Abstract
Despite major advances in early detection and prognosis, chemotherapy resistance is a major hurdle in the battle against breast cancer. Identifying predictive markers and understanding the mechanisms are key steps to overcoming chemoresistance. Methylation-controlled J protein (MCJ, also known as DNAJC15) is a negative regulator of mitochondrial respiration and has been associated with chemotherapeutic drug sensitivity in cancer cell lines. Here we show, in a retrospective study of a large cohort of breast cancer patients, that low MCJ expression in breast tumors predicts high risk of relapse in patients treated with chemotherapy; however, MCJ expression does not correlate with response to endocrine therapy. In a prospective study in breast cancer patients undergoing neoadjuvant therapy, low MCJ expression also correlates with poor clinical response to chemotherapy and decreased disease-free survival. Using MCJ-deficient mice, we demonstrate that lack of MCJ is sufficient to induce mammary tumor chemoresistance in vivo. Thus, loss of expression of this endogenous mitochondrial modulator in breast cancer promotes the development of chemoresistance.
The loss of MCJ, a negative regulator of mitochondrial respiration, correlates with poor response to chemotherapy in breast cancer patients.
Introduction
Breast cancer is the most prevalent cancer among women worldwide (1, 2). Improved diagnosis and therapies have enhanced survival for breast cancer relative to other cancers. However, despite the number of advances in screening, prevention, and treatment, many patients still present with or develop late-stage disease, resulting in increased mortality. The rate of breast cancer recurrence and death remains very high (over 40,000 annual deaths in the USA). Chemotherapy has been and still is one of the most effective and widely used means of treating breast cancer. However, the percentage of nonresponders and failures following an initial response remains high. Although the estrogen receptor/progesterone receptor–positive (ER/PR+) group of breast cancer patients are treated with endocrine therapy, the triple-negative breast cancer (TNBC), defined by their lack of expression of ER/PR and human epidermal growth factor receptor 2 (HER2), are dependent on treatment with chemotherapy. Patients with TNBC have poor prognosis due to inherent resistance of their cancers to chemotherapy agents, and patients with TNBC tend to have increased risk of recurrence (3). Herceptin alone is commonly used to treat the HER2+ breast cancer group, but efficacy is best when combined with chemotherapy. Thus, lack of response to chemotherapy will also affect the treatment of HER2+ patients. In addition, neoadjuvant therapy (short-term chemotherapy treatment prior to surgery) in breast cancer has become a standard therapeutic approach for patients with large locally advanced cancers to reduce the size of the primary tumor, allowing more patients to undergo breast conservation therapy (4, 5). However, only a fraction of the patients achieve a complete response following neoadjuvant therapy (5, 6). Moreover, the major problem in breast cancer treatment is the development of metastasis, since metastatic breast cancer cells are frequently resistant to almost any known therapy (7).
Understanding the mechanisms underlying the development of multidrug resistance is therefore critical to improving therapeutic outcomes in breast cancer. Moreover, determining a group of markers that can identify cancer patients who will or will not benefit from specific therapy is a challenge. Cancer cells can be resistant to chemotherapy before they are exposed to chemotherapeutic drugs (due to intrinsic characteristics of the cell), or they can evolve this resistance during the course of the chemotherapy (8). Among the different intracellular mechanisms of drug resistance in cancer cells is the increase in transmembrane drug efflux by the activation of ATP binding cassette (ABC) transporter proteins (9), with the most studied type being ABCB1 (MDR1) (10). Over the past 15 years, several types of inhibitors of ABCB1 activity were developed but, upon testing, proved to be either toxic or of limited efficacy (11–14). In addition to ABCB1, ABCC1 (MRP1) (15, 16) and ABCG2 (BCRP) (17) have also been involved in drug efflux. However, despite the large number of clinical studies on the expression of these transporters in breast cancer, no correlation between expression of these transporters and chemotherapy response has been demonstrated. Multidrug resistance, thus, remains a major challenge in cancer treatment. Interestingly, a recent study has shown that inhibition of mitochondrial respiration can overcome chemoresistance in melanoma cells (18). Moreover, metastatic cancer cells (frequently resistant to any chemotherapy treatment) were also shown to be more dependent on mitochondrial respiration (19). Thus, mitochondrial respiration may be an alternative target to overcome chemoresistance.
Methylation-controlled J protein (MCJ) is a member of the DnaJ protein family of cochaperones that contains a transmembrane domain and has an N-terminal region with no homology to any other known protein. We and others have recently shown that MCJ localizes to the mitochondrial inner membrane (20, 21). Moreover, we have shown that MCJ is one of the first endogenous inhibitors of complex I of the respiratory chain (20). Using MCJ-deficient mice, we have shown that loss of MCJ leads to increased complex I activity, mitochondrial membrane potential, and mitochondrial ATP production (20, 22). MCJ was initially identified as a gene negatively regulated by methylation at CpG islands in ovarian cancer (23, 24). Methylation of the MCJ gene at the CpG islands has also been reported in Wilm’s tumors, malignant brain tumors, and melanoma (25–28). In ovarian cancer patients, high levels of CpG island methylation in the MCJ gene (which is associated with a loss of MCJ expression) correlated with increased resistance to chemotherapy and poor overall survival (29). We have shown that MCJ is expressed in drug-sensitive breast cancer cell lines (e.g., MCF7 and MDA-MB-231) but is not detected in several multidrug-resistant breast cancer cell lines (e.g., MCF7/ADR and MD22) (30). Blocking MCJ expression in vitro in drug-sensitive breast cancer cells did not affect cell proliferation and expansion but increased resistance to doxorubicin and paclitaxel in part by promoting drug efflux (30). However, the role of MCJ in multidrug resistance in vivo and its impact on chemotherapy response in breast cancer patients remains unknown.
In this study, we show that the loss of MCJ leads to multidrug resistance both in mouse and human breast cancer patients. Using a mouse model of mammary tumors, we show for the first time that MCJ deficiency causes chemoresistance of the tumors in vivo. Importantly, retrospective and prospective studies in breast cancer patients indicate that low MCJ expression in breast tumor specifically predicts poor response to chemotherapy. Thus, MCJ, an endogenous break of mitochondrial respiration, emerges as a key regulator of chemotherapy response in breast cancer. Restoring MCJ function could therefore overcome multidrug resistance.
Results
MCJ deficiency causes in vivo chemoresistance of mammary tumors in mice.
MCJ is expressed in drug-sensitive breast cancer cell lines but not in multidrug-resistant breast cancer cell lines, and blocking MCJ expression in drug-sensitive breast cancer cells makes them refractory to multiple drugs without affecting their growth (30). To investigate the role of MCJ in breast cancer chemoresistance in vivo, we used MMTV-PyMT transgenic mice (MMTV-Py mice) as a mammary tumor mouse model. These mice express the polyomavirus middle T antigen in the mammary gland and readily develop tumors (31). Western blot analysis using tumor extracts identified MCJ as being present in both normal mammary glands and mammary tumors (Figure 1A). MMTV-Py mice were then crossed with MCJ KO mice (20) to obtain MCJ KO MMTV-Py mice. No major differences in the kinetics of tumor growth between MMTV mice and MCJ KO MMTV-Py mice were observed, as determined by overall tumor mass and survival (Figure 1B) or following individual tumors (Supplemental Figure 1A; supplemental material available online with this article; doi:10.1172/jci.insight.86873DS1). Histological analyses showed no obvious differences between tumors from MMTV-Py and MCJ KO MMTV-Py mice (Figure 1C). Thus, MCJ does not seem to affect tumor development and growth per se.
Figure 1. Increased chemoresistance in murine mammary tumors lacking MCJ.
(A) MCJ expression in normal mammary gland from a WT female mouse (N, C57BL/6, The Jackson Laboratory) or in mammary tumors isolated from 2 independent MMTV-PyMT mice (T1, T2) examined by Western blot analysis using whole cell extracts. (B) Kaplan-Meier survival curve of MMTV-PyMT (MMTV) and MCJ KO MMTV-PyMT (MCJ KO MMTV) mice (n = 5). Death was determined as the time mice needed to be euthanized due to enlarged tumors. P > 0.05 by log-rank test. (C) Histology (H&E) of mammary tumors from MMTV and MCJ KO MMTV mice at ×200 magnification. Boxes indicate enlarged sections. (D) Tumors from MMTV and MCJ KO MMTV were harvested and dissociated. Tumor cells were plated and treated with doxorubicin for 16 hours. Mean ± SD of cell survival relative to cells without doxorubicin is shown (n = 3). (E) MMTV and MCJ KO MMTV (n = 4) mice were treated with doxorubicin by i.p. administration every other day for 2 weeks. The size of a tumor over time is represented as a percentage relative to the initial size prior to the treatment. *P < 0.05 by unpaired t test.
To examine whether loss of MCJ also confers chemoresistance in vitro in primary MMTV tumor cells, tumors were harvested from MMTV-Py and MCJ KO MMTV-Py mice and were dissociated to obtain a suspension of cancer cells that were then plated in culture. Cancer cells from both genotypes grew at similar rates (data not shown). Cells were then treated with doxorubicin or medium alone for 16 hours, and cell survival was determined. MCJ KO breast cancer cells were more resistant to doxorubicin treatment relative to WT cancer cells (Figure 1D). We then examined whether the lack of MCJ affects chemotherapy response in vivo in these mice. After tumors reached a detectable and measurable size, both MMTV-Py mice and MCJ KO MMTV-Py mice were treated with doxorubicin every 2 days. Tumor size was measured over time. While the size of mammary tumors in MMTV-Py mice decreased with treatment, the size of the tumors in MCJ KO MMTV-Py mice continued to increase during treatment with doxorubicin (Figure 1E and Supplemental Figure 1B) Thus, loss of MCJ in mammary tumors causes in vivo chemoresistance in mice.
A prospective study with neoadjuvant therapy of breast cancer indicates that low MCJ expression is an indicator of poor pathological and clinical responses.
We next investigated the correlation between MCJ expression and response to neoadjuvant chemotherapy in breast cancer patients in a prospective study. Biopsies were collected from a total of 62 female breast cancer patients. Following pathological assessment, 58 samples were diagnosed as malignant (94%) and 4 as nonmalignant tumors (6%). According to cancer stage, 18/58 (31%) patients were treated with neoadjuvant therapy consisting of 3–4 cycles of anthracyclines (doxorubicin or epirubicin) within a 3-week interval each, followed by 3–4 cycles of taxanes (paclitaxel or docetaxel). Chemotherapy was then followed by surgery for removal of residual cancer. Samples from 15/18 patients (3 patients were lost to follow-up) were included in the present study. Table 1 shows the clinical characteristics of the patient cohort. Expression of MCJ protein in tissue sections of tumor biopsies was detected by staining with an MCJ-specific antibody (30). Differences in staining intensity among patients were clearly discernible, indicating variations in MCJ expression. Based on the staining intensity, patients could be divided into 3 groups: those with negative or low (Figure 2, A and B, patients #6 and #10); intermediate (Figure 2, D and E, patients #8 and #14); and high (Figure 2, G and H, patients #7 and #15) MCJ expression.
Table 1. Clinical characteristics of study participants (n = 15).

Figure 2. Variable levels of MCJ expression in primary breast cancer biopsies.
(A) Core needle biopsies from breast cancer patients were taken at the time of diagnosis. Sections 3–5 μm in thickness were stained with anti-MCJ mAb and counterstained with hematoxylin. Pictures depict variable intensity of staining among different patient samples; low (patients [Pts.] #6, #10; A and B), intermediate (Pts. #8, #14; D and E), or high (Pts. #7, #15; G and H). Original magnification ×400. (C, F, and I) Relative expression levels of MCJ mRNA isolated from core needle biopsies obtained at diagnosis were compared using qPCR. Each patient sample was run at least in duplicate, and the data shown represent the individual determinations for the indicated patients, as well as the mean ± SEM of the obtained values. Variable levels of MCJ mRNA are depicted from representative patients.
RNA was extracted from frozen tumor biopsies obtained at diagnosis, prior to neoadjuvant therapy, and MCJ mRNA levels were measured by quantitative PCR (qPCR). The levels of MCJ expression in the tumors (expressed as arbitrary units relative to the reference standard) varied between 0.4 and 12.3 units (Supplemental Figure 2). The expression levels of MCJ by qPCR correlated with the expression levels of MCJ protein detected by IHC (Figure 2, C, F, and I). Given the superior quantitative aspect of qPCR, we used these expression data for subsequent analysis.
Next, we assessed the correlation between the level of MCJ expression and the clinical characteristics of the patients. Within this cohort of patients, MCJ expression did not associate with age (Pearson R = –0.41), cancer stage (R = –0.59) or Nottingham grade (R = 0.57) (Table 2). There was also no significant correlation between the level of MCJ expression and breast tumor subtypes (Figure 3A). After neoadjuvant therapy, residual tumors and sentinel lymph nodes were surgically resected, and residual cancer burden (RCB) was determined. Based on the pathological evaluation, 53.3% of patients had RCB 0, 26.7% with RCB II, and 20% had RCB III (Table 3). Next, the correlation between RCB scores and the levels of MCJ mRNA expression was investigated. Samples from patients with RCB of 0, II, or III had means ± SEM of 5.4 ± 1.2, 2.6 ± 1.2, and 2.0 ± 0.6 units, respectively (Figure 3B). These results suggested a potential association between the low MCJ expression and poor response to neoadjuvant chemotherapy.
Table 2. MCJ expression in relation to clinical markers.

Figure 3. Low tumor MCJ expression predicts poor pathological and clinical responses to neoadjuvant therapy and correlates with short disease-free survival.
(A) Levels of MCJ mRNA according to breast tumor subtypes. Levels of expression are given in arbitrary units relative to the reference standard (n ≥ 3). (B) MCJ expression according to pathological response (residual cancer burden, RCB). The graph depicts MCJ expression at the time of diagnosis for each RCB patient group. Differences between the groups were not statistically significant (n ≥ 3, P = 0.14). (C) Association between MCJ mRNA levels and clinical response based on WHO criteria: cCR (clinical complete response), cPR (clinical partial response), and cSD (clinical stable disease). Differences between the groups were significant (mean ± SEM, n ≥ 3, P = 0.04, 1-way ANOVA). Post hoc Tukey’s test was significant only for cSD vs. cPR (*P < 0.05). (D) Association between MCJ expression and clinical response based on a simplified, 2-group criteria: NR, nonresponders; R, responders. Differences between NR and R groups were statistically significant (mean ± SEM, unpaired t test, n ≥ 5, *P = 0.01). (E). Comparison between cumulative relapse-free survival rates among patients with low (≤ 3.1) or high (> 3.1) MCJ expression units (log-rank [Mantel-Cox] test, n ≥ 6, *P = 0.0336).
Table 3. Clinical and Pathological responses (n = 15).

Furthermore, we examined the association between MCJ expression and clinical response to neoadjuvant therapy. Following WHO criteria, clinical responses in our patient cohort were classified as complete response (cCR; 3/15), partial response (cPR; 7/15), or stable disease (cSD; 5/15) (Table 3). MCJ expression was significantly lower in the cSD tumors when comparing the 3 patient groups by 1-way ANOVA test (P = 0.04). Post hoc Tukey’s multiple comparisons test revealed significant difference between cSD and cPR groups (P < 0.05) (Figure 3C). We further investigated the correlation between MCJ expression and overall clinical response to chemotherapy using a classification that organizes the patients into 2 groups: responders (patients with cCR and cPR) and nonresponders (patients with cSD). Accordingly, 10/15 patients (66.6%) were classified as responders and 5/15 patients (33.3%) as nonresponders (Table 3). MCJ expression level was significantly higher in responders (mean ± SEM of 5.2 ± 0.93) than nonresponders (1.42 ± 0.50; P = 0.01) (Figure 3D). These findings indicate that, based on clinical as well as pathological criteria, patients with breast tumors expressing low MCJ mRNA levels are most likely to respond poorly to neoadjuvant chemotherapy. Thus, MCJ could be a marker for responsiveness to chemotherapy in the neoadjuvant setting.
Analysis of receiver operating characteristic (ROC) AUC was used to obtain the optimal cutoff value for MCJ expression level as a predictor of overall clinical response (10 responders and 5 nonresponders). A cutoff threshold of 3.1 expression units was 90% sensitive and 100% specific for a clinical response. This was used to divide the cases into “high” (>3.1 units) and “low” (≤3.1 units) MCJ expression groups. Following neoadjuvant therapy, overall response was evaluated over a median follow-up period of 48 months. Relapse-free survival (RFS) was calculated based on relapse status, as 3/15 (20%) of patients had active disease on the latest clinical follow-up. RFS curves for the high (9/15) and low (6/15) MCJ expression groups (P = 0.03; log-rank [Mantel-Cox] test) are shown in Figure 3E. Together, these data indicate that the level of MCJ could be a predictor for disease-free survival in human breast cancer, with low levels of MCJ in the primary tumors being prognostic of short RFS.
Low MCJ expression predicts poor response to chemotherapy but not to endocrine therapy in breast cancer patients.
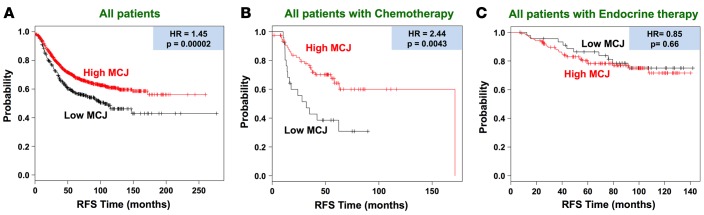
To further demonstrate the correlation between MCJ expression and chemotherapy response in a large cohort of patients, we used the Kaplan-Meier plotter (KM plotter) database for breast cancer that integrates gene expression (mRNA) and clinical data from 3 independent breast cancer repositories: cancer Biomedical Informatics Grid (caBIG), Gene Expression Omnibus (GEO), and The Cancer Genome Atlas (TCGA) (32). We first examined the association of MCJ expression using the DNAJC15 227808_at JetSet best probe set in the database with RFS in a total of 1,660 unclassified breast cancer patients. Patients whose breast cancers expressed low levels of MCJ (lowest quartile) exhibited worse prognosis than those patients whose tumors expressed high levels (upper 3 quartiles combined) of MCJ (hazard ratio [HR] = 1.45, P = 0.00002) (Figure 4A).
Figure 4. In large patient cohorts, low MCJ expression predicts poor survival after chemotherapy.
Kaplan-Meier curves for relapse-free survival (RFS) in breast cancer using the Kaplan-Meier Plotter (www.kmplot.com) database. “Low MCJ” indicates patients with tumors expressing MCJ mRNA in the lowest quartile, while “High MCJ” includes all other patients (MCJ expression in the top 3 quartiles). (A) Includes all patients regardless of therapy received (n = 1,660). (B) Includes all patients known to be treated with chemotherapy (n = 104). (C) Includes all patients known to have received endocrine therapy (n = 185). Hazard ratio (HR) and log-rank P values are shown.
We then addressed whether the association of low MCJ levels with overall poor prognosis was potentially mediated by the effect of MCJ on chemotherapy responses. We analyzed the relationship of MCJ expression with RFS restricting the analysis to patients who received chemotherapy and excluding patients with endocrine therapy (n = 104). Breast cancer patients with low–MCJ-expressing tumors had markedly shorter RFS than those patients with high–MCJ-expressing tumors (HR = 2.44, P = 0.0043) (Figure 4B). In contrast, when the analysis was restricted to patients who received endocrine therapy, but not chemotherapy (n = 185), no difference in RFS between low-MCJ and high-MCJ patients was detected (HR = 0.85, P = 0.66) (Figure 4C). Thus, accordingly with the results in our neoadjuvant therapy prospective study, results from these analyses further indicate that low MCJ expression is a predictive marker for poor response to chemotherapy.
MCJ as a marker for chemotherapy response in TNBC.
We further utilized the KM plotter database to investigate the relative expression of MCJ among clinical breast cancer subtypes (32). Interestingly, there was significantly lower expression of MCJ in the TNBC subtype (ER–, PR–, HER2–) relative to the Luminal A (P = 2.1 × 10–18), Luminal B (P = 7.1 × 10–7), HER2 (P = 5.9 × 10–11), ER+ (P = 6.2 × 10–11), and ER/PR+ (P = 3 × 10–4) subtypes (Figure 5A). There were only marginal differences in MCJ expression among the other subtypes. MCJ expression was not significantly correlated with patient age (Supplemental Figure 3A), tumor stage (Supplemental Figure 3B), or lymph node status (Supplemental Figure 3C). Thus, the overall MCJ expression is selectively reduced in TNBC tumors. We therefore examined the association of MCJ expression with RFS in the TNBC patients who had chemotherapy and observed significantly shorter relapse time in the low-MCJ group (HR = 2.70, P = 0.028) (Figure 5B). In patients with HER2+ breast tumors, low MCJ expression was not associated with significant differences in RFS (HR = 1.7, P = 0.22) (Figure 5C). As expected, there were only a few ER/PR+ breast cancer patients undergoing chemotherapy (data not shown). Together, these studies indicate that low MCJ expression could be used as a predictive marker for chemotherapy response in all breast cancer subtypes — particularly within the TNBC group, which is the group treated almost exclusively with chemotherapy.
Figure 5. Reduced expression of MCJ and the negative impact of low MCJ expression on survival is most evident in TNBC patients.
(A) MCJ expression was analyzed in breast cancers of the indicated subtypes using the Kaplan-Meier plotter database (integrating caBIG, GEO, and TCGA databases). A gradient of expression is shown across most classes, with statistical significance shown by letters at P < 0.01. Any comparison denoted with an “a” is not significantly different from another marked with an “a”. Similarly, “b” comparisons are not different from each other. However, “a”, “b”, and “c” are all different from each other. “ab” denotes a comparison not different from either “a” or “b” designation, but still denotes a difference from “c”. Further, a highly significant reduction in MCJ expression is seen in TNBC versus the other classes (P < 0.003). HER2+ER– (n = 162), ER+PR+ (n =74), luminal A (n = 908), luminal B (n = 436), ER+ (n = 844), TNBC (n = 384). (B and C) Kaplan-Meier curves were generated as in Figure 4, except that analyses were restricted to (B) triple-negative breast cancer (TNBC) patients known to have received chemotherapy (n = 53), and (C) HER2+ patients who received chemotherapy (n = 71).
MCJ promoter methylation associates with silencing gene expression in breast cancer.
No previous studies have reported methylation of the MCJ gene in breast cancer patients. We assessed MCJ gene methylation and expression data in breast tumors (n = 684) from the TCGA database to directly determine the relationship between DNA methylation at different locations and gene expression. Both CpG loci identified within the structural gene (cg27496116 and cg26053469) were highly methylated in most breast tumors and did not correlate with MCJ expression (Supplemental Figure 4, A and B). Analysis of the CpG loci within the human MCJ promoter showed that the 3 more distal CpG sites (>1 kb upstream of the transcriptional start site; Supplemental Figure 5A; cg11679069, cg13561081, and cg00131557) were highly methylated and not significantly correlated with DNAJC15 mRNA levels (Supplemental Figure 5, B–D). Two CpG loci 500–450 bp upstream of the transcription start site (TSS) (Supplemental Figure 6A; cg05035143 and cg12504148) were methylated in most tumors and had partial correlation with DNAJC15 mRNA levels (Supplemental Figure 6, B and C). Importantly, methylation of 3 CpG sites (cg14729962, cg09677945, and cg15988970) located in the proximal promoter (–200 to +1) (Figure 6A) revealed a strong inverse correlation with DNAJC15 mRNA levels and were unmethylated in most tumors (Figure 6, B–D). Thus, methylation in the proximal promoter contributes to the regulation of MCJ expression in breast tumors.
Figure 6. MCJ proximal promoter DNA methylation and gene expression in TCGA breast tumors.
(A) Line drawing of the MCJ gene promoter region with genomic position and transcription start site (TSS) in red. Vertical lines indicate CpG sites and are labeled to indicate CpGs plotted in B–D. (B–D) MCJ gene expression versus DNA methylation in TCGA breast tumors (n = 684) at promoter CpGs cg14729962 (B), cg09677945 (C), and cg15988970 (D) indicates increased expression associated with decreased DNA methylation (all P values < 2.2 × 10–16 by linear regression).
Discussion
Breast cancer is the second leading cause of cancer death among women. Chemotherapy remains one of the most effective and widely used means of treating breast cancer. While endocrine therapy is used to treat ER/PR+ breast cancer, and Herceptin in combination with standard chemotherapy is used to treat HER2+ breast cancer, chemotherapy is typically the only option for treatment of TNBC. While TNBC patients may respond initially, their rate of recurrence with multidrug resistance is very high. Chemotherapy is also used as the first line of neoadjuvant therapy prior to surgery or radiotherapy, with the goal of diminishing tumor size. In addition, independently of the type of the primary breast cancer, metastatic breast cancer is resistant to either endocrine therapy or Herceptin, leaving chemotherapy as the only alternative treatment, and the response is minimal. Therefore, understanding the mechanisms for development of chemoresistance, identifying biomarkers that predict chemotherapy response, and developing strategies that overcome chemoresistance are major goals that could have an impact on breast cancer therapy. Here, we demonstrate that low MCJ expression is a predictive marker for poor response to chemotherapy in human breast cancer patients. Additionally, loss of MCJ causes chemoresistance in a mammary tumor animal model, thus uncovering a mechanism that leads to chemoresistance in breast cancer.
A number of individual molecules that were identified in vitro as modulators of chemotherapy response failed validation as biomarkers by themselves in human breast cancer. For instance, despite the number of studies showing the expression of MDR1 (ABCB1) in multidrug-resistant breast cell lines, studies in a large cohort of breast cancer patients failed to show a clear correlation between MDR1 and overall response or survival (33). The idea that one molecule will not be able to predict prognosis or type of therapy, together with the emergence of gene expression patterns, stimulated the identification of groups of multiple genes as predictors. However, 2 gene expression signature tests have currently been incorporated into clinical diagnoses, but with limits in their applications. The 21-gene recurrence score assay (34) is limited for use to patients with ER+, node-negative early breast cancer to assess the 10-year risk of distant disease recurrence and to predict of the likelihood of response to adjuvant chemotherapy. The 18-gene vascular invasion assay is suitable for the prediction of tumor invasion and metastatic potential (35). No reliable gene expression test is currently available to predict chemotherapy response either in the context of neoadjuvant or adjuvant chemotherapy. Using 2 different approaches, we reveal here that MCJ is a predictive marker for chemotherapy response in human breast cancer. Our prospective study with neoadjuvant treatment of breast cancer patients shows that MCJ expression level was predictive of patients’ responses to the neoadjuvant chemotherapy. Additionally, in the follow-up analysis of these patients, data show that low MCJ expression predicts increased risk of recurrence. Using a more global unbiased approach in a large cohort of breast cancer patients, our studies also reveal that low MCJ expression in breast tumors predicts poor response to chemotherapy. In contrast, MCJ expression does not predict response to endocrine therapy, further showing the specificity of MCJ as a marker of chemotherapy response, which is consistent with its mechanism of action. While methylation of the MCJ gene has been correlated with chemoresistance in ovarian cancer (29), this is the first study examining the correlation between the expression of MCJ (instead of methylation of the gene) and the response to chemotherapy in cancer patients. Our results here indicate that MCJ could be a predictive marker for chemotherapy response in breast cancer, but it may also be a predictive marker for chemotherapy response in other types of cancers.
In addition to being a biomarker for chemoresistance, previous studies in ovarian cell lines and breast cancer cell lines suggested that loss of MCJ causes chemoresistance in vitro. Our study here shows for the first time to our knowledge that the loss of MCJ (MCJ-deficient mice) induces chemoresistance in vivo. Lack of MCJ does not seem to affect mammary tumor development or growth, but it decreases the response to chemotherapy in vivo. MCJ is an endogenous inhibitor of complex I of the mitochondrial respiratory chain (20), and loss of MCJ in macrophages (22), CD8 T cells (36), and both breast cancer cell lines and primary mammary tumor cells (data not shown) results in marked increase in mitochondrial respiration. While research has focused on the need to inhibit glycolysis as a means of preventing cancer growth, emerging evidence indicates that mitochondria are important targets in cancer (37–39). Recent studies have shown that multidrug resistance in melanoma can be overcome in vitro by blocking the mitochondrial ATP production (18). In addition, increased mitochondrial activity has also been shown to enhance metastatic dissemination (40), correlating with the poor chemotherapy response of metastatic cancer. Although there are a number of mechanisms by which mitochondrial respiration could contribute to chemoresistance in cancer, it is possible that ABC drug efflux transporters are highly dependent on mitochondria-derived ATP (instead of glycolysis) due to the dynamic ability of mitochondria to localize to regions of the cell.
Here, we also show that, in breast cancer patients, MCJ expression is low or undetectable in some breast tumors. Methylation is currently the best-characterized mechanism to regulate MCJ gene expression (23–28). However, the target methylation CpG sites within the MCJ gene responsible for its expression of MCJ gene are not fully defined and could be cell-type dependent. It was previously reported that loss of MCJ expression in drug-resistant ovarian cancer cell lines is dependent on methylation of CpG loci localized at the first exon (24). In contrast, using an unbiased approach to examine methylation and gene expression in breast cancer, our analyses indicate that methylation on 3 CpG loci localized within the proximal DNAJC15 promoter correlates with the loss of MCJ expression. The methylation at the proximal promoter could have a major impact in the binding of transcription factors that regulate MCJ transcription. In addition to methylation, we have recently reported that the expression of the mouse MCJ gene can be attenuated by Ikaros (41), a transcriptional regulator associated predominantly with the silencing of gene expression by promoting the formation of heterochromatin (42). Ikaros binding sites were identified in the proximal promoter of mouse MCJ gene (41), and they are also present in the human MCJ gene (data not shown). Thus, in addition to methylation, other mechanisms that regulate transcription of MCJ may be involved in the downregulation of MCJ expression in some of the breast tumors.
In summary, our study provides evidence that low expression of MCJ protein is associated with poor response to chemotherapy in human breast cancer, identifying MCJ as a new marker to predict chemotherapy response. Furthermore, we show that loss of MCJ is sufficient to cause chemoresistance in vivo. Thus, MCJ is a therapeutic target for breast cancer treatment. MCJ agonists could be used in combination with standard chemotherapeutic drugs to overcome chemoresistance, not only in primary cancers, but also in metastatic cancers that are highly refractory to any conventional therapies.
Methods
Mice and mammary tumor model studies
All mice used were female and 3–4 months of age. MCJ KO mice were previously described (20). MCJ KO mice were also crossed with the previously described MMTV-PyMT mice (31) (The Jackson Laboratory). MMTV-PyMT and MCJ KO MMTV-PyMT mice were fully (over 12 generations) backcrossed into FVB/N background and were maintained in the colony until their mammary tumors reached the maximum size institutionally approved, at which time they were euthanized. Survival curves were established based when the mice had to be euthanized according to the institutional regulations. Chemotherapy treatment was performed by i.p. injection of doxorubicin (2 mg/kg) or PBS as vehicle every other day for 12–14 days. Tumor size was determined using a caliper. Since MMTV-PyMT mice developed multifocal tumors, the size of the detectable tumor in the right or left thoracic mammary gland was followed, for precision only. Tumor size was measured prior to treatment and over time until the day of harvest. All mice were housed under sterile conditions at the animal care facility at the University of Vermont.
For in vitro studies, tumors were harvested from either MMTV-Py mice or MCJ KO MMTV-Py mice and dissociated using Mouse Tumor Dissociation Kits (Miltenyi Biotec). Tumor cells were plated and treated with doxorubicin (1 or 3 μM), and survival was determined by Trypan blue exclusion and cell count. Histological analysis (H&E staining) of the mammary tumors was performed with paraffin-embedded tumor sections.
Cell lines
MCF-7 and MDA-MB-231 human breast cancer cell lines were obtained from the American Type Culture Collection.
Western blot analysis
Mouse tumor cell extracts were analyzed for MCJ (20) and GAPDH (catalog sc-25778, Santa Cruz Biotechnology Inc.) as previously described (20).
Breast cancer prospective study
Breast cancer patients.
This prospective study was performed on a total of 62 patients with a suspicious breast mass that attended the Breast Care Clinic at Tawam Hospital (Al Ain, United Arab Emirates) from February 2010 to December 2012. Ultrasound-guided biopsies were obtained from every patient, and tumors were classified according to the American Joint Committee on Cancer (AJCC) to determine appropriate treatment. Patients with stage I and IV disease were excluded from the study. Prior to biopsy, an informed and written consent was obtained from every patient included in the study. Ethical approval for this study was obtained from the Al Ain Medical district human research ethics committee (protocol 08/13).
Neoadjuvant therapy.
All patients received neoadjuvant therapy consisting of 3–4 cycles of anthracyclines (doxorubicin or epirubicin) within a 3-week interval each, followed by 3–4 cycles of taxanes (paclitaxel or docetaxel). Additionally, patients with HER2+ tumors (3+ by IHC or FISH for equivocal results) received therapy with trastuzumab. All therapeutic modalities were according to protocols adopted by the Medical Oncology Department of Tawam Hospital and are in line with international guidelines.
Biopsy samples.
Biopsies were taken under local anesthesia with a 2% lidocaine solution using the core needle (inner diameter 1.6–2.4 mm) biopsy technique. Two biopsies per patient were collected and immediately snap-frozen in liquid nitrogen and processed for histological and RNA analysis. After completion of neoadjuvant chemotherapy, patients underwent surgical axillary node dissection and resection of the residual tumor. Pathology samples were processed following the standard protocol in the pathology laboratories (Tawam Hospital) and 2 of the authors (A. Albawardi and S. Almarzooqi) examined the H&E-stained slides from patients’ surgical tissues and reviewed the pathology reports.
IHC.
One of the biopsy samples from each patient was fixed in 10% formalin for 20 hours and then dehydrated and embedded in paraffin, following an established protocol (43). Sections 3–5 μm in thickness were cut on a microtome (Shandon AS325, Thermo Scientific), deparaffinized, and rehydrated. After blocking the endogenous peroxidase activity, antigen retrieval and protein block (DAKO) were performed. Sections were then incubated overnight at 4°C with anti-MCJ monoclonal antibody (BioMosaic), followed by HRP-conjugated polyclonal anti-mouse IgG secondary antibody (DAKO Envision + Dual Link System-HRP) and developed using DAB chromogen substrate. Slides were counterstained with hematoxylin and examined using an Olympus BX5 light microscope (Olympus America).
qPCR analysis.
Total RNA was extracted from a second biopsy of each patient using the TRIzol reagent protocol (Invitrogen) and repurified on Qiagen columns (RNeasy Plus Mini kit, Qiagen) as previously described (44). Similarly, total RNA was extracted from MCF-7 and MDA-MB-231 human breast cancer cell lines. The level of DNAJC15 gene expression was determined in both human breast cancer cell lines. Both cell lines were found to express DNAJC15, and based on 5 independent experiments, relative DNAJC15 expression in the 2 human cell lines differed by only 28% (mean ± SEM of 1.97 ± 0.23 for MDA-MB-231 vs. 1.42 ± 0.15 for MCF-7 cells). For calculating the fold-change in DNACJ15 expression in primary human tumors, the average of the two means was used as the reference standard. Total RNA concentration was determined by measuring absorbance at 260 nm on a Nanodrop 1000 spectrophotometer (Thermo Scientific). The purity of the RNA preparation was assessed based on the 260:280 nm absorbance ratio. cDNA was synthesized using Taqman reverse transcription reagents (Applied Biosystems) using the manufacturer’s protocol. For cDNA amplification, a GeneAmp PCR System 2700 (Applied Biosystems) was used. cDNA was amplified in TaqMan Universal Master Mix II (Applied Biosystems) using 40 ng of cDNA. The forward and reverse oligonucleotide primers used for MCJ (Hs01098150_m1) and HPRT (Hs02800695_m1) amplification were obtained from Applied Biosystems. The transcript level of the MCJ gene was normalized according to the ΔCq method to respective mRNA levels of the housekeeping gene HPRT. The expression of the MCJ gene is reported as fold-change in relation to the reference standard and is given in arbitrary units.
Tumor molecular subtypes.
Tumors were classified using the ER, PR, and HER2 molecular subtypes as proposed in the St. Gallen Consensus Report 2011 (45). According to these classifications, breast tumors are divided into 5 subgroups: luminal A (ER+, PR+, HER2–), luminal B (ER+, PR–, HER2–), luminal B-like (ER+, PR+/–, HER2+), HER2+ (ER–, PR–, HER2+), and basal or triple negative (ER–, PR–, HER2–).
Histological grading of the tumors.
Tumors were graded according to the Nottingham Combined Histologic Grade (Elston-Ellis modification of Scarff-Bloom-Richardson grading) system (46). This scoring system combines nuclear grade, tubule formation, and mitotic rate. Each of these features was given a score of 1–3 and then added to get a final total score ranging from 3–9. The final score determined the grade of the tumor as follows: grade-1 tumors (a score of 3–5), grade-2 tumors (a score of 6–7), and grade-3 tumors (a score of 8–9).
Evaluation of the pathological response.
Pathological response was assessed on resected specimens following the completion of neoadjuvant therapy. Pathological response was measured by calculating the RCB. The RCB of treated breast cancers is classified into 4 different categories according to established guidelines (47). (i) RCB 0 indicates no carcinoma in breast or lymph nodes. This group also included patients with residual in-situ ductal carcinoma and with no evidence of residual invasive disease. (ii) RCB I indicates partial response and minimal residual disease. (iii) RCB II indicates partial response and moderate residual disease. (iv) RCB III indicates chemoresistant, extensive residual disease. To calculate RCB, we used the MD Anderson Residual Cancer Burden Calculator, which is based on the assessment of primary tumor beds for residual tumor and lymph node status (48).
Evaluation of the clinical response.
After completion of neoadjuvant therapy, the evaluation of the patient’s response to chemotherapy was based on measuring the size of the residual tumor by ultrasound and MRI or physical examination by the breast surgeon according to standard procedures (49). Response to chemotherapy was determined following the WHO criteria (50, 51). Accordingly, the response of the target breast lesions was classified as cCR (disappearance of all target lesions), cPR (>50% decrease in the size of the lesions), clinical progressive disease (cPD; ≥25% increase in the size of the lesions), and SD (≤50% decrease and < 25% increase in the size of the lesions). Additionally, due to the small cohort of patients in our study, the clinical response was simplified, and patients with cCR and cPR were grouped as “responders”, while patients with cSD and cPD were grouped as “nonresponders”.
Analyses of publicly available datasets
RFS analysis and MCJ expression.
To explore potential correlations between the expression of MCJ in breast cancers and the survival of the corresponding patients, we used the KM plotter website (www.kmplot.com), which combines data from 3 data bases: caBIG, GEO, and TCGA (32). Expression levels were determined using primary breast cancer tumors prior to any treatment. For these analyses, we used the JetSet defined best probe set for DNAJC15 227808_at. Patients were split into the lower quartile of DNAJC15 expression compared with the upper 3 quartiles, and analyses were further restricted by ER/PR/HER2 status and treatment regimen (endocrine therapy or chemotherapy) as described in Results. Chemotherapy in these patients primarily included anthracyclines, cyclophosphamide, taxol, and — less frequently — 5-fluorouracil.
Methylation analysis.
Level 3 DNA methylation and normalized RNASeq data from invasive breast tumors was accessed from the TCGA data portal in October of 2013. Matched DNA methylation and normalized gene expression data were available from 684 tumors. Ten CpG sites in and near the DNAJC15 gene from the Illumina HumanMethylation450 array passed quality assessment and control procedures. They were available in the level-3 data (52) and included 8 promoter CpG sites and 2 CpG sites in the DNAJC15 gene body. Linear regression was used to test associations of DNAJC15 CpG methylation with DNAJC15 RNAseq expression.
Statistics.
Standard descriptive and inferential statistical analysis was conducted using SPSS statistical software (release 21; IBM SPSS Inc.). Univariate analyses were conducted using 1-way ANOVA and Pearson’s correlation coefficients. Multivariable modeling was conducted with multiple logistic regression using a forward stepwise conditional method for variable selection. No cases were excluded from the analysis, and missing values were not imputed. Colinearity was assessed using a correlation matrix of the variables included in the model. For in vitro studies and animal studies, statistical significance was determined by 2-tailed t test. Bars represent the mean with the SD or SEM. KM survival curves were analyzed using the log-rank (Mantel-Cox) test. All statistical tests were 2-sided. P < 0.05 was considered statistically significant.
Study approval.
Ethical approval for the human prospective study was obtained from the Al Ain Medical District Human Research Ethics Committee (protocol 08/13, Al Ain, United Arab Emirates). An informed and written consent was obtained from every patient included in the study. All animal procedures were approved by the University of Vermont Institutional Animal Care and Use Committee.
Author contributions
MJFC conceived the prospective clinical trial study, designed the experiments, coordinated the investigations, analyzed the data, and wrote the manuscript. IF provided patient samples and acquired patient clinical data. KJ acquired and analyzed data and wrote the manuscript. DPC conducted experiments, acquired and analyzed data, and wrote the manuscript. MAJ supervised patient treatment and wrote the manuscript. YAM conducted qPCR analysis of patient samples and acquired data. GB conducted histological studies and acquired data. SA and AA assessed patient specimens and determined the pathological response to treatment. MJH performed statistical analysis of the prospective patient trial. TSR conducted experiments and acquired data. HES and HET provided patient samples and helped in study design. AK provided patient samples. DEO conducted experiments and acquired data. BCC analyzed data and wrote the manuscript. JD analyzed data and wrote the manuscript. BKAR conceived the prospective clinical trial study, designed experiments, coordinated investigations, analyzed data, and wrote the manuscript. MR designed research studies, conducted experiments, acquired and analyzed data, and wrote the manuscript.
Supplementary Material
Acknowledgments
We wish to thank the nursing staff of the Tawam Hospital-Johns Hopkins Medicine Breast Care Center (Al Ain, United Arab Emirates) for their invaluable assistance in coordinating the collection of patient samples. We also thank Phani Gummadidala and Brian Silverstrim for technical support (University of Vermont). This work was funded by a grant from the Terry Fox Fund for Cancer Research (M.J. Fernández-Cabezudo and B.K. al-Ramadi), NIH grant R01 DE022772 (B.C. Christensen), NIH grant R21 CA127099 (M. Rincon), NIH grant R21 AI110016 (M. Rincon), and Lake Champlain Cancer Center Research Organization (LLCRO) (M. Rincon). Bioinformatic analyses were supported in part by the Biostatistics/Bioinformatics Shared Resources of Colorado’s NIH/NCI Cancer Center (support grant P30 CA046934).
Footnotes
Conflict of interest: M. Rincon and J. DeGregori are members of the scientific advisory board of Mitotherapeutix.
Reference information:JCI Insight. 2016;1(7):e86873. doi:10.1172/jci.insight.86873.
Contributor Information
Issam Faour, Email: ifaour@tawamhospital.ae.
Kenneth Jones, Email: KENNETH.L.JONES@ucdenver.edu.
Ghada Bashir, Email: ghadab@uaeu.ac.ae.
Saeeda Almarzooqi, Email: saeeda.almarzooqi@uaeu.ac.ae.
Alia Albawardi, Email: alia.albawardi@uaeu.ac.ae.
M. Jawad Hashim, Email: jhashim@uaeu.ac.ae.
Haytham El-Salhat, Email: elsalhat@hotmail.com.
Hakam El-Taji, Email: hakamtaji@yahoo.com.
Adnan Kassis, Email: aak196@yahoo.fr.
Dylan E. O’Sullivan, Email: dosull14run17epi@gmail.com.
Brock C. Christensen, Email: Brock.Christensen@Dartmouth.edu.
James DeGregori, Email: james.degregori@ucdenver.edu.
Basel K. al-Ramadi, Email: ramadi.b@uaeu.ac.ae.
Mercedes Rincon, Email: mrincon@uvm.edu.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R, et al. Cancer treatment and survivorship statistics, 2012. CA Cancer J Clin. 2012;62(4):220–241. doi: 10.3322/caac.21149. [DOI] [PubMed] [Google Scholar]
- 3.Wu J, Li S, Jia W, Su F. Response and prognosis of taxanes and anthracyclines neoadjuvant chemotherapy in patients with triple-negative breast cancer. J Cancer Res Clin Oncol. 2011;137(10):1505–1510. doi: 10.1007/s00432-011-1029-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thompson AM, Moulder-Thompson SL. Neoadjuvant treatment of breast cancer. Ann Oncol. 2012;23 Suppl 10:x231–x236. doi: 10.1093/annonc/mds324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Espie M. The management of breast cancer. Diagn Interv Imaging. 2014;95(7–8):753–757. doi: 10.1016/j.diii.2014.04.003. [DOI] [PubMed] [Google Scholar]
- 6.King TA, Morrow M. Surgical issues in patients with breast cancer receiving neoadjuvant chemotherapy. Nat Rev Clin Oncol. 2015;12(6):335–343. doi: 10.1038/nrclinonc.2015.63. [DOI] [PubMed] [Google Scholar]
- 7.Santa-Maria CA, Gradishar WJ. Changing treatment paradigms in metastatic breast cancer: lessons learned. JAMA Oncol. 2015;1(4):528–534; quiz 549. doi: 10.1001/jamaoncol.2015.1198. [DOI] [PubMed] [Google Scholar]
- 8.Kerbel RS, Kobayashi H, Graham CH. Intrinsic or acquired drug resistance and metastasis: are they linked phenotypes? J Cell Biochem. 1994;56(1):37–47. doi: 10.1002/jcb.240560108. [DOI] [PubMed] [Google Scholar]
- 9.Kovalev AA, Tsvetaeva DA, Grudinskaja TV. Role of ABC-cassette transporters (MDR1, MRP1, BCRP) in the development of primary acquired multiple drug resistance in patients with early metastatic breast cancer. Exp Oncol. 2013;35(4):287–290. [PubMed] [Google Scholar]
- 10.Wilson TR, Longley DB, Johnston PG. Chemoresistance in solid tumours. Ann Oncol. 2006;17(Suppl 10):x315–x324. doi: 10.1093/annonc/mdl280. [DOI] [PubMed] [Google Scholar]
- 11.Gottesman MM, Fojo T, Bates SE. Multidrug resistance in cancer: role of ATP-dependent transporters. Nat Rev Cancer. 2002;2(1):48–58. doi: 10.1038/nrc706. [DOI] [PubMed] [Google Scholar]
- 12.Krishna R, Mayer LD. Multidrug resistance (MDR) in cancer. Mechanisms, reversal using modulators of MDR and the role of MDR modulators in influencing the pharmacokinetics of anticancer drugs. Eur J Pharm Sci. 2000;11(4):265–283. doi: 10.1016/S0928-0987(00)00114-7. [DOI] [PubMed] [Google Scholar]
- 13.Munagala S, et al. Synthesis and evaluation of Strychnos alkaloids as MDR reversal agents for cancer cell eradication. Bioorg Med Chem. 2014;22(3):1148–1155. doi: 10.1016/j.bmc.2013.12.022. [DOI] [PubMed] [Google Scholar]
- 14.Palmeira A, Sousa E, Vasconcelos MH, Pinto MM. Three decades of P-gp inhibitors: skimming through several generations and scaffolds. Curr Med Chem. 2012;19(13):1946–2025. doi: 10.2174/092986712800167392. [DOI] [PubMed] [Google Scholar]
- 15.Cole SP, et al. Pharmacological characterization of multidrug resistant MRP-transfected human tumor cells. Cancer Res. 1994;54(22):5902–5910. [PubMed] [Google Scholar]
- 16.Cole SP, et al. Overexpression of a transporter gene in a multidrug-resistant human lung cancer cell line. Science. 1992;258(5088):1650–1654. doi: 10.1126/science.1360704. [DOI] [PubMed] [Google Scholar]
- 17.Doyle LA, Ross DD. Multidrug resistance mediated by the breast cancer resistance protein BCRP (ABCG2) Oncogene. 2003;22(47):7340–7358. doi: 10.1038/sj.onc.1206938. [DOI] [PubMed] [Google Scholar]
- 18.Roesch A, et al. Overcoming intrinsic multidrug resistance in melanoma by blocking the mitochondrial respiratory chain of slow-cycling JARID1B(high) cells. Cancer Cell. 2013;23(6):811–825. doi: 10.1016/j.ccr.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Porporato PE, et al. A mitochondrial switch promotes tumor metastasis. Cell Rep. 2014;8(3):754–766. doi: 10.1016/j.celrep.2014.06.043. [DOI] [PubMed] [Google Scholar]
- 20.Hatle K, et al. MCJ/DnaJC15, an endogenous mitochondrial repressor of the respiratory chain that controls metabolic alterations. Mol Cell Biol. 2013;33(11):2302–2314. doi: 10.1128/MCB.00189-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schusdziarra C, Blamowska M, Azem A, Hell K. Methylation-controlled J-protein MCJ acts in the import of proteins into human mitochondria. Hum Mol Genet. 2013;22(7):1348–1357. doi: 10.1093/hmg/dds541. [DOI] [PubMed] [Google Scholar]
- 22.Navasa N, et al. Regulation of oxidative stress by methylation-controlled J protein controls macrophage responses to inflammatory insults. J Infect Dis. 2015;211(1):135–145. doi: 10.1093/infdis/jiu389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shridhar V, et al. Loss of expression of a new member of the DNAJ protein family confers resistance to chemotherapeutic agents used in the treatment of ovarian cancer. Cancer Res. 2001;61(10):4258–4265. [PubMed] [Google Scholar]
- 24.Strathdee G, Davies BR, Vass JK, Siddiqui N, Brown R. Cell type-specific methylation of an intronic CpG island controls expression of the MCJ gene. Carcinogenesis. 2004;25(5):693–701. doi: 10.1093/carcin/bgh066. [DOI] [PubMed] [Google Scholar]
- 25.Ehrlich M, et al. Hypomethylation and hypermethylation of DNA in Wilms tumors. Oncogene. 2002;21(43):6694–6702. doi: 10.1038/sj.onc.1205890. [DOI] [PubMed] [Google Scholar]
- 26.Lau DT, Hesson LB, Norris MD, Marshall GM, Haber M, Ashton LJ. Prognostic significance of promoter DNA methylation in patients with childhood neuroblastoma. Clin Cancer Res. 2012;18(20):5690–5700. doi: 10.1158/1078-0432.CCR-12-0294. [DOI] [PubMed] [Google Scholar]
- 27.Lindsey JC, et al. Epigenetic inactivation of MCJ (DNAJD1) in malignant paediatric brain tumours. Int J Cancer. 2006;118(2):346–352. doi: 10.1002/ijc.21353. [DOI] [PubMed] [Google Scholar]
- 28.Muthusamy V, et al. Epigenetic silencing of novel tumor suppressors in malignant melanoma. Cancer Res. 2006;66(23):11187–11193. doi: 10.1158/0008-5472.CAN-06-1274. [DOI] [PubMed] [Google Scholar]
- 29.Strathdee G, Vass JK, Oien KA, Siddiqui N, Curto-Garcia J, Brown R. Demethylation of the MCJ gene in stage III/IV epithelial ovarian cancer and response to chemotherapy. Gynecol Oncol. 2005;97(3):898–903. doi: 10.1016/j.ygyno.2005.03.023. [DOI] [PubMed] [Google Scholar]
- 30.Hatle KM, et al. Methylation-controlled J protein promotes c-Jun degradation to prevent ABCB1 transporter expression. Mol Cell Biol. 2007;27(8):2952–2966. doi: 10.1128/MCB.01804-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guy CT, Cardiff RD, Muller WJ. Induction of mammary tumors by expression of polyomavirus middle T oncogene: a transgenic mouse model for metastatic disease. Mol Cell Biol. 1992;12(3):954–961. doi: 10.1128/MCB.12.3.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gyorffy B, Surowiak P, Budczies J, Lanczky A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PloS One. 2013;8(12):e86873. doi: 10.1371/journal.pone.0082241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu M, Ocana A, Tannock IF. Reversal of ATP-binding cassette drug transporter activity to modulate chemoresistance: why has it failed to provide clinical benefit? Cancer Metastasis Rev. 2013;32(1–2):211–227. doi: 10.1007/s10555-012-9402-8. [DOI] [PubMed] [Google Scholar]
- 34.Brufsky AM. Predictive and prognostic value of the 21-gene recurrence score in hormone receptor-positive, node-positive breast cancer. Am J Clin Oncol. 2014;37(4):404–410. doi: 10.1097/COC.0000000000000086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mannelqvist M, Wik E, Stefansson IM, Akslen LA. An 18-gene signature for vascular invasion is associated with aggressive features reduced survival in breast cancer. PloS One. 2014;9(6):e86873. doi: 10.1371/journal.pone.0098787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Champagne DP, et al. Fine-tuning of CD8 T cell mitochondrial respiration by MCJ/DnaJC15 distates protection to influenza virus. Immunity. doi: 10.1016/j.immuni.2016.02.018. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wolf DA. Is reliance on mitochondrial respiration a “chink in the armor” of therapy-resistant cancer? Cancer Cell. 2014;26(6):788–795. doi: 10.1016/j.ccell.2014.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Viale A, Corti D, Draetta GF. Tumors and mitochondrial respiration: a neglected connection. Cancer Res. 2015;75(18):3685–3686. doi: 10.1158/0008-5472.CAN-15-0491. [DOI] [PubMed] [Google Scholar]
- 39.Viale A, Draetta GF. Sugar? No thank you, just a deep breath of oxygen for cancer stem cells. Cell Metab. 2015;22(4):543–545. doi: 10.1016/j.cmet.2015.09.020. [DOI] [PubMed] [Google Scholar]
- 40.Rivadeneira DB, et al. Survivin promotes oxidative phosphorylation, subcellular mitochondrial repositioning, and tumor cell invasion. Sci Signal. 2015;8(389):e86873. doi: 10.1126/scisignal.aab1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Navasa N, et al. Ikaros mediates the DNA methylation-independent silencing of MCJ/DNAJC15 gene expression in macrophages. Sci Rep. 2015;5:e86873. doi: 10.1038/srep14692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.John LB, Ward AC. The Ikaros gene family: transcriptional regulators of hematopoiesis and immunity. Mol Immunol. 2011;48(9–10):1272–1278. doi: 10.1016/j.molimm.2011.03.006. [DOI] [PubMed] [Google Scholar]
- 43.al-Ramadi BK, Fernandez-Cabezudo MJ, El-Hasasna H, Al-Salam S, Bashir G, Chouaib S. Potent anti-tumor activity of systemically-administered IL2-expressing Salmonella correlates with decreased angiogenesis enhanced tumor apoptosis. Clin Immunol. 2009;130(1):89–97. doi: 10.1016/j.clim.2008.08.021. [DOI] [PubMed] [Google Scholar]
- 44.Kaimala S, et al. Salmonella-mediated tumor regression involves targeting of tumor myeloid suppressor cells causing a shift to M1-like phenotype reduction in suppressive capacity. Cancer Immunol Immunother. 2014;63(6):587–599. doi: 10.1007/s00262-014-1543-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Goldhirsch A, et al. Personalizing the treatment of women with early breast cancer: highlights of the St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2013. Ann Oncol. 2013;24(9):2206–2223. doi: 10.1093/annonc/mdt303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Edge S, Byrd DR, Compton CC, Fritz AG, Greene FL, Trotti A, eds. AJCC Cancer Staging Manual. 7th ed. New York, New York, USA: Springer-Verlag; 2010. [Google Scholar]
- 47.Symmans WF, et al. Measurement of residual breast cancer burden to predict survival after neoadjuvant chemotherapy. J Clin Oncol. 2007;25(28):4414–4422. doi: 10.1200/JCO.2007.10.6823. [DOI] [PubMed] [Google Scholar]
- 48.MD Anderson Cancer Center Residual Cancer Burden Calculator. [April 18, 2016];MD Anderson Web site. http://www3.mdanderson.org/app/medcalc/index.cfm?pagename=jsconvert3
- 49.Kuerer HM, et al. Clinical course of breast cancer patients with complete pathologic primary tumor and axillary lymph node response to doxorubicin-based neoadjuvant chemotherapy. J Clin Oncol. 1999;17(2):460–469. doi: 10.1200/JCO.1999.17.2.460. [DOI] [PubMed] [Google Scholar]
- 50.Therasse P. Measuring the clinical response. What does it mean? Eur J Cancer. 2002;38(14):1817–1823. doi: 10.1016/S0959-8049(02)00182-X. [DOI] [PubMed] [Google Scholar]
- 51.Khokher S, Qureshi MU, Chaudhry NA. Comparison of WHO and RECIST criteria for evaluation of clinical response to chemotherapy in patients with advanced breast cancer. Asian Pac J Cancer Prev. 2012;13(7):3213–3218. doi: 10.7314/APJCP.2012.13.7.3213. [DOI] [PubMed] [Google Scholar]
- 52.Cancer Genome Atlas Network Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.