Abstract
The ribonuclease XRN2 is an essential player in RNA metabolism. In Caenorhabditis elegans, XRN2 functions with PAXT-1, which shares a putative XRN2-binding domain (XTBD) with otherwise unrelated mammalian proteins. Here, we characterize structure and function of an XTBD – XRN2 complex. Although XTBD stably interconnects two XRN2 domains through numerous interacting residues, mutation of a single critical residue suffices to disrupt XTBD – XRN2 complexes in vitro, and recapitulates paxt-1 null mutant phenotypes in vivo. Demonstrating conservation of function, vertebrate XTBD-containing proteins bind XRN2 in vitro, and human CDKN2AIPNL (C2AIL) can substitute for PAXT-1 in vivo. In vertebrates, where three distinct XTBD-containing proteins exist, XRN2 may partition to distinct stable heterodimeric complexes, likely differing in subcellular localization or function. In C. elegans, complex formation with the unique PAXT-1 serves to preserve the stability of XRN2 in the absence of substrate.
Introduction
Ribonucleases (RNases) are key enzymes in RNA metabolism, central to both RNA processing and degradation. The nuclear 5’→3’ exoribonuclease XRN2 functions in ribosomal and small RNA processing1–6, transcriptional termination7,8, clearance of aberrant pre-mRNA9 and tRNA10, degradation of miRNAs11, and other pathways (reviewed in 12,13). Accordingly, XRN2 is conserved across eukaryotes, and essential for viability of yeast and worms14,15. The paralagous XRN1 plays a central role in mRNA degradation16, and both enzymes prefer 5’ monophosphorylated, single-stranded RNAs as substrates17–20.
In yeast, XRN2 (Rat1p) occurs in a complex with Rai1p (Rat1 interacting protein)18,21, which promotes XRN2 activity through unknown mechanisms21,22. However, the metazoan Rai1p orthologues Dom3z (DXO) do not bind to their respective XRN2s21. In C. elegans, XRN2 forms instead a complex with the novel protein PAXT-1 (Partner of XRN-Two)23. Loss of PAXT-1 causes a reduction in XRN2 levels, and, when worms are grown at a relatively high temperature of 26°C, developmental arrest, which can be prevented by increased xrn-2 gene dosage23. As paxt-1 mutations cause reduced XRN2 protein but not mRNA levels, PAXT-1 may stabilize the XRN2 protein23.
Although PAXT-1 is 375 amino acids long, its N-terminal 121 amino acids suffice for XRN2 co-immunoprecipitation, XRN2 accumulation, and animal viability23. This portion of PAXT-1 contains a predicted domain of unknown function, DUF3469, and although PAXT-1 is not well conserved outside nematodes, DUF3469 occurs also in unrelated vertebrate and ciliate but not yeast proteins23. Although it is not known whether these proteins bind XRN2, three of them, mammalian NKRF (NRF) and CDKN2AIP (CARF), and ciliate Tan1 were observed in XRN2 complexes3,24,25. Hence, DUF3469 may function as an XRN-Two-binding domain (XTBD)23.
Here, we sought to elucidate XTBD structure and function. We identify a core XTBD that binds XRN2, present its crystal structure in complex with XRN2, and identify a single XTBD amino acid that makes crucial and conserved contributions to complex formation in vitro and in vivo. We demonstrate that XTBDs are generic XRN2 binders in vitro, and that, despite limited sequence similarity, human CDKN2AIPNL (C2AIL) can substitute for C. elegans PAXT-1 in vivo. Finally, we provide evidence that PAXT-1 buffers cellular RNase activity by preserving XRN2 stability in the absence of substrate.
Results
Conserved Binding Interface of an XTBD – XRN2 Complex
Previously, we showed that the N-terminal amino acids 1–121 of C. elegans PAXT-1, which contain the XTB domain, suffice for XRN2 binding in vivo23. However, Pfam annotates the boundaries of XTBD as residues 7–93 (http://pfam.xfam.org/family/xtbd). Hence, we sought to determine the XTBD core element experimentally. Based on HHPred secondary structure predictions26 and disorder analysis by PSIPRED Protein Analysis Workbench27 and DisEMBL28, we decided to examine recombinant proteins comprising residues 1–121, 1–96, and 1–75, respectively. We found that all three polypeptides bound recombinant XRN2 equally well and comparably to full-length PAXT-1 (Supplementary Fig. 1a). Thus, the functional XTBD is 75 amino acids long, substantially smaller than previously annotated.
To obtain structural information on XTBD, we expressed and purified an XRN2 – PAXT-11–75 complex. To obtain high-quality diffracting protein crystals, we deleted a zinc-finger-containing loop (residues 258–294), a predicted disordered region in the middle of the protein (residues 418–531), and the glycine-rich C-terminus (residues 788–975) of XRN2, yielding XRN2ΔZLC. Following bacterial co-expression of XRN2ΔZLC and PAXT-11–75, purification and crystallization, we determined the structure of the 83 kDa complex at 2.85 Å resolution. The macromolecular complex crystallized in space group P212121 with six XTBD–XRN2ΔZLC heterodimers in the crystallographic asymmetric unit and a solvent content of ~64%. We performed molecular replacement and model building with the help of phased anomalous difference Fourier electron density maps obtained from seleno-methionine derivative crystals; Table 1 summarizes data collection and refinement statistics. The final XTBD–XRN2ΔZLC structure consists of residues 4–25, 35–148, 153–409, and 534–787 for XRN2ΔZLC (chain A) and 1–73 for XTBD (chain B), which show clear electron density.
Table 1. Data collection and refinement statistics.
| XTBD-XRN2ΔZLC | |
|---|---|
| Data collection | |
| Space group | P212121 |
| Cell dimensions | |
| a, b, c (Å) | 170.21, 200.77, 202.99 |
| α, β, γ (°) | 90, 90, 90 |
| Resolution (Å) | 50.00-2.84 (2.91-2.84) * |
| Rsym or Rmerge | 0.126 (1.261) |
| I / σI | 7.1 (0.75) |
| Completeness (%) | 94.6 (84.5) |
| Redundancy | 3.2 (3.1) |
| Refinement | |
| Resolution (Å) | 49.39-2.84 |
| No. reflections | 155484 |
| Rwork / Rfree | 0.185 / 0.238 |
| No. atoms | |
| Protein | 33362 |
| Ligand/ion | 135 |
| Water | 53 |
| B-factors | |
| Protein | 85.9 |
| Ligand/ion | 122.9 |
| Water | 56.4 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.011 |
| Bond angles (°) | 1.18 |
One crystal was used for for data collection and refinement.
Values in parentheses are for highest-resolution shell.
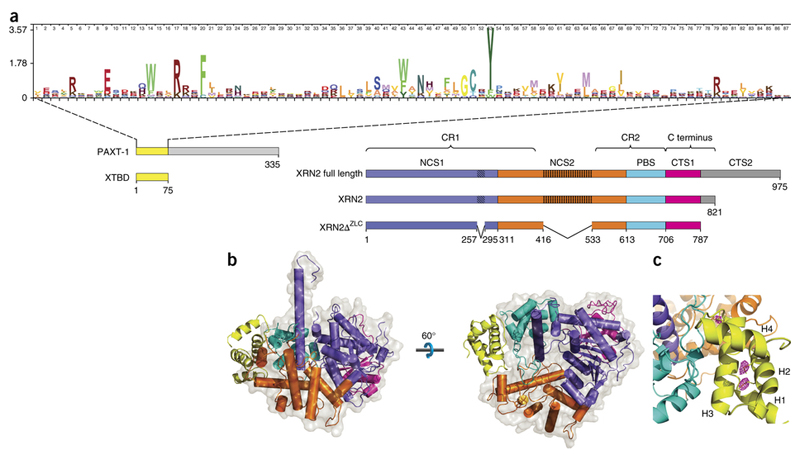
Previous work utilized sequence conservation to assign domains to XRN family proteins, namely CR1 (conserved region 1; residues 1–409) and CR2 (residues 543–713), which are connected by a non-conserved disordered region and extended by a poorly conserved C-terminus, respectively21 (Fig. 1a). To facilitate description and functional dissection of XRN2, we will use an alternative annotation system, namely a nuclease core segment 1 (NCS1; residues 1–310), a nuclease core segment 2 (NCS2; 311–612), a PAXT-1 binding segment (PBS; 613-705), a C-terminal segment 1 (CTS1; 706–787) and a C-terminal segment 2 (CTS2; 788–975) (Fig. 1a,b).
Figure 1.
Architecture of the XTBD – XRN2ΔZLC Complex. (a) Schematic of the PAXT-1 domain, XRN2 functional segments and protein constructs used in this study. Hatched and dotted areas within NCS1 and NCS2 mark the zinc-finger and disordered loop, respectively. Curly-brackets relate the functional segments to the previously assigned domains of XRN nucleases; i.e., conserved regions 1, 2 (CR1, CR2) and the C-terminus. The blow-up shows a sequence logo49 of XTBD. (b) Overall structure of the XTBD – XRN2ΔZLC complex as cartoon model with transparent surface for XRN2ΔZLC. Colors for XRN2ΔZLC and for XTBD correspond to those in (a). (c) Cartoon representation of XRN2 (blue, orange, cyan) in complex with the XTBD (yellow). Seleno-Methionine residues are shown as sticks with corresponding anomalous difference Fourier peaks in magenta (5 σ).
The structure of XRN2ΔZLC is highly similar to that of S. pombe Rat1 (Protein Data Bank (PDB) 3FQD 21) with a root-mean-square deviation (r.m.s.d.) of 1.46 Å over 580 aligned Cα atoms (53% sequence identity). It also superimposes very well onto the exonuclease cores of D. melanogaster and K. lactis XRN1s with r.m.s.d. values of 1.51 Å (503 aligned Cα atoms, 44% sequence identity, PDB 2Y35 20) and 1.36 Å (475 atoms, 42% identity, PDB 3PIE; ref. 29), respectively.
The crystal structure confirms PAXT-11-75 as the core XTBD. It binds to a large groove on XRN2, which is formed by NCS2 and PBS. XRN2 residues 645–681 form a long loop protruding from the globular core of the PBS and fold around the base of a large α-helix (residues 108–139), known as the tower domain21 (Fig. 1b and Supplementary Fig. 1b). XTBD folds into a globular four-helix bundle (H1–H4) connected by three loops (L1–L3) (Figure 1C). H1–H3 form an antiparallel helical array and H4 folds back on top of H2–H3 at a 90° angle. H1 is short, comprising only six residues, whereas helices H2–H4 are much longer (10–15 residues). The four-helical bundle is mainly stabilized by hydrophobic helix-helix interactions together with additional polar interactions between side chains located on neighboring helices. A peculiarity is L3, connecting H3 and H4, which is in a completely linear conformation (Supplementary Fig. 1c). Although a DALI search30 against PDB identified many structures with a topologically similar arrangement of three to four α- helices, either as single units or as part of a larger helical array, these differed substantially in their helix-to-helix angles and therefore represent only distant hits with low Z-scores (Z < 4.5) and rather large r.m.s.d. values (> 3.0 Å). Hence, it seems that the four-helical bundle of XTBD represents a structurally unique arrangement for XRN2 binding with no closely related protein structure present in the PDB.
XTBD Tyrosine 56 is Critical for Complex Formation
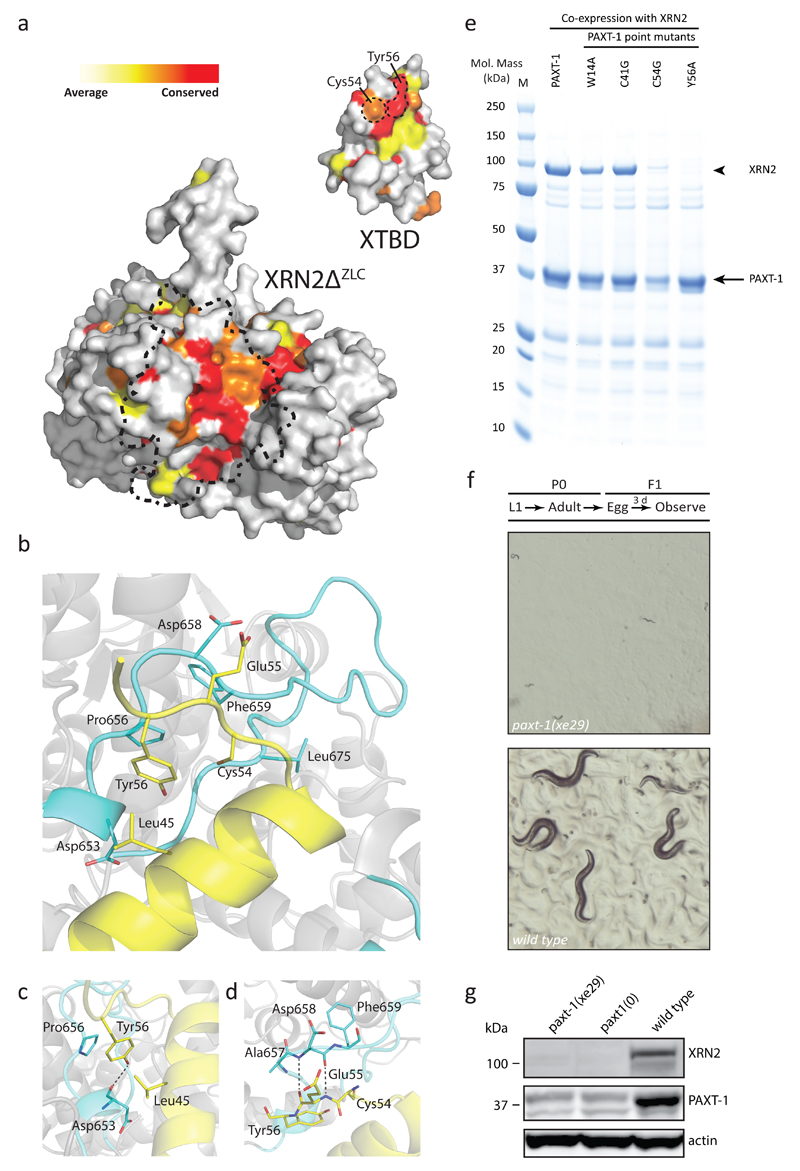
Crystal packing analysis suggests that the XTBD–XRN2ΔZLC complex exists as a single heterodimer and SEC MALS (size-exclusion chromatography coupled to multi-angle light scattering) experiments confirm this: The XTBD–XRN2ΔZLC complex elutes as monodisperse heterodimer with a measured mass of 80.9 kDa (calculated mass: 83.0 kDa) (Supplementary Fig. 2a). Analysis of the protein-protein interface by PISA31 and EPPIC32 reveals a solvent-accessible area on either protein of ~1000 Å2 upon complex formation. 24 residues of XTBD interact with 36 residues of XRN2ΔZLC in a hydrophobic and polar manner, and these interface residues are highly conserved as shown by ConSurf analysis33,34 of both proteins (Fig. 2a). The hydrophobic character of the interface is mainly formed by XTBD residues on L1, H3, and L3 (Trp14, Ile37, Cys41, Leu52, Cys54, Tyr56, Leu60) and contributes to interaction with XRN2ΔZLC residues Val553, Phe560, Pro591, Pro650, Ile652, Asp653, Pro656, Pro677, and Phe678 (Fig. 2b and Supplementary Fig. 2b). The formation of 13 hydrogen bonds and three salt bridges provides additional binding energy (Fig. 2c,d and Supplementary Fig. 2c). Although the XTBD - XRN2 interface is relatively small at ~1000 Å2, the complex is very stable and resists disruption by high salt washes (data not shown) and high dilution (Supplementary Fig. 2a).
Figure 2.
Tyr56 Is Critical for Stability of the Highly Conserved XTBD–XRN2ΔZLC Interface. (a) Surface representation of XRN2ΔZLC and XTBD structures highlighting conservation of the protein – protein interface (XTBD rotated 180° with respect to XRN2ΔZLC). Position-specific conservation scores computed by Consurf33,34 are displayed from white (weak to average conservation, Consurf levels 1-6) to red (most conserved, level 9). The dashed-dotted line on XRN2ΔZLC represents the projection of bound XTBD. Dashed lines on XTBD indicate residues analyzed in detail. (b-d) Representation of XTBD residues Cys54 and Tyr56 and their interaction with XRN2ΔZLC residues, zooming in on (c) interactions of the critical Tyr56 and (d) the side chain arrangement of Cys54 and Tyr56. Hydrogen bonds are indicated by gray dotted lines. (e) Gel showing results of Ni-NTA-mediated pull-down of His6-PAXT-1 and its point mutant variants, respectively, co-expressed with XRN2. (f) Stereomicroscropic images of C. elegans grown as indicated schematically. The paxt-1(xe29) allele encodes PAXT-1_Y56A and was obtained by genome editing of endogenous paxt-1. (g) Western blot of 100 µg lysate from worms of the indicated genotype grown at 26°C using antibodies specific to the indicated endogenous proteins.
To further dissect the XTBD–XRN2ΔZLC interaction, we leveraged the structural information to design PAXT-1 interface point mutant proteins, which we co-expressed with XRN2. We examined complex formation by purifying the PAXT-1 proteins through their His6-tags by Immobilized Metal Affinity Chromatography (IMAC). The PAXT-1 point mutations Trp14-to-Ala and Cys41-to-Gly, respectively did not compromise XRN2 co-purification (Fig. 2e; Supplementary Data Set 1). Less XRN2 co-purified with PAXT-1_Cys54-to-Gly but this mutation also decreased the amount of PAXT-1 purified (Fig. 2e and Supplementary Fig. 2e). Cys54-to-Gly may thus disrupt protein integrity, although we note that the level of PAXT-1_Cys54-to-Gly in bacterial lysates was comparable to that of wild-type PAXT-1 (Supplementary Figure 2e). By contrast, the Tyr56-to-Ala mutation completely abrogated interaction of XTBD with XRN2 without affecting PAXT-1 levels or purification (Fig. 2e and Supplementary Fig. 2e,f), highlighting Tyr56 as a key interface residue.
Since numerous side chains contribute to the total binding energy of the interface, loss of binding through a single point mutation appears surprising. However, it is explained by the crucial role of Tyr56 in shaping the unique linear conformation of the loop L3. With all seven residues of XTBD51-57 engaged in XRN2 binding (Supplementary Fig. 2b), L3 contributes 33% (338 Å2) buried surface area (BSA) and four hydrogen bonds to the XTBD – XRN2 interface, whereas the other 17 interacting residues together contribute the remaining 698 Å2 BSA and nine hydrogen bonds. Mutation of Tyr56 to alanine abrogates the key constraint for the 5157 conformation by eliminating the phenol ring that is normally sandwiched between Leu45 (XTBD, H3) and Pro656 (XRN2ΔZLC), forming a CH – π – interaction with Pro656 (XRN2ΔZLC) (Fig. 2c). Through its hydroxyl group, Tyr56 also forms a hydrogen bond (2.8 Å) with the backbone carbonyl of Asp653 (XRN2ΔZLC) (Fig. 2c). A second constraining residue, XTBD_Cys54, is in hydrophobic interaction with Pro656 (XRN2ΔZLC) and additionally forms a weak hydrogen bond (3.8 Å) with the backbone carbonyl of Leu675 (XRN2ΔZLC) (Supplementary Fig. 2c). Together, Cys54 and Tyr56 promote formation of an isolated β-bridge between backbone atoms of XTBD_Glu55 and XRN2_Asp658 (Fig. 2d). Mutation of Tyr56 to alanine and Cys54 to glycine removes the constraining side chains for this stretch and induces backbone flexibility, thus compromising all interactions between XTBD_51 – 57 and XRN2. The resulting loss of one third of BSA explains the severe consequences of these mutations. By contrast Trp14-to-Ala or Cys41-to-Gly have only limited effects on the BSA and thus no major effect on complex formation in vitro.
To validate the importance of XTBD_Tyr56 for XRN2 binding under physiological conditions, we utilized genome editing35,36 to create the Tyr56-to-Ala point mutation in endogenous PAXT-1. Strikingly, when we grew the resulting paxt-1(xe29) worms at 26°C, they arrested as L1 larvae, just like paxt1(0) animals (Fig. 2f). Also, just like in paxt-1(0) animals23, accumulation of XRN2 was reduced and PAXT-1 undetectable by western blotting (Fig. 2g and Supplementary Fig. 2d; Supplementary Data Set 1). Formally, we cannot exclude that destabilization of PAXT-1 through the Y56A mutation causes these effects. However, the protein is stable in vitro (Fig. 2e), as is a human XTBD protein with an analogous mutation in human cells (see below). Moreover, loss of XRN2 destabilizes PAXT-1 in vivo23. Hence, we propose that PAXT-1 depletion in vivo is a secondary consequence of its inability to bind XRN2. At any rate, our data identify Tyr56 as a critical residue for PAXT-1-mediated stabilization of XRN2 in vitro and in vivo, and explain the structural basis of this importance. They also explain the high degree of conservation of this residue: In XTBDs, this position is always held by residues with aromatic side chains (Tyr, Phe, Trp; Fig. 1a and Supplementary Fig. 3b), which can occupy the XRN2 Pro656 pocket via a stacking interaction.
XTBD Is a Generic XRN2 Adapter
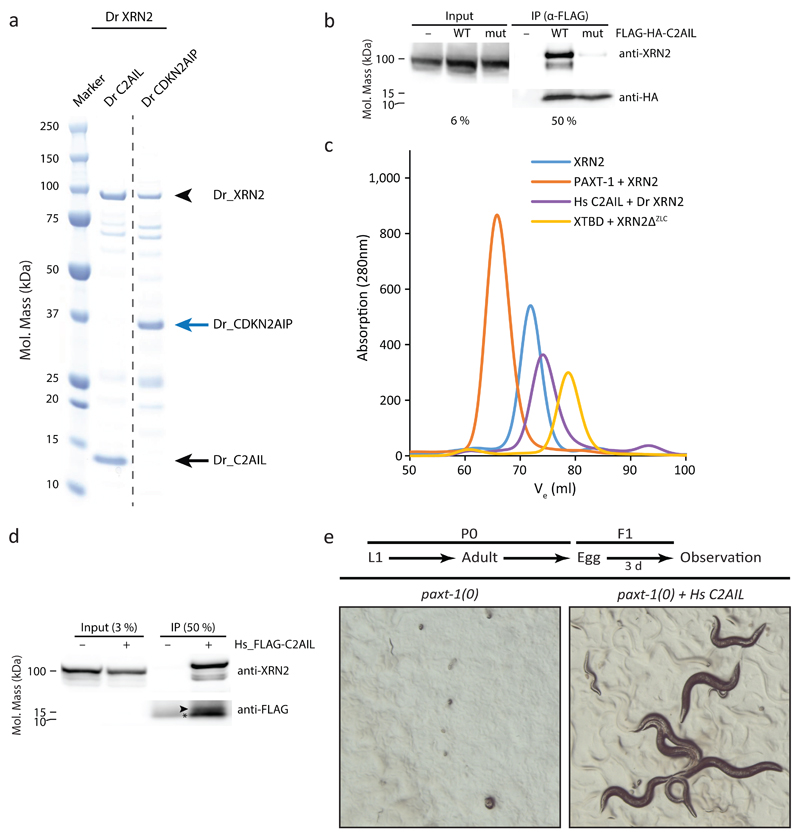
Although different XTBD-containing proteins occur in complexes with XRN2 3,23–25, these interactions were generally examined through co-immunoprecipitation, leaving it unclear whether they are direct and mediated by XTBDs. The fact that the interaction surfaces of both XRN2 and XTBD are highly conserved (Fig. 2a; Supplementary Figure 3b, c) supports a conserved and direct interaction. To test this notion experimentally, we produced recombinant XRN2 and the XTBD-containing proteins CDKN2AIP and CDKN2AIPNL (C2AIL in the following) from Danio rerio (zebrafish) in E. coli and determined their interactions. Whereas CDKN2AIP is a tumor suppressor that can activate p53 (ref. 37–39), C2AIL is as yet uncharacterized. When purifying the N-terminally-His6-tagged XTBD-proteins, XRN2 co-purification was readily detectable in both cases (Fig. 3a; Supplementary Data Set 1). Similarly, human XRN2 co-purified with human His6-C2AIL (data not shown).
Figure 3.
XTBD is a generic XRN2 binder. (a) Gel showing results of Ni-NTA-mediated pulldown of D. rerio His6-C2AIL and D. rerio His6-CDKN2AIP, respectively, each co-expressed with D. rerio XRN2 (aa 1-803) in E. coli. The dashed line indicates splicing of two non-adjacent lanes from a single gel. (b) Western blot of anti-FLAG immunoprecipitates and input lysates from HEK293T cells expressing wild-type transgenic human FLAG-HA-tagged C2AIL (FLAG-HA-C2AIL, “WT”), a Tyr82-to-Ala mutant variant thereof (“mut”), or GFP as mock control (“-“). (c) SEC elution profiles of C. elegans XRN2 alone (blue line), and complexes of PAXT-1 – XRN2 (orange), XTBD – XRN2ΔZLC (yellow) and H. sapiens C2AIL – D. rerio XRN2 (aa 1-803, purple line). (d) Western blot of anti-FLAG immunoprecipitates and input lysates from paxt-1(0) mutant C. elegans expressing human FLAG-tagged C2AIL (Hs_FLAG-C2AIL) from an integrated single-copy integrated transgene. ‘–‘ denotes non-transgenic wild-type animals used as negative control. (e) Stereomicroscropic images of paxt-1(0) mutant C. elegans carrying the Hs_FLAG-C2AIL encoding transgene or no transgene and grown as indicated by the scheme (above pictures).
To confirm specificity of the interaction between human C2AIL and XRN2, we mutated C2AIL_Tyr82, which corresponds to PAXT-1_Tyr56 (data not shown). We transfected human HEK293T cells with either wild-type or Tyr82-to-Ala mutant human FLAG-HA-C2AIL and immunoprecipitated the respective protein through an anti-FLAG antibody. Western blotting revealed robust co-immunoprecipitation of endogenous XRN2 with wild-type but not mutant FLAG-HA-C2AIL (Fig. 3b and Supplementary Data Set 1). Finally, D. rerio XRN2 formed a stable, chimeric complex with human C2AIL by SEC (Fig. 3c). We conclude that XTBD constitutes a generic XRN2-binding domain whose mode of interaction with XRN2 is conserved across animal phylogeny.
Human C2AIL Can Substitute for C. elegans PAXT-1 in vivo
To validate our insights into XTBD conservation from in vitro and cell-based studies in vivo, we expressed a codon-optimized and flag-tagged human c2ail single-copy transgene in paxt-1(0) mutant C. elegans. Endogenous C. elegans XRN2 co-immunoprecipitated with FLAG-Hs_C2AIL from worm lysates as confirmed by western blot (Fig. 3d and Supplementary Data Set 1) and mass spectrometry (data not shown). This confirms the formation of a chimeric complex in vivo.
Although PAXT-1 is dispensable at lower temperatures, paxt-1(0) animals arrest as L1-stage larvae when grown at 26°C, a phenotype that expression of PAXT-1_1-121 rescues23. Strikingly, the hs_c2ail transgenic worms similarly continue development into adulthood and go on to produce F2 progeny (Fig. 3e). Hence, not only the molecular but also the developmental function of PAXT-1 can be taken over by human C2AIL, despite only ~35% sequence identity between their XTBDs, but consistent with the good correlation of the C2AIL homology model with the XTBD structure (Supplementary Fig. 3a).
XTBD- or Substrate-Binding Stabilize XRN2
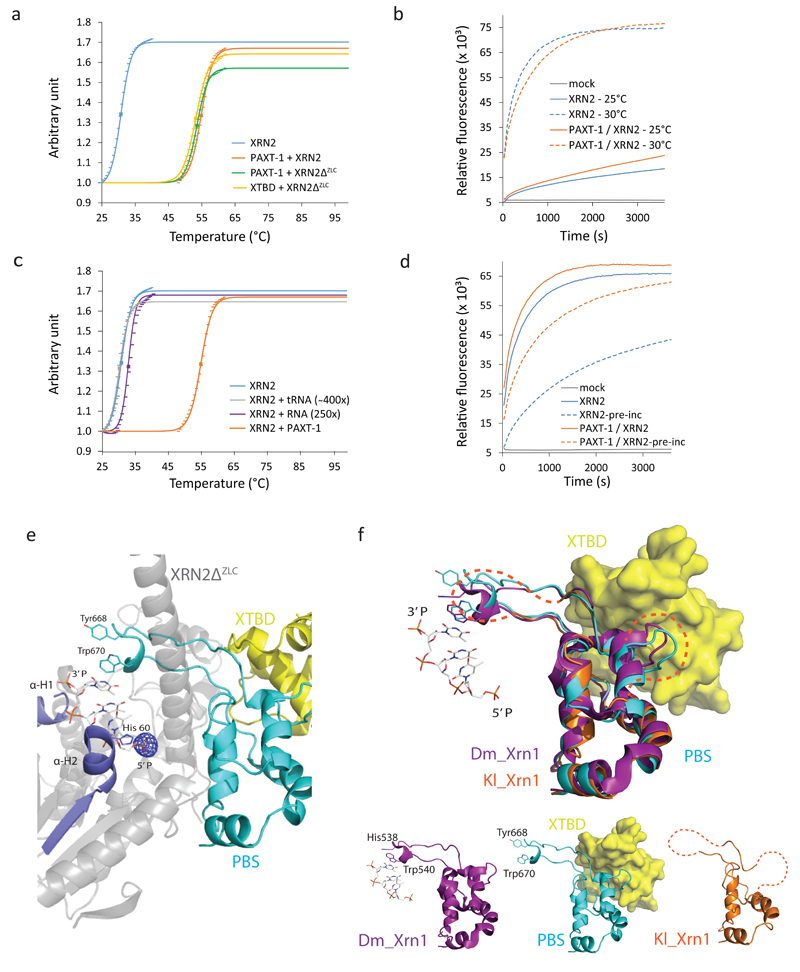
Previously we examined the influence of PAXT-1 on the enzymatic activity of XRN2 23 and found greater activity of the PAXT-1 – XRN2 complex than XRN2 alone over extended periods of time. However, initial velocities remained unchanged in the presence or absence of PAXT-1 23, and examination of Michaelis-Menten enzyme kinetics also failed to provide evidence for modulation of XRN2 activity by PAXT-1 (Supplementary Fig. 4a). Since XRN2 protein but not mRNA levels are reduced in C. elegans lacking PAXT-1 23, PAXT-1 thus appears to have a stabilizing rather than stimulatory effect on XRN2. To test this hypothesis experimentally, we monitored unfolding of recombinant XRN2 and its complex in vitro and determined the melting temperature Tm (Fig. 4a). We found a striking shift in Tm of 24°C, from Tm ≈ 31°C for XRN2 to Tm ≈ 55°C for the PAXT-1–XRN2 complex, demonstrating a pronounced stabilizing activity of PAXT-1 towards XRN2. The activity largely resides in the XTBD as the Tm values of PAXT-1 – XRN2, PAXT-1 – XRN2ΔZLC and XTBD – XRN2ΔZLC complexes differed little at 55°C, 54°C, and 53°C, respectively.
Figure 4.
XTBD or RNA substrate, respectively, stabilize XRN2 by promoting PBS folding. (a), (c) Graphs showing unfolding of (a) XRN2 alone and its indicated complexes, and (c) XRN2 in the presence of an RNA substrate or tRNA, respectively. Results for XRN2 alone and XRN2 – PAXT-1 from (a) are included for comparison. (a), (c) Data points of the sigmoidal part of each melt curve and their fits to the Boltzman equation, normalized to 1 and scaled, are indicated. Squares represent melting points (Tm). (b) Plot of activity over time of XRN2 and its complex, respectively, at 25°C and 30°C, respectively. ‘Mock’ is the negative control (no enzyme) at 30°C. (d) Plot of activity of XRN2 alone and its complex with or without preincubation at 30°C. (e) Representation of XRN2 substrate binding, inferred by superposition of XRN2ΔZLC (gray/red/cyan) in complex with XTBD (yellow) on D. melanogaster XRN1 in complex with substrate (PDB 2Y35 20). For the latter, only substrate is shown, in stick model. An mFo-DFc map of the XTBD–XRN2ΔZLC complex (calculated in absence of SO42-) is overlaid and displayed at the SO42- position only (5.5 σ). (f) Comparison of the PBSs of C. elegans XRN2 (cyan) in complex with XTBD (yellow, surface representation), D. melanogaster XRN1 (purple; PDB 2Y35 20) in complex with a tri-nucleotide substrate (stick-model), and K. lactis XRN1 (orange; PDB 3PIF 29) by superposition (top) or individual representation (bottom). Disordered regions within the K. lactis PBS are marked in orange dashed lines.
At 31°C, the Tm of XRN2 alone is close to the temperature at which paxt-1(0) larvae arrest development (26°C), suggesting that XRN2 destabilization in vivo may occur through its unfolding and subsequent degradation. To examine the consequences of temperature increases on XRN2 activity in vitro, we tested it at two temperatures within, or close to, the physiological range of C. elegans: 25°C, the default temperature of previous activity assays, and 30°C. From the melt curve, we estimated that 45% of XRN2 would be unfolded at 30°C. Nonetheless, and contrary to our expectation, we found that XRN2 activity was consistently, and over extended times, higher at 30°C than 25°C (Fig. 4b).
Although an increase in temperature accelerates enzymatic reaction, according to Arrhenius’ equation we would expect at most a modest effect with the narrow temperature window that we used. Hence, we considered that XRN2 might be unexpectedly stable under the conditions of the assay, possibly due to substrate binding. To test this, we examined XRN2 thermal stability in the absence and presence of the 30mer RNA substrate used for catalytic assays. This revealed a stabilizing effect of the substrate, which increased the Tm by 2°C to ~33°C (Fig. 4c). Although the effect was small relative to that seen for PAXT-1, it is above the 1°C threshold previously proposed to be relevant40, and it occurred reproducibly at two different substrate concentrations (data not shown). Moreover, it was highly specific, as inclusion of tRNA, which is not a good XRN2 substrate due to high secondary structure and paired 5’ terminal nucleotides18,20, did not increase thermal stability of XRN2. We conclude that substrate binding can stabilize XRN2.
To test if the stabilizing effect of substrate binding explained the unexpectedly robust activity of XRN2 at 30°C, we pre-incubated either XRN2 alone or a PAXT-1 – XRN2 complex in buffer for five minutes at this temperature before adding substrate. Whereas the PAXT-1 – XRN2 complex exhibited robust activity under these conditions, activity of XRN2 alone was compromised (Fig. 4d and Supplementary Fig. 4b). Hence, PAXT-1 can preserve activity of XRN2 in the absence of substrate.
XRN2 Stability Depends on Structural Integrity of the PBS
To understand the interaction of XRN2 with substrate, we superimposed the XRN1 – RNA substrate complex (PDB 2Y35; 20) onto the XTBD – XRN2 structure. This revealed a crucial role of the PBS-protruding loop in substrate binding, as its Trp670 base-stacks with three unpaired 5’-terminal nucleotides and His60 of α-helix 2 (Fig. 4e). Moreover, Trp670 in XRN2, like Trp540 in XRN1 20, acts in concert with the N-terminal α-helix 1 to restrict sterically access to the active site to single stranded RNA. A sulfate ion from the crystallization buffer of the XTBD – XRN2 complex occupies the precise position of the 5’ phosphate of the XRN1–RNA substrate complex (Fig. 4e), confirming conservation of the mode of substrate binding.
From the structures of the two complexes it is thus evident that both substrate and PAXT-1 bind to the PBS. To determine whether this might affect PBS structure, we compared the PBSs from these two complexes to that of K. lactis XRN1, which was crystallized in the absence of both substrate and additional proteins (PDB 3PIF; 29). Strikingly, whereas the PBS is fully structured in XRN2 – XTBD and Drosophila XRN1 – RNA complexes, it is heavily disordered in the structure of K. lactis XRN1 and the domain could not be entirely built in the crystallographic model (Fig. 4f). Hence, although not a proof, these structures are consistent with the notion that stabilization of XRN2 is achieved through maintenance of the integrity of the PBS, which both XTBD- and substrate-binding can accomplish (Fig. 4f).
Discussion
Based on the recurring identification of XTBD- (DUF3469-) containing proteins in complexes that also contain XRN2 3,23–25, we previously hypothesized that it mediate XRN2-binding23. Here, we have tested this notion and demonstrate that a core XTBD, substantially smaller than initially predicted, mediates binding to XRN2. Chimeric interactions between XTBD-containing proteins and XRN2 from different species in vivo and in vitro confirm the generality of this finding. Although XRN2 and its paralogue XRN1 are highly similar, including in the PAXT-1 binding groove (Supplementary Fig. 1e), binding of PAXT-1 to XRN1 is disfavored: the NCS2 of XRN1 extends further into the groove than that of XRN2, suggesting a steric clash with XTBD (Supplementary Fig. 1e). This is consistent with co-immunoprecipitation experiments, which revealed a specific interaction of PAXT-1 with XRN2, but not XRN1 23. Hence, XTBD is a generic and specific XRN2 binding domain. Interestingly, its binding to XRN2 is entirely distinct from that of Rai1p (Supplementary Figure 1d; 21 ), which may explain the distinct functions of the two proteins, promotion of XRN2 activity by Rai1p 21,22 versus XRN2 stabilization by PAXT-1 (23. and this study).
Although PAXT-1 is the only XTBD-containing protein in C. elegans, many vertebrates have three such predicted XTBD proteins, CDKN2AIP, C2AIL, and NKRF 23. Outside their XTBDs, these proteins exhibit little sequence homology, and their cellular functions appear distinct: Mammalian NKRF was identified as a transcriptional repressor41, CDKN2AIP appears to function as an activator of p5337–39, and C2AIL currently has no known function. However, the strong interactions that we observe between XTBDs and XRN2 imply that all of these proteins will exert at least part of their functions with or through XRN2. The ability to uncouple these proteins from XRN2 through a single point mutation now enables testing of this notion for each of these proteins.
The stable and heterodimeric binding between XRN2 and XTBD also implies that vertebrate XRN2 exist in multiple separate and non-interconvertible complexes, potentially diversifying XRN2 functions. Different binding partners may endow XRN2 with distinct catalytic activities, facilitate differential regulation of XRN2 through distinct subunits, or recruit XRN2 to distinct cellular locations. Experiments on human U-2 OS osteosarcoma cell lines rovide circumstantial evidence for the latter, as XRN2 immunofluorescence staining is observed in both nucleus and nucleoplasms, whereas NKRF locates to the nucleolus, from which the nucleoplasmic CDKN2AIP appears excluded42. As the phosphorylation status of NKRF may modulate the interaction between NKRF and XRN2 43, it will be interesting to see whether it also affects localization of NKRF and/or XRN2.
According to such a model of functional specialization of XRN2 complexes, XTBD serves to bridge XRN2 with specific protein domains that provide additional functionalities. As a complex of XRN2 with full-length PAXT-1 did not crystallize, we do not know what such functions could be in the case of the PAXT-1 C-terminal sequence, which is conserved across nematodes. Speculatively, it may modulate XRN2 activity or block access of specific substrates to the XRN2 active site. This is because the structure of the XRN2-XTBD complex implies that the PAXT-1 C-terminus would be located right in front of the XRN2 nuclease cleft, and because analysis by PSIPRED 27 and DisEMBL 28 suggests it to be structured (data not shown). Since we have thus far been unable to observe regulation of XRN2 activity by PAXT-1 in vitro, such functions might require specific substrates or conditions that remain to be identified. Alternatively, the PAXT-1 C-terminus might modulate PAXT-1 stability or binding to XRN2.
Supporting a more specialized role of the PAXT-1 C-terminus, it is dispensable for C. elegans viability23. Together with the fact that the heterologous human C2AIL can substitute for PAXT-1 in vivo, this then implies functions of XTBD beyond that of a passive XRN2 adapter. We propose stabilization of XRN2 by promoting folding of its PBS segment to be such a function. The Tm is a measure of a protein’s thermal stability and depends on numerous molecular parameters such as the amino acid composition, surface hydrophobicity, number of hydrogen bonds and labile secondary structures such as loops44. Upon XTBD-binding, 13 hydrogen bonds are formed, a hydrophobic patch covered and the flexibility of the PBS loop reduced. Decreased thermal stability of XRN2 in the absence of XTBD in vitro may then reflect decreased half-life in vivo45–47, as even partially unfolded proteins exhibit increased susceptibility to proteolysis47,48. This then explains how PAXT-1/XTBD binding increases XRN2 levels in vivo.
Although PAXT-1 is expressed throughout C. elegans development23, its loss causes defects only at specific stages: when paxt-1(0) mutant animals are shifted to the restrictive temperature of 26°C during the fourth larval (L4) stage, they will continue development through adulthood and produce F1 progeny, which, however, will arrest at the L1 stage23. This delayed effect does not simply reflect slow depletion of XRN2, as L1 stage arrest of F1s also occurs if parental animals were already shifted to 26°C during their L1 stage23. Moreover, although animals may conceivably be more dependent on XRN2 activity during some developmental stages than others, developmental arrest can be induced in virtually any stage upon direct inactivation of XRN2 15. Hence, we speculate that it is the stability of XRN2 that varies with development stage. Specifically, with substrate binding conferring some stability to XRN2, the stabilizing activity of PAXT-1 may be more important when substrate levels are low. Thus, a key function of PAXT-1 may be to preserve, rather than promote XRN2 activity, guarding enzyme not bound by substrate against decay to retain a constant pool of active protein.
Online Methods
Experimental replicates and sample sizes
Experiments were performed in experimental replicates as follows: Fig. 2e - n=3, Fig. 2f - n=3, Fig. 2g - n=3, Fig. 3a - n=2, Fig. 3b - n=2, Fig. 3d - n=2, Fig. 3e - n=3, Fig. 4a - n=3, Fig. 4b - n=2, Fig. 4c - n=2, Fig. 4d - n=3. For enzymatic and unfolding assays in Figure 4 and Supplementary Figure 4, replicate experiments were carried out on separate occasions but utilized the same sample preparations. All other experiments used biological replicates.
Animal experiments
Experiments were not blinded; no statistical calculations were performed.
Protein Expression and Purification
Expression constructs were cloned into a modified bis-cistronic pCOLADuet™ vector from Novagen. XTBD-containing constructs were cloned into the first multi-cloning site (MSC), and thus N-terminally extended by a Hexahistidine (His6) tag followed by a tobacco etch virus protease (TEV) cleavage site, whereas the XRN2 constructs in the second MSC remain untagged. All constructs were expressed in E. coli BL21 (DE3). Cells were lysed by sonication and the lysate cleared by centrifugation. Immobilized metal affinity chromatography (IMAC) slurry (Qiagen) was used to bind the His6-tag of the target protein. Next, protein was eluted through cleavage by TEV (home made)50. IMAC was repeated and the unbound fraction collected, diluted, and subjected to a Mono Q™ 5/50 GL (GE Healthcare) anion exchange column. Positive fractions were pooled, filtered with a Millex-GV 0.22 µm filter and loaded onto a Superdex™ 200 HiLoad™ 16/600 (GE Healthcare) column. Finally the target fractions were collected and concentrated using Millipore® Amicon® Ultra Centifugal Filter Concentrators with suitable pore size.
Incorporation of seleno-methionine was done as described by Molecular Dimensions’ SelenoMethionine media kit, using E.coli B834(DE3).
Crystallization, Data Collection, and Structure Solution
Protein crystallization was carried out at 20°C using the sitting-drop vapor diffusion method with a Phoenix robot (Art Robbins) dispensing 300 nl drops (100 nl crystallization buffer and 200 nl protein solution). XRN2ΔZLC–XTBD crystals, obtained in 2 M ammonium sulfate, 0.1 M Bis-TRIS pH 6.5 or 0.1 M TRIS pH 8.5 were harvested and flash-cooled in liquid nitrogen after cryoprotection with 3.2 M ammonium sulfate, 0.1 M Bis-TRIS pH 6.5 or 0.1 M TRIS pH 8.5. Highest resolution data was obtained with a seleno-methionine derivative crystal diffracting to 2.85 Å (λ=0.97796, T=100 K) at the SLS PX-II beamline (Villigen, Switzerland) and belonged to space group P212121 with six XRN2ΔZLC–XTBD heterodimers in the asymmetric unit. Diffraction data was integrated and scaled using the XDS program package51 and the structure was solved by the molecular replacement method with PHASER 52 using an XRN2 homology model consisting of the exonuclease core as search model. Phases from this solution were calculated and used for automatic model building with BUCCANEER 53. The XRN2ΔZLC–XTBD structure was further improved by the crystallographic simulated annealing routine followed by grouped B-factor refinement in PHENIX 54 and several rounds of manual rebuilding in COOT 55 and refinement in PHENIX and BUSTER (Bricogne G., Blanc E., Brandl M., Flensburg C., Keller P., Paciorek W., Roversi P, Sharff A., Smart O.S., Vonrhein C., Womack T.O. (2011). BUSTER version 2.11.5 Cambridge, United Kingdom: Global Phasing Ltd.; www.globalphasing.com/buster/). Non-crystallographic symmetry restraints were used throughout the structure solution and refinement process. NCS copies of XRN2ΔZLC and XTBD are highly similar with RMSD values between 0.456 to 0.597 Å (Cα) for XRN2ΔZLC (chain A related to all other XRN2ΔZLC chains present in the asymmetric unit) and 0.313 to 0.741 Å (Cα) for XTBD (chain B related to all other XTBD chains present in the asymmetric unit). Model building and validation was carried out with the help of anomalous difference Fourier electron density maps to locate seleno-methionine positions. The final structure was validated using COOT (Ramachandran plot statistics: Allowed 99.6%, outliers: 0.4%) and the wwPDB Validation Server (http://wwpdb-validation.wwpdb.org/validservice). Atomic coordinates and structure factors for the XRN2ΔZLC – XTBD complex have been deposited in the PDB with entry code 5FIR. Structural images for figures were prepared with PyMOL (www.pymol.org; Version 1.3r1).
Sequence Logo
The profile hidden Markov model (HMM) of DUF3469 from Pfam was exported and uploaded to Skylign.org 49. For concise representation, the following parameters were set: a) Alignment Processing to “Create HMM - remove mostly-empty columns”, b) Fragment Handling to “Alignment sequences are full length” and c) Letter Height to “Information Content - Above Background”. The resulting logo was exported without showing the gap parameters.
Thermal Shift Assay
In a 20 µl reaction, 2 µg of assayed protein was mixed with Protein Thermal Shift™ Dye and – Buffer from Applied Biosystems to a final concentration of 1x. In the presence of RNA substrate, however, 1 µg of protein was used instead to reduce the otherwise high concentrations of RNA. Purified yeast tRNA (Ambion™) or a 30mer XRN2 substrate, identical to that used in the kinetic assays (see below) were used when examining the effect of substrate binding. The reaction was prepared in a 96 well MicroAmp® optical microplate at 4°C and sealed with MicroAmp® Optical Adhesive Film. After centrifugation for 25 s at 1,000 g the plate was read in a StepOnePlus™ real-time PCR system using a melt curve program from 25 – 99°C. No normalizing quencher was used and reactions were run in triplicates. Raw data was exported and plotted using Microsoft Excel. The sigmoidal part of the curve was fit to the Boltzman equation (equation 1) using non-linear regression by maximizing R² and varying Tm and C, and the melting point Tm calculated as the inflection point. The resulting function was normalized to 1 and plotted (see below). Raw-data was also normalized to 1 and plotted as dashed lines. In addition, XRN2, XRN2 + tRNA, and XRN2 + RNA curves and values were re-scaled by a factor of 1.9, 1.3, and 1.8, respectively for concise representation.
| (equation 1) |
where Fpre is baseline fluorescence, Fpost is maximal fluorescence, T is temperature in °C, Tm is melting temperature in °C and C is enthalpy.
Real-time Exonuclease Activity Assay and Kinetic Analysis
The kinetic assay was performed as described in 56 with the following modifications: The RNA-DNA duplex substrate, where the RNA is FAM-labeled whereas the DNA carries a quencher, was diluted with fourfold excess of unlabeled 5’ phosphorylated let-7, which increased the dynamic range of the qPCR system. 300 fmol enzyme was assayed with varying concentrations of total substrate (sum of RNA-DNA duplex and let-7) in an excess from 5 to 200 fold. Each kinetic reaction was run in triplicates and gave 1 data point. After each data point was normalized to its baseline, the initial linear phase from 0 to 180 s was fitted by linear regression. The slope of this function equals the velocity at the corresponding substrate concentration [S]. Furthermore the average of three velocities was plotted over [S] and fitted to the Michaelis – Menten-model (equation 2) with the least-square method.
| (equation 2) |
where V is velocity, Vmax is maximal velocity, [S] is substrate concentration and KM is the Michaelis- Menten- constant.
A 50x substrate excess is used in Figure 4b. Preincubation time in Figure 4d is 5 min at 30°C.
Strains
Standard procedures were used to culture the Bristol N2 wild-type, mutant and transgenic strains 57. HW1091: paxt1-(xe5) is the null mutation described by 23. Generation of HW 1641: paxt-1(xe5) I; xeSi203[Ppaxt-1::C.elegans_co_human_C2AIL_FLAG_ operon_GFP_tbb-2-3'UTR, unc-119(+)] II and HW 1644: paxt-1(xe29[I:8597012-8597014 = TAT -> GCT, I:8597041 A -> T] (expressing PAXT-1_Y56A and containing a silent mutation to facilitate screening for genome editing events) are described in detail below.
Genome Editing of C. elegans Using CRISPR-Cas9
A suitable site for a 20 nt long sgRNA was identified by considering availability of a PAM site to create a double strand break on the paxt-1 gene. Two complementary oligonucleotides were hybridized and cloned by Gibson assembly58 into pIK111, a derivative of the PU6::sgRNA backbone59,60. Furthermore, a 100 nt DNA single stranded repair oligo was designed with the desired point mutation and a silent mutation at the sgRNA binding site. A CRISPR co-conversion mix, also containing Peft-3::Cas9::tbb-23’ UTR (ref. 36), the sgRNA and oligonucleotide to recreate the sqt-1(sc1) mutation (analogous to 35) was injected into N2 animals61. Roller progeny were cloned and their progeny were analyzed for desired mutations by PCR.
Rescue Experiments Using MosSCI
The paxt-1 promoter, H. sapiens C2AIL (codon-optimized + two artificial introns) coding sequence and tbb-2 3’ UTR sequences were cloned into pCFJ150 by Gateway cloning. An operon_GFP sequence between C2AIL and the tbb-2 3’UTR was included to permit visualization of transgene expression without altering the sequence of the encoded protein. The resulting plasmid was inserted into the ttTi5605 site by MosSCI (refs. 62,63), yielding xeIs203[Ppaxt-1::C.elegans_co_human_C2AIL_FLAG_operon_GFP_tbb-2-3'UTR,unc-119(+)] II animals. The integrants were outcrossed to wild-type N2 and crossed into the paxt-1(xe5) mutant. Rescue of the paxt-1(0) mutant phenotype was examined as reported23.
Cell Culture and Transient Transfection of HEK293T Cells
HEK293T cells were available in our lab, checked for mycoplasma contamination and grown in DMEM-F12 (Gibco™) supplemented with 10% (v/v) FCS and 25 U/ml Penicillin-Streptomycin (Gibco™) at 37°C. Prior to transfection (described in 64) using the PEI method, 2·106 cells were plated on a 6 cm dish per construct and grown for one day. Each transfection was run in 2 technical duplicates with plasmids derived from pIRESneo (Addgene plasmid #10822) to encode N-terminally FLAG-HA-tagged wild-type or Tyr82-to-Ala mutated hsa_C2AIL. pSD44 harboring GFP was used as mock control. It is modified from the pRRLSIN.cPPT.PGK-GFP.WPRE (Addgene plasmid #12252) backbone, containing the SV40 enhancer/early promoter driving expression of a puromycin selectable marker. Cells were harvested 72 h after transfection.
Western Blot and Co-Immunoprecipitation Experiments
Worm lysates were prepared by douncing harvested worms with 150 pestle strokes in extraction buffer (50 mM HEPES, pH 7.4, 50 mM KCl, 5 mM MgCl2, 0.1% w/v Triton X-100, 10% w/v Glycerol) finalized with 7 mg protease inhibitor (Roche complete EDTA-free), 2 µl 1 M DTT and 10 µl 100 mM PMSF per ml extraction buffer. The lysate was cleared by centrifugation at 16,000 g for 20 min and protein concentrations of the lysates were measured using Bradford reagent (Bio-Rad Protein Assay). Subsequently, 100 µg of lysate were loaded and separated by a Bis-TRIS SDS-PAGE 4 – 12% gradient gel (NuPAGE® Life Technologies). Proteins were then blotted on a PVDF membrane using standard procedures. Antibodies against XRN2 15, PAXT-1 23 and FLAG-tag (Monoclonal ANTI-FLAG® M2-Peroxidase, Sigma, Cat.-Nr. A8592) were used in a 1:500, 1:500 and 1:2000 dilution, respectively. Horseradish peroxidase-conjugated antibodies (GE Healthcare, Little Chalfont, UK) were used for detection, except for FLAG (see above). For IP experiments, 3 mg lysate was incubated with 40 µl magnetic anti-FLAG bead slurry (Sigma. Cat # M8823) in 1 ml finalized extraction buffer for 2 h on the rotating wheel at 4°C. Then beads were washed 4 times with 600 µl extraction buffer followed by 1 h elution on a vertical shaker (4°C) with 40 µl of 1 µg / µl FLAG peptide.
Immunoprecipitation of FLAG-HA-tagged Hs_C2AIL was performed using Anti-FLAG M2 Magnetic Beads (Sigma. Cat # M8823) as described64. For this, 2·106 transfected HEK293T cells per IP and construct were washed with 4 ml cold PBS, harvested and lysed in 500 µl lysis buffer (50 mM TRIS pH 7.5, 150 mM NaCl, 1% v/v TritonX-100, 1 mM EDTA, 1x EDTA-free Roche cOmplete Protease Inhibitor Cocktail) and incubated for 30 min on ice. Subsequently, the lysate was cleared by centrifugation at 16,000 g for 10 min. Per IP, 40 µl of bead slurry (see above) was washed twice with cold TBS (50 mM TRIS pH7.4, 150 mM NaCl), added to the lysate and filled to 1 ml final volume with TBS. After 2 h of incubation on the rotating wheel, beads were washed three times with 400 µl TBS and boiled at 70°C for 10 min in 40 µl SDS loading buffer for western blot analysis detecting the transgene with anti-HA antibody (1:1000 dilution; clone 3F10, Roche). Human XRN2 was detected as described23.
Microscopy
Stereoscopic images were taken with a M205 A stereo microscope (Leica, Solms, Germany).
Supplementary Material
Acknowledgements
We thank J. Keusch for technical support, D. Hess for mass spectrometry analysis, M. de la Mata for help with cell transfections, M. Zou for zebrafish lysate, and J.A. Chao and N. Thomä for critical comments on the manuscript. We are grateful to the staff of the Swiss Light Source, Paul Scherrer Institute, Villingen, where parts of the experiments were performed. The research leading to these results has received funding from the European Union Seventh Framework Programme (FP7/2007-2013) under grant agreement number 241985 (European Research Council “miRTurn”; to H.Gr.), the Swiss National Science Foundation (SNF 31003A_143313; to H.Gr.), and the Novartis Research Foundation through the Friedrich Miescher Institute (to I.K., H.Gu., H.Gr.).
Footnotes
Accession Codes
The atomic coordinates have been deposited at the Protein Data Bank, with accession codes 5FIR.
Author Contribution
H.R. and H.Gr. designed research. H.R. designed and performed in vitro and in vivo experiments. H.Gu. and H.R. did crystallographic structure determination and analyses. I.K. set up CRISPR, injected worms, and assisted in crossing worm lines. H.R., H.Gr. and H.Gu. wrote the manuscript. All authors supported editing the manuscript.
References
- 1.Petfalski E, Dandekar T, Henry Y, Tollervey D. Processing of the precursors to small nucleolar RNAs and rRNAs requires common components. Mol Cell Biol. 1998;18:1181–9. doi: 10.1128/mcb.18.3.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang M, Pestov DG. 5'-end surveillance by Xrn2 acts as a shared mechanism for mammalian pre-rRNA maturation and decay. Nucleic Acids Res. 2011;39:1811–22. doi: 10.1093/nar/gkq1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Couvillion MT, Bounova G, Purdom E, Speed TP, Collins K. A Tetrahymena Piwi Bound to Mature tRNA 3' Fragments Activates the Exonuclease Xrn2 for RNA Processing in the Nucleus. Mol Cell. 2012;48:509–20. doi: 10.1016/j.molcel.2012.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Geerlings TH, Vos JC, Raué HA. The final step in the formation of 25S rRNA in Saccharomyces cerevisiae is performed by 5'-->3' exonucleases. RNA. 2000;6:1698–703. doi: 10.1017/s1355838200001540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chanfreau G, Rotondo G, Legrain P, Jacquier A. Processing of a dicistronic small nucleolar RNA precursor by the RNA endonuclease Rnt1. EMBO J. 1998;17:3726–37. doi: 10.1093/emboj/17.13.3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zakrzewska-Placzek M, Souret FF, Sobczyk GJ, Green PJ, Kufel J. Arabidopsis thaliana XRN2 is required for primary cleavage in the pre-ribosomal RNA. Nucleic Acids Res. 2010;38:4487–502. doi: 10.1093/nar/gkq172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.West S, Gromak N, Proudfoot NJ. Human 5' --> 3' exonuclease Xrn2 promotes transcription termination at co-transcriptional cleavage sites. Nature. 2004;432:522–5. doi: 10.1038/nature03035. [DOI] [PubMed] [Google Scholar]
- 8.Kim M, et al. The yeast Rat1 exonuclease promotes transcription termination by RNA polymerase II. Nature. 2004;432:517–22. doi: 10.1038/nature03041. [DOI] [PubMed] [Google Scholar]
- 9.Davidson L, Kerr A, West S. Co-transcriptional degradation of aberrant pre-mRNA by Xrn2. EMBO J. 2012;31:2566–78. doi: 10.1038/emboj.2012.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chernyakov I, Whipple JM, Kotelawala L, Grayhack EJ, Phizicky EM. Degradation of several hypomodified mature tRNA species in Saccharomyces cerevisiae is mediated by Met22 and the 5'-3' exonucleases Rat1 and Xrn1. Genes Dev. 2008;22:1369–80. doi: 10.1101/gad.1654308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chatterjee S, Großhans H. Active turnover modulates mature microRNA activity in Caenorhabditis elegans. Nature. 2009;461:546–549. doi: 10.1038/nature08349. [DOI] [PubMed] [Google Scholar]
- 12.Miki TS, Großhans H. The multifunctional RNase XRN2. Biochem Soc Trans. 2013;41:825–30. doi: 10.1042/BST20130001. [DOI] [PubMed] [Google Scholar]
- 13.Nagarajan VK, Jones CI, Newbury SF, Green PJ. XRN 5'→3' exoribonucleases: Structure, mechanisms and functions. Biochim Biophys Acta. 2013;1829:590–603. doi: 10.1016/j.bbagrm.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Amberg DC, Goldstein AL, Cole CN. Isolation and characterization of RAT1: an essential gene of Saccharomyces cerevisiae required for the efficient nucleocytoplasmic trafficking of mRNA. Genes Dev. 1992;6:1173–89. doi: 10.1101/gad.6.7.1173. [DOI] [PubMed] [Google Scholar]
- 15.Miki TS, Rüegger S, Gaidatzis D, Stadler MB, Großhans H. Engineering of a conditional allele reveals multiple roles of XRN2 in Caenorhabditis elegans development and substrate specificity in microRNA turnover. Nucleic Acids Res. 2014;42:4056–67. doi: 10.1093/nar/gkt1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parker R, Sheth U. P bodies and the control of mRNA translation and degradation. Mol Cell. 2007;25:635–46. doi: 10.1016/j.molcel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 17.Kenna M, Stevens A, McCammon M, Douglas MG. An essential yeast gene with homology to the exonuclease-encoding XRN1/KEM1 gene also encodes a protein with exoribonuclease activity. Mol Cell Biol. 1993;13:341–50. doi: 10.1128/mcb.13.1.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stevens A, Poole TL. 5'-exonuclease-2 of Saccharomyces cerevisiae. Purification and features of ribonuclease activity with comparison to 5'-exonuclease-1. J Biol Chem. 1995;270:16063–9. doi: 10.1074/jbc.270.27.16063. [DOI] [PubMed] [Google Scholar]
- 19.Poole TL, Stevens A. Comparison of features of the RNase activity of 5'-exonuclease-1 and 5'-exonuclease-2 of Saccharomyces cerevisiae. Nucleic Acids Symp Ser. 1995:79–81. [PubMed] [Google Scholar]
- 20.Jinek M, Coyle SM, Doudna JA. Coupled 5' nucleotide recognition and processivity in XRN1-mediated mRNA decay. Mol Cell. 2011;41:600–8. doi: 10.1016/j.molcel.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xiang S, et al. Structure and function of the 5'-->3' exoribonuclease Rat1 and its activating partner Rai1. Nature. 2009;458:784–8. doi: 10.1038/nature07731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xue Y, et al. Saccharomyces cerevisiae RAI1 (YGL246c) is homologous to human DOM3Z and encodes a protein that binds the nuclear exoribonuclease Rat1p. Mol Cell Biol. 2000;20:4006–15. doi: 10.1128/mcb.20.11.4006-4015.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miki TS, Richter H, Rüegger S, Großhans H. PAXT-1 promotes XRN2 activity by stabilizing it through a conserved domain. Mol Cell. 2014;53:351–60. doi: 10.1016/j.molcel.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 24.Brannan K, et al. mRNA decapping factors and the exonuclease Xrn2 function in widespread premature termination of RNA polymerase II transcription. Mol Cell. 2012;46:311–24. doi: 10.1016/j.molcel.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Close P, et al. DBIRD complex integrates alternative mRNA splicing with RNA polymerase II transcript elongation. Nature. 2012;484:386–9. doi: 10.1038/nature10925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Söding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33:W244–8. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Buchan DW, Minneci F, Nugent TC, Bryson K, Jones DT. Scalable web services for the PSIPRED Protein Analysis Workbench. Nucleic Acids Res. 2013;41:W349–57. doi: 10.1093/nar/gkt381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Linding R, et al. Protein disorder prediction: implications for structural proteomics. Structure. 2003;11:1453–9. doi: 10.1016/j.str.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 29.Chang JH, Xiang S, Xiang K, Manley JL, Tong L. Structural and biochemical studies of the 5'→3' exoribonuclease Xrn1. Nat Struct Mol Biol. 2011;18:270–6. doi: 10.1038/nsmb.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Holm L, Rosenström P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–9. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–97. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 32.Duarte JM, Srebniak A, Schärer MA, Capitani G. Protein interface classification by evolutionary analysis. BMC Bioinformatics. 2012;13:334. doi: 10.1186/1471-2105-13-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ashkenazy H, Erez E, Martz E, Pupko T, Ben-Tal N. ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 2010;38:W529–33. doi: 10.1093/nar/gkq399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Celniker G, et al. ConSurf: Using Evolutionary Data to Raise Testable Hypotheses about Protein Function. Israel Journal Of Chemistry. 2013;53:199–206. [Google Scholar]
- 35.Arribere JA, et al. Efficient marker-free recovery of custom genetic modifications with CRISPR/Cas9 in Caenorhabditis elegans. Genetics. 2014;198:837–46. doi: 10.1534/genetics.114.169730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dickinson DJ, Ward JD, Reiner DJ, Goldstein B. Engineering the Caenorhabditis elegans genome using Cas9-triggered homologous recombination. Nat Methods. 2013;10:1028–34. doi: 10.1038/nmeth.2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cheung CT, Singh R, Kalra RS, Kaul SC, Wadhwa R. Collaborator of ARF (CARF) regulates proliferative fate of human cells by dose-dependent regulation of DNA damage signaling. J Biol Chem. 2014;289:18258–69. doi: 10.1074/jbc.M114.547208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hasan MK, et al. CARF is a novel protein that cooperates with mouse p19ARF (human p14ARF) in activating p53. J Biol Chem. 2002;277:37765–70. doi: 10.1074/jbc.M204177200. [DOI] [PubMed] [Google Scholar]
- 39.Hasan MK, et al. Alternative reading frame protein (ARF)-independent function of CARF (collaborator of ARF) involves its interactions with p53: evidence for a novel p53-activation pathway and its negative feedback control. Biochem J. 2004;380:605–10. doi: 10.1042/BJ20040337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Querol E, Perez-Pons JA, Mozo-Villarias A. Analysis of protein conformational characteristics related to thermostability. Protein Eng. 1996;9:265–71. doi: 10.1093/protein/9.3.265. [DOI] [PubMed] [Google Scholar]
- 41.Feng X, et al. Identification of a negative response element in the human inducible nitric-oxide synthase (hiNOS) promoter: The role of NF-kappa B-repressing factor (NRF) in basal repression of the hiNOS gene. Proc Natl Acad Sci U S A. 2002;99:14212–7. doi: 10.1073/pnas.212306199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Uhlén M, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 43.Rother S, et al. NF-κB-repressing factor phosphorylation regulates transcription elongation via its interactions with 5'→3' exoribonuclease 2 and negative elongation factor. FASEB J. 2015 doi: 10.1096/fj.15-270256. [DOI] [PubMed] [Google Scholar]
- 44.Vieille C, Zeikus GJ. Hyperthermophilic enzymes: sources, uses, and molecular mechanisms for thermostability. Microbiol Mol Biol Rev. 2001;65:1–43. doi: 10.1128/MMBR.65.1.1-43.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ghaemmaghami S, Oas TG. Quantitative protein stability measurement in vivo. Nat Struct Biol. 2001;8:879–82. doi: 10.1038/nsb1001-879. [DOI] [PubMed] [Google Scholar]
- 46.McLendon G, Radany E. Is protein turnover thermodynamically controlled? J Biol Chem. 1978;253:6335–7. [PubMed] [Google Scholar]
- 47.Parsell DA, Sauer RT. The structural stability of a protein is an important determinant of its proteolytic susceptibility in Escherichia coli. J Biol Chem. 1989;264:7590–5. [PubMed] [Google Scholar]
- 48.Kumar S, Tsai CJ, Nussinov R. Factors enhancing protein thermostability. Protein Eng. 2000;13:179–91. doi: 10.1093/protein/13.3.179. [DOI] [PubMed] [Google Scholar]
- 49.Wheeler TJ, Clements J, Finn RD. Skylign: a tool for creating informative, interactive logos representing sequence alignments and profile hidden Markov models. BMC Bioinformatics. 2014;15:7. doi: 10.1186/1471-2105-15-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Blommel PG, Fox BG. A combined approach to improving large-scale production of tobacco etch virus protease. Protein Expr Purif. 2007;55:53–68. doi: 10.1016/j.pep.2007.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr. 2010;66:125–32. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cowtan K. The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr D Biol Crystallogr. 2006;62:1002–11. doi: 10.1107/S0907444906022116. [DOI] [PubMed] [Google Scholar]
- 54.Afonine PV, et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr. 2012;68:352–67. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sinturel F, et al. Real-time fluorescence detection of exoribonucleases. RNA. 2009;15:2057–62. doi: 10.1261/rna.1670909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gibson DG, et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6:343–5. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 59.Friedland AE, et al. Heritable genome editing in C. elegans via a CRISPR-Cas9 system. Nat Methods. 2013;10:741–3. doi: 10.1038/nmeth.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Katic I, Großhans H. Targeted heritable mutation and gene conversion by Cas9-CRISPR in Caenorhabditis elegans. Genetics. 2013;195:1173–6. doi: 10.1534/genetics.113.155754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Katic I, Xu L, Ciosk R. CRISPR/Cas9 Genome Editing in Caenorhabditis elegans: Evaluation of Templates for Homology-Mediated Repair and Knock-Ins by Homology-Independent DNA Repair. G3 (Bethesda) 2015;5:1649–56. doi: 10.1534/g3.115.019273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Frøkjaer-Jensen C, et al. Single-copy insertion of transgenes in Caenorhabditis elegans. Nat Genet. 2008;40:1375–83. doi: 10.1038/ng.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Frøkjær-Jensen C, Davis MW, Ailion M, Jorgensen EM. Improved Mos1-mediated transgenesis in C. elegans. Nat Methods. 2012;9:117–8. doi: 10.1038/nmeth.1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.de la Mata M, et al. Potent degradation of neuronal miRNAs induced by highly complementary targets. EMBO Rep. 2015;16:500–11. doi: 10.15252/embr.201540078. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.