and the STRICT predicate filter was used to eliminate non-informative predicates. There were 5195 concepts and 17,058 predications in the predications graph and 239 distinct paths in the candidate graph. The algorithm terminated in less than one hour producing 10 subgraphs (and 153 singletons) at 2σ and 7 subgraphs (and 205 singletons) at 3σ.
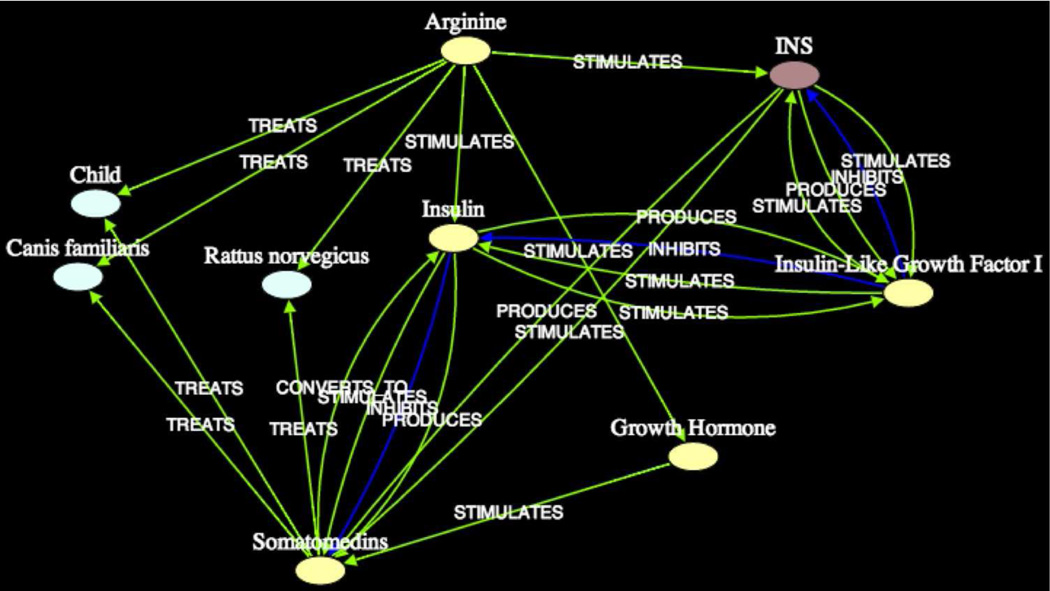
Figure 14 shows the intermediate Growth Hormone in subgraph5 at 3σ. The sequence of predications [Arginine STIMULATES Growth Hormone] and [Growth Hormone STIMULATES Somatomedins] is entirely correct and requires no further proof (in terms of rediscovery). Still, for verification, we confirmed in the article [PMID6394628] by Chew et al. that dietary Arginine stimulates the release of Growth Hormones. These Growth Hormones then stimulate the production of Somatomedin C (IGF1), which leads to cell growth and increased body size and muscle (i.e., protein synthesis), as noted in article [PMID7194347] by Clemmons et al. The same association is captured in subgraph6 at 2σ (not shown).
Figure 14.
Subgraph5 (k = 2, 3σ) Somatomedin C – Arginine
In subgraph5, several articles from which the seemingly spurious predication [Arginine TREATS Child] was extracted, upon investigation, were shown to actually discuss Glucagon and Insulin. This includes the article [PMID7204541] by Blethen et al. whose title is “Plasma somatomedins in children with hyper-insulinism.” Likewise, the article [PMID6205015] by Binoux et al. from which the predication [Arginine TREATS Rattus norvegicus] was extracted, discusses observations regarding Insulin-like Growth Factor 1 in the serum of rats. The article [PMID7007553] by Ashby et al. from which the same predication was extracted, discusses the effects of Progesterone and Insulin in rats, resulting from Glucose and Arginine stimulation. Based on these observations, it is reasonable to conclude that this subgraph captures the shared context of role of Insulin with Somatomedin C and Arginine.
Subgraph7 at 3σ (not shown) contains the concept Growth as an intermediate instead of Growth Hormone (similar to subgraph2 at 2σ, also not shown). The sequence of predications [IGF1 CAUSES Growth] and [Growth PRODUCES Somatomedins] is interesting because the article [PMID3748655] by van Buul-Offers et al. from which the predication [IGF1 CAUSES Growth] was extracted states that IGF1 “increases body length and weight, as well as the growth of several organs of Snell dwarf mice,” which is consistent with Swan-son’s report. The association between Malnutrition and Somatomedin production was found in the article [PMID7023246] by McCumbee et al., from which the predication [Growth PRODUCES Somatomedins], was extracted. No obvious association to Wound Healing was found using our methods. Table 4 shows that 3 out of 4 intermediates could be found using our approach.
Table 4.
Comparison of rediscoveries with other approaches for Somatomedin C - Arginine
| Scenario | Intermediate(s) | Cameron | Srinivasan [7] |
||
|---|---|---|---|---|---|
| S | P | S | P | ||
| Somatomedin C - Arginine | Growth Hormone | Y | 5 | Y | 1 |
| Body Weight | Y* | 7 | Y | 4 | |
| Malnutrition | Y* | 7 | - | - | |
| Wound Healing | - | - | Y | 4 | |
4.1.4. Remaining Experiments
This section briefly presents the results for the remaining 6 rediscoveries attempted.
Scenario 4
For the Indomethacin - Alzheimer’s Disease discovery [12] by Smalheiser and Swanson in 1995, there were 15 subgraphs at 2σ. Srinivasan found all 8 intermediates, while we only recovered 6 out of 8 intermediates from subgraphs 2, 3, 4, and 14 (shown in Table 5).
Table 5.
Comparison of rediscoveries with other approaches for Indomethacin - Alzhiemer’s Disease
| Scenario | Intermediate(s) | Cameron | Srinivasan [7] |
||
|---|---|---|---|---|---|
| S | P | S | P | ||
| Indomethacin – Alzheimer’s Disease |
Acetylcholine | Y | 4 | Y | 2 |
| Lipid Peroxidation | Y* | 2 | Y | 4 | |
| M2-muscarinic | - | - | Y | 3 | |
| Membrane Fluidity | - | - | Y | 10 | |
| Lymphocytes | Y* | 14 | Y | 4 | |
| Thyrotropin | Y | ZR-20 | Y | 9 | |
| T-lymphocytes (T- Cells) |
Y* | 3 | Y | 5 | |
Scenario 5
For Estrogen - Alzheimer’s Disease [13] by Smalheiser and Swanson in 1995, we found 3 out of 8 intermediates from 3 subgraphs at 2σ, as shown in Table 6. Srinivasan did not attempt this experiment.
Table 6.
Comparison of rediscoveries with other approaches for Estrogen - Alzheimer’s Disease
| Scenario | Intermediate(s) | Cameron | |
|---|---|---|---|
| S | P | ||
| Estrogen – Alzheimer’s Disease |
Antioxidant Activity | Y* | 4 |
| Alipoprotein E (ApoE) | Y* | 3 | |
| Calbindin D28k | Y | 4 | |
| Cathepsin D | - | - | |
| Cytochrome C oxidase | - | - | |
| Glutamate | - | - | |
| Receptor Polymorphism | - | - | |
| Superoxide Dismustase | - | - | |
Scenario 6
For Calcium-Independent PLA2 - Schizophrenia [14] by Smal-heiser and Swanson in 1997, our algorithm produced 10 subgraphs at 2σ, all of which were singletons. Here, our results are comparable to Srinivasan’s, except that we are able to retrieve the article [PMID7782894] by Kuo et al. deemed crucial to the discovery, through discovery browsing from singleton2. The seemingly innocuous singleton in subgraph2 (not shown), which states that [Phospholipase A2 INHIBITS Proteins] [Proteins CAUSES Schizophrenia] leads to the article [PMID7739414 ] by Berry, from which the predication [Proteins CAUSES Schizophrenia] was extracted. The article shows that the specific protein discussed was the selenium transport protein Selenoprotein P, as noted by Smalheiser. The article by Kuo is #4 in the search results of a MEDLINE search for Phospholipase A2 AND Selenium AND 1865:1997[DP].
Scenario 7
For Chlorpromazine - Cardiac Hypertrophy [29] by Wren et al. in 2002, there were 14 subgraphs at 2σ. The intermediate Isoproterenol was found in subgraph12 (as shown in Table 8). The article [PMID6165961] by Rossi et al. from which the predication [Chlorpromazine INHIBITS Isoproterenol] was extracted, together with the article [PMID203365] by Tsang et al. from which the predication [Isoproterenol CAUSES Cardiomegaly] was extracted, substantiated these predications. Subgraph5 contained the predication [Chlorpromazine INHIBITS Calcineurin] extracted from the article [PMID9001710] by Gong et al. and the predication [Calcineurin CAUSES Cardiac Hypertrophy] extracted from several articles, including [PMID9568714, PMID10679475, PMID11248077, PMID11773940, PMID10189350].
Table 8.
Comparison of rediscoveries with other approaches for Chlorpromazine - Cardia Hypertrophy
| Scenario | Intermediate(s) | Cameron | |
|---|---|---|---|
| S | P | ||
| Chlorpromazine – Cardiac Hypertrophy |
Calcineurin | Y | 5 |
| Isoproterenol | Y | 12 | |
Scenario 8
For Testosterone - Sleep [30] by Miller and Rindflesch in 2011, which articulates that “testosterone enhances sleep quality by inhibiting cortisol,” we found 11 subgraphs at 2σ and 10 subgraphs at 3σ. Cortisol (or Hydrocortisone) was found in subgraph7 at 3σ and also in subgraph11 at 2σ. The article [PMID8548511] by Kern et al. confirmed that [Hydrocortisone DISRUPTS Sleep], while the crucial article [PMID15841103] by Rubinow et al., noted by Miller, confirms that [Testosterone INHIBITS Hydrocortisone].
Scenario 9
For Diethylhexyl Phthalate (DEHP) - Sepsis [28] by Cairelli and Rindflesch in 2013, which articulates one possible mechanism for the obesity paradox [55], we did not find the intermediate PParGamma altogether. In our retrospective analysis, we found that the novel intermediate PParGamma was present in the predications graph, but not in the candidate graph. This is because no direct links between PParGamma and Sepsis existed in the candidate graph - consisting of paths of length 3 between DEHP and Sepsis. In the predications graph the predication, which states that [DEPH STIMULATES PParGamma] was present (extracted from [PMID22953781, PMID16326050]). We also noticed predications between PParGamma and Liver, Genes, STAT5A gene, etc. However, none of these concepts were linked directly to Sepsis. These observations suggest that the path length specified is perhaps too short. It also suggests that additional concepts, related to Sepsis (as terminals) may be necessary.
In summary, several approaches succeed in providing automation for finding intermediates. These approaches leverage keyword-based, concept-based relations-based, graph-based and hybrid techniques. Many also provide predicates between concepts, while more recent approaches are able to substantiate intermediates with provenance in MEDLINE. The main innovation of our approach is that we are able to retrieve and substantiate existing discoveries, on different thematic dimensions, using implicit and explicit semantics as suggested by Gordon and Dumais [32], not frequency, graph metrics or specificity. To the best of our knowledge, an approach that has rediscovered as many intermediates, with such degree of automation and substantiation has never been developed. In the next section the statistical evaluation is presented.
4.2. Statistical Evaluation
In the previous section, we showed that our context-driven, automatic subgraph creation method facilitated the rediscovery of 8 existing discoveries with their substantiation in MEDLINE. While these are encouraging results, one might argue that our experiments were biased since we knew the intermediates to be found in the first place. Hence, it was easy to find them in the subgraphs. A more important question is how interesting are subgraphs in general, such that an arbitrary domain expert might be motivated to explore them altogether? To address this question, we conducted a statistical evaluation, which uses association rarity to compute interestingness. If the interestingness score of the subgraphs across an entire experiment is low, then the rediscoveries were fortuitous and the associations that led to the rediscoveries were serendipitous, rather than systematic. While this not a complete loss, it is still less than ideal.
To perform this evaluation, for each path in each subgraph across the 8 rediscoveries (excluding singletons), a PubMed query was executed using the eUtils Web Service9. This was used to determine the number of documents that contain the association in MEDLINE, with the date restriction enforced.
For example, for the path [Arginine STIMULATES Growth Hormone], [Growth Hormone STIMULATES Somatomedins], the query “Arginine AND Growth Hormone AND Somatomedins AND 1865:1989/04[DP]” was composed, where Arginine, Growth Hormone, and Somatomedins represent an association. The rarity
| (10) |
of a set of associations across all subgraphs in an experiment E, is computed as the average of the association rarity, where f(A(pi)) is the frequency of a unique association A(pi) from path pi in MEDLINE. The interestingness
| (11) |
of an experiment E is computed as the normalized reciprocal of its rarity.
Table 10 shows the rarity and interestingness scores for each of the eight successful rediscoveries. For the Raynaud Syndrome - Dietary Fish Oils experiment, there were 10 unique intermediates/associations among the 4 subgraphs at 2σ; each of which had a zero-rarity in MEDLINE. This is not surprising, since Swanson noted in [1] that only four articles from the Raynaud literature overlapped with the Fish Oil literature by 1986. The rarity of these subgraphs is therefore 0.00, and the interestingness is 1 (meaning absolutely interesting).
Table 10.
Rarity and Interestingness score of the subgraphs in the rediscoveries
| Experiment | #Unique Associations |
MEDLINE Frequency |
r(E) | I(E) |
|---|---|---|---|---|
| Raynaud Syndrome – Dietary Fish Oils | 10 | 0 | 0.00 | 1.00 |
| Magnesium - Migraine | 48 | 27 | 0.56 | 0.64 |
| Somatomedin C – Arginine | 18 | 306 | 17.00 | 0.06 |
| Indomethacin – Alzhiemer’s Disease | 21 | 9 | 0.43 | 0.70 |
| Estrogen – Alzhiemer’s Disease | 42 | 36 | 0.86 | 0.54 |
| Calcium Independent PLA2 - Schizophrenia | 10 | 0 | 0.00 | 1.00 |
| Chlorpromazine – Cardiac Hypertrophy | 21 | 2 | 0.10 | 0.91 |
| Testosterone - Sleep | 61 | 654 | 10.72 | 0.09 |
| Average | 29 | 129 | 3.71 | 0.62 |
For Magnesium - Migraine there were 48 unique intermediates/associations, across a total of 27 documents (Table 10, row 3). The most commonly known intermediates were Hypertensive Disease (3), Individual (3), and Vascular Diseases (4) respectively. The overall rarity of the subgraphs in the experiment is therefore 27/48 = 0.56 and the interestingness is 0.64 (i.e., somewhat interesting).
For Somatomedin C - Arginine there were 18 unique intermediates/associations across a total of 306 documents (Table 10, row 4). The most commonly known intermediates were Child (16), Somatropin (63), and Growth Hormone (63). There were only two zero-rarity associations, which were from the intermediates Mus (0) and Falls (0). Clearly these are not interesting. Not surprisingly, the overall rarity score of these subgraphs is 306/18 = 17 and their interestingness is low (0.06). These high association frequencies suggest that perhaps the field is more well-studied. It also partially supports the observation by Gordon and Dumais [32] that while frequency of intermediates may be sufficient for finding novel intermediates in some cases, it may be insufficient to capture the related concepts that elucidate complex associations.
For Indomethacin – Alzheimers there were 21 unique associations across a total of 9 documents (Table 10, row 5). Hydrogen Peroxide (2), Interleukin-1 (2) and Free Radicals (3) were the most commonly known intermediates. The overall rarity score is 9/21 = 0.43 and the interestingness is 0.70 (i.e., quite interesting).
For Estrogen – Alzheimers there were 42 unique associations across a total of 36 documents (Table 10, row 6), among which 36 were zero-rarity associations. Metabolism (6), Dementia (10), and Senile dementia (10) were the most commonly known intermediates. The rarity score is 36/42 = 0.86 and the interestingness is 0.54.
For Calcium-Independent PLA2 – Schizophrenia there were 10 unique intermediates/associations (singletons described in Section 4.1.6), each of which was zero-rarity. Hence, the rarity of this subgraph is 0.00 and the interestingness was high (1.0).
For Chlorpromazine – Cardiac Hypertrophy there were 21 unique intermediates/associations across a total of 2 documents (Table 10, row 8) and 19 at zero-rarity. The most commonly known were Catecholamines (1) and Hypertensive disease (1). The rarity is therefore 2/21 = 0.10 and the interestingness is high (0.91).
For Testosterone – Sleep, there were 61 unique intermediates/associations across a total of 654 documents (Table 10, row 9) and 20 at zero-rarity. The most commonly known were Proteins (63), Symptoms (91), and Hormones (207). The overall rarity score is therefore 654/61 = 10.72 and the interestingness is low (0.09). This is not surprising, since these two domains (Testosterone and Sleep) are fairly well studied.
Across all 8 rediscoveries, the average rarity score is therefore 3.71 and the average interestingness is 0.62. This suggests that an association chosen at random from the rediscoveries is likely to be known to only approximately 4 documents in MEDLINE. Such a low rarity score suggests that the subgraphs themselves might be quite interesting to a domain expert. This is however not surprising, since most of the discoveries, at the time when made would have been inherently interesting situations and possibly not well studied in the literature. Testosterone – Sleep (2011) and Somatomedin C – Arginine (1990) are exceptional.
5. Discussion
This paper showed that the use of implicit and explicit semantics to find and elucidate associations among concepts along multiple thematic dimensions can be effective for LBD. Another important contribution is that domain scientists can infer relationships not explicitly stated in the subgraphs, based on meaningfully connected semantic predications. Our overall approach however, has several limitations. The first limitation is the assumption that the context of a semantic predication, expressed in terms of the distribution of MeSH descriptors is reliable for generating meaningful subgraphs. Not all MeSH descriptors assigned to an article are relevant to all its semantic predications, and hence the predication context vectors could be noisy. Ideally, direct mappings between semantic predications and MeSH descriptors could help resolve this discrepancy. Since, such mappings are unavailable our specification of context is subject to limitations of distributional semantics.
The second limitation is the degree of domain expertise still required for discovery browsing. Although impractical to eliminate, one improvement could be providing additional background knowledge to supplement the subgraphs where appropriate. In this way, assertional knowledge from the literature would be complemented with definitional knowledge from structured knowledge sources (though deep integration). Metrics for determining interesting neighboring concepts in background knowledge need to be developed for concepts in the subgraph to overcome this limitation.
Another limitation is the inability to systematically detect contradicting semantic predications. While the provenance of predications in MEDLINE allows domain experts to adjudicate, a method for resolving conflicting predications would be beneficial. We believe that temporal analysis of semantic predications could enable conflict resolution. However, since many unresolved paradoxes inherent in science itself are reported in the literature, it is unclear whether one might reliably detect and resolve such contradictions automatically, using temporal, statistical and/or semantic approaches.
The reliability of the statistical evaluation is also another limitation of our approach. Rare associations are generally interesting but not always. While alternative methods for conducting statistical evaluation for LBD have been discussed [56], it is cumbersome to coordinate cut-off dates for each predication across the rediscoveries. The suggested techniques are therefore impractical to implement. We use association rarity to indicate interestingness, similar to existing research [45, 36].
A number of technical limitations exist in our approach. The first technical limitation is the manual selection of a threshold for MeSH semantic similarity based on dice similarity. While dice is advantageous because it is easy to implement, other similarity metrics and more principled ways of computing the threshold should be explored. Likewise, the threshold for path relatedness, which is based on the second (and third) standard deviation from the mean of the Gaussian distribution, could be unreliable. Our results show that the data distributions only approximate to Gaussian. The p-values from the X2 test of the three Gaussian distributions in Figure 4 are indeed more than the 0.05 value normally considered reliable. To overcome this limitation, we anticipate that path relatedness could be recomputed relative to the minimum relatedness score. Torvik et al. [40] and Smalheiser et al. [42] implemented an approach based loosely on this idea, which normalized the distribution, using a mixture of Gaussian models.
Across some experiments, we utilized predicate filters to eliminate non-informative relationships (such as ASSOCIATED.WITH, INTERACTS.WITH, AFFECTS, etc). This is a compromise to achieve scalability. Ideally, the system should not require any predicate filters. In fact, the omission of some predicates may be responsible for low recall in some of our experiments. Still, given that most experiments terminated in less than one hour, higher recall may not be too costly for performance. With the emergence of big data infrastructure, the performance limitations of our clustering may be resolved using alternative platforms, such as Apache Spark.
The choice of HAC could be considered another limitation. HAC was selected because it is an unsupervised, deterministic clustering algorithm, for which the number of clusters does not have to be known or specified a priori. The time complexity of HAC is O(N2log N) in the best case. While approaches, such as those by Ramakrishnan et al. [45] and van der Eijk et al. [47] may be applicable for subgraph creation, it is unclear how they might be adapted to generate complex subgraphs along multiple thematic dimensions.
These and other limitations suggest the next steps in this research. In future, labels for subgraphs should be provided. This is a crucial task, since our approach is predicated on the idea that each subgraph captures a different thematic dimension of association between two concepts. Additionally, a comparative study using full text, against titles and abstracts, could be useful. Since, full text is only available on a limited scale, this is not a straightforward task.
6. Conclusion
Leveraging rich representations of textual content from scientific literature based on implicit and explicit context can provide effective means for literature-based discovery, as illustrated in this paper. These rich representations facilitated the rediscovery of 8 out of 9 well-known discoveries and their substantiation. Our approach is therefore an advancement of LBD research since it illustrates that notions of context and shared context can be important for making discoveries from scientific literature, which do not rely on statistical frequency, graph metrics or specificity. A beta-version of the Obvio web application, which showcases the rediscoveries, is available online for optional viewing (http://knoesis-hpco.cs.wright.edu/obvio/), along with various other resources (wiki page - http://wiki.knoesis.org/index.php/Obvio, video demo - http://bit.ly/obviodemo), which help put the contributions of this research into perspective. Further details about each experiment are also given in [54].
Table 7.
Comparison of rediscoveries with other approaches for Calcium-Independent PLA2 - Schizophrenia
| Scenario | Intermediate(s) | Cameron | Srinivasan [7] |
||
|---|---|---|---|---|---|
| S | P | S | P | ||
| Calcium-Independent PLA2 – Schizophrenia |
Oxidative Stress | Y* | 3 | Y | 3 |
| Selenium | Y* | 3 | - | - | |
| Vitamin E | Y* | 3 | - | - | |
Table 9.
Comparison of rediscoveries with other approaches for Testosterone - Sleep
| Scenario | Intermediate(s) | Cameron | Goodwin [21] |
||
|---|---|---|---|---|---|
| S | P | S | P | ||
| Testosterone - Sleep | Cortisol/Hydrocortisone | Y | 10 | Y | 4 |
Highlights.
Implicit and explicit semantics to capture context can be effective for LBD.
Multifaceted subgraphs can be captured using clustering based on shared context.
Discovery browsing using predicates and provenance in MEDLINE could enhance LBD.
Acknowledgments
This research was supported in part by the Intramural Research Program of the US National Institutes of Health, National Library of Medicine. We would especially like to thank Pavan Kapanipathi, Wenbo Wang and Shreyansh Bhatt for their insightful feedback on many aspects of this work. We also thank Swapnil Soni, Nishita Jaykumar, Vishnu Bompally, Gary A. Smith, Drashti Dave, Swapna Abhyankar, Mike Cairelli and Gaurish Anand, who contributed to other aspects of this work.
Appendix A. The Obvio Web Application.
This Appendix describes the Obvio web application (shown in Figure A.15) developed to showcase the rediscoveries. The system consists of 11 components, which can be used to explore subgraphs generated for closed discovery scenarios, using the following steps.
Figure A.15: Screenshot of the Obvio Web Application
Step 1
The user must first select a start term (A) using component 1. For example, the concept Chlorpromazine can be selected as an A-term.
Step 2
The user must then select the target term (C) using component 2. For example, the concept Cardiac Hypertrophy has been selected as the target term, for the given source.
Step 3
The user must then select the ‘Search’ button to submit the search request. Obvio retrieves the metadata for the search terms, which are then displayed in the ‘metadata panel’ immediately below search terms (component 3).
Step 4
The identifiers of the preprocessed subgraphs are shown in the ‘subgraph panel’ in component 4.
Step 5
The user must then select the identifier of a subgraph from the subgraph panel. The corresponding subgraph will be displayed in the ‘viewer’ (component 5).
Step 6
Interesting semantic predications may then be explored by clicking on the edge between concepts of interest in the viewer.
Step 7
The number of MEDLINE articles that contain the visualized semantic predications is shown the ‘Result Metadata Panel’ (component 6). The identifier for the MEDLINE article is also shown (currently shown, 2000 Feb 18). The title of the article is shown in component 7, while the date of publication is shown in component 8. The selected semantic predication is shown in component 9 (currently shown, Calcineurin-CAUSES-Cardiac Hypertrophy). The set of MEDLINE articles that contain the predication are also available for inspection in component 10. More importantly, the sentence from which the semantic predication was extracted will be highlighted.
Step 8
The user may also utilize the functionality from the ‘Filtering panel’ in component 11, to view different perspectives in the subgraphs based on semantic types and groups. Note that the original subgraph can be restored by clicking an arbitrary point in the viewer. Also, when any node in the subgraph has been selected, only the inlinks and outlinks connected to the selected node are displayed.
Footnotes
MeSH is a controlled vocabulary (or thesaurus) of biomedical terms, organized in a hierarchical structure - https://www.nlm.nih.gov/mesh/
SemRep – http://semrep.nlm.nih.gov/
Obvio video demo - http://bit.ly/obviodemo, Obvio Project page - http://wiki.knoesis.org/index.php/Obvio
Semantic Navigator – http://mor2.nlm.nih.gov:8000/perl/auth/semnav.pl
SemMedDB - http://skr3.nlm.nih.gov/SemMedDB/
PubMed – http://www.ncbi.nlm.nih.gov/pubmed
Kloeze, J. Prostaglandins, Proceedings of the 2nd Nobel Symposium, pp. 241–252 (BERT-STR(iM, S. and SAMUELSON, B., eds.) Almqvist and Wiksell, Stockholm, 1967.
Query: Alprostadil AND Raynaud AND 1865:1985/11[DP]. Confirmed in search result #12
eUtils Help - http://www.ncbi.nlm.nih.gov/books/NBK25500/
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest: None.
References
- 1.Swanson DR. Fish oil, raynaud’s syndrome, and undiscovered public knowledge. Perspect Biol Med. 1986;30(1):7–18. doi: 10.1353/pbm.1986.0087. [DOI] [PubMed] [Google Scholar]
- 2.Zahavi J, Hamilton W, O’Reilly M, Leyton J, Cotton L, Kakkar V. Plasma exchange and platelet function in raynauds phenomenon. Thromb Res. 1980;19(1–2):85–93. doi: 10.1016/0049-3848(80)90406-5. [DOI] [PubMed] [Google Scholar]
- 3.DiGiacomo RA, Kremer JM, Shah DM. Fish-oil dietary supplementation in patients with raynaud’s phenomenon: a double-blind, controlled, prospective study. Am J Med. 1989;2:158–164. doi: 10.1016/0002-9343(89)90261-1. [DOI] [PubMed] [Google Scholar]
- 4.Dyerberg J, Jorgensen KA. Marine oils and thrombogenesis. Prog Lipid Res. 1982;21(4):255–269. doi: 10.1016/0163-7827(82)90011-x. [DOI] [PubMed] [Google Scholar]
- 5.Bang HO, Dyerberg J. The lipid Metabolism in Greenlanders - Meddr Gronland. Man and Society; 1981. [Google Scholar]
- 6.Bang HO, Dyerberg J, Nielsen A. Plasma lipid and lipoprotein pattern in greenlandic west-coast eskimos. Lancet. 1971;1(7710):1143–1146. doi: 10.1016/s0140-6736(71)91658-8. [DOI] [PubMed] [Google Scholar]
- 7.Bang HO, Dyerberg J, Sinclair HM. The composition of the eskimo food in north western greenland. Am J Clin Nutr. 1980;33(12):2657–2661. doi: 10.1093/ajcn/33.12.2657. [DOI] [PubMed] [Google Scholar]
- 8.Bang HO, Dyerberg J. The bleeding tendency in greenland eskimos. Dan Med Bull. 1980;27:202–205. [PubMed] [Google Scholar]
- 9.Pringle R, Walder D, JP W. Blood viscosity and raynaud’s disease. Lancet. 1965;1(7395):1086–1088. doi: 10.1016/s0140-6736(65)92673-5. [DOI] [PubMed] [Google Scholar]
- 10.Moncada S. Biology and therapeutic potential of prostacyclin. Stroke; a journal of cerebral circulation. 1983;14(2):157–168. doi: 10.1161/01.str.14.2.157. [DOI] [PubMed] [Google Scholar]
- 11.Moncada S, Vane JR. Prostacyclin and its clinical applications. Annals of clinical research. 1984;16(5–6):241–252. [PubMed] [Google Scholar]
- 12.Smalheiser NR, Swanson DR. Indomethacin and Alzheimer’s disease. Neurology. 1996;46(2) doi: 10.1212/wnl.46.2.583. [DOI] [PubMed] [Google Scholar]
- 13.Smalheiser NR, Swanson DR. Linking estrogen to alzheimer’s disease: An informatics approach. Neurology. 1996;47(3):809–810. doi: 10.1212/wnl.47.3.809. [DOI] [PubMed] [Google Scholar]
- 14.Smalheiser NR, Swanson DR. Calcium-independent phospholipase a2 and schizophrenia. Arch Gen Psychiatry. 1998;55(8):752–753. doi: 10.1001/archpsyc.55.8.752. [DOI] [PubMed] [Google Scholar]
- 15.Swanson DR. Migraine and magnesium: eleven neglected connections. Perspect Biol Med. 1988;31(4):526–557. doi: 10.1353/pbm.1988.0009. [DOI] [PubMed] [Google Scholar]
- 16.Swanson DR. Somatomedin c and arginine: implicit connections between mutually isolated literatures. Perspectives in biology and medicine. 1990;33(2):157–186. doi: 10.1353/pbm.1990.0031. [DOI] [PubMed] [Google Scholar]
- 17.Weeber M, Vos R, Klein H, de Jong-van den Berg LTW, Aronson AR, Molema G. Case report: Generating hypotheses by discovering implicit associations in the literature: A case report of a search for new potential therapeutic uses for thalidomide. JAMIA. 2003;10(3):252–259. doi: 10.1197/jamia.M1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Srinivasan P, Libbus B. Mining medline for implicit links between dietary substances and diseases. Bioinformatics. 2004;20:290–296. doi: 10.1093/bioinformatics/bth914. [DOI] [PubMed] [Google Scholar]
- 19.Ahlers CB, Hristovski D, Kilicoglu H, Rindflesch TC. Using the literature-based discovery paradigm to investigate drug mechanisms. AMIA Annu Symp Proc. 2007:6–10. [PMC free article] [PubMed] [Google Scholar]
- 20.Yetisgen-Yildiz M, Pratt W. Using statistical and knowledge-based approaches for literature-based discovery. Journal of Biomedical Informatics. 2006;39(6):600–611. doi: 10.1016/j.jbi.2005.11.010. [DOI] [PubMed] [Google Scholar]
- 21.Hristovski D, Kastrin A, Peterlin B, Rindflesch TC. Combining semantic relations and dna microarray data for novel hypotheses generation. 2010 [PMC free article] [PubMed] [Google Scholar]
- 22.Hristovski D, Friedman C, Rindflesch TC, Peterlin B. Exploiting semantic relations for literature-based discovery. AMIA Annu Symp Proc. 2006:349–353. [PMC free article] [PubMed] [Google Scholar]
- 23.Hristovski D, Peterlin B, Mitchell JA, Humphrey SM. Improving literature based discovery support by genetic knowledge integration. Stud Health Technol Inform. 2003;95:68–73. [PubMed] [Google Scholar]
- 24.Hristovski D, Peterlin B, Mitchell JA, Humphrey SM. Using literature-based discovery to identify disease candidate genes. I J Medical Informatics. 2005;74(2–4):289–298. doi: 10.1016/j.ijmedinf.2004.04.024. [DOI] [PubMed] [Google Scholar]
- 25.Frijters R, van Vugt M, Smeets R, van Schaik RC, de Vlieg J, Alkema W. Literature mining for the discovery of hidden connections between drugs, genes and diseases. PLoS Computational Biology. 2010;6(9) doi: 10.1371/journal.pcbi.1000943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cameron D, Bodenreider O, Yalamanchili H, Danh T, Vallabhaneni S, Thirunarayan K, et al. A graph-based recovery and decomposition of swan-son’s hypothesis using semantic predications. Journal of Biomedical Informatics. 2013;46(2):238–251. doi: 10.1016/j.jbi.2012.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wilkowski B, Fiszman M, Miller CM, Hristovski D, Arabandi S, Rosemblat G, et al. Graph-based methods for discovery browsing with semantic predications. AMIA Annu Symp Proc. 2011 [PMC free article] [PubMed] [Google Scholar]
- 28.Cairelli MJ, Miller CM, Fiszman M, Workman TE, Rindflesch TC. Semantic medline for discovery browsing: Using semantic predications and the literature-based discovery paradigm to elucidate a mechanism for the obesity paradox. AMIA. 2013:164–173. [PMC free article] [PubMed] [Google Scholar]
- 29.Wren JD, Bekeredjian R, Stewart JA, Shohet RV, Garner HR. Knowledge discovery by automated identification and ranking of implicit relationships. Bioinformatics. 2004;20(3):389–398. doi: 10.1093/bioinformatics/btg421. [DOI] [PubMed] [Google Scholar]
- 30.Miller CM, Rindflesch TC, Fiszman M, Hristovski D, Shin D, Rosemblat G, et al. A closed literature-based discovery technique finds a mechanistic link between hypogonadism and diminished sleep quality in aging men. Sleep. 2012;35(2):279–285. doi: 10.5665/sleep.1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sheth AP, Ramakrishnan C, Thomas C. Semantics for the semantic web: The implicit, the formal and the powerful. Int J Semantic Web Inf Syst. 2005;1(1):1–18. [Google Scholar]
- 32.Gordon MD, Dumais ST. Using latent semantic indexing for literature based discovery. JASIS. 1998;49(8):674–685. [Google Scholar]
- 33.Gordon MD, Lindsay RK. Toward discovery support systems: A replication, re-examination, and extension of swanson’s work on literature-based discovery of a connection between raynaud’s and fish oil. JASIS. 1996;47(2):116–128. [Google Scholar]
- 34.Weeber M, Klein H, de Jong-van den Berg LTW, Vos R. Using concepts in literature-based discovery: Simulating swanson’s raynaud-fish oil and migraine-magnesium discoveries. JASIST. 2001;52(7):548–557. [Google Scholar]
- 35.Lindsay RK, Gordon MD. Literature-based discovery by lexical statistics. JASIS. 1999;50(7):574–587. [Google Scholar]
- 36.Srinivasan P. Text mining: Generating hypotheses from medline. JASIST. 2004;55(5):396–413. [Google Scholar]
- 37.Hristovski D, Stare J, Peterlin B, Dzeroski S. Supporting discovery in medicine by association rule mining in Medline and UMLS. Studies in health technology and informatics. 2001;84(Pt 2):1344–1348. [PubMed] [Google Scholar]
- 38.Pratt W, Yetisgen-Yildiz M. LitLinker: capturing connections across the biomedical literature; K-CAP ’03; NY, USA: ACM. Proceedings of the 2nd international conference on Knowledge capture; 2003. pp. 105–112. [Google Scholar]
- 39.Swanson DR, Smalheiser NR. An interactive system for finding complementary literatures: a stimulus to scientific discovery. Artificial Intelligence. 1997;91(2):183–203. [Google Scholar]
- 40.Torvik VI, Smalheiser NR. A quantitative model for linking two disparate sets of articles in medline. Bioinformatics. 2007;23(13):1658–1665. doi: 10.1093/bioinformatics/btm161. [DOI] [PubMed] [Google Scholar]
- 41.Wren JD. Extending the mutual information measure to rank inferred literature relationships. BMC Bioinformatics. 2004;5:145. doi: 10.1186/1471-2105-5-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smalheiser NR, Torvik VI, Zhou W. Arrowsmith two-node search interface: A tutorial on finding meaningful links between two disparate sets of articles in medline. Comput Methods Prog Biomed. 2009;94(2):190–197. doi: 10.1016/j.cmpb.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hu X, Li G, Yoo I, Zhang X, Xu X. A semantic-based approach for mining undiscovered public knowledge from biomedical literature. Granular Computing. 2005:22–27. [Google Scholar]
- 44.Rindflesch TC, Fiszman M. The interaction of domain knowledge and linguistic structure in natural language processing: interpreting hyper-nymic propositions in biomedical text. Journal of Biomedical Informatics. 2003;36(6):462–477. doi: 10.1016/j.jbi.2003.11.003. [DOI] [PubMed] [Google Scholar]
- 45.Ramakrishnan C, Milnor WH, Perry M, Sheth AP. Discovering informative connection subgraphs in multi-relational graphs. SIGKDD Explorations. 2005;7(2):56–63. [Google Scholar]
- 46.Goodwin JC, Cohen T, Rindflesch TC. Discovery by scent: Discovery browsing system based on the information foraging theory. BIBM Workshops. 2012:232–239. [Google Scholar]
- 47.van der Eijk CC, van Mulligen EM, Kors JA, Mons B, van den Berg J. Constructing an associative concept space for literature-based discovery. J Am Soc Inf Sci Technol. 2004;55(5):436–444. [Google Scholar]
- 48.Spangler S, Wilkins AD, Bachman BJ, Nagarajan M, Dayaram T, Haas P, et al. Automated hypothesis generation based on mining scientific literature; KDD ’14; New York, NY, USA. Proceedings of the 20th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining; 2014. pp. 1877–1886. [Google Scholar]
- 49.Zhang H, Fiszman M, Shin D, Wilkowski B, Rindflesch TC. Clustering cliques for graph-based summarization of the biomedical research literature. BMC Bioinformatics. 2013;14:182. doi: 10.1186/1471-2105-14-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang H, Fiszman M, Shin D, Miller CM, Rosemblat G, Rindflesch TC. Degree centrality for semantic abstraction summarization of therapeutic studies. Journal of Biomedical Informatics. 2011;44(5):830–838. doi: 10.1016/j.jbi.2011.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kilicoglu H, Shin D, Fiszman M, Rosemblat G, Rindflesch TC. Semmeddb: a pubmed-scale repository of biomedical semantic predications. Bioinformatics. 2012;28(23):3158–3160. doi: 10.1093/bioinformatics/bts591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Anyanwu K, Sheth AP. ρ-queries: enabling querying for semantic associations on the semantic web; WWW Budapest, Hungary. Proceedings of the Twelfth International World Wide Web Conference; 2003. May 20–24, pp. 690–699. [Google Scholar]
- 53.Wikipedia. http://en.wikipedia.org/wiki/reachability.
- 54.Cameron D. A context-driven subgraph model for literature-based discovery. Ph.D. thesis; Wright State University; 2014. URL http://knoesis.org/library/resource.php?id=2007. [Google Scholar]
- 55.Abhyankar S, Leishear K, Callaghan FM, Demner-Fushman D, McDonald CJ. Lower short- and long-term mortality associated with overweight and obesity in a large cohort study of adult intensive care unit patients. Crit Care. 2012;16(6):R235. doi: 10.1186/cc11903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yetisgen-Yildiz M, Pratt W. A new evaluation methodology for literature-based discovery systems. Journal of Biomedical Informatics. 2009;42(4):633–643. doi: 10.1016/j.jbi.2008.12.001. [DOI] [PubMed] [Google Scholar]