Abstract
In epilepsy, seizures and comorbidities (e.g., cognitive deficits and depression) occur when specific thresholds are crossed. These thresholds depend on the diathesis (or vulnerability) of a given individual. The diathesis is controlled by multiple genetic and environmental factors. Diathesis changes over multiple timescales: on a daily basis, and as part of the development/aging processes, etc. The diathesis–epilepsy model introduced here provides a conceptual framework to understand how past events (e.g., a very stressful event) can directly influence the occurrence of epilepsy and comorbidities later in life. Experimental evidence supports this model, and the existence of biomarkers predictive of a vulnerability state have led to the development of preventive therapeutic strategies. Epigenetic modifications could be a key determinant of diathesis. Their role is discussed.
The susceptibility of an individual to seizures and comorbidities depends on various genetic and environmental factors. Epigenetic modifications caused by certain events (e.g., traumatic brain injury) may alter one’s susceptibility to seizures and comorbidities.
A seizure is a type of activity that is endogenous to most brain neuronal networks. All it takes is to give the network a hard enough push. For example, an electroconvulsive shock will trigger a seizure in any “normal” human brain (Luttges and McGaugh 1967). Whether forced in “normal” tissue or spontaneously occurring in chronic “epileptic” networks, the dynamics of seizure with focal onset appear to follow universal rules across brain regions and species (Jirsa et al. 2014). In particular, seizures occur when brain activities cross a given threshold (Jirsa et al. 2014). In “normal” tissue, the threshold is high, and seizures are more difficult to trigger than in chronic epilepsy (Bankstahl et al. 2013). It can thus be hypothesized that the threshold is lowered in epilepsy (i.e., the probability to have a seizure is increased in patients with epilepsy as compared with the general population).
Then, the question we need to address is: What determines the threshold in a given individual? Many epilepsies can be tracked down to an original brain insult, such as a genetic mutation, early life stress, brain trauma, meningitis, etc. (Goldberg and Coulter 2013). This brain insult will induce changes in the circuitry, which may ultimately lower the threshold, and enable the occurrence of spontaneous seizures. Tracking down the mechanisms underlying the modification of the threshold is a very complex task because epilepsy is multifactorial, in the sense that a given insult will not result in a similar outcome in two individuals. The best examples can be found in human genetics. Mutations in familial forms of epilepsies are characterized by an incomplete penetrance and by different forms of epilepsy in family members carrying the same mutation (Depienne et al. 2010; Scheffer 2011). Likewise, brain insults, such as a traumatic brain injury or febrile seizures, only increase the likelihood of developing epilepsy later in life (i.e., not all affected individuals will develop spontaneous seizures) (Bolkvadze and Pitkanen 2012; Choy et al. 2014).
What then makes a given individual develop epilepsy and not another? Obviously, gene X environment interactions play a crucial role. This means that a brain insult (genetic or environmental) will result in epilepsy when specific gene X environment conditions are met. Genetics allowed us to identify mutations causally linked to a phenotype. However, carrying a mutation is not a sufficient condition to develop epilepsy. A seminal human genetic study showed that healthy individuals carry mutations usually associated with epilepsy, demonstrating that epilepsy is multifactorial/polygenic (Klassen et al. 2011). Hence, even with a mutation associated with epilepsy, seizures will happen only if adequate genetic background and/or environmental factors are present. This relates to the concept developed by Eve Marder that multiple solutions exist to produce a given type of neuronal network activity (Prinz et al. 2004). This means that a deleterious mutation can be counterbalanced if adequate homeostatic mechanisms can be triggered and a stable solution can be found by the system. Direct evidence for this concept has been provided experimentally, as crossing two different types of mutant mice (each with a different “epilepsy” gene) masked epilepsy in offspring, in effect raising the threshold for the occurrence of spontaneous seizures in the double mutant (Glasscock et al. 2007).
Importantly, if a brain insult can lower seizure threshold, the threshold can also be increased. The proof-of-concept that a threshold can be genetically manipulated has been provided in experimental models of sudden death in epilepsy (SUDEP) (Aiba 2015). In these models, a wave of spreading depolarization (SD) in the brain stem leads to death. SD is also a type of activity that is endogenous to most brain networks, and it is characterized by a threshold (El Houssaini et al. 2015). This threshold can be raised by an appropriate genetic manipulation, thus increasing the lifespan of SUDEP models (Aiba 2015), further revealing the polygenic nature of epilepsy and its consequences.
The previous considerations revolve around inherited mutations. To make things more complex, somatic mutations can occur during embryonic development (Poduri et al. 2013), affecting specific populations of neurons in specific brain areas. These mutations could be missed by common genetic analyses. In this case also, somatic mutations could result in a pathological state or counterbalance an inherited mutation. Somatic mutations may not be an epiphenomenon, as it is believed that they are common during cell division, although their functional outcome is not known (Frumkin et al. 2005; Lynch 2010).
These studies directly show that the genetic background can directly influence the threshold for pathological activities. Another main determinant is the environment, which includes the past history of an individual. Association/retrospective studies suggest that several risk factors increase the probability to develop epilepsy. For example, an episode of depression earlier in life can favor the development of epilepsy (Hoppe and Elger 2011; Hesdorffer et al. 2012). Hence, the past experience of individuals may prime them to develop epilepsy (Hoppe and Elger 2011; Hesdorffer et al. 2012). This issue is very difficult to investigate clinically and experimentally as past events may leave a mark in the system without giving rise to an obvious phenotype. However, it is a parameter as important to consider as the genetic background.
What kind of trace can be left in neuronal networks by environmental events? Obvious candidates include cell death, chronic inflammation, or oxidative stress, which can be relatively easily identified. A more subtle change, but more drastic in terms of functional consequences, can be brought about by epigenetic reprogramming. Broadly speaking, epigenetics refers to different ways to dynamically regulate gene expression with touching the DNA code.
Thus, the past of an individual—past meaning both the genetic background and factors acting on one’s life—is a key determinant for the development of epilepsy. The same argument holds for comorbidities associated with epilepsy, including depression and cognitive deficits that affect a substantial number of patients with epilepsy. Among patients with temporal lobe epilepsy (TLE), 14%–31% develop depression (Adams et al. 2008; Sanchez-Gistau et al. 2010) and 30% cognitive deficits (Jones et al. 2007). Comorbidities are often considered as more detrimental by patients for their quality of life than seizures themselves (Hermann et al. 2000; Bell et al. 2011). The general mechanisms underlying depression and cognitive deficits remain unknown. Arguably, as for seizures, multiple paths may lead to these comorbidities (i.e., there may be multiple overlapping mechanisms). It can be assumed that some reorganization in the circuitry is responsible for their expression (Kleen et al. 2012). Because epilepsy is associated with network reorganizations, it is natural to propose that such reorganizations enable the expression of comorbidities, although causality has never been clearly shown. A different, but complementary, hypothesis would be to take into account past events that may have altered the circuitry (with no obvious expression of a specific phenotype) before epileptogenesis, and primed the system for the development of comorbidities, even perhaps independent of epilepsy.
Stress is the most studied event in relationship to pathologies (McEwen 2013). Past stressful events can change the epigenetics landscape, thus inducing a functional reorganization of networks (Klengel and Binder 2015). There is also a large body of literature linking stress, epilepsy, and depression. But here, the focus will be on the influence of past stressful events occurring before epileptogenesis on the subsequent development of epilepsy and comorbidities (Koutsogiannopoulos et al. 2009; Lanteaume et al. 2009), and the role of possible epigenetic reprogramming.
INFLUENCE OF PAST STRESSFUL EVENTS: THE DIATHESIS–EPILEPSY MODEL
The diathesis–stress model provides a conceptual framework to understand the occurrence of depression. Diathesis (or predisposition) is the intrinsic vulnerability of an individual, which depends on multiple genetic X environmental factors. Diathesis changes during one’s life, and cannot be easily measured objectively. Even if it remains mostly abstract, this metavariable is useful to model how an organism will respond to a challenge (internal or external). Another useful metavariable is the allostatic load (the accumulation of unresolved stress that wears an organ when an individual is exposed to internal or external challenging events). The diathesis–stress model posits that if diathesis and stress cross a certain threshold, a psychiatric disorder will develop (Monroe and Simons 1991). In epilepsy, the psychological burden of seizures (helplessness, social stigma) can produce a chronic state of stress, increasing the allostatic load, thus favoring the emergence of depression in vulnerable patients (Hoppe and Elger 2011). The same concept may be extended to the occurrence of cognitive deficits after epilepsy onset. Because diathesis can be different from one individual to another, the diathesis–stress model can account for the fact that not all patients with epilepsy develop depression and cognitive deficit comorbidities.
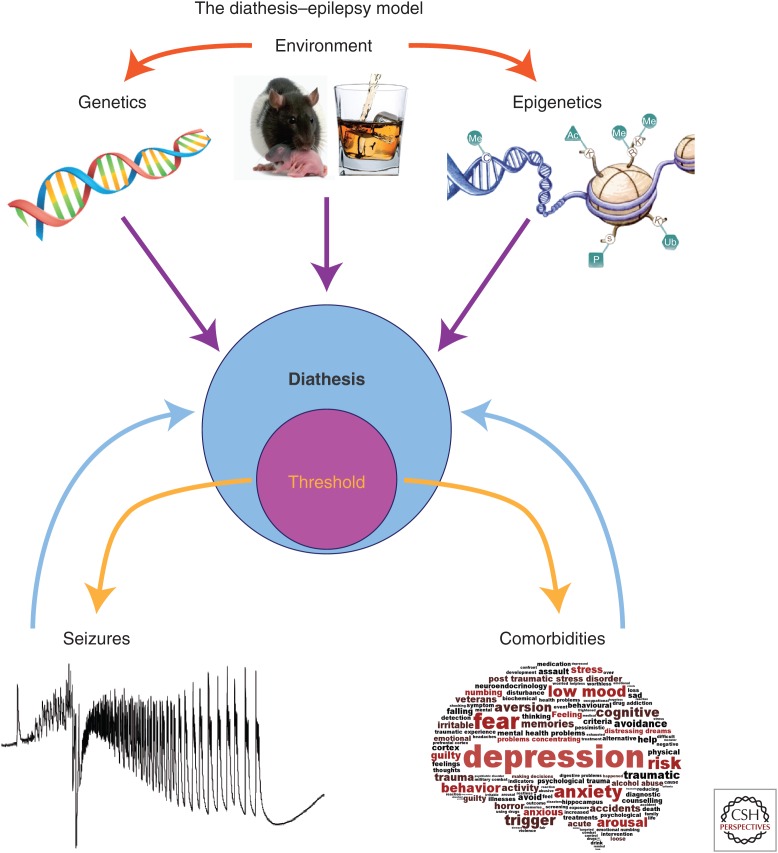
The diathesis–stress model considers the accumulation of stress as a trigger of the pathology, given the predisposition of the individual. A complementary extension of the diathesis–stress scenario is introduced here: the so-called diathesis–epilepsy model (Fig. 1). In this model, a past history of stress would increase the allostatic load of some individuals, changing their diathesis and predisposing them to epilepsy and comorbidities. The brain insult would here act as the trigger for epilepsy and/or comorbidities. Such scheme could account for the fact that only some individuals develop epilepsy after a given brain insult, and also why some develop comorbidities after epilepsy onset (Fig. 2). In other words, the epileptogenic insult would act as a stressor, enabling threshold crossing in predisposed individuals. Numerous clinical data support this model. For example, it is well established that depression favors the occurrence of epilepsy later in life (Hoppe and Elger 2011). In this case, the stressful events triggered a phenotype (depression) that could be easily identified. However, the accumulation of stressful events during one’s life may leave some trace, without obvious phenotypic traits in some individuals (those with a high susceptibility). Indeed, a link between an initial very stressful event (death of relative or unemployment) and the development of morbidities, including “emotional vulnerability,” has been reported in TLE patients (Lanteaume et al. 2009). In rodents, early-life stress can result in epilepsy (Dube et al. 2015), probably because the activated stress system can interfere with a proper maturation of neuronal networks (Chen and Baram 2015).
Figure 1.
The diathesis–epilepsy model. Diathesis is the vulnerability of an individual, here to seizures and comorbidities (e.g., depression and cognitive deficits). The vulnerability to epilepsy and comorbidities does not necessarily imply similar mechanisms, although a cross talk is likely. Seizures and comorbidities occur when a certain threshold is crossed (the threshold is part of diathesis, as it is determined by it). Numerous factors control diathesis: our genetic background, environmental factors, and epigenetic modifications. Environmental factors can change our genetic background (e.g., mutations) and our epigenetic landscape (e.g., maternal care, exposure to psychoactive substances). Epigenetics covers the multiple ways to change gene expression (e.g., changing the code of histone tails, the methylation state of the DNA, etc.). Epigenetic modifications occur naturally during the night/day cycle, aging etc., but also following environmental challenges (e.g., intense stress). Decreasing the thresholds will increase the probability of the occurrence of seizures and comorbidities. Once the latter occur, they could feedback negatively onto diathesis, further increasing vulnerability.
Figure 2.
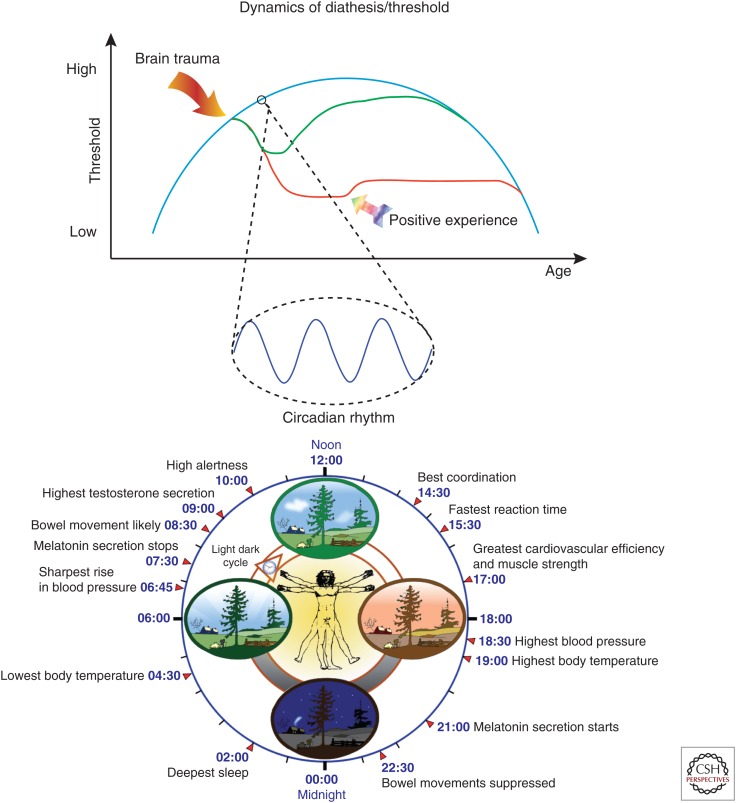
Dynamics of diathesis. The blue curve schematizes the inverted curve of epilepsy incidence as a function of age. It can be proposed that seizure threshold reflects this curve during one’s life (the immature and aging brains are more susceptible to seizures, albeit for different reasons). If everything was governed by genetic factors, the threshold at a given time point would entirely be determined by the genetic background, which would impose a different diathesis in individuals. At a smaller timescale, the threshold normally oscillates in a circadian-dependent manner, following the cycling of genes and proteins, which may occur in all brain regions. Other cycles can be added (e.g., menstrual for catamenial epilepsy, and perhaps even seasonal). Life events also have the potential to alter diathesis (e.g., after a brain trauma), which could cause cell death, inflammatory processes, epigenetic modifications etc. As a result, the threshold for seizure would be lowered more or less permanently in a vulnerable population (red curve), while it may recover in a less vulnerable population (green curve). The threshold for comorbidities would also be affected (not represented—but it does not have to follow the same curves for the vulnerable and nonvulnerable populations). These alterations can be more or less permanent, as homeostatic mechanisms may restore previous diathesis or as positive life experiences (winning the lottery, publishing in Nature) may increase the different thresholds. Diathesis is dynamical by nature, constantly changing during one’s life. Identifying diathesis in individuals is very difficult, but the identification of biomarkers would open the way to preventive treatments, as shown in experimental studies (see text).
TESTING THE DIATHESIS–EPILEPSY MODEL
Assessing diathesis objectively is very difficult in humans in the absence of an obvious phenotype (such as depression). However, it can be performed experimentally in rodents. Social defeat (as a first hit) produces a vulnerability state to depression in >40% of social defeat exposed Sprague–Dawley rats (Blugeot et al. 2011). Low levels of serum brain derived neurotrophic factor (BDNF) measured after recovery from social defeat identify the vulnerable population. At this stage, neither nonvulnerable nor vulnerable animals display depression-like behavior. When exposed to chronic mild stress (as a second hit), only vulnerable animals (those with persistent low serum BDNF levels) display depression-like behavior (Blugeot et al. 2011), in keeping with the diathesis–stress model. After social defeat, the diathesis is clearly different in the two groups exposed to the same type of stressful event. It can also be concluded that laboratory animals were not biologically equivalent as they reacted differently to the first stressful event, also probably reflecting a different diathesis. Indeed, future vulnerable and nonvulnerable animals (i.e., before social defeat) could already be distinguished based on different electroencephalogram (EEG) properties in specific frequency bands (D Claverie, C Becker, A Ghestem, et al., unpubl.), indicating different internal states.
Using a similar experimental procedure, it became possible to test the diathesis–epilepsy model (Becker et al. 2015). Rats exposed to social defeat separated into two groups of animals, vulnerable and nonvulnerable, based on their serum BDNF levels. Vulnerable animals had a lower threshold to reach status epilepticus and had considerably accelerated epileptogenesis as compared with nonvulnerable animals. Hence, a past, unresolved history of stress, changed diathesis in some animals rendering them more susceptible to the development of epilepsy.
Going a step further, the presence of cognitive deficits and depression was assessed in animals with spontaneous seizures (Becker et al. 2015). A depression-like profile and severe cognitive deficits were found in the vulnerable population only (with low serum BDNF levels after the first hit). Importantly, animals exposed to status epilepticus only, displayed neither depression nor cognitive deficits once epileptic. This shows that, in these experimental conditions, the brain insult triggering epileptogenesis (status epilepticus) acted as a revelator of comorbidities in a vulnerable population.
Together, these results validate the diathesis–epilepsy model. A past history of unresolved stress may sensitize some individuals to epilepsy and comorbidities. Importantly, in a well-controlled laboratory environment, epilepsy by itself does not necessarily result in comorbidities (Becker et al. 2015). The latter statement appears to be at odds with some, but not all, studies reporting depression-like behavior and cognitive deficits in experimental models of epilepsy, including work from our own group (Lenck-Santini and Holmes 2008; Chauviere et al. 2009; Inostroza et al. 2011, 2012, 2013; Sankar and Mazarati 2012; Tyler et al. 2012). Why do comorbidities occur in some laboratories and not others? Different strains, species, and models could explain such discrepancies. A complementary explanation could be that diathesis levels, which include genetic, environmental, and epigenetic factors, vary between animals across laboratories. This may directly come from the breeder (Langer et al. 2011) or other environmental factors. For example, unresolved stress/long-term deleterious consequences may occur in some animals postnatally, as a function of the dam’s behavior (Francis et al. 2003). It is also difficult to predict the effect of housing social animals like rodents in individual cages, without social contacts. We partially removed the latter confounding factor in our latest study, allowing animals to explore their environment outside their cage at least 1 h/day (Becker et al. 2015). Although, with epilepsy, animals were quiet and not aggressive (as is often witnessed when animals are maintained in individual cages with scant social contact).
Finally, the state of vulnerability to depression could be reversed using a BDNF mimetic (Becker et al. 2015). Hence, here also, the threshold can be manipulated in the other direction, to return to “normal” diathesis levels. However, the treatment did not normalize the progression of epileptogenesis, suggesting that the biological processes controlling the vulnerability to depression and vulnerability to epilepsy are not identical (Becker et al. 2015).
Thus, the diathesis–epilepsy model provides a useful conceptual framework to understand why epilepsy and associated comorbidities develop in some individuals only (although this is one possibility among many others). In such a model, the threshold to seizures and comorbidities can be included within the diathesis (Fig. 1). The identification of potential biomarkers of vulnerability (serum BDNF and EEG patterns) renders possible prospective studies in humans. How can we explain such vulnerability? As mentioned before, it all comes down to genetics X environmental factors (in their broadest sense). Epigenetics constitute a useful link between genetics and environment.
EPIGENETICS AS A KEY PHYSIOLOGICAL PROCESS
The definition used by the National Institutes of Health (NIH) for the Roadmap Epigenomics Project is used here: “For purposes of this program, epigenetics refers to both heritable changes in gene activity and expression (in the progeny of cells or of individuals) and also stable, long-term alterations in the transcriptional potential of a cell that are not necessarily heritable” (see www.roadmapepigenomics.org/overview). Basically, epigenetics can include anything that can regulate gene expression independently on gene sequences (i.e., DNA methylation, histone modification, and micro/long noncoding RNAs) (Kobow and Blumcke 2014). In the case of histone modification (as an example), numerous enzymes exist that can transfer acetyl groups, methyl groups, or phosphorus (among others) to the histone tails, which influence gene transcription (Fig. 1). In addition to these “writing” molecules, there are “erasers,” which remove these signals from the histone tails, and of course there are “readers,” which inform about the code left on the histones (Borrelli et al. 2008). Hence, there are multiple enzymes that can change the epigenetic landscape and influence the way genes will be expressed or not.
It is important to first highlight that epigenetic modifications are, first and foremost, a physiological process. Epigenetic modifications occur naturally across time. Thus, during brain development, specific sets of genes are activated under the control of epigenetic mechanisms, while other sets of genes are regulated/expressed during aging (Jobe et al. 2012; Lardenoije et al. 2015). But epigenetic modifications also occur on a much faster timescale. This has been well established in the suprachiasmatic nucleus (SCN). Our biology is tightly controlled by circadian rhythms. The core time-keeping mechanism in the SCN includes transcriptional/translational feedback loops, with two transcriptional activators CLOCK and BMAL1, and two transcription repressors PER and CRY (Sahar and Sassone-Corsi 2013). At the beginning of the light phase, CLOCK and BMAL1 activate the transcription of the different per and cry genes, which proteins PER and CRY repress CLOCK–BMAL1 transcription. Because clock genes control numerous transcription factors, 10% to 20% of the genes expressed in cells oscillate during the night/day cycle (Aguilar-Arnal et al. 2013). Such circadian rhythm induces a daily remapping of SCN neuronal network at the gene/protein level, which allows switching between different functional/behavioral states (e.g., wakefulness, feeding, activity, etc.). CLOCK is itself a histone acetyltransferase, and the enzyme SIRT1 is a NAD+-dependent deacetylase (i.e., a “writer” and an “eraser,” respectively). There is thus a tight link between metabolism, epigenetics, and circadian rhythm (Orozco-Solis and Sassone-Corsi 2014). Because clock proteins PER2 and BMAL1 show rhythmic behavior in 18 out of 20 forebrain regions, including the hippocampus (Harbour et al. 2013, 2014), one can expect a circadian epigenetic regulation of genes in these regions. Hence, epigenetics controls gene expression during the night and day cycle. In a pathological context, the circadian clock can be reprogrammed, resulting in a different regulation of genes based on epigenetics mechanisms (Asher and Sassone-Corsi 2015). The outcome is that genes and proteins display different levels of expression according to the time of the day as a function of the requirements of the system in which they occur, and that such dynamic regulation may be affected/reprogrammed in pathologies. These issues remain to be tested in the temporal lobe regions in controls and experimental models of epilepsy.
EPIGENETICS IN EPILEPSY AND COMORBIDITIES
In the previous section, epigenetics is considered as a phenomenon that influences all instants of our life, as part of a normal time-dependent programmed process. But, environmental factors can also leave a profound epigenetic trace, affecting the way genes are expressed. Some are also part of our everyday life as certain memory and learning processes depend on epigenetic mechanisms (Graff and Tsai 2013). But some environmental factors can also have deleterious consequences via epigenetic reprogramming. This has been well established during development (Bale 2015). For example, maternal care (licking/grooming) plays a key role in determining the sensitivity to stress, via an epigenetic mechanism, which remains stable once the pups reached adulthood, but which can be reversed with chromatin-modifying drugs (Szyf 2009a,b).
Epigenetics is a relatively new but rapidly expanding field in epilepsy. Many epigenetic modifications have been identified already (Kobow and Blumcke 2014). But interpreting them is a difficult task, as changing the expression of genes and proteins with epigenetics will increase dramatically the number of possibilities to build functional and dysfunctional cells and networks. In addition, there is no clear separation between genetics and epigenetics, as mutations associated with the occurrence of epilepsy can involve direct/indirect epigenetic factors (e.g., ARX and MeCP2) (Amir et al. 1999; Poeta et al. 2013; Kobow and Blumcke 2014). Hence, epigenetic mechanisms may be central in certain forms of inherited epilepsies or pathologies associated with epilepsy.
In acquired forms of epilepsies, epigenetics could also play a key role. The brain insult triggering epileptogenesis (e.g., status epilepticus or traumatic brain injury) may alter the epigenetic landscape, change diathesis, and lower seizure threshold (Fig. 2). Numerous genes are up- or down-regulated following a brain insult and in chronic epileptogenic regions (Becker et al. 2002; Gorter et al. 2006). Epigenetic mechanisms may underlie such changes in gene expression (Qureshi and Mehler 2010). One typical example is BDNF, which is up-regulated in epilepsy via a decreased Bdnf DNA methylation (Ryley Parrish et al. 2013). In such a context of wide modifications at the epigenome level, it is not surprising to find chromatin-modifying agents among antiepileptic drugs. For example, valproic acid has several modes of action, including its action as a histone deacetylase inhibitor (Monti et al. 2009).
Because epigenetic mechanisms can simultaneously alter the expression of multiple genes, designing repair or antiepileptogenesis strategies is a complex task. A good example is provided by the neuron-restrictive silencer factor (NRSF), also known as repressor element-1-silencing factor (REST). NRSF acts both as a transcription and epigenetic regulator, and it plays a key role during development (Bithell 2011). After status epilepticus, NRSF is up-regulated (Palm et al. 1998). Because NRSF can potentially regulate hundreds of genes (Roopra et al. 2001; Johnson et al. 2007), it is an interesting candidate for large-scale changes of gene expression, with proepileptogenic consequences. Indeed, the down-regulation of the HCN1 protein found in experimental epilepsy (Jung et al. 2007; Marcelin et al. 2009) involves an epigenetic mechanism because of NRSF up-regulation (McClelland et al. 2011). Using an in vivo treatment with decoy deoxynucleotides against NRSF, HCN1 expression and function was restored (McClelland et al. 2011). Because NRSF controls a specific set of genes during epileptogenesis (McClelland et al. 2014), blocking its action in vivo had disease-modifying effects slowing down, but not preventing, epileptogenesis (McClelland et al. 2011). Thus, despite its widespread effects (McClelland et al. 2014), NRSF is one among potentially other factors that drive epileptogenesis. It is important to note that some of the modified genes may be restoring homeostasis in the network. Hence, by preventing the overall action of NRSF, “beneficial” effects may have been prevented. As mentioned above, polyepigenomic mechanisms may be at play, and targeted (as opposed to widespread) actions may be necessary to obtain antiepileptogenesis or antiepileptic effects.
Finally, to make things even more complex, epilepsy is characterized by time-dependent modifications following the initial brain insult (including inherited mutations) that led to epilepsy. Some epigenetic mechanisms found during a specific time period may not be valid at other time points. One key aspect of epigenetics is its dynamic nature, which naturally occurs from brain development to aging (Fig. 2).
EPIGENETICS AND THE DIATHESIS–EPILEPSY MODEL
Epigenetics provide a useful framework to study the impact of past events on the development of epilepsy and its comorbidities. Past events (e.g., stressful situations [Becker et al. 2015] and exposure to psychoactive drugs during development [Silva et al. 2013]), may have left an epigenetic mark, not sufficient to produce a phenotype by itself, but increasing diathesis, making these individuals more vulnerable (in effect bringing them close to threshold). The occurrence of a brain insult would thus act as a trigger and enable the development of epilepsy and/or comorbidities. It is likely that diathesis constantly changes in both directions as a function of various positive/negative life events (Fig. 2). But diathesis remains a key determinant for the development of epilepsy and comorbidities, which occur in some individuals and not others as a function of their genetic background and epigenomic landscape at any given time point.
Perhaps the best, albeit speculative, illustration of this concept (and its complexity) is the fact that seizures show a circadian pattern in mesial TLE and experimental models (Quigg et al. 1998). Because seizure threshold is also regulated in a circadian manner in a BMAL1-dependent manner (Gerstner et al. 2014), one can propose, as a working hypothesis, that normal epigenetic-circadian mechanisms in addition to the modifications brought about by past life experiences and proepileptogenic brain insults, bring neuronal networks close to seizure threshold at specific time points during the night and day cycle. Thus, diathesis is likely regulated across multiple timescales (Fig. 2).
CONCLUDING REMARKS
Our genetic background is a key determinant of our vulnerability to epilepsy and comorbidities. Such a background can be “finely tuned” by epigenetic mechanisms. The latter are physiological processes that change the protein landscape across multiple timescales (hours to decades), thus constantly changing our diathesis and threshold to epilepsy and comorbidities. Environmental factors can also dynamically modulate our diathesis, without necessarily affecting the threshold (i.e., they would act as priming factors). Other hits could be necessary to affect the threshold. It is important to note that thresholds for epilepsies and comorbidities may be different, involving distinct underlying mechanisms, although some intersection is expected. Of course, seizures and comorbidities can feed back onto diathesis and threshold (usually increasing vulnerability further). It is also important to note that diathesis can also be decreased (naturally with positive life experiences or pharmacologically). In conclusion, seizures and comorbidities occur in an ever-changing (dynamic) neural environment, critically depending on past experiences, which should be taken into consideration to understand their origin and plan future therapeutic strategies.
ACKNOWLEDGMENTS
The author acknowledges INSERM and the European Union’s Seventh Framework Programme (FP7/2007-2013) under Grant agreement No. 602102 (EPITARGET), and Dr. P. P. Quilichini for her help with the manuscript and figures.
Footnotes
Editors: Gregory L. Holmes and Jeffrey L. Noebels
Additional Perspectives on Epilepsy: The Biology of a Spectrum Disorder available at www.perspectivesinmedicine.org
REFERENCES
- Adams SJ, O’Brien TJ, Lloyd J, Kilpatrick CJ, Salzberg MR, Velakoulis D. 2008. Neuropsychiatric morbidity in focal epilepsy. Br J Psychiatry 192: 464–469. [DOI] [PubMed] [Google Scholar]
- Aguilar-Arnal L, Hakim O, Patel VR, Baldi P, Hager GL, Sassone-Corsi P. 2013. Cycles in spatial and temporal chromosomal organization driven by the circadian clock. Nat Struct Mol Biol 20: 1206–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aiba I, Noebels JL. 2015. Spreading depolarization in the brainstem mediates sudden cardiorespiratory arrest in mouse SUDEP models. Sci Transl Med 7: 282ra46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amir RE, Van dVI, Wan M, Tran CQ, Francke U, Zoghbi HY. 1999. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet 23: 185–188. [DOI] [PubMed] [Google Scholar]
- Asher G, Sassone-Corsi P. 2015. Time for food: The intimate interplay between nutrition, metabolism, and the circadian clock. Cell 161: 84–92. [DOI] [PubMed] [Google Scholar]
- Bale TL. 2015. Epigenetic and transgenerational reprogramming of brain development. Nat Rev Neurosci 16: 332–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankstahl M, Bankstahl JP, Loscher W. 2013. Pilocarpine-induced epilepsy in mice alters seizure thresholds and the efficacy of antiepileptic drugs in the 6-Hertz psychomotor seizure model. Epilepsy Res 107: 205–216. [DOI] [PubMed] [Google Scholar]
- Becker AJ, Chen J, Paus S, Normann S, Beck H, Elger CE, Wiestler OD, Blumcke I. 2002. Transcriptional profiling in human epilepsy: Expression array and single cell real-time qRT-PCR analysis reveal distinct cellular gene regulation. NeuroReport 13: 1327–1333. [DOI] [PubMed] [Google Scholar]
- Becker C, Bouvier E, Ghestem A, Siyoucef S, Claverie D, Camus F, Bartolomei F, Benoliel JJ, Bernard C. 2015. Predicting and treating stress-induced vulnerability to epilepsy and depression. Ann Neurol 78: 128–136. [DOI] [PubMed] [Google Scholar]
- Bell B, Lin JJ, Seidenberg M, Hermann B. 2011. The neurobiology of cognitive disorders in temporal lobe epilepsy. Nat Rev Neurol 7: 154–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bithell A. 2011. REST: Transcriptional and epigenetic regulator. Epigenomics 3: 47–58. [DOI] [PubMed] [Google Scholar]
- Blugeot A, Rivat C, Bouvier E, Molet J, Mouchard A, Zeau B, Bernard C, Benoliel JJ, Becker C. 2011. Vulnerability to depression: From brain neuroplasticity to identification of biomarkers. J Neurosci 31: 12889–12899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolkvadze T, Pitkanen A. 2012. Development of post-traumatic epilepsy after controlled cortical impact and lateral fluid-percussion-induced brain injury in the mouse. J Neurotrauma 29: 789–812. [DOI] [PubMed] [Google Scholar]
- Borrelli E, Nestler EJ, Allis CD, Sassone-Corsi P. 2008. Decoding the epigenetic language of neuronal plasticity. Neuron 60: 961–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauviere L, Rafrafi N, Thinus-Blanc C, Bartolomei F, Esclapez M, Bernard C. 2009. Early deficits in spatial memory and θ rhythm in experimental temporal lobe epilepsy. J Neurosci 29: 5402–5410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Baram TZ. 2015. Toward understanding how early-life stress reprograms cognitive and emotional brain networks. Neuropsychopharmacology 10.1038/npp.2015.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy M, Dube CM, Patterson K, Barnes SR, Maras P, Blood AB, Hasso AN, Obenaus A, Baram TZ. 2014. A novel, noninvasive, predictive epilepsy biomarker with clinical potential. J Neurosci 34: 8672–8684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depienne C, Trouillard O, Gourfinkel-An I, Saint-Martin C, Bouteiller D, Graber D, Barthez-Carpentier MA, Gautier A, Villeneuve N, Dravet C, et al. 2010. Mechanisms for variable expressivity of inherited SCN1A mutations causing Dravet syndrome. J Med Genet 47: 404–410. [DOI] [PubMed] [Google Scholar]
- Dube CM, Molet J, Singh-Taylor A, Ivy A, Maras PM, Baram TZ. 2015. Hyper-excitability and epilepsy generated by chronic early-life stress. Neurobiol Stress 2: 10–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Houssaini K, Ivanov AI, Bernard C, Jirsa VK. 2015. Seizures, refractory status epilepticus, and depolarization block as endogenous brain activities. Phys Rev E 91: 010701. [DOI] [PubMed] [Google Scholar]
- Francis DD, Szegda K, Campbell G, Martin WD, Insel TR. 2003. Epigenetic sources of behavioral differences in mice. Nat Neurosci 6: 445–446. [DOI] [PubMed] [Google Scholar]
- Frumkin D, Wasserstrom A, Kaplan S, Feige U, Shapiro E. 2005. Genomic variability within an organism exposes its cell lineage tree. PLoS Comput Biol 1: e50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstner JR, Smith GG, Lenz O, Perron IJ, Buono RJ, Ferraro TN. 2014. BMAL1 controls the diurnal rhythm and set point for electrical seizure threshold in mice. Front Syst Neurosci 8: 121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasscock E, Qian J, Yoo JW, Noebels JL. 2007. Masking epilepsy by combining two epilepsy genes. Nat Neurosci 10: 1554–1558. [DOI] [PubMed] [Google Scholar]
- Goldberg EM, Coulter DA. 2013. Mechanisms of epileptogenesis: A convergence on neural circuit dysfunction. Nat Rev Neurosci 14: 337–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorter JA, van Vliet EA, Aronica E, Breit T, Rauwerda H, Lopes da Silva FH, Wadman WJ. 2006. Potential new antiepileptogenic targets indicated by microarray analysis in a rat model for temporal lobe epilepsy. J Neurosci 26: 11083–11110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graff J, Tsai LH. 2013. Histone acetylation: Molecular mnemonics on the chromatin. Nat Rev Neurosci 14: 97–111. [DOI] [PubMed] [Google Scholar]
- Harbour VL, Weigl Y, Robinson B, Amir S. 2013. Comprehensive mapping of regional expression of the clock protein PERIOD2 in rat forebrain across the 24-h day. PLoS ONE 8: e76391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbour VL, Weigl Y, Robinson B, Amir S. 2014. Phase differences in expression of circadian clock genes in the central nucleus of the amygdala, dentate gyrus, and suprachiasmatic nucleus in the rat. PLoS ONE 9: e103309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann BP, Seidenberg M, Bell B. 2000. Psychiatric comorbidity in chronic epilepsy: Identification, consequences, and treatment of major depression. Epilepsia 41: S31–S41. [DOI] [PubMed] [Google Scholar]
- Hesdorffer DC, Ishihara L, Mynepalli L, Webb DJ, Weil J, Hauser WA. 2012. Epilepsy, suicidality, and psychiatric disorders: A bidirectional association. Ann Neurol 72: 184–191. [DOI] [PubMed] [Google Scholar]
- Hoppe C, Elger CE. 2011. Depression in epilepsy: A critical review from a clinical perspective. Nat Rev Neurol 7: 462–472. [DOI] [PubMed] [Google Scholar]
- Inostroza M, Cid E, Brotons-Mas J, Gal B, Aivar P, Uzcategui YG, Sandi C, Menendez de la Prida L. 2011. Hippocampal-dependent spatial memory in the water maze is preserved in an experimental model of temporal lobe epilepsy in rats. PLoS ONE 6: e22372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inostroza M, Cid E, Menendez de la Prida L, Sandi C. 2012. Different emotional disturbances in two experimental models of temporal lobe epilepsy in rats. PLoS ONE 7: e38959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inostroza M, Brotons-Mas JR, Laurent F, Cid E, de la Prida LM. 2013. Specific impairment of “what-where-when” episodic-like memory in experimental models of temporal lobe epilepsy. J Neurosci 33: 17749–17762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jirsa VK, Stacey WC, Quilichini PP, Ivanov AI, Bernard C. 2014. On the nature of seizure dynamics. Brain 137: 2210–2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jobe EM, McQuate AL, Zhao X. 2012. Crosstalk among epigenetic pathways regulates neurogenesis. Front Neurosci 6: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DS, Mortazavi A, Myers RM, Wold B. 2007. Genome-wide mapping of in vivo protein–DNA interactions. Science 316: 1497–1502. [DOI] [PubMed] [Google Scholar]
- Jones JE, Bell B, Fine J, Rutecki P, Seidenberg M, Hermann B. 2007. A controlled prospective investigation of psychiatric comorbidity in temporal lobe epilepsy. Epilepsia 48: 2357–2360. [DOI] [PubMed] [Google Scholar]
- Jung S, Jones TD, Lugo JN Jr, Sheerin AH, Miller JW, D’Ambrosio R, Anderson AE, Poolos NP. 2007. Progressive dendritic HCN channelopathy during epileptogenesis in the rat pilocarpine model of epilepsy. J Neurosci 27: 13012–13021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klassen T, Davis C, Goldman A, Burgess D, Chen T, Wheeler D, McPherson J, Bourquin T, Lewis L, Villasana D, et al. 2011. Exome sequencing of ion channel genes reveals complex profiles confounding personal risk assessment in epilepsy. Cell 145: 1036–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleen JK, Scott RC, Lenck-Santini PP, Holmes GL. 2012. Cognitive and behavioral co-morbidities of epilepsy. In Jasper’s basic mechanisms of the epilepsies (ed. Noebels JL, Avoli M, Rogawski MA, Olsen RW, Delgado-Escueta AV). National Center for Biotechnology Information, Bethesda, MD. [PubMed] [Google Scholar]
- Klengel T, Binder EB. 2015. Epigenetics of stress-related psychiatric disorders and gene X environment interactions. Neuron 86: 1343–1357. [DOI] [PubMed] [Google Scholar]
- Kobow K, Blumcke I. 2014. Epigenetic mechanisms in epilepsy. Prog Brain Res 213: 279–316. [DOI] [PubMed] [Google Scholar]
- Koutsogiannopoulos S, Adelson F, Lee V, Andermann F. 2009. Stressors at the onset of adult epilepsy: Implications for practice. Epileptic Disord 11: 42–47. [DOI] [PubMed] [Google Scholar]
- Langer M, Brandt C, Loscher W. 2011. Marked strain and substrain differences in induction of status epilepticus and subsequent development of neurodegeneration, epilepsy, and behavioral alterations in rats [corrected]. Epilepsy Res 96: 207–224. [DOI] [PubMed] [Google Scholar]
- Lanteaume L, Bartolomei F, Bastien-Toniazzo M. 2009. How do cognition, emotion, and epileptogenesis meet? A study of emotional cognitive bias in temporal lobe epilepsy. Epilepsy Behav 15: 218–224. [DOI] [PubMed] [Google Scholar]
- Lardenoije R, Iatrou A, Kenis G, Kompotis K, Steinbusch HW, Mastroeni D, Coleman P, Lemere CA, Hof PR, van den Hove DL, et al. 2015. The epigenetics of aging and neurodegeneration. Prog Neurobiol 131: 21–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenck-Santini PP, Holmes GL. 2008. Altered phase precession and compression of temporal sequences by place cells in epileptic rats. J Neurosci 28: 5053–5062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luttges MW, McGaugh JL. 1967. Permanence of retrograde amnesia produced by electroconvulsive shock. Science 156: 408–410. [DOI] [PubMed] [Google Scholar]
- Lynch M. 2010. Evolution of the mutation rate. Trend Genet 26: 345–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcelin B, Chauviere L, Becker A, Migliore M, Esclapez M, Bernard C. 2009. h channel-dependent deficit of θ oscillation resonance and phase shift in temporal lobe epilepsy. Neurobiol Dis 33: 436–447. [DOI] [PubMed] [Google Scholar]
- McClelland S, Flynn C, Dube C, Richichi C, Zha Q, Ghestem A, Esclapez M, Bernard C, Baram TZ. 2011. Neuron-restrictive silencer factor-mediated hyperpolarization-activated cyclic nucleotide gated channelopathy in experimental temporal lobe epilepsy. Ann Neurol 70: 454–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClelland S, Brennan GP, Dube C, Rajpara S, Iyer S, Richichi C, Bernard C, Baram TZ. 2014. The transcription factor NRSF contributes to epileptogenesis by selective repression of a subset of target genes. eLife 3: e01267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen BS. 2013. The brain on stress: Toward an integrative approach to brain, body, and behavior. Perspect Psychol Sci 8: 673–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monroe SM, Simons AD. 1991. Diathesis-stress theories in the context of life stress research: Implications for the depressive disorders. Psychol Bull 110: 406–425. [DOI] [PubMed] [Google Scholar]
- Monti B, Polazzi E, Contestabile A. 2009. Biochemical, molecular and epigenetic mechanisms of valproic acid neuroprotection. Curr Mol Pharmacol 2: 95–109. [DOI] [PubMed] [Google Scholar]
- Orozco-Solis R, Sassone-Corsi P. 2014. Epigenetic control and the circadian clock: Linking metabolism to neuronal responses. Neuroscience 264: 76–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm K, Belluardo N, Metsis M, Timmusk T. 1998. Neuronal expression of zinc finger transcription factor REST/NRSF/XBR gene. J Neurosci 18: 1280–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poduri A, Evrony GD, Cai X, Walsh CA. 2013. Somatic mutation, genomic variation, and neurological disease. Science 341: 1237758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poeta L, Fusco F, Drongitis D, Shoubridge C, Manganelli G, Filosa S, Paciolla M, Courtney M, Collombat P, Lioi MB, et al. 2013. A regulatory path associated with X-linked intellectual disability and epilepsy links KDM5C to the polyalanine expansions in ARX. Am J Hum Genet 92: 114–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prinz AA, Bucher D, Marder E. 2004. Similar network activity from disparate circuit parameters. Nat Neurosci 7: 1345–1352. [DOI] [PubMed] [Google Scholar]
- Quigg M, Straume M, Menaker M, Bertram EH III. 1998. Temporal distribution of partial seizures: Comparison of an animal model with human partial epilepsy. Ann Neurol 43: 748–755. [DOI] [PubMed] [Google Scholar]
- Qureshi IA, Mehler MF. 2010. Epigenetic mechanisms underlying human epileptic disorders and the process of epileptogenesis. Neurobiol Dis 39: 53–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roopra A, Huang Y, Dingledine R. 2001. Neurological disease: Listening to gene silencers. Mol Interv 1: 219–228. [PubMed] [Google Scholar]
- Ryley Parrish R, Albertson AJ, Buckingham SC, Hablitz JJ, Mascia KL, Davis Haselden W, Lubin FD. 2013. Status epilepticus triggers early and late alterations in brain-derived neurotrophic factor and NMDA glutamate receptor Grin2b DNA methylation levels in the hippocampus. Neuroscience 248: 602–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahar S, Sassone-Corsi P. 2013. The epigenetic language of circadian clocks. Handb Exp Pharmacol 217: 29–44. [DOI] [PubMed] [Google Scholar]
- Sanchez-Gistau V, Pintor L, Sugranyes G, Bailles E, Carreno M, Donaire A, Boget T, Setoain X, Bargallo N, Rumia J. 2010. Prevalence of interictal psychiatric disorders in patients with refractory temporal and extratemporal lobe epilepsy in Spain. A comparative study. Epilepsia 51: 1309–1313. [DOI] [PubMed] [Google Scholar]
- Sankar R, Mazarati A. 2012. Neurobiology of depression as a comorbidity of epilepsy. In Jasper’s basic mechanisms of the epilepsies (ed. Noebels JL, Avoli M, Rogawski MA, Olsen RW, Delgado-Escueta AV). National Center for Biotechnology Information, Bethesda, MD. [PubMed] [Google Scholar]
- Scheffer IE. 2011. Does genotype determine phenotype? Sodium channel mutations in Dravet syndrome and GEFS+. Neurology 76: 588–589. [DOI] [PubMed] [Google Scholar]
- Silva CG, Metin C, Fazeli W, Machado NJ, Darmopil S, Launay PS, Ghestem A, Nesa MP, Bassot E, Szabo E, et al. 2013. Adenosine receptor antagonists including caffeine alter fetal brain development in mice. Sci Transl Med 5: 197ra104. [DOI] [PubMed] [Google Scholar]
- Szyf M. 2009a. The early life environment and the epigenome. Biochim Biophys Acta 1790: 878–885. [DOI] [PubMed] [Google Scholar]
- Szyf M. 2009b. Epigenetics, DNA methylation, and chromatin-modifying drugs. Annu Rev Pharmacol Toxicol 49: 243–263. [DOI] [PubMed] [Google Scholar]
- Tyler AL, Mahoney JM, Richard GR, Holmes GL, Lenck-Santini PP, Scott RC. 2012. Functional network changes in hippocampal CA1 after status epilepticus predict spatial memory deficits in rats. J Neurosci 32: 11365–11376. [DOI] [PMC free article] [PubMed] [Google Scholar]