Abstract
Genetically encoded unnatural amino acids provide powerful strategies for modulating the molecular functions of proteins in mammalian cells. However this approach has not been coupled to genome-wide measurements, because efficient unnatural amino acid incorporation is limited to transient expression settings that lead to very heterogeneous expression. We demonstrate that stable \ integration of the Methanosarcina mazei pyrrolysyl-tRNA synthetase (PylS)/tRNAPylCUA pair (and its derivatives) into the mammalian genome enables efficient, homogeneous unnatural amino acid incorporation into target proteins in diverse mammalian cells, and we reveal the distinct transcriptional responses of ES cells and MEFs to amber suppression. Genetically encoding Nε-acetyl-lysine in place of six lysine residues in histone H3 enables deposition of pre-acetylated histones into cellular chromatin, via a pathway that is orthogonal to enzymatic modification. Upon synthetically encoding lysine-acetylation at natural modification sites we determine the consequences of acetylation at specific amino acids in histones on gene expression.
Introduction
Genetic code expansion enables the site-specific incorporation of 'designer' amino acids into proteins produced in cells1. Numerous useful unnatural amino acids have been incorporated, facilitating new approaches to study outstanding problems in biology.2–5
Genetic code expansion uses orthogonal aminoacyl-tRNA synthetase/tRNACUA pairs that direct the incorporation of unnatural amino acids in response to an amber stop codon introduced at a desired site into a gene of interest1. The pyrrolysyl-tRNA synthetase (PylRS, encoded by PylS)/tRNAPylCUA (encoded by PylT) pair from Methanosarcina species (commonly M. mazei, as used here, or M. barkeri) has emerged as a particularly versatile system for genetic code expansion1. This pair has been evolved to direct the incorporation of numerous structurally and functionally diverse, designer amino acids, and has been developed for unnatural amino acid incorporation in diverse hosts, including Esherichia coli, Saccharomyces cerevisiae, Caenorhabditis elegans, Drosophila melanogaster and mammalian cells1.
Current methods for incorporating unnatural amino acids in mammalian cells are primarily based on transient transfection and/or transient expression6–9, limiting the scope of most, unnatural amino acid mutagenesis experiments to cell lines that can be efficiently transfected. Since transient expression (including viral transduction7) experiments lead to heterogeneous expression levels, current approaches severely limit the ability of investigators to couple precise perturbations - that may be effected by unnatural amino acid mutagenesis - to global genomic, epigenomic, transcriptomic, metabolomic or proteomic measurements. An ideal method for introducing unnatural amino acids into cells would express PylS, PylT and a gene of interest containing an amber codon from an integrated locus, facilitating uniform levels of unnatural amino acid incorporation for all cells in a clonal population. The multiple copies of PylT commonly required for efficient unnatural amino acid incorporation in mammalian cells8 are too large and too repetitive for retroviral packaging10 and cannot be straightforwardly installed in diverse cell types.
PiggyBac transgenesis enables the rapid and efficient integration of large and complex sequences into the genome of mammalian cells and many other hosts11. Here we demonstrate that PiggyBac transposon mediated integration of an optimized PylS/PylT cassette, containing multiple copies of PylT, enables the generation of stable lines that show robust, efficient and uniform unnatural amino acid incorporation.
Histone proteins, which package DNA in the eukaryotic nucleus, are densely post-translationally modified12, and understanding the in vivo functions of these modifications is of intense interest. N-epsilon acetyl-lysine (AcK), and other post-translational modifications, and their non-removable or selectively-removable analogs (including lysine methylation13, 14, tri-fluoroacetyl-lysine15, crotonyl-lysine16, 17, butyryl-lysine, propionyl-lysine16), have been genetically encoded into recombinant histones expressed in E. coli, using an acetyl-lysine-tRNA synthetase (AcKRS)/tRNAPylCUA pair18 (or other PylRS/tRNAPylCUA derivatives); this approach - along with other powerful strategies to synthetically install chromatin modifications19 - allows scientists to address the mechanistic consequences of specific modifications on chromatin structure and function in vitro. In vitro experiments can provide a clear link between molecular cause and effect20–23, but are abstracted from the appreciable complexity of the cellular environment. In contrast, in vivo experiments typically provide a wealth of correlative information about changes of chromatin state in a native context24, but it is commonly impossible to infer causation from these experiments.
Manipulation of cellular chromatin modifications would provide a route to understanding causation and mechanism. However, it is very challenging to selectively perturb a modification at a particular site on a histone. Manipulations of enzymes that modify histones have pleiotropic effects, and may affect the modification of non-histone proteins as much as histone proteins25, 26, confounding efforts to assign causation to a particular modification state. Modifying enzymes may be recruited to genomic loci by synthetically tethering them to DNA binding modules (such as catalytically inactive Cas9 variants) 27. The locus specific consequences observed in these experiments may result from post-translational modification of many sites on the nucleosome and other chromatin bound proteins, and do not provide a causative link between a modification at a specific site in a protein and its consequences.
The direct genetic encoding of post-translationally modified amino acids (and their non-removable analogs) into histones in mammalian cells would provide a route to study the specific consequences of defined modifications in histones, without the pleiotropic effects that may be mediated by modifying enzymes. Here we demonstrate that AcK can be genetically programmed into defined sites in histones and deposited into cellular chromatin. This approach reveals genes that can be regulated by the site-specific acetylation of a histone.
Results
PiggyBac integration of the orthogonal PylRS/tRNACUA pair
We developed an approach for integration of a PylS and PylT expression cassette8 embedded in a PiggyBac targeting vector (Fig. 1a) into the mammalian genome. A resistance marker was expressed from an internal ribosome entry site in the PylS transcript, to enable the selection of integrants that express PylS and PylT. By delivering the gene of interest containing the stop codon on an analogous vector, we doubled the number of PylT genes introduced.
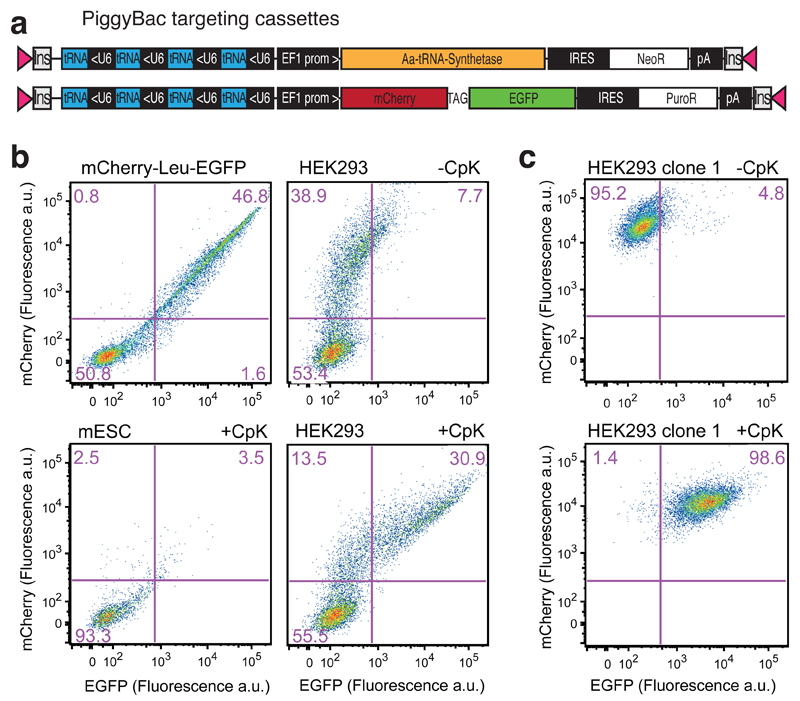
Figure 1.
Transient transfection leads to heterogeneous levels of an unnatural amino acid containing protein, with many cells untransfected, this problem is solved by PiggyBac integration of the unnatural amino acid incorporation machinery. a PiggyBac targeting vectors used in transient transfection and stable cell line generation, Insulators (Ins), inverted terminal repeats (pink triangles). Amber codon between mCherry and EGFP is indicated as ‘TAG’ b FACS analysis of transiently transfected HEK293 and mouse ES cells (mESC), 48 h after transfection and addition of 0.5 mM CpK. Cells were gated for live and single cell population. c Representative FACS analysis of a HEK293 cell line with the unnatural amino acid incorporation machinery integrated, cultured 48 hours with or without addition of 0.5 mM CpK.
Transient transfection experiments with the PiggyBac constructs confirmed the efficient, amino-acid dependent incorporation of Nε-[(tert-butoxy)carbonyl]-l-lysine (BocK) or Nε-[((2-methylcycloprop-2-en-1-yl)methoxy)carbonyl]-l-lysine (CpK), known PylRS substrates13, 28, in response to the amber stop codon in mCherry-TAG-EGFP, in HEK293 cells and mouse embryonic stem cells (ESC) (Fig. 1b, Supplementary Fig 1); in both cell lines, the majority of mCherry positive cells were also EGFP positive.
However, flow cytometry revealed several drawbacks of transient transfection for unnatural amino acid incorporation. The low transfection efficiency of some cell lines, such as ESCs, leads to many cells not receiving the unnatural amino acid incorporation machinery and/or gene of interest. For cells that are transfected, there is great variability in stop codon read through and unnatural amino acid incorporation; this results from variability in both the expression level of the gene of interest and the expression level of the unnatural amino acid incorporation machinery (Fig. 1b,Supplementary Fig. 1) consistent with the copy number of PylT being a major determinant of unnatural amino acid incorporation efficiency8).
To address the limitations of transient transfection approaches we created stable cell lines for unnatural amino acid incorporation (Supplementary Fig. 2). We co-transfected HEK293 cells with the two targeting vectors (Fig. 1a) and a plasmid carrying the PiggyBac transposase gene. After selection, with Puromycin (5 μg mL-1) and G418 (1,000 μg mL-1), we isolated single clones with strong mCherry fluorescence by FACS. The clonal cell population responded uniformly to the addition of 0.5 mM CpK (Fig.1c, Supplementary Fig. 2,). Additional experiments demonstrate the generality of the approach across different cell lines (Supplementary Fig. 3).
Efficient unnatural amino acid incorporation in mouse ESC
Integrating the unnatural amino acid incorporation machinery in mouse embryonic stem cells would provide new approaches to study stem cell function and developmental biology29 and may provide access to differentiated cell lines for unnatural amino acid incorporation.
We transfected an ES cell line (E14), with the 4xPylT/PylS, 4xPylT/sfGFP150TAG (Fig. 2a) and PiggyBac transposase, and selected stable integrants with Puromycin (5 μg mL-1) and G418 (1,000 μg mL-1) from 3 to 7 days after transfection. More than half of the cells in the resulting polyclonal pool expressed sfGFP when BocK or CpK were added, whereas we observed no sfGFP fluorescence in the absence of unnatural amino acid (Supplementary Fig. 4). We derived individual clones by FACS that maintained robust unnatural amino acid dependent expression, consistent with high-fidelity incorporation of the unnatural amino acid in response to the amber stop codon, and exhibited a defined and homogeneous level of sfGFP expression (Fig. 2b and Supplementary Fig. 4). We also generated E14 cell lines expressing 4xPylT/PylS and 4xPylT/mCherry-TAG-EGFP with the same protocol, and observed, near-quantitative, uniform and dose-dependent incorporation of BocK and CpK (Supplementary Fig. 5). These experiments demonstrate that the unnatural amino acid incorporation machinery can be stably integrated into ES cells and used to direct the site-specific incorporation of unnatural amino acids.
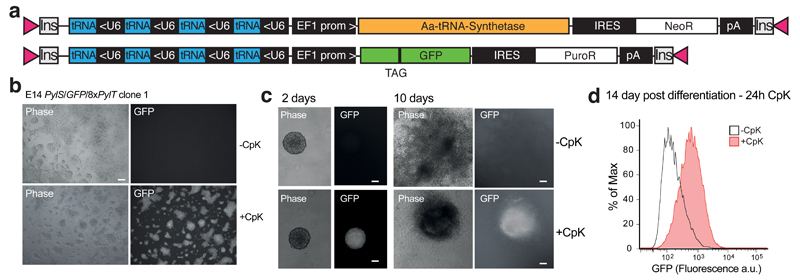
Figure 2.
PiggyBac-mediated generation and differentiation of mouse embryonic stem cell lines for unnatural amino acid mutagenesis. a PiggyBac targeting vectors used to generate stable mouse embryonic stem cell (ESC) lines from an E14 line, containing bidirectional EF1 and U6 expression cassette, internal ribosome entry site (IRES) and resistance markers (NeoR, PuroR), Insulators (Ins) and inverted terminal repeats (pink triangles). b Representative images of a clone grown in the presence or absence of 0.2 mM CpK for 48 hours. Scale bar: 100 µm. c Embryoid body differentiation protocol to produce beating cardiomyocyte aggregates in the presence (bottom) or absence (top) of 0.2 mM CpK. Scale bar: 100 µm. d FACS analysis of cells differentiated via embryoid body protocol in the absence of CpK and incubated with 0.5 mM CpK for 24 h.
To demonstrate the differentiation potential of ES cells containing the unnatural amino acid incorporation machinery, we performed a well-established embryoid body (EB) differentiation protocol, in the presence of CpK (0.2 mM), to produce cardiac myocytes from a cell line bearing 4xPylT/PylS and 4xPylT/sfGFP150TAG. After 7 days the EBs were smaller than those grown in the absence of CpK (Fig. 2c), but nonetheless formed beating cardiomyocyte assemblies (Supplementary Video 1). The differentiated tissue retained the propensity for unnatural amino acid incorporation, albeit at a lower efficiency, as judged by the appearance of sfGFP fluorescence upon addition of CpK to cardiac myocytes that were differentiated in the absence of unnatural amino acid (Fig. 2d). Manipulating mouse ESC may provide a facile in vitro strategy to derive differentiated cell types that can site-specifically incorporate unnatural amino acids.
Cell-specific transcription responses to amber suppression
We leveraged the ability to generate stable cell lines to define the general and cell-type specific effects of amber suppression on gene expression. In two mouse ESC (E14) and two MEF cell lines clones, in which 4xPylT/PylS and 4xPylT/sfGFP150TAG were integrated, we observed 1302 common and significant (P <0.005) and 36 common, and significant (P <0.005) changes in gene expression upon addition of 0.2 mM CpK (48 h), respectively (Fig. 3a,b, Supplementary Fig. 6)‥ Only 11 genes identified in ESCs were also dysregulated in MEF cells (Supplementary Fig. 6). These observations suggest that, in contrast to ES cells - which are known to be plastic30 - amber suppression has a minimal effect on gene expression in MEFs.
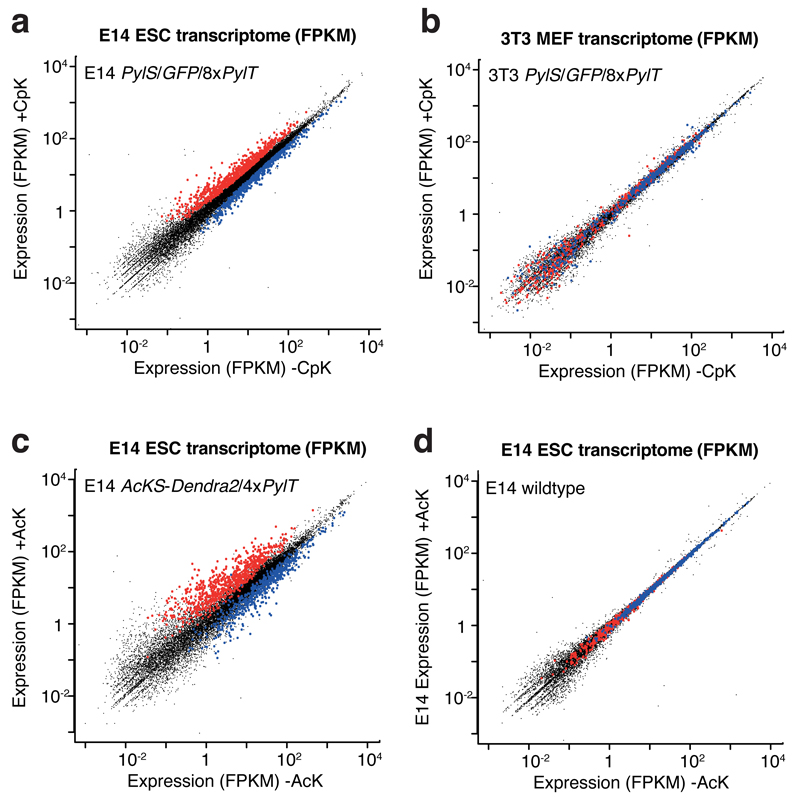
Figure 3.
RNA-Seq analysis of E14 ESC cell lines incorporating the unnatural amino acids CpK or AcK. a RNA-Seq analysis of E14 cell line bearing PylS/4xPylT and GFP150TAG/4xPylT. Whole transcriptome fragments per kilobase of exon per million fragments mapped (FPKM) values in the presence versus absence of 0.2 mM CpK for 48h are plotted. Significantly (P <0.005) up- and downregulated genes from two biological replicates are colored in red and blue, respectively. Further analysis and biological replicates are shown in Supplementary Fig. 6. b RNA-Seq analysis of 3T3 mouse embryonic fibroblast (MEF) cell line bearing PylS/4xPylT and GFP150TAG/4xPylT. Whole transcriptome FPKM values in the presence versus absence of 0.2 mM CpK for 48 h are plotted. Up- and downregulated gene sets defined in ESC (a) are colored in red and blue, respectively‥ c RNA-Seq analysis of E14 cell line bearing AcKS-TAGDendra2/4xPylT. Whole transcriptome FPKM values in the presence versus absence of 10 mM AcK for 24h are plotted. Significantly up- and downregulated genes from (a) are colored in red and blue, respectively. d RNA-Seq analysis of wild type E14 cell line, in the presence and absence of 10 mM AcK for 24h.
Strikingly, the 1302 genes that were mis-regulated in the PylS/sfGFP150TAG/8xPylT E14 lines upon addition of CpK were similarly perturbed in the 4xPylT/AcKS cell line, in the presence of Ack (compare Figs. 3a and 3c,d see also Supplementary Fig. 6). This correlation suggests that the transcriptional changes we observe in ES cells are an effect of amber suppression, and may be independent of the specific unnatural amino acid incorporated at the amber stop codon.
Intriguingly, many mis-regulated genes were not stem-cell specific, and the majority of stem-cell specific genes were unaffected (Supplementary Fig. 7), although known pluripotency factors, including Pou5f1, Nanog, Sox231, were downregulated 1.5 to 2-fold (Supplementary Fig. 7). Upregulated genes fell into a variety of categories related to development (via gene ontology)32 (Supplementary Fig. 7), and there was a significant enrichment of primary metabolic enzymes amongst the downregulated genes, suggesting that amber suppression may have a repressive effect on primary metabolism in mouse ESC, through an unknown mechanism (Supplementary Fig. 7). Notably, we do not observe upregulation of genes involved in protein folding stress responses, suggesting that amber suppression does not cause a large number of proteins to be aberrantly elongated and misfolded, consistent with previous observations in mammalian cells33.
Encoded site-specific histone acetylation in cells
We developed a two-step strategy to generate a panel of cell lines with well matched unnatural amino acid incorporation efficiency, that incorporate an unnatural amino acid at distinct positions in a target protein or proteins (Supplementary Fig. 8). We used this strategy to generate a series of histone H3 variants in which acetyl-lysine is co-translationally incorporated (Fig. 4a, Supplementary Fig. 8). In the first step, we co-transfected E14 cells with the 4xPylT/AcKS cassette and PiggyBac transposase, and selected with 10 μg mL-1 Puromycin. We isolated a single 4xPylT/AcKS clone by FACS, using a Dendra2 fluorescent reporter fused in-frame behind the AcKS ORF, separated by an amber codon, as a measure for efficient incorporation of acetyl-lysine at amber codons (Fig. 4a). Incorporation was dose-dependent, and maximal at 10 mM AcK (Supplementary Fig. 9). In the second step, we co-transfected an E14 AcKS/4xPylT cell line with the PiggyBac transposase plasmid, and a 4xPylT/histone H3.2(XXTAG)-HA or 4xPylT/histone H3.3(XXTAG)-HA cassette, where XX indicates the position in the histone gene targeted for conversion of a lysine codon to TAG. We selected with 10 μg mL-1 Puromycin and 2 mg mL-1 G418 and isolated polyclonal pools that are closely related to the parental cell line and to each other.
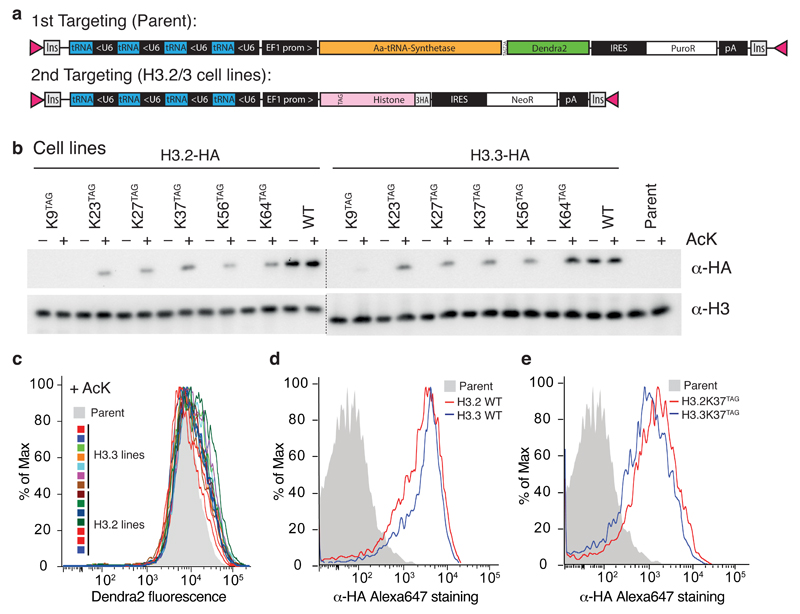
Figure 4.
Site-specific incorporation of acetyl-lysine into the histone H3 in ESC. a PiggyBac targeting vectors used to generate stable mouse ESC lines, Insulators (Ins) , inverted terminal repeats (pink triangles). b Western blot panel of histone cell lines bearing AcKS-TAGDendra2/4xPylT and H3.2, H3.2(XXTAG)-HA, H3.3 or H3.3 (XXTAG) - designated WT or KXXTAG in the figure, respectively - with a C-terminal triple HA-tag and 4xPylT. Synthetic histone expression is detected in acid-extracted chromatin by western blot against HA tag, an endogenous histone H3 loading control is shown. Expression of histone H3.2 and H3.3 genes containing TAG codons is dependent on addition of AcK (10 mM for 24 hours). c Amber suppression efficiency in all cell lines from panel b, as measured by FACS analysis of Dendra2 fluorescence, in the presence of 10 mM AcK d, e Expression of HA-tagged Histone H3 without (d) and with amber codon (e) in single cells as measured by fluorescent staining with anti-HA Alexa647 antibody and FACS analysis. Data for the entire panel of H3 variants is shown in Supplementary Fig. 10.
Western blot against a C-terminal HA tag demonstrates AcK-dependent read through of the amber codon in H3(XXTAG)-HA containing cell lines (Fig. 4b). Although H3.3-HA was expressed continuously whereas expression of H3.3(XXTAG)-HA was induced by addition of AcK for 24 h, the levels of H3.3 produced from H3.3(XXTAG)-HA reached 50 to 100% of the H3.3-HA control in most positions (Fig. 4b). All resulting cell lines showed a defined and comparable level of acetyl-lysine incorporation at an amber stop codon (Fig. 4c), and histone expression levels were comparable between H3.2 and H3.3 pairs with the TAG codon at the same position (Fig. 4d,e, Supplementary Fig. 10). However, there was some variability in the level of acetyl lysine incorporation at distinct sites in histone H3 (Fig. 4b), consistent with context effects in amber suppression efficiency 34, and we observed minimal incorporation for H3.3(K9TAG)-HA.
Genetically directed acetylation in chromatin
We focused on detecting H3.3 acetylation in chromatin, since, unlike H3.2, H3.3 undergoes dynamic, replication independent, deposition35. H3.3-HA from each expressed H3.3(XXTAG)-HA variant was deposited into chromatin, as judged by anti-H3.3 and anti-HA immunoblot (Supplementary Fig. 11). The amount of transgenic H3.3 produced in the 24 h window following addition of AcK to cells was a fraction of the total H3.3 (Supplementary Fig. 11); this is observed in many histone transgenesis experiments and is a consequence of the large number of endogenous histone genes and tight control of cellular histone protein levels36. We directly detected K37ac and K56ac on histone H3.3-HA in chromatin-extracted histones (Supplementary Fig. 11), as well as in immunoprecipitated soluble and nucleosomal H3.3-HA (Supplementary Fig. 11). We also detected K27ac and K64ac on HA-tagged histones immunoprecipitated from nucleosomal histones (Supplementary Fig. 11). Notably, we only detected K37ac, K56ac and K64ac from transgenic H3.3-HA when acetylation was genetically encoded at these sites, showing that genetically encoded acetylation produces higher acetylation levels than endogenous enzymes at these sites. In contrast, we detected K27ac on all transgenic H3.3-HA, consistent with enzymes in mouse ESCs efficiently acetylating K27. Transient expression experiments in HEK293T confirm that H3K27ac can be specifically incorporated into chromatin and detected using the anti H3K27ac antibody (Supplementary Fig. 11). We note that less K27ac was detected with the anti K27ac antibody when the histones were pre-acetylated at K56, suggesting a possible negative crosstalk of acetylations on histone H3. We did not detect K9ac or K23ac on the tagged histone species, either because the antibodies used did not provide the necessary sensitivity (despite their apparent ability to detect the abundant K9ac, K23ac acetylation, respectively, on the endogenous histones), and/or because the encoded acetylation at those sites was rapidly lost through deacetylation. In the future, it will be interesting to investigate genetically encoding non-hydrolyzable analogs of post-translational modifications.
Consistent with the role of K56ac in histone deposition in human cells37, the majority of H3.3 K56ac-HA is in the chromatin-bound fraction (Supplementary Fig. 11). The K56ac signal resulting from H3.3(K56TAG)-HA expression was easily detectable even in the presence of the bulk endogenous histones, suggesting that the encoded acetylation is retained in H3.3K56ac-HA, and that endogenous levels of K56ac are extremely low38, 39.
Encoded H3 acetylation regulates transcription
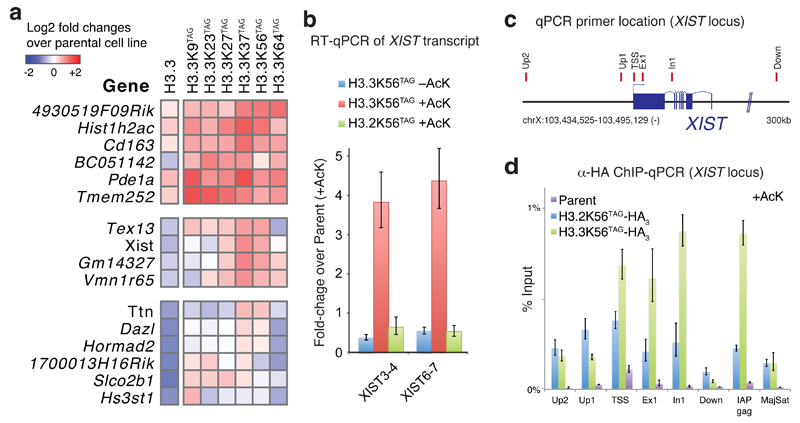
To investigate changes in gene expression resulting from genetically encoding acetylation in H3.3 we carried out mRNA-Seq on all H3.3 cell lines. We identified sixteen candidates genes in H3.3(XXTAG)-HA cell lines (Fig. 5a) that showed 1) significant (P <0.005) up-regulation over the AcKS/4xPylT parental cell line (which provides a control for the effects of amber suppression with AcK), 2) significant (P <0.005) and more than 2-fold up-regulation over the AcKS/H3.3/8xPylT cell line (which provides a control for amber suppression and the effects of expressing the non-acetlyated histone- in the H3.3(XXTAG)-HA cell lines). As H3.3 with encoded acetylation provides a fraction of H3 in the cell, the genes identified may be a subset of those regulated by H3 acetylation. We compared the expression levels in all H3.3(XXTAG)-HA cell lines and the H3.3-HA control cell line to the parental cell line (Fig.5a, see Supplementary Fig. 12 for absolute expression counts and a wider range of controls). Notably, a subset of genes identified, including TEX13, XIST, GM14327, VMN165, were particularly strongly induced in H3.3 (K37TAG)-HA and H3.3 (K56TAG)-HA cell lines, for which we have confirmed the site-specific acetylation on H3.3 in chromatin (). We thus focused on the XIST (X-inactive specific transcript) gene, which gives rise to the non-coding Xist RNA required for X inactivation during development40. We confirmed by RT-qPCR that XIST expression in the H3.3 K27TAG, K37TAG and K56TAG cell lines were higher than in the control H3.3 cell line, in the presence of 10mM AcK (Fig. 5b, Supplementary Fig. 12 ). Furthermore, we did not observe XIST induction in H3.2 K56TAG cell lines, in the presence of AcK (Fig.5b). XIST induction increased with H3.3 K56Ac expression levels (Supplementary Fig.12). The proteins expressed from H3.3-HA, H3.3 (K37TAG)-HA, and H3.3 (K56TAG)-HA were specifically enriched at the transcription start site, and coding region of the XIST gene, as judged by anti-HA ChIP (Fig. 5c,d, Supplementary Fig. 13).
Figure 5.
Genetically encoded, site-specific histone acetylation in chromatin regulates gene expression. a RNA-Seq analysis of E14 ESC cell lines bearing AcKS-Dendra2/4xPylT and H3.3(XXTAG)/4xPylT. Heatmap of genes that were found to be significantly ( P <0.005) upregulated in at least one H3.3(XXTAG) cell line, with respect to both the Parental AcKS-Dendra2/4xPylT and the matched control AcKS-Dendra2/H3.3/8xPylT cell lines. b Reverse transcription-quantitative PCR (RT-qPCR) analysis of noncoding XIST RNA expression in H3.3(56TAG) cell lines, with and without addition of 10 mM AcK (24 h) to the medium, and a matched H3.2(56TAG) control cell line, showing that XIST upregulation is dependent on K56 acetylation of histone H3.3. Error bars indicate 95% confidence interval of three technical replicates. c locations of qPCR primers used in (d). d. ChIP assay validating variant-specific incorporation of H3.3K56TAG-HA3, in the presence of 10 mM AcK for 24 h, at the XIST locus at primer sites tiling the XIST locus. ChIP assay was performed using α-HA magnetic beads. Error bars indicate 95% confidence interval of three technical replicates.
Discussion
The ability to rapidly generate stable cell lines with defined amber suppression efficiency will be broadly enabling for unnatural amino acid mutagenesis. We anticipate that our approach for creating cell populations that have well matched amber suppression efficiency and express different mutants of proteins of interest will prove particularly useful. A key application of stable cell lines for unnatural amino acid mutagenesis will be in the coupling of precise molecular, spatial and temporal perturbations, effected by site-specific unnatural amino acid incorporation, to measurements of cell-, organism- or ecosystem- wide consequences. Such approaches will require careful control for the effects of amber suppression itself, and we have demonstrated that these effects are cell-type specific and require direct measurement in control experiments.
The approach we have reported for genetically installing post-translational modifications, (and by extension their non-hydrolyzable analogs) into chromatin, and a recent approach that used protein trans-splicing to assemble a ubiquitinated histone in chromatin within isolated cell nuclei36, are orthogonal to enzymatic modification. Orthogonal routes to chromatin modification allow the consequences of post-translational modifications at defined sites on a histone to be defined, without the pleiotropic effects of modifying enzymes at other sites on histones or on other proteins. In the future, orthogonal routes to chromatin modification may be extended to facilitate the site-specific and orthogonal modification of a histone deposited at defined genomic loci. Given the potential of histone modifications to control epigenetic phenomena we anticipate that orthogonal approaches to chromatin modification may facilitate insight into whether specific modifications cause epigenetic change. Orthogonal approaches to chromatin modification may form a basis for ‘synthetic epigenetics’ in which precise (natural or unnatural) modification states are programmed into cells to control cell and organism fate.
Online Methods
Antibodies
Following primary antibodies were used for western blotting: anti-GFP (sc-9996 , Santa Cruz Biotechnologies), anti-mCherry (ABE3523, SourceBiosciences), anti-FLAG M2 (SIGMA), a-Actin (#4967, Cell Signaling Technology), a-H3 (ab1791, Abcam), a-H3K56ac (07-677, Millipore), a-H3K37ac (61587, ActiveMotif). a-H3K64ac (ActiveMotif 39545, gift from Robert Schneider), a-H3K27ac (ab4729, Abcam).
ESC culture
E14 ESC41 were acquired from Babraham Institute, Cambridge, UK. ESCs were cultured under standard conditions (10% CO2, 90% Humidity, 37°C) in KO-DMEM (LifeTechnologies #10829-018), 2 mM Glutamax (Gibco), 0.1 mM Non-essential amino acids (Gibco), 15% ES grade FBS (Gibco), 0.1 mM beta-mercaptoethanol, and leukemia inhibitory factor (LIF, Millipore ESG1107). ESCs were routinely tested for mycoplasma.
Transient transfection
Cells were transfected with TransIT-293 (HEK293) or TransIT-2020 (all other cell types) according to the manufacturers protocol. Amino acid was added at transfection and cells were incubated for 24-48 hours.
Stable cell line generation
Cells were transfected in a 6-well plate with TransIT-293 (HEK293) or TransIT-2020 (all other cell types) with 2.5 μg Plasmid DNA, PiggyBac targeting plasmid(s) and Super PiggyBac Transposase Plasmid (SBI) at a ratio of 5:1. After 48 hours, cells were split 1:6 into a 6-well plate each and selection antibiotic was added. Optimal antibiotic concentrations (5-10 μg mL-1 Puromycin, 500-2 mg mL-1 G418) were experimentally determined. Cells were grown for at least 7 days under selection for polyclonal pools or FACS sorted into wells of single cells after 3-5 days of selection for clonal cell lines. Experiments were performed at early passages (< 10 for polyclonal pools, < 20 for clonal cell lines) Antibiotic resistance was periodically confirmed in long-term cultures.
Western blotting of nucleocytoplasmic extracts
Cells were seeded in a 24 plate (2x105 per well) and incubated with or without amino acid at indicated concentration for the indicated time. Cells were washed with PBS and lysed by adding ice-cold 100 μL RIPA (Sigma) supplemented with Protease Inhibitor Cocktail (Roche). After 10 min incubation on ice, lysates were cleared by centrifugation (10 min x 24k r.p.m., 4 °C) and 20 μL 6× SDS sample buffer was added, and samples were incubated at 95 °C for 10 min. 10 μL were loaded per lane on a NuPAGE gel. Proteins were transferred using an iBlot onto a Nitrocellulose membrane, blocked with 5% milk powder/TBST and incubated with primary followed by secondary antibodies (Bio-Rad Goat Anti-Rabbit IgG (H+L)-HRP Conjugate #1721019; Goat Anti-Mouse IgG (H + L)-HRP Conjugate #1706516). Primary antibody was typically incubated over night at 4°C in 5% Milk/TBST, whereas secondary was incubated in TBST for 1h at RT. HRP signal was visualised using Illuminata Forte (Millipore) substrate and a ChemiDoc digital imager (Bio-Rad). For acetylation-specific antibodies, milk was replaced with purified BSA (SIGMA A9418). Acetylation-specific antibodies were used at 1:500 to 1:5,000 dilutions with consistent results.
FACS analysis of live and fixed cells
Cells were trypsinised and triturated to dissociate aggregates, and taken up in full medium before centrifugation. Cells centrifuged at 500g x 5 min, washed in PBS/5%FBS, and resuspended in PBS for analysis. For fixations, cell suspension was treated with 1% Paraformaldehyde for 10 min at RT and quenched with full medium, followed by centrifugation (500g x 5 min), and resuspension in PBS for analysis. For immunofluorescent staining, cells were permeabilised in PBS/5%FBS/0.01% Triton X-100 and incubated with fluorescent dye-conjugated antibody for 1 hour followed by 3 washes with PBS and centrifugation (500g x 5 min), and resuspended in PBS for analysis.
Western blotting of chromatin extracts
Cells were grown in 24 well plate with or without amino acid at indicated concentration for the indicated time. Cells were washed with PBS and lysed by adding ice-cold 500 μL Lysis buffer (1 % Triton X-100 in PBS) supplemented with Protease Inhibitor Cocktail (Roche). After 10 min incubation on ice, nuclei were pelleted by centrifugation (10 min x 10k r.p.m., 4 °C), and resuspended well in 100μL 0.4 N H2SO4. Suspension was rocked at RT for 1h and insoluble material was removed by centrifugation (10 min x 24,000 r.p.m., 4 °C). Supernatant was neutralised with 6× SDS sample buffer/1M Tris base. SDS page and western blotting was performed.
Analysis of nucleosomal histones
Cells were trypsinised from two T125 tissue culture plates (~2x107 cells) and washed twice with DMEM/10%FBS and PBS. For isolating nuclei, cells were resuspended in ice-cold hypotonic buffer (10 mM HEPES pH 7.5, 20 mM NaCl, 0.01% Triton X-100). Nuclei were pelleted, washed with hypotonic lysis buffer and pelleted again, and resuspended in digestion buffer (10 mM HEPES pH 7.5, 20 mM NaCl, 0.01% Triton X-100). MNase was added and nuclei were incubated at 37 °C for 10 min. Nuclei were lysed in RIPA buffer supplemented with 10mM EDTA and insoluble material was removed by centrifugation. Supernatant was used for immunprecipitation of tagged histone H3 using anti-HA magnetic beads (Pierce # 88836). Digestion to mononucleosomes was confirmed by purification of a DNA sample and agarose gel electrophoresis.
RNA-Seq
RNA-Seq was carried out from two biological replicates for each clone/condition, in two independent clones for ESC and MEF cells. RNEasy Kit (Qiagen) was used to isolate total RNA from one 6-well plate per replicate. RNA was quality controlled using a Bioanalyzer (RIN>9). Library generation and sequencing was carried out using the Illumina Truseq Kit at Bejing Genomics Institute (BGI) Hong Kong. Reads were aligned using TopHat2/Bowtie2 to the mouse genome (build mm10). RPKM and differential expression values were calculated using CuffLinks/CuffDiff. Data analysis was performed using R and cummeRbund42. cummeRbund was used to confirm sample quality and comparable dispersion across samples. Gene ontology analysis was performed using geneontology.org 32. Statistically significant changes (P <0.005) were extracted from CuffDiff2, and are based on a Poisson model distribution corrected for both count uncertainty and count overdispersion43.
RT-qPCR
Total RNA was reverse transcribed using the Superscript III Kit (Life Technologies) and random d(T)20. qPCR was carried out using Power SYBR Green Master Mix (Life Technologies) and IDT PrimeTime Primers (Xist 3–4, Mm.PT.58.10791082; Xist 6–7,Mm.PT.58.43610994; Actb, Mm.PT.58.33540333).
ChIP-qPCR
ChIP was performed essentially as described by the Zhao lab44, 45. ES cells were trypsinized, resuspended in full medium, pelleted and washed twice with ice-cold PBS before flash-freezing. Frozen cells were resuspended in MNase digestion buffer and protocol was carried out as described using anti-HA magnetic beads (Pierce # 88836).
Plasmid Sequences
pPiggyBac 4xU6-PylT(U25C)/EF1-MmPylS-IRES-Puro
ACTCTTCCTTTTTCAATATTATTGAAGCATTTATCAGGGTTATTGTCTCATGAGCGGATACATATTTGAATGTATTTAGAAAAATAAACAAATAGGGGTTCCGCGCACATTTCCCCGAAAAGTGCCACCTAAATTGTAAGCGTTAATATTTTGTTAAAATTCGCGTTAAATTTTTGTTAAATCAGCTCATTTTTTAACCAATAGGCCGAAATCGGCAAAATCCCTTATAAATCAAAAGAATAGACCGAGATAGGGTTGAGTGTTGTTCCAGTTTGGAACAAGAGTCCACTATTAAAGAACGTGGACTCCAACGTCAAAGGGCGAAAAACCGTCTATCAGGGCGATGGCCCACTACGTGAACCATCACCCTAATCAAGTTTTTTGGGGTCGAGGTGCCGTAAAGCACTAAATCGGAACCCTAAAGGGAGCCCCCGATTTAGAGCTTGACGGGGAAAGCCGGCGAACGTGGCGAGAAAGGAAGGGAAGAAAGCGAAAGGAGCGGGCGCTAGGGCGCTGGCAAGTGTAGCGGTCACGCTGCGCGTAACCACCACACCCGCCGCGCTTAATGCGCCGCTACAGGGCGCGTCCCATTCGCCATTCAGGCTGCGCAACTGTTGGGAAGGGCGATCGGTGCGGGCCTCTTCGCTATTACGCCAGCTGGCGAAAGGGGGATGTGCTGCAAGGCGATTAAGTTGGGTAACGCCAGGGTTTTCCCAGTCACGACGTTGTAAAACGACGGCCAGTGAGCGCGCCTCGTTCATTCACGTTTTTGAACCCGTGGAGGACGGGCAGACTCGCGGTGCAAATGTGTTTTACAGCGTGATGGAGCAGATGAAGATGCTCGACACGCTGCAGAACACGCAGCTAGATTAACCCTAGAAAGATAATCATATTGTGACGTACGTTAAAGATAATCATGCGTAAAATTGACGCATGTGTTTTATCGGTCTGTATATCGAGGTTTATTTATTAATTTGAATAGATATTAAGTTTTATTATATTTACACTTACATACTAATAATAAATTCAACAAACAATTTATTTATGTTTATTTATTTATTAAAAAAAAACAAAAACTCAAAATTTCTTCTATAAAGTAACAAAACTTTTATGAGGGACAGCCCCCCCCCAAAGCCCCCAGGGATGTAATTACGTCCCTCCCCCGCTAGGGGGCAGCAGCGAGCCGCCCGGGGCTCCGCTCCGGTCCGGCGCTCCCCCCGCATCCCCGAGCCGGCAGCGTGCGGGGACAGCCCGGGCACGGGGAAGGTGGCACGGGATCGCTTTCCTCTGAACGCTTCTCGCTGCTCTTTGAGCCTGCAGACACCTGGGGGGATACGGGGAAAAGGCCTCCAAGGCCACTAGGAAAAACCGCACTTGTCCGGAAACCCCGGGAATCTAACCCGGCTGAACGGATTTAGAGTCCGTTCGATCTACATGATCAGGTTTCCGGTGTTTCGTCCTTTCCACAAGATATATAAAGCCAAGAAATCGAAATACTTTCAAGTTACGGTAAGCATATGATAGTCCATTTTAAAACATAATTTTAAAACTGCAAACTACCCAAGAAATTATTACTTTCTACGTCACGTATTTTGTACTAATATCTTTGTGTTTACAGTCAAATTAATTCTAATTATCTCTCTAACAGCCTTGTATCGTATATGCAAATATGAAGGAATCATGGGAAATAGGCCCTCTTCCTGCCCAACTAGGAAAAACCGCACTTGTCCGGAAACCCCGGGAATCTAACCCGGCTGAACGGATTTAGAGTCCGTTCGATCTACATGATCAGGTTTCCGGTGTTTCGTCCTTTCCACAAGATATATAAAGCCAAGAAATCGAAATACTTTCAAGTTACGGTAAGCATATGATAGTCCATTTTAAAACATAATTTTAAAACTGCAAACTACCCAAGAAATTATTACTTTCTACGTCACGTATTTTGTACTAATATCTTTGTGTTTACAGTCAAATTAATTCTAATTATCTCTCTAACAGCCTTGTATCGTATATGCAAATATGAAGGAATCATGGGAAATAGGCCCTCTTCCTGCCCAACTAGGAAAAACCGCACTTGTCCGGAAACCCCGGGAATCTAACCCGGCTGAACGGATTTAGAGTCCGTTCGATCTACATGATCAGGTTTCCGGTGTTTCGTCCTTTCCACAAGATATATAAAGCCAAGAAATCGAAATACTTTCAAGTTACGGTAAGCATATGATAGTCCATTTTAAAACATAATTTTAAAACTGCAAACTACCCAAGAAATTATTACTTTCTACGTCACGTATTTTGTACTAATATCTTTGTGTTTACAGTCAAATTAATTCTAATTATCTCTCTAACAGCCTTGTATCGTATATGCAAATATGAAGGAATCATGGGAAATAGGCCCTCTTCCTGCCCAACTAGGAAAAACCGCACTTGTCCGGAAACCCCGGGAATCTAACCCGGCTGAACGGATTTAGAGTCCGTTCGATCTACATGATCAGGTTTCCGGTGTTTCGTCCTTTCCACAAGATATATAAAGCCAAGAAATCGAAATACTTTCAAGTTACGGTAAGCATATGATAGTCCATTTTAAAACATAATTTTAAAACTGCAAACTACCCAAGAAATTATTACTTTCTACGTCACGTATTTTGTACTAATATCTTTGTGTTTACAGTCAAATTAATTCTAATTATCTCTCTAACAGCCTTGTATCGTATATGCAAATATGAAGGAATCATGGGAAATAGGCCCTCTTCCTGCCCAACTAGTGGATCTGCGATCGCTCCGGTGCCCGTCAGTGGGCAGAGCGCACATCGCCCACAGTCCCCGAGAAGTTGGGGGGAGGGGTCGGCAATTGAACGGGTGCCTAGAGAAGGTGGCGCGGGGTAAACTGGGAAAGTGATGTCGTGTACTGGCTCCGCCTTTTTCCCGAGGGTGGGGGAGAACCGTATATAAGTGCAGTAGTCGCCGTGAACGTTCTTTTTCGCAACGGGTTTGCCGCCAGAACACAGCTGAAGCTTCGAGGGGCTCGCATCTCTCCTTCACGCGCCCGCCGCCCTACCTGAGGCCGCCATCCACGCCGGTTGAGTCGCGTTCTGCCGCCTCCCGCCTGTGGTGCCTCCTGAACTGCGTCCGCCGTCTAGGTAAGTTTAAAGCTCAGGTCGAGACCGGGCCTTTGTCCGGCGCTCCCTTGGAGCCTACCTAGACTCAGCCGGCTCTCCACGCTTTGCCTGACCCTGCTTGCTCAACTCTACGTCTTTGTTTCGTTTTCTGTTCTGCGCCGTTACAGATCCAAGCTGTGACCGGCGCCTACTCTAGAGCTAGCGTTTAAACTTAAGAAGCTTGCCACCATGGACTACAAGGACGACGACGACAAGATGGACAAGAAGCCCCTGAACACCCTGATCAGCGCCACAGGACTGTGGATGTCCAGAACCGGCACCATCCACAAGATCAAGCACCACGAGGTGTCCCGGTCCAAAATCTACATCGAGATGGCCTGCGGCGATCACCTGGTCGTCAACAACAGCAGAAGCAGCCGGACAGCCAGAGCCCTGCGGCACCACAAGTACAGAAAGACCTGCAAGCGGTGCAGAGTGTCCGACGAGGACCTGAACAAGTTCCTGACCAAGGCCAACGAGGACCAGACCAGCGTGAAAGTGAAGGTGGTGTCCGCCCCCACCCGGACCAAGAAAGCCATGCCCAAGAGCGTGGCCAGAGCCCCCAAGCCCCTGGAAAACACCGAAGCCGCTCAGGCCCAGCCCAGCGGCAGCAAGTTCAGCCCCGCCATCCCCGTGTCTACCCAGGAAAGCGTCAGCGTCCCCGCCAGCGTGTCCACCAGCATCTCTAGCATCTCAACCGGCGCCACAGCTTCTGCCCTGGTCAAGGGCAACACCAACCCCATCACCAGCATGTCTGCCCCTGTGCAGGCCTCTGCCCCAGCCCTGACCAAGTCCCAGACCGACCGGCTGGAAGTGCTCCTGAACCCCAAGGACGAGATCAGCCTGAACAGCGGCAAGCCCTTCCGGGAGCTGGAAAGCGAGCTGCTGAGCCGGCGGAAGAAGGACCTCCAGCAAATCTACGCCGAGGAACGGGAGAACTACCTGGGCAAGCTGGAAAGAGAGATCACCCGGTTCTTCGTGGACCGGGGCTTCCTGGAAATCAAGAGCCCCATCCTGATCCCCCTGGAGTACATCGAGCGGATGGGCATCGACAACGACACCGAGCTGAGCAAGCAGATTTTCCGGGTGGACAAGAACTTCTGCCTGCGGCCCATGCTGGCCCCCAACCTGTACAACTACCTGCGGAAACTGGATCGCGCTCTGCCCGACCCCATCAAGATTTTCGAGATCGGCCCCTGCTACCGGAAAGAGAGCGACGGCAAAGAGCACCTGGAAGAGTTTACAATGCTGAACTTTTGCCAGATGGGCAGCGGCTGCACCAGAGAGAACCTGGAATCCATCATCACCGACTTTCTGAACCACCTGGGGATCGACTTCAAGATCGTGGGCGACAGCTGCATGGTGTACGGCGACACCCTGGACGTGATGCACGGCGACCTGGAACTGTCTAGCGCCGTCGTGGGACCCATCCCTCTGGACCGGGAGTGGGGCATCGATAAGCCCTGGATCGGAGCCGGCTTCGGCCTGGAACGGCTGCTGAAAGTCAAGCACGACTTTAAGAACATCAAGCGGGCTGCCAGAAGCGAGAGCTACTACAACGGCATCAGCACCAACCTGTGATGAGGATCCGCGGCCGCGCCCCTCTCCCTCCCCCCCCCCTAACGTTACTGGCCGAAGCCGCTTGGAATAAGGCCGGTGTGCGTTTGTCTATATGTTATTTTCCACCATATTGCCGTCTTTTGGCAATGTGAGGGCCCGGAAACCTGGCCCTGTCTTCTTGACGAGCATTCCTAGGGGTCTTTCCCCTCTCGCCAAAGGAATGCAAGGTCTGTTGAATGTCGTGAAGGAAGCAGTTCCTCTGGAAGCTTCTTGAAGACAAACAACGTCTGTAGCGACCCTTTGCAGGCAGCGGAACCCCCCACCTGGCGACAGGTGCCTCTGCGGCCAAAAGCCACGTGTATAAGATACACCTGCAAAGGCGGCACAACCCCAGTGCCACGTTGTGAGTTGGATAGTTGTGGAAAGAGTCAAATGGCTCTCCTCAAGCGTATTCAACAAGGGGCTGAAGGATGCCCAGAAGGTACCCCATTGTATGGGATCTGATCTGGGGCCTCGGTGCACATGCTTTACATGTGTTTAGTCGAGGTTAAAAAAACGTCTAGGCCCCCCGAACCACGGGGACGTGGTTTTCCTTTGAAAAACACGATGATAATATGGCCACAACCATGGCGTCCGGAATGACCGAGTACAAGCCCACGGTGCGCCTCGCCACCCGCGACGACGTCCCCAGGGCCGTACGCACCCTCGCCGCCGCGTTCGCCGACTACCCCGCCACGCGCCACACCGTCGATCCGGACCGCCACATCGAGCGGGTCACCGAGCTGCAAGAACTCTTCCTCACGCGCGTCGGGCTCGACATCGGCAAGGTGTGGGTCGCGGACGACGGCGCCGCGGTGGCGGTCTGGACCACGCCGGAGAGCGTCGAAGCGGGGGCGGTGTTCGCCGAGATCGGCCCGCGCATGGCCGAGTTGAGCGGTTCCCGGCTGGCCGCGCAGCAACAGATGGAAGGCCTCCTGGCGCCGCACCGGCCCAAGGAGCCCGCGTGGTTCCTGGCCACCGTCGGCGTCTCGCCCGACCACCAGGGCAAGGGTCTGGGCAGCGCCGTCGTGCTCCCCGGAGTGGAGGCGGCCGAGCGCGCCGGGGTGCCCGCCTTCCTGGAGACCTCCGCGCCCCGCAACCTCCCCTTCTACGAGCGGCTCGGCTTCACCGTCACCGCCGACGTCGAGGTGCCCGAAGGACCGCGCACCTGGTGCATGACCCGCAAGCCCGGTGCCTAGGTCGACAATCAACCTCTGGATTACAAAATTTGTGAAAGATTGACTGGTATTCTTAACTATGTTGCTCCTTTTACGCTATGTGGATACGCTGCTTTAATGCCTTTGTATCATGTTAACTTGTTTATTGCAGCTTATAATGGTTACAAATAAAGCAATAGCATCACAAATTTCACAAATAAAGCATTTTTTTCACTGCATTCTAGTTGTGGTTTGTCCAAACTCATCAATGTATCTTATCATGTCTGGAATTGACTCAAATGATGTCAATTAGTCTATCAGAAGCTATCTGGTCTCCCTTCCGGGGGACAAGACATCCCTGTTTAATATTTAAACAGCAGTGTTCCCAAACTGGGTTCTTATATCCCTTGCTCTGGTCAACCAGGTTGCAGGGTTTCCTGTCCTCACAGGAACGAAGTCCCTAAAGAAACAGTGGCAGCCAGGTTTAGCCCCGGAATTGACTGGATTCCTTTTTTAGGGCCCATTGGTATGGCTTTTTCCCCGTATCCCCCCAGGTGTCTGCAGGCTCAAAGAGCAGCGAGAAGCGTTCAGAGGAAAGCGATCCCGTGCCACCTTCCCCGTGCCCGGGCTGTCCCCGCACGCTGCCGGCTCGGGGATGCGGGGGGAGCGCCGGACCGGAGCGGAGCCCCGGGCGGCTCGCTGCTGCCCCCTAGCGGGGGAGGGACGTAATTACATCCCTGGGGGCTTTGGGGGGGGGCTGTCCCTGATATCTATAACAAGAAAATATATATATAATAAGTTATCACGTAAGTAGAACATGAAATAACAATATAATTATCGTATGAGTTAAATCTTAAAAGTCACGTAAAAGATAATCATGCGTCATTTTGACTCACGCGGTCGTTATAGTTCAAAATCAGTGACACTTACCGCATTGACAAGCACGCCTCACGGGAGCTCCAAGCGGCGACTGAGATGTCCTAAATGCACAGCGACGGATTCGCGCTATTTAGAAAGAGAGAGCAATATTTCAAGAATGCATGCGTCAATTTTACGCAGACTATCTTTCTAGGGTTAATCTAGCTGCATCAGGATCATATCGTCGGGTCTTTTTTCCGGCTCAGTCATCGCCCAAGCTGGCGCTATCTGGGCATCGGGGAGGAAGAAGCCCGTGCCTTTTCCCGCGAGGTTGAAGCGGCATGGAAAGAGTTTGCCGAGGATGACTGCTGCTGCATTGACGTTGAGCGAAAACGCACGTTTACCATGATGATTCGGGAAGGTGTGGCCATGCACGCCTTTAACGGTGAACTGTTCGTTCAGGCCACCTGGGATACCAGTTCGTCGCGGCTTTTCCGGACACAGTTCCGGATGGTCAGCCCGAAGCGCATCAGCAACCCGAACAATACCGGCGACAGCCGGAACTGCCGTGCCGGTGTGCAGATTAATGACAGCGGTGCGGCGCTGGGATATTACGTCAGCGAGGACGGGTATCCTGGCTGGATGCCGCAGAAATGGACATGGATACCCCGTGAGTTACCCGGCGGGCGCGCTTGGCGTAATCATGGTCATAGCTGTTTCCTGTGTGAAATTGTTATCCGCTCACAATTCCACACAACATACGAGCCGGAAGCATAAAGTGTAAAGCCTGGGGTGCCTAATGAGTGAGCTAACTCACATTAATTGCGTTGCGCTCACTGCCCGCTTTCCAGTCGGGAAACCTGTCGTGCCAGCTGCATTAATGAATCGGCCAACGCGCGGGGAGAGGCGGTTTGCGTATTGGGCGCTCTTCCGCTTCCTCGCTCACTGACTCGCTGCGCTCGGTCGTTCGGCTGCGGCGAGCGGTATCAGCTCACTCAAAGGCGGTAATACGGTTATCCACAGAATCAGGGGATAACGCAGGAAAGAACATGTGAGCAAAAGGCCAGCAAAAGGCCAGGAACCGTAAAAAGGCCGCGTTGCTGGCGTTTTTCCATAGGCTCCGCCCCCCTGACGAGCATCACAAAAATCGACGCTCAAGTCAGAGGTGGCGAAACCCGACAGGACTATAAAGATACCAGGCGTTTCCCCCTGGAAGCTCCCTCGTGCGCTCTCCTGTTCCGACCCTGCCGCTTACCGGATACCTGTCCGCCTTTCTCCCTTCGGGAAGCGTGGCGCTTTCTCATAGCTCACGCTGTAGGTATCTCAGTTCGGTGTAGGTCGTTCGCTCCAAGCTGGGCTGTGTGCACGAACCCCCCGTTCAGCCCGACCGCTGCGCCTTATCCGGTAACTATCGTCTTGAGTCCAACCCGGTAAGACACGACTTATCGCCACTGGCAGCAGCCACTGGTAACAGGATTAGCAGAGCGAGGTATGTAGGCGGTGCTACAGAGTTCTTGAAGTGGTGGCCTAACTACGGCTACACTAGAAGGACAGTATTTGGTATCTGCGCTCTGCTGAAGCCAGTTACCTTCGGAAAAAGAGTTGGTAGCTCTTGATCCGGCAAACAAACCACCGCTGGTAGCGGTGGTTTTTTTGTTTGCAAGCAGCAGATTACGCGCAGAAAAAAAGGATCTCAAGAAGATCCTTTGATCTTTTCTACGGGGTCTGACGCTCAGTGGAACGAAAACTCACGTTAAGGGATTTTGGTCATGAGATTATCAAAAAGGATCTTCACCTAGATCCTTTTAAATTAAAAATGAAGTTTTAAATCAATCTAAAGTATATATGAGTAAACTTGGTCTGACAGTTACCAATGCTTAATCAGTGAGGCACCTATCTCAGCGATCTGTCTATTTCGTTCATCCATAGTTGCCTGACTCCCCGTCGTGTAGATAACTACGATACGGGAGGGCTTACCATCTGGCCCCAGTGCTGCAATGATACCGCGAGACCCACGCTCACCGGCTCCAGATTTATCAGCAATAAACCAGCCAGCCGGAAGGGCCGAGCGCAGAAGTGGTCCTGCAACTTTATCCGCCTCCATCCAGTCTATTAATTGTTGCCGGGAAGCTAGAGTAAGTAGTTCGCCAGTTAATAGTTTGCGCAACGTTGTTGCCATTGCTACAGGCATCGTGGTGTCACGCTCGTCGTTTGGTATGGCTTCATTCAGCTCCGGTTCCCAACGATCAAGGCGAGTTACATGATCCCCCATGTTGTGCAAAAAAGCGGTTAGCTCCTTCGGTCCTCCGATCGTTGTCAGAAGTAAGTTGGCCGCAGTGTTATCACTCATGGTTATGGCAGCACTGCATAATTCTCTTACTGTCATGCCATCCGTAAGATGCTTTTCTGTGACTGGTGAGTACTCAACCAAGTCATTCTGAGAATAGTGTATGCGGCGACCGAGTTGCTCTTGCCCGGCGTCAATACGGGATAATACCGCGCCACATAGCAGAACTTTAAAAGTGCTCATCATTGGAAAACGTTCTTCGGGGCGAAAACTCTCAAGGATCTTACCGCTGTTGAGATCCAGTTCGATGTAACCCACTCGTGCACCCAACTGATCTTCAGCATCTTTTACTTTCACCAGCGTTTCTGGGTGAGCAAAAACAGGAAGGCAAAATGCCGCAAAAAAGGGAATAAGGGCGACACGGAAATGTTGAATACTCAT
pPiggyBac 4xU6-PylT(U25C)/EF1-sfGFP(150TAG)-IRES-Neo
ACTCTTCCTTTTTCAATATTATTGAAGCATTTATCAGGGTTATTGTCTCATGAGCGGATACATATTTGAATGTATTTAGAAAAATAAACAAATAGGGGTTCCGCGCACATTTCCCCGAAAAGTGCCACCTAAATTGTAAGCGTTAATATTTTGTTAAAATTCGCGTTAAATTTTTGTTAAATCAGCTCATTTTTTAACCAATAGGCCGAAATCGGCAAAATCCCTTATAAATCAAAAGAATAGACCGAGATAGGGTTGAGTGTTGTTCCAGTTTGGAACAAGAGTCCACTATTAAAGAACGTGGACTCCAACGTCAAAGGGCGAAAAACCGTCTATCAGGGCGATGGCCCACTACGTGAACCATCACCCTAATCAAGTTTTTTGGGGTCGAGGTGCCGTAAAGCACTAAATCGGAACCCTAAAGGGAGCCCCCGATTTAGAGCTTGACGGGGAAAGCCGGCGAACGTGGCGAGAAAGGAAGGGAAGAAAGCGAAAGGAGCGGGCGCTAGGGCGCTGGCAAGTGTAGCGGTCACGCTGCGCGTAACCACCACACCCGCCGCGCTTAATGCGCCGCTACAGGGCGCGTCCCATTCGCCATTCAGGCTGCGCAACTGTTGGGAAGGGCGATCGGTGCGGGCCTCTTCGCTATTACGCCAGCTGGCGAAAGGGGGATGTGCTGCAAGGCGATTAAGTTGGGTAACGCCAGGGTTTTCCCAGTCACGACGTTGTAAAACGACGGCCAGTGAGCGCGCCTCGTTCATTCACGTTTTTGAACCCGTGGAGGACGGGCAGACTCGCGGTGCAAATGTGTTTTACAGCGTGATGGAGCAGATGAAGATGCTCGACACGCTGCAGAACACGCAGCTAGATTAACCCTAGAAAGATAATCATATTGTGACGTACGTTAAAGATAATCATGCGTAAAATTGACGCATGTGTTTTATCGGTCTGTATATCGAGGTTTATTTATTAATTTGAATAGATATTAAGTTTTATTATATTTACACTTACATACTAATAATAAATTCAACAAACAATTTATTTATGTTTATTTATTTATTAAAAAAAAACAAAAACTCAAAATTTCTTCTATAAAGTAACAAAACTTTTATGAGGGACAGCCCCCCCCCAAAGCCCCCAGGGATGTAATTACGTCCCTCCCCCGCTAGGGGGCAGCAGCGAGCCGCCCGGGGCTCCGCTCCGGTCCGGCGCTCCCCCCGCATCCCCGAGCCGGCAGCGTGCGGGGACAGCCCGGGCACGGGGAAGGTGGCACGGGATCGCTTTCCTCTGAACGCTTCTCGCTGCTCTTTGAGCCTGCAGACACCTGGGGGGATACGGGGAAAAGGCCTCCAAGGCCACTAGGAAAAACCGCACTTGTCCGGAAACCCCGGGAATCTAACCCGGCTGAACGGATTTAGAGTCCGTTCGATCTACATGATCAGGTTTCCGGTGTTTCGTCCTTTCCACAAGATATATAAAGCCAAGAAATCGAAATACTTTCAAGTTACGGTAAGCATATGATAGTCCATTTTAAAACATAATTTTAAAACTGCAAACTACCCAAGAAATTATTACTTTCTACGTCACGTATTTTGTACTAATATCTTTGTGTTTACAGTCAAATTAATTCTAATTATCTCTCTAACAGCCTTGTATCGTATATGCAAATATGAAGGAATCATGGGAAATAGGCCCTCTTCCTGCCCAACTAGGAAAAACCGCACTTGTCCGGAAACCCCGGGAATCTAACCCGGCTGAACGGATTTAGAGTCCGTTCGATCTACATGATCAGGTTTCCGGTGTTTCGTCCTTTCCACAAGATATATAAAGCCAAGAAATCGAAATACTTTCAAGTTACGGTAAGCATATGATAGTCCATTTTAAAACATAATTTTAAAACTGCAAACTACCCAAGAAATTATTACTTTCTACGTCACGTATTTTGTACTAATATCTTTGTGTTTACAGTCAAATTAATTCTAATTATCTCTCTAACAGCCTTGTATCGTATATGCAAATATGAAGGAATCATGGGAAATAGGCCCTCTTCCTGCCCAACTAGGAAAAACCGCACTTGTCCGGAAACCCCGGGAATCTAACCCGGCTGAACGGATTTAGAGTCCGTTCGATCTACATGATCAGGTTTCCGGTGTTTCGTCCTTTCCACAAGATATATAAAGCCAAGAAATCGAAATACTTTCAAGTTACGGTAAGCATATGATAGTCCATTTTAAAACATAATTTTAAAACTGCAAACTACCCAAGAAATTATTACTTTCTACGTCACGTATTTTGTACTAATATCTTTGTGTTTACAGTCAAATTAATTCTAATTATCTCTCTAACAGCCTTGTATCGTATATGCAAATATGAAGGAATCATGGGAAATAGGCCCTCTTCCTGCCCAACTAGGAAAAACCGCACTTGTCCGGAAACCCCGGGAATCTAACCCGGCTGAACGGATTTAGAGTCCGTTCGATCTACATGATCAGGTTTCCGGTGTTTCGTCCTTTCCACAAGATATATAAAGCCAAGAAATCGAAATACTTTCAAGTTACGGTAAGCATATGATAGTCCATTTTAAAACATAATTTTAAAACTGCAAACTACCCAAGAAATTATTACTTTCTACGTCACGTATTTTGTACTAATATCTTTGTGTTTACAGTCAAATTAATTCTAATTATCTCTCTAACAGCCTTGTATCGTATATGCAAATATGAAGGAATCATGGGAAATAGGCCCTCTTCCTGCCCAACTAGTGGATCTGCGATCGCTCCGGTGCCCGTCAGTGGGCAGAGCGCACATCGCCCACAGTCCCCGAGAAGTTGGGGGGAGGGGTCGGCAATTGAACGGGTGCCTAGAGAAGGTGGCGCGGGGTAAACTGGGAAAGTGATGTCGTGTACTGGCTCCGCCTTTTTCCCGAGGGTGGGGGAGAACCGTATATAAGTGCAGTAGTCGCCGTGAACGTTCTTTTTCGCAACGGGTTTGCCGCCAGAACACAGCTGAAGCTTCGAGGGGCTCGCATCTCTCCTTCACGCGCCCGCCGCCCTACCTGAGGCCGCCATCCACGCCGGTTGAGTCGCGTTCTGCCGCCTCCCGCCTGTGGTGCCTCCTGAACTGCGTCCGCCGTCTAGGTAAGTTTAAAGCTCAGGTCGAGACCGGGCCTTTGTCCGGCGCTCCCTTGGAGCCTACCTAGACTCAGCCGGCTCTCCACGCTTTGCCTGACCCTGCTTGCTCAACTCTACGTCTTTGTTTCGTTTTCTGTTCTGCGCCGTTACAGATCCAAGCTGTGACCGGCGCCTACTCTAGAGCTAGCGCCACCATGGTGTCCAAGGGCGAGGAACTGTTCACCGGCGTGGTGCCCATCCTGGTGGAACTGGATGGCGACGTGAACGGCCACAAGTTCTCTGTGCGGGGAGAGGGCGAAGGCGACGCCACAAATGGCAAGCTGACCCTGAAGTTCATCTGCACCACCGGCAAGCTGCCCGTGCCTTGGCCTACCCTCGTGACCACACTGACCTACGGCGTGCAGTGCTTCAGCAGATACCCCGACCATATGAAGCGGCACGACTTCTTCAAGAGCGCCATGCCCGAGGGCTACGTGCAGGAACGGACCATCAGCTTCAAGGACGACGGCACCTACAAGACCAGAGCCGAAGTGAAGTTCGAGGGCGACACCCTCGTGAACCGGATCGAGCTGAAGGGCATCGATTTCAAAGAGGACGGCAACATCCTGGGCCACAAGCTGGAGTACAACTTCAACAGCCACTAGGTGTACATCACCGCCGACAAGCAGAAGAACGGCATCAAGGCCAACTTCAAGATCCGGCACAACGTGGAAGATGGCAGCGTGCAGCTGGCCGACCACTACCAGCAGAACACCCCCATCGGAGATGGCCCCGTGCTGCTGCCCGACAACCACTACCTGAGCACCCAGAGCGTGCTGAGCAAGGACCCCAACGAGAAGCGGGACCACATGGTGCTGCTGGAATTCGTGACCGCCGCTGGCATCACCCACGGCATGGACGAGCTGTACAAGGGCAGCCACCATCACCATCACCATTAATAATAAGGCCGCGCCCCTCTCCCTCCCCCCCCCCTAACGTTACTGGCCGAAGCCGCTTGGAATAAGGCCGGTGTGCGTTTGTCTATATGTTATTTTCCACCATATTGCCGTCTTTTGGCAATGTGAGGGCCCGGAAACCTGGCCCTGTCTTCTTGACGAGCATTCCTAGGGGTCTTTCCCCTCTCGCCAAAGGAATGCAAGGTCTGTTGAATGTCGTGAAGGAAGCAGTTCCTCTGGAAGCTTCTTGAAGACAAACAACGTCTGTAGCGACCCTTTGCAGGCAGCGGAACCCCCCACCTGGCGACAGGTGCCTCTGCGGCCAAAAGCCACGTGTATAAGATACACCTGCAAAGGCGGCACAACCCCAGTGCCACGTTGTGAGTTGGATAGTTGTGGAAAGAGTCAAATGGCTCTCCTCAAGCGTATTCAACAAGGGGCTGAAGGATGCCCAGAAGGTACCCCATTGTATGGGATCTGATCTGGGGCCTCGGTGCACATGCTTTACATGTGTTTAGTCGAGGTTAAAAAAACGTCTAGGCCCCCCGAACCACGGGGACGTGGTTTTCCTTTGAAAAACACGATGATAATATGGCCACAACCATGGCGTCCGGAATGACCGAGTACAAGCCCACGGTGCGCCTCGCCACCCGCGACGACGTCCCCAGGGCCGTACGCACCCTCGCCGCCGCGTTCGCCGACTACCCCGCCACGCGCCACACCGTCGATCCGGACCGCCACATCGAGCGGGTCACCGAGCTGCAAGAACTCTTCCTCACGCGCGTCGGGCTCGACATCGGCAAGGTGTGGGTCGCGGACGACGGCGCCGCGGTGGCGGTCTGGACCACGCCGGAGAGCGTCGAAGCGGGGGCGGTGTTCGCCGAGATCGGCCCGCGCATGGCCGAGTTGAGCGGTTCCCGGCTGGCCGCGCAGCAACAGATGGAAGGCCTCCTGGCGCCGCACCGGCCCAAGGAGCCCGCGTGGTTCCTGGCCACCGTCGGCGTCTCGCCCGACCACCAGGGCAAGGGTCTGGGCAGCGCCGTCGTGCTCCCCGGAGTGGAGGCGGCCGAGCGCGCCGGGGTGCCCGCCTTCCTGGAGACCTCCGCGCCCCGCAACCTCCCCTTCTACGAGCGGCTCGGCTTCACCGTCACCGCCGACGTCGAGGTGCCCGAAGGACCGCGCACCTGGTGCATGACCCGCAAGCCCGGTGCCTAGGTCGACAATCAACCTCTGGATTACAAAATTTGTGAAAGATTGACTGGTATTCTTAACTATGTTGCTCCTTTTACGCTATGTGGATACGCTGCTTTAATGCCTTTGTATCATGCGTTAACTAAACTTGTTTATTGCAGCTTATAATGGTTACAAATAAAGCAATAGCATCACAAATTTCACAAATAAAGCATTTTTTTCACTGCATTCTAGTTGTGGTTTGTCCAAACTCATCAATGTATCTTATCATGTCTGGAATTGACTCAAATGATGTCAATTAGTCTATCAGAAGCTCATCTGGTCTCCCTTCCGGGGGACAAGACATCCCTGTTTAATATTTAAACAGCAGTGTTCCCAAACTGGGTTCTTATATCCCTTGCTCTGGTCAACCAGGTTGCAGGGTTTCCTGTCCTCACAGGAACGAAGTCCCTAAAGAAACAGTGGCAGCCAGGTTTAGCCCCGGAATTGACTGGATTCCTTTTTTAGGGCCCATTGGTATGGCTTTTTCCCCGTATCCCCCCAGGTGTCTGCAGGCTCAAAGAGCAGCGAGAAGCGTTCAGAGGAAAGCGATCCCGTGCCACCTTCCCCGTGCCCGGGCTGTCCCCGCACGCTGCCGGCTCGGGGATGCGGGGGGAGCGCCGGACCGGAGCGGAGCCCCGGGCGGCTCGCTGCTGCCCCCTAGCGGGGGAGGGACGTAATTACATCCCTGGGGGCTTTGGGGGGGGGCTGTCCCTGATATCTATAACAAGAAAATATATATATAATAAGTTATCACGTAAGTAGAACATGAAATAACAATATAATTATCGTATGAGTTAAATCTTAAAAGTCACGTAAAAGATAATCATGCGTCATTTTGACTCACGCGGTCGTTATAGTTCAAAATCAGTGACACTTACCGCATTGACAAGCACGCCTCACGGGAGCTCCAAGCGGCGACTGAGATGTCCTAAATGCACAGCGACGGATTCGCGCTATTTAGAAAGAGAGAGCAATATTTCAAGAATGCATGCGTCAATTTTACGCAGACTATCTTTCTAGGGTTAATCTAGCTGCATCAGGATCATATCGTCGGGTCTTTTTTCCGGCTCAGTCATCGCCCAAGCTGGCGCTATCTGGGCATCGGGGAGGAAGAAGCCCGTGCCTTTTCCCGCGAGGTTGAAGCGGCATGGAAAGAGTTTGCCGAGGATGACTGCTGCTGCATTGACGTTGAGCGAAAACGCACGTTTACCATGATGATTCGGGAAGGTGTGGCCATGCACGCCTTTAACGGTGAACTGTTCGTTCAGGCCACCTGGGATACCAGTTCGTCGCGGCTTTTCCGGACACAGTTCCGGATGGTCAGCCCGAAGCGCATCAGCAACCCGAACAATACCGGCGACAGCCGGAACTGCCGTGCCGGTGTGCAGATTAATGACAGCGGTGCGGCGCTGGGATATTACGTCAGCGAGGACGGGTATCCTGGCTGGATGCCGCAGAAATGGACATGGATACCCCGTGAGTTACCCGGCGGGCGCGCTTGGCGTAATCATGGTCATAGCTGTTTCCTGTGTGAAATTGTTATCCGCTCACAATTCCACACAACATACGAGCCGGAAGCATAAAGTGTAAAGCCTGGGGTGCCTAATGAGTGAGCTAACTCACATTAATTGCGTTGCGCTCACTGCCCGCTTTCCAGTCGGGAAACCTGTCGTGCCAGCTGCATTAATGAATCGGCCAACGCGCGGGGAGAGGCGGTTTGCGTATTGGGCGCTCTTCCGCTTCCTCGCTCACTGACTCGCTGCGCTCGGTCGTTCGGCTGCGGCGAGCGGTATCAGCTCACTCAAAGGCGGTAATACGGTTATCCACAGAATCAGGGGATAACGCAGGAAAGAACATGTGAGCAAAAGGCCAGCAAAAGGCCAGGAACCGTAAAAAGGCCGCGTTGCTGGCGTTTTTCCATAGGCTCCGCCCCCCTGACGAGCATCACAAAAATCGACGCTCAAGTCAGAGGTGGCGAAACCCGACAGGACTATAAAGATACCAGGCGTTTCCCCCTGGAAGCTCCCTCGTGCGCTCTCCTGTTCCGACCCTGCCGCTTACCGGATACCTGTCCGCCTTTCTCCCTTCGGGAAGCGTGGCGCTTTCTCATAGCTCACGCTGTAGGTATCTCAGTTCGGTGTAGGTCGTTCGCTCCAAGCTGGGCTGTGTGCACGAACCCCCCGTTCAGCCCGACCGCTGCGCCTTATCCGGTAACTATCGTCTTGAGTCCAACCCGGTAAGACACGACTTATCGCCACTGGCAGCAGCCACTGGTAACAGGATTAGCAGAGCGAGGTATGTAGGCGGTGCTACAGAGTTCTTGAAGTGGTGGCCTAACTACGGCTACACTAGAAGGACAGTATTTGGTATCTGCGCTCTGCTGAAGCCAGTTACCTTCGGAAAAAGAGTTGGTAGCTCTTGATCCGGCAAACAAACCACCGCTGGTAGCGGTGGTTTTTTTGTTTGCAAGCAGCAGATTACGCGCAGAAAAAAAGGATCTCAAGAAGATCCTTTGATCTTTTCTACGGGGTCTGACGCTCAGTGGAACGAAAACTCACGTTAAGGGATTTTGGTCATGAGATTATCAAAAAGGATCTTCACCTAGATCCTTTTAAATTAAAAATGAAGTTTTAAATCAATCTAAAGTATATATGAGTAAACTTGGTCTGACAGTTACCAATGCTTAATCAGTGAGGCACCTATCTCAGCGATCTGTCTATTTCGTTCATCCATAGTTGCCTGACTCCCCGTCGTGTAGATAACTACGATACGGGAGGGCTTACCATCTGGCCCCAGTGCTGCAATGATACCGCGAGACCCACGCTCACCGGCTCCAGATTTATCAGCAATAAACCAGCCAGCCGGAAGGGCCGAGCGCAGAAGTGGTCCTGCAACTTTATCCGCCTCCATCCAGTCTATTAATTGTTGCCGGGAAGCTAGAGTAAGTAGTTCGCCAGTTAATAGTTTGCGCAACGTTGTTGCCATTGCTACAGGCATCGTGGTGTCACGCTCGTCGTTTGGTATGGCTTCATTCAGCTCCGGTTCCCAACGATCAAGGCGAGTTACATGATCCCCCATGTTGTGCAAAAAAGCGGTTAGCTCCTTCGGTCCTCCGATCGTTGTCAGAAGTAAGTTGGCCGCAGTGTTATCACTCATGGTTATGGCAGCACTGCATAATTCTCTTACTGTCATGCCATCCGTAAGATGCTTTTCTGTGACTGGTGAGTACTCAACCAAGTCATTCTGAGAATAGTGTATGCGGCGACCGAGTTGCTCTTGCCCGGCGTCAATACGGGATAATACCGCGCCACATAGCAGAACTTTAAAAGTGCTCATCATTGGAAAACGTTCTTCGGGGCGAAAACTCTCAAGGATCTTACCGCTGTTGAGATCCAGTTCGATGTAACCCACTCGTGCACCCAACTGATCTTCAGCATCTTTTACTTTCACCAGCGTTTCTGGGTGAGCAAAAACAGGAAGGCAAAATGCCGCAAAAAAGGGAATAAGGGCGACACGGAAATGTTGAATACTCAT
Supplementary Material
Editorial summary.
Stable integration of genes that facilitate the incorporation of unnatural amino acids into amber stop codon in genes of interest, allows targeted integration of acetyl-lysine into histone H3.3 and the investigation of its effect in mouse embryonic stem cells.
Acknowledgements
This work was supported the Medical Research Council, UK (U105181009 and UPA0241008, to JWC). S.J.E. was supported by EMBO ALTF 1232-2011 and Herchel Smith Fund. O.S.W. was funded through the Ph.D. program in Chemical Biology and Molecular Medicine, Cambridge. We are grateful to T. Elliott (MRC-LMB) for CpK, and to the MRC-LMB FACS facility (M. Daly, F. Zhang, V. Romashova) for support, and J. Sale for helpful comments on the manuscript.
Footnotes
Please note a slash”/” between synthetases and tRNAs eg “PylRS/tRNACUA is standard nomenclature in the field, it has a specific meaning that is not amibguoous. Please do not edit this nomenclature to and, or a hyphen in copy editing.
Accession Codes
RNA-Seq data is available at the NCBI Gene Expression Omnibus under accession number GSE73823.
Author contributions
S.J.E. and J.W.C. conceived the experimental strategy, analysed the data and wrote the paper. S.J.E. performed most experiments and analysed and interpreted the RNA seq data. O.S.W. and R.J.E. performed qPCR experiment and analysis.
Competing financial interests
The authors declare no competing financial interest.
References
- 1.Chin JW. Expanding and Reprogramming the Genetic Code of Cells and Animals. Annu Rev Biochem. 2014;83:379–408. doi: 10.1146/annurev-biochem-060713-035737. [DOI] [PubMed] [Google Scholar]
- 2.Davis L, Chin JW. Designer proteins: applications of genetic code expansion in cell biology. Nature Rev Mol Cell Biol. 2012;13:168–182. doi: 10.1038/nrm3286. [DOI] [PubMed] [Google Scholar]
- 3.Tsai Y-H, Essig S, James JR, Lang K, Chin JW. Selective, rapid and optically switchable regulation of protein function in live mammalian cells. Nature Chemistry. 2015;7:554–561. doi: 10.1038/nchem.2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baker AS, Deiters A. Optical control of protein function through unnatural amino acid mutagenesis and other optogenetic approaches. ACS chemical biology. 2014;9:1398–1407. doi: 10.1021/cb500176x. [DOI] [PubMed] [Google Scholar]
- 5.Lang K, Chin JW. Cellular incorporation of unnatural amino acids and bioorthogonal labeling of proteins. Chem Rev. 2014;114:4764. doi: 10.1021/cr400355w. [DOI] [PubMed] [Google Scholar]
- 6.Gautier A, Deiters A, Chin JW. Light-Activated Kinases Enable Temporal Dissection of Signaling Networks in Living Cells. Journal of the American Chemical Society. 2011;133:2124–2127. doi: 10.1021/ja1109979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chatterjee A, Xiao H, Bollong M, Ai HW, Schultz PG. Efficient viral delivery system for unnatural amino acid mutagenesis in mammalian cells. Proc Natl Acad Sci U S A. 2013;110:11803–11808. doi: 10.1073/pnas.1309584110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schmied WH, Elsasser SJ, Uttamapinant C, Chin JW. Efficient multisite unnatural amino acid incorporation in mammalian cells via optimized pyrrolysyl tRNA synthetase/tRNA expression and engineered eRF1. J Am Chem Soc. 2014;136:15577–15583. doi: 10.1021/ja5069728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xiao H, et al. Genetic incorporation of multiple unnatural amino acids into proteins in mammalian cells. Angew Chem Int Ed Engl. 2013;52:14080–14083. doi: 10.1002/anie.201308137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shen B, et al. Genetically encoding unnatural amino acids in neural stem cells and optically reporting voltage-sensitive domain changes in differentiated neurons. Stem cells. 2011;29:1231–1240. doi: 10.1002/stem.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ding S, et al. Efficient transposition of the piggyBac (PB) transposon in mammalian cells and mice. Cell. 2005;122:473–483. doi: 10.1016/j.cell.2005.07.013. [DOI] [PubMed] [Google Scholar]
- 12.Huang H, Sabari BR, Garcia BA, Allis CD, Zhao Y. SnapShot: histone modifications. Cell. 2014;159:458–458 e451. doi: 10.1016/j.cell.2014.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nguyen DP, Garcia Alai MM, Kapadnis PB, Neumann H, Chin JW. Genetically Encoding Nε-Methyl- l-lysine in Recombinant Histones. Journal of the American Chemical Society. 2009;131:14194–14195. doi: 10.1021/ja906603s. [DOI] [PubMed] [Google Scholar]
- 14.Groff D, Chen PR, Peters FB, Schultz PG. A Genetically Encoded ε-N-Methyl Lysine in Mammalian Cells. ChemBioChem. 2010;11:1066–1068. doi: 10.1002/cbic.200900690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hancock SM, Uprety R, Deiters A, Chin JW. Expanding the Genetic Code of Yeast for Incorporation of Diverse Unnatural Amino Acids via a Pyrrolysyl-tRNA Synthetase/tRNA Pair. Journal of the American Chemical Society. 2010;132:14819–14824. doi: 10.1021/ja104609m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gattner MJ, Vrabel M, Carell T. Synthesis of epsilon-N-propionyl-, epsilon-N-butyryl-, and epsilon-N-crotonyl-lysine containing histone H3 using the pyrrolysine system. Chemical communications. 2013;49:379–381. doi: 10.1039/c2cc37836a. [DOI] [PubMed] [Google Scholar]
- 17.Kim CH, Kang M, Kim HJ, Chatterjee A, Schultz PG. Site-specific incorporation of epsilon-N-crotonyllysine into histones. Angew Chem Int Ed Engl. 2012;51:7246–7249. doi: 10.1002/anie.201203349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Neumann H, Peak-Chew SY, Chin JW. Genetically encoding Nε-acetyllysine in recombinant proteins. Nature Chemical Biology. 2008;4:232–234. doi: 10.1038/nchembio.73. [DOI] [PubMed] [Google Scholar]
- 19.Muller MM, Muir TW. Histones: at the crossroads of peptide and protein chemistry. Chemical reviews. 2015;115:2296–2349. doi: 10.1021/cr5003529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tropberger P, et al. Regulation of transcription through acetylation of H3K122 on the lateral surface of the histone octamer. Cell. 2013;152:859–872. doi: 10.1016/j.cell.2013.01.032. [DOI] [PubMed] [Google Scholar]
- 21.Neumann H, et al. A method for genetically installing site-specific acetylation in recombinant histones defines the effects of H3 K56 acetylation. Molecular cell. 2009;36:153–163. doi: 10.1016/j.molcel.2009.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Di Cerbo V, et al. Acetylation of histone H3 at lysine 64 regulates nucleosome dynamics and facilitates transcription. eLife. 2014;3:e01632. doi: 10.7554/eLife.01632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nguyen UT, et al. Accelerated chromatin biochemistry using DNA-barcoded nucleosome libraries. Nat Methods. 2014;11:834–840. doi: 10.1038/nmeth.3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Henikoff S, Shilatifard A. Histone modification: cause or cog? Trends in genetics : TIG. 2011;27:389–396. doi: 10.1016/j.tig.2011.06.006. [DOI] [PubMed] [Google Scholar]
- 25.Choudhary C, et al. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 26.Zhao S, et al. Regulation of cellular metabolism by protein lysine acetylation. Science. 2010;327:1000–1004. doi: 10.1126/science.1179689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hathaway NA. Dynamics and memory of heterochromatin in living cells. Cell. 2012;149:1447–1460. doi: 10.1016/j.cell.2012.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Elliott TS, et al. Proteome labeling and protein identification in specific tissues and at specific developmental stages in an animal. Nature biotechnology. 2014;32:465–472. doi: 10.1038/nbt.2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Keller G. Embryonic stem cell differentiation: emergence of a new era in biology and medicine. Genes & development. 2005;19:1129–1155. doi: 10.1101/gad.1303605. [DOI] [PubMed] [Google Scholar]
- 30.Streets AM, et al. Microfluidic single-cell whole-transcriptome sequencing. Proc Natl Acad Sci U S A. 2014;111:7048–7053. doi: 10.1073/pnas.1402030111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ng HH, Surani MA. The transcriptional and signalling networks of pluripotency. Nature cell biology. 2011;13:490–496. doi: 10.1038/ncb0511-490. [DOI] [PubMed] [Google Scholar]
- 32.Ashburner M, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nature genetics. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Uttamapinant C, et al. Genetic code expansion enables live-cell and super-resolution imaging of site-specifically labeled cellular proteins. J Am Chem Soc. 2015;137:4602–4605. doi: 10.1021/ja512838z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dabrowski M, Bukowy-Bieryllo Z, Zietkiewicz E. Translational readthrough potential of natural termination codons in eucaryotes - The impact of RNA sequence. RNA Biol. 2015;12:950–958. doi: 10.1080/15476286.2015.1068497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Elsaesser SJ, Goldberg AD, Allis CD. New functions for an old variant: no substitute for histone H3.3. Current opinion in genetics & development. 2010;20:110–117. doi: 10.1016/j.gde.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.David Y, Vila-Perello M, Verma S, Muir TW. Chemical tagging and customizing of cellular chromatin states using ultrafast trans-splicing inteins. Nat Chem. 2015;7:394–402. doi: 10.1038/nchem.2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Das C, Lucia MS, Hansen KC, Tyler JK. CBP/p300-mediated acetylation of histone H3 on lysine 56. Nature. 2009;459:113–117. doi: 10.1038/nature07861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Drogaris P, et al. Histone deacetylase inhibitors globally enhance h3/h4 tail acetylation without affecting h3 lysine 56 acetylation. Scientific reports. 2012;2:220. doi: 10.1038/srep00220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tan Y, Xue Y, Song C, Grunstein M. Acetylated histone H3K56 interacts with Oct4 to promote mouse embryonic stem cell pluripotency. Proc Natl Acad Sci U S A. 2013;110:11493–11498. doi: 10.1073/pnas.1309914110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chow JC, Yen Z, Ziesche SM, Brown CJ. Silencing of the mammalian X chromosome. Annual review of genomics and human genetics. 2005;6:69–92. doi: 10.1146/annurev.genom.6.080604.162350. [DOI] [PubMed] [Google Scholar]
- 41.Hooper M, Hardy K, Handyside A, Hunter S, Monk M. HPRT-deficient (Lesch-Nyhan) mouse embryos derived from germline colonization by cultured cells. Nature. 1987;326:292–295. doi: 10.1038/326292a0. [DOI] [PubMed] [Google Scholar]
- 42.Trapnell C, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nature protocols. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Trapnell C, et al. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nature Biotechnology. 2013;31:46–+. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cuddapah S, et al. Native chromatin preparation and Illumina/Solexa library construction. Cold Spring Harbor protocols. 2009;2009 doi: 10.1101/pdb.prot5237. pdb prot5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cuddapah S, et al. Global analysis of the insulator binding protein CTCF in chromatin barrier regions reveals demarcation of active and repressive domains. Genome research. 2009;19:24–32. doi: 10.1101/gr.082800.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.