Abstract
Measuring precise concentrations of proteins can provide insights into biological processes. Here, we use efficient protein extraction and sample fractionation and state-of-the-art quantitative mass spectrometry techniques to generate a comprehensive, condition-dependent protein abundance map of Escherichia coli. We measure cellular protein concentrations for 55% of predicted E. coli genes (>2300 proteins) under 22 different experimental conditions and identify methylation and N-terminal protein acetylations previously not known to be prevalent in bacteria. We uncover system-wide proteome allocation, expression regulation, and post-translational adaptations. These data provide a valuable resource for the systems biology and broader E. coli research communities.
Introduction
Transcriptome analyses have provided valuable insights in gene regulation. However, transcriptome data do not capture post-transcriptional processes, such as protein turnover, and therefore do not provide a complete picture of the expression state1,2. Thus, to understand how biological systems function, there is a need to complement these transcript-based insights with quantitative protein information.
Recent developments in mass spectrometry-based proteomics have enabled absolute protein levels to be measured on a system-wide level in microbes3–5 and mammalian cell lines4,6. However, because in-depth protein quantification requires extensive sample fractionation, proteome studies have so far been limited to a few samples, conditions3,7,8 or cellular compartments9,10. Post-translational modifications have also been broadly characterized in E. coli, but these too have been restricted to one or few conditions and have relied on enrichment techniques to identify respective specific modification11–15. In contrast, studies that investigated the E. coli proteome across multiple conditions were limited in terms of protein coverage16,17, or with regards to absolute quantification12.
Here, we quantify proteins across 22 experimental conditions. By reducing sample fractionation to a few high-quality fractions and using high-resolution mass spectrometry, we could double sample throughput without compromising on proteome coverage. Using an efficient protein extraction method, we obtain quantitative information also on membrane and ribosomal proteins that are notoriously difficult to extract quantitatively18. Overall, we determine protein abundance levels for approximately 55% of the predicted E. coli genes (>2300 proteins). This not only doubles the number of proteins absolutely quantified in E. coli3, but also provides the most comprehensive condition-dependent protein abundance map for any organism to date. In addition, we identify eleven (3 novel) different types of post-translational modifications (PTMs) including 318 novel PTMs, predominately Nα-acetylations and methylations, which were not previously reported in E. coli. We also uncover growth-rate dependent proteome re-arrangements, providing fundamental insights in global resource allocation.
Results
Experimental design
We grew E. coli BW2511319 under 22 different growth conditions in biological triplicates. These conditions included (i) growth on minimal media with excess of different carbon and energy sources, (ii) growth in glucose-limited chemostat cultures with varying growth rates, (iii) growth on glucose excess with different stress conditions, (iv) growth on complex medium, and (v) one and three days into stationary phase. Additionally, to enable use of the generated data also for other E. coli strains, we determined protein abundances under glucose and LB growth conditions also for two other frequently used strains; MG165520 and NCM372221.
Generation of condition-dependent proteome profiles
Quantitative proteome analyses were carried out using a combination of recently developed mass spectrometry (MS) based strategies4,5,22 and an efficient protein extraction method, which together allowed for system-wide accurate quantification of protein levels across a large number of conditions (Fig. 1). First, aliquots of all samples taken from the different conditions were subjected to shotgun LC-MS analysis to identify as many peptides as possible and to determine their condition-dependent intensities by label-free quantification. Towards maximizing the number of quantified proteins, we optimized protein extraction, sample pre-fractionation and LC parameters (Supplementary Fig. 1-3) and combined the data of two independent large-scale LC-MS analyses using different samples and experimental parameters (Supplementary Fig. 4).
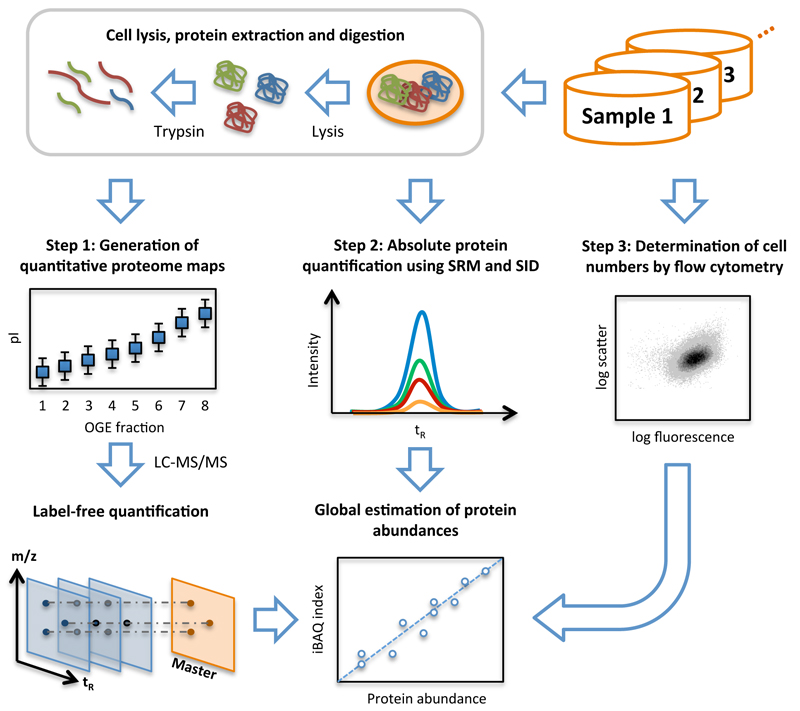
Figure 1. Workflow of system-wide protein abundance determination.
The workflow comprised three steps. First, cells of the various samples were lysed, proteins extracted and proteolyzed using trypsin. The peptide mixtures were either further fractionated using OFFGEL electrophoresis (OGE) or directly analyzed in biological triplicates by shotgun LC-MS/MS and quantified by label-free quantification. Second, the cellular concentrations of 41 proteins covering all components of the glycolysis pathway were determined across all samples by selected-reaction monitoring (SRM) and stable isotope dilution (SID). Therefore, for each protein, heavy labeled reference peptides (selected from the shotgun LC-MS/MS experiment) were synthetized. After spiking known amounts of these references into each sample, absolute quantities were determined for the corresponding proteins by SRM. Third, the numbers of cells taken for LC-MS/MS analyses were determined for each sample by flow cytometry. With this information, the protein concentrations determined by SRM/SID could be transformed to protein copies/cell and a quantitative model was built to translate MS-intensities of all quantified proteins to cellular abundance estimates using the Intensity Based Absolute Quantification (iBAQ) approach4.
Second, we accurately quantified a sub-set of identified proteins to establish a “calibration” for the determined MS-intensities of all identified proteins. Here, we selected 41 proteins, which we expected to be expressed at different abundances. Specifically, we selected the enzymes and iso-enzymes of the glycolytic pathway (including proteins with hypothetical function), tricarboxylic acid cycle enzymes and a few other proteins (Supplementary Table 1). These proteins’ concentrations were determined in each sample using stable isotope dilution (SID) and selected reaction monitoring (SRM) LC-MS/MS analysis23,24 (Supplementary Table 2&3). The concentration range of the 41 proteins covered more than four orders of magnitudes ranging from around 92’000 (Mdh, on acetate medium) to only 2 (YbhA, 3 days into stationary phase) copies per cell.
To determine the concentrations of proteins that we did not quantify with synthetic peptides, we used summed precursor MS-intensities originating from the respective protein, and a quantitative model established for each sample using the absolutely quantified proteins (cf. 4,25). We observed good correlation (r2> 0.8) and low median error rates (determined by bootstrapping5) between measured and estimated abundances being below 60% and 100% for the unfractionated (dataset 2) and Off-Gel electrophoresis (OGE)-fractionated (dataset 1) samples, respectively (Supplementary Fig. 4 and 5). Finally, together with the cell numbers determined from flow cytometric analyses (Step 3) and condition-dependent cell volumes26, accurate protein abundances per cell and per cell volume were calculated (Supplementary Table 4-6).
We determined absolute quantities for 2359 proteins across all conditions reflecting around 55% of the predicted ORFs and >95% of the proteome mass27,28. The dataset is an unbiased representation of the E. coli proteome – including very hydrophobic proteins – with highly reproducible and accurate protein concentration determined for 22 growth conditions (seeSupplementary Note 1, Supplementary Fig. 6-9 and Supplementary Tables 4-8). The high correlation coefficients of absolute protein levels observed with previously published small datasets comprising a few single conditions confirm the high quality of our dataset (Supplementary Fig. 8).
To test the applicability of our dataset to other E. coli strains, we determined absolute protein levels for two additional, commonly used E. coli strains (MG1655 and NCM3722) at two conditions and compared the levels with the data from BW25113 (Supplementary Fig. 10 and Supplementary Table 9). We found highly similar protein levels, with the exception of proteins of the flagella assembly apparatus that are particularly high in MG165529. This indicates that the data acquired for BW25113 is to a significant extend also valid for other E. coli strains.
Growth rate-dependent changes in protein abundance
Seminal studies in bacterial physiology uncovered that the mass fractions of cellular macromolecules (i.e. protein, RNA, DNA) are a function of growth rate, irrespective of the composition of the growth medium30–32. It was further found that the amounts of ribosomal proteins increased relative to the total protein amount with increasing growth rate33,34. Recently, using E. coli strains subjected to gradual carbon and nitrogen limitation, as well as gradual ribosome inhibition by chloramphenicol, it was found that the proteome undergoes growth-rate and limitation-dependent re-arrangements17.
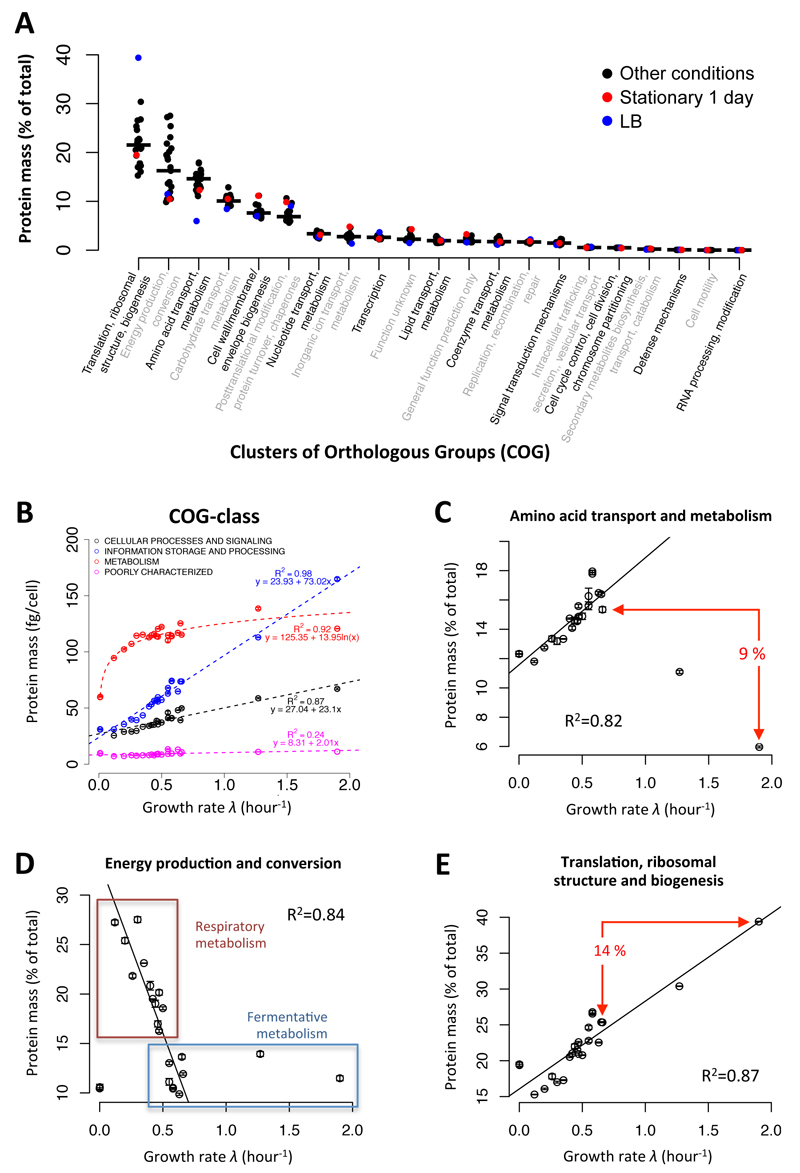
Here, we further explore this idea and investigate protein resource allocation across conditions. We found that few cellular processes - as defined by COG classification (Clusters of Orthologous Groups35,36, Supplementary Table 10 and 11) - make up most of the proteome mass (Fig. 2A), with six COG processes comprising around 80% of the total proteome. Combining the masses of proteins assigned to each of the four main COG classes and correlating the combined masses with growth rates (Fig. 2B and Supplementary Table 12), we found that proteins involved in “metabolism” showed a logarithmic increase in abundance with growth rate, ”cellular processes and signaling“, and ”information storage and processing“ (containing e.g. ribosomal proteins) a linear increase (Fig. 2B) and the levels of poorly characterized proteins stayed constant. Similar growth-rate dependent trends could be found in the 21 different functional COG-categories (Supplementary Fig. 11). Thus, extending the study of Hwa and colleagues17 to a large range of different growth conditions, our data demonstrates that the abundance of many functional processes strongly correlates with growth rate; also when the proteins of certain functional COG-categories are correlated against the growth rate (cf. Fig. 2C-E).
Figure 2. Fractions of protein mass in different COG processes.
(A) The fractions of total protein masses assigned to the 20 COG categories35, respectively, are indicated for each environmental condition. The median values (black line) as well as the values for 1-day stationary phase (red) and LB (blue) conditions are shown. (B) Correlation of growth rate and absolute mass (Da) of all proteins assigned to the four major COG-classes. (C) COG process of “amino acid transport and metabolism”; the difference in relative protein mass between the fastest rich (LB) and fastest minimal medium (42°C glucose) growth condition is indicated. (D) COG process of “energy production and conversion”; the boxes indicate conditions with a respiratory or fermentative metabolism, respectively, as major source for ATP production. (E) Same as in (C) for the COG process of “translation, ribosomal structure and biogenesis”. In sub-figures C-E, the squared Pearson correlation coefficients (R2) are calculated for all conditions indicated with a black circle and a robust linear regression was applied (see supplemental text for details). Error bars show the standard deviations between triplicate measurements.
However, we also noted that in some conditions the fraction of the proteins of certain metabolic COG categories deviated from the growth-rate correlation, suggesting an altered demand for proteome resources. This was, for instance, the case when comparing conditions where amino acids were present or absent in the growth medium (Fig. 2C), or between conditions with respiratory versus fermentative metabolism (Fig. 2D). In the first case, the COG category of “amino acid transport and metabolism” was approximately 9% lower on the LB medium condition compared to the fastest growth condition without amino acids present. In the second case, on average about 10% of the protein mass was invested for energy generation on fermentative carbon sources (COG category “energy production and conversion”), while substrates that largely rely on respiration invested 15-30% of their total protein mass in energy regeneration (Fig. 2D). Increased allocation of protein resources in these metabolic processes was accompanied by lower allocation in proteins connected with “translation, ribosomal structure and biogenesis” (Fig. 2E). Since it has been suggested that the amount of ribosomes determines the cellular growth rate37, these observations propose that the investments required for metabolic processes of amino acid biosynthesis or energy metabolism under specific conditions constrain the possible investments in ribosomes, and thus can be considered growth-limiting factors.
Role of transcriptional regulation in resource allocation
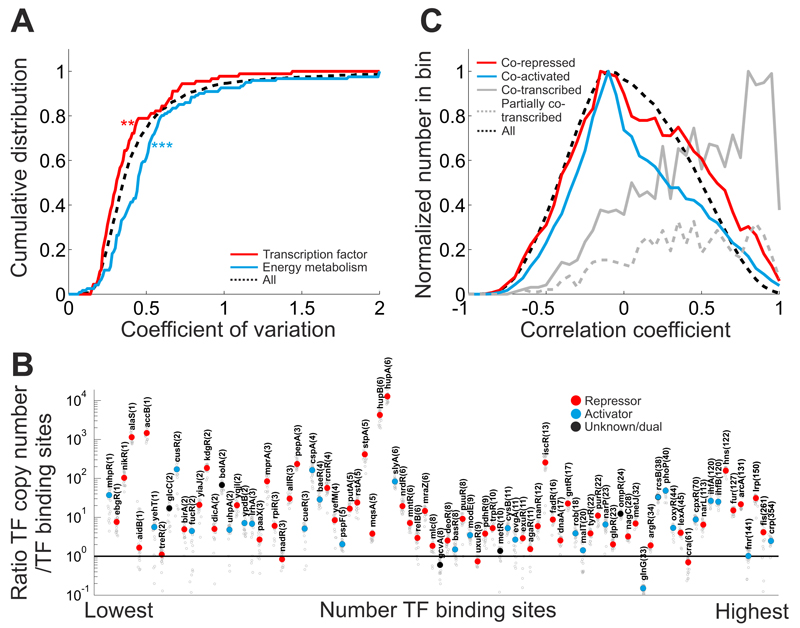
Next, we aimed to identify those cellular processes, which rely on transcriptional regulation for the adaptation to different conditions. Therefore, we determined the concentration variability of each detected protein, across conditions by calculating the coefficient of variation (CV). Here, we found different median variabilities in different COG categories (Supplementary Fig. 12). For instance, consistent with the fact that many of the tested conditions are carbon source changes, proteins belonging to the COG-categories “carbohydrate transport and metabolism” and “energy conversion and metabolism” were highly variable across conditions compared to the rest of the proteome. In contrast, proteins belonging to the COG category “transcription” exhibited significantly lower variability across conditions. In particular, the 90 reliably quantified transcription factors revealed significantly less variability across conditions than the rest of the proteome (Fig. 3A). Thus, proteins belonging to the COG category “transcription” may be subject to posttranslational regulation instead of transcriptional regulation.
Figure 3. Role of transcriptional regulatory network in determining proteome resource allocation.
(A) Cumulative distribution of the coefficient of variation (CV) for two representative protein classes (red: 90 transcription factors, blue: proteins belonging to the COG category “energy conversion and metabolism”) compared to the whole detected proteome (black dashed lines). Protein concentrations were calculated from protein copy numbers and cell volumes for all conditions. For each protein, the coefficient of variation was calculated as its relative standard deviation across conditions using only conditions, in which this protein was reliably quantified (relative error of quantification < 30%). Only proteins, for which more than 50% of the conditions yielded reliable quantification were used. Transcription factors have a lower median CV than the rest of the proteome (one-sided Wilcoxon rank sum test, p-value 0.012), whereas proteins of energy metabolism have a higher median CV (one-sided Wilcoxon rank sum test, p-value 2.17·10-5). (B) Relationship between transcription factor copy number and corresponding number of chromosomal binding sites per cell. Transcription factors were sorted according to the number of reported binding sites (based on RegulonDB40). Transcription factor copy numbers were normalized for the number of proteins in the active transcription factor complex (i.e. considering eventual multimerization of the transcription factors). The number of chromosomal binding sites was adjusted to account for growth-rate dependent differences in DNA content (as described in48). Small grey circles: TF/binding site ratio for each condition. Large circles: median ratio across all conditions. Transcription factors are marked as repressors (red) or activators (blue) if more than 50% of their binding sites are reported as repressing or activating, respectively. TFs with predominantly dual, or unknown, effect are marked in black. The number of distinct TF binding sites in the chromosome is shown in brackets after TF names. Note that HupA/B (HU complex) with a median ratio of >104 also play a histone-related role as part of the nucleoid complex49, and the observed high HupA/B copy numbers are likely reflecting HU’s role in the structural integrity of the chromosome. (C) Distribution of protein cross-correlations across conditions. Cross-correlation was calculated as pairwise Pearson correlation coefficient between proteins across all 22 conditions. Distribution for proteins whose genes are targeted by at least one common repressor or activator are shown as red and blue line, respectively. Grey line: protein pairs, which share at least one transcription unit (=co-transcribed). Grey dashed line: fraction of co- transcribed protein pairs, which also have non-overlapping transcription units (=partially co- transcribed). Black dashed line: cross-correlation of all detected protein pairs.
Despite the mostly low variation across conditions for individual transcription factors, the overall range in copy numbers between transcription factors was very large (from approx. 10 to >10000 copies per cell). To test whether these differences are related to the number of the transcription factors’ binding sites on the chromosome, we determined the ratio between transcription factor copy number and the number of reported chromosomal binding sites (Fig. 3B). While some extreme outliers exist (cf. caption of Fig. 3), we found that most transcription factors had only a median ratio around 10, with some of the global regulators, such as Cra, Fnr and Crp, having even lower ratios between 1 and 2. Since transcription factors also bind unspecifically to DNA38, which further reduces the number of free transcription factors, such low ratios make it unlikely that all available binding sites are actually occupied by the respective transcription factor at a given time point, which in turn may cause considerable competition between different binding sites for a relatively scarce transcription factor. Recently, it was found that such competition is used to establish the hierarchy of sugars co-utilization39, and the generally low ratio between TF copy number and binding sites suggests that similar hierarchical regulation may extend to other transcriptional regulators.
We next investigated the extent to which the topology of the transcriptional regulatory network can explain the expression of proteins across conditions. Therefore, we calculated the pairwise Pearson correlation between all proteins and compared the correlation coefficients of co-activated/co-repressed proteins (i.e. proteins sharing at least one transcriptional repressor or activator; as reported in RegulonDB40) with those of the rest of the protein pairs (Fig. 3C). Here, across conditions, we found that co-transcribed proteins (i.e. proteins from the same operon) have a clear bias towards strong positive correlations. Co-transcribed protein pairs with weak correlation had additional, non-overlapping transcription units (Fig. 3C, grey dashed line). The strong bias for strong positive correlations in co-transcribed proteins suggests that differential posttranscriptional regulation of gene expression within operons plays a limited role in E. coli. In contrast, we found that co-activated/co-repressed proteins (i.e. proteins that are regulated by the same transcription factors) show weak correlations. This finding suggests that in different conditions distinct subsets of a transcription factor’s targets are activated or repressed, which makes the topology of the transcriptional regulatory network a poor predictor of protein expression across conditions.
Distribution of protein mass between peri- and cytoplasm
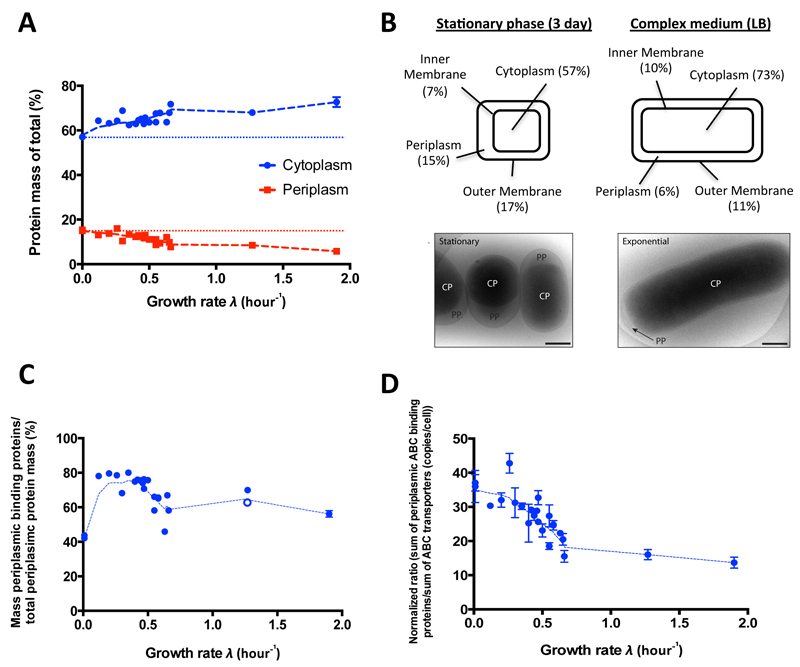
Next, we investigated the condition-specific localization of protein mass between different cellular compartments. 1174 of the measured proteins had a compartmental localization assigned representing between 76% and 83% of the total protein mass at the different conditions (Supplementary Table 13). Generally, we found that the protein mass fraction of the cytosolic proteins significantly increased with growth rate (Fig. 4A), while correspondingly the mass fraction of periplasmic proteins significantly decreased, even when considering geometric alterations resulting from increased cell volumes at faster growth rates (Supplementary Fig. 13). At the stationary phase conditions, periplasmic proteins accounted for 15% of the expressed protein mass, while on LB medium only 6%. On an absolute level, the mass of all periplasmic proteins per cell was higher in slowly growing E. coli cells (despite their smaller size) compared to their fast growing counterparts (Supplementary Table 14). Further, we found that the relative mass of proteins associated with the inner membrane increased, while the relative mass of proteins located at the outer membrane decreased at faster cell growth (Supplementary Table 14).
Figure 4. Condition-dependent distribution of protein mass in different cellular compartments.
(A) Mass fractions of proteins annotated as periplasmic (red) and cytoplasmic (blue) to total protein mass as a function of growth rate. Compartmental localization was done according to UniProt/gene ontology50. The trend lines (Lowess curves, dashed lines) are indicated. (B) Upper panel: Volume fractions of different cell compartments at the 3-day stationary phase and LB growth condition as hypothesized on the basis of protein mass fraction assuming constant volumetric protein concentrations across conditions. Lower panel: Cryo-electron microscopy analysis of E. coli cells grown to 3-day stationary phase after a glucose culture (left), or grown on LB-medium (right) confirms this hypothesis. Scale bars represent 500 nm. (C) Distributions of the mass fractions of periplasmic ABC binding proteins to all annotated periplasmic proteins by growth rate. Notably, we discarded one extremely abundant protein (OppA) for the Glycerol-AA growth condition (λ=1.27) and show the results with (filled blue circle) and without (unfilled blue circle) this outlier. The corresponding trend line (Lowess curve, dashed line) excluding this outlier is indicated. (D) Same as in (C) for the average ratio of all periplasmic ABC binding proteins to their corresponding ABC transporters (in copies per cell). Error bars show the standard deviations between triplicate measurements. Linear regression line slopes were significant from zero (p<0.0001) for all plots.
Taking these identified distributions of the protein mass together and assuming constant protein concentrations across cellular compartments suggested that the volume fractions between cytoplasm and periplasm change as a function of growth rate (Fig. 4B – upper panel), with the cytoplasm assuming higher and the periplasm lower volume fractions at high growth rates. To test this, we generated cryo-electron microscopy images of cells grown on LB medium and in stationary phase. We indeed found a significantly reduced periplasmic space at the fast growth rate condition (Fig. 4B – lower panel and Supplementary Fig. 14) consistent with the observed significant decrease in protein mass in the periplasmic space.
To investigate the growth rate-dependent distribution between cyto- and periplasmic proteins, we focused on protein classes that constituted a large fraction of the periplasmic proteome. We found that periplasmic binding proteins with ABC transporter functions were significantly enriched in the periplasm covering up to 80% of the total protein mass of the periplasm. Notably, the mass of periplasmic ABC transporter binding proteins in the periplasm decreased with increasing growth rates, explaining a large part of the observed reduction of the protein mass in the periplasm in fast growing cells (Fig. 4C). Focusing on stoichiometries between the periplasmic binding proteins and their membrane-bound counterparts, we found a high excess of periplasmic binding proteins compared to their ABC transporters of up to >100 fold at low growth rates (Supplementary Table 15) and we found that these stoichiometries (with some exceptions, Supplementary Table 16) decreased significantly with increasing growth rates (Fig. 4D).
Thus, at lower growth rates cells apparently increase the abundance of the periplasmic proteins and binding proteins and express higher numbers of binding proteins as compared to the respective ABC transporters. Eventually, these measures allow cells to increase the efficiency of nutrient uptake in less favorable conditions.
Post-translational modifications
We performed a global and unrestricted protein modification search41,42 to identify the most frequent post-translational modifications (PTMs) in our protein dataset (Supplementary Fig. 15). We identified 11 different types of PTMs (Table 1, Supplementary Table 17) and confirmed many known lysine acetylation and phosphorylation sites from previous studies focusing on single PTMs11,14,43. We also found that certain PTMs are enriched in specific protein classes and pathways (Table 1), and modify proteins of different expression levels (Fig. 5A) and that proteins can carry different types of PTMs at the same residue (Supplementary Table 18).
Table 1. Overview of identified post-translational modifications.
| Protein modification | Unique sites identified | Unique modified proteins | Known sitesa | Selected enriched KEGG pathways/SwissProt-Keywords foundb |
|---|---|---|---|---|
| Acetyl (K) | 61 | 44 | 25c | glycolysis / gluconeogenesis, citrate cycle (TCA cycle), pyruvate metabolism, ribosome, acetylation, phosphoprotein |
| Accetyl (Protein N-term) | 32 | 31d | 1e | nucelotide binding, atp-binding, acetylation, protein transport |
| Dimethy (K) | 14 | 14 | ||
| Dimethy (R) | 2 | 2 | ||
| Formyl (Protein N-term) | 24 | 24 | phosphoprotein, cytoplasm, pyridoxal phosphate, homodimer, transferase | |
| Methyl (K) | 84 | 64 | acetylation, phosphoprotein, methylated amino acid, periplasm, ribosome, ABC transporters, RNA degradation | |
| Methyl (R) | 67 | 55 | acetylation, protein biosynthesis, cytoplasm, homodimer, phosphoprotein, citrate cycle (TCA cycle), ribosome | |
| Phospho (S/T) | 24 | 21 | 8f | metall binding, phosphoprotein, magnesium, manganese |
| Succinyl (K) | 17 | 15 | 3g | DNA binding, periplasm, heterodimer |
| Trimethy (K) | 14 | 13 | protein biosynthesis, acetylation | |
| Trimethy (R) | 16 | 16 | protein biosynthesis |
Known sites from recent large-scale studies
Benjamini probability <0.05
Welnert, B. T. et al, Accetyl-phosphate is a critical determinant of lysine acetylation in E, coll. Mol Cell 51, 265-272 (2013). The largest dateset (52) was used for comparision.
Two acetlyated N-terminal (+/- methlonline) were identified for protein sufa
Smith, V. F., Schwartz, B. L., Randall, L. L., and Smith, R. D, (1996) Electrospray mass spectrometric Investigation of the chaperone SecB. Protein Scl, 5, 488–494
Macek B. et al. Phosphoproteome analysis of E. coli reveals evolutionary conservation of bacterial Ser/Thr/Tyr phosphorylation, Molecular &Amp; Cellular Proteomks 2008;7:299–307.
Colak, G. et al. Identification of Iysine succinylation substrates and the succinylation regulatory enzyme CobB in Escherichla coll. Mol Cell Proteomks 12, 3509–3520 (2013).
Figure 5. Identification and quantification of post-translational modifications (PTMs).
(A) Distribution of the corresponding protein abundances (in copies per cell, glucose medium) for the different PTMs identified. (B) Number of all identified N-terminal amino acids (protein N-terminus) carrying a Nα-acetylation. (C) Bar chart displaying summed modification abundances per protein for all quantified PTMs with increasing growth rate. The corresponding Lowess curve is indicated as dashed line. Of note, the linear regression line slope was significant from zero (p<0.0127). (D) Bar chart displaying the number of identified N-terminal protein acetylations for wild-type and three mutant strains lacking the three known N-acetyltransferases annotated in the E. coli genome. The mean value and the calculated significance (t-Test (two-tailed distribution, two-sample assuming equal variance), p-value <0.01 (**)) are indicated.
Notably, we identified a large number of PTMs, in particular protein methylation and N-terminal protein acetylation sites (Table 1). We found 31 proteins that were acetylated at the N-terminus. While a majority of eukaryotic proteins are N-terminally acetylated, so far Nα-acetylation in bacteria has been considered extremely rare44,45 and its function remains unclear46. We found that Nα-acetylation mostly occurred on N-terminal serine, alanine, methionine and threonine (Fig. 5B). Furthermore, mainly caused by a decreasing Nα-acetylation with growth rate (Supplementary Fig. 16), we found the highest total PTM abundances per protein to anticorrelate with growth rate (Fig. 5C and Supplementary Table 19). This together with the high number of identified Nα-acetylations suggest that N-terminal Nα- acetylation might also have physiological relevance in bacteria.
To further investigate into this, we analyzed Nα-acetylations in three mutant strains each lacking one of the three known E. coli Nα-acetyltransferases (NATs), originally assigned to only single target proteins. We found that only in the ribosomal-protein-alanine acetyltransferase rimJ mutant the number of Nα-acetylations significantly decreased (Fig. 5D and Supplementary Table 20-21). We further found that the decrease could be mainly ascribed to serine and threonine residues that did not get Nα-acetylated in the rimJ mutant (Supplementary Fig. 17), which are the Nα-acetylations that we found to increase at slow growth rates in the wild-type (Supplementary Fig. 18 and Supplementary Table 22). This finding suggests that RimJ is not only involved in the Nα-acetylation of its known target protein (RpsE), but might play a wider role in Nα-acetylation of other proteins with N-terminal serine and threonine residues, and this in a growth rate-dependent manner.
Discussion
In this work we determined absolute copy numbers for >2300 proteins mapped across 22 growth conditions and covering the full dynamic range from ~1 to more than 100 000 copies per cells. With this protein and condition coverage, we extended proteomic analyses of microbes to the level of transcriptomics, enabling large-scale biological discovery also on the proteome level. Furthermore, we present the first global dataset on methylation and Nα-acetylation in bacteria, and provide indication that these posttranslational modifications might have physiological relevance also in E. coli.
The generated protein abundance data will allow researchers of the systems biology community to develop quantitative models on certain biological subsystems, a task that requires precise knowledge on protein abundances. Furthermore, the data will also enable global computational studies, particularly drawing on the broad protein- and condition-coverage achieved. Finally, the data will also become a valuable resource for the broader E. coli community for novel discoveries.
Currently, proteomics analyses as done here can yet only be done in dedicated labs, where expertise ranging from sample handling via mass spectrometry to downstream bioinformatics analyses is present. However, we expect as technology advances quantitative proteomics technologies will become more broadly accessible – eventually through service companies – for a broader range of researchers. Still, for the scientific community significant challenges lie ahead, specifically those related to the elucidation of the second half of the – until now – obscure proteome, and the investigation of the identified novel types of posttranslational modifications.
Online methods
Strains and plasmids
The Escherichia coli K-12 strain BW25113 (genotype: F-, Δ(araD-araB)567, ΔlacZ4787(∷rrnB-3), λ-, rph-1, Δ(rhaD-rhaB)568, hsdR514)19 was used to generate the proteome map for all 22 conditions. Mutant strains with either the rimL, rimJ or rimI gene deleted were taken from the KEIO collection19. Correctness of the deletions were checked by PCR. Additionally, the proteome for the glucose and LB condition was also determined for the strains MG1655 (genotype: F-, λ-, rph-1)20 and NCM3722 (genotype: F+)21.
Media
LB-medium was prepared as follows: Five grams of yeast extract (BD), 10 g Tryptone (BD) and 10 g NaCl were dissolved in one liter of water and the mixture sterilized by autoclaving. LB-plates were produced by adding 20 g agar (BD) to the LB-medium mixture before autoclaving. M9 minimal medium without carbon source was prepared in the following way: To 700 ml of water, 200 ml of 5x base salt solution (211 mM Na2HPO4, 110 mM KH2PO4, 42.8 mM NaCl, 56.7 mM (NH4)2SO4, in H2O, autoclaved), 10 ml of trace elements (0.63 mM ZnSO4, 0.7 mM CuCl2, 0.71 mM MnSO4, 0.76 mM CoCl2, in H2O, autoclaved), 1 ml 0.1 M CaCl2 solution, in H2O, autoclaved, 1 ml 1 M MgSO4 solution, in H2O, autoclaved, 2 ml of 500x thiamine solution (1.4 mM, in H2O, filter sterilized) and 0.6 ml 0.1 M FeCl3 solution (in H2O, filter sterilized) were added. The resulting solution was filled up to 1 liter with water. All chemicals used were obtained from Sigma-Aldrich unless indicated otherwise. To prepare M9 minimal medium with a specific amount of carbon source, aqueous stock solutions were used. Aqueous stock solutions were prepared for every carbon source, adjusted to pH 7 by titration with 1 M sodium hydroxide or fuming hydrochloric acid. M9 minimal medium was complemented with carbon source by mixing appropriate amounts of carbon source free M9 minimal medium and carbon source stock solutions. The medium was always filtrated after preparation (Steritop-GP 500ml, Millipore). The following carbon sources and concentrations were used: acetate (sodium acetate, 3.5 g/L), fumarate (disodium fumarate, 2.8 g/L), galactose (2.3 g/L), glucose (5 g/L), glucosamine (2.1 g/L), glycerol (2.2 g/L), pyruvate (sodium pyruvate, 3.3 g/L), succinate (disodium succinate hexahydrate, 5.7 g/L), fructose (5 g/L), mannose (5 g/L), xylose (5 g/L). For chemostat growth only 1 g/L of glucose was used. Glucose minimal medium for the cells grown with osmotic stress was supplemented with NaCl to a concentration of 50 mM, for the cells grown with pH stress, fuming hydrochloric acid was titrated to the medium until a pH of 6 was reached. The glycerol + amino acid medium was made by supplementing the media with glycerol to a concentration of 2.2 g/L, and complete CSM mixture (ForMedium) and the following individual amino acids: alanine, asparagine, cysteine, glutamate, glycine, proline and serine to final concentrations of alanine 1.0 mg/L (0.01mM), adenine 10.2 mg/L (0.1mM), arginine 51.1 mg/L (0.3mM), asparagine 1.6 mg/L (0.01mM), aspartic acid 81.8 mg/L (0.6mM), cysteine 1.2 mg/L (0.01mM), glutamate 15.2 mg/L (0.1mM), glutamine 13.9 mg/L (0.1mM), glycine 0.4 mg/L (0.01mM), histidine 20.5 mg/L (0.1mM), isoleucine 51.1 mg/L (0.4mM), leucine 102.3 mg/L (0.8mM), lysine 51.1 mg/L (0.4mM), methionine 20.5 mg/L (0.14mM), phenylalanine 51.1 mg/L (0.3mM), proline 5.2 mg/L (0.05mM), serine 9.2 mg/L (0.1mM), threonine 102.3 mg/L (0.9mM), tryptophan 51.1 mg/L (0.3mM), tyrosine 51.1 mg/L (0.3mM), valine 143.2 mg/L (1.2mM), uracil 20.5 mg/L (0.2mM). An overview about the used growth conditions can be found in Supplementary Table 23-24.
Cultivation
Cells taken from -80°C stocks were streaked out on LB-agar plates. The cells were grown on the plate overnight and kept at 4°C for a maximum of three weeks. For the preculture, a single colony was picked from a plate and grown overnight in 50 ml M9 glucose medium in a 500 ml unbaffled wide- neck Erlenmeyer flask covered by a 38 mm silicone sponge closure (BellCo glass) at 37°C, 300 rpm and 5 cm shaking diameter (ISF-4-V shaker, Kühner). For the batch cultures, the cells from a preculture were re-inoculated into 50 ml of the appropriate pre-warmed medium in a 500 ml unbaffled wide-neck Erlenmeyer flask covered by a 38 mm silicone sponge closure (BellCo glass) and grown at 37°C, orbital shaking at 300 rpm and 5 cm shaking diameter (ISF-4-V, Kühner). To ensure steady state growth, the cells were first grown to exponential phase and then passaged into a second shake- flask containing fresh medium ensuring the cells had undergone at least 10 divisions under the respective condition and were thus in a steady state. The cells undergoing temperature stress were grown at 42°C. Cells grown in a chemostat were inoculated from a preculture to an OD of 0.1 and allowed to grow in batch mode to an OD of around 0.8 before dilution (rates: 0.12, 0.2, 0.35, 0.5) was started51. Starved cells were continuously shaken after reaching stationary phase for either 1 or 3 days.
Determination of cell counts and growth rates
For all shake flask batch cultures, cell counts were determined over time using an Accuri® C6 Flow Cytometer (BD Biosciences). Samples used for flow cytometric analysis were diluted with M9 minimal medium to an OD600 value of around 0.001, corresponding to a cell density of approximately 106 cells/ml. The instrument settings were the following: Flow rate: medium, FSC-H: 106, SSC-H: 105: all log scale. Analysis of the data was done with CFlow plus analysis (Version 1.0.264.15). The growth rate of the cultures was determined from the cell counts over time at cell concentrations from 105 cells/ml to 109 cells/ml. The growth rate was calculated from at least four consecutive measurements.
Sample preparation
Samples for proteome analyses were taken from cells that were grown until they reached 10 divisions in exponential state, collected by centrifugation at 20,000g at 4°C, washed twice with 2 ml ice-cold PBS buffer, harvested by centrifugation at 20,000g and pellet was snap frozen in liquid nitrogen and stored at -80°C until further processing. Cells were resuspended either in 100µl lysis buffer 1 (100 mM ammoniumbicarbonate, 2% sodium deoxycholate) for the dataset 2 (see Supplementary Fig. 4 for details) or 100µl lysis buffer 2 (100 mM ammoniumbicarbonate, 8M urea, 0.1% RapiGest™) for the dataset 1. The cells were disrupted by strong vortexing for 3 x 30 seconds followed by indirect sonication (100% amplitude, 0.5 cycle, 3 × 10 s) in a VialTweeter (Hielscher). A small aliquot of the supernatant was taken to determine the protein concentration of each sample using a BCA assay (Thermo Fisher Scientific). Proteins obtained from the different samples were reduced with 5mM TCEP for 60 (15)min at 37 (99)°C for dataset 1 (2), respectively, and alkylated with 10mM iodoacetamide for 30min in the dark at 25°C. After quenching the reaction with 12 mM N-acetyl- cysteine, the proteins were proteolyzed for 4h at 37°C using sequencing-grade Lys-C (Wako Chemicals) at 1/200 w/w. Then, the samples were diluted with 100mM ammoniumbicarbonate buffer to a final sodium deoxycholate (urea) concentration of 1% (1.6M) for dataset 2 (1) samples, respectively, and further digested by incubation with sequencing-grade modified trypsin (1/50, w/w; Promega, Madison, Wisconsin) over night at 37°C. The samples were acidified with 2M HCl to a final concentration of 50mM, incubated for 15min at 37°C and the precipitated detergent removed by centrifugation at 10,000g for 15min. Subsequently, an aliquot of the heavy reference peptide mix (see Supplementary Table 1 for details) were spiked into each sample at a concentration of 200/20 fmol of heavy reference peptides per 1µg of total endogenous protein mass. All peptide samples were then desalted by C18 reversed-phase spin columns according to the manufacturer’s instructions (Macrospin, Harvard Apparatus), separated in aliquots of 150 ug peptides, dried under vacuum and stored at -80ºC until further use. For LC-MS analysis, samples were solubilized in solvent A (98% water, 2% acetonitrile, 0.15% formic acid) at a concentration of 0.5 ug/ul and 4 ul were injected per LC-MS run. All samples of dataset 2 were prepared in biological triplicates.
Off-Gel electrophoresis
150 ug of dried peptides of each sample were solubilized in 1800 µl Off-Gel electrophoresis buffer, respectively, according to the manufacturer`s instructions (3100 OFFGEL Fractionator, Agilent Technologies). Then, all 19 peptide mixtures were separated on a 12cm pH 3-10 IPG strip (GE Healthcare), respectively, using a protocol of 1h rehydration at maximum 500V, 50μA and 200mW. Peptides were separated at maximum 8000V, 100μA and 300mW until 20kVh was reached. Subsequently, the 12 fractions were combined to 4 final fractions (F1-F4) using the following pooling scheme; (F1) 1-3, (F2) 4-6, (F3) 7-9 and (F4) 10-12. The pooled fractions were subsequently desalted using C18 reversed-phase columns according to the manufacturer’s instructions (Microspin, Harvard Apparatus), dried under vacuum and subjected to LC-MS/MS analysis. For the initial comparison of different fractionation schemes (Supplementary Fig. 1), the following additional fractions pooling scheme was employed; 1, 2, 3, 4, 5-6, 7-8, 9-10 and 11-12.
LC-MS/MS analysis
Two independent LC-MS experiments were carried out comprising samples with and without OGE- fractionation, respectively. The fractionated samples (dataset 1) were analyzed using a previously described µRPLC-MS system9 with some modifications. The hybrid Orbitrap-Velos mass spectrometer was interfaced to a nano electrospray ion source coupled online to an Easy-nLC system (all ThermoScientific). 1µg of peptides were separated on a RP-LC column (75 µm x 20 cm) packed in-house with C18 resin (Magic C18 AQ 3 µm; Michrom BioResources) using a linear gradient from 95% solvent A (98% water, 2% acetonitrile, 0.15% formic acid) and 5% solvent B (98% acetonitrile, 2% water, 0.15% formic acid) to 30% solvent B over 120 min at a flow rate of 0.2 µl/min. Each survey scan acquired in the Orbitrap at 60,000 FWHM was followed by 10 MS/MS scans of the most intense precursor ions in the linear ion trap. Preview mode was enabled and dynamic exclusion was set for 60 sec. Charge state screening was employed to select for ions with at least two charges and rejecting ions with undetermined charge state. The normalized collision energy was set to 32%, and one microscan was acquired for each spectrum. The unfractionated samples (dataset 2) were analyzed on a hybrid Orbitrap-Elite mass spectrometer connected online to an Easy-nLC 1000 system (both Thermo Scientific). Peptide separation was performed on a (75 µm x 45 cm) packed in-house with C18 resin (Reprosil-AQ Pur, Dr. Maisch 1.9 µm) using a linear gradient from 95% solvent A and 5% solvent B to 30% solvent B over 180 min at a flow rate of 0.2 µl/min. For MS1, 10E6 ions were accumulated in the Orbitrap cell over a maximum time of 300 ms and scanned at a resolution of 120,000 FWHM (at 400 m/z) followed by 10 MS/MS scans of the most intense precursor ions in the Orbitrap acquired at a target setting of 50,000 ions, accumulation time of 100 ms and a resolution of 15,000 FWHM (at 400 m/z). The normalized collision energy was set to 35%, and one microscan was acquired for each spectrum. A list comprising names of all samples and LC-MS runs included in this study is shown in Supplementary Table 25.
Protein identification and label-free quantification
The acquired raw-files were imported into the Progenesis LC-MS software (v4.0, Nonlinear Dynamics Limited), which was used to extract peptide precursor ion intensities across all samples applying the default parameters. The generated mgf-files were searched using MASCOT against a decoy database (consisting of forward and reverse protein sequences) of the predicted proteome from E.coli (UniProt, download date: 2012/07/20). The database consists of 4431 E. coli proteins as well as known contaminants such as porcine trypsin, human keratins and high abundant bovine serum proteins (Uniprot), resulting in a total of 10388 protein sequences. The search criteria were set as follows: full tryptic specificity was required (cleavage after lysine or arginine residues, unless followed by proline); 2 missed cleavages were allowed; carbamidomethylation (C) was set as fixed modification; oxidation (M) was applied as variable modifications; mass tolerance of 10 ppm (precursor) and 0.6 (0.02 for the HCD dataset) Da (fragments). The database search results were filtered using the ion score to set the false discovery rate (FDR) to 1% on the peptide and protein level, respectively, based on the number of reverse protein sequence hits in the datasets. The relative quantitative data obtained were normalized and statistically analyzed using our in-house software script SafeQuant52 (see also Supplementary Note 2).
Absolute quantification of glycolytic proteins by targeted LC-MS
41 proteins covering all enzymes and iso-enzymes of the glycolytic pathway were selected for absolute quantification by selected reaction monitoring (SRM) and stable isotope dilution (SID) (Supplementary Table 1). For each protein, one heavy reference peptide was synthesized matching the sequence of the endogenous peptide with the highest precursor ion MS-intensity determined in the label-free quantification experiment. Peptides containing missed cleavages or a glutamine at the N- terminus were excluded. Based on Top3 quantification25,53, the proteins were ranked according to their expected cellular abundance and grouped into two groups containing either high or low abundant proteins. According to this, a standard mixture comprising all 41 heavy reference peptide was generated containing 10/1 pmol/ul of peptides matching to high/low concentrated proteins (Supplementary Table 1). To generated the SRM assays, an aliquot of this mixture containing 500/50 fmol of each reference peptide was analyzed by shotgun LC-MS/MS using HCD fragmentation, database searched by Mascot applying the same settings as above with two changes; isotopically labeled arginine (+10 Da) and lysine (+8 Da) were added as variable modifications and the mass tolerance for MS2 fragments was set to 0.02 Da. The resulting dat-file was imported to Skyline version 1.4 (https://brendanx-uw1.gs.washington.edu/labkey/project/home/software/Skyline/begin. view) to generate a spectral library and select the best transitions for each peptide. After collision energy optimization, the 424 transitions (up to six transitions per peptide) were scheduled into time segments of 10 minutes and the final transition list (Supplementary Table 26) imported to a triple quadrupole mass spectrometer (TSQ Vantage) connected to an electrospray ionsoure (both ThermoFisher Scientific). Peptide separation was carried out using an nEasy-LC systems (ThermoFisher Scientific) equipped with a RP-HPLC column (75 μm x 20 cm) packed in-house with C18 resin (Magic C18 AQ 3 μm; Michrom BioResources) using a linear gradient from 95% solvent A (0.15% formic acid, 2% acetonitrile) and 5% solvent B (98% acetonitrile, 0.15% formic acid) to 35% solvent B over 90 minutes at a flow rate of 0.2 μl/min. Each sample was analyzed in duplicate. All raw-files were imported into Skyline for protein quantification. Based on the number of cells counted by FACS for each sample, absolute abundances for the selected proteins (in copies/cell) could be calculated across all samples in both data sets (Supplementary Table 2-3).
Proteome-wide estimation of protein abundances
The absolute protein concentrations determined for 41 glycolytic proteins were aligned with the summed protein intensities as provided by the Progenesis LC-MS software (v4.0, Nonlinear Dynamics Limited) divided by the number of expected tryptic peptides as recently specified4,25. The thus generated models were applied to estimate absolute protein levels for all quantified proteins in the CID and HCD dataset, respectively, and the expected errors were calculated by bootstrapping25 (Supplementary Fig. 5). To control for variations in protein extraction efficiency, which was lower for stationary phase samples, we used the total protein mass per cell (that is the summed masses of all quantified proteins) accurately determined in triplicates for the glucose experiment by our LC-MS approach (Supplementary Fig. 5A) and, assuming that the volumetric protein concentration is condition independent54, we adjusted the total protein mass per cell for each condition according to the precisely measured cellular volumes (Supplementary Table 23 and Supplementary Note 3) determined previously26. Due to the higher number of quantified membrane proteins, higher number of growth conditions included and the analysis in biological triplicates (Supplementary Fig. 4), protein quantities obtained from data set 2 were employed for all quantitative analysis carry out in this study. Data generated in data set 1 was only included in the qualitative analysis of identified protein modifications illustrated in Table 1 and Fig. 5A and B.
To assess the technical and biological variability of our label-free protein quantification approach, we performed duplicate SRM and shotgun LC-MS analyses of three independent biological samples grown in glucose media and chemostat µ=0.5 and correlated the protein abundances determined by our data analysis pipeline (Supplementary Fig. 7). Besides, stoichiometries were determined for quantified components of protein complexes with known subunit composition (Supplementary Table 27).
Analysis of post-translational modifications
The extensive LC-MS dataset generated also allowed us to search for different post-translational modifications at various positions. To get an overview of the potential modification present in our dataset, we first carried out an Open Modification Search. Therefore, a spectral library was compiled from the MS data obtained from the glucose condition sample sequentially applying the software tools X!Tandem (TPP v4.6)55 and Peptide Prophet (TPP v4.6)56 followed by Liberator and DeLiberator (v.1.46 and v 0.19)57. The search parameters of the protein sequence database search tool X!Tandem were set as follows: full tryptic specificity (cleavage after lysine or arginine residues unless followed by proline), up to two missed cleavages, carbamidomethyl (C) as fixed modification, oxidation (M) as a variable modification, 10 ppm precursor mass tolerance, 0.6 Da fragment mass tolerance, screening a target-decoy UniProtKB/SwissProt E.coli (UniProt, download date: 2012/07/20) protein sequence database. X!Tandem search results were processed using PeptideProphet and a consensus spectrum target-decoy spectral library was created using Liberator and DeLiberator applying default parameters. Next all MS/MS spectra from all samples were screened against this spectral library in an Open Modification Search using QuickMod (v.1.03)42. The search parameters of QuickMod were set to: fragment mass tolerance 0.06 Da, modification mass tolerance 150 Da, False Discovery Rate cutoff 0.01, while default values were used for all other parameters.
To verify the modifications detected above and extend the modification search space to lysine and arginine modifications that alter tryptic cleavage and therefore are missed by the QuickMod search tool, we re-searched all acquired MS/MS-scans against the E. coli protein database using Mascot and allowing additional variable modifications. Specifically, the following five sets of variable modifications were included: (1) acetyl (protein N-term and K); (2) phospho (S,T,Y); (3) mono-, di- and tri-methylation (K); (4) mono-, di- and tri-methylation (R) and (5) formyl (protein N-term) and succinyl (K). All other parameters were set as described above. All peptide spectrum matches (PSM) identifying modified peptides were extracted and, for each modification and site, the false discovery rate adjusted to less than 1%, respectively, as described above. If the same modification was identified at multiple sites in the same peptide, the position of the modification determined in the PSM with the highest Mascot Ion Score was selected.
Protein modifications were quantified using label-free quantification as described above (see Supplementary Table 22. The local error rate for all identified and quantified peptides carrying a modification was set to 1% and to control for protein regulations, all calculated ratios of modified peptides were normalized by the ratios of their corresponding proteins. All filtering and statistical analysis steps were carried out using our in-house software tool SafeQuant52.
Electron microscopy
For cryo-electron microscopy, cells were taken from the culture by centrifugation at 1500g for 1 min. The pellet was resuspended in 20 µl of the supernatant after which 2.5 µl of this suspension was attached to glow-discharged 200 mesh Quantifoil R3.5/1 grids inside a vitrobot (FEI, the Netherlands) of which the chamber was set to room temperature and 100% humidity. After blotting for 10 s, the grids were plunge-frozen into liquid nitrogen-cooled liquid ethane. The complete procedure from culture to frozen samples maximally took 3 min. The frozen grids were then transferred into a FEI Tecnai20 transmission electron microscope running at 200 kV and imaged with a cooled slow-scan charge-coupled device camera (Ultrascan 4000; Gatan) using the low-dose procedure. Measurements on the periplasm and cytoplasm were performed in ImageJ and the results are illustrated in detail in Supplementary Fig. 14 and Supplementary Table 28-29.
Supplementary Material
Acknowledgements
Funding is acknowledged from NWO (VIDI grant to M.H.), Dupont (Dupont Young Professorship Award to M.H.), the Swiss National Science Foundation (31003A_132428/1 to M.B.) and the Commission of the European Communities through the PROSPECTS consortium (EU FP7 project 201648) (R.A.), the PROMYS consortium (EU H2020 project 613745) (M.H.) and for a Marie Curie IEF grant (330150) (Ke.K.), and the European Research Council (ERC-2008-AdG 233226) (R.A.) Further, the authors would like to thank Jakub Radzikowski for performing a number of experiments. We also like to thank Dirk Bumann and Samuel Marguerat for critical reading of the manuscript.
Footnotes
Accession codes
All mass spectrometry raw data files have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository47 with the dataset identifier PXD000498 and DOI 10.6019/PXD000498.
Author contributions
SV and BV performed all batch cultivations. KaK performed the chemostat cultures and did the TF-based analysis. KeK performed the electron microscopy analyses. LC prepared samples for dataset 1. AS prepared samples for dataset 2. AS performed all shotgun LC-MS analyses. MB carried out all targeted LC-MS analyses. AS, EA and MB analyzed MS data. AS and MH wrote the manuscript with input from KaK, EA and RA. AS, RA, and MH conceived the study.
Competing financial interest statement
The authors declare that they have no competing interests as defined by Nature Publishing Group, or other interests that might be perceived to influence the results and/or discussion reported in this paper.
References
- 1.Marguerat S, et al. Quantitative analysis of fission yeast transcriptomes and proteomes in proliferating and quiescent cells. Cell. 2012;151:671–683. doi: 10.1016/j.cell.2012.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee MV, et al. A dynamic model of proteome changes reveals new roles for transcript alteration in yeast. Mol Syst Biol. 2011;7:514. doi: 10.1038/msb.2011.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ishihama Y, et al. Protein abundance profiling of the Escherichia coli cytosol. BMC Genomics. 2008;9:102. doi: 10.1186/1471-2164-9-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schwanhäusser B, et al. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 5.Malmstrom J, et al. Proteome-wide cellular protein concentrations of the human pathogen Leptospira interrogans. Nature. 2009;460:762–765. doi: 10.1038/nature08184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beck M, et al. The quantitative proteome of a human cell line. Mol Syst Biol. 2011;7:549. doi: 10.1038/msb.2011.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krug K, et al. Deep Coverage of the Escherichia coli Proteome Enables the Assessment of False Discovery Rates in Simple Proteogenomic Experiments. Mol Cell Proteomics. 2013;12:3420–3430. doi: 10.1074/mcp.M113.029165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li G-W, Burkhardt D, Gross C, Weissman JS. Quantifying absolute protein synthesis rates reveals principles underlying allocation of cellular resources. Cell. 2014;157:624–635. doi: 10.1016/j.cell.2014.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maddalo G, et al. Systematic analysis of native membrane protein complexes in Escherichia coli. J Proteome Res. 2011;10:1848–1859. doi: 10.1021/pr101105c. [DOI] [PubMed] [Google Scholar]
- 10.Masuda T, Saito N, Tomita M, Ishihama Y. Unbiased quantitation of Escherichia coli membrane proteome using phase transfer surfactants. Mol Cell Proteomics. 2009;8:2770–2777. doi: 10.1074/mcp.M900240-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Macek B, et al. Phosphoproteome analysis of E. coli reveals evolutionary conservation of bacterial Ser/Thr/Tyr phosphorylation. Mol Cell Proteomics. 2008;7:299–307. doi: 10.1074/mcp.M700311-MCP200. [DOI] [PubMed] [Google Scholar]
- 12.Soares NC, Spät P, Krug K, Macek B. Global Dynamics of the Escherichia coli Proteome and Phosphoproteome During Growth in Minimal Medium. J Proteome Res. 2013;12:2611–2621. doi: 10.1021/pr3011843. [DOI] [PubMed] [Google Scholar]
- 13.Zhang J, et al. Lysine acetylation is a highly abundant and evolutionarily conserved modification in Escherichia coli. Mol Cell Proteomics. 2009;8:215–225. doi: 10.1074/mcp.M800187-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weinert BT, et al. Acetyl-phosphate is a critical determinant of lysine acetylation in E. coli. Mol Cell. 2013;51:265–272. doi: 10.1016/j.molcel.2013.06.003. [DOI] [PubMed] [Google Scholar]
- 15.Colak G, et al. Identification of lysine succinylation substrates and the succinylation regulatory enzyme CobB in Escherichia coli. Mol Cell Proteomics. 2013;12:3509–3520. doi: 10.1074/mcp.M113.031567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ishii N, et al. Multiple High-Throughput Analyses Monitor the Response of E. coli to Perturbations. Science. 2007;316:593–597. doi: 10.1126/science.1132067. [DOI] [PubMed] [Google Scholar]
- 17.Scott M, Klumpp S, Mateescu EM, Hwa T. Emergence of robust growth laws from optimal regulation of ribosome synthesis. Mol Syst Biol. 2014;10:747. doi: 10.15252/msb.20145379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Savas JN, Stein BD, Wu CC, Yates JR. Mass spectrometry accelerates membrane protein analysis. Trends Biochem Sci. 2011;36:388–396. doi: 10.1016/j.tibs.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baba T, et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2 doi: 10.1038/msb4100050. 2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blattner FR, et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 21.Soupene E, et al. Physiological studies of Escherichia coli strain MG1655: growth defects and apparent cross-regulation of gene expression. J Bacteriol. 2003;185:5611–5626. doi: 10.1128/JB.185.18.5611-5626.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmidt A, et al. Absolute quantification of microbial proteomes at different states by directed mass spectrometry. Mol Syst Biol. 2011;7:510. doi: 10.1038/msb.2011.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Picotti P, Bodenmiller B, Mueller LN, Domon B, Aebersold R. Full dynamic range proteome analysis of S. cerevisiae by targeted proteomics. Cell. 2009;138:795–806. doi: 10.1016/j.cell.2009.05.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gillette MA, Carr SA. Quantitative analysis of peptides and proteins in biomedicine by targeted mass spectrometry. Nat Meth. 2013;10:28–34. doi: 10.1038/nmeth.2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ahrné E, Molzahn L, Glatter T, Schmidt A. Critical assessment of proteome-wide label-free absolute abundance estimation strategies. PROTEOMICS. 2013;13:2567–2578. doi: 10.1002/pmic.201300135. [DOI] [PubMed] [Google Scholar]
- 26.Volkmer B, Heinemann M. Condition-dependent cell volume and concentration of Escherichia coli to facilitate data conversion for systems biology modeling. PLoS ONE. 2011;6:e23126. doi: 10.1371/journal.pone.0023126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Akhtar MK, Jones PR. Construction of a synthetic YdbK-dependent pyruvate:H2 pathway in Escherichia coli BL21(DE3) Metab Eng. 2009;11:139–147. doi: 10.1016/j.ymben.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 28.Stouthamer AH. A theoretical study on the amount of ATP required for synthesis of microbial cell material. Antonie Van Leeuwenhoek. 1973;39:545–565. doi: 10.1007/BF02578899. [DOI] [PubMed] [Google Scholar]
- 29.Wood TK, González Barrios AF, Herzberg M, Lee J. Motility influences biofilm architecture in Escherichia coli. Appl Microbiol Biotechnol. 2006;72:361–367. doi: 10.1007/s00253-005-0263-8. [DOI] [PubMed] [Google Scholar]
- 30.SCHAECHTER M, MAALOE O, KJELDGAARD NO. Dependency on medium and temperature of cell size and chemical composition during balanced grown of Salmonella typhimurium. J Gen Microbiol. 1958;19:592–606. doi: 10.1099/00221287-19-3-592. [DOI] [PubMed] [Google Scholar]
- 31.Brunschede H, Dove TL, Bremer H. Establishment of exponential growth after a nutritional shift-up in Escherichia coli B/r: accumulation of deoxyribonucleic acid, ribonucleic acid, and protein. J Bacteriol. 1977;129:1020–1033. doi: 10.1128/jb.129.2.1020-1033.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shahab N, Flett F, Oliver SG, Butler PR. Growth rate control of protein and nucleic acid content in Streptomyces coelicolor A3(2) and Escherichia coli B/r. Microbiology (Reading, Engl) 1996;142 ( Pt 8):1927–1935. doi: 10.1099/13500872-142-8-1927. [DOI] [PubMed] [Google Scholar]
- 33.Gausing K. Regulation of ribosome production in Escherichia coli: synthesis and stability of ribosomal RNA and of ribosomal protein messenger RNA at different growth rates. J Mol Biol. 1977;115:335–354. doi: 10.1016/0022-2836(77)90158-9. [DOI] [PubMed] [Google Scholar]
- 34.Ehrenberg M, Bremer H, Dennis PP. Medium-dependent control of the bacterial growth rate. Biochimie. 2013;95:643–658. doi: 10.1016/j.biochi.2012.11.012. [DOI] [PubMed] [Google Scholar]
- 35.Tatusov RL, et al. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Galperin MY, Makarova KS, Wolf YI, Koonin EV. Expanded microbial genome coverage and improved protein family annotation in the COG database. Nucleic Acids Res. 2015;43:D261–9. doi: 10.1093/nar/gku1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koch AL. Why can't a cell grow infinitely fast? Can J Microbiol. 1988;34:421–426. doi: 10.1139/m88-074. [DOI] [PubMed] [Google Scholar]
- 38.Hammar P, et al. The lac repressor displays facilitated diffusion in living cells. Science. 2012;336:1595–1598. doi: 10.1126/science.1221648. [DOI] [PubMed] [Google Scholar]
- 39.Aidelberg G, et al. Hierarchy of non-glucose sugars in Escherichia coli. BMC Syst Biol. 2014;8:133. doi: 10.1186/s12918-014-0133-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Salgado H, et al. RegulonDB v8.0: omics data sets, evolutionary conservation, regulatory phrases, cross-validated gold standards and more. Nucleic Acids Res. 2013;41:D203–13. doi: 10.1093/nar/gks1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ahrné E, Müller M, Lisacek F. Unrestricted identification of modified proteins using MS/MS. PROTEOMICS. 2010;10:671–686. doi: 10.1002/pmic.200900502. [DOI] [PubMed] [Google Scholar]
- 42.Ahrné E, Nikitin F, Lisacek F, Müller M. QuickMod: A tool for open modification spectrum library searches. J Proteome Res. 2011;10:2913–2921. doi: 10.1021/pr200152g. [DOI] [PubMed] [Google Scholar]
- 43.Hu LI, Lima BP, Wolfe AJ. Bacterial protein acetylation: the dawning of a new age. Mol Microbiol. 2010;77:15–21. doi: 10.1111/j.1365-2958.2010.07204.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jones JD, O'Connor CD. Protein acetylation in prokaryotes. PROTEOMICS. 2011;11:3012–3022. doi: 10.1002/pmic.201000812. [DOI] [PubMed] [Google Scholar]
- 45.Bonissone S, Gupta N, Romine M, Bradshaw RA, Pevzner PA. N-terminal Protein Processing: A Comparative Proteogenomic Analysis. Mol Cell Proteomics. 2013;12:14–28. doi: 10.1074/mcp.M112.019075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Soppa J. Protein acetylation in archaea, bacteria, and eukaryotes. Archaea. 2010;2010 doi: 10.1155/2010/820681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vizcaíno JA, et al. The PRoteomics IDEntifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res. 2013;41:D1063–9. doi: 10.1093/nar/gks1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dennis PP, Bremer H. Macromolecular composition during steady-state growth of Escherichia coli B-r. J Bacteriol. 1974;119:270–281. doi: 10.1128/jb.119.1.270-281.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dorman CJ. Nucleoid-associated proteins and bacterial physiology. Adv Appl Microbiol. 2009;67:47–64. doi: 10.1016/S0065-2164(08)01002-2. [DOI] [PubMed] [Google Scholar]
- 50.Apweiler R, et al. UniProt: the Universal Protein knowledgebase. Nucleic Acids Res. 2004;32:D115–9. doi: 10.1093/nar/gkh131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nanchen A, Schicker A, Sauer U. Nonlinear dependency of intracellular fluxes on growth rate in miniaturized continuous cultures of Escherichia coli. Appl Environ Microbiol. 2006;72:1164–1172. doi: 10.1128/AEM.72.2.1164-1172.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Glatter T. Large-scale quantitative assessment of different in-solution protein digestion protocols reveals superior cleavage efficiency of tandem Lys-C/trypsin proteolysis over trypsin digestion. J Proteome Res. 2012;11:5145–5156. doi: 10.1021/pr300273g. [DOI] [PubMed] [Google Scholar]
- 53.Silva JC, Gorenstein MV, Li G-Z, Vissers JPC, Geromanos SJ. Absolute quantification of proteins by LCMSE: a virtue of parallel MS acquisition. Mol Cell Proteomics. 2006;5:144–156. doi: 10.1074/mcp.M500230-MCP200. [DOI] [PubMed] [Google Scholar]
- 54.Klumpp S, Zhang Z, Hwa T. Growth rate-dependent global effects on gene expression in bacteria. Cell. 2009;139:1366–1375. doi: 10.1016/j.cell.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Craig R, Beavis R C. TANDEM: matching proteins with tandem mass spectra. Bioinformatics. 2004;20:1466–1467. doi: 10.1093/bioinformatics/bth092. [DOI] [PubMed] [Google Scholar]
- 56.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 57.Ahrné E, et al. An improved method for the construction of decoy peptide MS/MS spectra suitable for the accurate estimation of false discovery rates. PROTEOMICS. 2011;11:4085–4095. doi: 10.1002/pmic.201000665. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.