Abstract
Mouse double minute 2 (MDM2) is a critical negative regulator of the tumor suppressor p53, playing a key role in controlling its transcriptional activity, protein stability, and nuclear localization. MDM2 expression is up-regulated in numerous cancers, resulting in a loss of p53-dependent activities, such as apoptosis and cell-cycle arrest. Genetic amplification and inheritance of MDM2 promoter single-nucleotide polymorphisms (SNPs) are the two best-studied mechanisms for up-regulating MDM2 activity. This article provides an overview of these events in human cancer, highlighting the frequent occurrence of MDM2 amplification in sarcoma and the role of SNP309 and SNP285 in regulating MDM2 expression and cancer risk. The availability of large-scale genomic profiling datasets, like those from The Cancer Genome Atlas Research Network, have provided the opportunity to evaluate the consequences of MDM2 amplification and SNP inheritance across high-quality tumor samples from diverse cancer indications.
MDM2, a key regulator of the tumor suppressor p53, is functionally overexpressed in numerous cancers. This is primarily caused by amplification of the MDM2 gene, but some tumors achieve comparable levels of MDM2 overexpression via unknown mechanisms.
P53 is now clearly recognized as the most frequently mutated gene in human cancer. Genetic inactivation of this tumor suppressor and transcription factor occurs in nearly half of all human tumors (Kandoth et al. 2013), serving as a testament to the strong selective growth advantage that this molecular event provides. Indeed, it has been hypothesized that tumor survival itself is not possible in the context of an intact p53 pathway (Junttila and Evan 2009), as the stressed environments in which tumors develop would otherwise unleash p53, resulting in cell-cycle arrest and, in many cases, apoptosis. Under this scenario, tumors showing wild-type p53 must abrogate p53 function in some other way, and it has been proposed that many of these tumors are impaired in their ability to stimulate apoptosis (Tovar et al. 2006; Junttila and Evan 2009). One small but revealing fraction of these p53WT tumors is thought to diminish p53 function by directly or indirectly increasing the expression of p53’s key negative regulator, a gene called mouse double minute 2 (MDM2).
It is informative to view MDM2 from a historical perspective. MDM2 was discovered in a screen for genes amplified in a spontaneously transformed mouse cell line (Cahilly-Snyder et al. 1987). In subsequent functional studies, MDM2 was shown to be capable of inducing tumorigenicity when overexpressed in this and other rodent cells, implying that MDM2 could act as an oncogene in these contexts (Fakharzadeh et al. 1991; Finlay 1993). In an independent line of investigation, a protein known only as p90 (named for its apparent molecular weight by migration in SDS-PAGE) was discovered to bind p53 (Hinds et al. 1990). These two parallel story lines were linked when protein sequencing revealed p90 to be MDM2, suggesting the possibility that MDM2 might regulate or mediate the function of p53 (Momand et al. 1992). This hypothesis was borne out when MDM2 was shown to be capable of inhibiting p53-mediated transcriptional activation of a plasmid containing a p53 response element (Momand et al. 1992; Oliner et al. 1993). This inhibitory function was shown to be conferred through the ability of MDM2 to conceal the p53 activation domain from the cellular transcription machinery (Oliner et al. 1993). Additionally, MDM2 was observed to abrogate p53 function by two other mechanisms: (1) targeting p53 for ubiquitin-mediated degradation, and (2) exporting p53 from the nucleus to the cytoplasm (Haupt et al. 1997; Honda et al. 1997; Kubbutat et al. 1997; Roth et al. 1998).
In vivo confirmation that MDM2 antagonizes p53 function came from mouse knockout studies in which p53 deletion rescued the embryonic lethality of MDM2-null mice (Jones et al. 1995; Montes de Oca Luna et al. 1995). Additional support for an in vivo relationship between these two proteins emerged from murine tumor studies in which MDM2 expression was experimentally increased or decreased. Transgenic overexpression of MDM2, under the control of its native promoter (Jones et al. 1998) or a heterologous promoter (Lundgren et al. 1997), induced spontaneous tumor formation. Conversely, mice displaying reduced MDM2 expression, caused by allelic haploinsufficiency (Alt et al. 2003; Terzian et al. 2007) or hypomorphism (Mendrysa et al. 2006), developed fewer tumors than their normal counterparts when crossed into genetically engineered tumor models.
The role of MDM2 as a bona fide human oncogene was cemented when it was discovered to be genetically amplified in more than a third of human sarcoma samples (Oliner et al. 1992). Subsequent studies revealed MDM2 amplification in a range of tumor types outside of sarcoma (Momand et al. 1998). These somatic changes are not the only reported tumor-predisposing genetic alterations in MDM2. Indeed, Bond et al. (2004, 2005) published the discovery of a germline single-nucleotide polymorphism (SNP) in the MDM2 promoter that increased MDM2 expression, enhanced cancer risk, and accelerated tumorigenesis. Together, MDM2 amplification and promoter polymorphism represent the two most intensely studied alterations in this gene, and this review will explore the consequences of these alterations on MDM2 expression and tumorigenesis.
MDM2 AMPLIFICATION
Early cytogenetic studies characterizing chromosomal abnormalities in soft-tissue sarcomas identified recurrent aberrations associated within the 12q13-14 locus (Turc-Carel et al. 1986; Meltzer et al. 1991). On cloning the MDM2 gene, its locus was mapped to the same region on chromosome 12 (Oliner et al. 1992). Building on the earlier evidence for aberrations at this locus, a diverse panel of sarcoma samples was profiled for genetic alterations in MDM2, identifying MDM2 amplification in a significant number (17 of 47) of samples (Oliner et al. 1992). Analysis of broader panels of sarcoma samples confirmed these initial findings, highlighting the presence of MDM2 amplification in many histological subtypes of sarcoma, including osteosarcoma, liposarcoma, lipoma, leiomyosarcoma, rhabdomyosarcoma, malignant schwannoma, fibrosarcoma, hemangiopericytoma, and malignant fibrous histiocytoma (Forus et al. 1993; Ladanyi et al. 1993; Leach et al. 1993; Cordon-Cardo et al. 1994; Florenes et al. 1994; Nakayama et al. 1995; Miller et al. 1996; Patterson et al. 1997). In addition, MDM2 amplification was shown to associate with overexpression of both RNA (Oliner et al. 1992; Florenes et al. 1994) and protein (Oliner et al. 1992; Patterson et al. 1997), and to occur in a mutually exclusive manner with p53 mutation (Leach et al. 1993; Florenes et al. 1994; Miller et al. 1996). Together, these findings supported the developing hypothesis that elevated MDM2 expression via gene amplification represented an alternative mechanism to p53 mutation to inactivate the p53-signaling pathway and promote tumor progression in sarcoma.
Although MDM2 amplification is found in many histological subtypes of sarcoma, two subtypes in particular, well-differentiated and dedifferentiated liposarcomas, show exceptionally high MDM2 amplification rates (>90%) (Coindre et al. 2010). Both of these subtypes display relatively simple genomic profiles, harboring supernumerary rings or marker chromosomes containing the 12q13-15 amplicon, including MDM2 and cyclin-dependent kinase 4 (CDK4) (Chibon et al. 2002; Coindre et al. 2003, 2010). Given its prevalence in these two subtypes, MDM2 amplification is frequently evaluated as an ancillary means for the differential diagnosis of well-differentiated and dedifferentiated liposarcomas from closely related adipocytic neoplasms (Binh et al. 2005; Shimada et al. 2006; Coindre et al. 2010; Lokka et al. 2014).
The high frequency of amplification and overexpression of MDM2 in sarcoma, together with the fact that these events tend to occur in a mutually exclusive manner with p53 mutations, support the idea that MDM2 represents an attractive therapeutic target in this setting. Nutlins, the first drug-like MDM2 inhibitors, are a class of cis-imidazole small molecules that bind to the hydrophobic p53-binding pocket of MDM2 and prevent its interaction with p53 (Vassilev et al. 2004). Disruption of the interaction of the two proteins results in the stabilization of p53 and activation of the downstream p53-dependent transcriptional program, inducing cell-cycle arrest and/or programmed cell death (Vassilev et al. 2004; Tovar et al. 2006). In vitro studies evaluating the ability of nutlin-3 to induce apoptosis in a small number of MDM2-amplified sarcoma cell lines reported clear induction of Annexin-V (Tovar et al. 2006) and TUNEL (Muller et al. 2007) staining following treatment with nutlin-3. These findings translated to the in vivo setting in which daily administration of nutlin-3 induced extensive tumor regression in established MDM2-amplified osteosarcoma xenograft models (Tovar et al. 2006). Taken together, these results suggest that (1) the p53-signaling pathway downstream from MDM2 is structurally intact, but functionally inactivated by virtue of MDM2 overexpression in the MDM2-amplified sarcoma setting, and (2) that MDM2 antagonists might be expected to show significant clinical activity in this setting.
More recent studies have characterized the activity of nutlin-3 in primary human sarcoma tissue biopsies (Pishas et al. 2014). Here, the investigators evaluated the ability of MDM2 amplification status to serve as a predictive biomarker to identify those patients most likely to respond to the MDM2 antagonist. Immunohistochemical analysis of 24 explant sarcoma tissues treated with nutlin-3 identified three biopsies (one each of myxoid liposarcoma, Ewing sarcoma, and chondrosarcoma) with robust apoptotic responses (activated caspase-3). Surprisingly, neither MDM2 amplification nor expression showed significant correlation with apoptotic response to nutlin-3. In fact, of the six biopsies deemed to be MDM2 amplified, none showed a robust apoptotic response to nutlin-3 treatment. Furthermore, of the three biopsies that elicited the strongest apoptotic responses, none displayed evidence of MDM2 amplification or overexpression. The investigators then performed a global transcriptomic analysis in an effort to identify biomarkers with better predictive strength. The results suggested that the promoter methylation state of GADD45A (growth arrest and DNA-damage-inducible, α), a p53 target gene involved in coordinating DNA repair and cell-cycle regulation, could serve as a potential predictive biomarker of nutlin-3 efficacy in sarcomas. In contrast to the aforementioned studies performed in MDM2-amplified cell lines and tumor xenograft models in which MDM2 amplification predisposed cells to undergo apoptosis, these results highlighted that MDM2 amplification may not be the ideal predictive biomarker of response to MDM2 antagonists in sarcoma.
A proof-of-mechanism clinical trial with RG7112, an MDM2 antagonist and member of the nutlin family, was performed in chemotherapy-naïve well-differentiated and dedifferentiated MDM2-amplified liposarcoma (Ray-Coquard et al. 2012). Patients received a 1440 mg/m2 dose of RG7112 for 10 days of a 28-day cycle (up to three cycles), followed by surgical resection. The primary endpoint of this study was the assessment of MDM2 inhibition and p53 reactivation as measured by changes in a panel of exploratory biomarkers, including p53, p21, MDM2, Ki-67, MIC-1, and apoptosis (TUNEL staining). Most patients showed changes in the measured endpoints in line with expectations for MDM2 inhibition and p53 reactivation, suggesting that RG7112 was pharmacologically active on its intended target. Changes in exploratory biomarkers included increases in apoptosis, as measured by TUNEL staining, and reduced proliferative capacity, as measured by Ki-67 expression. Furthermore, MDM2 messenger RNA (mRNA) expression and p53 and p21 protein expression were elevated in tumor biopsies, along with elevated levels of MIC-1 expression in patient blood. Somewhat disappointingly, the best response evaluation criteria in solid tumors (RECIST) to RG7112 was a confirmed partial response in one patient, with 14 patients showing stable disease and five patients displaying disease progression. Considering that these patients were primarily amplified for MDM2, one might have expected more profound effects with RG7112 in this setting. However, patients were limited to one to three treatment cycles of RG7112, which might have prevented a full assessment of its efficacy potential.
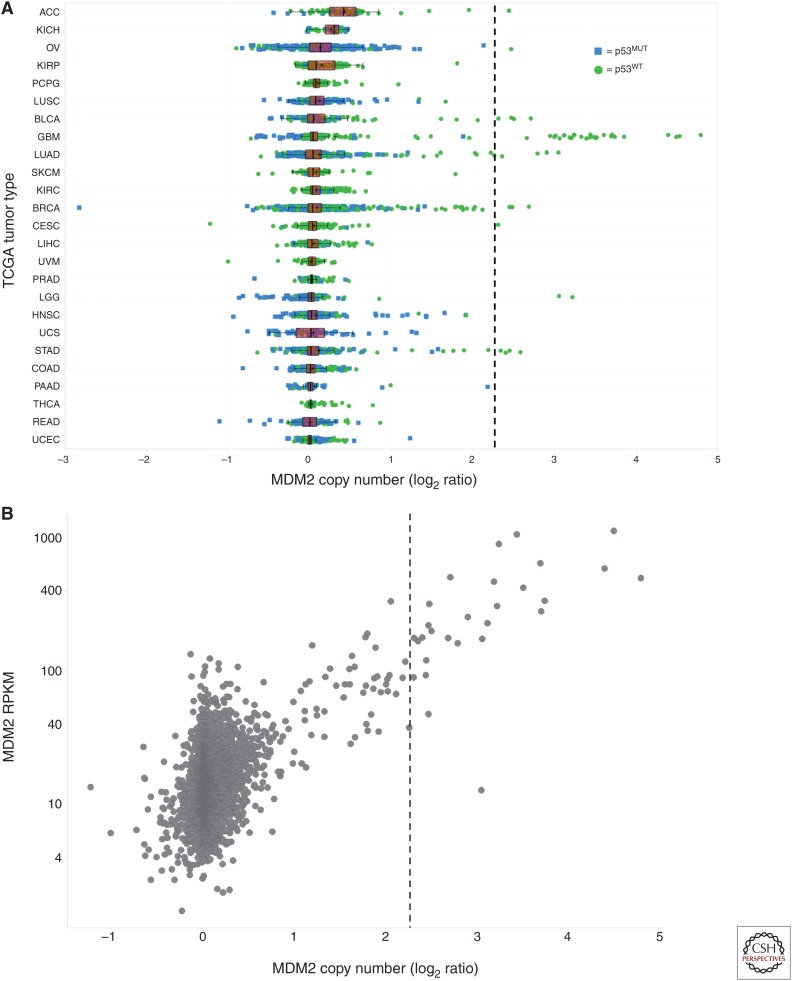
Numerous studies have reported evidence for MDM2 amplification outside the sarcoma setting, including pan-cancer analyses, which have identified the MDM2 gene as being located in one of the most statistically significant “peak” regions of recurrent amplification in the cancer genome (Beroukhim et al. 2010; Zack et al. 2013). More focused efforts have reported infrequent copy number (CN) gains in MDM2 across diverse tumor types, including malignant glioma (Reifenberger et al. 1993), breast carcinoma (Quesnel et al. 1994; McCann et al. 1995; Choschzick et al. 2010), ovarian carcinoma (Foulkes et al. 1995; Courjal et al. 1996; Mayr et al. 2006), urothelial carcinoma (Habuchi et al. 1994; Lianes et al. 1994), lung carcinoma (Marchetti et al. 1995; Higashiyama et al. 1997; Dworakowska et al. 2004; Job et al. 2010; Zhao et al. 2014), and others. To gain a better understanding for the propensity of MDM2 amplification across diverse cancer indications, The Cancer Genome Atlas (TCGA) data (The Cancer Genome Atlas Research Network 2015) were mined for evidence of primary tumor samples harboring amplification of the MDM2 locus. Functionally relevant MDM2 amplification was defined as the CN above which MDM2 amplification and p53 mutation were mutually exclusive, a phenomenon observed at MDM2 log2 CN ratio ≥2.25, or 9.5 copies per cell (Saiki et al. 2015). Evidence of MDM2 amplification was found in ∼1.2% of all TCGA primary tumor samples (Table 1, Fig. 1A), although amplification rates were not uniform across tumor types. Tumor types harboring MDM2 amplification in >2% of primary tumor samples included sarcoma (18.7%), glioblastoma multiforme (7.2%), bladder urothelial carcinoma (2.9%), and cholangiocarcinoma (2.8%) (Table 1, Fig. 1A). Furthermore, a strong correlation was observed between MDM2 amplification and elevated MDM2 expression (Fig. 1B).
Table 1.
Annual U.S. incidence of MDM2-amplified tumorsa
| Tumor type | MDM2 amplificationb (TCGAc) | Annual incidenced (U.S.) | Projected annual U.S. incidence with MDM2 amplification |
|---|---|---|---|
| Sarcoma | 18.7% (48/257) | 11,930 | 2231 |
| Liposarcoma | 78.6% (44/56) | 5000e | 3930 |
| Glioblastoma multiforme | 7.2% (42/581) | 11,390f | 820 |
| Bladder urothelial carcinoma | 2.9% (12/412) | 74,000 | 2146 |
| Cholangiocarcinoma | 2.8% (1/36) | 3000 | 84 |
| Stomach adenocarcinoma | 1.6% (7/442) | 24,590 | 393 |
| Testicular germ cell tumors | 1.3% (2/150) | 8430 | 110 |
| Adrenocortical carcinoma | 1.1% (1/90) | 200 | 2 |
| Lung adenocarcinoma | 0.9% (5/528) | 66,700g | 600 |
| Brain lower grade glioma | 0.4% (2/513) | 3000f,h | 12 |
| Breast invasive carcinoma | 0.4% (4/1097) | 234,190 | 937 |
| Ovarian serous cystadenocarcinoma | 0.3% (2/583) | 13,190i | 40 |
| Cervical squamous carcinoma | 0.3% (1/295) | 12,900 | 39 |
| Lung squamous cell carcinoma | 0.2% (1/501) | 57,170g | 114 |
| Uterine corpus endometrial carcinoma | 0% (0/544) | 54,870 | - |
| Kidney carcinomak | 0% (0/886) | 61,560 | - |
| Colorectal adenocarcinomal | 0% (0/628) | 132,700 | - |
| Head and neck squamous cell carcinoma | 0% (0/522) | 42,440 | - |
| Thyroid carcinoma | 0% (0/504) | 62,450 | - |
| Prostate adenocarcinoma | 0% (0/498) | 220,800 | - |
| Liver hepatocellular carcinoma | 0% (0/370) | 35,660 | - |
| Esophageal carcinoma | 0% (0/184) | 16,980 | - |
| Pancreatic adenocarcinoma | 0% (0/184) | 48,960 | - |
| Pheochromocytoma and paraganglioma | 0% (0/175) | 2850j | - |
| Thymoma | 0% (0/123) | 530 | - |
| Skin cutaneous melanoma | 0% (0/105) | 73,870 | - |
| Mesothelioma | 0% (0/87) | 3000 | - |
| Uveal melanoma | 0% (0/80) | 2580 | - |
| Uterine carcinosarcoma | 0% (0/56) | 1600 | - |
| Lymphoid neoplasm diffuse large B-cell lymphoma | 0% (0/48) | 23,950 | - |
| Total | 1.2% (128/10,479) | 7528–9227 |
aData adapted and updated from Saiki et al. (2015).
bBased on MDM2 log2 CN ratio ≥2.25.
cThe occurrence of MDM2 amplification was derived from tumor sample data generated by the TCGA Research Network (cancergenome.nih.gov).
dExcept as noted, estimates of annual U.S. incidence were taken from the American Cancer Society (2015).
eData from Schwartz (2014).
fData from Dolecek et al. (2012).
gData from Lung Cancer Research Foundation (2015).
hData from Pouratian et al. (2007).
iData from Levanon et al. (2008).
jData from Young and Kaplan (2014).
kData for renal clear cell carcinoma (0/531), papillary cell carcinoma (0/289), and chromophobe carcinoma (0/66) were combined.
lData for colon adenocarcinoma (0/463) and rectum adenocarcinoma (0/165) were combined.
Figure 1.
(A) Mouse double minute 2 (MDM2) copy number (CN) and p53 mutation status for The Cancer Genome Atlas (TCGA) primary tumor samples. Tumor samples have been annotated as p53 wild type (WT) or mutant (MUT) by color and shape. Samples have been grouped by tumor type: adrenocortical carcinoma (ACC), bladder urothelial carcinoma (BLCA), breast invasive carcinoma (BRCA), cervical squamous cell carcinoma (CESC), colon adenocarcinoma (COAD), glioblastoma multiforme (GBM), head and neck squamous cell carcinoma (HNSC), kidney chromophobe (KICH), kidney renal clear cell carcinoma (KIRC), kidney renal papillary cell carcinoma (KIRP), lower grade glioma (LGG). Liver hepatocellular carcinoma (LIHC), lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC), ovarian carcinoma (OV), pancreatic adenocarcinoma (PAAD), pheochromocytoma and paraganglioma (PCPG), prostate adenocarcinoma (PRAD), rectum adenocarcinoma (READ), skin cutaneous melanoma (SKCM), stomach adenocarcinoma (STAD), thyroid carcinoma (THCA), uterine corpus endometrial carcinoma (UCEC), uterine carcinoma (UCS), and uveal melanoma (UVM). (B) Scatterplot of MDM2 CN (log2 CN ratio) versus expression (reads per kilobase of transcript per million [RPKM]) for p53WT TCGA primary tumor samples. Functionally relevant MDM2 amplification was defined as the CN above which MDM2 amplification and p53 mutation were mutually exclusive (dashed line at MDM2 log2 CN ratio ≥2.25 in A and B).
MDM2 OVEREXPRESSION
Although MDM2 amplification is one important means by which MDM2 expression can be up-regulated, other mechanisms for overexpressing MDM2 have been proposed. One such mechanism that has received significant attention involves germline inheritance of a SNP, SNP309, in the first intron of the MDM2 promoter (Bond et al. 2004). Most SNP309 studies have focused on the G and T alleles at this position, which together comprise the vast majority of genotypes (Bond et al. 2004). The G/G genotype enhances the binding affinity of the Sp1 transcription factor to the MDM2 promoter by 1.2-fold in surface plasmon resonance assays (Knappskog et al. 2011) and twofold in electromobility shift assays (Bond et al. 2004), and it increases MDM2 promoter activity by ∼1.5-fold in transcriptional reporter assays (Bond et al. 2004), all compared with the T/T genotype. Cultured cells with the G/G genotype were shown to express much higher average MDM2 mRNA levels than cells bearing the T/T genotype (Bond et al. 2004), but a closer look at this result revealed that much of this difference was driven by a single G/G-genotype cell line (CCF-STTG1) reported to harbor an elevated MDM2 CN, ranging from 18.7 to 52 copies per cell (He et al. 1994; Cancer Cell Line Encyclopedia 2015). The G/G and T/T cell line sets used in this expression comparison were also quite small in number (four lines per genotype), were unmatched for tumor type, and were of mixed p53 mutational status, raising the possibility that the observed MDM2 expression differences were unrelated to the SNP309 genotype. A separate study comparing basal transcription from the SNP309 G and T alleles in a G/T heterozygous cell line revealed 1.48-fold greater expression of the G allele than the T allele (Hu et al. 2007a). Additionally, mice in which the endogenous MDM2 intron 1 was replaced with human intron 1 (with either SNP309 G/G or T/T) showed higher MDM2 transcription in the G/G genotype than the T/T genotype in some tissues (Post et al. 2010).
The most compelling results from the initial report describing the discovery of SNP309 involve epidemiological comparisons suggesting that humans who inherit the G/G genotype suffer accelerated tumor formation (Bond et al. 2004). However, data from subsequent studies conflict as to the relevance of MDM2 SNP309 to human cancer risk. For example, a combined analysis of 11 breast cancer studies, five colorectal cancer studies, and seven lung cancer studies involving 11,633 cases and 12,907 controls found that the SNP309 G/G variant does not impact the risk of breast or colorectal cancer, but it does confer a modestly increased risk in lung cancer (Wilkening et al. 2007). A breast cancer meta-analysis of 12,986 cases and 12,993 controls reported that MDM2 SNP309 status was a risk factor in Chinese populations, but not in non-Chinese populations (Economopoulos and Sergentanis 2010). A meta-analysis of 14,770 cases and 14,524 controls covering 25 studies across multiple tumor types and ethnicities found that SNP309 was a low-penetrance susceptibility tumor marker (Hu et al. 2007b). In four of these 25 studies, p53 mutational status was profiled, and no differences in SNP309-associated risk were seen when comparing p53WT and p53Mutant tumors. Studies in lung cancer (Lind et al. 2006) and gastric cancer (Ohmiya et al. 2006) showed increased risk in the G/G genotype in the p53Mutant group. This is a counterintuitive result, given that there should be no selective oncogenic advantage conferred by mutationally inactivating p53 if SNP309-induced MDM2 overexpression has already shut down the p53 pathway. A similarly puzzling result came from a study in which the T/T genotype, but not the G/G or T/G genotypes, increased breast cancer risk in the p53Mutant population relative to the p53WT population (Boersma et al. 2006). In contrast, a colorectal cancer study showed that somatic p53 mutation abolishes the ability of the SNP309 G-allele to accelerate tumorigenesis (Menin et al. 2006), which is in line with model-based prediction.
A potential explanation for the inconsistent relationship between SNP309 genotype and cancer risk emerged with the finding that an activated estrogen receptor (ER) signaling pathway may be required for SNP309 to accelerate tumor formation (Bond et al. 2006; Bond and Levine 2007). Like Sp1, the ER binds a region of the MDM2 promoter that includes SNP309 (Okumura et al. 2002; Kinyamu and Archer 2003; Phelps et al. 2003), and estrogen up-regulates MDM2 transcription to a greater extent (1.28-fold higher) in the context of the SNP309 G allele than the T allele (Hu et al. 2007a). Studies of diffuse large B-cell lymphoma, soft-tissue sarcoma, breast cancer, and colorectal cancer have shown an earlier age of onset in females, but not males, carrying the SNP309 G allele, an effect accentuated or seen exclusively in women under the average age of menopause (age 51) or whose tumors, in the case of breast cancer, are scored as highly ER positive (reviewed in Bond and Levine 2007). An analysis of cancer patients with Li–Fraumeni syndrome (inherited p53 mutation carriers) showed that female gender and SNP309 G genotype cooperate synergistically to accelerate tumorigenesis (Atwal et al. 2008). To avoid tumor-type biases, this study excluded gender-specific cancers, focusing only on soft-tissue sarcomas, osteosarcomas, and leukemias.
The discovery of an additional modifier of MDM2 SNP309 G, a nearby MDM2 intronic polymorphism called SNP285 C, provided the potential to further reconcile the apparently conflicting results seen in epidemiological studies linking SNP309 G with increased cancer risk (Paulin et al. 2008; Knappskog et al. 2011). The SNP285 C allele is only observed on the SNP309 G haplotype, indicating that these two SNPs are in linkage disequilibrium (Paulin et al. 2008; Knappskog et al. 2011). The SNP285 C/SNP309 G combination haplotype is present in 6%–8% of Caucasians, but is absent in Asian and African American populations (Knappskog et al. 2011, 2014). In a survey of healthy controls from Norway, the Netherlands, the United Kingdom, and Finland, SNP285 C/SNP309 G constituted a minority (11%) of SNP309 G haplotypes (Knappskog et al. 2011). SNP285 C functionally opposes SNP309 G and it does so by neutralizing the SNP309 G–mediated increase in Sp1 affinity for the MDM2 promoter (Knappskog et al. 2011). In fact, SNP285 C more effectively attenuates Sp1-binding affinity than SNP309 G enhances it, thus providing SNP285 C/SNP309 G a net reduction in Sp1 binding relative to wild type (SNP285 G/SNP309 T) and conferring a potential cancer-protective function. The above findings imply that the SNP285 C/SNP309 G subset of SNP309 G alleles might be at least partially responsible for the lack of observable association between SNP309 G and cancer risk in some studies involving Caucasians (Hu et al. 2007b; Economopoulos and Sergentanis 2010). Indeed, in Caucasians with breast cancer, exclusion of SNP285 C/SNP309 G carriers enabled identification of an association between SNP309 G and cancer risk, a link that was not discernable in the full SNP309 G population (Knappskog et al. 2011). In ovarian cancer, removing SNP285 C/SNP309 G carriers strengthened an already statistically significant SNP309 G–mediated increase in cancer risk (Knappskog et al. 2011). This is a counterintuitive result, as the vast majority of ovarian cancers are p53Mutant (The Cancer Genome Atlas Research Network 2015) and should, therefore, be unaffected by SNPs that modulate MDM2-expression levels. Together, the published data around SNPs 285 and 309 present a conflicting picture as to the importance of these polymorphisms in modulating cancer risk.
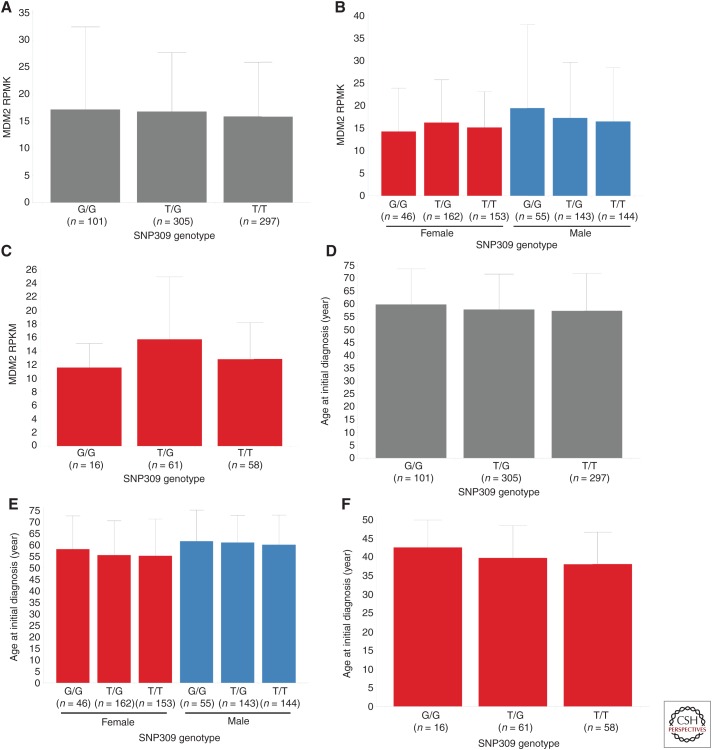
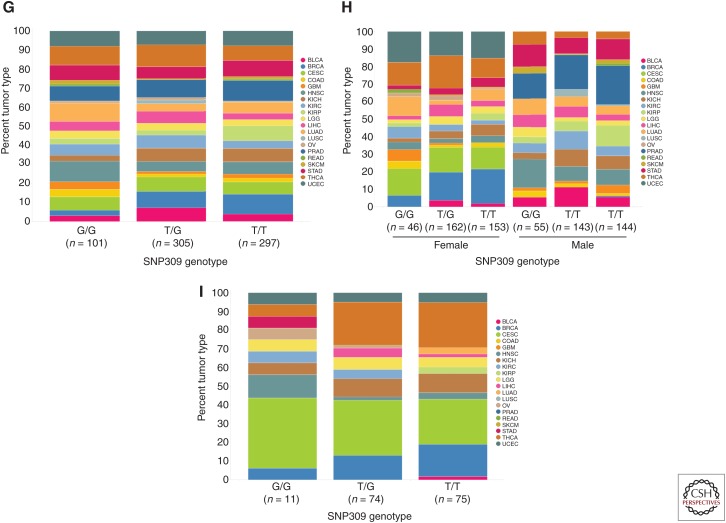
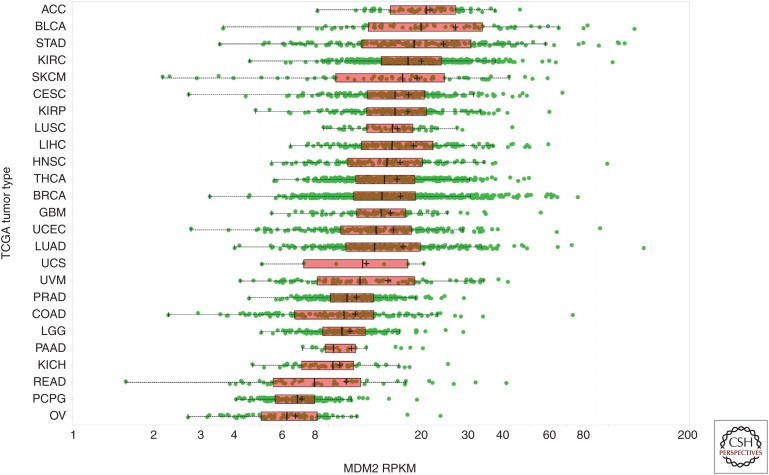
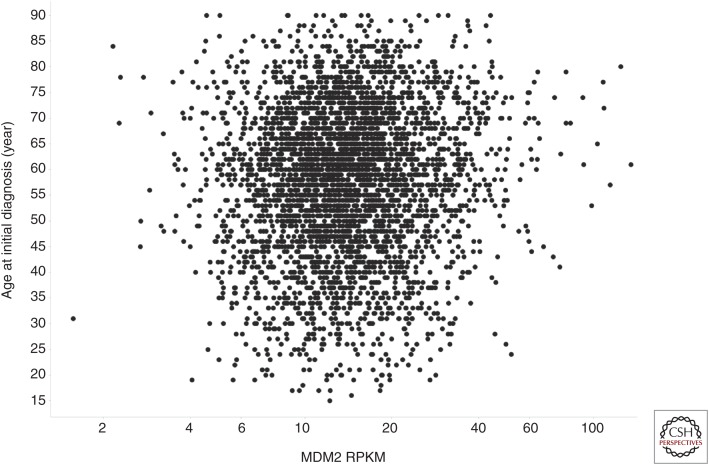
The emergence of large-scale genomic profiling performed under standardized conditions has afforded the ability to further evaluate whether MDM2 promoter SNPs play a role in human cancer development. The concept that the SNP309 G allele predisposes people to higher cancer risk is predicated on the idea that the G allele enhances MDM2 expression. To test the validity of this proposed relationship, TCGA data (The Cancer Genome Atlas Research Network 2015) were mined via ArrayStudio (Omicsoft TCGAL, and 2015 Q1 release, Cary, NC) and the Cancer Genomics Hub (Wilks et al. 2014) to search for an association between the SNP309 G allele and increased MDM2 expression. To avoid any possibility of elevated MDM2 CN confounding the analysis, only tumors with MDM2 log2 CN ratio <1 (i.e., less than four copies per cell) were included. A pan-cancer analysis of p53WT TCGA tumors with MDM2 log2 CN ratio <1 revealed no differences in MDM2 expression in tumors harboring the SNP309 G/G, G/T, and T/T genotypes (Fig. 2A). Similarly, no relationship between the SNP309 G allele and MDM2 expression was observed when subdividing the samples by gender (Fig. 2B) or when restricting the analysis to women under the average age of menopause (Fig. 2C). Further analysis of these same tumor subsets similarly failed to expose any association between SNP309 status and age of initial cancer diagnosis (Fig. 2D–F). It is formally possible that these negative findings (Fig. 2A–F) could be attributable to differences in tumor-type composition across the SNP309 genotypes present in TCGA (Fig. 2G–I). However, this seems unlikely given that mean MDM2 expression did not differ markedly across tumor types (Fig. 3; mean RPKM 15.4 ± 4.9), suggesting that tumor type is not a strong modifier of MDM2 mRNA levels. Last, in an analysis agnostic to MDM2 promoter SNP status, no relationship was seen between MDM2 mRNA level and age of initial cancer diagnosis (Fig. 4). Together, these data suggest that (1) MDM2 SNP309 does not alter MDM2 expression or modulate cancer risk across a large swath of human tumors, and (2) MDM2 expression itself is not a determinant of cancer risk in these tumors. Nevertheless, these results cannot rule out the possibility that MDM2 promoter SNPs might confer subtle changes in cancer risk in narrow populations of cancers.
Figure 2.
Relationship between single-nucleotide polymorphism (SNP)309 genotype and mouse double minute 2 (MDM2) expression (reads per kilobase of transcript per million [RPKM]) for (A) p53WT The Cancer Genome Atlas (TCGA) primary tumor samples containing MDM2 log2 copy number (CN) ratio <1.0, (B) grouped by gender, or (C) limited to female patients with initial age of diagnosis <51 (premenopausal). (D-F) Relationships between SNP309 genotype and age at initial diagnosis (years) for the same corresponding groups. Mean and standard deviation for MDM2 expression or age at initial diagnosis within each genotype have been shown. With one exception, no significant differences were observed in MDM2 expression or age at initial diagnosis when comparing either the G/G and T/G populations or G/G and T/T populations (Student's two-tailed t-test assuming unequal variances). Although a statistically significant difference was observed in MDM2 expression when comparing the G/G and T/G populations in premenopausal females (C, p-value = 0.006), the direction of the difference was counter to that expected based on previous reports of the G/G genotype correlating with elevated MDM2-expression levels. Furthermore, MDM2 expression did not trend with G allele dosage. (G-I) Tumor-type composition for each of the SNP309 genotype populations for the corresponding groups analyzed in (A-F), respectively (see Fig. 1 for tumor-type abbreviations).
Figure 3.
Mouse double minute 2 (MDM2) expression (reads per kilobase of transcript per million [RPKM]) in The Cancer Genome Atlas (TCGA) p53WT primary tumor samples with MDM2 log2 copy number (CN) ratio <1.0, grouped by tumor type (see Figure 1 for tumor-type abbreviations).
Figure 4.
Scatter plot of mouse double minute 2 (MDM2) expression (reads per kilobase of transcript per million [RPKM]) versus age at initial diagnosis (years) for p53WT The Cancer Genome Atlas (TCGA) primary tumor samples with MDM2 log2 copy number (CN) ratio <1.0.
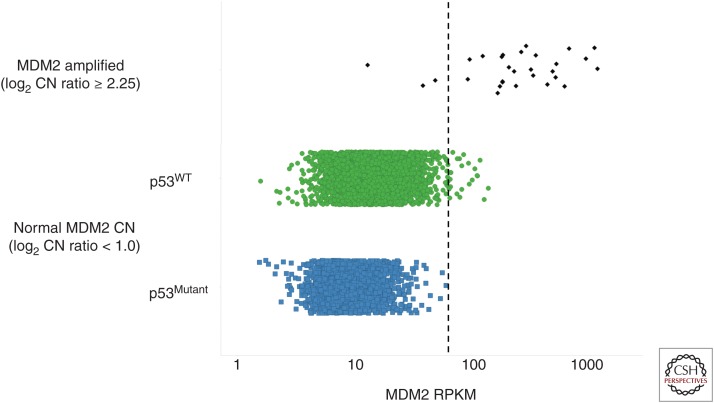
As discussed above, MDM2 amplification is associated with elevated MDM2 mRNA expression (Fig. 1B), and it is reasonable to assume that this level of MDM2 overexpression would be required to functionally inactivate p53. To determine whether comparable levels of MDM2 overexpression are achievable by other means, we mined TCGA data (The Cancer Genome Atlas Research Network 2015) in search of tumors with similarly elevated MDM2 levels but normal MDM2 CN (log2 CN ratio <1). We reasoned that functionally elevated MDM2 expression would be mutually exclusive with p53 mutation and, therefore, set an expression threshold that excluded all p53Mutant tumors (Fig. 5). As expected, the vast majority of MDM2-amplified tumors also exceeded this expression threshold (Fig. 5). Based on this analysis, only a small fraction of all tumors with normal MDM2 CN (∼0.5%) reached MDM2-expression levels similar to those achieved by MDM2 amplification. Together, the results discussed herein implicate MDM2 amplification as the dominant mechanism through which human tumors raise MDM2 levels to abrogate p53. Even so, the rate of MDM2 amplification pales in comparison to other mechanisms for inactivating the p53 pathway, such as mutation of p53 itself.
Figure 5.
Dot plot of mouse double minute 2 (MDM2) expression (reads per kilobase of transcript per million [RPKM]) in The Cancer Genome Atlas (TCGA) primary tumor samples grouped by MDM2 amplified samples (log2 copy number [CN] ratio ≥2.25) or p53WT and p53Mutant samples with normal MDM2 CN (log2 CN ratio <1.0). The threshold for functionally elevated MDM2 expression as defined by mutual exclusivity with p53-mutated samples has been indicated by the dashed line.
ACKNOWLEDGMENTS
We thank Henry Bigelow for his contributions to the analysis of the TCGA Whole Genome Sequencing data for evaluation of SNP309 status, and Rick Kendall, Angela Coxon, and Paul Hughes for their critical review of the manuscript.
Footnotes
Editors: Guillermina Lozano and Arnold J. Levine
Additional Perspectives on The p53 Protein available at www.perspectivesinmedicine.org
REFERENCES
- Alt JR, Greiner TC, Cleveland JL, Eischen CM. 2003. Mdm2 haplo-insufficiency profoundly inhibits Myc-induced lymphomagenesis. EMBO J 22: 1442–1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atwal GS, Rabadan R, Lozano G, Strong LC, Ruijs MW, Schmidt MK, van’t Veer LJ, Nevanlinna H, Tommiska J, Aittomaki K, et al. 2008. An information-theoretic analysis of genetics, gender and age in cancer patients. PloS ONE 3: e1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, Barretina J, Boehm JS, Dobson J, Urashima M, et al. 2010. The landscape of somatic copy-number alteration across human cancers. Nature 463: 899–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binh MB, Sastre-Garau X, Guillou L, de Pinieux G, Terrier P, Lagace R, Aurias A, Hostein I, Coindre JM. 2005. MDM2 and CDK4 immunostainings are useful adjuncts in diagnosing well-differentiated and dedifferentiated liposarcoma subtypes: A comparative analysis of 559 soft tissue neoplasms with genetic data. Am J Surg Pathol 29: 1340–1347. [DOI] [PubMed] [Google Scholar]
- Boersma BJ, Howe TM, Goodman JE, Yfantis HG, Lee DH, Chanock SJ, Ambs S. 2006. Association of breast cancer outcome with status of p53 and MDM2 SNP309. J Natl Cancer Inst 98: 911–919. [DOI] [PubMed] [Google Scholar]
- Bond GL, Levine AJ. 2007. A single nucleotide polymorphism in the p53 pathway interacts with gender, environmental stresses and tumor genetics to influence cancer in humans. Oncogene 26: 1317–1323. [DOI] [PubMed] [Google Scholar]
- Bond GL, Hu W, Bond EE, Robins H, Lutzker SG, Arva NC, Bargonetti J, Bartel F, Taubert H, Wuerl P, et al. 2004. A single nucleotide polymorphism in the MDM2 promoter attenuates the p53 tumor suppressor pathway and accelerates tumor formation in humans. Cell 119: 591–602. [DOI] [PubMed] [Google Scholar]
- Bond GL, Hu W, Levine A. 2005. A single nucleotide polymorphism in the MDM2 gene: From a molecular and cellular explanation to clinical effect. Cancer Res 65: 5481–5484. [DOI] [PubMed] [Google Scholar]
- Bond GL, Hirshfield KM, Kirchhoff T, Alexe G, Bond EE, Robins H, Bartel F, Taubert H, Wuerl P, Hait W, et al. 2006. MDM2 SNP309 accelerates tumor formation in a gender-specific and hormone-dependent manner. Cancer Res 66: 5104–5110. [DOI] [PubMed] [Google Scholar]
- Cahilly-Snyder L, Yang-Feng T, Francke U, George DL. 1987. Molecular analysis and chromosomal mapping of amplified genes isolated from a transformed mouse 3T3 cell line. Somat Cell Mol Genet 13: 235–244. [DOI] [PubMed] [Google Scholar]
- Chibon F, Mariani O, Derre J, Malinge S, Coindre JM, Guillou L, Lagace R, Aurias A. 2002. A subgroup of malignant fibrous histiocytomas is associated with genetic changes similar to those of well-differentiated liposarcomas. Cancer Genet Cytogenet 139: 24–29. [DOI] [PubMed] [Google Scholar]
- Choschzick M, Heilenkotter U, Lebeau A, Jaenicke F, Terracciano L, Bokemeyer C, Sauter G, Simon R. 2010. MDM2 amplification is an independent prognostic feature of node-negative, estrogen receptor-positive early-stage breast cancer. Cancer Biomark 8: 53–60. [DOI] [PubMed] [Google Scholar]
- Coindre JM, Mariani O, Chibon F, Mairal A, De Saint Aubain Somerhausen N, Favre-Guillevin E, Bui NB, Stoeckle E, Hostein I, Aurias A. 2003. Most malignant fibrous histiocytomas developed in the retroperitoneum are dedifferentiated liposarcomas: A review of 25 cases initially diagnosed as malignant fibrous histiocytoma. Mod Pathol 16: 256–262. [DOI] [PubMed] [Google Scholar]
- Coindre JM, Pedeutour F, Aurias A. 2010. Well-differentiated and dedifferentiated liposarcomas. Virchows Archiv 456: 167–179. [DOI] [PubMed] [Google Scholar]
- Cordon-Cardo C, Latres E, Drobnjak M, Oliva MR, Pollack D, Woodruff JM, Marechal V, Chen J, Brennan MF, Levine AJ. 1994. Molecular abnormalities of mdm2 and p53 genes in adult soft tissue sarcomas. Cancer Res 54: 794–799. [PubMed] [Google Scholar]
- Courjal F, Cuny M, Rodriguez C, Louason G, Speiser P, Katsaros D, Tanner MM, Zeillinger R, Theillet C. 1996. DNA amplifications at 20q13 and MDM2 define distinct subsets of evolved breast and ovarian tumours. Br J Cancer 74: 1984–1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolecek TA, Propp JM, Stroup NE, Kruchko C. 2012. CBTRUS Statistical Report: Primary brain and central nervous system tumors diagnosed in the United States in 2005–2009. Neuro Oncol 14: v1–v49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dworakowska D, Jassem E, Jassem J, Peters B, Dziadziuszko R, Zylicz M, Jakobkiewicz-Banecka J, Kobierska-Gulida G, Szymanowska A, Skokowski J, et al. 2004. MDM2 gene amplification: A new independent factor of adverse prognosis in non-small cell lung cancer (NSCLC). Lung Cancer 43: 285–295. [DOI] [PubMed] [Google Scholar]
- Economopoulos KP, Sergentanis TN. 2010. Differential effects of MDM2 SNP309 polymorphism on breast cancer risk along with race: A meta-analysis. Breast Cancer Res Treat 120: 211–216. [DOI] [PubMed] [Google Scholar]
- Fakharzadeh SS, Trusko SP, George DL. 1991. Tumorigenic potential associated with enhanced expression of a gene that is amplified in a mouse tumor cell line. EMBO J 10: 1565–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlay CA. 1993. The mdm-2 oncogene can overcome wild-type p53 suppression of transformed cell growth. Mol Cell Biol 13: 301–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florenes VA, Maelandsmo GM, Forus A, Andreassen A, Myklebost O, Fodstad O. 1994. MDM2 gene amplification and transcript levels in human sarcomas: Relationship to TP53 gene status. J Natl Cancer Inst 86: 1297–1302. [DOI] [PubMed] [Google Scholar]
- Forus A, Florenes VA, Maelandsmo GM, Meltzer PS, Fodstad O, Myklebost O. 1993. Mapping of amplification units in the q13-14 region of chromosome 12 in human sarcomas: Some amplica do not include MDM2. Cell Growth Differ 4: 1065–1070. [PubMed] [Google Scholar]
- Foulkes WD, Stamp GW, Afzal S, Lalani N, McFarlane CP, Trowsdale J, Campbell IG. 1995. MDM2 overexpression is rare in ovarian carcinoma irrespective of TP53 mutation status. Br J Cancer 72: 883–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habuchi T, Kinoshita H, Yamada H, Kakehi Y, Ogawa O, Wu WJ, Takahashi R, Sugiyama T, Yoshida O. 1994. Oncogene amplification in urothelial cancers with p53 gene mutation or MDM2 amplification. J Natl Cancer Inst 86: 1331–1335. [DOI] [PubMed] [Google Scholar]
- Haupt Y, Maya R, Kazaz A, Oren M. 1997. Mdm2 promotes the rapid degradation of p53. Nature 387: 296–299. [DOI] [PubMed] [Google Scholar]
- He J, Reifenberger G, Liu L, Collins VP, James CD. 1994. Analysis of glioma cell lines for amplification and overexpression of MDM2. Genes Chromosomes Cancer 11: 91–96. [DOI] [PubMed] [Google Scholar]
- Higashiyama M, Doi O, Kodama K, Yokouchi H, Kasugai T, Ishiguro S, Takami K, Nakayama T, Nishisho I. 1997. MDM2 gene amplification and expression in non-small-cell lung cancer: Immunohistochemical expression of its protein is a favourable prognostic marker in patients without p53 protein accumulation. Br J Cancer 75: 1302–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinds PW, Finlay CA, Quartin RS, Baker SJ, Fearon ER, Vogelstein B, Levine AJ. 1990. Mutant p53 DNA clones from human colon carcinomas cooperate with ras in transforming primary rat cells: A comparison of the “hot spot” mutant phenotypes. Cell Growth Differ 1: 571–580. [PubMed] [Google Scholar]
- Honda R, Tanaka H, Yasuda H. 1997. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett 420: 25–27. [DOI] [PubMed] [Google Scholar]
- Hu W, Feng Z, Ma L, Wagner J, Rice JJ, Stolovitzky G, Levine AJ. 2007a. A single nucleotide polymorphism in the MDM2 gene disrupts the oscillation of p53 and MDM2 levels in cells. Cancer Res 67: 2757–2765. [DOI] [PubMed] [Google Scholar]
- Hu Z, Jin G, Wang L, Chen F, Wang X, Shen H. 2007b. MDM2 promoter polymorphism SNP309 contributes to tumor susceptibility: Evidence from 21 case-control studies. Cancer Epidemiol Biomarkers Prev 16: 2717–2723. [DOI] [PubMed] [Google Scholar]
- Job B, Bernheim A, Beau-Faller M, Camilleri-Broet S, Girard P, Hofman P, Mazieres J, Toujani S, Lacroix L, Laffaire J, et al. 2010. Genomic aberrations in lung adenocarcinoma in never smokers. PloS ONE 5: e15145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones SN, Roe AE, Donehower LA, Bradley A. 1995. Rescue of embryonic lethality in Mdm2-deficient mice by absence of p53. Nature 378: 206–208. [DOI] [PubMed] [Google Scholar]
- Jones SN, Hancock AR, Vogel H, Donehower LA, Bradley A. 1998. Overexpression of Mdm2 in mice reveals a p53-independent role for Mdm2 in tumorigenesis. Proc Natl Acad Sci 95: 15608–15612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junttila MR, Evan GI. 2009. p53—A Jack of all trades but master of none. Nat Rev Cancer 9: 821–829. [DOI] [PubMed] [Google Scholar]
- Kandoth C, McLellan MD, Vandin F, Ye K, Niu B, Lu C, Xie M, Zhang Q, McMichael JF, Wyczalkowski MA, et al. 2013. Mutational landscape and significance across 12 major cancer types. Nature 502: 333–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinyamu HK, Archer TK. 2003. Estrogen receptor-dependent proteasomal degradation of the glucocorticoid receptor is coupled to an increase in mdm2 protein expression. Mol Cell Biol 23: 5867–5881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knappskog S, Bjornslett M, Myklebust LM, Huijts PE, Vreeswijk MP, Edvardsen H, Guo Y, Zhang X, Yang M, Ylisaukko-Oja SK, et al. 2011. The MDM2 promoter SNP285C/309G haplotype diminishes Sp1 transcription factor binding and reduces risk for breast and ovarian cancer in Caucasians. Cancer Cell 19: 273–282. [DOI] [PubMed] [Google Scholar]
- Knappskog S, Gansmo LB, Dibirova K, Metspalu A, Cybulski C, Peterlongo P, Aaltonen L, Vatten L, Romundstad P, Hveem K, et al. 2014. Population distribution and ancestry of the cancer protective MDM2 SNP285 (rs117039649). Oncotarget 5: 8223–8234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubbutat MH, Jones SN, Vousden KH. 1997. Regulation of p53 stability by Mdm2. Nature 387: 299–303. [DOI] [PubMed] [Google Scholar]
- Ladanyi M, Cha C, Lewis R, Jhanwar SC, Huvos AG, Healey JH. 1993. MDM2 gene amplification in metastatic osteosarcoma. Cancer Res 53: 16–18. [PubMed] [Google Scholar]
- Leach FS, Tokino T, Meltzer P, Burrell M, Oliner JD, Smith S, Hill DE, Sidransky D, Kinzler KW, Vogelstein B. 1993. p53 Mutation and MDM2 amplification in human soft tissue sarcomas. Cancer Res 53: 2231–2234. [PubMed] [Google Scholar]
- Levanon K, Crum C, Drapkin R. 2008. New insights into the pathogenesis of serous ovarian cancer and its clinical impact. J Clin Oncol 26: 5284–5293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lianes P, Orlow I, Zhang ZF, Oliva MR, Sarkis AS, Reuter VE, Cordon-Cardo C. 1994. Altered patterns of MDM2 and TP53 expression in human bladder cancer. J Natl Cancer Inst 86: 1325–1330. [DOI] [PubMed] [Google Scholar]
- Lind H, Zienolddiny S, Ekstrom PO, Skaug V, Haugen A. 2006. Association of a functional polymorphism in the promoter of the MDM2 gene with risk of nonsmall cell lung cancer. Int J Cancer 119: 718–721. [DOI] [PubMed] [Google Scholar]
- Lokka S, Scheel AH, Dango S, Schmitz K, Hesterberg R, Ruschoff J, Schildhaus HU. 2014. Challenging dedifferentiated liposarcoma identified by MDM2-amplification, a report of two cases. BMC Clin Pathol 14: 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundgren K, Montes de Oca Luna R, McNeill YB, Emerick EP, Spencer B, Barfield CR, Lozano G, Rosenberg MP, Finlay CA. 1997. Targeted expression of MDM2 uncouples S phase from mitosis and inhibits mammary gland development independent of p53. Genes Dev 11: 714–725. [DOI] [PubMed] [Google Scholar]
- Marchetti A, Buttitta F, Pellegrini S, Merlo G, Chella A, Angeletti CA, Bevilacqua G. 1995. mdm2 gene amplification and overexpression in non-small cell lung carcinomas with accumulation of the p53 protein in the absence of p53 gene mutations. Diagn Mol Pathol 4: 93–97. [DOI] [PubMed] [Google Scholar]
- Mayr D, Kanitz V, Anderegg B, Luthardt B, Engel J, Lohrs U, Amann G, Diebold J. 2006. Analysis of gene amplification and prognostic markers in ovarian cancer using comparative genomic hybridization for microarrays and immunohistochemical analysis for tissue microarrays. Am J Clin Pathol 126: 101–109. [DOI] [PubMed] [Google Scholar]
- McCann AH, Kirley A, Carney DN, Corbally N, Magee HM, Keating G, Dervan PA. 1995. Amplification of the MDM2 gene in human breast cancer and its association with MDM2 and p53 protein status. Br J Cancer 71: 981–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meltzer PS, Jankowski SA, Dal Cin P, Sandberg AA, Paz IB, Coccia MA. 1991. Identification and cloning of a novel amplified DNA sequence in human malignant fibrous histiocytoma derived from a region of chromosome 12 frequently rearranged in soft tissue tumors. Cell Growth Differ 2: 495–501. [PubMed] [Google Scholar]
- Mendrysa SM, O’Leary KA, McElwee MK, Michalowski J, Eisenman RN, Powell DA, Perry ME. 2006. Tumor suppression and normal aging in mice with constitutively high p53 activity. Genes Dev 20: 16–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menin C, Scaini MC, De Salvo GL, Biscuola M, Quaggio M, Esposito G, Belluco C, Montagna M, Agata S, D’Andrea E, et al. 2006. Association between MDM2-SNP309 and age at colorectal cancer diagnosis according to p53 mutation status. J Natl Cancer Inst 98: 285–288. [DOI] [PubMed] [Google Scholar]
- Miller CW, Aslo A, Won A, Tan M, Lampkin B, Koeffler HP. 1996. Alterations of the p53, Rb and MDM2 genes in osteosarcoma. J Cancer Res Clin Oncol 122: 559–565. [DOI] [PubMed] [Google Scholar]
- Momand J, Zambetti GP, Olson DC, George D, Levine AJ. 1992. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell 69: 1237–1245. [DOI] [PubMed] [Google Scholar]
- Momand J, Jung D, Wilczynski S, Niland J. 1998. The MDM2 gene amplification database. Nucleic Acids Res 26: 3453–3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montes de Oca Luna R, Wagner DS, Lozano G. 1995. Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature 378: 203–206. [DOI] [PubMed] [Google Scholar]
- Muller CR, Paulsen EB, Noordhuis P, Pedeutour F, Saeter G, Myklebost O. 2007. Potential for treatment of liposarcomas with the MDM2 antagonist Nutlin-3A. Int J Cancer 121: 199–205. [DOI] [PubMed] [Google Scholar]
- Nakayama T, Toguchida J, Wadayama B, Kanoe H, Kotoura Y, Sasaki MS. 1995. MDM2 gene amplification in bone and soft-tissue tumors: Association with tumor progression in differentiated adipose-tissue tumors. Int J Cancer 64: 342–346. [DOI] [PubMed] [Google Scholar]
- Ohmiya N, Taguchi A, Mabuchi N, Itoh A, Hirooka Y, Niwa Y, Goto H. 2006. MDM2 promoter polymorphism is associated with both an increased susceptibility to gastric carcinoma and poor prognosis. J Clin Oncol 24: 4434–4440. [DOI] [PubMed] [Google Scholar]
- Okumura N, Saji S, Eguchi H, Nakashima S, Saji S, Hayashi S. 2002. Distinct promoter usage of mdm2 gene in human breast cancer. Oncol Reps 9: 557–563. [PubMed] [Google Scholar]
- Oliner JD, Kinzler KW, Meltzer PS, George DL, Vogelstein B. 1992. Amplification of a gene encoding a p53-associated protein in human sarcomas. Nature 358: 80–83. [DOI] [PubMed] [Google Scholar]
- Oliner JD, Pietenpol JA, Thiagalingam S, Gyuris J, Kinzler KW, Vogelstein B. 1993. Oncoprotein MDM2 conceals the activation domain of tumour suppressor p53. Nature 362: 857–860. [DOI] [PubMed] [Google Scholar]
- Patterson H, Barnes D, Gill S, Spicer J, Fisher C, Thomas M, Grimer R, Fletcher C, Gusterson B, Cooper C. 1997. Amplification and over-expression of the MDM2 gene in human soft tissue tumours. Sarcoma 1: 17–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulin FE, O’Neill M, McGregor G, Cassidy A, Ashfield A, Ali CW, Munro AJ, Baker L, Purdie CA, Lane DP, et al. 2008. MDM2 SNP309 is associated with high grade node positive breast tumours and is in linkage disequilibrium with a novel MDM2 intron 1 polymorphism. BMC Cancer 8: 281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelps M, Darley M, Primrose JN, Blaydes JP. 2003. p53-independent activation of the hdm2-P2 promoter through multiple transcription factor response elements results in elevated hdm2 expression in estrogen receptor α-positive breast cancer cells. Cancer Res 63: 2616–2623. [PubMed] [Google Scholar]
- Pishas KI, Neuhaus SJ, Clayer MT, Schreiber AW, Lawrence DM, Perugini M, Whitfield RJ, Farshid G, Manavis J, Chryssidis S, et al. 2014. Nutlin-3a efficacy in sarcoma predicted by transcriptomic and epigenetic profiling. Cancer Res 74: 921–931. [DOI] [PubMed] [Google Scholar]
- Post SM, Quintas-Cardama A, Pant V, Iwakuma T, Hamir A, Jackson JG, Maccio DR, Bond GL, Johnson DG, Levine AJ, et al. 2010. A high-frequency regulatory polymorphism in the p53 pathway accelerates tumor development. Cancer Cell 18: 220–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouratian N, Asthagiri A, Jagannathan J, Shaffrey ME, Schiff D. 2007. Surgery insight: The role of surgery in the management of low-grade gliomas. Nat Clin Pract Neuro 3: 628–639. [DOI] [PubMed] [Google Scholar]
- Quesnel B, Preudhomme C, Fournier J, Fenaux P, Peyrat JP. 1994. MDM2 gene amplification in human breast cancer. Eur J Cancer 30A: 982–984. [DOI] [PubMed] [Google Scholar]
- Ray-Coquard I, Blay JY, Italiano A, Le Cesne A, Penel N, Zhi J, Heil F, Rueger R, Graves B, Ding M, et al. 2012. Effect of the MDM2 antagonist RG7112 on the P53 pathway in patients with MDM2-amplified, well-differentiated or dedifferentiated liposarcoma: An exploratory proof-of-mechanism study. Lancet Oncol 13: 1133–1140. [DOI] [PubMed] [Google Scholar]
- Reifenberger G, Liu L, Ichimura K, Schmidt EE, Collins VP. 1993. Amplification and overexpression of the MDM2 gene in a subset of human malignant gliomas without p53 mutations. Cancer Res 53: 2736–2739. [PubMed] [Google Scholar]
- Roth J, Dobbelstein M, Freedman DA, Shenk T, Levine AJ. 1998. Nucleo-cytoplasmic shuttling of the hdm2 oncoprotein regulates the levels of the p53 protein via a pathway used by the human immunodeficiency virus rev protein. EMBO J 17: 554–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiki AY, Caenepeel S, Cosgrove E, Su C, Boedigheimer M, Oliner JD. 2015. Identifying the determinants of response to MDM2 inhibition. Oncotarget 6: 7701–7712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada S, Ishizawa T, Ishizawa K, Matsumura T, Hasegawa T, Hirose T. 2006. The value of MDM2 and CDK4 amplification levels using real-time polymerase chain reaction for the differential diagnosis of liposarcomas and their histologic mimickers. Hum Pathol 37: 1123–1129. [DOI] [PubMed] [Google Scholar]
- Terzian T, Wang Y, Van Pelt CS, Box NF, Travis EL, Lozano G. 2007. Haploinsufficiency of Mdm2 and Mdm4 in tumorigenesis and development. Mol Cell Biol 27: 5479–5485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tovar C, Rosinski J, Filipovic Z, Higgins B, Kolinsky K, Hilton H, Zhao X, Vu BT, Qing W, Packman K, et al. 2006. Small-molecule MDM2 antagonists reveal aberrant p53 signaling in cancer: Implications for therapy. Proc Natl Acad Sci 103: 1888–1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turc-Carel C, Limon J, Dal Cin P, Rao U, Karakousis C, Sandberg AA. 1986. Cytogenetic studies of adipose tissue tumors. II: Recurrent reciprocal translocation t(12;16) (q13;p11) in myxoid liposarcomas. Cancer Genet Cytogenet 23: 291–299. [DOI] [PubMed] [Google Scholar]
- Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, Kong N, Kammlott U, Lukacs C, Klein C, et al. 2004. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science 303: 844–848. [DOI] [PubMed] [Google Scholar]
- Wilkening S, Bermejo JL, Hemminki K. 2007. MDM2 SNP309 and cancer risk: A combined analysis. Carcinogenesis 28: 2262–2267. [DOI] [PubMed] [Google Scholar]
- Wilks C, Cline MS, Weiler E, Diehkans M, Craft B, Martin C, Murphy D, Pierce H, Black J, Nelson D, et al. 2014. The Cancer Genomics Hub (CGHub): Overcoming cancer through the power of torrential data. Database (Oxford) 10.1093/database/bau093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zack TI, Schumacher SE, Carter SL, Cherniack AD, Saksena G, Tabak B, Lawrence MS, Zhang CZ, Wala J, Mermel CH, et al. 2013. Pan-cancer patterns of somatic copy number alteration. Nat Genet 45: 1134–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Yang J, Chen Z, Gao Z, Zhou F, Li X, Li J, Shi S, Feng X, Sun N, et al. 2014. Identification of somatic alterations in stage I lung adenocarcinomas by next-generation sequencing. Genes Chromosomes Cancer 53: 289–298. [DOI] [PubMed] [Google Scholar]