Phenotypic diversity between laboratory-domesticated bacterial strains is a common problem and often results in the failed reproduction of published data. However, researchers rarely compare such strains to elucidate the underlying mutation(s). In this study, we tested one of the best-studied V. cholerae isolates, O1 El Tor strain C6706 (a patient isolate from Peru), with respect to two main phenotypes: natural competence for transformation and type VI secretion. We recently demonstrated that the two phenotypes are coregulated and specifically induced upon the growth of pandemic V. cholerae O1 El Tor strains on chitinous surfaces. We provide evidence that of seven C6706 strains collected from different laboratories, four were impaired in the tested phenotypes due to a mutation in a QS gene. Collectively, our data indicate that the circulation of such a mutated wild-type strain of C6706 might have had important consequences for QS-related data.
KEYWORDS: Vibrio cholerae, luxO mutation, natural competence for transformation, quorum sensing, type VI secretion system
ABSTRACT
Vibrio cholerae, the causative agent of cholera, is a model organism for studying virulence regulation, biofilm formation, horizontal gene transfer, and the cell-to-cell communication known as quorum sensing (QS). As in any research field, discrepancies between data from diverse laboratories are sometimes observed for V. cholerae. Such discrepancies are often caused by the use of diverse patient or environmental isolates. In this study, we investigated the inability of a few laboratories to reproduce high levels of natural transformation, a mode of horizontal gene transfer that is specifically induced on chitinous surfaces. This irreproducibility was mostly related to one specific isolate of V. cholerae: the O1 El Tor C6706 strain. C6706 was previously described as QS proficient, an important prerequisite for the induction of natural competence for transformation. To elucidate the underlying problem, we collected seven isolates of the same C6706 strain from different research laboratories in North America and Europe and compared their phenotypes. Importantly, we observed a split response with respect to QS-related gene expression, including chitin-induced natural competence and type VI secretion (T6S). While approximately half of the strains behaved as reported for several other O1 El Tor pandemic isolates that are commonly studied in the laboratory, the other half were significantly impaired in QS-related expression patterns. This impairment was caused by a mutation in a QS-related gene (luxO). We conclude that the circulation of such QS-impaired wild-type strains is responsible for masking several important phenotypes of V. cholerae, including natural competence for transformation and T6S.
IMPORTANCE Phenotypic diversity between laboratory-domesticated bacterial strains is a common problem and often results in the failed reproduction of published data. However, researchers rarely compare such strains to elucidate the underlying mutation(s). In this study, we tested one of the best-studied V. cholerae isolates, O1 El Tor strain C6706 (a patient isolate from Peru), with respect to two main phenotypes: natural competence for transformation and type VI secretion. We recently demonstrated that the two phenotypes are coregulated and specifically induced upon the growth of pandemic V. cholerae O1 El Tor strains on chitinous surfaces. We provide evidence that of seven C6706 strains collected from different laboratories, four were impaired in the tested phenotypes due to a mutation in a QS gene. Collectively, our data indicate that the circulation of such a mutated wild-type strain of C6706 might have had important consequences for QS-related data.
INTRODUCTION
Although Vibrio cholerae, the causative agent of cholera, has been studied for more than a century, we still lack important information needed to fully understand its environmental lifestyle, its transmission to humans, and its full pathogenic potential. In the context of pathogenesis, a plethora of studies have provided important information about its major virulence factors (e.g., the cholera toxin [Ctx] and the toxin-coregulated pilus [TCP]) (1–4). However, the identification of additional virulence factors to explain, for instance, the mild diarrhea caused by ctx-negative V. cholerae strains, as elucidated in an infant rabbit model of cholera, is still important (5). One such putative virulence factor is the recently discovered type VI secretion system (T6SS) of V. cholerae (6). The T6SS is a molecular spear used to transport toxic effectors to other Gram-negative bacteria or eukaryotes in a contact-dependent manner (7). The result of this intoxication is the killing of the adjacent cell if it does not exert immunity against the toxic effectors, as is the case for the attacker’s siblings (i.e., a kin-discrimination mechanism).
The T6SS has primarily been studied in two nonpandemic isolates of V. cholerae (strain V52, an isolate from Sudan, and strain 2740-80, a nontoxigenic isolate from Florida) that harbor a constitutively active T6SS. The cues leading to the production of this system in the pandemic O1 El Tor strains, however, remained largely unknown. In this context, we recently showed that the T6SS of several pandemic V. cholerae O1 El Tor isolates is induced upon growth on chitin (8), which is one of the primary niches of the pathogen in its natural aquatic habitat (9). In particular, we demonstrated that the T6SS is part of the chitin-induced natural competence regulon and is therefore coregulated with the DNA-uptake machinery of V. cholerae (8, 10).
Natural competence for transformation is a widespread mode of horizontal gene transfer that is used by many prokaryotes to incorporate new genetic material into their own genomes (11, 12). Such genetic material is acquired through the uptake of external DNA via competence-induced DNA-uptake machineries (10). The sophisticated regulatory network that drives natural competence in V. cholerae has been studied for more than a decade (13), first by us and more recently also by others (reviewed in reference 14). Briefly, upon growth on chitin, the bacterium produces the main regulator of transformation, TfoX, which subsequently leads to the production of the type IV pilus part of the DNA-uptake machinery (15) (including the major pilin subunit PilA; Fig. 1). However, TfoX alone is not sufficient to allow DNA uptake to occur, as the induction of the second part of the DNA-uptake machinery (e.g., the protein ComEA, which pulls the DNA into the periplasm [16, 17], and the inner membrane transporter ComEC [15]) requires additional input from the quorum-sensing (QS) circuitry (18) (Fig. 1). This input occurs via the master regulator of QS, HapR, which itself is produced only at a high cell density (HCD) (for a review, see reference 19). Hence, HapR acts as a positive coactivator of chitin-induced natural competence. Additionally, HapR also represses the gene that encodes a nuclease (dns) (Fig. 1), which, if not repressed, has a major impact on natural transformation through the degradation of external and periplasmic DNA (16, 20, 21). Notably, the two input signals (e.g., HCD signaled through HapR and chitin signaled through TfoX) merge in the production of the QS- and TfoX-dependent regulator QstR (22), which is ultimately required for the production of the pilus-unrelated part of the DNA-uptake machinery (15–17) and for the induction of the T6SS (8) (Fig. 1).
FIG 1 .
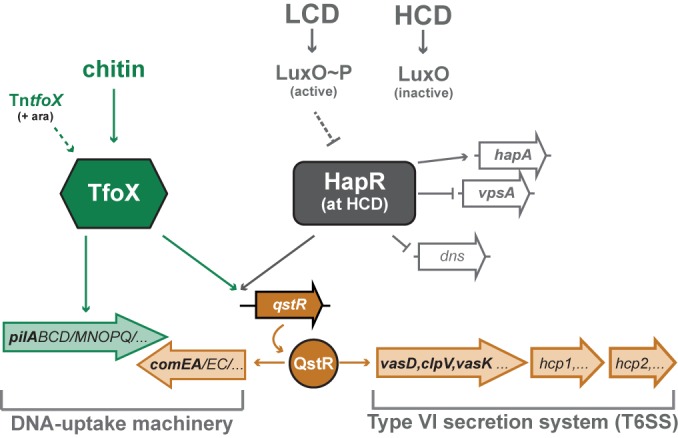
TfoX- and quorum-sensing (QS)-dependent regulation of the DNA-uptake machinery and the type VI secretion system (T6SS) in V. cholerae. The scheme shows the TfoX- and QS-dependent regulation of the competence regulon in V. cholerae, which includes genes encoding the DNA-uptake machinery and the T6SS. The activation of most genes requires dual input from chitin and a high cell density (HCD; compared with low cell density, LCD), which results in the production of the transformation regulator TfoX and the main regulator of QS, HapR, respectively. The signals of both of these proteins merge in the expression of qstR, which encodes the QS- and TfoX-dependent transcription factor QstR. TfoX, HapR, and QstR are required for the production of the essential parts of the DNA-uptake machinery and the T6SS (shown by orange arrows), whereas the type IV pilus part of the DNA-uptake machinery relies solely on the activation by TfoX (green arrow). QS-impaired C6706 mutant strains possess reduced HapR levels and, accordingly, reduced expression of the QstR-regulated genes. Natural transformation and T6SS-mediated interbacterial killing are therefore vastly impaired. The genes whose expression levels were measured in this study are in bold. LuxO~P, phosphorylated LuxO.
Chitin-induced natural competence for transformation is conserved among V. cholerae strains as well as noncholera Vibrio species (such as V. vulnificus [23], V. fischeri [24], and V. parahaemolyticus [25]). However, despite this conservation and several reports that used our previously published protocol (26–28) or derivatives of it to genetically modify Vibrio strains, we frequently obtain requests from researchers who are unable to use chitin-induced natural transformation as a tool (especially for strain C6706; see below). The nontransformability of QS-defective strains, such as the first sequenced strain of V. cholerae N16961 (29), which contains an authentic frameshift mutation within hapR, was reported early on (13). The primary cause for the lack of natural transformation in this and other QS-defective strains is the absence of dns repression (Fig. 1), which results in constitutively high nuclease activity (20). Consistent with these data is a recent report that identified another transformation-inhibitory nuclease in a horizontally acquired integrative and conjugative element (ICE), which rendered such ICE-carrying strains similarly nontransformable (30). However, despite the fact that the well-studied V. cholerae C6706 strain (31), an O1 El Tor patient isolate from Peru, does not contain such an ICE and that this strain has been described as QS proficient and naturally transformable (13), several researchers have reported to us its nontransformability. Here, we followed up on this nontransformability by testing seven C6706 isolates obtained from different laboratories located in North America and Europe. We show that approximately half of these wild-type (WT) strains contain the same compromising mutation within luxO, resulting in impaired QS behavior and, consequently, in low natural transformability and T6SS activity.
RESULTS
The majority of C6706 strains are severely impaired in their natural transformability.
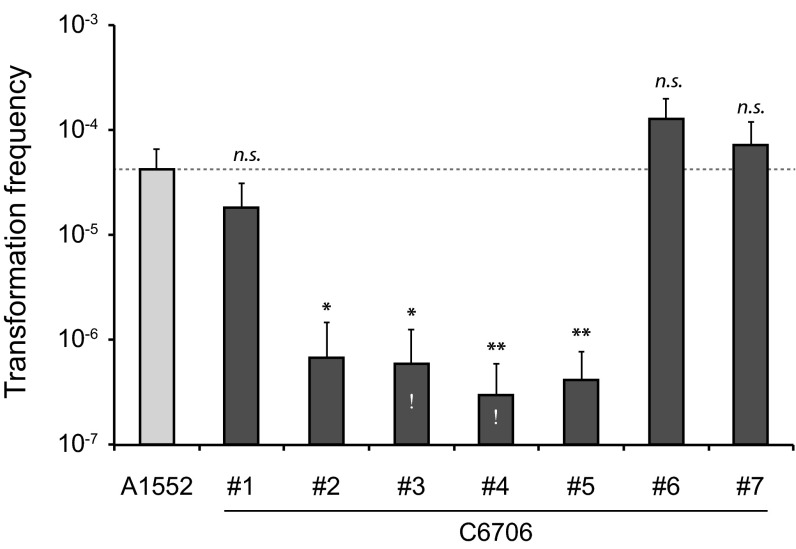
In 2005, it was shown for the first time that the human pathogen V. cholerae could enter a state of natural competence and that this phenotype depends on the presence of chitin (13). That study and follow-up studies showed that many patient isolates of V. cholerae, as well as environmental samples, are naturally transformable in a chitin-dependent manner (8, 13, 32, 33). However, frequent concerns exist in the field with respect to the transformability of O1 El Tor pandemic strain C6706 (personal communications from several researchers to M.B.). We therefore asked seven principal investigators working on diverse aspects of V. cholerae to share their C6706 strains with us. First, we tested these seven samples in a well-established chitin-dependent transformation assay (26) and compared the transformation frequencies to those of our main laboratory strain, the QS- and competence-proficient V. cholerae O1 El Tor A1552 strain. As presented in Fig. 2, our data confirmed that four of these seven wild-type C6706 strains were transformable only sparsely compared with the remaining three C6706 samples and the A1552 control strain. This bipartite response indicates that the nontransformability of such C6706 strains was not caused by an improper following of published protocols but rather by genetic differences between circulating C6706 strains.
FIG 2 .
The seven samples of strain C6706 are transformation variable. The seven representatives of strain C6706 (and strain A1552 as a control) were grown on chitin flakes and scored for natural transformability. The data represent the average transformation frequencies of at least three biological replicates (±SD), and the dashed line shows the value for the A1552 control strain. If no transformants were recovered in a subset of the independent experiments, the detection limit value was used for calculations (indicated by the white exclamation mark). Statistically significant differences between the results from the different C6706 strains and the A1552 control strain were determined by Student’s t test (*, P < 0.05; **, P < 0.01; n.s., not significant).
The majority of C6706 strains produce lowered hapR transcript levels and changed levels of expression of HapR-regulated genes.
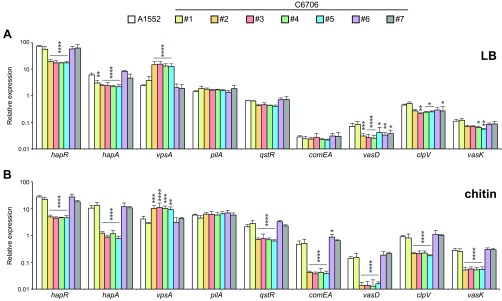
To elucidate whether the impairment of natural transformation was caused by a lack of chitin induction or by a problem in the QS circuit, we tested the seven C6706 strains for the expression of several QS-related and QS-unrelated genes, first in the absence of chitin (at HCD). In particular, we first monitored the transcript levels of hapR by quantitative reverse transcription-PCR (qRT-PCR) because the gene encodes an important coregulator of natural transformation and T6SS (Fig. 1). Interestingly, the hapR transcript levels were reduced in the same C6706 isolates (isolates 2 to 5) that also showed low transformability (Fig. 3A). These low hapR transcript levels were mirrored in lowered expression of hapA (for which HapR acts as an activator [34]) (Fig. 1) and in higher expression of the Vibrio polysaccharide synthesis gene vpsA (for which HapR acts as a repressor [35, 36]) (Fig. 1) than in the three highly transformable C6706 samples (sample 1, sample 6, and sample 7) and the A1552 control strain (Fig. 3A). The expression levels of competence-related genes did not differ between the samples under such chitin-independent HCD conditions (Fig. 3A), consistent with the fact that the competence regulon is not induced in LB medium in pandemic O1 El Tor strains.
FIG 3 .
Two different patterns exist for the expression of QS-responsive genes in the seven C6706 samples. Data represent the relative expression levels of hapR and selected HapR-regulated genes (see Fig. 1 for details) as measured by qRT-PCR. Different V. cholerae C6706 strains (strain 1 to strain 7) were cultured in liquid LB medium to an HCD (A) or were statically grown on chitinous surfaces (B). Strain A1552 served as the positive control. The data represent the means (±SD) of the results of three independent biological experiments. Statistical differences between the A1552 control strain and the indicated C6706 strains (strain 1 to strain 7) were determined using two-way ANOVA. Only significantly different values are indicated: *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
When the same strains underwent a chitin-induced expression analysis, a similar QS-dependent pattern became obvious. As highlighted in Fig. 3B, the same differences in the hapR transcript levels were observed upon growth on chitinous surfaces, as described for LB-grown bacteria (Fig. 3A). The low levels of the HapR regulator were again reflected in the changed levels of expression of hapA and vpsA (Fig. 3B). Notably, while expression of the pilA QS-independent competence gene (Fig. 1) was induced in all of the tested strains upon growth on chitin (Fig. 3), the expression of the chitin- (TfoX-) and QS-coregulated competence genes was severely reduced in the same subset of QS-impaired C6706 strains, which were almost nontransformable (Fig. 2 and 3B). The affected genes were qstR, which itself requires induction by TfoX and HapR (22) (Fig. 1), and all of the tested QstR-dependent genes that encode either parts of the DNA-uptake machinery (e.g., comEA) or components of the T6SS (e.g., vasD, clpV, and vasK) (Fig. 3B). Notably, this striking difference in competence gene expression was observed even though we have previously demonstrated that chitin-attached bacteria show heterogeneity with respect to competence expression (18). We therefore conclude that the low HapR level produced in 4 of 7 of the C6706 isolates is not sufficient to properly induce the competence regulon, which includes the T6SS.
QS-impaired wild-type C6706 strains show reduced TfoX-induced interbacterial killing.
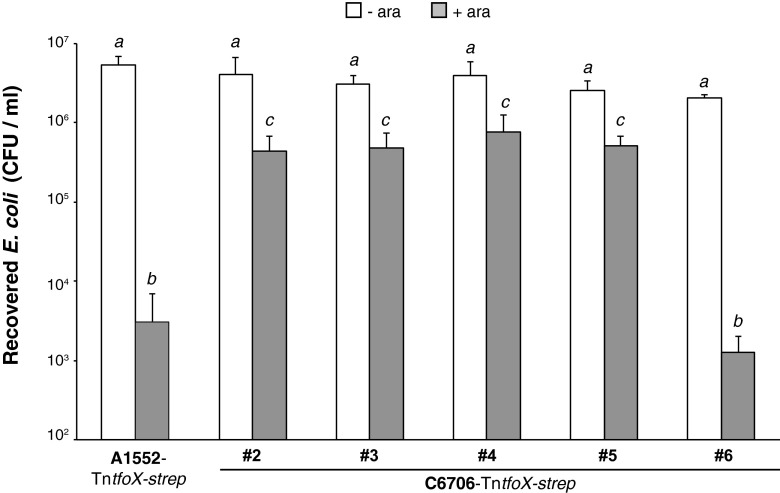
Because we found decreased HapR levels in approximately half of the seven C6706 strains compared with the rest of the C6706 strains and the A1552 control, we considered whether this decrease was also reflected in a reduced ability to kill other Gram-negative bacteria, such as Escherichia coli, by means of T6SS-mediated attack. To answer this question, we introduced a transposon carrying an arabinose-inducible copy of tfoX (TntfoX-strep [18, 37]) into the chromosome of a representative set of C6706 strains, which allowed us to induce TfoX through the provision of the inducer (e.g., in a chitin-independent manner, as the latter condition does not support the growth of E. coli). We mixed these V. cholerae strains with an arabinose-non-degrading E. coli strain (37) and scored the recoverability of the E. coli prey after 4 h of coincubation. As shown in Fig. 4, QS-impaired C6706 strains 2 to 5 reduced the E. coli numbers only slightly, whereas the QS-proficient C6706 sample (sample 6) significantly reduced the prey population. This phenotype therefore reflects the expression analysis.
FIG 4 .
TfoX-induced interbacterial killing is vastly reduced in QS-impaired C6706 isolates. Data represent results of a killing assay of the indicated V. cholerae predator strains performed with E. coli as prey. Strain A1552 served as a positive control. All of the strains contained the TntfoX-strep transposon, which permits the induction of tfoX through the addition of arabinose (18, 37). The surviving prey were quantified as CFU per milliliter (y axis) after coculturing with the indicated V. cholerae strains on plain (-ara) or arabinose-containing (+ara) LB agar plates was performed. The averages of results from three biological replicates (±SD) are shown. Statistical differences were determined using two-way ANOVA. Data labeled with the same character (a, b, or c) are not significantly different. Differences between data labeled with different characters are significant (P < 0.0001 for comparisons between data labeled with b and data labeled with c).
QS-impaired wild-type C6706 strains possess a mutation in the QS gene luxO.
To elucidate the cause of the QS impairment and the low hapR transcript (and HapR protein) levels in a subset of the C6706 samples but not in the second half or in other O1 El Tor isolates, we sequenced two important QS genes: hapR itself and the gene encoding LuxO, the upstream-acting regulator of the hapR transcript (19). The rationale behind performing the latter was that at a low cell density (LCD), LuxO is in its phosphorylated and therefore active form. Phosphorylated LuxO (LuxO~P) indirectly (via the regulation of small RNAs) leads to the degradation of hapR mRNA (19) (Fig. 1). Importantly, and as shown in Table 1, all four of the QS-impaired C6706 strains contained the same G-to-A mutation in luxO, which resulted in an amino acid change from glycine to serine at position 333 of the protein (according to reference 38; annotated as glycine 319 initially [29]). None of the other three QS-proficient C6706 samples harbored this mutation, nor did any of the other O1 El Tor, O1 classical, or O37 strains (seven, two, and three isolates, respectively) which were tested at the same time (Table 1).
TABLE 1 .
Sequenced luxO and hapR genes in commonly studied V. cholerae strains
| V. cholerae strain (serogroup) | Description of luxO genea | Description of hapR genea |
|---|---|---|
| A1552 (O1) | Wild-type sequence | Wild-type hapR sequence (without frameshift mutation, as is the case for N16961) |
| C6706#1 (O1) | Wild-type sequence | Wild-type sequence |
| C6706#2 (O1) | Mutant luxO (change of amino acid G333Sb) | Wild-type sequence |
| C6706#3 (O1) | Mutant luxO (change of amino acid G333Sb) | Wild-type sequence |
| C6706#4 (O1) | Mutant luxO (change of amino acid G333Sb) | Wild-type sequence |
| C6706#5 (O1) | Mutant luxO (change of amino acid G333Sb) | Wild-type sequence |
| C6706#6 (O1) | Wild-type sequence | Wild-type sequence |
| C6706#7 (O1) | Wild-type sequence | Wild-type sequence |
| N16961 (O1) | Wild-type sequence | Frameshift mutation in hapR confirmed (29) |
| N16961rep (O1) | Wild-type sequence | Wild-type sequence (frameshift mutation of strain N16961 repaired) |
| C6709 (O1) | Wild-type sequence | Wild-type sequence |
| E7946 (O1) | Wild-type sequence | Wild-type sequence |
| DRC-193A (O1) | Wild-type sequence | Wild-type sequence |
| P27459 (O1) | Wild-type sequence | Wild-type sequence |
| O395 (O1 classical) | Wild-type sequence | Frameshift mutation in hapR confirmed (39) |
| 569B (O1 classical) | Wild-type sequence | Mutation in hapR (T169G leading to amino acid change Y57D) |
| V52 (O37) | Wild-type sequence | hapR mutation confirmed (premature stop codon) (43) |
| ATCC 25872 (O37) | Wild-type sequence | Wild-type sequence |
| ATCC 25873 (O37) | Wild-type sequence | Wild-type sequence |
DISCUSSION
Here, we provide evidence for the circulation of a QS-impaired wild-type version of V. cholerae strain C6706. Four of seven tested C6706 samples obtained from international laboratories were severely affected with respect to their natural transformability and, accordingly, also with respect to their T6SS production and activity. Using qRT-PCR, we showed that the low transformability perfectly correlated with low levels of hapR transcripts and a changed expression pattern of several HapR-regulated genes. Accordingly, HapR-/QstR-coregulated competence genes (e.g., those encoding the DNA-uptake machinery and the T6SS) were not induced upon growth on chitin, explaining the compromised transformation and T6SS responses in these C6706 samples. Sequencing the hapR and luxO genes of the C6706 isolates and other O1 and non-O1 V. cholerae strains showed that the QS-impaired C6706 strains contained a mutation in luxO. Interestingly, the exact same mutation was recently described for a luxU mutant derivative of strain C6706 (38). In their study, Jung et al. provided evidence that the G333S amino acid change mimics the active form of LuxO (38). This change therefore explains why the mutated C6706 strains consistently had lower hapR transcript and protein levels than the nonmutated C6706 strains and non-C6706 control strain A1552.
An earlier study reported frequent mutations in the hapR gene of V. cholerae (39), and we speculated that such mutations are overrepresented in culture collections due to a sampling bias (40). Importantly, none of these O1 El Tor and classical isolates of V. cholerae contained the exact same mutation in hapR (39), excluding the clonal expansion of one successful mutant strain. Interestingly, however, Joelsson and colleagues mentioned in their study that the HapR protein level was reduced in strain C6706 compared with that in several other O1 serogroup strains (39), possibly caused by the here-described mutation of several wild-type C6706 strains. In this context, it should be noted that a recent study on V. fischeri showed that luxO mutations are frequently isolated from cultures in prolonged stationary phase (41). Importantly, the authors describe the isolation of a plethora of different luxO mutant alleles, all of which mimic the gene encoding a constitutively active LuxO protein (41). In our study, however, we found the exact same mutation in four different C6706 strains obtained from different laboratories, and this mutant allele of luxO exactly matches a previously reported mutation in a C6706-derived luxU mutant (38). Thus, it appears rather unlikely that the mutation arose independently in those five different strains. Instead, it can be assumed that the mutated C6706 strain was circulated among different laboratories. It is therefore of prime importance for any group studying QS-related phenotypes, such as the QS network, virulence expression, biofilm formation, natural competence for transformation, and T6SS production, in strain C6706 to ensure that the wild-type laboratory stock(s) (some laboratories seem to have more than one stock of the wild-type C6706 strain) and mutants thereof from other laboratories do not contain the previously reported (38) and here-described (for the WT) luxO mutation. Indeed, as we show in this study, for natural transformation and T6SS production/interbacterial killing, this mutation masks important QS-dependent phenotypes and therefore leads to the irreproducibility of such features.
It is unnecessary to mention that mutations can occur at any time. It is important that the presence of such mutations in commonly used strains is communicated within the scientific community in a timely manner to avoid unnecessary investments of time and resources using flawed reagents. Indeed, we have received numerous inquiries with respect to the nontransformability of C6706. The current study solved this mystery, as we revealed a QS-impairing mutation in a circulating C6706 strain. Our recommendation for researchers who work with V. cholerae C6706 is therefore to take measures to ensure that the luxO gene is not mutated in their laboratory stock.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The V. cholerae strains used in this study are listed in Table 2. E. coli strain TOP10-TnKan (37) served as prey in the interbacterial killing assay (see below). Bacteria were grown in liquid LB medium under shaking conditions or on LB agar plates (1.5% agar) unless otherwise stated. The temperature was kept at room temperature, 30°C, or 37°C. Half-concentrated defined artificial seawater (0.5× DASW [13]) was used for the chitin-induced natural transformation experiments. Antibiotics and other supplements were added at the following concentrations: kanamycin at 75 µg/ml and l-arabinose at 0.2%.
TABLE 2 .
Bacterial strains (V. cholerae) and plasmids used in this study
| Strain or plasmid | Genotype/descriptiona | Internal strain no. | Reference(s) or source |
|---|---|---|---|
| Strains | |||
| A1552 (WT) | Wild type, O1 El Tor Inaba; Rifr | MB_1 | 44 |
| A1552-TntfoX-strep | A1552 containing mini-Tn7-araC-PBAD-tfoX-strep; Rifr, Gentr | MB_3420 | 37 |
| C6706 (strain #1–7) | V. cholerae O1 El Tor strain C6706; isolated in 1991, Peru; Strr (31) | MB_1144 (#1), MB_1990 (#2), MB_2599 (#3), MB_3087 (#4), MB_3594 (#5), MB_3601 (#6), MB_4242 (#7) | Obtained from diverse laboratories in North America and Europe |
| C6706#2-TntfoX-strep | C6706#2 containing mini-Tn7-araC-PBAD-tfoX-strep; Strr, Gentr | MB_4146 | This study |
| C6706#3-TntfoX-strep | C6706#3 containing mini-Tn7-araC-PBAD-tfoX-strep; Strr, Gentr | MB_4148 | This study |
| C6706#4-TntfoX-strep | C6706#4 containing mini-Tn7-araC-PBAD-tfoX-strep; Strr, Gentr | MB_4150 | This study |
| C6706#5-TntfoX-strep | C6706#5 containing mini-Tn7-araC-PBAD-tfoX-strep; Strr, Gentr | MB_4152 | This study |
| C6706#6-TntfoX-strep | C6706#6 containing mini-Tn7-araC-PBAD-tfoX-strep; Strr, Gentr | MB_4154 | This study |
| N16961 | N16961, hapR frameshift; Strr | MB_2 | 29 |
| N16961-rep | N16961, hapR frameshift repaired (TransFLP); Strr | MB_2254 | 45 |
| C6709 | V. cholerae O1 El Tor Inaba; isolated in 1991, Peru; Strr | MB_1503 | 46, 47 |
| E7946 | V. cholerae strain El Tor Ogawa; isolated in 1978, Bahrain; Strr | MB_2600 | 48, 49 |
| ATCC 25872 | V. cholerae non-O1 (O37); isolated in 1965, Czechoslovakia; Strr | MB_276 | 50, 51 |
| ATCC 25873 | V. cholerae non-O1 (O37); isolated in 1965, Czechoslovakia; Strr | MB_277 | 50, 51 |
| DRC-193A | V. cholerae O1; patient isolate from 2011 (isolated at the Institut National de Recherche Biomédicale; Democratic Republic of the Congo); ctxAB+ tcp+ hapR+; Strr | MB_1954 | 8 |
| P27459 | V. cholerae O1 El Tor Inaba; isolated in 1976, Bangladesh; Strr | MB_1504 | 47, 52 |
| V52 | V. cholerae non-O1 (O37); Isolated in 1968, Sudan; Strr | MB_1510 | 47, 53 |
| O395 | V. cholerae O1 classical (Ogawa); Strr | MB_1147 | 54 |
| 569B | V. cholerae O1 classical (Inaba); Strr | MB_1148 | 51 |
| Plasmids | |||
| pUX-BF13 | oriR6K, helper plasmid with Tn7 transposition function; Ampr | MB_457 | 55 |
| pGP704-mTntfoX-strep | pGP704 with mini-Tn7 carrying araC and PBAD-driven tfoX-strep; Ampr, Gentr | MB_3664 | 37 |
Amp, ampicillin; Gent, gentamicin; Rif, rifampin; Str, streptomycin.
For the selection of V. cholerae after triparental mating performed with E. coli donor strains, thiosulfate citrate bile salts sucrose (TCBS) agar plates were used. The plates were prepared following the standard protocol provided by the manufacturer (Sigma-Aldrich/Fluka, Buchs, Switzerland).
Natural transformation assays on chitin surfaces.
The natural transformability of the diverse V. cholerae strains was tested through an established transformation assay performed using chitin flakes (26, 27). The genomic DNA of strain A1552-lacZ-Kan (26) served as the transforming material. The frequencies were calculated as the number of kanamycin-resistant transformants divided by the total number of CFUs. The averages (±standard deviation [SD] as shown by the error bar) of results of four independent biological replicates are indicated in the figure. For calculation purposes, the value was set to the detection limit for the experiments that resulted in the absence of transformants (e.g., values below the detection limit of the assay). Significant differences between V. cholerae strain A1552 and the seven isolates of strain C6706 were evaluated with Student’s t test on log-transformed data (42).
Gene expression analysis by quantitative reverse transcription-PCR (qRT-PCR).
For the LB growth conditions, the bacteria were grown at 30°C for 6 h under shaking conditions in liquid LB medium to reach a high cell density and processed as previously reported (8, 18). RNA that was extracted from chitin-grown bacteria was obtained by growing V. cholerae on chitin flakes (Sigma-Aldrich, Switzerland) (26). After 22 h of static incubation on chitin surfaces (for each strain in quadruplicate), the samples were centrifuged for 3 min, the supernatant was removed, and the pellet was resuspended in 1 ml of Tri Reagent (Sigma-Aldrich, Switzerland). After vortex mixing was performed to ensure homogenization, the samples were again centrifuged to remove residual chitin flakes. The supernatant was transferred to a new tube, shock-frozen on dry ice, and stored at −80°C.
The expression of representative genes was analyzed by quantitative reverse transcription-PCR (qRT-PCR) as previously described (18). The transcript levels of the indicated genes were normalized to the expression of gyrA to obtain the relative expression values. All of the experiments were performed three independent times, and averages (±SD) of results of all of the biological replicates are provided. Statistical analyses were based on two-way analysis of variance (ANOVA), which was performed using GraphPad Prism version 7 for Mac (GraphPad Software, San Diego, CA, USA).
Interbacterial killing assay.
The interbacterial killing assay was performed as previously described (8) using E. coli strain TOP10-Kan (37) as the prey and the indicated V. cholerae strains as predators. The bacteria were grown in the absence (−ara) or presence (+ara) of 0.2% arabinose to induce the chromosomal copy on tfoX (harbored on transposon TntfoX-strep [18, 37]). Coincubation occurred at 37°C for 4 h with a ratio of predator to prey of 10:1. Recovered E. coli cells were enumerated through serial dilution followed by the counting of CFU per milliliter. Three independent experiments were performed, and averages (±SD) of results of these biological replicates are given in the figure. Significant differences were determined using two-way ANOVA (GraphPad Prism).
ACKNOWLEDGMENTS
We acknowledge all of the researchers who provided us with their published C6706 strains or other V. cholerae strains. For obvious reasons, we keep their names anonymous. We thank Lisa Metzger for scientific input and Tiziana Scrignari for technical assistance.
Funding Statement
The Swiss National Science Foundation provided funding for this work (grants 31003A_143356 and 31003A_162551).
REFERENCES
- 1.De SN. 1959. Enterotoxicity of bacteria-free culture-filtrate of Vibrio cholerae. Nature 183:1533–1534. doi: 10.1038/1831533a0. [DOI] [PubMed] [Google Scholar]
- 2.Holmgren J. 1981. Actions of cholera toxin and the prevention and treatment of cholera. Nature 292:413–417. doi: 10.1038/292413a0. [DOI] [PubMed] [Google Scholar]
- 3.Taylor RK, Miller VL, Furlong DB, Mekalanos JJ. 1987. Use of phoA gene fusions to identify a pilus colonization factor coordinately regulated with cholera toxin. Proc Natl Acad Sci U S A 84:2833–2837. doi: 10.1073/pnas.84.9.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nelson EJ, Harris JB, Morris JG Jr, Calderwood SB, Camilli A. 2009. Cholera transmission: the host, pathogen and bacteriophage dynamic. Nat Rev Microbiol 7:693–702. doi: 10.1038/nrmicro2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ritchie JM, Rui H, Bronson RT, Waldor MK. 2010. Back to the future: studying cholera pathogenesis using infant rabbits. mBio 1:e00047-10. doi: 10.1128/mBio.00047-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pukatzki S, Ma AT, Sturtevant D, Krastins B, Sarracino D, Nelson WC, Heidelberg JF, Mekalanos JJ. 2006. Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc Natl Acad Sci U S A 103:1528–1533. doi: 10.1073/pnas.0510322103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ho BT, Dong TG, Mekalanos JJ. 2014. A view to a kill: the bacterial type VI secretion system. Cell Host Microbe 15:9–21. doi: 10.1016/j.chom.2013.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Borgeaud S, Metzger LC, Scrignari T, Blokesch M. 2015. The type VI secretion system of Vibrio cholerae fosters horizontal gene transfer. Science 347:63–67. doi: 10.1126/science.1260064. [DOI] [PubMed] [Google Scholar]
- 9.Pruzzo C, Vezzulli L, Colwell RR. 2008. Global impact of Vibrio cholerae interactions with chitin. Environ Microbiol 10:1400–1410. doi: 10.1111/j.1462-2920.2007.01559.x. [DOI] [PubMed] [Google Scholar]
- 10.Matthey N, Blokesch M. 2016. The DNA-uptake process of naturally competent Vibrio cholerae. Trends Microbiol 24:98–110. doi: 10.1016/j.tim.2015.10.008. [DOI] [PubMed] [Google Scholar]
- 11.Seitz P, Blokesch M. 2013. Cues and regulatory pathways involved in natural competence and transformation in pathogenic and environmental gram-negative bacteria. FEMS Microbiol Rev 37:336–363. doi: 10.1111/j.1574-6976.2012.00353.x. [DOI] [PubMed] [Google Scholar]
- 12.Johnston C, Martin B, Fichant G, Polard P, Claverys JP. 2014. Bacterial transformation: distribution, shared mechanisms and divergent control. Nat Rev Microbiol 12:181–196. doi: 10.1038/nrmicro3199. [DOI] [PubMed] [Google Scholar]
- 13.Meibom KL, Blokesch M, Dolganov NA, Wu C-Y, Schoolnik GK. 2005. Chitin induces natural competence in Vibrio cholerae. Science 310:1824–1827. doi: 10.1126/science.1120096. [DOI] [PubMed] [Google Scholar]
- 14.Metzger LC, Blokesch M. 2016. Regulation of competence-mediated horizontal gene transfer in the natural habitat of Vibrio cholerae. Curr Opin Microbiol 30:1–7. doi: 10.1016/j.mib.2015.10.007. [DOI] [PubMed] [Google Scholar]
- 15.Seitz P, Blokesch M. 2013. DNA-uptake machinery of naturally competent Vibrio cholerae. Proc Natl Acad Sci U S A 110:17987–17992. doi: 10.1073/pnas.1315647110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seitz P, Blokesch M. 2014. DNA transport across the outer and inner membranes of naturally transformable Vibrio cholerae is spatially but not temporally coupled. mBio 5:e01409-14. doi: 10.1128/mBio.01409-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seitz P, Pezeshgi Modarres H, Borgeaud S, Bulushev RD, Steinbock LJ, Radenovic A, Dal Peraro M, Blokesch M. 2014. ComEA is essential for the transfer of external DNA into the periplasm in naturally transformable Vibrio cholerae Cells. PLoS Genet 10:e1004066. doi: 10.1371/journal.pgen.1004066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lo Scrudato M, Blokesch M. 2012. The regulatory network of natural competence and transformation of Vibrio cholerae. PLoS Genet 8:e1002778. doi: 10.1371/journal.pgen.1002778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rutherford ST, Bassler BL. 2012. Bacterial quorum sensing: its role in virulence and possibilities for its control. Cold Spring Harb Perspect Med 2:a012427. doi: 10.1101/cshperspect.a012427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blokesch M, Schoolnik GK. 2008. The extracellular nuclease Dns and its role in natural transformation of Vibrio cholerae. J Bacteriol 190:7232–7240. doi: 10.1128/JB.00959-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Suckow G, Seitz P, Blokesch M. 2011. Quorum sensing contributes to natural transformation of Vibrio cholerae in a species-specific manner. J Bacteriol 193:4914–4924. doi: 10.1128/JB.05396-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lo Scrudato M, Blokesch M. 2013. A transcriptional regulator linking quorum sensing and chitin induction to render Vibrio cholerae naturally transformable. Nucleic Acids Res 41:3644–3658. doi: 10.1093/nar/gkt041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gulig PA, Tucker MS, Thiaville PC, Joseph JL, Brown RN. 2009. User friendly cloning coupled with chitin-based natural transformation enables rapid mutagenesis of Vibrio vulnificus. Appl Environ Microbiol 75:4936–4949. doi: 10.1128/AEM.02564-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pollack-Berti A, Wollenberg MS, Ruby EG. 2010. Natural transformation of Vibrio fischeri requires tfoX and tfoY. Environ Microbiol 12:2302–2311. doi: 10.1111/j.1462-2920.2010.02250.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen Y, Dai J, Morris JG Jr, Johnson JA. 2010. Genetic analysis of the capsule polysaccharide (K antigen) and exopolysaccharide genes in pandemic Vibrio parahaemolyticus O3:K6. BMC Microbiol 10:274. doi: 10.1186/1471-2180-10-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marvig RL, Blokesch M. 2010. Natural transformation of Vibrio cholerae as a tool-optimizing the procedure. BMC Microbiol 10:155. doi: 10.1186/1471-2180-10-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.De Souza Silva O, Blokesch M. 2010. Genetic manipulation of Vibrio cholerae by combining natural transformation with FLP recombination. Plasmid 64:186–195. doi: 10.1016/j.plasmid.2010.08.001. [DOI] [PubMed] [Google Scholar]
- 28.Blokesch M. 2012. TransFLP—a method to genetically modify V. cholerae based on natural transformation and FLP-recombination. J Vis Exp 68:e3761. doi: 10.3791/3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heidelberg JF, Eisen JA, Nelson WC, Clayton RA, Gwinn ML, Dodson RJ, Haft DH, Hickey EK, Peterson JD, Umayam L, Gill SR, Nelson KE, Read TD, Tettelin H, Richardson D, Ermolaeva MD, Vamathevan J, Bass S, Qin H, Dragoi I, Sellers P, McDonald L, Utterback T, Fleishmann RD, Nierman WC, White O. 2000. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406:477–483. doi: 10.1038/35020000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dalia AB, Seed KD, Calderwood SB, Camilli A. 2015. A globally distributed mobile genetic element inhibits natural transformation of Vibrio cholerae. Proc Natl Acad Sci U S A 112:10485–10490. doi: 10.1073/pnas.1509097112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thelin KH, Taylor RK. 1996. Toxin-coregulated pilus, but not mannose-sensitive hemagglutinin, is required for colonization by Vibrio cholerae O1 El Tor biotype and O139 strains. Infect Immun 64:2853–2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blokesch M, Schoolnik GK. 2007. Serogroup conversion of Vibrio cholerae in aquatic reservoirs. PLoS Pathog 3:e81. doi: 10.1371/journal.ppat.0030081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miller MC, Keymer DP, Avelar A, Boehm AB, Schoolnik GK. 2007. Detection and transformation of genome segments that differ within a coastal population of Vibrio cholerae strains. Appl Environ Microbiol 73:3695–3704. doi: 10.1128/AEM.02735-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jobling MG, Holmes RK. 1997. Characterization of hapR, a positive regulator of the Vibrio cholerae HA/protease gene hap, and its identification as a functional homologue of the Vibrio harveyi luxR gene. Mol Microbiol 26:1023–1034. doi: 10.1046/j.1365-2958.1997.6402011.x. [DOI] [PubMed] [Google Scholar]
- 35.Zhu J, Mekalanos JJ. 2003. Quorum sensing-dependent biofilms enhance colonization in Vibrio cholerae. Dev Cell 5:647–656. doi: 10.1016/S1534-5807(03)00295-8. [DOI] [PubMed] [Google Scholar]
- 36.Hammer BK, Bassler BL. 2003. Quorum sensing controls biofilm formation in Vibrio cholerae. Mol Microbiol 50:101–104. doi: 10.1046/j.1365-2958.2003.03688.x. [DOI] [PubMed] [Google Scholar]
- 37.Metzger LC, Stutzmann S, Scrignari T, Van der Henst C, Matthey N, Blokesch M. 2016. Independent regulation of type VI secretion in Vibrio cholerae by TfoX and TfoY. Cell Rep 15:951–958. doi: 10.1016/j.celrep.2016.03.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jung SA, Chapman CA, Ng WL. 2015. Quadruple quorum-sensing inputs control Vibrio cholerae virulence and maintain system robustness. PLoS Pathog 11:e1004837. doi: 10.1371/journal.ppat.1004837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Joelsson A, Liu Z, Zhu J. 2006. Genetic and phenotypic diversity of quorum-sensing systems in clinical and environmental isolates of Vibrio cholerae. Infect Immun 74:1141–1147. doi: 10.1128/IAI.74.2.1141-1147.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Blokesch M. 2012. A quorum sensing-mediated switch contributes to natural transformation of Vibrio cholerae. Mob Genet Elements 2:224–227. doi: 10.4161/mge.22284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kimbrough JH, Stabb EV. 2015. Antisocial luxO mutants provide a stationary-phase survival advantage in Vibrio fischeri ES114. J Bacteriol 198:673–687. doi: 10.1128/JB.00807-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Keene ON. 1995. The log transformation is special. Stat Med 14:811–819. doi: 10.1002/sim.4780140810. [DOI] [PubMed] [Google Scholar]
- 43.Chun J, Grim CJ, Hasan NA, Lee JH, Choi SY, Haley BJ, Taviani E, Jeon YS, Kim DW, Brettin TS, Bruce DC, Challacombe JF, Detter JC, Han CS, Munk AC, Chertkov O, Meincke L, Saunders E, Walters RA, Huq A, Nair GB, Colwell RR. 2009. Comparative genomics reveals mechanism for short-term and long-term clonal transitions in pandemic Vibrio cholerae. Proc Natl Acad Sci U S A 106:15442–15447. doi: 10.1073/pnas.0907787106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yildiz FH, Schoolnik GK. 1998. Role of rpoS in stress survival and virulence of Vibrio cholerae. J Bacteriol 180:773–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kühn J, Finger F, Bertuzzo E, Borgeaud S, Gatto M, Rinaldo A, Blokesch M. 2014. Glucose- but not rice-based oral rehydration therapy enhances the production of virulence determinants in the human pathogen Vibrio cholerae. PLoS Negl Trop Dis 8:e3347. doi: 10.1371/journal.pntd.0003347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wachsmuth IK, Evins GM, Fields PI, Olsvik O, Popovic T, Bopp CA, Wells JG, Carrillo C, Blake PA. 1993. The molecular epidemiology of cholera in Latin America. J Infect Dis 167:621–626. doi: 10.1093/infdis/167.3.621. [DOI] [PubMed] [Google Scholar]
- 47.Nesper J, Kraiss A, Schild S, Blass J, Klose KE, Bockemühl J, Reidl J. 2002. Comparative and genetic analyses of the putative Vibrio cholerae lipopolysaccharide core oligosaccharide biosynthesis (wav) gene cluster. Infect Immun 70:2419–2433. doi: 10.1128/IAI.70.5.2419-2433.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miller VL, DiRita VJ, Mekalanos JJ. 1989. Identification of toxS, a regulatory gene whose product enhances ToxR-mediated activation of the cholera toxin promoter. J Bacteriol 171:1288–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lazinski DW, Camilli A. 2013. Homopolymer tail-mediated ligation PCR: a streamlined and highly efficient method for DNA cloning and library construction. BioTechniques 54:25–34. doi: 10.2144/000113981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Aldova E, Laznickova K, Stepankova E, Lietava J. 1968. Isolation of nonagglutinable vibrios from an enteritis outbreak in Czechoslovakia. J Infect Dis 118:25–31. doi: 10.1093/infdis/118.1.25. [DOI] [PubMed] [Google Scholar]
- 51.Felsenfeld O, Stegherr-Barrios A, Aldová E, Holmes J, Parrott MW. 1970. In vitro and in vivo studies of streptomycin-dependent cholera vibrios. Appl Microbiol 19:463–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pearson GD, Woods A, Chiang SL, Mekalanos JJ. 1993. CTX genetic element encodes a site-specific recombination system and an intestinal colonization factor. Proc Natl Acad Sci U S A 90:3750–3754. doi: 10.1073/pnas.90.8.3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bik EM, Gouw RD, Mooi FR. 1996. DNA fingerprinting of Vibrio cholerae strains with a novel insertion sequence element: a tool to identify epidemic strains. J Clin Microbiol 34:1453–1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mekalanos JJ. 1983. Duplication and amplification of toxin genes in Vibrio cholerae. Cell 35:253–263. doi: 10.1016/0092-8674(83)90228-3. [DOI] [PubMed] [Google Scholar]
- 55.Bao Y, Lies DP, Fu H, Roberts GP. 1991. An improved Tn7-based system for the single-copy insertion of cloned genes into chromosomes of gram-negative bacteria. Gene 109:167–168. doi: 10.1016/0378-1119(91)90604-A. [DOI] [PubMed] [Google Scholar]