Abstract
Reconstructing the complete evolutionary history of extant life on our planet will be one of the most fundamental accomplishments of scientific endeavor, akin to the completion of the periodic table, which revolutionized chemistry. The road to this goal is via comparative genomics because genomes are our most comprehensive and objective evolutionary documents. The genomes of plant and animal species have been systematically targeted over the past decade to provide coverage of the tree of life. However, multicellular organisms only emerged in the last 550 million years of more than three billion years of biological evolution and thus comprise a small fraction of total biological diversity. The bulk of biodiversity, both past and present, is microbial. We have only scratched the surface in our understanding of the microbial world, as most microorganisms cannot be readily grown in the laboratory and remain unknown to science. Ground-breaking, culture-independent molecular techniques developed over the past 30 years have opened the door to this so-called microbial dark matter with an accelerating momentum driven by exponential increases in sequencing capacity. We are on the verge of obtaining representative genomes across all life for the first time. However, historical use of morphology, biochemical properties, behavioral traits, and single-marker genes to infer organismal relationships mean that the existing highly incomplete tree is riddled with taxonomic errors. Concerted efforts are now needed to synthesize and integrate the burgeoning genomic data resources into a coherent universal tree of life and genome-based taxonomy.
Uncultured microorganisms represent the majority (>80%) of microbial diversity. Newly developed culture-independent methods to sequence microbial genomes will allow the development of a more complete, objective tree of life.
SETTING THE STAGE FOR A TAXONOMIC CLASSIFICATION BASED ON EVOLUTIONARY RELATIONSHIPS
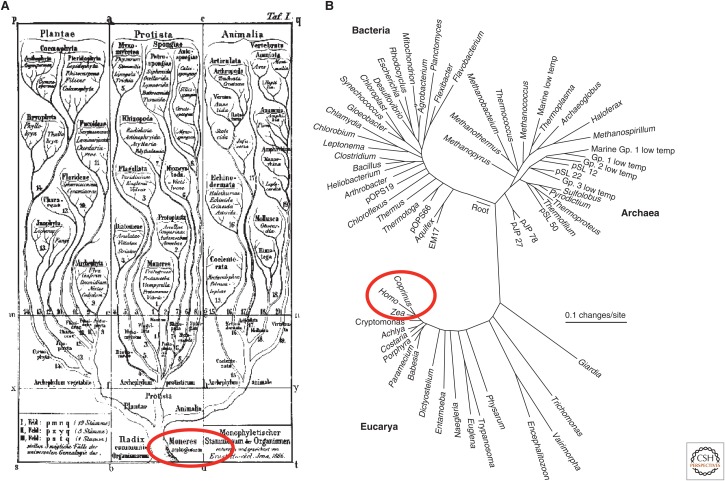
Closely following on from Darwin’s thesis that all life forms on our planet arose from a common ancestor (Darwin 1859), have been biologists’ attempts to classify life naturalistically according to evolution. Initially and understandably, phenotype (morphology, development, etc.) was the primary basis for inferring relationships between organisms resulting in a schema that lumped all microbial life (which had been discovered 200 years earlier through the advent of the microscope) into a single “primitive” kingdom at the base of the tree (Fig 1A) (Haeckel 1866). The discovery of the structure of DNA in the latter half of the 20th century and its role as the heritable blueprint of life led to the proposal that genes are a more objective basis than phenotype for inferring evolutionary (phylogenetic) relationships (Zuckerkandl and Pauling 1965). Carl Woese ran with this idea focusing on comparison of universally conserved components of the protein manufacturing machinery in the cell, the ribosomal RNA (rRNA) genes, which he correctly reasoned would produce an objective tree of life. His first trees and all subsequent efforts turned the phenotype-based tree on its head; instead of microorganisms occupying a lowly corner of the tree, all multicellular life clustered together in a corner of one of three newly described primary lines of descent (Fig. 1B) (Woese and Fox 1977). Small subunit ribosomal RNA- (16S rRNA)-based classification of bacteria and archaea was enthusiastically embraced by microbiologists following Woese’s discoveries, in large part because natural relationships between microbes are virtually undetectable using phenotypic properties (Stanier and Van Niel 1962). Thirty years on, 16S rRNA sequences form the basis of microbial classification; however, vast numbers of discrepancies exist between taxonomy and phylogeny with many currently defined taxa not forming evolutionarily coherent (monophyletic) groups. A conspicuous case in point is the genus Clostridium, which is superficially united by a common morphology and ability to produce endospores, but represents dozens of phylogenetically distinct groups within the phylum Firmicutes (Yutin and Galperin 2013). This greatly impedes our ability to understand the ecology and evolution of ecosystems, such as mammalian guts, where clostridia are important functional populations. The large number of unresolved taxonomic errors is due to a combination of historical artifacts (phenotypic classification) and limitations with rRNA gene trees such as poor phylogenetic resolution, inadequate reference sequences, and sequencing artifacts (notably chimeras, as discussed below). Genome trees inferred that using multiple marker genes offer greater phylogenetic resolution than 16S rRNA and other single-marker gene trees (Ciccarelli et al. 2006; Lang et al. 2013) and are, therefore, a more reliable basis for taxonomic classification. However, publicly available genome sequences are still far from representative of microbial diversity as a whole, as revealed by the development of culture-independent methods.
Figure 1.
Two representations of the tree of life. The first (A) based on phenotypic comparisons resulting in lumping of microorganisms into a single undifferentiated mass at the base of the tree (circled in red), and the second (B) based on genotypic (rRNA genes) comparisons revealing that most diversity (including the Eucarya) is actually microbial with multicellular life forms only emerging relatively recently in evolutionary history (circled in red). (Panel A is reproduced from Haekcel 1866, and is freely available in the public domain and free of known restrictions under copyright law. Panel B is based on Figure 2 in Barns et al. 1996.)
MOST MICROBIAL DIVERSITY IS UNCULTURED BUT HAS RECENTLY BECOME READILY ACCESSIBLE GENOMICALLY
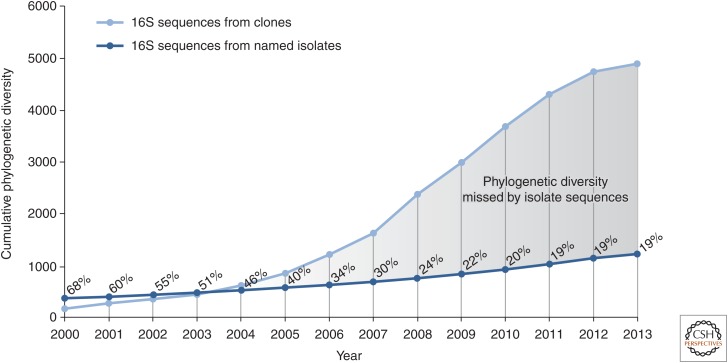
Culture-independent molecular techniques founded on 16S rRNA in the mid-1980s (Olsen et al. 1986) highlighted our ignorance of most of the tree of life by crudely outlining its borders. This was achieved by sequencing 16S rRNA genes from bulk DNAs extracted directly from environmental sources (Pace 1997). This type of microbial community profiling has improved with increased sequencing and computing capacity. The startling conclusion from more than two decades of such culture-independent environmental sequence surveys is that >80% of microbial evolutionary diversity is represented by uncultured microorganisms distributed across upward of 100 major lines of descent within the Bacteria and Archaea (Harris et al. 2013) and that the amount of recognized microbial dark matter is still increasing (Fig. 2).
Figure 2.
Increase in recognized phylogenetic diversity as measured by rRNA sequence novelty (y-axis) over time (x-axis). Recognized diversity has increased by an order of magnitude in the past 10 years, most of which (∼80%) is a result of newly identified uncultured lineages (microbial dark matter). Note that the apparent leveling off of novel phylogenetic diversity discovery in 2013 is likely a consequence of the recent move from full-length 16S rRNA sequencing to partial gene sequencing on next-generation sequencing platforms, which are not included in this estimate (adapted from Rinke et al. 2013).
The task to obtain representative genomic coverage of all recognized microbial diversity, estimated conservatively to represent hundreds of thousands of species (Curtis and Sloan 2005), is daunting. However, two promising culture-independent approaches have emerged to achieve this goal. The first is metagenomics, the application of high-throughput sequencing of DNA extracted directly from environmental samples, and the second is single-cell genomics, the physical separation and amplification of cells before sequencing. Initially, it was unfeasible to extract genomes of individual populations from metagenomic data (a bioinformatic process called binning) because of insufficient sequencing depth and inadequate binning tools. Only in some instances could complete or near-complete genomes be reconstructed from environmental sequences, and these were typically of dominant populations with minimal genomic heterogeneity (Tyson et al. 2004; Garcia Martin et al. 2006; Elkins et al. 2008). In contrast, most populations in early metagenomic studies remained unidentified, being represented by a smattering of anonymous sequence data. Single-cell genomics overcomes this problem by separating environmental samples into their component populations up front, most efficiently via cell sorting, thereby avoiding the need for binning and allowing sequencing resources to be focused on individual genomes (Lasken and McLean 2014). The potential of this approach for articulating microbial dark matter was shown in a recent study describing 200 single-cell genomes representing 29 major uncultured bacterial and archaeal lineages that challenge established boundaries between the three domains of life. These include an archaeal-type purine synthesis in bacteria and complete sigma factors in Archaea similar to those in Bacteria (Rinke et al. 2013). Despite this promising start, we need thousands of microbial dark matter genomes, rather than hundreds, to gain a comprehensive picture of microbial evolution and ecology. Technical challenges associated with single-cell genomics, including the need for whole-genome amplification and the high potential for contamination, currently preclude this level of scale-up. Moreover, the average completeness of a single-cell genome is currently only 35% (Parks et al. 2015) because of large biases in genome coverage introduced during the whole-genome amplification step (Lasken and McLean 2014). Recently, a new and effective binning strategy based on differing relative abundance patterns of populations between related microbial communities has been developed by multiple groups (Albertsen et al. 2013; Alneberg et al. 2013; Sharon and Banfield 2013; Imelfort et al. 2014; Kang et al. 2014; Nielsen et al. 2014). This approach uses differential sequencing coverage of a given population between metagenomic data sets as a proxy of its relative abundance, and leverages the much higher genome coverage afforded by modern sequencers, which produce tens of billions of sequence base pairs per run. Unlike single-cell genomics, differential coverage binning will scale to produce tens of thousands of population genomes in a short time frame. For example, ∼50 Gbp of sequence data (a single lane of HiSeq Illumina data) from more than three related environmental samples will typically produce 50 to 100 high-quality population bins (>80% complete, <10% contaminated) (Imelfort et al. 2014; Parks et al. 2015). Furthermore, the method does not require specialized equipment beyond access to high-throughput sequencing and high-performance computing, and therefore is being rapidly adopted by research groups worldwide. It is entirely feasible that >500,000 population genomes will be generated in the next few years providing the necessary volume of data to adequately cover all of the major lines of descent in the bacterial and archaeal domains. This will form the basis of a comprehensive genome-based classification framework of microbial diversity.
RECONCILING TAXONOMY WITH PHYLOGENY
Although phylogenetic inference is an objective approximation of evolutionary history, microbial systematics—the classification of microorganisms into hierarchical taxonomic groups (nominally domain, phylum, class, order, family, genus, species) based on phylogenetic inference (or other criteria), is a largely subjective human construct to help organize information. Few would argue the natural evolutionary distinction between Bacteria and Archaea so the rank of Domain (Kingdom) is not especially controversial, with the exception of the placement of the Eucarya (Spang et al. 2015). However, the circumscription of all ranks subordinate to Domain is often fiercely debated between taxonomic “lumpers” and “splitters” (Endersby 2009), which unfortunately diminishes the enterprise in the eyes of other scientists as they consider it subjective and arbitrary. Microbial systematics has been divorced from evolutionary theory for much of its history (Doolittle 2015), so the process of reconciling taxonomy with phylogeny will serve to unite the two. This involves two main tasks.
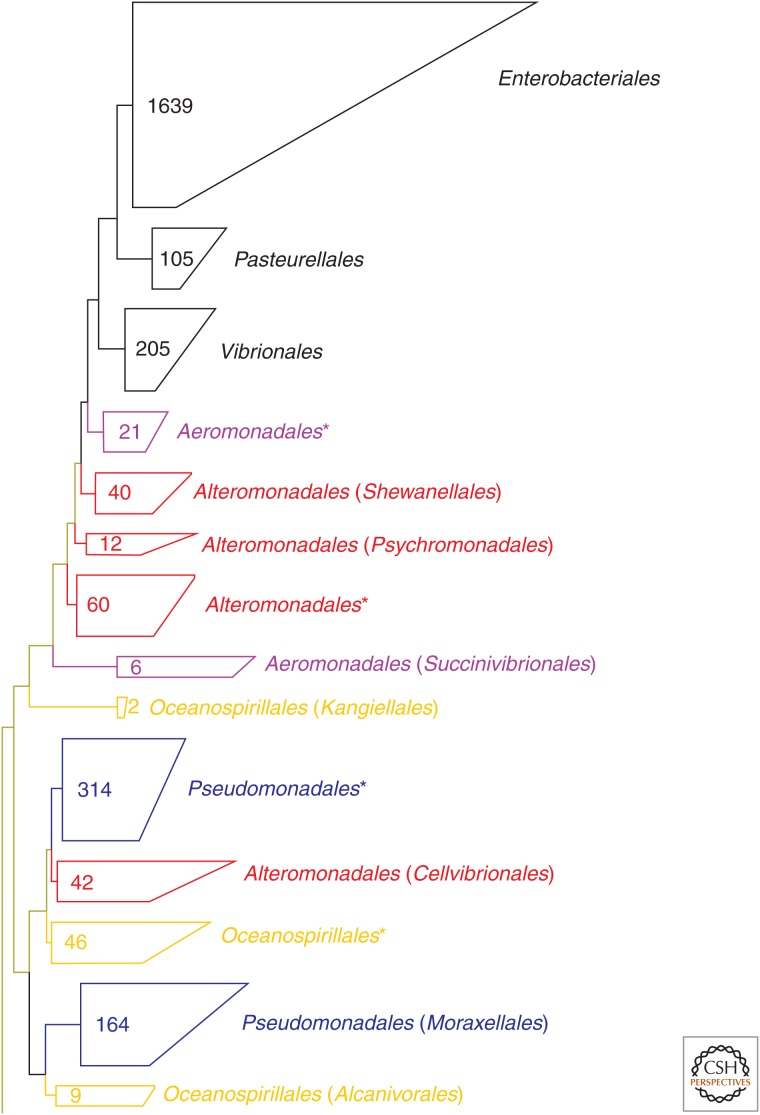
The first is to identify polyphyletic taxa at all ranks (phylum to species) in a phylogenetic tree and to reclassify them according to a standard set of rules. Figure 3 is a genome-based reconstruction of the Gammaproteobacteria highlighting some of the polyphyletic orders in this class. In this tree, the order Alteromonadales comprises four distinct lines of descent. To correct this conflict between phylogeny and taxonomy, the group containing the type genus after which the order was named, Alteromonas, is first identified (starred in tree) and retains the order name. The other groups can then be renamed after the oldest validly described genus in each group (shown in parentheses in Fig. 3), in this case Shewanellales after Shewanella, Psychromonadales after Psychromonas, and Cellvibrionales after Cellvibrio. The three other polyphyletic orders in Figure 3, Aeromonadales, Oceanospirillales, and Pseudomonadales, can be similarly corrected. This same approach can be used for all taxonomic ranks above genus, and if no validly named isolate exists for a given group, which is indeed the case for the majority of branches because of microbial dark matter, groups can be named after environmental 16S rRNA sequences or population genomes, with the caveat that the name should be unique in the taxonomy (McDonald et al. 2012). Using this approach, we found that 85% of 635 named isolates belonging to the class Clostridia required reassignment at one or more ranks to reconcile existing taxonomic classifications with a genome-based phylogeny (Table 1).
Figure 3.
A tree of isolate genomes belonging to the class Gammaproteobacteria inferred from a concatenated alignment of 83 single-copy marker genes. Lineages are collapsed at the order level showing that some are monophyletic (in black), but many others are polyphyletic (colored), the most extreme case in this instance being the Alteromonadales. Renaming of orders to remove polyphyly is shown in parentheses. Numbers inside collapsed groups indicate the number of genomes comprising a given order.
Table 1.
Extent of taxonomic reassignments required to reconcile existing taxonomy with genome-based phylogeny for members of the class Clostridia
| No. ranks requiring reassignment | No. changes/635 | % of total |
|---|---|---|
| 0 | 92 | 14.5 |
| 1 | 123 | 19.4 |
| 2 | 309 | 48.7 |
| 3 | 111 | 17.5 |
Only changes in three ranks are included: order, family, and genus.
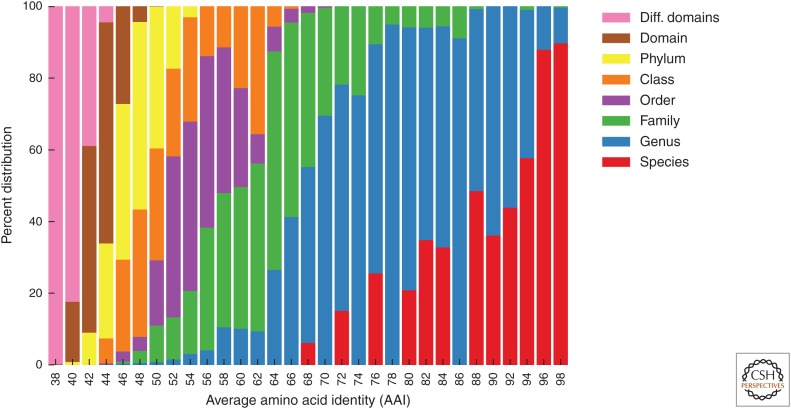
The second task concerns the unevenness of rank assignments according to sequence-based metrics. Konstantinidis and Tiedje proposed the average amino acid identity (AAI) of shared genes between two genomes as a measure of relatedness (Konstantinidis and Tiedje 2005). They plotted taxonomic ranks as a function of AAI for 175 fully sequenced strains revealing a high degree of overlap between different ranks, even nonadjacent ranks. We repeated this analysis with nearly 6000 genomes and the current National Center for Biotechnology Information (NCBI) taxonomy with much the same result—up to five different ranks overlapped at a given AAI and the range of AAIs for each rank often exceeded 20% (Fig. 4). Recently, Yarza et al. (2014) proposed using 16S rRNA gene sequence identity thresholds to rationally circumscribe different taxonomic ranks. However, like AAIs, this does not take into account different evolutionary tempos across the tree of life, which can result in an uneven application of taxonomic ranks as fast-evolving lineages will have lower sequence identities than their slower evolving counterparts for the same divergence times. Ideally, ancestors belonging to the same rank should have been contemporaries in the past. Therefore, defining and normalizing rank distributions using estimated evolutionary divergences is a more naturalistic approach for assigning ranks. Although accurately estimating divergence times has proven challenging (Arbogast et al. 2002), branch lengths in a genome-based phylogeny may provide a suitable approximation if these lengths are appropriately normalized to take into account varying evolutionary rates.
Figure 4.
A histogram of pairwise average amino acid identities (AAI) between 290 archaeal and 5582 bacterial reference genomes colored by rank affiliation showing the uneveness of current taxonomic classifications. Extreme outliers include isolates classified in the same species (red) with only 68% AAI (Bdellovibrio bacteriovorus) and others classified in different genera (green) sharing as high as 94% AAI (e.g., Tannerella and Coprobacter) (adapted from Konstantinidis and Tiedje 2005).
TRANSITIONING FROM A 16S rRNA- TO GENOME-BASED TAXONOMY
To date, 16S rRNA has been the most widely used gene for inferring evolutionary relationships between microorganisms and serves as the primary basis for microbial taxonomy. Its high degree of sequence conservation (Woese 1987) has the dual benefits of providing a domain-level overview of microbial diversity, and enables culture-independent environmental surveys using near universal polymerase chain reaction (PCR) priming sites. The public repository of 16S rRNA gene sequences number in the millions and it is, therefore, also the most comprehensively sequenced marker gene available to us. However, this same sequence conservation also leads to chimera formation in environmental surveys. Chimeras are PCR-induced artifacts whereby an incompletely synthesized 16S rRNA amplicon acts as a primer on a different template producing a hybrid molecule. Such mispriming only requires short stretches of sequence identity between the 3′-end of the incomplete template and homologous region of the foreign template, which abounds in 16S rRNA because of its high degree of conservation. Moreover, very similar chimeras can be formed in independent studies of similar habitats in which parent sequences are present in approximately similar ratios (e.g., Bacteroides–Clostridium chimeras produced in human gut surveys) (Haas et al. 2011). Chimeras have been recognized as a problem associated with PCR-based surveys since the 1990s and alarms have been raised about the growing number of undetected chimeras in the public databases (Hugenholtz and Huber 2003; Ashelford et al. 2005). Environmental sequences now dominate the public databases in both number and phylogenetic diversity coverage, unlike the early 2000s when they still represented a minority of available data (Fig. 1). It is very likely that the problem of undetected chimeras has increased accordingly as suggested by the poor overlap between different chimera-detection tools (Haas et al. 2011). All such tools detect chimeric sequences by comparison to a reference set of 16S rRNA sequences. If the reference set is compromised by undetected chimeras, detection of new chimeric sequences is also compromised. In short, chimeras are a recognized but underestimated problem in 16S rRNA data sets that artificially inflate diversity estimates and introduce noise into phylogenetic trees, ultimately compromising taxonomic classifications based on these trees.
Concatenated protein marker gene trees derived from isolate and population genomes are much less susceptible to chimeric artifacts and provided completeness and contamination checks used for the latter (Parks et al. 2015). Combined with the higher phylogenetic resolution afforded by concatenated markers, such trees are the logical choice for a phylogenetically reconciled taxonomy. However, the 16S rRNA database is currently two orders of magnitude larger than the genome database and significant efforts have been invested into curating 16S rRNA trees for taxonomic classification (Kim et al. 2012; McDonald et al. 2012; Quast et al. 2012; Cole et al. 2013). Therefore, and despite ambiguities arising from chimeric sequences, efforts should be made to incorporate 16S rRNA-based classifications into genome-based taxonomy to provide taxonomic continuity in the literature. For microbial isolates, the process of connecting 16S rRNA to genome sequences to allow transfer of taxonomic information should be straightforward. However, for many isolates, 16S rRNA genes were sequenced before their genomes, introducing the risk of connecting different taxa between trees. Sequencing of type material is very important in this context as it provides unambiguous signposts in phylogenetic trees, not only for transfer of taxonomic information, but also to enable correction of the numerous polyphyly errors that presently exist in microbial taxonomy (Fig. 3; Table 1) (Kyrpides et al. 2014). For most microbial diversity, however, we are reliant on single-cell or population genomes to connect 16S rRNA-based classifications to their corresponding locations in genome trees. Unfortunately, rRNA genes are often difficult to recover in genomes derived from metagenomic data. This is because of the high conservation of these genes, which often results in chimeric assembly, and the fact that the rRNA operon is typically the largest repeat in a microbial genome if present in greater than one copy. This confounds differential coverage binning because the rRNA genes do not bin with the rest of the genome as a result of having a higher coverage if present in the assembly as a collapsed repeat. Development of new methods to obtain full-length non-chimeric 16S rRNA genes for population genomes should be a priority as this will enable a smooth transition to a genome-based microbial taxonomy. Moreover, 16S rRNA is likely to remain a valuable tool in the microbiologist’s toolbox for the foreseeable future, for example, 16S rRNA-targeted fluorescence in situ hybridization (Amann et al. 2001), so it cannot be easily ignored as we transition to a genome-based taxonomy.
APPLICATIONS FOR A REPRESENTATIVE GENOME TREE AND GENOME-BASED TAXONOMY
One important and largely open question is the extent to which lateral gene transfer (LGT) plays a role in microbial evolution and ecology. This topic was first broached in a holistic manner with the availability of the first microbial genome sequences. Phylogenetic analysis of numerous gene families revealed a high level of inferred discordance with the 16S rRNA-based tree over evolutionary timescales, raising the question as to whether the universal tree of life would be better represented as a network because of LGT blurring organismal boundaries (Doolittle 1999). Despite the inference that LGT is a relatively common phenomenon, a comparison of 56 genomes selected to maximize 16S rRNA-defined phylogenetic diversity with randomly selected genome data sets of the same size showed that tree-based selection resulted in higher rates of discovery of novel gene families, gene fusions, and operon arrangements (Wu et al. 2009). These findings argue against LGT overriding vertical inheritance. However, few would argue against LGT being a potent evolutionary force, the most public face of which is transfer of antibiotic resistance between bacteria (Arber 2014). An analysis of microbial isolate genomes to identify recently laterally transferred genes (>99% nucleotide identity) in humans and the environment points to ecology being a more important driver of LGT than phylogeny, particularly in humans (Smillie et al. 2011). This analysis only considered isolate genomes, however, which do not adequately represent the ecosystems from which they were obtained. Population genomics provides the means to obtain a representative sampling of the component members of a given ecosystem, and a genome-based taxonomy then becomes the essential framework for determining the frequency and mode of LGT between organisms of varying evolutionary relatedness (Beiko et al. 2005).
Current concepts on many topics have likely been oversimplified by limited and highly skewed sampling of microbial diversity, and stand to be overhauled by microbial dark matter genome sequencing. Recent examples include the discovery of nonphotosynthetic basal Cyanobacteria (Di Rienzi et al. 2013; Soo et al. 2014) and a large radiation of candidate bacterial phyla with unusual ribosome structures and biogenesis mechanisms that may actually be more representative of the “average” bacterium than our current preconceptions (Rinke et al. 2013; Brown et al. 2015). Similarly, categorization of the bacterial cell envelope as either single (monoderm) or double (diderm) layer structures, based on studies of mostly Firmicutes and Proteobacteria, will become more sophisticated with greater representation of candidate phyla (Sutcliffe 2010). In summary, our knowledge of the microbial world is set to expand dramatically in the coming years as microbial dark matter is rapidly illuminated through genome sequencing. Coupling these advances to a systematized genome-based classification will serve to organize this valuable resource into a coherent framework that will greatly facilitate analysis and communication.
Footnotes
Editor: Howard Ochman
Additional Perspectives on Microbial Evolution available at www.cshperspectives.org
REFERENCES
- Albertsen M, Hugenholtz P, Skarshewski A, Nielsen KL, Tyson GW, Nielsen PH. 2013. Genome sequences of rare, uncultured bacteria obtained by differential coverage binning of multiple metagenomes. Nat Biotechnol 31: 533–538. [DOI] [PubMed] [Google Scholar]
- Alneberg J, Bjarnason BS, de Bruijn I, Schirmer M, Quick J, Ijaz UZ, Lahti L, Loman NJ, Andersson AF, Quince C. 2013. CONCOCT: Clustering cONtigs on COverage and ComposiTion. arXiv:1312.4038. [DOI] [PubMed] [Google Scholar]
- Amann R, Fuchs BM, Behrens S. 2001. The identification of microorganisms by fluorescence in situ hybridisation. Curr Opin Biotechnol 12: 231–236. [DOI] [PubMed] [Google Scholar]
- Arber W. 2014. Horizontal gene transfer among bacteria and its role in biological evolution. Life (Basel) 4: 217–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbogast BS, Edwards SV, Wakeley J, Beerli P, Slowinski JB. 2002. Estimating divergence times from molecular data on phylogenetic and population genetic timescales. Annu Rev Ecol Syst 33: 707–740. [Google Scholar]
- Ashelford KE, Chuzhanova NA, Fry JC, Jones AJ, Weightman AJ. 2005. At least 1 in 20 16S rRNA sequence records currently held in public repositories is estimated to contain substantial anomalies. Appl Environ Microbiol 71: 7724–7736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beiko RG, Harlow TJ, Ragan MA. 2005. Highways of gene sharing in prokaryotes. Proc Natl Acad Sci 102: 14332–14337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CT, Hug LA, Thomas BC, Sharon I, Castelle CJ, Singh A, Wilkins MJ, Wrighton KC, Williams KH, Banfield JF. 2015. Unusual biology across a group comprising more than 15% of domain bacteria. Nature 523: 208–211. [DOI] [PubMed] [Google Scholar]
- Ciccarelli FD, Doerks T, von Mering C, Creevey CJ, Snel B, Bork P. 2006. Toward automatic reconstruction of a highly resolved tree of life. Science 311: 1283–1287. [DOI] [PubMed] [Google Scholar]
- Cole JR, Wang Q, Fish JA, Chai B, McGarrell DM, Sun Y, Brown CT, Porras-Alfaro A, Kuske CR, Tiedje JM. 2013. Ribosomal Database Project: Data and tools for high throughput rRNA analysis. Nucleic Acids Res 42: D633–D642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis TP, Sloan WT. 2005. Microbiology. Exploring microbial diversity—A vast below. Science 309: 1331–1333. [DOI] [PubMed] [Google Scholar]
- Darwin CR. 1859. On the origin of species by means of natural selection, or the preservation of favoured races in the struggle for life, 1st ed. John Murray, London. [PMC free article] [PubMed] [Google Scholar]
- Di Rienzi SC, Sharon I, Wrighton KC, Koren O, Hug LA, Thomas BC, Goodrich JK, Bell JT, Spector TD, Banfield JF, et al. 2013. The human gut and groundwater harbor non-photosynthetic bacteria belonging to a new candidate phylum sibling to cyanobacteria. eLife 2: e01102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doolittle WF. 1999. Phylogenetic classification and the universal tree. Science 284: 2124–2129. [DOI] [PubMed] [Google Scholar]
- Doolittle WF. 2015. Rethinking the tree of life. Microbe Mag, Oct, www.microbemagazine.org. [Google Scholar]
- Elkins JG, Podar M, Graham DE, Makarova KS, Wolf Y, Randau L, Hedlund BP, Brochier-Armanet C, Kunin V, Anderson I, et al. 2008. A korarchaeal genome reveals insights into the evolution of the Archaea. Proc Natl Acad Sci 105: 8102–8107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endersby J. 2009. Lumpers and splitters: Darwin, Hooker, and the search for order. Science 326: 1496–1499. [DOI] [PubMed] [Google Scholar]
- García Martín H, Ivanova N, Kunin V, Warnecke F, Barry KW, McHardy AC, Yeates C, He S, Salamov AA, Szeto E, et al. 2006. Metagenomic analysis of two enhanced biological phosphorus removal (EBPR) sludge communities. Nat Biotechnol 24: 1263–1269. [DOI] [PubMed] [Google Scholar]
- Haas BJ, Gevers D, Earl AM, Feldgarden M, Ward DV, Giannoukos G, Ciulla D, Tabbaa D, Highlander SK, Sodergren E, et al. 2011. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res 21: 494–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeckel H. 1866. Generelle morphologie der organismen: Allgemeine grundzüge der organischen Formen-Wissenschaft, mechanisch begründet durch die von Charles Darwin reformirte descendenztheorie, von Ernst Haeckel. Reimer, Berlin. [Google Scholar]
- Harris JK, Caporaso JG, Walker JJ, Spear JR, Gold NJ, Robertson CE, Hugenholtz P, Goodrich J, McDonald D, Knights D, et al. 2013. Phylogenetic stratigraphy in the Guerrero Negro hypersaline microbial mat. ISME J 7: 50–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugenholtz P, Huber T. 2003. Chimeric 16S rDNA sequences of diverse origin are accumulating in the public databases. Int J Syst Evol Microbiol 53: 289–293. [DOI] [PubMed] [Google Scholar]
- Imelfort M, Parks D, Woodcroft BJ, Dennis P, Hugenholtz P, Tyson GW. 2014. GroopM: An automated tool for the recovery of population genomes from related metagenomes. PeerJ 2: e603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang DD, Froula J, Egan R, Wang Z. 2014. MetaBAT: Metagenome binning based on abundance and tetranucleotide frequency. Presented at the Ninth Annual DOE Joint Genome Institute User Meeting Walnut Creek, CA. [Google Scholar]
- Kim OS, Cho YJ, Lee K, Yoon SH, Kim M, Na H, Park SC, Jeon YS, Lee JH, Yi H, et al. 2012. Introducing EzTaxon-e: A prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol 62: 716–721. [DOI] [PubMed] [Google Scholar]
- Konstantinidis KT, Tiedje JM. 2005. Towards a genome-based taxonomy for prokaryotes. J Bacteriol 187: 6258–6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyrpides NC, Hugenholtz P, Eisen JA, Woyke T, Göker M, Parker CT, Amann R, Beck BJ, Chain PS, Chun J, et al. 2014. Genomic encyclopedia of bacteria and archaea: Sequencing a myriad of type strains. PLoS Biol 12: e1001920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang JM, Darling AE, Eisen JA. 2013. Phylogeny of bacterial and archaeal genomes using conserved genes: Supertrees and supermatrices. PLoS ONE 8: e62510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasken RS, McLean JS. 2014. Recent advances in genomic DNA sequencing of microbial species from single cells. Nat Rev Genet 15: 577–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, Andersen GL, Knight R, Hugenholtz P. 2012. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J 6: 610–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen HB, Almeida M, Juncker AS, Rasmussen S, Li J, Sunagawa S, Plichta DR, Gautier L, Pedersen AG, Le Chatelier E, et al. 2014. Identification and assembly of genomes and genetic elements in complex metagenomic samples without using reference genomes. Nat Biotechnol 32: 822–828. [DOI] [PubMed] [Google Scholar]
- Olsen GJ, Lane DJ, Giovannoni SJ, Pace NR, Stahl DA. 1986. Microbial ecology and evolution: A ribosomal RNA approach. Annu Rev Microbiol 40: 337–365. [DOI] [PubMed] [Google Scholar]
- Pace NR. 1997. A molecular view of microbial diversity and the biosphere. Science 276: 734–740. [DOI] [PubMed] [Google Scholar]
- Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. 2015. CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res 25: 1043–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO. 2012. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res 41: D590–D596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinke C, Schwientek P, Sczyrba A, Ivanova NN, Anderson IJ, Cheng JF, Darling A, Malfatti S, Swan BK, et al. 2013. Insights into the phylogeny and coding potential of microbial dark matter. Nature 499: 431–437. [DOI] [PubMed] [Google Scholar]
- Sharon I, Banfield JF. 2013. Microbiology. Genomes from metagenomics. Science 342: 1057–1058. [DOI] [PubMed] [Google Scholar]
- Smillie CS, Smith MB, Friedman J, Cordero OX, David LA, Alm EJ. 2011. Ecology drives a global network of gene exchange connecting the human microbiome. Nature 480: 241–244. [DOI] [PubMed] [Google Scholar]
- Soo RM, Skennerton CT, Sekiguchi Y, Imelfort M, Paech SJ, Dennis PG, Sheen JA, Parks DH, Tyson GW, Hugenholtz P. 2014. An expanded genomic representation of the phylum Cyanobacteria. Genome Biol Evol 6: 1031–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spang A, Saw JH, Jørgensen SL, Zaremba-Niedzwiedzka K, Martijn J, Lind AE, van Eijk R, Schleper C, Guy L, Ettema TJ. 2015. Complex archaea that bridge the gap between prokaryotes and eukaryotes. Nature 521: 173–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanier RY, Van Niel CB. 1962. The concept of a bacterium. Arch Mikrobiol 42: 17–35. [DOI] [PubMed] [Google Scholar]
- Sutcliffe IC. 2010. A phylum level perspective on bacterial cell envelope architecture. Trends Microbiol 18: 464–470. [DOI] [PubMed] [Google Scholar]
- Tyson GW, Chapman J, Hugenholtz P, Allen EE, Ram RJ, Richardson PM, Solovyev VV, Rubin EM, Rokhsar DS, Banfield JF. 2004. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature 428: 37–43. [DOI] [PubMed] [Google Scholar]
- Woese CR. 1987. Bacterial evolution. Microbiol Rev 51: 221–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese CR, Fox GE. 1977. Phylogenetic structure of the prokaryotic domain: The primary kingdoms. Proc Natl Acad Sci 71: 5088–5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D, Hugenholtz P, Mavromatis K, Pukall R, Dalin E, Ivanova NN, Kunin V, Goodwin L, Wu M, Tindall BJ, et al. 2009. A phylogeny-driven genomic encyclopaedia of Bacteria and Archaea. Nature 462: 1056–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarza P, Yilmaz P, Pruesse E, Glöckner FO, Ludwig W, Schleifer KH, Whitman WB, Euzéby J, Amann R, Rosselló-Móra R. 2014. Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat Rev Microbiol 12: 635–645. [DOI] [PubMed] [Google Scholar]
- Yutin N, Galperin MY. 2013. A genomic update on clostridial phylogeny: Gram-negative spore formers and other misplaced clostridia. Environ Microbiol 15: 2631–2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuckerkandl E, Pauling L. 1965. Molecules as documents of evolutionary history. J Theor Biol 8: 357–366. [DOI] [PubMed] [Google Scholar]