Abstract
Aminoglycosides are natural or semisynthetic antibiotics derived from actinomycetes. They were among the first antibiotics to be introduced for routine clinical use and several examples have been approved for use in humans. They found widespread use as first-line agents in the early days of antimicrobial chemotherapy, but were eventually replaced in the 1980s with cephalosporins, carbapenems, and fluoroquinolones. Aminoglycosides synergize with a variety of other antibacterial classes, which, in combination with the continued increase in the rise of multidrug-resistant bacteria and the potential to improve the safety and efficacy of the class through optimized dosing regimens, has led to a renewed interest in these broad-spectrum and rapidly bactericidal antibacterials.
The continued increase in multidrug-resistant bacteria has led to renewed interest in legacy aminoglycosides (e.g., streptomycin) and efforts to develop novel aminoglycosides (e.g., plazomicin).
Aminoglycosides are potent, broad-spectrum antibiotics that act through inhibition of protein synthesis. The class has been a cornerstone of antibacterial chemotherapy since streptomycin (Fig. 1) was first isolated from Streptomyces griseus and introduced into clinical use in 1944. Several other members of the class (Fig. 1) were introduced over the intervening years including neomycin (1949, S. fradiae), kanamycin (1957, S. kanamyceticus), gentamicin (1963, Micromonospora purpurea), netilmicin (1967, derived from sisomicin), tobramycin (1967, S. tenebrarius), and amikacin (1972, derived from kanamycin). A shift away from systemic use of the class began in the 1980s with the availability of the third-generation cephalosporins, carbapenems, and fluoroquinolones, which were perceived to be less toxic and/or provide broader coverage than the aminoglycosides. However, increasing resistance to these classes of drugs, combined with more extensive knowledge of the basis of aminoglycoside resistance, has led to renewed interest in the legacy aminoglycosides and the development of novel aminoglycosides such as arbekacin and plazomicin (Fig. 4). These latter agents were designed to overcome common aminoglycoside resistance mechanisms thereby maintaining potency against multidrug-resistant (MDR) pathogens.
Figure 1.
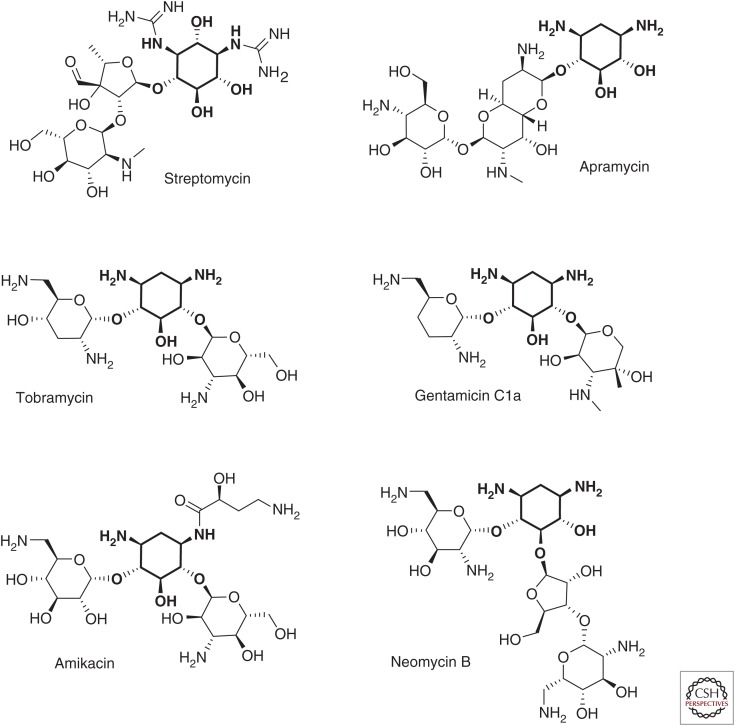
Structures of representative aminoglycosides, including the atypical aminoglycosides streptomycin and apramycin, 4,6-substituted AGs tobramycin, gentamcin, and amikacin, and the 4,5-substituted AG neomycin. The deoxystreptamine or streptidine rings are in bold.
Figure 4.
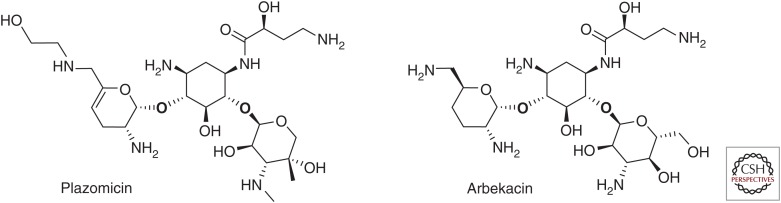
Structures of plazomicin and arbekacin.
Additionally, improved dosing schemes have been developed that attempt to reduce aminoglycoside toxicity while maintaining efficacy. Specifically, clinical studies have reported a lower incidence of nephrotoxicity with once-daily dosing (Nicolau et al. 1995). These data are consistent with the concept that higher doses given less often should reduce the risk of toxicity while maintaining and possibly enhancing efficacy (Drusano et al. 2007). The advantages of once-daily dosing of aminoglycosides are now widely accepted and, for many infection types, this dosing schedule has become the standard of care (Avent et al. 2011). Additionally, inhaled delivery of aminoglycosides has become an area of renewed interest because it allows for greater local exposure within the lungs with reduced systemic toxicity. Inhaled tobramycin is available in the European Union (EU) and the United States for the treatment of patients with chronic Pseudomonas aeruginosa lung infection associated with cystic fibrosis (CF), and inhaled amikacin and arbekacin are in development for potential use in CF and acute respiratory tract infections.
SPECTRUM OF ACTIVITY
Aminoglycosides are active against various Gram-positive and Gram-negative organisms. Aminoglycosides are particularly potent against members of the Enterobacteriaceae family, including Escherichia coli, Klebsiella pneumoniae and K. oxytoca, Enterobacter cloacae and E. aerogenes, Providencia spp., Proteus spp., Morganella spp., and Serratia spp. (Ristuccia and Cunha 1985; Aggen et al. 2010; Landman et al. 2010). Furthermore, aminoglycosides are active against Yersinia pestis (Heine 2015) and Francisella tularensis (Ikäheimo et al. 2000), the causative agents of plague and tularemia, respectively. The class also has good activity against Staphylococcus aureus, including methicillin-resistant and vancomycin-intermediate and -resistant isolates, P. aeruginosa and to a lesser extent Acinetobacter baumannii (Ristuccia and Cunha 1985; Karlowsky et al. 2003; Aggen et al. 2010; Landman et al. 2011). Many Mycobacterium spp. are also susceptible to aminoglycosides including Mycobacterium tuberculosis, M. fortuitum, M. chelonae, and M. avium (Swenson et al. 1985; Ho et al. 1997).
Active electron transport is required for aminoglycoside uptake into cells, so the class inherently lacks activity against anaerobic bacteria (Kislak 1972; Martin et al. 1972; Ramirez and Tolmasky 2010). Aminoglycosides are also inactive against most Burkholderia spp. and Stenotrophomonas spp. as well as Streptococcus spp. and Enterococcus spp. (Brogden et al. 1976; Vakulenko and Mobashery 2003; Brooke 2012; Podnecky et al. 2015).
Contemporary large-scale surveillance programs provide an understanding of current aminoglycoside susceptibility among important pathogens associated with common infection types. A recent surveillance study of Gram-negative organisms isolated from patients hospitalized in intensive care units (ICUs) in the United States and the EU found that amikacin and gentamicin showed good activity against key Gram-negative pathogens (Sader et al. 2014). In the United States, 99.5% and 87.9% of E. coli isolates were susceptible to amikacin and gentamicin, respectively, according to the Clinical and Laboratory Standards Institute (CLSI) criteria. Likewise, 97.3% and 87.2% of E. coli isolates from the EU were susceptible to amikacin and gentamicin, respectively, according to the European Committee on Antimicrobial Susceptibility Testing (EUCAST) criteria. Among Klebsiella spp., 94.8% and 92.7% of U.S. isolates and 90.5% and 83.3% of EU isolates were susceptible to amikacin and gentamicin, respectively. Amikacin was one of the few agents that retained activity against P. aeruginosa (97.3% susceptibility in the United States and 84.9% in the EU according to CLSI and EUCAST criteria, respectively). Similarly, aminoglycoside susceptibility rates were high among isolates collected from U.S. medical centers in 2012 (Sader et al. 2015). In this study, CLSI-based susceptibility rates were 99.0%, 88.2%, and 86.3% among E. coli and 88.2%, 89.2%, and 82.4% against K. pneumoniae for amikacin, gentamicin, and tobramycin, respectively. However, activity was reduced among K. pneumoniae carbapenemase (KPC)-producing K. pneumoniae, which are often resistant to multiple classes of drugs, for amikacin (42.5% susceptible), gentamicin (50.0% susceptible), and tobramycin (25.0% susceptible). Amikacin was the most active agent tested against P. aeruginosa (98% susceptible), followed closely by tobramycin (90% susceptible) and gentamicin (88.0% susceptible). Broad potency was also observed against S. aureus (96.0%, 95.0%, and 76.0% for amikacin, gentamicin, and tobramycin, respectively), but these compounds were less active against A. baumannii (≤58.0% susceptible).
The broad-spectrum activity of aminoglycosides is enhanced in vitro through synergy with other classes of antimicrobials. This phenomenon, in which the combined effect of two antimicrobial agents is greater than the sum of their individual effects, is particularly well characterized between aminoglycosides and cell-wall-active agents such as β-lactams. In vitro synergy between aminoglycosides and β-lactams has been observed in both Gram-negative and -positive organisms, including wild-type and MDR isolates, using a variety of methodologies (Eliopoulos and Eliopoulos 1988). These in vitro observations have helped to stimulate the use of aminoglycoside-containing combination therapy in the treatment of a number of infection types (see below).
MECHANISM OF ACTION
Aminoglycosides inhibit protein synthesis by binding, with high affinity, to the A-site on the 16S ribosomal RNA of the 30S ribosome (Kotra et al. 2000). Although aminoglycoside class members have a different specificity for different regions on the A-site, all alter its conformation. As a result of this interaction, the antibiotic promotes mistranslation by inducing codon misreading on delivery of the aminoacyl transfer RNA. This results in error prone protein synthesis, allowing for incorrect amino acids to assemble into a polypeptide that is subsequently released to cause damage to the cell membrane and elsewhere (Davis et al. 1986; Mingeot-Leclercq et al. 1999; Ramirez and Tolmasky 2010; Wilson 2014). Some aminoglycosides can also impact protein synthesis by blocking elongation or by directly inhibiting initiation (Davis 1987; Kotra et al. 2000; Wilson 2014). The exact mechanism of binding and the subsequent downstream effects varies by chemical structure, but all aminoglycosides are rapidly bactericidal (Davis 1987; Mingeot-Leclercq et al. 1999) and typically produce a prolonged postantibiotic effect (PAE) (Zhanel et al. 1991) The PAE has been shown to be directly related to the length of time that the bacteria take to recover from the inhibition of protein synthesis (Stubbings et al. 2006). It is hypothesized that this is dependent on the eventual disassociation of the antibiotic from its target and exit from the cell.
Aminoglycosides are characterized by a core structure of amino sugars connected via glycosidic linkages to a dibasic aminocyclitol, which is most commonly 2-deoxystreptamine (Mingeot-Leclercq et al. 1999). Aminoglycosides are broadly classified into four subclasses based on the identity of the aminocyclitol moiety: (1) no deoxystreptamine (e.g., streptomycin, which has a streptidine ring); (2) a mono-substituted deoxystreptamine ring (e.g., apramycin); (3) a 4,5-di-substituted deoxystreptamine ring (e.g., neomycin, ribostamycin); or (4) a 4,6-di-substituted deoxystreptamine ring (e.g., gentamicin, amikacin, tobramycin, and plazomicin) (Magnet and Blanchard 2005; Wachino and Arakawa 2012). Examples of each subclass are shown in Figure 1. The core structure is decorated with a variety of amino and hydroxyl substitutions that have a direct influence on the mechanisms of action and susceptibility to various aminoglycoside-modifying enzymes (AMEs) associated with each of the aminoglycosides.
Aminoglycoside entry into bacterial cells is comprised of three distinct stages, the first of which increases permeability of the bacterial membrane, whereas the second and third are energy-dependent. The first stage involves electrostatic binding of the polycationic aminoglycoside to the negatively charged components of the bacterial membrane, such as the phospholipids and teichoic acids of Gram-positive organisms and the phospholipids and lipopolysaccharide (LPS) of Gram-negative organisms, followed by displacement of magnesium ions (Davis 1987; Taber et al. 1987; Ramirez and Tolmasky 2010). These cations are responsible for cross bridging and stabilization of the lipid components of the bacterial membrane and their removal leads to disruption of the outer membrane, enhanced permeability, and initiation of aminoglycoside uptake (Hancock et al. 1981, 1991; Hancock 1984; Ramirez and Tolmasky 2010). This phenomenon facilitates entry into the cytoplasm via a slow, energy-dependent, electron-transport-mediated process (Kislak 1972; Martin et al. 1972; Davis 1987; Taber et al. 1987; Ramirez and Tolmasky 2010). Inhibition of protein synthesis and mistranslation of proteins occurs once aminoglycoside molecules access the cytoplasm. These mistranslated proteins insert into and cause damage to the cytoplasmic membrane itself and facilitate subsequent aminoglycoside entry (Nichols and Young 1985; Davis et al. 1986). This then leads to rapid uptake of additional aminoglycoside molecules into the cytoplasm, increased inhibition of protein synthesis, mistranslation, and accelerated cell death (Davis et al. 1986; Davis 1987; Taber et al. 1987; Ramirez and Tolmasky 2010).
MECHANISMS OF AMINOGLYCOSIDE RESISTANCE
Aminoglycoside resistance takes many different forms including enzymatic modification, target site modification via an enzyme or chromosomal mutation, and efflux. Each of these mechanisms has varying effects on different members of the class and often multiple mechanisms are involved in any given resistant isolate. Resistance to aminoglycosides via target site mutations has not been observed because nearly all prokaryotes, with the exception of Mycobacterium spp. (Bercovier et al. 1986) and Borrelia spp. (Schwartz et al. 1992), encode multiple copies of rRNA. Although contemporary large-scale surveillance programs provide an understanding of phenotypic aminoglycoside resistance among important pathogens, these studies have generally not focused on the epidemiology of specific resistance mechanisms (Jones et al. 2014; Sader et al. 2015).
Enzymatic Drug Modification
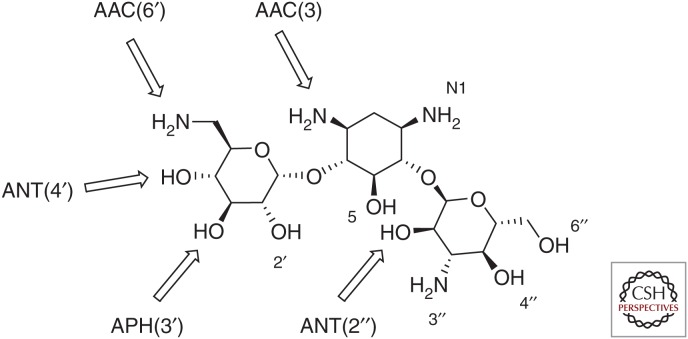
AMEs are often found on plasmids containing multiple resistance elements, including other AMEs or β-lactamases. The mobility of these enzymes might be tied to their origins, which has been hypothesized to be via horizontal gene transfer from the actinomycetes responsible for the natural production of aminoglycosides (Shaw et al. 1993; Ramirez and Tolmasky 2010). More than 100 AMEs have been described and are broadly categorized into three groups based on their ability to acetylate, phosphorylate, or adenylate amino or hydroxyl groups found at various positions around the aminoglycoside core scaffold (Ramirez and Tolmasky 2010). These modifications decrease the binding affinity of the drug for its target and lead to a loss in antibacterial potency (Llano-Sotelo et al. 2002). These three families of AMEs include aminoglycoside N-acetyltransferases (abbreviated AACs), aminoglycoside O-nucleotidyltransferases (ANTs), and aminoglycoside O-phosphotransferases (APHs). The classes are further divided into subtypes according to the position on the aminoglycoside that the enzyme modifies followed by a Roman numeral and, in some cases, a letter when multiple enzymes exist that modify the same position (Shaw et al. 1993; Ramirez and Tolmasky 2010). The major sites of aminoglycoside modification for the most common AMEs are shown for kanamycin A in Figure 2.
Figure 2.
Sites of chemical modification by representative aminoglycoside-modifying enzymes (AMEs) on kanamycin A.
Aminoglycoside Acetyltransferases
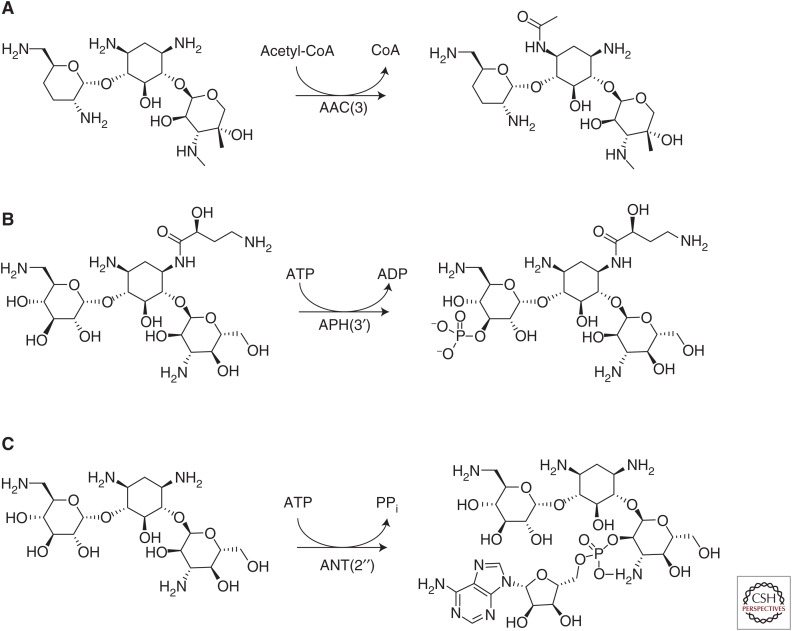
The aminoglycoside acetyltransferases or AACs comprise the largest group of AMEs. They are part of the GCN5-related N-acetyltransferase (GNAT) superfamily of ∼10,000 described proteins (Ramirez and Tolmasky 2010). These enzymes acetylate amino groups found at various positions on the aminoglycoside scaffold in an acetyl-CoA-dependent reaction (Fig. 3A). There are four main subclasses of this group of enzymes whose nomenclature is derived from the specific amino group that is modified. Specifically, AAC(1) and AAC(3) target the amino groups found at position 1 and 3, respectively, of the 2-deoxystreptamine ring, whereas AAC(2′) and AAC(6′) target amino groups found at the 2′ and 6′ position of the 2,6-dideoxy-2,6-diaminoglucose ring (Mingeot-Leclercq et al. 1999; Wright and Thompson 1999; Azucena and Mobashery 2001; Magnet and Blanchard 2005).
Figure 3.
Examples of aminoglycoside modification by AMEs. (A) An example of chemical modification of gentamicin catalyzed by the aminoglycoside acetyltransferase AAC(3). (B) An example of chemical modification catalyzed by the aminoglycoside phosphotransferase APH(3′) on amikacin. (C) Adenylation of the 2″ hydroxyl of kanamycin A catalyzed by the aminoglycoside nucleotidyltransferase ANT(2″).
A representative of the AAC family (AAC(6′)-IV) was the first AME found in bacteria to be described in the literature (Okamoto and Suzuki 1965). Since then, >70 members of the AAC family have been described. Frequently observed class members found among Gram-negative bacteria include the AAC(6′)-1 enzyme that leads to amikacin, netilmicin, and tobramycin resistance, AAC(3)-IIa, which is responsible for resistance to gentamicin, tobramycin, and netilmicin, and AAC(3)-I, which modifies gentamicin (Shaw et al. 1993; Castanheira et al. 2015). Less common are the AAC(6′)-APH(2″) hybrid enzyme responsible for high-level aminoglycoside resistance in Enterococcus faecalis (Culebras and Martínez 1999), the chromosomally encoded AAC(6′)-Ii enzyme responsible for intrinsic resistance to aminoglycosides among E. faecium (Costa et al. 1993) and the chromosomal AAC(2′) enzyme found in Mycobacterium spp. and Providencia stuartii (Rather et al. 1993; Aínsa et al. 1997; Macinga and Rather 1999; Ramirez and Tolmasky 2010). Last, a variant of AAC(6′)-Ib that has acquired the ability to modify fluoroquinolones without significantly altering its activity against aminoglycosides has been identified in clinical isolates of Gram-negative bacteria (Robicsek et al. 2006; Strahilevitz et al. 2009).
Aminoglycoside Phosphotransferases
The second largest group of AMEs is the APHs. This structurally diverse group of enzymes acts like kinases in that they catalyze the ATP-dependent phosphorylation of hydroxyl groups found on aminoglycosides (Fig. 3B). APH enzymes are functionally and structurally similar to the serine–threonine and tyrosine kinases found in eukaryotes (Wright and Thompson 1999). The modifications made by these enzymes lower binding affinity to the target by decreasing the hydrogen bonding potential of aminoglycoside hydroxyl groups with important rRNA residues. Most of the >30 described APH enzymes belong to the APH(3′) subfamily (Kim and Mobashery 2005), although variants that target the 2″ hydroxyl also exist (Ramirez and Tolmasky 2010). These enzymes are found in diverse groups of Gram-negative bacteria, although APH(3′)-IIIa was discovered in S. aureus and Enterococcus spp. All members of the family lead to kanamycin and neomycin resistance with various members of the family also able to modify a variety of other aminoglycosides, including amikacin and gentamicin B (Shaw et al. 1993).
Aminoglycoside Nucleotidyltransferases
The final group of AMEs is the ANTs. These enzymes act by adding AMP from an ATP donor to hydroxyl groups at the 2″, 3″, 4′, 6, and 9 positions (Fig. 3C). The most clinically relevant members of the class include ANT(2″) and ANT(4′) (Kotra et al. 2000; Magnet and Blanchard 2005), which were first described in K. pneumoniae and S. aureus, respectively (Benveniste and Davies 1971; Le Goffic et al. 1976). ANT(2″) broadly effects the activity of 4,6-di-substituted aminoglycosides (Gates 1988), whereas ANT(4′) targets kanamycin A, B, and C, gentamicin A, amikacin, tobramycin, and neomycin B and C. Other members of this class include ANT(3″), ANT(6), and ANT(9), which confer resistance to streptomycin and spectinomycin (Hollingshead and Vapnek 1985; Murphy 1985; Ounissi et al. 1990).
16S rRNA Methylation
Target site modification leading to aminoglycoside resistance occurs via the action of 16S rRNA methyltransferases (RMTs). These enzymes modify specific rRNA nucleotide residues in a manner that blocks aminoglycosides from effectively binding to their target (Beauclerk and Cundliffe 1987; Cundliffe 1989; Wachino and Arakawa 2012). There are two general classes of RMTs that are characterized by the specific nucleotide residues that they modify. These include enzymes that render bacteria resistant to 4,6-di-substituted aminoglycosides via methylation of the N7 position of nucleotide G1405 (Thompson et al. 1985; Beauclerk and Cundliffe 1987) and those that affect both 4,6- and 4,5-di-substituted aminoglycosides through methylation of the N1 position of nucleotide A1408 (Skeggs et al. 1985; Beauclerk and Cundliffe 1987; Mingeot-Leclercq et al. 1999).
The first clinical case of a pathogen with an RMT as a mechanism of aminoglycoside resistance was reported in a P. aeruginosa isolate from Japan in 2003 (Yokoyama et al. 2003). This isolate contained a plasmid-encoded RMT, named RmtA for ribosomal methyltransferase A. Subsequently, several additional plasmid-borne RMTs, encoded by the genes armA, rmtB1, rmtB2, rmtC, rmtD, rmtD2, rmtE, rmtF, rmtG, and rmtH, have emerged in clinical isolates that show high-level resistance to multiple aminoglycosides (Yokoyama et al. 2003; Doi et al. 2004; Yan et al. 2004; Yamane et al. 2005; Wachino et al. 2006; Wachino and Arakawa 2012). These enzymes modify the G1405 nucleotide and, thus, impact the activity of all 4,6-di-substituted aminoglycosides (i.e., amikacin, gentamicin, and tobramycin). In 2007, the enzyme NpmA was discovered encoded on a plasmid in an aminoglycoside-resistant E. coli clinical isolate, also from Japan (Wachino et al. 2007). This methyltransferase modifies the A1408 nucleotide and, thus, impacts 4,6- and 4,5-di-substituted as well as monosubstituted aminoglycosides, conferring pan-aminoglycoside resistance. To date, there has been only one additional report of this enzyme in a clinical isolate (Al Sheikh et al. 2014).
Efflux-Mediated Resistance
Several members of the resistance–nodulation–division (RND) family of efflux systems have been shown to be involved in intrinsic aminoglycoside resistance in various pathogens (Aires et al. 1999; Westbrock-Wadman et al. 1999; Rosenberg et al. 2000; Magnet et al. 2001; Hocquet et al. 2003; Islam et al. 2004). In the opportunistic pathogen P. aeruginosa, intrinsic low-level resistance to aminoglycosides, tetracycline and erythromycin, is mediated by the expression of the multiple efflux (Mex) XY-OprM system. MexXY orthologs are found in other species of bacteria. For example, members of the Burkholderia cenopacia complex are often intrinsically resistant to aminoglycosides via RND efflux pumps (Buroni et al. 2009). A homologous transporter in E. coli, AcrD, participates in efflux of aminoglycosides (Rosenberg et al. 2000) as does the Acinetobacter drug efflux (Ade) ABC efflux system in A. baumannii (Magnet et al. 2001) and the major facilitator superfamily (MFS) in Mycobacteria spp.
Molecular Epidemiology of Aminoglycoside Resistance Mechanisms
Comprehensive molecular data regarding the current prevalence of specific aminoglycoside resistance mechanisms among common pathogens is sparse. One recent study evaluated the aminoglycoside resistance mechanisms found among 200 Gram-negative bacilli isolates selected at random from an extensive culture collection (Castanheira et al. 2015). Ninety-nine Enterobacteriaceae (91.9% AME-positive), 49 A. baumannii (79.6% AME-positive), and 52 P. aeruginosa (63.5% AME positive) with a variety of aminoglycoside resistance profiles were included. A diversity of AME genes were identified with the most prevalent being aac(6′)-Ib (n = 75), ant(3′)-Ia (n = 51), and aac(3)-IIa (n = 45), and with 26 isolates harboring more than one AME gene. In addition, 21 of the isolates were found to carry an RMT including 12 Enterobacteriaceae, seven A. baumannii, and two P. aeruginosa. Many of these isolates were also resistant to other common antibiotics used to treat Gram-negative infections, highlighting the ability of these resistance mechanisms to spread through conjugation of plasmids and nonreplicative transposons among bacteria (Courvalin 1994; Waters 1999; Dzidic and Bedeković 2003; Feizabadi et al. 2004).
AMEs are commonly found in association with other key resistance elements, such as carbapenemases and extended spectrum β-lactamases (ESBLs). Among 50 carbapenem-resistant K. pneumoniae clinical isolates from two U.S. medical centers (80% possessing KPC-2, 10% possessing KPC-3 with all KPC+ isolates also expressing TEM-1 and SHV-12), 98% of isolates expressed at least one AME (Almaghrabi et al. 2014). Specifically, 98% were positive for aac(6′)-Ib, 56% positive for aph(3′)-Ia, 38% positive for aac(3′)-IV, and 2% positive for ant(2″)-Ia. Overall, 40%, 98%, and 16% of the strains were nonsusceptible to gentamicin, tobramycin, and amikacin, respectively. Plazomicin, a novel aminoglycoside in clinical development that was designed to evade AME-based resistance, had MICs that ranged from 0.25 to 1 µg/mL against this set of isolates. AME characterization in 330 aminoglycoside-resistant clinical Enterobacteriaceae isolates from Spain revealed the presence of aph(3″)-Ib and ant(3″)-Ia genes in 65.4% and 37.5% of the isolates, consistent with 92% phenotypic resistance to streptomycin found in this strain collection (Miró et al. 2013). These isolates were resistant to other aminoglycosides to varying degrees, including gentamicin (18.4%), tobramycin (16.9%), and amikacin (1.5%), indicating the presence of other AMEs; aph(3′)-Ia was found in 13.9% of isolates, aac(3)-IIa in 12.4%, aac(6′)-Ib in 4.2%, ant(2″)-Ia in 3.6%, and aph(3″)-IIa 1.2% (Miró et al. 2013). Underscoring the association of AMEs with resistance elements to other key antibiotic classes, many of these isolates also produced ESBLs and were resistant to the fluoroquinolones.
Similar to the AMEs, comprehensive data describing the prevalence and molecular epidemiology of RMTs is lacking. A recent literature review described reports of widespread, global dissemination of genes encoding RMTs among isolates from both human and livestock sources (Wachino and Arakawa 2012). Despite the widespread detection of RMTs across many regions, the prevalence appears to vary by region with the highest rates reported in Asia. A SENTRY surveillance study from the Asia-Pacific region (2007–2008) detected genes encoding RMTs in 6.9% (China), 10.5% (India), 1.5% (Hong Kong), 6.1% (Korea), and 5.0% (Taiwan) of Enterobacteriaceae isolates (Bell et al. 2010). Lower rates (≤1.3%) of RMTs among Enterobacteriaceae have been reported from single institution or local studies from European medical centers (Wachino and Arakawa 2012). Similar to the association between AMEs and other key resistance mechanisms, an association between RMTs and specific β-lactamases has been described. Seventy-six percent of the RMT containing isolates described in the SENTRY surveillance study above also possessed a CTX-M ESBL (Bell et al. 2010), and RMTs are frequently found in association with the New Delhi metallo-β-lactamase (NDM) (Berçot et al. 2011; Livermore et al. 2011; Mushtaq et al. 2011; Poirel et al. 2014).
AGENTS IN DEVELOPMENT
Plazomicin is a new aminoglycoside that was specifically engineered to be resistant to the action of the AMEs that are prevalent in key Gram-negative pathogens (Armstrong and Miller 2010). It is synthesized from a sisomicin scaffold that is intrinsically refractory to modification by APH(3′)-III, -VI, and -VII and ANT(4′), which confer amikacin resistance because of an absence of the 3′- and 4′-OH groups. Modification at the N-1 position via addition of a hydroxylaminobutyric acid substituent sterically hinders the action of the AAC(3), ANT(2″), and APH(2″) enzymes, which confer resistance to gentamicin and tobramycin. Finally, addition of a hydroxyethyl substituent at the 6′ position inhibits the action of the AAC(6′) enzymes, which confer resistance to a broad range of agents, including amikacin, tobramycin, and gentamicin (Fig. 4). Importantly, these modifications to sisomicin do not reduce intrinsic potency as has been associated with previous efforts to protect the 6′ position and, as predicted, lead to improved activity against Enterobacteriaceae (MIC90 ≤2 mg/L) that are resistant to currently available aminoglycosides (Nagabhushan et al. 1982; Aggen et al. 2010). Plazomicin retains vulnerability to modification by AAC(2′)-I, a chromosomal AME found in P. stuartii and some mycobacterial species. However, this enzyme is rare, has not been found on a mobile element and has not been shown to have clinical relevance in Mycobacterium spp. In addition, plazomicin, like all 4,6-linked aminoglycosides, is inactive against isolates that produce RMTs. As described above, this mechanism of resistance frequently travels on mobile genetic elements with NDM-positive Enterobacteriaceae, and, thus, plazomicin is not active against many isolates harboring this enzyme (Berçot et al. 2011; Livermore et al. 2011; Mushtaq et al. 2011; Poirel et al. 2014).
Similar to plazomicin, arbekacin, a semi-synthetic derivative of dibekacin, was specifically engineered to overcome the action of a subset of clinically important AMEs (Fig. 4). Arbekacin is stable to the action of AMEs commonly found in methicillin-resistant S. aureus (MRSA), such as APH, ANT, and AAC and possesses potent activity against this clinically important pathogen (Matsumoto 2014). Although arbekacin has been approved for use in Japan since 1990 for the treatment of sepsis and pneumonia caused by MRSA, this molecule also retains potent activity against key Gram-negative pathogens, including MDR strains, and has more recently gained attention as a potential therapy for infections caused by these organisms. In a recent surveillance study of isolates from hospitalized patients with pneumonia, arbekacin was the most potent aminoglycoside tested against ESBL-producing E. coli and was slightly more active than amikacin and tobramycin against ESBL- and KPC-expressing K. pneumoniae. Similarly, arbekacin possessed greater activity against P. aeruginosa and Acinetobacter spp., including MDR isolates, than the other aminoglycosides tested (amikacin, gentamicin, and tobramcin) (Sader et al. 2015). Because of its broad-spectrum in vitro activity against both MDR Gram-positive and Gram-negative pathogens, arbekacin is currently under development as an inhalational agent for the treatment of mechanically ventilated patients with bacterial pneumonia (clinicaltrials.gov/ct2/show/NCT02459158) and is under study at Walter Reed Army Medical Hospital for the treatment of patients with infections caused by MDR organisms that have limited treatment options (clinicaltrials.gov/ct2/show/NCT01659515).
PHARMACOKINETICS AND PHARMACODYNAMICS
Aminoglycosides are poorly absorbed via the gastrointestinal (GI) tract and are, thus, administered via the intravenous or intramuscular route (Ramirez and Tolmasky 2010; Craig 2011). The volume of distribution for members of the aminoglycoside class approaches total body volume, indicating a broad distribution into tissues, including the lung (Simon et al. 1973). This feature has led to extensive use of aminoglycosides as part of combination regimens for the treatment of pneumonia. Aminoglycosides are rapidly cleared through the urinary tract, which also makes these drugs ideal for the treatment of urinary tract infections (Lode et al. 1976; Ramirez and Tolmasky 2010; Craig 2011).
The relationship between aminoglycoside pharmacokinetics (PKs) and pharmacodynamics (PDs) has been studied extensively in mice. The PK/PD variable that is most often correlated with efficacy of aminoglycosides is the ratio of area under the concentration–time curve (AUC) to MIC, although peak concentration also appears to play a role. The magnitude of the PK/PD target for aminoglycosides is not as well defined as it is for other antibiotic classes because of, until recently, the lack of development of new aminoglycosides. Available data suggests that significant variations in the PK/PD target exist between species and body site of infection. For example, an AUC/MIC target of 100 was reportedly associated with a 1- to 2-log10 kill in the mouse neutropenic thigh infection model with amikacin and K. pneumoniae (Craig 2011), whereas this same group reported better efficacy at the same dose level and with the same strain in the mouse lung infection model (Craig et al. 1991). These results were potentially a result of a longer measured PAE in the lung compared with the thigh (Craig et al. 1991).
AUC/MIC thresholds have been correlated to efficacy in patients in whom extensive PK sampling was also conducted. These case reports and reviews describe a variety of different ways that the AUC/MIC ratio might be used to predict outcomes in patients. Smith et al. (2001) reviewed 23 patients with intra-abdominal infections (n = 16) or lower respiratory infections (n = 7) treated with tobramycin infused >30 min to achieve a Cmax of between 4 and 10 mg/L. A PK/PD model constructed from these patients data showed that improved efficacy was observed when an AUC/MIC ratio of ≥110 was achieved compared with AUC/MIC ratios of <110 (80% clinical cure rate compared with 47%). Other investigators have found similar correlations between exposure and efficacy. Jacobs (2001) describes the need to achieve an AUC/MIC ratio of 25 for less severe infections or in immune competent patients and a ratio of at least 100 in patients with severe infections and/or those that are immune-compromised. Zelenitsky et al. (2003) found significantly improved outcomes in patients with bacteremia treated with gentamicin when AUC/MIC ratios exceeded 70 compared with AUC/MIC ratios <70 (90% cure vs. 45.5%, respectively). However, a much stronger correlation with outcome was noted for Cmax/MIC with 84% and 90% clinical cure rates for Cmax/MIC values of 4.8 or 8 compared with 0% clinical cure for Cmax/MIC < 2.9.
There is substantial evidence that administering larger doses of aminoglycosides less frequently may be associated with improved outcomes compared with providing the same total daily dose over more frequent dosing schedules (Barclay et al. 1999). This dosing approach, known as once daily or extended interval dosing, takes advantage of three features of aminoglycosides—concentration-dependent killing, rapid elimination, and a prolonged PAE. Larger doses are thought to increase target body site concentrations to improve PD while minimizing potential toxicity through allowance for a period of time in which there is little or no drug in circulation. The PAE properties of the class allow for an extended period of killing after the drug is cleared from the body and before the next dose is administered. A number of meta-analyses of results from clinical trials comparing once versus multiple daily administration of aminoglycosides have been published (Barclay et al. 1999). Overall, the results of these studies suggest that once daily dosing is associated with reduced nephrotoxicity and equal, if not slightly improved, efficacy.
Therapeutic drug management (TDM), the clinical practice of measuring drug concentrations at designated intervals for use in optimizing individualized dosage regimens, has further improved the safety profile of aminoglycosides. Older studies using multiple daily doses of these drugs have shown nephrotoxicity rates of 10% to 20% (Humes et al. 1982; Moore et al. 1984), whereas lower rates of nephrotoxicity ranging from 0% to 14% have been reported with once-daily dosing regimens with dose adjustment guided by the use of TDM (Prins et al. 1993; Nicolau et al. 1995; Murry et al. 1999; Rybak et al. 1999; Buijk et al. 2002). The advantages of once-daily or extended interval dosing of aminoglycosides combined with the use of TDM are now widely accepted and, for many infection types, this dosing approach has become the standard of care (Avent et al. 2011).
CLINICAL USES OF AMINOGLYCOSIDES
The spectrum of activity, rapid bactericidal activity, and favorable chemical and pharmacokinetic properties of aminoglycosides make them a clinically useful class of drugs. Aminoglycosides are used as single agents and in combination with other antibiotics in both empirical and definitive therapy for a broad range of indications (Avent et al. 2011; Jackson et al. 2013).
In patients with serious infections caused by Gram-negative pathogens, the receipt of empiric combination therapy containing at least one antimicrobial agent to which the pathogen is susceptible leads to lower mortality and improved outcomes (Tamma et al. 2012). In addition to helping ensure that the pathogen is adequately covered by at least one active drug, the use of empiric aminoglycoside-β-lactam combination therapy has also been theorized to contribute to improved outcomes by taking advantage of the in vitro synergy observed between these classes and to prevent the emergence of resistance (Pankuch et al. 2010; Le et al. 2011). Although clinical data to support the latter two theoretical benefits of combination therapy are conflicting (Tamma et al. 2012), aminoglycosides are often combined with β-lactams for the empirical treatment of severe sepsis and certain nosocomial infections in patients with a high risk of mortality or when there is concern that the causative pathogen may be resistant to more commonly used agents (American Thoracic Society; Infectious Diseases Society of America 2005; Dellinger et al. 2013). More recently, aminoglycosides have increasingly become important components of therapy for patients infected with MDR pathogens, such as carbapenem-resistant Enterobacteriaceae (CRE), for whom few treatment options remain. In a retrospective cohort study of 50 cases of sepsis caused by carbapenem- and colistin-resistant K. pneumoniae, the receipt of gentamicin as part of definitive therapy was associated with lower mortality compared with the receipt of non-gentamicin-containing regimens (Gonzalez-Padilla et al. 2015). The novel aminoglycoside plazomicin (see section on Agents in Development) is currently under phase 3 study for the treatment of serious infections caused by CRE (clinicaltrials.gov/ct2/show/NCT01970371).
Aminoglycosides are also an important component of combination therapy for multidrug-resistant tuberculosis (MDR-TB) and certain non-tuberculous mycobacterial (NTM) infections. Current MDR-TB treatment guidelines recommend inclusion of one of the following agents during the intensive phase of therapy: amikacin, kanamycin, streptomycin, or capreomycin, a cyclic peptide antibiotic that is often considered as an aminoglycoside because of its mechanism of action. Each of these agents possesses potent bactericidal activity against M. tuberculosis (Ho et al. 1997) and the choice of agent depends on previous injectable use (if any) and the likelihood of resistance. A meta-analysis including 32 studies with >9000 treatment episodes did not reveal any clear differences in efficacy among the available agents (World Health Organization 2011). Similar to treatment of MDR-TB, combination therapy for patients with fibrocavitary, severe nodular/bronchiectatic or macrolide-resistant lung disease because of the M. avium complex generally includes amikacin or streptomycin (Griffith et al. 2007). Among the rapidly growing mycobacteria, amikacin is the preferred agent for infections because of M. fortuitum or M. abscessus, whereas tobramycin is the most active agent against M. chelonae (Griffith et al. 2007).
Aminoglycosides remain the preferred therapy for certain zoonotic infections such as plague and tularemia. Although streptomycin has traditionally been the agent of choice for these infection types, gentamicin is now widely used because of the broader availability of this agent as well as data suggesting similar efficacy to streptomycin (Mwengee et al. 2006; Snowden and Stovall 2011). Aminoglycosides are bactericidal against these organisms and the use of bacteriostatic agents, such as doxycycline or chloramphenicol has led to treatment failures (Dennis et al. 2001; Snowden and Stovall 2011).
Inhaled tobramycin therapy in CF patients with chronic lung infection caused by P. aeruginosa has been shown to improve respiratory function, decrease hospitalizations, and reduce systemic antibiotic use (Ramsey et al. 1999), and has contributed to a significant increase in survival for these patients (Sawicki et al. 2012). Inhaled aminoglycosides, alone or as part of combination therapy, are currently under evaluation as adjunctive agents for treatment of additional respiratory infection types including chronic lung infections associated with non-CF bronchiectasis and refractory NTM infections of the lung, and for the prevention and/or treatment of ventilator associated infections, tracheobronchitis (VAT), and pneumonia (VAP). Regarding the latter indication, although definitive data from large randomized trials is not available, a number of small studies focused on the prevention or treatment of VAP have provided encouraging results. A meta-analysis of eight comparative trials of prophylactic aerosolized antibiotics (four of which used inhaled tobramycin or gentamicin) found that ICU-acquired pneumonia was less common in the group of patients that received antibiotic prophylaxis compared with those who received no prophylaxis (Falagas et al. 2006). Similarly, a meta-analysis of five comparative trials of adjunctive aerosolized antibiotics in the treatment of VAP, each of which evaluated an inhaled aminoglycoside versus placebo or no therapy, revealed that administration of aerosolized antibiotics was associated with better treatment success compared with control in both the intent-to-treat and clinically evaluable populations (Ioannidou et al. 2007). The relative inefficiency of drug delivery through the ventilator circuit is a significant challenge with the use of aerosolized antibiotics for the treatment of ventilator-associated infections. To overcome this issue, BAY41-6551, an investigational drug–device combination of amikacin specifically formulated for inhalation, has been developed. This drug–device combination is currently under phase 3 study as adjunctive therapy in intubated and mechanically ventilated patients with Gram-negative pneumonia (clinicaltrials.gov/ct2/show/NCT01799993 and clinicaltrials.gov/ct2/show/results/NCT00805168).
Because of their poor oral bioavailability, aminoglycosides are a key element of oropharyngeal or gut decolonization/decontamination regimens, including those targeting MDR pathogens. The purpose of these decontamination regimens is to eradicate potential pathogens from the oropharynx and digestive tract of patients at risk for nosocomial or postoperative infections. Selective digestive decontamination (SDD) consists of the oropharyngeal and gastric administration of non-absorbable antibiotics lacking anaerobic activity (often a polymyxin, an aminoglycoside, and amphotericin) along with a short course of systemic antibiotic therapy, whereas selective oropharyngeal decontamination (SOD) consists of application of non-absorbable antibiotics to the oropharynx alone. More than 50 randomized studies and 10 meta-analyses of SDD/SOD have been published. Overall, these data suggest that SDD/SOD are associated with improved survival in ICU patients (Price et al. 2014) and SDD is associated with a reduction in the rate of postoperative infection, including anastomotic leakage, in patients undergoing elective GI surgery (Abis et al. 2013; Roos et al. 2013). Despite these successes in the use of SOD and SDD to improve patient outcomes in the setting of low levels of antibiotic resistance, controversy remains regarding their effectiveness in the setting of high levels of antibiotic resistance as well as their impact on antibiotic resistance. No relationship between the use of SDD or SOD and the development of antimicrobial resistance has been shown in individual studies or meta-analyses in the setting of low antibiotic resistance (Daneman et al. 2013; Plantinga and Bonten 2015) and an international multicenter study of the effects of SDD and SOD on ICU-level antibiotic resistance in countries with higher levels of resistance is currently ongoing (clinicaltrials.gov/ct2/show/NCT02208154).
The ability of paromomycin to bind to eukaryotic ribosomes has led to the use of this agent in the treatment of protozoal infections. Like other aminoglycosides, oral paromomycin is poorly absorbed and may be used for the treatment of noninvasive amebiasis, cryptosporidiosis, trichomoniasis, and giardiasis in patients in whom other agents are contraindicated (Stover et al. 2012). More recently, this agent has been used to treat both cutaneous and visceral leishmaniasis. Topical paromomycin yielded a significantly higher cure rate compared with control therapy in patients with cutaneous leishmaniasis caused by Leishmania major (Ben Salah et al. 2013). Intramuscular paromomycin monotherapy was noninferior to standard amphotericin B therapy in a randomized control trial in patients with visceral disease in India (Sundar et al. 2007) and short-course combination regimens containing this agent were also noninferior to standard therapy with fewer adverse events (Sundar et al. 2011).
CONCLUSIONS
The aminoglycosides are a critical component of the current antibacterial arsenal. Their broad spectrum of activity, rapid bactericidal action, and favorable chemical and pharmacokinetic properties make them a clinically useful class of drugs across numerous infection types, including certain protozoal infections. The use of aminoglycosides waned as a result of the emergence of other classes of broad-spectrum agents with improved safety profiles, but the emergence of MDR pathogens has led to renewed interest in this class of drugs. Improved understanding of the drivers of toxicity and efficacy has led to the implementation of optimized dosing regimens that improve safety while maintaining efficacy. Increased understanding of key aminoglycoside resistance mechanisms combined with innovative medicinal chemistry approaches have led to the synthesis and development of novel agents specifically designed to evade resistance while maintaining potency against fully susceptible isolates. Given the dearth of new agents in the antibiotic pipeline and the ever-increasing specter of resistance, further optimization of the aminoglycoside scaffold to generate new agents with superior potency against MDR pathogens as well as an improved safety profile is warranted.
Footnotes
Editors: Lynn L. Silver and Karen Bush
Additional Perspectives on Antibiotics and Antibiotic Resistance available at www.perspectivesinmedicine.org
REFERENCES
- Abis GS, Stockmann HB, van Egmond M, Bonjer HJ, Vandenbroucke-Grauls CM, Oosterling SJ. 2013. Selective decontamination of the digestive tract in gastrointestinal surgery: Useful in infection prevention? A systematic review. J Gastrointest Surg 17: 2172–2178. [DOI] [PubMed] [Google Scholar]
- Aggen JB, Armstrong ES, Goldblum AA, Dozzo P, Linsell MS, Gliedt MJ, Hildebrandt DJ, Feeney LA, Kubo A, Matias RD, et al. 2010. Synthesis and spectrum of the neoglycoside ACHN-490. Antimicrob Agents Chemother 54: 4636–4642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aínsa JA, Pérez E, Pelicic V, Berthet FX, Gicquel B, Martín C. 1997. Aminoglycoside 2′-N-acetyltransferase genes are universally present in mycobacteria: Characterization of the aac(2′)-Ic gene from Mycobacterium tuberculosis and the aac(2′)-Id gene from Mycobacterium smegmatis. Mol Microbiol 24: 431–441. [DOI] [PubMed] [Google Scholar]
- Aires JR, Köhler T, Nikaido H, Plésiat P. 1999. Involvement of an active efflux system in the natural resistance of Pseudomonas aeruginosa to aminoglycosides. Antimicrob Agents Chemother 43: 2624–2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almaghrabi R, Clancy CJ, Doi Y, Hao B, Chen L, Shields RK, Press EG, Iovine NM, Townsend BM, Wagener MM, et al. 2014. Carbapenem-resistant Klebsiella pneumoniae strains exhibit diversity in aminoglycoside-modifying enzymes, which exert differing effects on plazomicin and other agents. Antimicrob Agents Chemother 58: 4443–4451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al Sheikh YA, Marie MA, John J, Krishnappa LG, Dabwab KH. 2014. Prevalence of 16S rRNA methylase genes among β-lactamase-producing Enterobacteriaceae clinical isolates in Saudi Arabia. Libyan J Med 9: 24432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- American Thoracic Society; Infectious Diseases Society of America. 2005. Guidelines for the management of adults with hospital-acquired, ventilator-associated, and healthcare-associated pneumonia. Am J Respir Crit Care Med 171: 388–416. [DOI] [PubMed] [Google Scholar]
- Armstrong ES, Miller GH. 2010. Combating evolution with intelligent design: The neoglycoside ACHN-490. Curr Opin Microbiol 13: 565–573. [DOI] [PubMed] [Google Scholar]
- Avent ML, Rogers BA, Cheng AC, Paterson DL. 2011. Current use of aminoglycosides: Indications, pharmacokinetics and monitoring for toxicity. Intern Med J 41: 441–449. [DOI] [PubMed] [Google Scholar]
- Azucena E, Mobashery S. 2001. Aminoglycoside-modifying enzymes: Mechanisms of catalytic processes and inhibition. Drug Resist Updat 4: 106–117. [DOI] [PubMed] [Google Scholar]
- Barclay ML, Kirkpatrick CM, Begg EJ. 1999. Once daily aminoglycoside therapy. Is it less toxic than multiple daily doses and how should it be monitored? Clin Pharmacokinet 36: 89–98. [DOI] [PubMed] [Google Scholar]
- Beauclerk AA, Cundliffe E. 1987. Sites of action of two ribosomal RNA methylases responsible for resistance to aminoglycosides. J Mol Biol 193: 661–671. [DOI] [PubMed] [Google Scholar]
- Bell J, Andersson P, Jones R, Turnidge J. 2010. 16S rRNA methylase containing Enterobacteriaceae in the SENTRY Asia-Pacific region frequently harbour plasmid-mediated quinolone resistance CTXM types. 20th European Congress of Clinical Microbiology and Infectious Diseases (ECCMID), Abstract O559 Vienna, April 10–13. [Google Scholar]
- Ben Salah A, Ben Messaoud N, Guedri E, Zaatour A, Ben Alaya N, Bettaieb J, Gharbi A, Belhadj Hamida N, Boukthir A, Chlif S, et al. 2013. Topical paromomycin with or without gentamicin for cutaneous leishmaniasis. N Engl J Med 368: 524–532. [DOI] [PubMed] [Google Scholar]
- Benveniste R, Davies J. 1971. R-factor mediated gentamicin resistance: A new enzyme which modifies aminoglycoside antibiotics. FEBS Lett 14: 293–296. [DOI] [PubMed] [Google Scholar]
- Berçot B, Poirel L, Nordmann P. 2011. Updated multiplex polymerase chain reaction for detection of 16S rRNA methylases: High prevalence among NDM-1 producers. Diagn Microbiol Infect Dis 71: 442–445. [DOI] [PubMed] [Google Scholar]
- Bercovier H, Kafri O, Sela S. 1986. Mycobacteria possess a surprisingly small number of ribosomal RNA genes in relation to the size of their genome. Biochem Biophys Res Commun 136: 1136–1141. [DOI] [PubMed] [Google Scholar]
- Brogden RN, Pinder RM, Sawyer PR, Speight TM, Avery GS. 1976. Tobramycin: A review of its antibacterial and pharmacokinetic properties and therapeutic use. Drugs 12: 166–200. [DOI] [PubMed] [Google Scholar]
- Brooke JS. 2012. Stenotrophomonas maltophilia: An emerging global opportunistic pathogen. Clin Microbiol Rev 25: 2–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buijk SE, Mouton JW, Gyssens IC, Verbrugh HA, Bruining HA. 2002. Experience with a once-daily dosing program of aminoglycosides in critically ill patients. Intensive Care Med 28: 936–942. [DOI] [PubMed] [Google Scholar]
- Buroni S, Pasca MR, Flannagan RS, Bazzini S, Milano A, Bertani I, Venturi V, Valvano MA, Riccardi G. 2009. Assessment of three resistance-nodulation-cell division drug efflux transporters of Burkholderia cenocepacia in intrinsic antibiotic resistance. BMC Microbiol 9: 200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castanheira M, Costello SE, Jones RN, Mendes RE. 2015. Prevalence of aminoglycoside resistance genes among contemporary Gram-negative resistant isolates collected worldwide. 25th European Congress of Clinical Microbriology and Infectious Diseases (ECCMID), Abstract O011 Copenhagen, April 25–28. [Google Scholar]
- Cattoir V, Nordmann P. 2009. Plasmid-mediated quinolone resistance in Gram-negative bacterial species: An update. Curr Med Chem 16: 1028–1046. [DOI] [PubMed] [Google Scholar]
- Costa Y, Galimand M, Leclercq R, Duval J, Courvalin P. 1993. Characterization of the chromosomal aac(6′)-Ii gene specific for Enterococcus faecium. Antimicrob Agents Chemother 37: 1896–1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courvalin P. 1994. Transfer of antibiotic resistance genes between Gram-positive and Gram-negative bacteria. Antimicrob Agents Chemother 38: 1447–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig WA. 2011. Optimizing aminoglycoside use. Crit Care Clin 27: 107–121. [DOI] [PubMed] [Google Scholar]
- Craig WA, Redington J, Ebert SC. 1991. Pharmacodynamics of amikacin in vitro and in mouse thigh and lung infections. J Antimicrob Chemother 27: 29–40. [DOI] [PubMed] [Google Scholar]
- Culebras E, Martínez JL. 1999. Aminoglycoside resistance mediated by the bifunctional enzyme 6′-N-aminoglycoside acetyltransferase-2″-O-aminoglycoside phosphotransferase. Front Biosci 4: D1–D8. [DOI] [PubMed] [Google Scholar]
- Cundliffe E. 1989. How antibiotic-producing organisms avoid suicide. Annu Rev Microbiol 43: 207–233. [DOI] [PubMed] [Google Scholar]
- Daneman N, Sarwar S, Fowler RA, Cuthbertson BH, Group SCS. 2013. Effect of selective decontamination on antimicrobial resistance in intensive care units: A systematic review and meta-analysis. Lancet Infect Dis 13: 328–341. [DOI] [PubMed] [Google Scholar]
- Davis BD. 1987. Mechanism of bactericidal action of aminoglycosides. Microbiol Rev 51: 341–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis BD, Chen LL, Tai PC. 1986. Misread protein creates membrane channels: An essential step in the bactericidal action of aminoglycosides. Proc Natl Acad Sci 83: 6164–6168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellinger RP, Levy MM, Rhodes A, Annane D, Gerlach H, Opal SM, Sevransky JE, Sprung CL, Douglas IS, Jaeschke R, et al. 2013. Surviving sepsis campaign: International guidelines for management of severe sepsis and septic shock: 2012. Crit Care Med 41: 580–637. [DOI] [PubMed] [Google Scholar]
- Dennis DT, Inglesby TV, Henderson DA, Bartlett JG, Ascher MS, Eitzen E, Fine AD, Friedlander AM, Hauer J, Layton M, et al. 2001. Tularemia as a biological weapon: Medical and public health management. JAMA 285: 2763–2773. [DOI] [PubMed] [Google Scholar]
- Doi Y, Yokoyama K, Yamane K, Wachino J, Shibata N, Yagi T, Shibayama K, Kato H, Arakawa Y. 2004. Plasmid-mediated 16S rRNA methylase in Serratia marcescens conferring high-level resistance to aminoglycosides. Antimicrob Agents Chemother 48: 491–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drusano GL, Ambrose PG, Bhavnani SM, Bertino JS, Nafziger AN, Louie A. 2007. Back to the future: Using aminoglycosides again and how to dose them optimally. Clin Infect Dis 45: 753–760. [DOI] [PubMed] [Google Scholar]
- Dzidic S, Bedeković V. 2003. Horizontal gene transfer-emerging multidrug resistance in hospital bacteria. Acta Pharmacol Sin 24: 519–526. [PubMed] [Google Scholar]
- Eliopoulos GM, Eliopoulos CT. 1988. Antibiotic combinations: Should they be tested? Clin Microbiol Rev 1: 139–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falagas ME, Siempos II, Bliziotis IA, Michalopoulos A. 2006. Administration of antibiotics via the respiratory tract for the prevention of ICU-acquired pneumonia: A meta-analysis of comparative trials. Crit Care 10: R123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feizabadi MM, Asadi S, Zohari M, Gharavi S, Etemadi G. 2004. Genetic characterization of high-level gentamicin-resistant strains of Enterococcus faecalis in Iran. Can J Microbiol 50: 869–872. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Padilla M, Torre-Cisneros J, Rivera-Espinar F, Pontes-Moreno A, López-Cerero L, Pascual A, Natera C, Rodríguez M, Salcedo I, Rodríguez-López F, et al. 2015. Gentamicin therapy for sepsis due to carbapenem-resistant and colistin-resistant Klebsiella pneumoniae. J Antimicrob Chemother 70: 905–913. [DOI] [PubMed] [Google Scholar]
- Griffith DE, Aksamit T, Brown-Elliott BA, Catanzaro A, Daley C, Gordin F, Holland SM, Horsburgh R, Huitt G, Iademarco MF, et al. 2007. An official ATS/IDSA statement: Diagnosis, treatment, and prevention of nontuberculous mycobacterial diseases. Am J Respir Crit Care Med 175: 367–416. [DOI] [PubMed] [Google Scholar]
- Hancock RE. 1984. Alterations in outer membrane permeability. Annu Rev Microbiol 38: 237–264. [DOI] [PubMed] [Google Scholar]
- Hancock RE, Raffle VJ, Nicas TI. 1981. Involvement of the outer membrane in gentamicin and streptomycin uptake and killing in Pseudomonas aeruginosa. Antimicrob Agents Chemother 19: 777–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock RE, Farmer SW, Li ZS, Poole K. 1991. Interaction of aminoglycosides with the outer membranes and purified lipopolysaccharide and OmpF porin of Escherichia coli. Antimicrob Agents Chemother 35: 1309–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho YI, Chan CY, Cheng AF. 1997. In-vitro activities of aminoglycoside-aminocyclitols against mycobacteria. J Antimicrob Chemother 40: 27–32. [DOI] [PubMed] [Google Scholar]
- Hocquet D, Vogne C, El Garch F, Vejux A, Gotoh N, Lee A, Lomovskaya O, Plésiat P. 2003. MexXY-OprM efflux pump is necessary for adaptive resistance of Pseudomonas aeruginosa to aminoglycosides. Antimicrob Agents Chemother 47: 1371–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingshead S, Vapnek D. 1985. Nucleotide sequence analysis of a gene encoding a streptomycin/spectinomycin adenylyltransferase. Plasmid 13: 17–30. [DOI] [PubMed] [Google Scholar]
- Humes HD, Weinberg JM, Knauss TC. 1982. Clinical and pathophysiologic aspects of aminoglycoside nephrotoxicity. Am J Kidney Dis 2: 5–29. [DOI] [PubMed] [Google Scholar]
- Ikäheimo I, Syrjälä H, Karhukorpi J, Schildt R, Koskela M. 2000. In vitro antibiotic susceptibility of Francisella tularensis isolated from humans and animals. J Antimicrob Chemother 46: 287–290. [DOI] [PubMed] [Google Scholar]
- Ioannidou E, Siempos II, Falagas ME. 2007. Administration of antimicrobials via the respiratory tract for the treatment of patients with nosocomial pneumonia: A meta-analysis. J Antimicrob Chemother 60: 1216–1226. [DOI] [PubMed] [Google Scholar]
- Islam S, Jalal S, Wretlind B. 2004. Expression of the MexXY efflux pump in amikacin-resistant isolates of Pseudomonas aeruginosa. Clin Microbiol Infect 10: 877–883. [DOI] [PubMed] [Google Scholar]
- Jackson J, Chen C, Buising K. 2013. Aminoglycosides: How should we use them in the 21st century? Curr Opin Infect Dis 26: 516–525. [DOI] [PubMed] [Google Scholar]
- Jones RN, Flonta M, Gurler N, Cepparulo M, Mendes RE, Castanheira M. 2014. Resistance surveillance program report for selected European nations (2011). Diagn Microbiol Infect Dis 78: 429–436. [DOI] [PubMed] [Google Scholar]
- Karlowsky JA, Draghi DC, Jones ME, Thornsberry C, Friedland IR, Sahm DF. 2003. Surveillance for antimicrobial susceptibility among clinical isolates of Pseudomonas aeruginosa and Acinetobacter baumannii from hospitalized patients in the United States, 1998 to 2001. Antimicrob Agents Chemother 47: 1681–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C, Mobashery S. 2005. Phosphoryl transfer by aminoglycoside 3′-phosphotransferases and manifestation of antibiotic resistance. Bioorg Chem 33: 149–158. [DOI] [PubMed] [Google Scholar]
- Kislak JW. 1972. The susceptibility of Bacteroides fragilis to 24 antibiotics. J Infect Dis 125: 295–299. [DOI] [PubMed] [Google Scholar]
- Kotra LP, Haddad J, Mobashery S. 2000. Aminoglycosides: Perspectives on mechanisms of action and resistance and strategies to counter resistance. Antimicrob Agents Chemother 44: 3249–3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landman D, Babu E, Shah N, Kelly P, Bäcker M, Bratu S, Quale J. 2010. Activity of a novel aminoglycoside, ACHN-490, against clinical isolates of Escherichia coli and Klebsiella pneumoniae from New York City. J Antimicrob Chemother 65: 2123–2127. [DOI] [PubMed] [Google Scholar]
- Landman D, Kelly P, Bäcker M, Babu E, Shah N, Bratu S, Quale J. 2011. Antimicrobial activity of a novel aminoglycoside, ACHN-490, against Acinetobacter baumannii and Pseudomonas aeruginosa from New York City. J Antimicrob Chemother 66: 332–334. [DOI] [PubMed] [Google Scholar]
- Le J, McKee B, Srisupha-Olarn W, Burgess DS. 2011. In vitro activity of carbapenems alone and in combination with amikacin against KPC-producing Klebsiella pneumoniae. J Clin Med Res 3: 106–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Goffic F, Baca B, Soussy CJ, Dublanchet A, Duval J. 1976. ANT(4′)I: A new aminoglycoside nucleotidyltransferase found in “staphylococcus aureus.” Ann Microbiol (Paris) 127: 391–399 (author’s transl.). [PubMed] [Google Scholar]
- Livermore DM, Mushtaq S, Warner M, Zhang JC, Maharjan S, Doumith M, Woodford N. 2011. Activity of aminoglycosides, including ACHN-490, against carbapenem-resistant Enterobacteriaceae isolates. J Antimicrob Chemother 66: 48–53. [DOI] [PubMed] [Google Scholar]
- Llano-Sotelo B, Azucena EF, Kotra LP, Mobashery S, Chow CS. 2002. Aminoglycosides modified by resistance enzymes display diminished binding to the bacterial ribosomal aminoacyl-tRNA site. Chem Biol 9: 455–463. [DOI] [PubMed] [Google Scholar]
- Lode H, Grunert K, Koeppe P, Langmaack H. 1976. Pharmacokinetic and clinical studies with amikacin, a new aminoglycoside antibiotic. J Infect Dis 134: S316–S322. [DOI] [PubMed] [Google Scholar]
- Macinga DR, Rather PN. 1999. The chromosomal 2′-N-acetyltransferase of Providencia stuartii: Physiological functions and genetic regulation. Front Biosci 4: D132–D140. [DOI] [PubMed] [Google Scholar]
- Magnet S, Blanchard JS. 2005. Molecular insights into aminoglycoside action and resistance. Chem Rev 105: 477–498. [DOI] [PubMed] [Google Scholar]
- Magnet S, Courvalin P, Lambert T. 2001. Resistance-nodulation-cell division-type efflux pump involved in aminoglycoside resistance in Acinetobacter baumannii strain BM4454. Antimicrob Agents Chemother 45: 3375–3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin WJ, Gardner M, Washington JA. 1972. In vitro antimicrobial susceptibility of anaerobic bacteria isolated from clinical specimens. Antimicrob Agents Chemother 1: 148–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto T. 2014. Arbekacin: Another novel agent for treating infections due to methicillin-resistant Staphylococcus aureus and multidrug-resistant Gram-negative pathogens. Clin Pharmacol 6: 139–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mingeot-Leclercq MP, Glupczynski Y, Tulkens PM. 1999. Aminoglycosides: Activity and resistance. Antimicrob Agents Chemother 43: 727–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miró E, Grünbaum F, Gómez L, Rivera A, Mirelis B, Coll P, Navarro F. 2013. Characterization of aminoglycoside-modifying enzymes in enterobacteriaceae clinical strains and characterization of the plasmids implicated in their diffusion. Microb Drug Resist 19: 94–99. [DOI] [PubMed] [Google Scholar]
- Moore RD, Smith CR, Lipsky JJ, Mellits ED, Lietman PS. 1984. Risk factors for nephrotoxicity in patients treated with aminoglycosides. Ann Intern Med 100: 352–357. [DOI] [PubMed] [Google Scholar]
- Murphy E. 1985. Nucleotide sequence of a spectinomycin adenyltransferase AAD(9) determinant from Staphylococcus aureus and its relationship to AAD(3″)(9). Mol Gen Genet 200: 33–39. [DOI] [PubMed] [Google Scholar]
- Murry KR, McKinnon PS, Mitrzyk B, Rybak MJ. 1999. Pharmacodynamic characterization of nephrotoxicity associated with once-daily aminoglycoside. Pharmacotherapy 19: 1252–1260. [DOI] [PubMed] [Google Scholar]
- Mushtaq S, Irfan S, Sarma JB, Doumith M, Pike R, Pitout J, Livermore DM, Woodford N. 2011. Phylogenetic diversity of Escherichia coli strains producing NDM-type carbapenemases. J Antimicrob Chemother 66: 2002–2005. [DOI] [PubMed] [Google Scholar]
- Mwengee W, Butler T, Mgema S, Mhina G, Almasi Y, Bradley C, Formanik JB, Rochester CG. 2006. Treatment of plague with gentamicin or doxycycline in a randomized clinical trial in Tanzania. Clin Infect Dis 42: 614–621. [DOI] [PubMed] [Google Scholar]
- Nagabhushan T, Miller G, Weinstein M. 1982. Structure–activity relationships in aminoglycoside-aminocyclitol antibiotics. In The aminoglycosides: Microbiology, clinical use and toxicology (ed. Whelton A, Neu HC), pp. 3–27. Marcel Dekker, New York. [Google Scholar]
- Nichols WW, Young SN. 1985. Respiration-dependent uptake of dihydrostreptomycin by Escherichia coli. Its irreversible nature and lack of evidence for a uniport process. Biochem J 228: 505–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolau DP, Belliveau PP, Nightingale CH, Quintiliani R, Freeman CD. 1995. Implementation of a once-daily aminoglycoside program in a large community-teaching hospital. Hosp Pharm 30: 674–676, 679–680. [PubMed] [Google Scholar]
- Okamoto S, Suzuki Y. 1965. Chloramphenicol-, dihydrostreptomycin-, and kanamycin-inactivating enzymes from multiple drug-resistant Escherichia coli carrying episome “R”. Nature 208: 1301–1303. [DOI] [PubMed] [Google Scholar]
- Ounissi H, Derlot E, Carlier C, Courvalin P. 1990. Gene homogeneity for aminoglycoside-modifying enzymes in Gram-positive cocci. Antimicrob Agents Chemother 34: 2164–2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankuch GA, Seifert H, Appelbaum PC. 2010. Activity of doripenem with and without levofloxacin, amikacin, and colistin against Pseudomonas aeruginosa and Acinetobacter baumannii. Diagn Microbiol Infect Dis 67: 191–197. [DOI] [PubMed] [Google Scholar]
- Plantinga NL, Bonten MJ. 2015. Selective decontamination and antibiotic resistance in ICUs. Crit Care 19: 259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podnecky NL, Rhodes KA, Schweizer HP. 2015. Efflux pump-mediated drug resistance in Burkholderia. Front Microbiol 6: 305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirel L, Savov E, Nazli A, Trifonova A, Todorova I, Gergova I, Nordmann P. 2014. Outbreak caused by NDM-1- and RmtB-producing Escherichia coli in Bulgaria. Antimicrob Agents Chemother 58: 2472–2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price R, MacLennan G, Glen J; SuDDICU Collaboration. 2014. Selective digestive or oropharyngeal decontamination and topical oropharyngeal chlorhexidine for prevention of death in general intensive care: Systematic review and network meta-analysis. BMJ 348: g2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prins JM, Büller HR, Kuijper EJ, Tange RA, Speelman P. 1993. Once versus thrice daily gentamicin in patients with serious infections. Lancet 341: 335–339. [DOI] [PubMed] [Google Scholar]
- Ramirez MS, Tolmasky ME. 2010. Aminoglycoside modifying enzymes. Drug Resist Updat 13: 151–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey BW, Pepe MS, Quan JM, Otto KL, Montgomery AB, Williams-Warren J, Vasiljev-K M, Borowitz D, Bowman CM, Marshall BC, et al. 1999. Intermittent administration of inhaled tobramycin in patients with cystic fibrosis. Cystic Fibrosis Inhaled Tobramycin Study Group. N Engl J Med 340: 23–30. [DOI] [PubMed] [Google Scholar]
- Rather PN, Orosz E, Shaw KJ, Hare R, Miller G. 1993. Characterization and transcriptional regulation of the 2′-N-acetyltransferase gene from Providencia stuartii. J Bacteriol 175: 6492–6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ristuccia AM, Cunha BA. 1985. An overview of amikacin. Ther Drug Monit 7: 12–25. [DOI] [PubMed] [Google Scholar]
- Robicsek A, Strahilevitz J, Jacoby GA, Macielag M, Abbanat D, Park CH, Bush K, Hooper DC. 2006. Fluoroquinolone-modifying enzyme: A new adaptation of a common aminoglycoside acetyltransferase. Nat Med 12: 83–88. [DOI] [PubMed] [Google Scholar]
- Roos D, Dijksman LM, Tijssen JG, Gouma DJ, Gerhards MF, Oudemans-van Straaten HM. 2013. Systematic review of perioperative selective decontamination of the digestive tract in elective gastrointestinal surgery. Br J Surg 100: 1579–1588. [DOI] [PubMed] [Google Scholar]
- Rosenberg EY, Ma D, Nikaido H. 2000. AcrD of Escherichia coli is an aminoglycoside efflux pump. J Bacteriol 182: 1754–1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybak MJ, Abate BJ, Kang SL, Ruffing MJ, Lerner SA, Drusano GL. 1999. Prospective evaluation of the effect of an aminoglycoside dosing regimen on rates of observed nephrotoxicity and ototoxicity. Antimicrob Agents Chemother 43: 1549–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sader HS, Farrell DJ, Flamm RK, Jones RN. 2014. Antimicrobial susceptibility of Gram-negative organisms isolated from patients hospitalised with pneumonia in U.S. and European hospitals: Results from the SENTRY Antimicrobial Surveillance Program, 2009–2012. Int J Antimicrob Agents 43: 328–334. [DOI] [PubMed] [Google Scholar]
- Sader HS, Rhomberg PR, Farrell DJ, Jones RN. 2015. Arbekacin activity against contemporary clinical bacteria isolated from patients hospitalized with pneumonia. Antimicrob Agents Chemother 59: 3263–3270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawicki GS, Signorovitch JE, Zhang J, Latremouille-Viau D, von Wartburg M, Wu EQ, Shi L. 2012. Reduced mortality in cystic fibrosis patients treated with tobramycin inhalation solution. Pediatr Pulmonol 47: 44–52. [DOI] [PubMed] [Google Scholar]
- Schwartz JJ, Gazumyan A, Schwartz I. 1992. rRNA gene organization in the Lyme disease spirochete, Borrelia burgdorferi. J Bacteriol 174: 3757–3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw KJ, Rather PN, Hare RS, Miller GH. 1993. Molecular genetics of aminoglycoside resistance genes and familial relationships of the aminoglycoside-modifying enzymes. Microbiol Rev 57: 138–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon VK, Mösinger EU, Malerczy V. 1973. Pharmacokinetic studies of tobramycin and gentamicin. Antimicrob Agents Chemother 3: 445–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skeggs PA, Thompson J, Cundliffe E. 1985. Methylation of 16S ribosomal RNA and resistance to aminoglycoside antibiotics in clones of Streptomyces lividans carrying DNA from Streptomyces tenjimariensis. Mol Gen Genet 200: 415–421. [DOI] [PubMed] [Google Scholar]
- Snowden J, Stovall S. 2011. Tularemia: Retrospective review of 10 years’ experience in Arkansas. Clin Pediatr (Phila) 50: 64–68. [DOI] [PubMed] [Google Scholar]
- Stover KR, Riche DM, Gandy CL, Henderson H. 2012. What would we do without metronidazole? Am J Med Sci 343: 316–319. [DOI] [PubMed] [Google Scholar]
- Strahilevitz J, Jacoby GA, Hooper DC, Robicsek A. 2009. Plasmid-mediated quinolone resistance: A multifaceted threat. Clin Microbiol Rev 22: 664–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubbings W, Bostock J, Ingham E, Chopra I. 2006. Mechanisms of the post-antibiotic effects induced by rifampicin and gentamicin in Escherichia coli. J Antimicrob Chemother 58: 444–448. [DOI] [PubMed] [Google Scholar]
- Sundar S, Jha TK, Thakur CP, Sinha PK, Bhattacharya SK. 2007. Injectable paromomycin for visceral leishmaniasis in India. N Engl J Med 356: 2571–2581. [DOI] [PubMed] [Google Scholar]
- Sundar S, Sinha PK, Rai M, Verma DK, Nawin K, Alam S, Chakravarty J, Vaillant M, Verma N, Pandey K, et al. 2011. Comparison of short-course multidrug treatment with standard therapy for visceral leishmaniasis in India: An open-label, non-inferiority, randomised controlled trial. Lancet 377: 477–486. [DOI] [PubMed] [Google Scholar]
- Swenson JM, Wallace RJ, Silcox VA, Thornsberry C. 1985. Antimicrobial susceptibility of five subgroups of Mycobacterium fortuitum and Mycobacterium chelonae. Antimicrob Agents Chemother 28: 807–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taber HW, Mueller JP, Miller PF, Arrow AS. 1987. Bacterial uptake of aminoglycoside antibiotics. Microbiol Rev 51: 439–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamma PD, Cosgrove SE, Maragakis LL. 2012. Combination therapy for treatment of infections with Gram-negative bacteria. Clin Microbiol Rev 25: 450–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J, Skeggs PA, Cundliffe E. 1985. Methylation of 16S ribosomal RNA and resistance to the aminoglycoside antibiotics gentamicin and kanamycin determined by DNA from the gentamicin-producer, Micromonospora purpurea. Mol Gen Genet 201: 168–173. [DOI] [PubMed] [Google Scholar]
- Vakulenko SB, Mobashery S. 2003. Versatility of aminoglycosides and prospects for their future. Clin Microbiol Rev 16: 430–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wachino J, Arakawa Y. 2012. Exogenously acquired 16S rRNA methyltransferases found in aminoglycoside-resistant pathogenic Gram-negative bacteria: An update. Drug Resist Updat 15: 133–148. [DOI] [PubMed] [Google Scholar]
- Wachino J, Yamane K, Shibayama K, Kurokawa H, Shibata N, Suzuki S, Doi Y, Kimura K, Ike Y, Arakawa Y. 2006. Novel plasmid-mediated 16S rRNA methylase, RmtC, found in a Proteus mirabilis isolate demonstrating extraordinary high-level resistance against various aminoglycosides. Antimicrob Agents Chemother 50: 178–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wachino J, Shibayama K, Kurokawa H, Kimura K, Yamane K, Suzuki S, Shibata N, Ike Y, Arakawa Y. 2007. Novel plasmid-mediated 16S rRNA m1A1408 methyltransferase, NpmA, found in a clinically isolated Escherichia coli strain resistant to structurally diverse aminoglycosides. Antimicrob Agents Chemother 51: 4401–4409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters VL. 1999. Conjugative transfer in the dissemination of β-lactam and aminoglycoside resistance. Front Biosci 4: D433–D456. [DOI] [PubMed] [Google Scholar]
- Westbrock-Wadman S, Sherman DR, Hickey MJ, Coulter SN, Zhu YQ, Warrener P, Nguyen LY, Shawar RM, Folger KR, Stover CK. 1999. Characterization of a Pseudomonas aeruginosa efflux pump contributing to aminoglycoside impermeability. Antimicrob Agents Chemother 43: 2975–2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson DN. 2014. Ribosome-targeting antibiotics and mechanisms of bacterial resistance. Nat Rev Microbiol 12: 35–48. [DOI] [PubMed] [Google Scholar]
- World Health Organization. 2011. Guidelines for the programmatic management of drug-resistant tuberculosis: 2011 update. World Health Organization, Geneva. [PubMed] [Google Scholar]
- Wright GD, Thompson PR. 1999. Aminoglycoside phosphotransferases: Proteins, structure, and mechanism. Front Biosci 4: D9–D21. [DOI] [PubMed] [Google Scholar]
- Yamane K, Wachino J, Doi Y, Kurokawa H, Arakawa Y. 2005. Global spread of multiple aminoglycoside resistance genes. Emerg Infect Dis 11: 951–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan JJ, Wu JJ, Ko WC, Tsai SH, Chuang CL, Wu HM, Lu YJ, Li JD. 2004. Plasmid-mediated 16S rRNA methylases conferring high-level aminoglycoside resistance in Escherichia coli and Klebsiella pneumoniae isolates from two Taiwanese hospitals. J Antimicrob Chemother 54: 1007–1012. [DOI] [PubMed] [Google Scholar]
- Yokoyama K, Doi Y, Yamane K, Kurokawa H, Shibata N, Shibayama K, Yagi T, Kato H, Arakawa Y. 2003. Acquisition of 16S rRNA methylase gene in Pseudomonas aeruginosa. Lancet 362: 1888–1893. [DOI] [PubMed] [Google Scholar]
- Zhanel GG, Hoban DJ, Harding GK. 1991. The postantibiotic effect: A review of in vitro and in vivo data. DICP 25: 153–163. [DOI] [PubMed] [Google Scholar]